Abstract
Background
Pancreatic patient–derived organoids (PDOs) are a well-established model for studying pancreatic ductal adenocarcinoma (PDAC) carcinogenesis and are potential predictors of clinical responses to chemotherapy. Oncolytic virotherapy is envisioned as a novel treatment modality for pancreatic cancer, and candidate viruses are being tested in clinical trials. Here, we explore the feasibility of using PDOs as a screening platform for the oncolytic adenovirus (OA) response.
Methods
Organoids were established from healthy pancreas and PDAC tissues and assessed for infectivity, oncoselectivity, and patient-dependent sensitivity to OA. Antitumour effects were studied in vivo in organoid xenografts. Further evaluation of oncolytic responses was conducted in organoids derived from orthotopic models or metastastic tissues.
Findings
Oncolytic adenoviruses display good selectivity, with replication only in organoids derived from PDAC tumours. Furthermore, responses of PDOs to a set of OAs reveal individual differences in cytotoxicity as well as in synergism with standard chemotherapy. Adenoviral cytotoxicity in PDOs is predictive of antitumour efficacy in a subcutaneous xenograft setting. Organoids from orthotopic tumours and metastases in nude mice mirror the viral preference of PDOs, indicating that PDO sensitivity to OAs could be informative about responses in both primary tumours and metastatic foci.
Interpretation
Our data imply that pancreatic PDOs can serve as predictive tools for screening for sensitivity to OA.
Keywords: Pancreatic ductal adenocarcinoma (PDAC), Patient-derived organoids (PDO), Oncolytic adenovirus (OA), Orthotopic tumours
Research in context.
Evidence before this study
A variety of oncolytic viruses engineered against cancer are currently in clinical trials, but heterogeneous responses in patients are observed. Currently, oncolytic virotherapies are preclinically tested in vitro in cancer cell lines and in vivo in mouse xenografts.
Added value of this study
We now show that patient-derived organoids (PDOs) could serve as a meaningful platform for preclinical studies of virotherapy and help tailor engineered viruses for future development. Our data suggest that virotherapy testing in PDOs could be a useful tool to inform about treatment selection for patients with pancreatic cancer in the future. OA activity was validated in pancreatic cancer organoids, highlighting patient-specific adenoviral sensitivity. The inter-patient variability in PDO responses was predictive of the antitumour efficacy in xenografts. Moreover, we observed similar viral preference in both primary tumours and metastasis in nude mice, mirroring results with patient PDOs. Finally, screening PDOs for response to both OAs and chemotherapy revealed enhanced sensitivity in combination treatments.
Implications of all the available evidence
We propose that PDOs can be used as a suitable platform to identify personalized viral therapies, alone or in combination with other therapies; this has the potential to greatly improve clinical benefits for patients with pancreatic cancer.
Alt-text: Unlabelled box
1. Introduction
Pancreatic ductal adenocarcinoma (PDAC) is a neoplastic malignancy with a very dismal prognosis, caused in part by a lack of specific symptoms and typically late diagnoses. However, the intrinsic biology of PDACs also gives it a remarkable resistance to most conventional treatments. This results in a 5-year overall survival rate of less than 7%, with almost all survivors being the patients who had undergone surgical resection [1,2].
Model systems based on traditional cell lines, xenograft models, or genetically engineered mouse models (GEMM) have substantially contributed to improving our current knowledge of PDAC [3]. Recently, patient-derived organoids (PDOs) from normal and neoplastic tissues have been proposed as a valuable model to study PDAC carcinogenesis. PDAC organoids from resected tumours recapitulate histo-architecture and phenotype heterogeneity while retaining molecular features of primary malignancies. Importantly, the relatively short time required to establish organoid cultures from surgical specimens minimizes potential culture-induced genetic drift [4], [5], [6].
One emerging strategy for treating PDAC is using oncolytic virotherapies [7,8]. Several viral derivatives are currently being tested in clinical trials, either as a single agent or (in most cases) in combination with standard treatments or novel compounds [9]. To date, clinical studies on oncolytic virotherapy have shown highly variable response rates, which might however be due to the heterogeneous nature of PDAC tumours. Such variability highlights the need of building robust systems that can help to find the best treatment options based either on a viral or a pharmacoviral approach for each specific patient. In this light, PDAC organoids are now being researched as a valuable tool for drug screening and for predicting clinical responses to treatments [5,10,11].
In the current work, we explored the feasibility of using PDAC organoid models as a platform to screen for selectivity and potency of OA anticancer therapies as a single agent or in combination with standard chemotherapeutics.
2. Materials and methods
2.1. Human specimens
Samples from PDAC and adjacent normal pancreatic parenchyma were obtained after surgery at Hospital Clinic, Barcelona, or at the University Medical Centre Utrecht Hospital. Experiments with human samples were approved by all necessary ethical committees. Written informed consent was obtained from all patients prior to acquisition of samples in accordance with the Declaration of Helsinki. Samples were confirmed to be tumour or normal based on pathologist assessment.
2.2. Organoid generation and maintenance
Tissues were processed following established protocols [5,12]. Briefly, fresh tissues were minced in small fragments and digested with collagenase and dispase II at 0.125 mg/ml in wash medium (DMEM 1% FBS, 1% penicillin/streptomycin) with gentle shaking at 37 °C. When ducts became visible, digestion was stopped by adding cold wash medium and centrifuged 300 g for 5 min. The cell pellet was plated in matrigel and grown in complete medium (advanced DMEM/F12 supplemented with: 1% penicillin/streptomycin, 1% glutaMAX, 1% HEPES 10 mM, N-acetylcysteine (10 mM), nicotinamide (1.25 mM), B27 (final concentration 1 ×), FGF-10 (100 ng/ml), gastrin I (100 nM), hEGF (50 ng/ml), A83-01 (500 nM), primocin (50 µg/ml), mNoggin conditioned medium (10% final volume), R-spondin1 conditioned medium (10% final volume), and Wnt3a-conditioned medium (50% final volume)). The remaining undigested tissue was treated for 10 min at 37 °C with the dissociating agent TrypLE Select (Gibco) and further processed as described above. After organoid generation, growth medium was changed every 4 days, and organoids were split by mechanical disaggregation when the appropriate confluence was reached. Organoids from orthotopic tumour xenografts and metastases in Foxn1nu/nu mice (ENVIGO, RRID: MGI:2174843) were generated with the same protocol.
2.3. Adenoviruses
Adenoviruses used in the study and previously described were: AdGFPLuc, a replication–defective adenovirus that expresses the green fluorescent protein (GFP) and the luciferase (luc) genes under the control of the cytomegalovirus promoter (CMV) [13]. The replication-competent adenoviruses: AdNuPARE1A, ICOVIR15, ICOVIR15-miR99b, ICOVIR15-miR485, and Adwt-E [14], [15], [16]. AdNuPARE1A incorporates an uPAR minimal promoter and 3 × SPS sequences recognizing the binding domain of the CSL transcription factor characteristic of Notch-responsive genes regulating E1A transcription [15]. ICOVIR15 presents a 24-bp deletion in the E1A region and incorporates E2F-responsive elements to redirect E1A transcription toward deregulation of the Rb/p16 pathway. The fibre protein contains an RGD motif inserted in the HI-loop region [14]. ICOVIR15-miR99b and ICOVIR15-miR485 contain the miR-99b or miR-485 genomic sequences, respectively, under the CMV promoter next to the R-ITR in the ICOVIR15 backbone [16]. Adwt-E additionally expresses enhanced-GFP (E) [16].
AdNuPARE1A-E and ICOVIR15-E were generated in the current study. The E gene was introduced downstream of the adenoviral fibre at the adenoviral genomes (pAdNuPARE1A or pICOVIR15), and viruses were produced as previously reported [16].
2.4. KRAS mutation analysis
Genomic DNA was extracted from PDAC organoids using the Blood DNA Isolation Mini Kit (Norgen Biotek) following manufacturer's protocol. The mutational status of KRAS was analysed by PCR amplification of exon 2 with the indicated primers (Table S1). PCR products were Sanger sequenced by GeneWiz® (RRID:SCR_003177) and mutations were analysed using Unipro UGENE software (RRID:SCR_005579), the NBLAST platform (RRID:SCR_015884) and Benchling (RRID:SCR_013955). Allele frequencies were calculated using the Mutation Quantifier Tool of Mutation Surveyor Software v5.1.2 (Softgenetics®, RRID:SCR_001247).
2.5. Histological analysis of tumour tissues and organoids
Tumour tissues were fixed overnight in 4% paraformaldehyde (PFA), embedded in paraffin, and sectioned at 5 µm for haematoxylin and eosin (H&E) staining and for immunohistochemistry (IHC).
Organoids were harvested for inclusion following the protocol in Broutier et al., 2016 [12]. Briefly, wells were washed with 1 ml of PBS, Matrigel drops were collected in 15 mL centrifuge tubes containing 10 ml of cold PBS on ice, and organoids were settled by gravity and washed. Subsequently, 4% PFA (4 ml) was added to the organoids and incubated for 30 min. Fixative was removed, samples were washed with PBS for 10 min 3 ×, and organoids were resuspended in 2% agar for paraffin embedding and sectioned at 5 µm.
For haematoxylin and eosin (H&E), tissue or organoid sections were deparaffinized, hydrated, and stained following standard protocols at Biobank of Hospital Clínic (RRID:SCR_004530). For immunohistochemistry (IHC), hydrated samples were incubated in sodium citrate buffer (pH 6.0, 10 mM) at 95° for 5 min under pressure. Sections were blocked with PBS-Triton 0.3% + 10% FBS + 1% BSA for 90 min, washed 3 × with PBS, and incubated overnight with primary antibodies against cytokeratin 19 (Anti-CK19, Abcam 52,625, RRID:AB_2,281,020, diluted 1:500 in PBS 0.1% BSA) or E1A (Santa Cruz Biotechnology sc-25 RRID:AB_626,645, diluted 1:200 in PBS 0.1% BSA). Endogenous peroxidases were blocked with Dual Endogenous Enzyme Block (Dako) for 10 min. Reactions were developed with Dako EnVision + Dual Link System-HRP (DAB+) (Dako). Sections were counterstained with Harris haematoxylin (Panreac), and samples visualized using a NIKON e600 inverted microscope.
2.6. mRNA quantification
Total RNA was extracted from patient tissues and organoids using miRNeasy Mini Kit (Qiagen). Samples were treated with TURBO-DNA free Kit (Invitrogen) and quantified with a Nanodrop 1000 spectrophotometer (RRID:SCR_018035). RNA (500 ng) was retrotranscribed to cDNA using PrimeScript™ RT Reagent Kit (Takara). Quantitative PCR (qPCR) was performed using specific primers (Table S1) and lightCycler® 480 SYBR Green I Master (Roche) in a Via ViiA 7 Real-Time PCR System (Applied Biosystem). Relative gene expression levels were quantified using the delta CT method, normalized to the HPRT gene and represented as 2ΔCt.
2.7. EGFP detection
IDIT4 organoids were infected with AdGFPLuc or Adwt-E at 50,000 pfu/well. IDIT1 and IDIS0 organoids were infected with 5000 pfu/well of Adwt-E, AdNuPARmE1A-E, or ICOVIR15-E. EGFP fluorescence was visualized in the Olympus IX51 microscope (at a wavelength of 480 nm). EGFP signals of individual organoids at each timepoint were quantified using the ImageJ software (RRID:SCR_003070) and expressed as mean% of stained area ± SEM.
2.8. Viral yield
IDIT4 organoids were infected with AdGFPLuc or Adwt-E at 50,000 pfu/well for 5 days. Infected cells were collected and centrifuged, and the cell pellet was resuspended in DMEM/F12 medium. Cells underwent three freeze / thaw cycles. Lysates were centrifuged at 600 g for 5 min, and medium containing viral particles was harvested (passage 1, P1). Fifteen µL of this medium were used to infect IDIT4 organoids for 5 days (passage 2, P2). This procedure was repeated to generate passage 3 (P3). Adenoviral genomes were extracted from media using Blood DNA Isolation Mini Kit (Norgen Biotek), following the manufacturer's protocol, and quantified by qPCR using primer sets for hexon (Table S1). The number of viral genomes in the different samples was determined by the interpolation of the crossing–threshold with a standard curve of a known concentration.
2.9. Viral entry
IDIT1, IDIT5, and IDIT6 organoids were enzymatically disaggregated by incubating with TrypLE™ Select (Gibco) at 37 °C for 10 min. Organoids were counted, and 50,000 cells were collected by centrifugation. Pelleted organoids were resuspended in 20 μl of F12 medium and incubated either with AdNuPARmE1A or ICOVIR15 at a dose of 5 pfu/cell for 30 min at 37 °C. Following incubation, organoids were washed twice with PBS followed by centrifugation. Organoid pellets were then resuspended in 200 μl of PBS, and DNA was extracted using the Blood DNA Isolation Mini Kit (Norgen Biotek) following the manufacturer's protocol. The amount of adenoviral and cellular genomes was quantified by qPCR using primer sets for hexon and albumin genes, respectively (Table S1). The number of viral genomes per cell in the different samples was determined by the interpolation of the crossing–threshold with a standard curve of known concentrations.
2.10. Cytotoxicity assays
After organoid dissociation, 5000 cells/well were incubated in triplicate with the different viruses at the indicated doses for 30 min at 37 °C. Tubes were transferred to ice, and the medium containing infected organoids (20% of final volume) was supplemented with Matrigel (80% of final volume), mixed, and seeded in 10 μl drops in triplicate in 96 multi-well plates. Once the Matrigel solidified, fresh growth medium was added, and cultures were incubated for 5 days at 37 °C. For the combined treatments, drugs were added to the medium at the following concentration: nab-paclitaxel (nP, Abraxane) (100 nM), and gemcitabine (2.5 ng/ml) alone or with the adenovirus AdNuPARmE1A (at 5000 pfu/well).
Cell viability was measured at 5 days after treatment, with an adapted colorimetric MTT assay (Affymetrix, USB® Products) as described in [17]. Briefly, MTT solution at 0.75 mg/ml was added for 1 h at 37 °C and replaced with 40 µl of 2% SDS for 2 h to solubilize Matrigel. Finally, DMSO was added to dissolve crystal precipitates. Absorbance was measured at 550 nm in a spectrophotometer (Synergy HT – Biotek). Results are expressed as a percentage of viability relative to non-infected control cells (100%).
Cytotoxicity was also measured by a clonogenic assay. Five days after treatment, organoids were harvested, mechanically disaggregated, and split in a new 96 multi-well plate at a 1:2 dilution. The following day, viable organoids showing a round shape, a visible epithelial layer, and a diameter ≥ 20 μm were counted under a microscope (Olympus IX51). Results are represented as the number of viable organoids/well, with non-infected organoids (MOCK) as internal control.
2.11. In vivo antitumoural activity
Animal procedures met the guidelines of European Community Directive 86/609/EEC, were approved by the Local Ethical Committee, and were performed by trained, certified personnel. Subcutaneous tumours were generated by injecting a suspension of IDIT1 organoids (500,000 cells/tumour) with Matrigel (Bioscience) at a 1:1 ratio, in each flank of Foxn1nu/nu mice (RRID: MGI:3,848,172; ENVIGO). Tumours were monitored twice a week, and volumes were calculated using the formula V = D × d2 × π ÷ 6. When the tumour mean size reached 100 mm3, mice were randomly allocated to treatment groups and intravenously injected with a single dose of saline or 5 × 1010 vp/animal (using AdNuPARmE1A or ICOVIR15).
2.12. In vivo metastatic model
Organoids (500,000 cells/animal) were resuspended in PBS and Matrigel (ratio 1:1, for each tumour). The orthotopic engraftment was performed as previously described [18,19]. Briefly, 100 µl of the cell suspension was injected to the tail region of the pancreas with a 29 G syringe. Animals were monitored once a week by tumour palpation, and mice were euthanized 2–3 months after implantation. Primary tumours and metastases were then collected and processed for organoid generation and/or histological analysis.
2.13. Organoid karyotyping
Confluent organoids were treated with 0.1 µg/ml colcemid (Gibco KaryoMAX Colcemid Solution) for 24 h. Organoids were then dissociated to single cells by incubation with TrypLE Select (Gibco) at 37 °C for 10 min. Cells were washed with PBS, incubated with prewarmed KCl (0.0075 M) (hypotonic solution) for 40 min, and fixed with methanol:glacial acetic acid (3:1). Samples were dropped onto microscope slides, mounted with ProlongTM Gold Antifade Reagent with DAPI (Invitrogen), and visualized under a Nikon Eclipse 50i fluorescence microscope. Metaphases were captured and counted using the Isis software. A minimum of 15 metaphases per sample were counted.
2.14. Statistical analysis
Experimental data are presented as mean ± SEM of at least three independent experiments. Statistical analyses were performed using GraphPad Prism software v5.0a (RRID:SCR_002798). Statistical differences were evaluated using a two-tailed non-parametric Mann–Whitney test, or a Student's T-test, and p < 0.05 was taken as the level of significance.
Differences in the in vivo tumour growth were analysed using the R v.2.14.1 software (RRID:SCR_001905) and by applying a linear mixed-effect model using the lme4 package (RRID:SCR_015654). Statistical differences were evaluated using a multiple comparison using Tukey contrast; p < 0.05 was taken as level of significance.
3. Results
3.1. Adenoviruses infect and replicate in PDOs
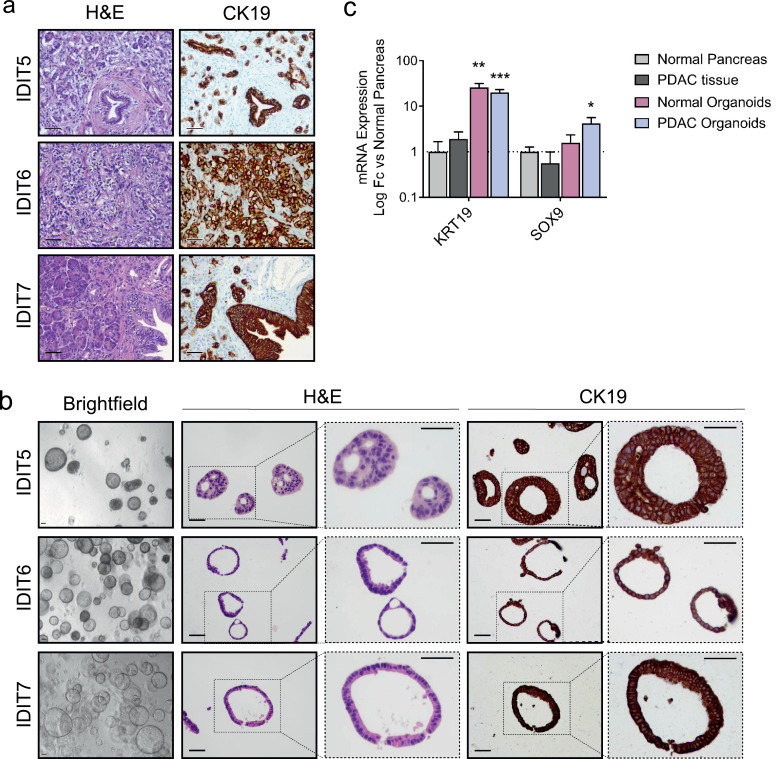
PDOs were established from normal or tumour tissue collected at the time of surgical resection according to published protocols [5]. Histological evaluation of tumour organoids revealed phenotypic characteristics of patient specimens. H&E staining of whole-mount organoids showed morphologic similarities to primary tumours, such as dysplastic cell organization and nuclear irregularities (Fig. 1a and b). CK19 immunohistochemical staining showed strong immunoreactivity in the cytoplasm and cell membrane of ductal cells in both primary tumours and established organoids (Fig. 1a and b). Confirmation of the ductal origin of organoids was evidenced by an enriched expression of the ductal markers KRT19 and SOX9 (Fig. 1c).
Fig. 1.
Human PDAC-derived organoids maintain characteristics of their tumour-of-origin and are enriched in ductal markers. (a) Representative histological sections of human pancreatic tumours. Left, H&E staining; right, CK19 IHC staining (scale bar 50 µm). (b) PDAC-derived organoids from matching tumours. Left, brightfield images (scale bar 50 µm); centre, H&E staining; right, CK19 IHC staining (scale bar 50 µm). (c) qRT-PCR analysis of ductal lineage markers (KRT19 and SOX9) from PDAC organoids (n = 7), normal pancreas organoids (n = 4), tumour tissue-of-origin (n = 4), and adjacent normal pancreas (n = 6). Data are represented as mean ± SEM; *p < 0.5, **p < 0.01, ***p < 0.05, non-parametric Mann–Whitney U test.
The tumourigenic origin of the established organoids was further confirmed through the identification of oncogenic mutations in the KRAS codon 12 in 7 out of 7 tumour organoids, in line with reported mutation frequencies at this hotspot (Table 1). One of the seven organoids (IDIT3) was homozygous for the mutant allele. No mutations were detected in the KRAS gene of normal organoids (Fig. S1).
Table 1.
KRAS allele frequency for the organoids used in the study. The allele frequency was estimated with the “Mutation Quantifier Tool” of Mutation Surveyor Software v5.1.2 (Softgenetics®).
| Organoid ID | KRAS status | Mutation | AA change | WT allele ratio | Mut allele ratio |
|---|---|---|---|---|---|
| IDIT1 | Mut | 12:25398285(-): G/C | G12R | 43.3% | 56.7% |
| IDIT2 | Mut | 12:25398285(-): G/A | G12R | 58.4% | 41.6% |
| IDIT3 | Mut | 12:25398284(-): G/T | G12V | 0% | 100% |
| IDIT4 | Mut | 12:25398284(-): G/A | G12D | 44.6% | 55.4% |
| IDIT5 | Mut | 12:25398284(-): G/A | G12D | 53% | 47% |
| IDIT6 | Mut | 12:25398284(-): G/A | G12D | 44.4% | 56.6% |
| IDIT7 | Mut | 12:25398284(-): G/T | G12V | 52.7/% | 47.3% |
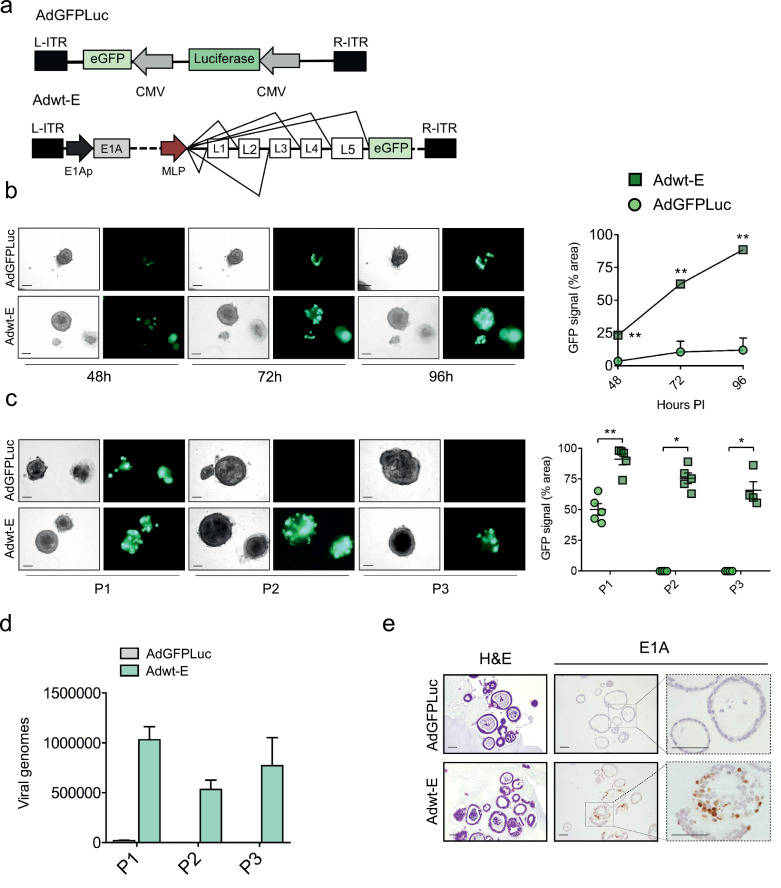
To investigate whether adenoviruses could infect and replicate in PDOs, tumour organoids were exposed to 5 × 105 pfu/well of the reporter adenovirus (AdGFPLuc) or the replication-competent adenovirus engineered with EGFP (Adwt-E) (Fig. 2a). Outer layers of organoid cultures showed susceptibility to adenoviral infection, as shown by GFP expression, in line with previous evidence [20] (Figs. 2b and c). Notably, whereas GFP signal was stable at all time-points analysed after AdGFPLuc infection, the fluorescence signal observed after Adwt-E infection significantly increased over time. GFP expression observed in the inner layers of organoids infected with Adwt-E suggested replication and virus spreading in PDOs (Fig. 2b). Evidence of viral replication was further demonstrated by incubating naïve organoids with cell extracts from Adwt-E or AdGFPLuc-infected organoids for consecutive passages. GFP expression in P2 and P3 passages was only observed in cultures that had been treated with the replication-competent virus (Fig. 2c). Additional confirmation came from quantification of the viral yield in each of the three passages. Viral genomes ranging between 5 × 105–1 × 106 were detected in Adwt-E organoids, whereas only 1 × 104 viral genomes were measured in the first passage of AdGFPLuc infected cultures, coinciding with the input dose (Fig. 2d). Positive E1A immunostaining confirmed viral replication of Adwt-E, showing virus spreading within organoids, while no staining was detected in the AdGFPLuc-infected PDOs (Fig. 2e).
Fig. 2.
Adenoviruses efficiently replicate in PDAC organoids. (a) Schematic representation of adenoviruses used in the experiment: the reporter non-replicative adenovirus AdGFPLuc and the replicative-competent Adwt-E, both expressing GFP. (b) Time-course GFP expression in tumour organoids infected with 5 × 105 pfu/well of AdGFPLuc or Adwt-E. Left panel shows representative images of IDIT4 organoids at the indicated time-points after infection (scale bar 100 μm). Right panel represents GFP quantification. Values are expressed as percentage of fluorescent area at each time point, normalized by total area of single organoids. Data are represented as mean ± SEM, (n > 3); **p < 0.01, ***p < 0.05 Non-parametric Mann-Whitney U Test. (c) Adenoviral propagation through passages. Organoids were infected with 5 × 105 pfu/well of AdGFPLuc or Adwt-E and 5 days later (P1) were harvested, followed by three cycles of freeze and thaw. Supernatants were used to infect fresh organoids and 5 days later (P2) the same procedure was repeated to generate P3. GFP expression was visualized at P1, P2 and P3 in organoids infected with Adwt-E, but only at P1 in AdGFPLuc infected organoids. Left panel shows representative images of organoids at P1, P2, P3 (scale bar 100 μm). Right panel represents GFP quantification at each passage. Values are expressed as percentage of fluorescent area for each time point, normalized by total area of single organoids. Data are represented as mean ± SEM, n > 3; * p < 0.05, ** p < 0.01, Non-parametric Mann-Whitney U Test. (d) Quantification of viral genomes content in P1, P2 and P3 organoid passages infected with AdGFPLuc or Adwt-E (n = 3). e. Histological sections of IDIT4 organoids infected with AdGFPLuc or Adwt-E. H&E (left panel), E1A immunostaining (right panel), scale bar 50 μm.
3.2. Oncoselective adenoviruses display viral attenuation in organoids from healthy pancreas
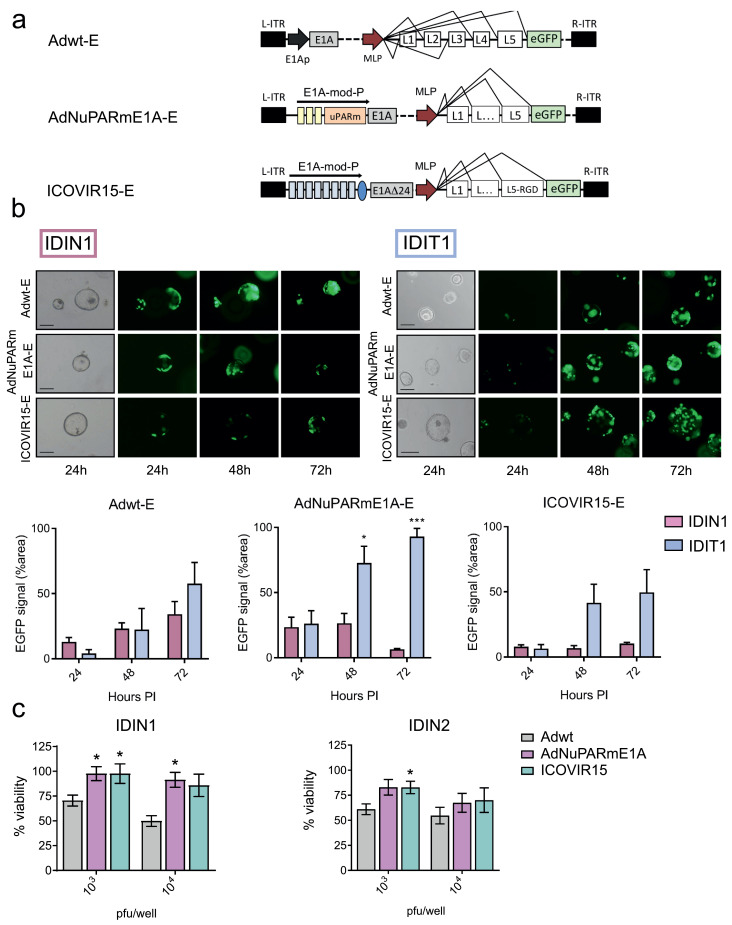
Next, we investigated the potential of using PDOs for testing oncolytic adenovirus selectivity. To this end, normal and tumour organoids from PDAC patients were infected with the oncolytic adenoviruses AdNuPARmE1A-E or ICOVIR15-E and the control, non-selective virus, Adwt-E (Fig. 3a). GFP expression was monitored at 24 h, 48 h, and 72 h after infection in individual organoids. GFP expression in PDOs from PDAC tumours similarly increased with time, independently of the virus used, according to viral replication and spread within the organoid (Fig. 3b). Notably, whereas GFP expression expanded in normal organoids exposed to Adwt-E, infection with oncolytic adenoviruses displayed similar GFP expression at all time points assessed, indicating the lack of replication of either AdNuPARmE1A-E or ICOVIR15-E in normal tissue (Fig. 3b). To further assess the use of normal organoids to screen for the tumour-specificity of oncolytic adenoviruses, we tested the cytotoxic effects of Adwt, AdNuPARmE1A, or ICOVIR15 in IDIN1 and IDN2 normal organoids, established from independent patients. In IDIN1, oncolytic viruses did not trigger cytotoxicity at the doses tested, whereas Adwt resulted in 30% and 50% organoid lethality. A similar tendency was observed in IDIN2, although some lethality appeared with AdNuPARmE1A and ICOVIR15 (Fig. 3c). These different behaviours could be due to the fact that normal organoids from adjacent tumours may be in an intermediate state between tumour and healthy tissue, despite not having any genetic alterations nor displaying a normal appearance [21]. In this scenario, oncolytic viruses might be able to trigger a certain degree of cytotoxicity. Altogether, these results support the oncoselective activity of AdNuPARmE1A-E and ICOVIR15-E,and validate PDOs as a system for studying oncoselectivity.
Fig. 3.
Oncolytic adenoviruses selectively replicate in PDAC derived organoids. (a) Schematic representation of Adwt-E, AdNuPARmE1A-E and ICOVIR15-E adenoviruses. Yellow boxes in AdNuPARmE1A-E represent SPS sequences sensitive to NOTCH activation. Blue boxes in ICOVIR15-E correspond to E2F-responsive elements. (b) Time-course GFP expression in IDIN1 (n = 3) normal organoids and IDIT1 (n = 3) tumour organoids, infected with 5 × 103 pfu/well of either adenoviruses. (Upper panels). Representative images of GFP expression in IDIT1 and IDIN1 at 24 h, 48 h, and 72 h after infection. (Lower panels). GFP quantification of organoids cultures infected with the different viruses. Values are expressed as percentage of fluorescent area for each time point, normalized for the area of single organoids. Data are represented as mean ± SEM, n ≥ 3; * p < 0.05, **p < 0.01, Student's t-test. (c) In vitro cytotoxicity of Adwt, AdNuPARmE1A and ICOVIR15 in normal pancreas organoids (IDIN1 and IDIN2). The indicated viruses have the same backbone as described in (a) but lacked the GFP gene, as shown in Fig. 4a. IDIN1 and IDIN2 organoids derived from two different patients were infected with the indicated doses and 5 days later, cell viability was assessed by MTT assay. Data are represented as mean ± SEM, n = 5; *p < 0.05, non-parametric Mann-Whitney U Test.
3.3. PDOs from PDAC exhibit different sensitivities to oncolytic adenoviruses
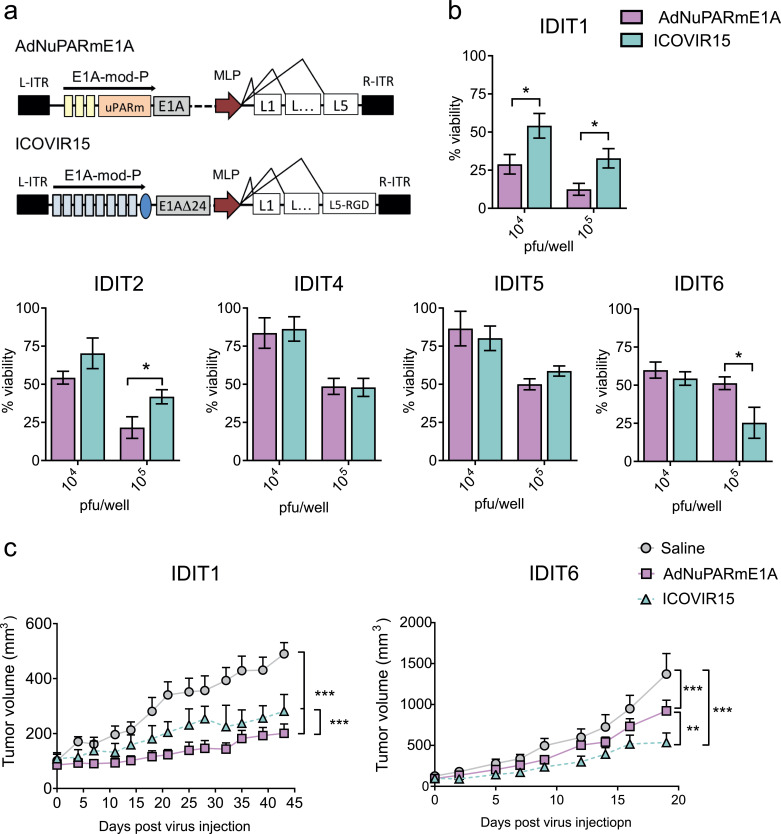
To determine whether the organoid model could be used to screen oncolytic adenoviruses, we tested the sensitivity of tumour organoids derived from five patients (IDIT1, IDIT2, IDIT4, IDIT5, and IDIT6) to two different oncolytic adenoviruses, AdNuPARmE1A and ICOVIR15 (Fig. 4a). Organoids were exposed to different doses of viral particles for 5 days. Cell viability at each dose and time point was measured by an adapted MTT assay [17]. Both AdNuPARmE1A and ICOVIR15 induced cytotoxicity to the cultures in a dose-dependent manner. We observed a heterogeneous response in terms of sensitivity to viral oncolysis, with ID50 values that ranged from 1 × 103 for the most sensitive (IDIT1), to 1 × 105 for the less sensitive organoids (IDIT4 and IDIT5) (Fig. 4b and Fig. S2a). Individual preference for a single virus could be observed in 3 out of 5 tumour organoids. The IDIT1 sample displayed a better response to AdNuPARmE1A, with an ID50 of 1 × 103 pfu/well, making it 10-fold more potent than ICOVIR15 (ID50 of about 1 × 104 pfu/well) (Fig. 4b. Fig S2). IDIT2 also had a 10-fold increased sensitivity for AdNuPARmE1A, although it was less permissive to oncolysis than IDIT1 (ID50 of 1 × 104 pfu/well for IDIT2 versus of 1 × 103 pfu/well for IDIT1). AdNuPARmE1A and ICOVIR15 triggered similar cytotoxicity levels in IDIT4 and IDIT5 organoids, which were the most resistant to oncolytic therapy, with an ID50 to either virus of 1 × 105 pfu/well. Interestingly, ICOVIR15 generated a better response in IDIT6 organoids (Fig. 4b). The different sensitivity of organoids to oncolysis, and to AdNuPARmE1A or ICOVIR15, was further confirmed with an organoid-formation assay (Fig. S2b).
Fig. 4.
PDAC organoids display heterogeneity in sensitivity to oncolytic adenoviruses. (a) Schematic representation of the oncolytic adenoviruses. (b) In vitro cytotoxicity of AdNuPARmE1A and ICOVIR15 in a battery of PDAC organoids. Organoids were infected with the indicated doses and 5 days later, cell viability was assessed by MTT assay. Data are represented as mean ± SEM, n ≥ 4; *p < 0.05, **p < 0.01, non-parametric Mann-Whitney U test. (c) In vivo antitumour effects of oncolytic adenovirus in IDIT1 and IDIT6 subcutaneous tumours. Tumour-bearing mice were intravenously injected with 5 × 1010 vp/mice of AdNuPARmE1A, ICOVIR15 or saline solution, and tumour growth was monitored. n ≥ 6 tumours/group, **p < 0.01, ***p < 0.005, Tukey range test for multiple comparison.
To investigate whether oncolytic efficacy in organoids could predict the in vivo response to viruses, IDIT1 or IDIT6 organoids were subcutaneously injected into the flanks of nude mice. When tumours reached 100 mm3, animals received a systemic dose of saline solution or ICOVIR15 or AdNuPARmE1A (at 5 × 1010 viral particles (vp)/animal). Treatment with either virus strongly inhibited tumour growth, but the anti-tumour effect was significantly higher in animals receiving the AdNuPARmE1A virus in IDIT1 tumours, and in those receiving ICOVIR15 in IDIT6 tumours, consistent with the in vitro data (Fig. 4c).
To gain insight into the preferential sensitivity of IDIT1/IDIT2 for AdNuPARmE1A, or of IDIT6 for ICOVIR15, we examined their engineered viral genomes to identify candidate genes that could act as potential biomarkers. AdNuPARmE1A was engineered to control the E1A gene, the first gene expressed in the adenovirus replication cycle, by the uPAR gene promoter and NOTCH-responsive elements. Thus, the transcriptional activity of uPAR and NOTCH pathway genes in organoids could reflect the regulation of E1A, since these genes share regulatory sequences with AdNuPARmE1A [15,22]. We quantified the mRNA content of the uPAR, HES1, and NOTCH1 genes from the five organoid lines. We observed that IDIT1 and IDIT2 expressed the highest content of all three genes, whereas IDIT6 had the lowest expression of these genes of all organoids (Fig. S3a). These results suggest that the regulatory sequences from the AdNuPARmE1A controlling the E1A gene are highly activated in IDIT1/IDIT2, thereby favouring adenoviral replication in these organoids, whereas such activation is diminished in IDIT6. Adenoviruses initiate their infection by the recognition of the fibre protein with the host cellular receptors coxsackie adenovirus receptor (CAR) and integrins. Particularly, binding to integrins ανβ5 is required for Ad5 entry in cells expressing very low CAR [23]. ICOVIR15 was engineered with a fibre protein containing an RGD motif to further facilitate viral entry by binding to integrins. To understand potential differences in viral response in organoids, we analysed the expression of the RGD-recognizing integrins αν, β3, and β5. However, their expression levels did not reveal any profile that would explain an enhanced viral entrance of ICOVIR15 in IDIT6 organoids (Fig. S3b).
To further explore potential differences in viral entry and explain the heterogeneous sensitivity of organoids to oncolytic viruses, we quantified the number of viral genomes at 4 h after infection in the high-sensitive IDIT1 organoids, the low-sensitive IDIT5 organoids, and IDIT6 organoids, which exhibit low sensitivity for AdNuPARmE1A but high for ICOVIR15. We observed that the highest number of viral genomes was detected in IDIT1 organoids, with both viruses having very similar entrance capacity. IDIT5 and IDIT6 organoids showed reduced but similar entry for the two viruses (Fig. S4).
The similarities in viral entry observed between the two viruses also suggests that the in vivo tumour targeting by either virus in IDIT1 and IDIT6 xenografts would be comparable and that, therefore, the different antitumor response to ICOVIR15 and AdNuPARmE1A mainly depends on organoids’ features. Nevertheless, an in vivo scenario is much more complex, particularly upon systemic administration, where hepatic viral clearance rapidly removes the virus from the circulation. Thus, it could be speculated that the differences in the fibre protein of the two viruses could distinctly impact viral sequestration by the liver. However, viral circulation kinetics of ICOVIR15 and AdNuPARmE1A are most likely comparable since ablation of CAR and integrin binding have been shown not to affect liver tropism [24].
Thus, the increased sensitivity of IDIT1 organoids to AdNuPARmE1A might be explained by a combination of a high viral entry together with an active transcription of E1A, favoured by the uPAR and Notch-responsive elements. In contrast, the poor activity of the Notch pathway in IDIT6 organoids could impair AdNuPARE1A replication in this model.
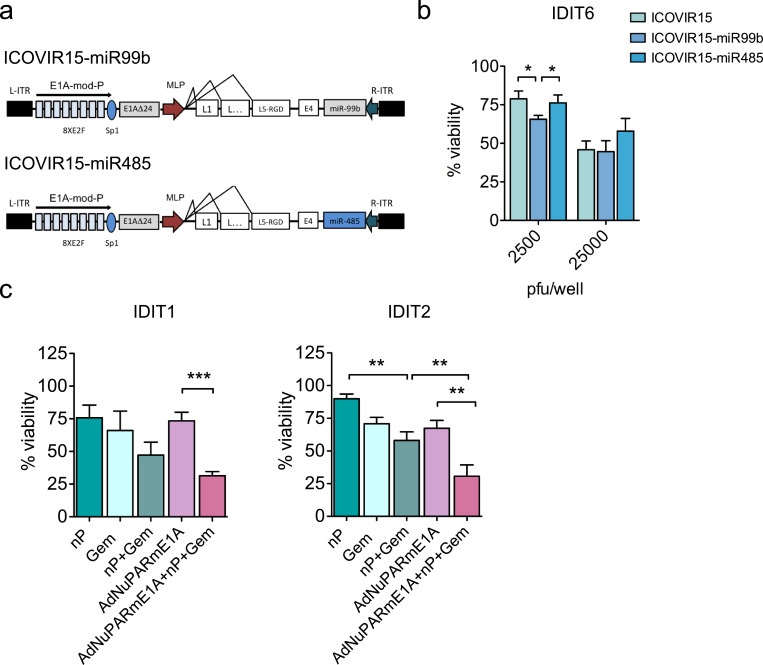
Previous work from our group has shown that ICOVIR15-miR-99b and ICOVIR-15-miR-485 exert enhanced antitumour responses in PANC-1 xenografts due to the role of miR-99b and miR-485 in regulating transcriptional repressors of viral proteins [16]. As IDIT6 organoids were sensitive to the parental ICOVIR-15, we explored the response of IDIT6 organoids to ICOVIR15-miR-99b or ICOVIR-15 miR-485; we observed statistically significant higher oncolysis with ICOVIR-miR99b (Figs. 5a and b). Note that the enhanced effect was only observed at low doses of infection yet is still relevant, as in virotherapy treatments, neoplastic cells may receive a repertoire of viral loads, but all cells infected with high or low viral loads will contribute to the final antitumour effect.
Fig. 5.
Organoids identify individual response to combined therapies or armed oncolytic adenoviruses. (a) Schematic representation of ICOVIR15-miR99b and ICOVIR15-miR485 oncolytic adenoviruses. (b) IDIT6 organoids were infected with parental ICOVIR15, ICOVIR15-miR99b, or ICOVIR15-miR485 at the indicated doses. Cytotoxicity was assessed 5 days later by MTT assay. Data are represented as mean ± SEM, n ≥ 4; *p < 0.05, non-parametric Mann–Whitney U test. (c) Combination treatments. IDIT1 and IDIT2 organoids were treated with 100 nM nab-paclitaxel and 2.5 ng/ml of gemcitabine, or AdNuPARmE1A (5000 pfu/well), or all in combination; after 5 days, cell viability was assessed by MTT assay. Data are represented as mean ± SEM; n ≥ 4, **p < 0.01, ***p < 0.005, non-parametric Mann–Whitney U test.
To ascertain the benefit of combining viral oncolysis and standard-of-care chemotherapeutic treatments, IDIT1 and IDIT2 organoids were exposed to nab-paclitaxel plus gemcitabine with or without AdNuPARmE1A, or AdNuPARmE1A alone, for 5 days. We observed differences in sensitivity to nab-paclitaxel plus gemcitabine, and the combined treatment with AdNuPARmE1A, in both organoid lines. Remarkably, in both PDOs, the highest cytotoxicity was achieved by combining nab-paclitaxel, gemcitabine, and the oncolytic virus (Fig. 5c). These results provide evidence of the feasibility of testing mixed viro-chemotherapy approaches in organoid models.
3.4. Organoids from orthotopic tumours and metastatic foci in mice mirror responses of PDO to oncolytic viruses
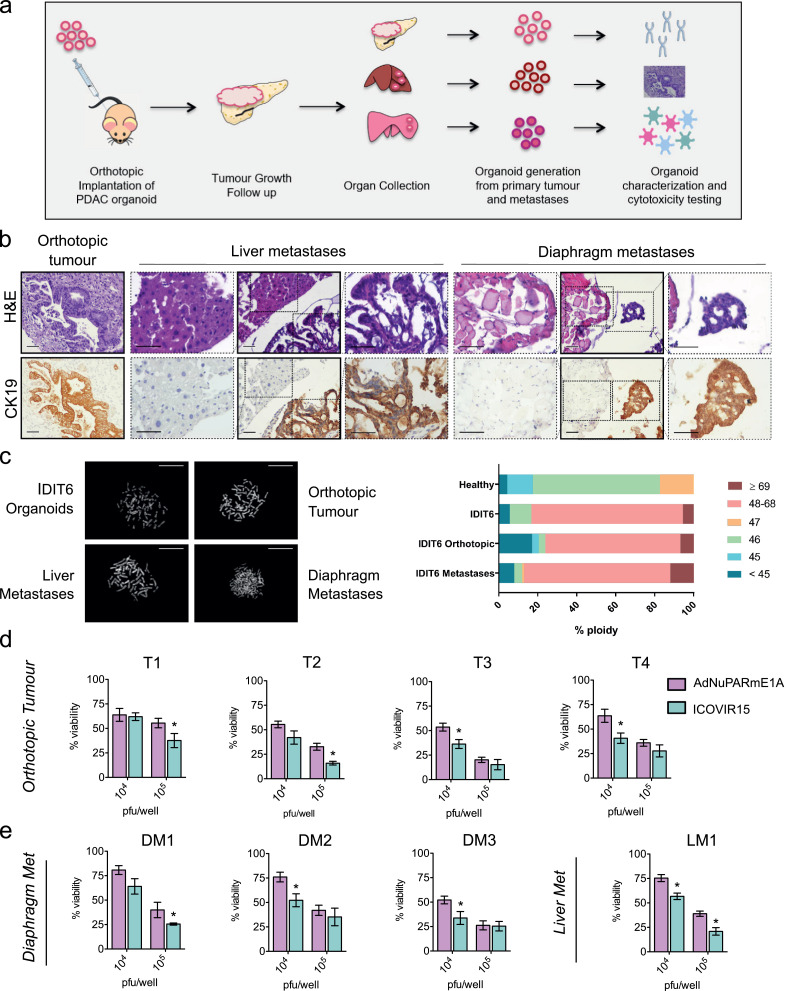
To test whether a candidate OA used on PDOs would reflect its activity both in primary tumours and metastatic foci, we generated orthotopic tumours from IDIT6 organoids. At three months (or sooner) following implantation, all animals developed distant metastases (Fig. 6a). Organoid engraftment generated pancreatic tumours with glandular structures that resembled human PDAC precursor lesions (e.g., pancreatic intraepithelial neoplasia, hPanIN), which were also identified in the metastatic foci (Fig. 6b). Karyotype analysis revealed that organoids from orthotopic tumours had a percentage of aneuploidy similar to IDIT6 PDOs, whereas organoids generated from metastases were highly aneuploid. In stark contrast, organoids derived from healthy pancreatic tissue mainly exhibited diploid karyotypes (Fig. 6c).
Fig. 6.
Primary tumours and metastases-derived organoids from PDOs implanted in nude mice mirror PDOs sensitivity to adenoviruses. (a) Schematic representation of the experiment. Six nude mice were orthotopically implanted with 5 × 105 cells. Tumour growth was followed by palpation, and after 2–3 months, animals were euthanized and organs were collected. New organoids were generated from the orthotopic tumours and their metastases and then further analysed. (b) Histological sections obtained from primary tumours and metastases stained for H&E (left) and CK19 (right); scale bar, 50 μm. (c) Karyotyping of IDIT6 original organoids and its derived orthotopic tumour and metastases. Metaphases were induced in the different lines via colcemid treatment (0.1 μg/ml) for 24 h. A minimum of 15 metaphases per sample was counted. Left panel, representative images of the different samples. Right panel, the percentage of ploidy in each group of samples. (d, e) Orthotopic tumours and metastases derived from IDIT6 organoids from four different mice were infected at the indicated doses with AdNuPARmE1A or ICOVIR15. After five days, cytotoxicity was analysed by MTT assay. Data are represented as mean ± SEM; n ≥ 5; *p < 0.05, non-parametric Mann–Whitney U test.
We examined the therapeutic sensitivity to AdNuPARmE1A or ICOVIR15 in organoids established from IDIT6 primary tumours of four independent mice: one liver metastasis and three diaphragmatic metastases. An analogous experiment was conducted on IDIT1-derived organoids from two primary tumours and a diaphragm metastasis. ICOVIR15 displayed enhanced cytotoxicity as compared to AdNuPARmE1A in organoids from all IDIT6-derived primary tumours and their metastatic foci, mirroring what we had observed in vitro in IDIT6 PDOs. (Fig. 6d and e). In contrast, organoids from primary tumours or metastases derived from IDIT1-derived organoids showed increased sensitivity to AdNuPARmE1A, again mirroring the response of their corresponding PDOs (Fig. S5).
These results show that oncolytic virotherapy acts in a similar manner in primary tumours and in metastases, despite increased genomic instability of the latter. Thus, tumour evolution does not seem to interfere with viral activity. These findings suggest that the predictive ability of PDOs to identify highly active oncolytic adenoviruses would facilitate the selection of successful treatments that could eradicate both primary tumours and metastases.
Discussion
Tumour organoids have been proposed as valuable tools for personalized drug testing [10,25,26]. Treatments in oncology are expanding, and novel therapies are continuously under development. Oncolytic viruses gained large interest in recent years, reaching late phases of clinical investigation [27], [28], [29], [30]. In the future, personalised virotherapy approaches have the potential to be used for identifying optimal treatment for individual patients. Three-dimensional cell cultures, such as spheroids or organotypic tumour slices, have been studied as an alternative to traditional 2D cultures to screen for viral therapies [31]. However, the inability to recapitulate the histological architecture in the spheroids, as well as the lack of long survival periods of organotypic cultures, have limited their use as screening platforms. Here we show that patient-derived organoids (PDOs) from PDAC are a promising model for evaluating both selectivity and sensitivity to oncolytic adenoviral therapies. We demonstrate for the first time that adenoviruses can replicate and expand in organoids, and that PDOs from individual patients display different sensitivities to oncolytic adenoviruses. This interpatient diversity in the response to adenoviruses aligned with differential expression of genes encoding cellular factors that regulate viral activity. For instance, the Notch-sensitive uPAR-regulated oncolytic adenovirus AdNuPARmE1A was more effective in PDOs with upregulated Notch pathway signalling and uPAR transcriptional activity. The observed variability in viral entry might also explain (at least in part) the differential responses of organoids to oncolysis. However, we could not associate this variability to the differential expression of membrane integrins in organoids, suggesting that other aspects besides viral internalisation might play a role. In fact, upon integrin receptor activation by viral RGD domains, a transient contribution of the endocytic machinery takes place to package a virus into clathrin-coated vesicles. Endosomal escape, cytoplasmic traffic, and transport at the nuclear pore are processes required to successfully deliver a viral genome into the nucleus of a cell. All of these are very tightly regulated functions, and it could be speculated that a variable degree of efficiency in each process might translate into differences in viral entry in organoids. This high level of biological complexity underscores the need for a personalised and easy-to-use tool for determining treatment responses, such as the one proposed here. Specifically, our findings open the possibility of using organoids as platforms to study viral tropism. This can contribute to better define tumour-specific targeting ligands in patient-derived models that encompass the full spectrum of the disease, generating targeted viruses with potentially improved therapeutic value.
The heterogeneous nature of cancers further underscores the need of using combination therapies. Our results validate the benefit of combining AdNuPARmE1A with the standard chemotherapeutic regimens in pancreatic cancer (such as gemcitabine and nab-paclitaxel) in sensitive PDOs. We recapitulated in organoids the previously reported synergistic effects in pancreatic cancer cell lines and PDX models [15,22]. These data suggest that organoids could serve to test the value of combined treatments for regimens of viruses and chemotherapeutics that have not been tested in vivo yet, in a personalised manner.
The tumour microenvironment shapes the response to anticancer therapies; thus, the lack of stromal components represents a strong limitation in current PDO systems. Until now, PDXs have been proposed for such screenings, but they are too time-consuming to support personalised therapy for patients with pancreatic cancer, especially considering the rapid disease progression at later stages. Moreover, the absence of an intact murine immune system limits the study of immunotherapies. This is why the development of PDX models in humanized mice with a fully competent human immune system is very attractive for evaluating the full potential of oncolytic virotherapy [32]. However, as these systems are very complex, the development of 3D co-cultures including PDOs, fibroblasts, and/or immune cells to model tumour complexity can open new avenues for evaluation of virotherapy responses alone or in combination treatments with chemotherapy, immunotherapeutic agents, or fibroblast-targeting drugs [33], [34], [35].
Notably, testing the OAs AdNuPARmE1A and ICOVIR15 in organoids derived from primary tumours or metastases in nude mice showed similar responses and sensitivity to PDOs. However, this might not be the case for all OAs, since preferential killing of high-metastatic versus low-metastatic cells has been reported for the replication-selective adenovirus vector OBP-401, in which E1A transcription is driven by the human telomerase reverse transcriptase (hTERT) [36]. Ideally, to elucidate the optimal adenoviral activity in metastatic nodules, OA screening should be performed in PDOs derived from patient metastases. However, in the absence of metastatic biopsies, our results provide an indication that OA screening in PDOs derived from patient primary tumours still may reflect their activity in metastatic foci. Of note, the relatively short time required to generate organoids and to assess OA response in PDOs suggest that a therapeutic candidate could be selected within a clinically meaningful time frame.
In conclusion, these results suggest that testing sensitivity to oncolytic adenoviruses alone or in combination with chemotherapy in PDOs is a reliable in vitro cancer model for the study of patient-specific responses. Considering the variety of oncolytic viruses under development and the upcoming armed viruses, with more potent antitumour activity, PDOs may represent an easy-to-handle platform to help make virotherapy-based, personalised treatment decisions.
Declaration of Competing Interest
SFB and HC declare that they hold a patent (WO2015/173425). All other authors declare no conflict of interest.
Acknowledgments
Acknowledgements
We thank Jordi Camps and Elena Asensio for the help with organoid karyotyping. This work was developed at the Centro Esther Koplowitz, Barcelona, Spain. We are indebted to the Banc de Tumors-Biobank core facility of the Hospital-Clínic-IDIBAPS for technical help (work supported by the Xarxa de Bancs de Tumors de Catalunya - XBTC). We also acknowledge the support of CERCA Programme/Generalitat de Catalunya.
Funding sources
GR and MRR are recipients of an FPI predoctoral contract (BES-2015-071612 and BES-2012-053726 respectively) from MINECO, and AM was recipient of an FPU predoctoral contract from Ministry of Education, Spain. This work was supported by grants to CF from the Spanish Ministry of Economia y Competitividad BIO2014-57716-C2-R, BIO2017-89754-C2-2R with partial support from the Generalitat de Catalunya SGR17/861. CIBERER and CIBERehd are initiatives of the ISCIII. The CF group was partially financed by the Instituto de Salud Carlos III (IIS10/00014) and co-financed by Fondo Europeo de Desarrollo Regional (FEDER); it also acknowledges the support of COST Action BM1204 EUPancreas and the Spanish Adenovirus Network (AdenoNet, BIO2015-68990-REDT).
Role of the funding source
The funders had no role in study design, data collection, interpretation, or the decision to submit the work for publication
Author contributions
CF and GR conceived and designed the experiments. GR conducted most of the in vitro and in vivo experiments. MR-R contributed with the generation of adenoviral genomes. AM-B established organoids cultures in the lab. SB trained AM-B and helped to establish organoids generation. SB and HC provided some organoid samples. SP helped with organoid karyotyping and characterization. MC and HB helped with histological analysis. CF and SS-C provided surgical specimens. CF supervised the project and wrote the manuscript, with input from all other authors. HC designed experiments and revised the manuscript.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2020.102786.
Appendix. Supplementary materials
References
- 1.Kleeff J., Korc M., Apte M., La Vecchia C., Johnson C.D., Biankin A.V. Pancreatic cancer. Nat Rev Dis Prim. 2016;2:16022. doi: 10.1038/nrdp.2016.22. [DOI] [PubMed] [Google Scholar]
- 2.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69:7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 3.Kersten K., Visser K.E., Miltenburg M.H., Jonkers J. Genetically engineered mouse models in oncology research and cancer medicine. EMBO Mol Med. 2017;9:137–153. doi: 10.15252/emmm.201606857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boj S.F., Hwang C.-.I., Baker L.A., Engle D.D., Tuveson D.A., Clevers H. Model organoids provide new research opportunities for ductal pancreatic cancer. Mol Cell Oncol. 2016;3 doi: 10.1080/23723556.2015.1014757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boj S.F., Hwang C.-.I., Baker L.A., Chio C., II, Engle D.D., Corbo V. Organoid models of human and mouse ductal pancreatic cancer. Cell. 2015;160:324–338. doi: 10.1016/j.cell.2014.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tuveson D., Clevers H. Cancer modeling meets human organoid technology. Science (80-) 2019;364:952–955. doi: 10.1126/science.aaw6985. [DOI] [PubMed] [Google Scholar]
- 7.Hajda J., Lehmann M., Krebs O., Kieser M., Geletneky K., Jäger D. A non-controlled, single arm, open label, phase II study of intravenous and intratumoral administration of ParvOryx in patients with metastatic, inoperable pancreatic cancer: parvOryx02 protocol. BMC Cancer. 2017;17:576. doi: 10.1186/s12885-017-3604-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sato-Dahlman M., Wirth K., Yamamoto M. Role of Gene Therapy in Pancreatic Cancer—A Review. Cancers (Basel) 2018;10:103. doi: 10.3390/cancers10040103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eissa I., Bustos-Villalobos I., Ichinose T., Matsumura S., Naoe Y., Miyajima N. The current status and future prospects of oncolytic viruses in clinical trials against melanoma, glioma, pancreatic, and breast cancers. Cancers (Basel) 2018;10:356. doi: 10.3390/cancers10100356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang L., Holtzinger A., Jagan I., BeGora M., Lohse I., Ngai N. Ductal pancreatic cancer modeling and drug screening using human pluripotent stem cell– and patient-derived tumor organoids. Nat Med. 2015;21:1364–1371. doi: 10.1038/nm.3973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tiriac H., Belleau P., Engle D.D., Plenker D., Deschênes A., Somerville T.D.D. Organoid profiling identifies common responders to chemotherapy in pancreatic cancer. Cancer Discov. 2018;8:1112–1129. doi: 10.1158/2159-8290.CD-18-0349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Broutier L., Andersson-Rolf A., Hindley C.J., Boj S.F., Clevers H., Koo B.-.K. Culture and establishment of self-renewing human and mouse adult liver and pancreas 3D organoids and their genetic manipulation. Nat Protoc. 2016;11:1724–1743. doi: 10.1038/nprot.2016.097. [DOI] [PubMed] [Google Scholar]
- 13.Bayo-Puxan N., Cascallo M., Gros A., Huch M., Fillat C., Alemany R. Role of the putative heparan sulfate glycosaminoglycan-binding site of the adenovirus type 5 fiber shaft on liver detargeting and knob-mediated retargeting. J Gen Virol. 2006;87:2487–2495. doi: 10.1099/vir.0.81889-0. [DOI] [PubMed] [Google Scholar]
- 14.Rojas J.J., Guedan S., Searle P.F., Martinez-Quintanilla J., Gil-Hoyos R., Alcayaga-Miranda F. Minimal RB-responsive E1A promoter modification to attain potency, selectivity, and transgene-arming capacity in oncolytic adenoviruses. Mol Ther. 2010;18:1960–1971. doi: 10.1038/mt.2010.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mato-Berciano A., Raimondi G., Maliandi M.V., Alemany R., Montoliu L., Fillat C. A NOTCH-sensitive uPAR-regulated oncolytic adenovirus effectively suppresses pancreatic tumor growth and triggers synergistic anticancer effects with gemcitabine and nab-paclitaxel. Oncotarget. 2017;8:22700–22715. doi: 10.18632/oncotarget.15169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rovira-Rigau M., Raimondi G., Marín M.Á., Gironella M., Alemany R., Fillat C. Bioselection reveals miR-99b and miR-485 as enhancers of adenoviral oncolysis in pancreatic cancer. Mol Ther. 2019;27:230–243. doi: 10.1016/j.ymthe.2018.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grabinger T., Luks L., Kostadinova F., Zimberlin C., Medema J.P., Leist M. Ex vivo culture of intestinal crypt organoids as a model system for assessing cell death induction in intestinal epithelial cells and enteropathy. Cell Death Dis. 2014;5:e1228. doi: 10.1038/cddis.2014.183. –e1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jose A., Sobrevals L., Ivorra A., Fillat C. Irreversible electroporation shows efficacy against pancreatic carcinoma without systemic toxicity in mouse models. Cancer Lett. 2012;317:16–23. doi: 10.1016/j.canlet.2011.11.004. [DOI] [PubMed] [Google Scholar]
- 19.Kim M.P., Evans D.B., Wang H., Abbruzzese J.L., Fleming J.B., Gallick G.E. Generation of orthotopic and heterotopic human pancreatic cancer xenografts in immunodeficient mice. Nat Protoc. 2009;4:1670–1680. doi: 10.1038/nprot.2009.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang N., Zhang H., Zhang B.-.Q., Liu W., Zhang Z., Qiao M. Adenovirus-Mediated Efficient Gene Transfer into Cultured Three-Dimensional Organoids. PLoS ONE. 2014;9:e93608. doi: 10.1371/journal.pone.0093608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aran D., Camarda R., Odegaard J., Paik H., Oskotsky B., Krings G. Comprehensive analysis of normal adjacent to tumor transcriptomes. Nat Commun. 2017;8:1077. doi: 10.1038/s41467-017-01027-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maliandi M V., Mato-Berciano A., Sobrevals L., Roue G., Jose A., Fillat C. AduPARE1A and gemcitabine combined treatment trigger synergistic antitumor effects in pancreatic cancer through NF-kappaB mediated uPAR activation. Mol Cancer. 2015;14:146. doi: 10.1186/s12943-015-0413-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lyle C., McCormick F. Integrin alphavbeta5 is a primary receptor for adenovirus in CAR-negative cells. Virol J. 2010;7:148. doi: 10.1186/1743-422X-7-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martin K., Brie A., Saulnier P., Perricaudet M., Yeh P., Vigne E. Simultaneous CAR- and alpha V integrin-binding ablation fails to reduce Ad5 liver tropism. Mol Ther. 2003;8:485–494. doi: 10.1016/s1525-0016(03)00182-5. [DOI] [PubMed] [Google Scholar]
- 25.Broutier L., Mastrogiovanni G., Verstegen M.M.A., Francies H.E., Gavarró L.M., Bradshaw C.R. Human primary liver cancer–derived organoid cultures for disease modeling and drug screening. Nat Med. 2017;23:1424–1435. doi: 10.1038/nm.4438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Picco G., Garnett M.J. A road map for precision cancer medicine using personalized models. Cancer Discov. 2017;7:456–458. doi: 10.1158/2159-8290.CD-17-0268. [DOI] [PubMed] [Google Scholar]
- 27.Hirooka Y., Kasuya H., Ishikawa T., Kawashima H., Ohno E., Villalobos I.B. A Phase I clinical trial of EUS-guided intratumoral injection of the oncolytic virus, HF10 for unresectable locally advanced pancreatic cancer. BMC Cancer. 2018;18:596. doi: 10.1186/s12885-018-4453-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ribas A., Dummer R., Puzanov I., VanderWalde A., Andtbacka R.H.I., Michielin O. Oncolytic virotherapy promotes intratumoral T cell infiltration and improves Anti-PD-1 Immunotherapy. Cell. 2017;170:1109–1119. doi: 10.1016/j.cell.2017.08.027. .e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lang F.F., Conrad C., Gomez-Manzano C., Yung W.K.A., Sawaya R., Weinberg J.S. Phase I study of DNX-2401 (Delta-24-RGD) oncolytic adenovirus: replication and immunotherapeutic effects in recurrent malignant glioma. J Clin Oncol. 2018;36:1419–1427. doi: 10.1200/JCO.2017.75.8219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chesney J., Awasthi S., Curti B., Hutchins L., Linette G., Triozzi P. Phase IIIb safety results from an expanded-access protocol of talimogene laherparepvec for patients with unresected, stage IIIB–IVM1c melanoma. Melanoma Res. 2018;28:44–51. doi: 10.1097/CMR.0000000000000399. [DOI] [PubMed] [Google Scholar]
- 31.Kloker L.D., Yurttas C., Lauer U.M. Three-dimensional tumor cell cultures employed in virotherapy research. Oncolytic Virother. 2018;7:79–93. doi: 10.2147/OV.S165479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Choi Y., Lee S., Kim K., Kim S.-.H., Chung Y.-.J., Lee C. Studying cancer immunotherapy using patient-derived xenografts (PDXs) in humanized mice. Exp Mol Med. 2018;50:99. doi: 10.1038/s12276-018-0115-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Neal J.T., Li X., Zhu J., Giangarra V., Grzeskowiak C.L., Ju J. Organoid modeling of the tumor immune microenvironment. Cell. 2018;175 doi: 10.1016/j.cell.2018.11.021. 1972–1988.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Öhlund D., Handly-Santana A., Biffi G., Elyada E., Almeida A.S., Ponz-Sarvise M. Distinct populations of inflammatory fibroblasts and myofibroblasts in pancreatic cancer. J Exp Med. 2017;214:579–596. doi: 10.1084/jem.20162024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tsai S., McOlash L., Palen K., Johnson B., Duris C., Yang Q. Development of primary human pancreatic cancer organoids, matched stromal and immune cells and 3D tumor microenvironment models. BMC Cancer. 2018;18:335. doi: 10.1186/s12885-018-4238-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yano S., Takehara K., Kishimoto H., Tazawa H., Urata Y., Kagawa S. OBP-401-GFP telomerase-dependent adenovirus illuminates and kills high-metastatic more effectively than low-metastatic triple-negative breast cancer in vitro. Cancer Gene Ther. 2017;24:45–47. doi: 10.1038/cgt.2016.67. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.