Abstract
The 2019 novel severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) outbreak has caused a large number of deaths, with thousands of confirmed cases worldwide. The present study followed computational approaches to identify B- and T-cell epitopes for the spike (S) glycoprotein of SARS-CoV-2 by its interactions with the human leukocyte antigen alleles. We identified 24 peptide stretches on the SARS-CoV-2 S protein that are well conserved among the reported strains. The S protein structure further validated the presence of predicted peptides on the surface, of which 20 are surface exposed and predicted to have reasonable epitope binding efficiency. The work could be useful for understanding the immunodominant regions in the surface protein of SARS-CoV-2 and could potentially help in designing some peptide-based diagnostics. Also, identified T-cell epitopes might be considered for incorporation in vaccine designs.
Keywords: SARS-CoV-2, Spike protein, Epitopes, Diagnostics
Highlights
-
•
Determination of variability and average solvent accessibility.
-
•
Identification of the B- and T-cell epitopes for spike glycoprotein of SARS-CoV-2.
-
•
Interactions of B and T cell epitopes with the human leukocyte antigen alleles.
1. Introduction
Emerging severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) is a recent pandemic and has been declared as a public health emergency by the World Health Organization (WHO, 2020b). The disease rapidly spread across the globe and caused havoc to humanity (Wu and McGoogan, 2020). By the start of May, SARS-CoV-2 had spread to 215 countries and infected over 3,862,676 people (WHO, 2020a). The WHO is continuously monitoring and updating health-related plans to curtail the disease spread. The absence of a specific treatment and vaccine worsens the situation and threatens the world.
The International Committee on Taxonomy of Viruses (ICTV), classified SARS-CoV-2 under the family Coronaviridae of order Nidovirales. The genomic sequence of SARS-CoV-2 isolated from the bronchoalveolar lavage fluid of a patient from Wuhan, China showed a length of 29,903 nucleotides [GenBank accession number NC_045512] (Wu et al., 2020). SARS-CoV-2 contains a positive-sense single-stranded RNA with 5ˊ and 3ˊ untranslated region. The genome codes for ORF1a, ORF1b, Spike (S), ORF3a, ORF3b, Envelope (E), Membrane (M), ORF6, ORF7a, ORF7b, ORF8, ORF9b, ORF14, Nucleocapsid (N), and ORF10 from 5ˊ to 3ˊ (Wu et al., 2020; Zhu et al., 2020).
The S glycoprotein forms a homotrimer and mediates viral entry into host cells. The S protein is a potential target for therapeutic and vaccine design against SARS-CoV-2 infection in humans (Li, 2016; Tortorici et al., 2019). The S glycoprotein comprises two functional subunits: the S1 subunit is responsible for binding to the host cell receptor and the S2 subunit is responsible for fusion of the virus with the cell membrane. Usually in CoVs, S is cleaved at the boundary between S1 and S2 subunits, which remain non-covalently bound in the prefusion conformation, to activate the protein for membrane fusion via extensive irreversible conformational changes (Burkard et al., 2014; Park et al., 2016; Walls et al., 2017). Setting it apart from other SARS-CoVs, it is found that the S glycoprotein of SARS-CoV-2 harbors a furin cleavage site at the boundary between the S1/S2 subunits (Walls et al., 2020). By now, it is evident that SARS-CoV-2 S uses angiotensin-converting enzyme 2 (ACE2) receptor-mediated entry into cells. Some studies suggest similar binding affinities to human ACE2 with the S protein of SARS-CoV-2 and SARS-CoV (Letko et al., 2020; Walls et al., 2020). However, some suggest that SARS-CoV-2 binds ACE2 with higher affinity than SARS-CoV (Tai et al., 2020; Wang et al., 2020; Wrapp et al., 2020).
As the situation worsens, there is a growing need for the development of suitable therapeutics, vaccines, and other diagnostics against SARS-CoV-2 for effective disease management strategies. Vaccines and diagnostic assays based on peptides have become increasingly substantial and indispensable for their advantages over conventional methods (Li et al., 2014; Mohanraj et al., 2017). The present study aimed to locate appropriate epitopes within a particular protein antigen that can elicit an immune response and could be selected for the synthesis of an immunogenic peptide. Using a computational approach, the S glycoprotein of SARS-CoV-2 was explored to identify various immunodominant epitopes for the development of diagnostics and vaccines. Besides, the results could also help us to understand the SARS-CoV-2 surface protein response towards T- and B-cells.
2. Materials & methods
2.1. Collection of the targeted protein sequence
The amino acid sequences (n = 98) of S protein available at the time of study on targeted SARS-CoV-2 were downloaded from the National Centre for Biotechnological Information (NCBI) database.
2.2. Identification of potential peptides
To identify an immunodominant region, it is of extreme importance to select the conserved region within the S protein of SARS-CoV-2. All the sequences were compared among themselves for variability using the protein variability server by the Shannon method (Garcia-Boronat et al., 2008). The average solvent accessibility (ASA) profile was predicted for each sequence using the SABLE server (Adamczak et al., 2004). BepiPred 1.0 Linear Epitope Prediction module incorporated in Immune Epitope Database (IEDB) was used to predict potential epitopes within the S protein (Haste Andersen et al., 2006; Larsen et al., 2006; Ponomarenko and Bourne, 2007; Vita et al., 2019). The FASTA sequence of the targeted protein was used as an input for all the default parameters.
2.3. Identification of B-cell epitopes
We used two web-based tools for B-cell epitope prediction: the IEDB and ABCpred servers (Saha and Raghava, 2006). S protein structure from the protein data bank (PDB, 6VSB) was analyzed for linear and discontinuous B-cell epitopes using the ElliPro module on the IEDB server with default settings (Ponomarenko et al., 2008; Wrapp et al., 2020). Also, the ABCpred server was used to detect B-cell epitopes using the artificial neural network (ann) method.
2.4. Identification of T-cell epitopes
T-cell epitopes with a binding affinity towards major histocompatibility complex (MHC)-I and MHC-II alleles were selected to boost up both cytotoxic T-cell and helper T-cell mediated immune response. IEDB server was used to predict the MHC-I and MHC-II binding epitopes for the targeted protein. The reference set of alleles was used for predicting the MHC-I and MHC-II T-cell epitopes (Karosiene et al., 2012; Nielsen et al., 2007; Nielsen et al., 2003; Peters and Sette, 2005; Sturniolo et al., 1999).
3. Results and discussion
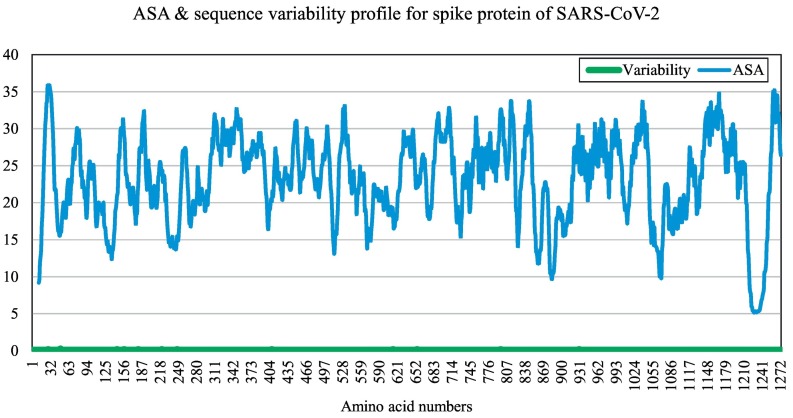
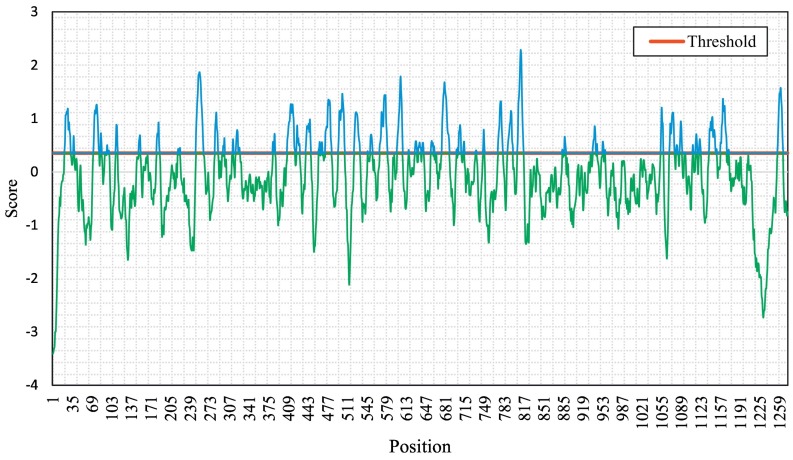
In our study, we targeted the S glycoprotein of SARS-CoV-2 as it is present outside the virus and interacts with the host receptor. At the time of the study, there were 98 sequences available for the targeted protein of SARS-CoV-2. The S glycoprotein sequence is 1273 amino acids long, except for that of the virus isolated from Kerala (India), which is a 1272 amino acid long S glycoprotein (GenBank accession number MT012098). Our interest here was to determine conserved regions first and then determine surface-exposed regions, which are potential epitopes to generate an immune response. We found that sequences among all the S proteins in the analysis are least variable and highly conserved, as shown in Fig. 1 . However, we found that there were 12 point mutations in the amino acid sequences collected. The mutated sites identified were as follows: positions 247 and 614 for sequence MT007544 (Australia), positions 145 and 408 for sequence MT012098 (India), position 49 for sequence MT027064 (USA), position 221 for sequence MT039890 (South Korea), position 28 for sequence MT049951 (China), position 797 for sequence MT093571 (Sweden), position 157 for sequence MT159716 (USA), positions 655 and 930 for sequence MT163720 (USA), and position 181 for sequence MT184910 (USA). Regions having a high ASA value are more surface exposed compared to others. We identified a total of 24 peptides of varying lengths, which were selected based on high ASA values (Table 1 ). The potential epitope regions were predicted using the sequence of the S protein of SARS-CoV-2 that showed the least variability (GenBank accession number NC_045512). The potential epitopes are represented by blue peaks, while green-colored slopes represent non-epitopic regions (Fig. 2 ).
Fig. 1.
Profiles of average solvent accessibility (blue) in % and amino acid sequence variability (green) in numbers of the 98 SARS-CoV-2 protein plotted against amino acid numbers. High ASA value means the solvent accessibility score is relatively higher for that region and it is more surface exposed with respect to its neighbours. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Table 1.
Conserved region selected based on protein variability, average solvent accessibility and antibody epitope prediction using BepiPred 1.0 Linear Epitope Prediction module of IEDB selected for further analysis.
| Sl. No. | Start | End | Length | Peptide |
|---|---|---|---|---|
| 1 | 21 | 38 | 18 | RTQLPPAYTNSFTRGVYY |
| 2 | 69 | 81 | 13 | HVSGTNGTKRFDN |
| 3 | 144 | 155 | 12 | YYHKNNKSWMES |
| 4 | 178 | 191 | 14 | DLEGKQGNFKNLRE |
| 5 | 249 | 261 | 13 | LTPGDSSSGWTAG |
| 6 | 278 | 287 | 10 | KYNENGTITD |
| 7 | 314 | 325 | 12 | QTSNFRVQPTES |
| 8 | 407 | 428 | 22 | VRQIAPGQTGKIADYNYKLPDD |
| 9 | 437 | 450 | 14 | NSNNLDSKVGGNYN |
| 10 | 461 | 485 | 25 | LKPFERDISTEIYQAGSTPCNGVEG |
| 11 | 493 | 506 | 14 | QSYGFQPTNGVGYQ |
| 12 | 521 | 533 | 13 | PATVCGPKKSTNL |
| 13 | 567 | 581 | 15 | RDIADTTDAVRDPQT |
| 14 | 597 | 607 | 11 | VITPGTNTSNQ |
| 15 | 625 | 648 | 24 | HADQLTPTWRVYSTGSNVFQTRAG |
| 16 | 654 | 661 | 8 | EHVNNSYE |
| 17 | 673 | 691 | 19 | SYQTQTNSPRRARSVASQS |
| 18 | 700 | 713 | 16 | GAENSVAYSNNSIA |
| 19 | 768 | 780 | 13 | TGIAVEQDKNTQE |
| 20 | 788 | 799 | 14 | IYKTPPIKDFGG |
| 21 | 805 | 816 | 12 | ILPDPSKPSKRS |
| 22 | 1134 | 1150 | 17 | NNTVYDPLQPELDSFKE |
| 23 | 1153 | 1171 | 19 | DKYFKNHTSPDVDLGDISG |
| 24 | 1255 | 1267 | 13 | KFDEDDSEPVLKG |
Fig. 2.
Graphical representation of B-cell linear epitopes of spike protein of SARS-CoV-2. B-cell linear epitopes predicted using BepiPred 1.0 module incorporated in IEDB server using default threshold value (0.35).
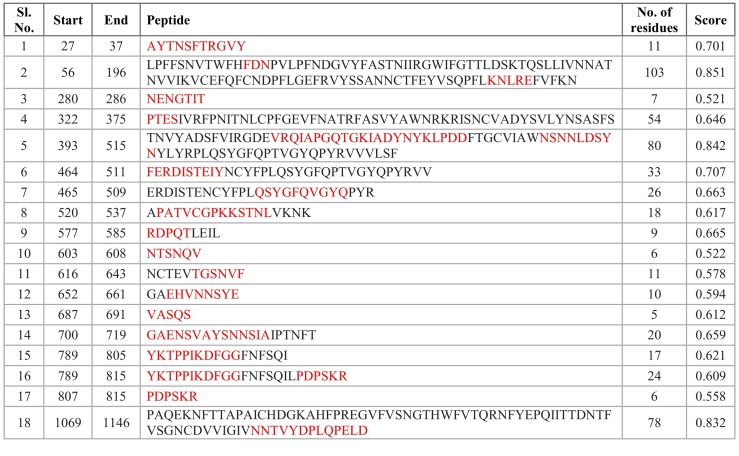
The existence of B-cell linear and discontinuous (conformational) epitopes within the identified segments could help us to identify the peptides, which can elicit an immune response (Purcell et al., 2007). We identified 18 linear epitopes, predicted by ElliPro (IEDB), which contained regions from 19 of our selected peptides (highlighted in red in Table 2 ). These identified B-cell linear epitopes were placed based on their positional value and scores. Epitopes with high scores have more potential for antibody binding. Five of our selected peptides (peptide numbers 3, 5, 19, 23, and 24 in Table 1) were not considered as potential linear B-cell epitopes. Some parts of our identified epitopes were in accordance with epitopes recognized in an earlier study (Ahmed et al., 2020), which further supports the credibility of our identified epitopes.
Table 2.
IEDB ElliPro predicted linear epitopes for spike protein of SARS-CoV-2. Sequences that match our selected peptides are marked in red.
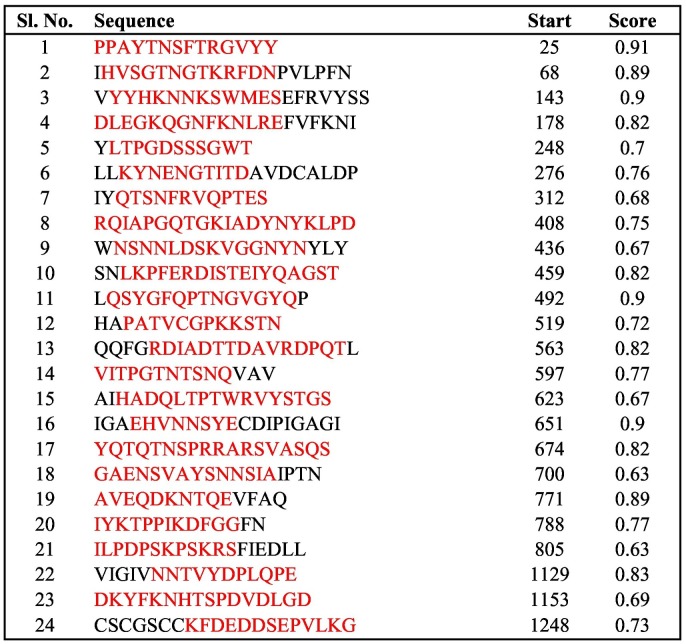
Using the same module, B-cell discontinuous epitopes were predicted, which gave 16 epitope regions that contained regions from 18 of our selected peptides (highlighted in red in Table S1). Six peptides (peptide numbers 3, 5, 14, 19, 23, and 24 in Table 1) were not predicted as discontinuous B-cell epitopes. To further confirm, we used the ABCpred server to detect B-cell epitopes, with a default threshold of 0.51. It identified various epitopes with different lengths and scores. Out of those, the regions that contained our selected peptides are highlighted in red in Table 3 . A high score represents good binding affinity with epitopes; most of our peptides scored more than 0.7 and were predicted as linear B-cell epitopes.
Table 3.
ABCpred determination of B-cell binding affinities. Note that high score indicates good binding affinity.
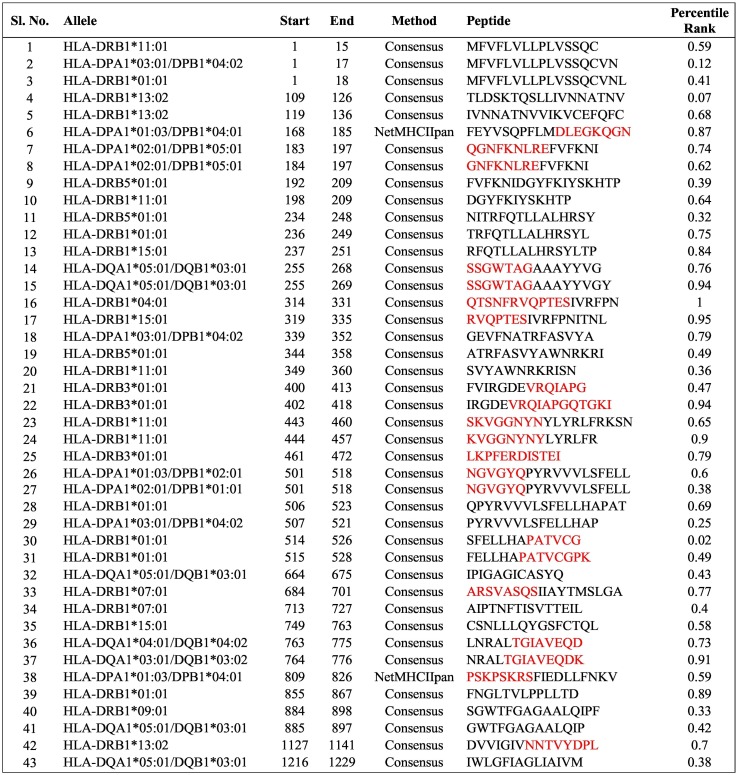
We used the IEDB server to determine the binding affinity for the human leucocyte antigen (HLA). As recommended by the IEDB server, reference HLA allele sets were used for the prediction of MHC-I and MHC-II T-cell epitopes, as they provide comprehensive coverage of the population. All the predictions were made using IEDB recommended procedures. The list of binding affinities for MHC-I T-cell epitopes is given in Table S2, where low rank represents high binding affinity. Similarly, the list of binding affinities for MHC-II T-cell epitopes are given in Table 4 . Regions from our selected peptides are highlighted in red. The epitopes with rank <1% for very high binding affinity were selected. We also observed that some of the peptides we identified as potential B-cell epitopes were present as T-cell epitopes with good binding affinities.
Table 4.
IEDB prediction of binding affinity with MHC-II alleles, peptides with percentile rank less than 1.00 are shown here. The binding affinity is considered higher for low percentile rank. Sequences that match our selected peptides are marked in red.
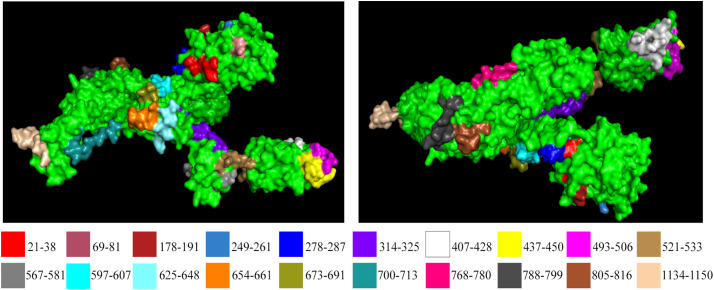
Overall, it was found that the regions identified in Table 1 not only had good B-cell and T-cell affinities, but the majority of them had also overlapped with discontinuous epitopes (Table S1). The peptide segments identified from the set of 98 sequences of the SARS-CoV-2 S glycoprotein appear to hold reasonable potential to act as immunogens. Peptide-based diagnostics and vaccines have previously been proposed against virus outbreaks (Dey et al., 2017; Ichihashi et al., 2011; Navalkar et al., 2015; Oany et al., 2014; Zhao et al., 2009). The availability of a 3D structure (6VSB) of the SARS-CoV-2 S glycoprotein provided an opportunity to inspect the predicted peptides. Placement of the peptide segments identified by ASA and conserved sequence analysis on the S glycoprotein showed that 20 of the regions we identified lie on the surface (Fig. 3 ). In order to limit recognition and evade the immune response of the host, coronaviruses use conformational masking and glycan shielding (Walls et al., 2019; Xiong et al., 2018). SARS-CoV-2 S trimer also exists in multiple distinct conformational states, which is necessary for receptor engagement, leading to the initiation of fusogenic conformational changes (Walls et al., 2020). The considerable number of peptides at the surface region of the S glycoprotein allows for the potential use of those peptide regions as immunogens. Binding to the ACE2 receptor is a critical initial step for the SARS-CoV-2 in entering target cells. Recent studies have also pointed out the vital role of ACE2 in mediating the entry of SARS-CoV-2 (Hoffmann et al., 2020). Receptor binding motif (RBM) is part of the receptor-binding domain (RBD) of SARS-CoV-2, which contains most of the contacting residue for ACE-2 binding (Lan et al., 2020). It was observed that some of our identified peptides from Table 1 (peptide no. 7–12) fall in the regions of RBD (amino acid no. 319–540) and RBM (amino acid no. 438–506), which makes them potential peptide regions to be used.
Fig. 3.
Our selected peptides are highlighted on spike protein of SARS-CoV-2 protein structure downloaded from PDB (ID: 6VSB).
The emergence of new viral diseases like SARS-CoV-2 represents a substantial global disease burden. Over the past few months, there have been increased research efforts for the design and development of diagnostics and vaccines for SARS-CoV-2. Some related analyses have been reported in distinct, parallel studies (Baruah and Bose, 2020; Bhattacharya et al., 2020; Grifoni et al., 2020). Our study leverages the available resources and computational methods and adds to the ongoing research focused on the development of diagnostics and vaccines against SARS-CoV-2. Other than already existing ones, we have identified a further number of peptides, which adds to the library of peptides that are likely to be recognized by human immune responses. Facilitated by high mutation rates, traditional vaccines based on antibody-mediated protection are often poor inducers of T-cell responses and can have limited success (Rosendahl Huber et al., 2014). Peptide-based sensitive and rapid diagnostic kits are considered a better alternative to the conventional serological tests, including whole antigenic protein (Mohanraj et al., 2017). In our study, we predicted both B-cell and T-cell epitopes for conferring immunity in different ways. We speculate that the identified epitopes with considerably good epitope binding efficiency have the potential to be an immunodominant peptide. The study could help us to use the predicted peptide as an immunogen for the development of diagnostics and vaccines against SARS-CoV-2.
4. Conclusion
In the present study, peptide segments were identified on S proteins for the development of diagnostics and vaccines against SARS-CoV-2. The recent availability of 3D data on 2019-CoV S glycoprotein has helped the search. SARS-CoV-2, being an RNA virus, has a high mutation rate and undergoes active recombination (Yi, 2020). Although the peptides identified are ideal candidates as immunogens for the development of peptide-based diagnostics and vaccines, more refinement and lab trials are essential steps that are yet to be undertaken for early development before the identified epitopes are rendered obsolete.
Declaration of Competing Interest
The authors declare no conflict of interest.
Acknowledgement
We thank Dr. Monika Koul and Dr. Nitin Chaudhary for proof reading the text and their valuable suggestions.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.meegid.2020.104382.
Appendix A. Supplementary data
Supplementary material
References
- Adamczak R., Porollo A., Meller J. Accurate prediction of solvent accessibility using neural networks-based regression. Proteins. 2004;56(4):753–767. doi: 10.1002/prot.20176. [DOI] [PubMed] [Google Scholar]
- Ahmed S.F., Quadeer A.A., McKay M.R. Preliminary identification of potential vaccine targets for the COVID-19 Coronavirus (SARS-CoV-2) based on SARS-CoV immunological studies. Viruses. 2020;12(3) doi: 10.3390/v12030254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baruah V., Bose S. Immunoinformatics-aided identification of T cell and B cell epitopes in the surface glycoprotein of 2019-nCoV. J. Med. Virol. 2020;92(5):495–500. doi: 10.1002/jmv.25698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya M., Sharma A.R., Patra P., Ghosh P., Sharma G., Patra B.C., Lee S.S., Chakraborty C. Development of epitope-based peptide vaccine against novel coronavirus 2019 (SARS-COV-2): Immunoinformatics approach. J. Med. Virol. 2020 doi: 10.1002/jmv.25736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkard C., Verheije M.H., Wicht O., van Kasteren S.I., van Kuppeveld F.J., Haagmans B.L., Pelkmans L., Rottier P.J., Bosch B.J., de Haan C.A. Coronavirus cell entry occurs through the endo−/lysosomal pathway in a proteolysis-dependent manner. PLoS Pathog. 2014;10(11) doi: 10.1371/journal.ppat.1004502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey S., Nandy A., Basak S.C., Nandy P., Das S. A bioinformatics approach to designing a Zika virus vaccine. Comput. Biol. Chem. 2017;68:143–152. doi: 10.1016/j.compbiolchem.2017.03.002. [DOI] [PubMed] [Google Scholar]
- Garcia-Boronat M., Diez-Rivero C.M., Reinherz E.L., Reche P.A. PVS: a web server for protein sequence variability analysis tuned to facilitate conserved epitope discovery. Nucleic Acids Res. 2008;36:W35–W41. doi: 10.1093/nar/gkn211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grifoni A., Sidney J., Zhang Y., Scheuermann R.H., Peters B., Sette A. A sequence homology and Bioinformatic approach can predict candidate targets for immune responses to SARS-CoV-2. Cell Host Microbe. 2020;27(4):671–680. doi: 10.1016/j.chom.2020.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haste Andersen P., Nielsen M., Lund O. Prediction of residues in discontinuous B-cell epitopes using protein 3D structures. Protein Sci. 2006;15(11):2558–2567. doi: 10.1110/ps.062405906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Kruger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A., Muller M.A., Drosten C., Pohlmann S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181(2):271–280. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichihashi T., Yoshida R., Sugimoto C., Takada A., Kajino K. Cross-protective peptide vaccine against influenza A viruses developed in HLA-A*2402 human immunity model. PLoS One. 2011;6(9) doi: 10.1371/journal.pone.0024626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karosiene E., Lundegaard C., Lund O., Nielsen M. NetMHCcons: a consensus method for the major histocompatibility complex class I predictions. Immunogenetics. 2012;64(3):177–186. doi: 10.1007/s00251-011-0579-8. [DOI] [PubMed] [Google Scholar]
- Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L., Wang X. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581(7807):215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- Larsen J.E., Lund O., Nielsen M. Improved method for predicting linear B-cell epitopes. Immunome Res. 2006;2 doi: 10.1186/1745-7580-2-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letko M., Marzi A., Munster V. Functional Assessment of Cell Entry and Receptor Usage for SARS-CoV-2 and Other Lineage B Betacoronaviruses. Nat. Microbiol. 2020;5(4):562–569. doi: 10.1038/s41564-020-0688-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F. Structure, function, and evolution of Coronavirus spike proteins. Annu. Rev. Virol. 2016;3(1):237–261. doi: 10.1146/annurev-virology-110615-042301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Joshi M.D., Singhania S., Ramsey K.H., Murthy A.K. Peptide vaccine: Progress and challenges. Vaccines. 2014;2(3):515–536. doi: 10.3390/vaccines2030515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohanraj U., Chander S., Chavan Y.G. Peptide based viral detection systems for effective diagnosis of common viral infections in India. Curr. Protein Pept. Sci. 2017;18(9):939–945. doi: 10.2174/1389203717666160724205226. [DOI] [PubMed] [Google Scholar]
- Navalkar K.A., Johnston S.A., Stafford P. Peptide based diagnostics: are random-sequence peptides more useful than tiling proteome sequences? J. Immunol. Methods. 2015;417:10–21. doi: 10.1016/j.jim.2014.12.002. [DOI] [PubMed] [Google Scholar]
- Nielsen M., Lundegaard C., Worning P., Lauemoller S.L., Lamberth K., Buus S., Brunak S., Lund O. Reliable prediction of T-cell epitopes using neural networks with novel sequence representations. Protein Sci. 2003;12(5):1007–1017. doi: 10.1110/ps.0239403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen M., Lundegaard C., Lund O. Prediction of MHC class II binding affinity using SMM-align, a novel stabilization matrix alignment method. BMC Bioinformatics. 2007;8 doi: 10.1186/1471-2105-8-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oany A.R., Emran A.A., Jyoti T.P. Design of an epitope-based peptide vaccine against spike protein of human coronavirus: an in silico approach. Drug Des. Devel. Ther. 2014;8:1139–1149. doi: 10.2147/DDDT.S67861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J.E., Li K., Barlan A., Fehr A.R., Perlman S., McCray P.B., Jr., Gallagher T. Proteolytic processing of Middle East respiratory syndrome coronavirus spikes expands virus tropism. Proc. Natl. Acad. Sci. U. S. A. 2016;113(43):12262–12267. doi: 10.1073/pnas.1608147113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters B., Sette A. Generating quantitative models describing the sequence specificity of biological processes with the stabilized matrix method. BMC Bioinformatics. 2005;6 doi: 10.1186/1471-2105-6-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponomarenko J., Bui H.H., Li W., Fusseder N., Bourne P.E., Sette A., Peters B. ElliPro: a new structure-based tool for the prediction of antibody epitopes. BMC Bioinformatics. 2008;9 doi: 10.1186/1471-2105-9-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponomarenko J.V., Bourne P.E. Antibody-protein interactions: benchmark datasets and prediction tools evaluation. BMC Struct. Biol. 2007;7 doi: 10.1186/1472-6807-7-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell A.W., McCluskey J., Rossjohn J. More than one reason to rethink the use of peptides in vaccine design. Nat. Rev. Drug Discov. 2007;6(5):404–414. doi: 10.1038/nrd2224. [DOI] [PubMed] [Google Scholar]
- Rosendahl Huber S., van Beek J., de Jonge J., Luytjes W., van Baarle D. T cell responses to viral infections - opportunities for peptide vaccination. Front. Immunol. 2014;5 doi: 10.3389/fimmu.2014.00171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha S., Raghava G.P. Prediction of continuous B-cell epitopes in an antigen using recurrent neural network. Proteins. 2006;65(1):40–48. doi: 10.1002/prot.21078. [DOI] [PubMed] [Google Scholar]
- Sturniolo T., Bono E., Ding J., Raddrizzani L., Tuereci O., Sahin U., Braxenthaler M., Gallazzi F., Protti M.P., Sinigaglia F., Hammer J. Generation of tissue-specific and promiscuous HLA ligand databases using DNA microarrays and virtual HLA class II matrices. Nat. Biotechnol. 1999;17(6):555–561. doi: 10.1038/9858. [DOI] [PubMed] [Google Scholar]
- Tai W., He L., Zhang X., Pu J., Voronin D., Jiang S., Zhou Y., Du L. Characterization of the receptor-binding domain (RBD) of 2019 novel coronavirus: implication for development of RBD protein as a viral attachment inhibitor and vaccine. Cell. Mol. Immunol. 2020 doi: 10.1038/s41423-020-0400-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tortorici M.A., Walls A.C., Lang Y., Wang C., Li Z., Koerhuis D., Boons G.J., Bosch B.J., Rey F.A., de Groot R.J., Veesler D. Structural basis for human coronavirus attachment to sialic acid receptors. Nat. Struct. Mol. Biol. 2019;26(6):481–489. doi: 10.1038/s41594-019-0233-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vita R., Mahajan S., Overton J.A., Dhanda S.K., Martini S., Cantrell J.R., Wheeler D.K., Sette A., Peters B. The immune epitope database (IEDB): 2018 update. Nucleic Acids Res. 2019;47(D1):D339–D343. doi: 10.1093/nar/gky1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A.C., Tortorici M.A., Snijder J., Xiong X., Bosch B.J., Rey F.A., Veesler D. Tectonic conformational changes of a coronavirus spike glycoprotein promote membrane fusion. Proc. Natl. Acad. Sci. U. S. A. 2017;114(42):11157–11162. doi: 10.1073/pnas.1708727114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A.C., Xiong X., Park Y.J., Tortorici M.A., Snijder J., Quispe J., Cameroni E., Gopal R., Dai M., Lanzavecchia A., Zambon M., Rey F.A., Corti D., Veesler D. Unexpected receptor functional mimicry elucidates activation of Coronavirus fusion. Cell. 2019;176(5):1026–1039. doi: 10.1016/j.cell.2018.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181(2):281–292. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Zhang Y., Wu L., Niu S., Song C., Zhang Z., Lu G., Qiao C., Hu Y., Yuen K.Y., Wang Q., Zhou H., Yan J., Qi J. Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell. 2020;181(4):894–904. doi: 10.1016/j.cell.2020.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO . 2020. Coronavirus disease 2019 (COVID-19) Situation Report - 71. [Google Scholar]
- WHO . 2020. WHO Director-General's opening remarks at the media briefing on COVID-19 - 11 March 2020. [Google Scholar]
- Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367(6483):1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G., Hu Y., Tao Z.W., Tian J.H., Pei Y.Y., Yuan M.L., Zhang Y.L., Dai F.H., Liu Y., Wang Q.M., Zheng J.J., Xu L., Holmes E.C., Zhang Y.Z. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579(7798):265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z., McGoogan J.M. Characteristics of and important lessons from the Coronavirus disease 2019 (COVID-19) outbreak in China. JAMA. 2020 doi: 10.1001/jama.2020.2648. [DOI] [PubMed] [Google Scholar]
- Xiong X., Tortorici M.A., Snijder J., Yoshioka C., Walls A.C., Li W., McGuire A.T., Rey F.A., Bosch B.J., Veesler D. Glycan Shield and fusion activation of a deltacoronavirus spike glycoprotein fine-tuned for Enteric infections. J. Virol. 2018;92(4) doi: 10.1128/JVI.01628-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi H. 2019 novel coronavirus is undergoing active recombination. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao K., Liu Q., Yu R., Li Z., Li J., Zhu H., Wu X., Tan F., Wang J., Tang X. Screening of specific diagnostic peptides of swine hepatitis E virus. Virol. J. 2009;6 doi: 10.1186/1743-422X-6-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P., Zhan F., Ma X., Wang D., Xu W., Wu G., Gao G.F., Tan W., China Novel Coronavirus I., Research T. A Novel Coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382(8):727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material