Abstract
Despite remarkable responses to cancer immunotherapy in a subset of patients, many patients remain resistant to therapies. It is now clear that elevated levels of tumor-infiltrating T cells as well as a systemic anti-tumor immune response are requirements for successful immunotherapies. However, the tumor microenvironment imposes an additional resistance mechanism to immunotherapy. We have developed a practical and improved strategy for cancer immunotherapy using an oncolytic virus and anti-OX40. This strategy takes advantage of a preexisting T cell immune repertoire in vivo, removing the need to know about present tumor antigens. We have shown in this study that the replication-deficient oncolytic Sindbis virus vector expressing interleukin-12 (IL-12) (SV.IL12) activates immune-mediated tumor killing by inducing OX40 expression on CD4 T cells, allowing the full potential of the agonistic anti-OX40 antibody. The combination of SV.IL12 with anti-OX40 markedly changes the transcriptome signature and metabolic program of T cells, driving the development of highly activated terminally differentiated effector T cells. These metabolically reprogrammed T cells demonstrate enhanced tumor infiltration capacity as well as anti-tumor activity capable of overcoming the repressive tumor microenvironment. Our findings identify SV.IL12 in combination with anti-OX40 to be a novel and potent therapeutic strategy that can cure multiple types of low-immunogenic solid tumors.
Keywords: oncolytic virus anti-OX40 combination therapy, anti-OX40, anti-tumor immunity, Sindbis, alpha-virus immunotherapy, cancer immunity
Graphical Abstract
Despite remarkable responses to cancer immunotherapy, many patients remain resistant to therapies. The harsh tumor microenvironment blocks tumor penetration, impairing the cancer killing functions of the immune system. In this study, the authors show that the combination of an oncolytic virus and anti-OX40 antibody can successfully cure multiple tumors by overcoming this resistance.
Introduction
Immune checkpoint modulation has shown remarkable promise in treating advanced cancers.1, 2, 3 Although high response rates with immune checkpoint blockade have been documented in patients with highly immunogenic tumors, the proportion of patients who respond to treatment is often still low. Major challenges to overcome are the lack of T cell infiltration into the tumor microenvironment as well as the immunosuppressive nature of the tumor, which inhibits the intratumoral immune response.4,5 To improve response rates, combining immune checkpoint inhibitory antibodies with oncolytic virus (OV) therapy has become an attractive and promising novel strategy in patients.6, 7, 8, 9 OVs can selectively infect and replicate in tumor cells, resulting in tumor cell lysis. Besides this direct virus-mediated anti-tumor activity, OVs also have the potential to induce an anti-tumor response indirectly by activating immune cells to help them target and kill cancer cells. However, major challenges are still ahead, as most OVs have to be administered intratumorally, which limits their application.10, 11, 12, 13 Furthermore, the therapeutic efficacy of an OV is dependent on its ability to directly infect and kill tumor cells, thus narrowing the spectrum of OV therapy.
It has recently been appreciated that the metabolic landscape of the tumor microenvironment may represent an additional resistance mechanism to immunotherapy.14, 15, 16, 17, 18 T cell effector responses are energetically demanding, and T cells undergo substantial metabolic reprogramming during activation.19 However, due to the metabolic deregulation of tumor cells, the tumor microenvironment becomes restricted by oxygen and nutrients. Thus, even with a strong immunotherapy, such as with anti-PD-1 or anti-CTLA4 antibodies, once recruited to the tumor site, actively infiltrating anti-tumor T cells from the periphery experience metabolic stress and become dysfunctional. New immunomodulatory strategies are therefore needed to overcome these resistances. Besides the B7 co-inhibitory receptors (e.g., CTLA4 and PD-1), the tumor necrosis factor receptor (TNFR) superfamily contains many other immune checkpoints, which could become the next generation of immune modulators. One of these receptors is OX40, a co-stimulatory receptor that is expressed on activated T cells. Signaling through OX40 promotes survival signals in effector T cells through nuclear factor κB (NF-κB) and nuclear factor of activated T cells (NFAT) activation that enhance the expression of molecules such as survivin, cyclin A, Bcl-2, and cytokines.20 Furthermore, OX40 signaling impairs immunosuppressive regulatory T cell function through direct inhibition of Foxp3 expression.21 Thus, OX40 stimulation has the potential to improve anti-tumor therapy by prolonging survival of pre-existing tumor-specific effector T cells as well as enhancing T cell activation through depletion of immunosuppressive immune cells.
In this study, we investigated the therapeutic efficacy of a replication-deficient oncolytic viral vector called Sindbis virus (SV). Because SV is a blood-borne pathogen, vectors from this virus can be administered in the bloodstream via the intravenous (i.v.) and intraperitoneal (i.p.) routes, which greatly facilitates their delivery.22 Furthermore, SV was genetically modified to be replication-defective by splitting its genome and deleting the packaging signal to block viral assembly after viral replication.23 We show that SV expressing the pro-inflammatory cytokine interleukin-12 (IL-12; SV.IL12) activates T cells as well as enhances the expression of OX40 on CD4 T effector cells and, therefore, potentiates efficacy of the agonistic anti-OX40 antibody therapy. Our data indicate that combination of SV.IL12 and anti-OX40 activates tumor immunity against low-immunogenic tumors through the metabolic rewiring of T cells into highly activated effector cells. This is in line with studies on other TNFRs, such as 41-BB, which reported a metabolic reprogramming of T cells after 41-BB stimulation.24 Furthermore, SV.IL12 in combination with anti-OX40 induces a marked immune cell infiltration into the tumor microenvironment. Considering that tumors tend to quickly escape the immune response by mutating or losing the expression of drug targets or tumor antigens targeted by the immune response, our treatment approach reduces the risk of developing tumor resistances and offers an attractive and safe strategy to change the immunogenic phenotype of various cancers without prior knowledge of tumor antigens.
Results
SV-Expressing IL-12 Enhances the Expression of OX40 on CD4 T Cells
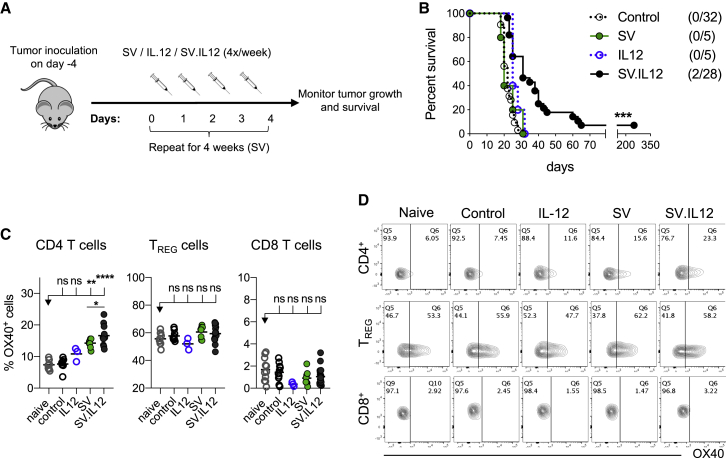
Previous studies from our laboratory demonstrated that replication-deficient SV.IL12 elicits a therapeutic efficacy in immune-deficient severe combined immunodeficiency (SCID) mice bearing human ovarian carcinoma tumors.25 Furthermore, Granot et al.25 demonstrated that the therapeutic efficacy of SV.IL12 was natural killer (NK) cell mediated and enhanced by the presence of IL-12. Due to the fact that IL-12 also plays an essential role in regulating the adaptive immune response,26 we investigated the therapeutic effect of SV.IL12 in immune-competent tumor-bearing mice (colon cancer; CT26). To exploit SV.IL12 for cancer therapy, tumor cells were i.p. implanted, and after tumor establishment (4 days after tumor cell injection [day 0]), we i.p. injected SV, SV.IL12, or IL-12 on 4 consecutive days (days 1, 2, 3, and 4) for a total of 4 weeks (Figures 1A and 1B). While untreated (control), SV-treated, and IL-12-treated mice succumb to cancer after 3 weeks, treatment with SV.IL12 slightly prolonged survival time, with an overall long-term survival rate of 7.1%. These data suggest that SV-expressing IL-12 is needed to induce the observed therapeutic efficacy. Shortly after i.p. injection, SV infects macrophages in mediastinal lymph nodes where T cells get subsequently activated (Figure S1). Even though SV.IL12-infected cells secrete significant amounts of IL-12, as observed in in vitro experiments (Figure S2A), i.p. injection of SV.IL12 did not significantly change levels of plasma IL-12 in mice (Figure S2B), thus suggesting that IL-12 produced by SV acts locally and stimulates transduced macrophages (Figure S1) that present tumor antigens to corresponding T cells and activates them further. That shapes the subsequent anti-tumor immune response, such as promoting the differentiation into T helper (Th)1 cells as well as increasing interferon (IFN)γ production (Figures S2C and S2D).27, 28, 29, 30 After 1 week of treatment, we analyzed the T cell response for their expression of inhibitory and activation markers. We observed that OX40 was markedly upregulated on CD4 T cells during SV.IL12 treatment, which was mostly among the effector CD4 T cells and less on the regulatory T cells (Figures 1C and 1D). Interestingly, SV treatment also induced OX40 upregulation on CD4 T cells, but to a lesser extent (Figures 1C and 1D). On the basis of the results above and previous studies that reported a beneficial effect of anti-OX40 in cancer treatment,20 we hypothesized that the agonistic anti-OX40 antibody could augment the therapeutic efficacy of SV.IL12.
Figure 1.
SV.IL12 Induces a Modest Therapeutic Efficacy and Increases OX40 Expression on CD4 T Cells
(A) Treatment schema. BALB/c mice were given intraperitoneal (i.p.) injections of SV, IL-12 (50 ng), or SV.IL12 at various times after injection of 7 × 104 CT.26.Fluc on day 0. (B) Survival plots of control and treated mice bearing CT26.Fluc tumors. Statistical significance between SV.IL12 and all other groups was determined with the Mantel-Cox test. Results are representatives of at least two independent experiments. (C and D) CT26 tumor-bearing mice were treated with SV, IL-12 (50 ng), or SV.IL12 on 4 consecutive days (days 1, 2, 3, and 4). On day 7, spleens were excised and a single-cell suspension was stained and analyzed by flow cytometry. As controls, naive and untreated (control) tumor-bearing mice were used. (C) Percentage of OX40 expression by CD4 T cells (left), regulatory T cells (TREG; middle), and CD8 T cells (right). (D) Representative flow cytometry plots indicating OX40 staining in different T cell subsets. Bars represent means and each symbol represents an individual mouse. Statistical significance was determined with the Kruskal-Wallis test followed by the Dunns’ test or the Mann-Whitney test. Results are representatives of at least two independent experiments.
Intraperitoneal Delivery of SV.IL12 and Anti-OX40 Antibody Cures Established Cancers
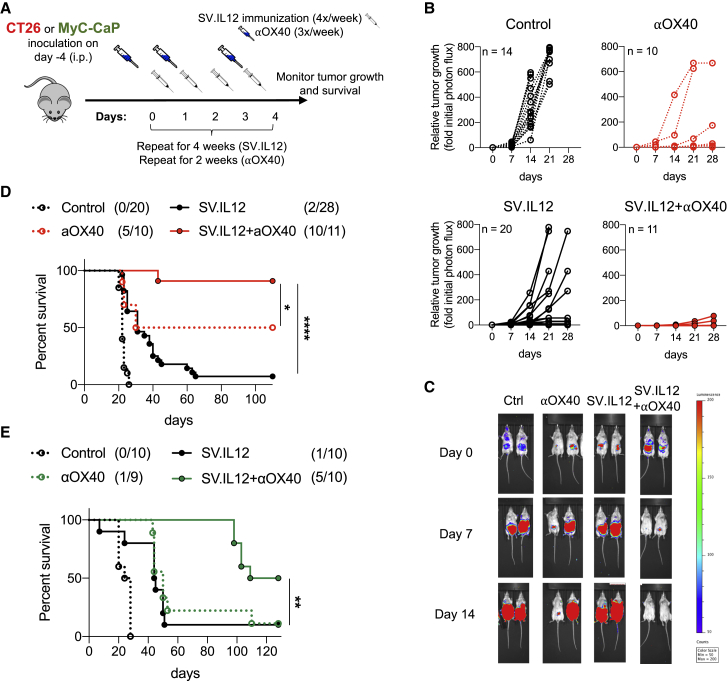
Similar to many other OVs, SV can directly infect cancer cells and provide a local immune response in the tumor microenvironment.22,31 However, as shown in previous publications, SV infectivity is not required for inducing a strong therapeutic efficacy, as SV also enters peripheral lymphoid organs, which induces a systemic response.32,33 To investigate whether the oncolytic activity of SV.IL12 in combination with anti-OX40 is required for successful anti-cancer therapy, SV non-susceptible (colon cancer; CT26) and susceptible (prostate cancer; MyC-CaP) tumor cell lines were used in this study (Figure S3).32,34 Immunocompetent female BALB/c and male FVB/NJ mice were implanted with either CT26 or MyC-CaP tumor cell lines, which expressed the firefly luciferase (Fluc) protein, respectively. This allowed us to monitor tumor growth in vivo using noninvasive bioluminescent imaging. Once tumors become established (day 0), mice were treated with SV.IL12 in combination with anti-OX40. SV.IL12 was i.p. injected on 4 consecutive days (days 1, 2, 3, and 4) for a total of 4 weeks (Figure 2A). Anti-OX40 was injected three times a week (days 0, 2, and 4) for a total of 2 weeks. In both tumor models, all untreated animals experienced progressive tumor growth and succumbed to cancer on week 3 (Figure 2; Figure S4). Mice bearing CT26.Fluc or MyC-CaP.Fluc tumors showed some delay in tumor growth when treated with i.p. injected SV.IL12 or anti-OX40 alone but with only a moderate effect on long-term survival (Figure 2; Figure S4). However, the combination of SV.IL12 with anti-OX40 resulted in complete regression of tumors in both tumor models (Figure 2; Figure S4). Tumors occasionally did recur in mice treated with combination therapy after treatment was completed, resulting in a long-term survival rate of 91.6% and 50% in the CT26 and MyC-CaP tumor models, respectively. In conclusion, combination of SV.IL12 with anti-OX40 elicits a strong therapeutic efficacy against two distinct solid tumors. Furthermore, these findings confirm that the oncolytic activity of SV is not required to induce a robust and effective anti-tumor response. Due to the fact that anti-OX40 monotherapy already resulted in a 20%–50% survival rate, we wanted to investigate whether the addition of SV.IL12 would allow us to reduce treatment frequencies while still maintaining the strong therapeutic efficacy of combination therapy. This is especially important for lowering risks of adverse events as well as being more convenient for patients in clinics. Interestingly, therapeutic efficacy in the CT26.Fluc tumor model was maintained with only one injection per week of SV.IL12 and anti-OX40 (Figure S5). This is in contrast with MyC-CaP.Fluc tumor-bearing mice for which the full treatment regimen was required (data not shown). Thus, in the following experiments mice bearing CT26 tumors were treated with one injection a week whereas MyC-CaP tumor-bearing mice received the full treatment regimen.
Figure 2.
SV.IL12 in Combination with Anti-OX40 Antibody Cures Established Tumors In Vivo
(A) Experimental protocol for the prostate and colon cancer model used in (B)–(E). FVB/NJ or BALB/c mice were given an i.p. injection of SV.IL12 and/or anti-OX40 at various times after injection of 105 MyC-CaP.Fluc or 7 × 104 CT26.Fluc cells on day 0, respectively. (B) CT26.Fluc tumor growth curves are shown as fold changes relative to the luminescence on day 0 of the same mouse. Each line represents an individual mouse. Left graphs: control (n = 14) (top) and SV.IL12 (n = 20) (bottom). Right graphs: anti-OX40 (n = 10) (top) and SV.IL12+anti-OX40 (n = 11) (bottom). (C) Representative bioluminescence images of control and treated CT26.Fluc-bearing mice. (D and E) Survival plots of control and treated mice bearing peritoneally disseminated CT26.Fluc (D) or MyC-CaP.Fluc (E) tumors. Statistical significances between SV.IL12+anti-OX40 and anti-OX40 or SV.IL12 were determined with the Mantel-Cox test. Results are representatives of at least two independent experiments. αOX40/aOX40, anti-OX40.
Combination Therapy Markedly Changes the Transcriptome Signature of T Cells
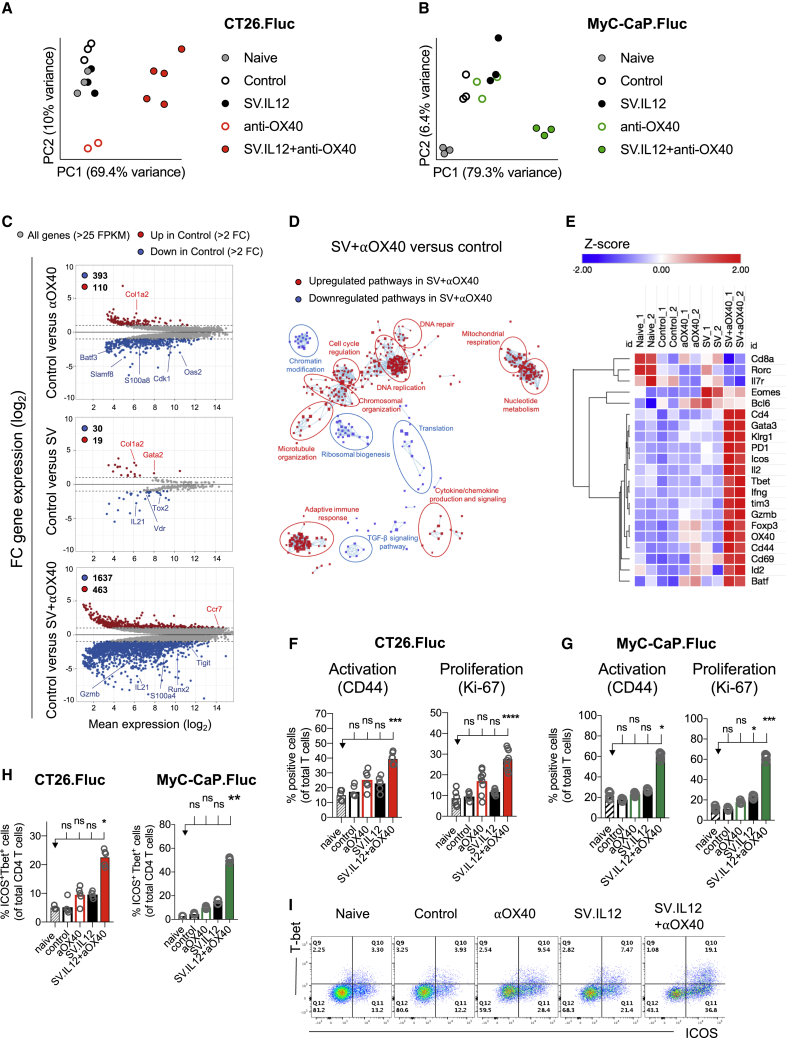
Due to the fact that OX40 is specifically expressed on T cells, which are crucial players during anti-tumor responses, we assessed the requirement of T cells during SV.IL12 with anti-OX40 treatment. The presence of both CD4 and CD8 T cells was required for eliciting the observed therapeutic efficacy, as mice treated with the corresponding depleting antibodies were unable to control tumor growth (Figure S6). To better understand the impact of the combination therapy on T cells, RNA sequencing was performed on isolated T cells from spleens derived from naive, control, as well as anti-OX40 and SV with or without anti-OX40-treated mice on day 7. Principal-component analysis (PCA) of normalized reads showed a distinct segregation between combined therapy and all other groups in both tumor models (Figures 3A and 3B). These data suggest that combined therapy induces a distinct T cell response in the periphery independently of the tumor model and mouse strain, suggesting an indirect and immunity-driven role of SV vectors with a negligible direct effect of the vector in tumors (Figure 3C). Indeed, gene expression profiles of control versus anti-OX40, SV, or SV in combination with anti-OX40 were clearly distinct, with the most differentially expressed genes (DEGs) in the latter one (503, 49, and 2,100 DEGs, respectively) (Figure 3C). Of 2,100 DEGs in control versus combination therapy, more genes were downregulated than upregulated (1,637 versus 463) >2-fold. Similarly, in control versus anti-OX40 as well as control versus SV more DEGs were downregulated than upregulated (393 versus 110 and 30 versus 19, respectively).
Figure 3.
Combination Therapy Markedly Changes the Transcriptome Signature of T Cells Favoring Effector T Cells with a Th1-type Phenotype
(A–E) RNA sequencing of T cells isolated from spleens derived from untreated tumor-bearing mice (control) compared with mice treated with SV.IL12 and/or anti-OX40 on day 7. (A and B) Principal component analysis (PCA) of normalized read counts from the CT26.Fluc (A) and MyC-CaP.Fluc tumor model (B). (C) MA plots of differentially expressed genes (DEGs; >2-fold) in T cells of control versus anti-OX40-treated mice (top graph), SV.IL12-treated mice (middle graph), or SV.IL12+anti-OX40-treated mice (bottom graph) in the CT.26 model. Significantly (p < 0.05) upregulated and downregulated DEGs are depicted in red or blue, respectively. (D) Pathway and network analysis based on DEGs in T cells isolated from CT26.Fluc-bearing mice treated with combination therapy. Downregulated (blue) and upregulated (red) pathways are shown, respectively. (E) Heatmap analysis of selected genes based on normalized read counts linked to T cell differentiation and activation as well as T cell lineage transcription factors. (F–I) Tumor-bearing mice were treated with SV.IL12 and/or anti-OX40. As controls, naive and untreated (control) tumor-bearing mice were used. On day 7, spleens were excised and a single-cell suspension was stained and analyzed by flow cytometry. (F and G) Percentage of CD44 or Ki-67 expression by T cells from the CT26.Fluc (F) or MyC-CaP.Fluc (G) tumor model. (H and I) Percentage of ICOS and T-bet expression by CD4 T cells (H); representative flow cytometry plots (I). Bars represent means and each symbol represents an individual mouse. Statistical significance was determined with the Kruskal-Wallis test followed by the Dunn’s test. Results are representatives of at least two independent experiments (F–I).
We performed unbiased pathway enrichment and network analyses of DEGs from control versus combination therapy to determine biological processes in T cells that are influenced by this treatment (Figure 3D). Although both upregulated and downregulated DEGs were included in the analysis, the vast majority of pathways were upregulated in T cells treated with combination therapy with the exception of four clusters (transforming growth factor β [TGF-β] signaling, ribosomal biogenesis, translation, and chromatin modification). The upregulated pathways were dominated by DNA replication, chromosomal organization, and cell cycle regulation, but they also included various metabolic and immunological processes, such as mitochondrial respiration, nucleotide metabolism, and adaptive immune responses.
Combination Therapy Enhances Systemic T Cell Responses, Favoring Th1-like ICOS CD4 T Cells
As T cells from combination therapy express a marked change in their transcriptome signature compared with all other groups, markers for T cell differentiation and activation (e.g., PD-1, ICOS, OX40, TIM3, KLRG1, IL-7 receptor [IL-7R]) as well as T cell lineage transcription factors (e.g., EOMES, TBET, GATA3, BCL6, RORC, FOXP3) were analyzed (Figure 3E). Only T cells from combined therapy expressed the gene signature of terminally differentiated effector T cells, which are characterized by high expression of the killer cell lectin-like receptor G1 (KLRG1) and low expression of IL-7R.35 Furthermore, genes encoding products associated with the differentiation and function of effector cells, such as Batf, Id2, Tbet, Gzmb, and Ifng, were also highly expressed in T cells isolated from mice treated with combined therapy compared with all other groups. The enhancement of effector T cells in combined therapy was confirmed by flow cytometry in both tumor models, as judged by the increased expression of the activation and proliferation markers CD44 and Ki-67, respectively (Figures 3F and 3G; Figure S7). Interestingly, CD4 T cells also expressed a marked anti-tumor effector phenotype (ICOS+T-bet+) that was on average 2- to 3-fold higher during combined therapy compared with SV.IL12 or anti-OX40 treatment (Figures 3H and 3I). Previous studies reported a correlation between expansion of ICOS+T-bet+ CD4 T cells and clinical benefit in cancer patients who received anti-CTLA4 therapy.3,36,37 In summary, SV.IL12 in combination with anti-OX40 induces a marked systemic T cell response and favors the differentiation of terminal effector T cells. Furthermore, combined therapy induces a sustained increase in the frequency of ICOS+T-bet+ CD4 T cells, which has also been reported to be elevated during successful anti-CTLA4 cancer therapy.
CD4 and CD8 T Cells Are Metabolically Reprogrammed in Mice Treated with SV.IL12 and anti-OX40
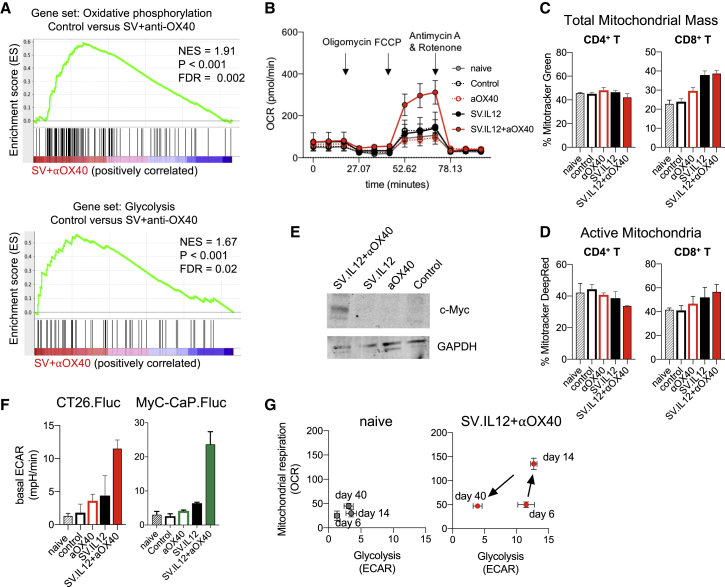
The tumor microenvironment can be a very challenging milieu for an effector T cell, as it is characterized by hypoxia, acidosis, and low levels of nutrient sources such as glucose and glutamine.14,15,38 Even when T cell activation and initiation of effector function are allowed, T cells may be unable to generate the bioenergetic intermediates necessary to carry out effector function in the tumor microenvironment. Thus, providing a metabolic support for T cells is crucial for the success of cancer treatments, as previously reported.39, 40, 41, 42 To test whether SV.IL12 in combination with anti-OX40 influences the metabolic state of T cells, gene set enrichment analysis (GSEA) of the RNA sequencing data was performed between T cells from combined therapy and control. GSEA analysis showed significantly higher expression of genes involved in oxidative phosphorylation and glycolysis pathways during combination therapy (Figure 4A). To confirm GSEA analysis, peripheral T cells from both tumor models were metabolically profiled using Seahorse analysis on day 7 (Figure 4B; Figures S8A and S8B). Oxidative phosphorylation and glycolytic profiles in T cells from naive, control, and mice treated with SV.IL12 and/or anti-OX40 were determined by measuring the oxygen consumption rate (OCR) and the extracellular acidification rate (ECAR), respectively. The basal OCR was enhanced in T cells from combined therapy and SV.IL12 treatment, but only the former harbored a dramatic increase in spare respiratory capacity in the CT26 model (Figure 4B; Figure S8A). This was in contrast to T cells from combined therapy in the MyC-CaP.Fluc tumor model, which expressed 3.75-fold higher basal OCR with no spare respiratory capacity (Figure S8B). The reason for this discrepancy between the two models might be the differences in the number of treatments, as MyC-CaP.Fluc-bearing mice receive 3 and 4 times more injections of anti-OX40 and SV.IL12, respectively.
Figure 4.
SV.IL12 in Combination with Anti-OX40 Promotes Metabolic Reprogramming of T Cells
Tumor-bearing mice were left untreated or treated with SV.IL12 and/or anti-OX40. T cells were isolated from spleens on day 7 or otherwise indicated. (A) Selected gene set enrichment analysis (GSEA) of oxidative phosphorylation and glycolysis pathways based on DEGs in control versus SV+anti-OX40. (B) Mitochondrial respiration was assessed by measuring the median values of oxygen consumption rates (OCRs) in T cells of indicated groups using an extracellular flux analyzer. Oligomycin, FCCP, antimycin A, and rotenone were injected as indicated to identify energetic mitochondrial phenotypes. (C and D) MitoTracker Green (C) and MitoTracker Deep Red FM (D) staining of T cells from indicated groups using flow cytometry. (E) Western blot of c-Myc protein expression in T cells of control or mice treated with anti-OX40, SV.IL12, or SV.IL12+anti-OX40. GAPDH (bottom) is the loading control. (F) Baseline extracellular acidification rates (ECARs) in T cells of indicated groups derived from the CT26.Fluc and MyC-CaP.Fluc tumor models. (G) Energy profile (OCRs versus ECARs) of T cells from naive or CT26.Fluc-bearing mice treated with SV.IL12+anti-OX40 on days 7, 14, and 40. Error bars indicate SEM. Results are representatives of at least two independent experiments (B–G).
Analysis of mitochondrial mass (Figure 4C; Figure S8C) and activity (Figure 4D; Figure S8C), using flow cytometry with the mitochondrial stain MitoTracker Green and Deep Red, respectively, revealed that SV.IL12 with or without anti-OX40 induced higher mitochondrial mass and activity in CD8 T cells but not in CD4 T cells. These data suggest that the observed increase in basal OCR was mainly driven by CD8 T cells. Interestingly, a slight decrease of active mitochondria occurred in CD4 T cells from mice treated with combined therapy, which might explain the increase in spare respiratory capacity in this group. To test whether the reduction of active mitochondria in CD4 T cells is associated with a switch toward glycolysis, the master regulator for glycolysis c-MYC and basal ECAR were measured in T cells from all groups. Indeed, the addition of anti-OX40 to SV.IL12 induced elevated protein expression of c-MYC as well as basal ECAR (Figures 4E and 4F). T cells from naive and control as well as SV.IL12- or anti-OX40-treated mice showed no signs of elevations. Collectively, these findings reveal that SV.IL12 induces enhanced oxidative phosphorylation in CD8 T cells, whereas the addition of anti-OX40 to SV.IL12 is needed to push CD4 T cells toward glycolysis by increasing the protein expression of c-MYC.
To determine the kinetics of peripheral T cell metabolism during the course of treatment with SV.IL12 and anti-OX40, OCR and ECAR were measured on days 7, 14, and 40 in CT26.Fluc-bearing mice (Figure 4G). As shown above, T cells on day 7 shifted toward a glycolytic state, which is associated with the initial effector phase. Two weeks into the treatment, T cells switched to a highly energetic state utilizing both metabolic pathways, oxidative phosphorylation and glycolysis, as reported for highly activated T cells.43 Once tumors were fully rejected and mice were tumor-free for a month, T cells returned to a more quiescent state, such as naive cells. Interestingly, T cells from MyC-CaP.Fluc-bearing mice switched to a highly energetic state early on during treatment (day 7) and remained in this metabolic phenotype 2 weeks after treatment had stopped (Figure S8D). The reason for this discrepancy might be the differences in the number of treatments applied in both tumor models, as MyC-CaP.Fluc-bearing mice received 3 and 4 times more anti-OX40 and SV.IL12, respectively. T cells from control as well as anti-OX40- or SV.IL12-treated mice in both tumor models remained in a quiescent state during the course of treatment (Figure S8E). In summary, SV.IL12 in combination with anti-OX40 metabolically rewires T cells to an energetic state using both metabolic pathways, oxidative phosphorylation and glycolysis. This phenotype does not occur in SV.IL12- or anti-OX40-treated mice, which succumb to cancer. Thus, the changed metabolic state of T cells correlates with an efficient anti-tumor response and better survival rate.
Metabolic Reprogrammed T Cells in SV.IL12 with Anti-OX40-Treated Mice Display Enhanced CD4-Mediated Cytokine Production and Anti-tumor Activity
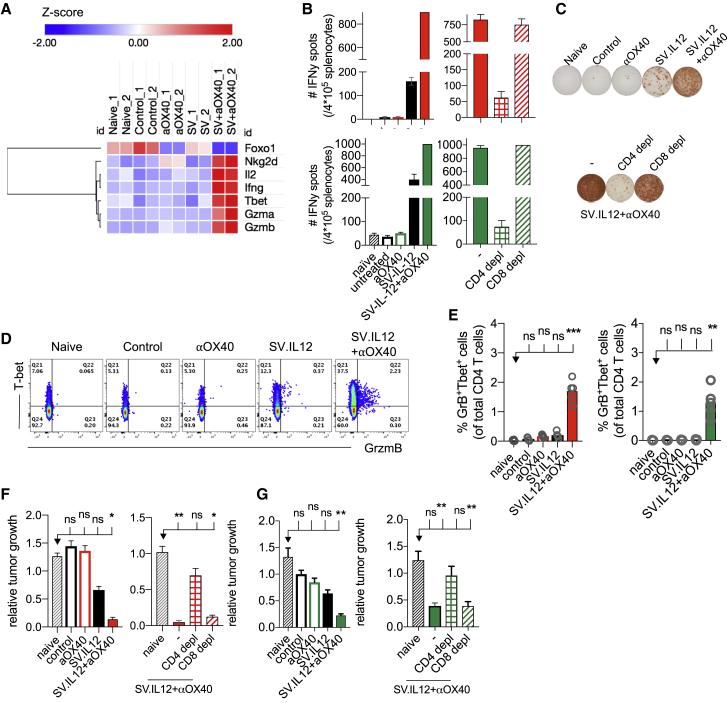
To test whether metabolic reprogrammed T cells in combined therapy possess enhanced effector functions, cytokine production and cytotoxicity were analyzed in T cells isolated from spleens on day 7. Genes encoding pro-inflammatory cytokines Ifng and Il2 were upregulated in T cells from mice treated with SV in combination with anti-OX40 (Figure 5A). ELISPOT (enzyme-linked immunospot) analysis of IFNγ by splenocytes confirmed RNA sequencing data, showing the strongest IFNγ secretion in mice treated with combined therapy in both tumor models (Figures 5B and 5C). Splenocytes from SV.IL12-treated mice also produced IFNγ but to a lesser extent. Interestingly, the main producers of IFNγ were CD4 T cells, as depletion of CD4 T cells but not CD8 T cells abolished IFNγ secretion in splenocytes from mice treated with combined therapy.
Figure 5.
Reprogrammed T Cells in SV+Anti-OX40-Treated Mice Display Enhanced CD4-Mediated Cytokine Production and Anti-tumor Activity
Tumor-bearing mice were left untreated or treated with SV.IL12 and/or anti-OX40. Spleens were excised on day 7 for further analysis. (A) Heatmap analysis of selected genes based on normalized read counts linked to cytokine expression and enhanced anti-tumor activity. RNA sequencing was performed on isolated T cells from CT26.Fluc tumor-bearing mice. (B and C) Interferon-γ (IFNγ) enzyme-linked immunospot analysis of splenocytes from control and treated mice as indicated (n = 5–10 mice per group). Additionally, IFNγ enzyme-linked immunospot analysis was measured in splenocytes depleted of CD4 or CD8 T cells in the CT26.Fluc (top) and MyC-CaP.Fluc (bottom) tumor models. (D and E) Percentage of T-bet and granzyme B (GrB) expression by CD4 T cells using flow cytometry. Representative flow cytometry plots from indicated groups are shown in (D) and statistical significances shown in (E). (F and G) Cytotoxic activity of T cells from control and treated mice (n = 5–10 mice per group). T cells were co-cultured at an effector-to-target cell ratio of 10:1 with either CT26.Fluc (F) or MyC-CaP.Fluc (G) tumor cell lines for 2 days. Additionally, T cells were depleted of CD4 or CD8 T cells and co-cultured as previously described. Cytotoxic activity was assessed based on viability of tumor cells, which was determined by measuring the luciferase activity and is shown relative to naive T cells. Bars represent means ± SEM (B, F, and G), and each symbol represents an individual mouse (E). Statistical significance was determined with the Kruskal-Wallis test followed by the Dunn’s test. Results are representatives of at least two independent experiments.
In addition, RNA levels of the cytotoxic proteases granzyme A and B were upregulated in mice treated with combination therapy compared with all other groups (Figure 5A). Protein expression of granzyme B correlated with RNA levels as measured by flow cytometry in both tumor models (Figures 5D and 5E; Figures S9A–S9D). Furthermore, granzyme B-positive cells were detected in CD8 as well as CD4 T cells, suggesting the presence of cytotoxic CD4 T cells in mice treated with combined therapy.44, 45, 46 Upregulation of granzyme B was associated with downregulation of the transcription factor Foxo1, which is known to control granzyme transcription through repression of the transcription factor T-bet (Figure 5A).47 Last, the enhanced cytotoxic potential of T cells from combined therapy was also supported by elevated expression of the NKG2D receptor, which has been shown to trigger T cell receptor (TCR)-independent cytotoxicity in activated T cells (Figure 5A; Figures S9E and S9F).48
Having observed upregulation of granzymes and cytotoxic receptors in combination therapy, the function of T cells was investigated using an ex vivo tumor growth assay. Splenocytes obtained from all groups were co-cultured at an effector-to-target cell ratio of 10:1 with either CT26.Fluc (Figure 5F) or MyC-CaP.Fluc (Figure 5G) tumor cell lines. The anti-tumor activity of splenocytes was determined by measuring the luciferase activity of cell lines, which correlates with tumor growth. Tumor growth was markedly reduced when co-cultured with splenocytes from mice receiving combined therapy compared with splenocytes from naive, control, and mice treated with anti-OX40 in both tumor models. The anti-tumor activity of splenocytes from mice treated with SV.IL12 alone was weaker than that from combined therapy. Surprisingly, tumor growth inhibition was mediated by CD4 T cells, as depletion of CD4 T cells but not CD8 T cells abolished the inhibitory effect on tumor cells. Taken together, these results clearly show that T cells from combined therapy elicit enhanced anti-tumor and functional activity, such as granzyme B and IFNγ production driven by CD4 T cells.
Mice Treated with SV.IL12 in Combination with Anti-OX40 Display Enhanced T Cell Migration and Intratumoral T Cell Immunity
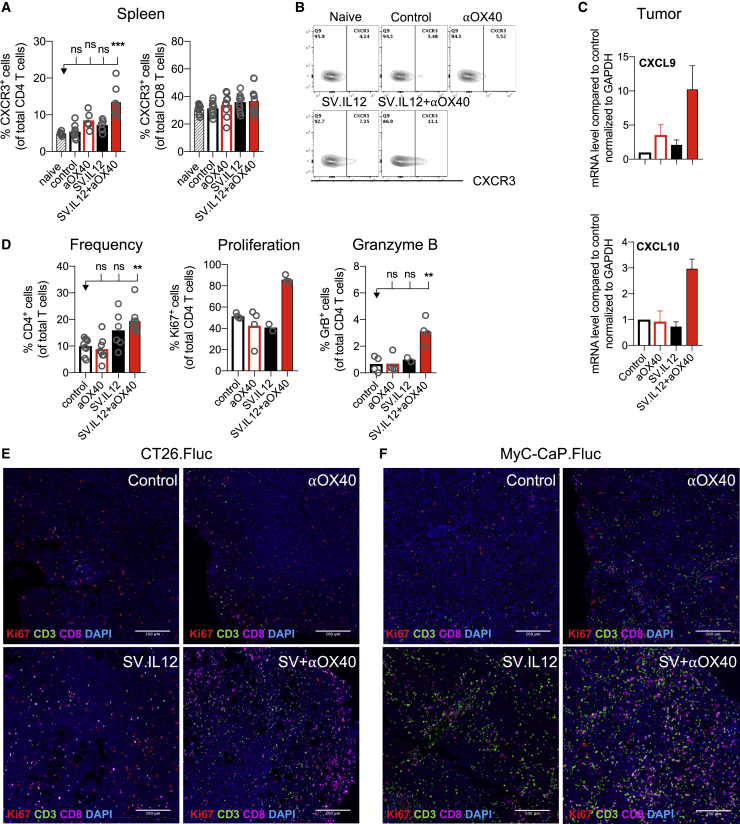
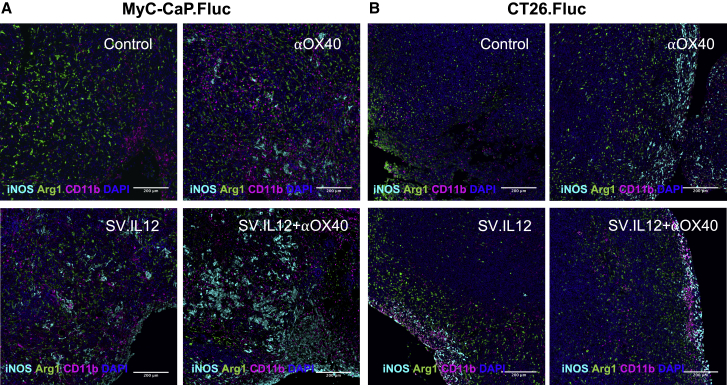
Only a minority of the total of treated patients respond to current immunotherapy, and the presence of tumor-infiltrating lymphocytes (TILs) has been shown to be one of the main factors that influence the responsiveness toward various therapies in multiple cancers.49,50 Due to the fact that SV-elicited anti-tumor responses do not necessarily require direct infection of the tumor or intratumoral injection, we wondered whether SV.IL12 therapy in combination with anti-OX40 could nevertheless alter the local tumor microenvironment and favor intratumoral immunity. To assess whether SV.IL12 in combination with anti-OX40 induces T cell infiltration into the tumor, the chemokine receptor CXCR3 on peripheral T cells was analyzed after 1 week of treatment. We observed that in the CT26.Fluc model CXCR3 levels were significantly upregulated on CD4 T cells during combination therapy compared with all other groups, and CXCR3 levels remained elevated during the course of treatment (Figures 6A and 6B; Figure S10A). In contrast, CXCR3 expression on CD8 T cells only appeared later on in treatment, suggesting that CD4 T cells are first recruited to the inflamed site followed by CD8 T cells (Figure S10A). MyC-CaP.Fluc tumor-bearing mice showed elevated levels of CXCR3 on CD4 and CD8 T cells after 1 week of combination treatment (Figures S10B and S10C). Furthermore, SV.IL12 treatment also increased CXCR3 expression on T cells but to a lesser extent. The reason for this discrepancy between the two models might be the differences in the number of treatments, as MyC-CaP.Fluc-bearing mice receive 3 and 4 times more injections of anti-OX40 and SV.IL12, respectively. Cells expressing CXCR3 follow the gradient of their ligands CXCL9, CXCL10, and CXCL1.51 Indeed, combination therapy also enhanced the production of CXCL9 and CXCL10 in the tumor microenvironment, as judged by real-time PCR, suggesting that CXCR3-positive T cells migrate to the tumor site (Figure 6C). Treatment of SV.IL12 or anti-OX40 alone did not alter the expression of these ligands. In line with these observations, an overall increase in T cells was observed in CT26.Fluc and MyC-CaP.Fluc peritoneally disseminated tumors from mice treated with combined therapy compared with control and anti-OX40-treated mice (Figures 6E and 6F). SV.IL12-treated mice also showed enhanced T cell infiltration but to a lesser extent. Furthermore, dissecting CD4 and CD8 T cells by flow cytometry revealed that combination therapy increases the proportion of CD4 T cells in CT26.Fluc tumors, which is consistent with the elevated CXCR3 expression on peripheral CD4 T cells (Figure 6D). These results clearly show that SV.IL12 in combination with anti-OX40 alters the tumor microenvironment by facilitating T cell infiltration via modulation of the CXCR3/CXCL9–11 axis. Not only did combination therapy increase T cell infiltration in both tumor models, but CD4 as well as CD8 T cells also demonstrated enhanced functional activity in the tumor, as judged by the Ki-67 and granzyme B expression (Figure 6D; Figures S10D and S11). In line with these results, a decrease in proliferation, as judged by the expression of Ki-67 in tumor cells, was observed in CT26.Fluc and MyC-CaP.Fluc tumor cells when treated with combined therapy compared with all other treatments (Figures 6E and 6F). These results suggest that the presence of activated T cells in the tumor microenvironment exert anti-tumor activity, which inhibits tumor growth. Besides from T cell activation, we also observed enhanced inducible nitric oxide synthase (iNOS) production in MyC-CaP.Fluc tumors treated with combination therapy compared with control or anti-OX40 treatment (Figure 7). SV.IL12 treatment alone also induced iNOS production but to a lesser extent. Interestingly, the amount of iNOS inversely correlated with ariginase1 production, suggesting a repolarization of tumor-associated macrophages from the M2-like (pro-tumor) into the M1-like (anti-tumor) phenotype during combination therapy. These trends were only observed in MyC-CaP.Fluc and not in CT26.Fluc tumors, which might be a consequence of SV directly infecting MyC-CaP cells.
Figure 6.
Mice Treated with SV.IL12 in Combination with Anti-OX40 Display Enhanced T Cell Migration and Intratumoral T Cell Activity
(A and B) CT26.Fluc-bearing mice were left untreated or were treated with SV.IL12 and/or anti-OX40. On day 7, spleens were excised and a single-cell suspension was stained and analyzed by flow cytometry. (A) Percentages of CXCR3 expression by CD4 (left graph) and CD8 (right graph) T cells are shown. Representative flow cytometry plots are shown in (B). (C–F) Tumors were harvested after 2 weeks of treatment from control and treated mice. (C) Intratumoral gene expression of CXCL9 (top) and CXCL10 (bottom) analyzed by real-time PCR. Data are normalized to GAPDH. (D) Intratumoral T cell immune responses from indicated groups were assessed by flow cytometry. Percentages of CD4 expression by T cells (left graph), Ki-67 expression (middle graph), and granzyme B expression (right graph) by CD4 T cells are shown. (E and F) Multiplex immunofluorescence staining of tumors isolated from CT26.Fluc (E) and MyC-CaP.Fluc (F) tumor-bearing mice. Representative images of T cell infiltration are shown for control as well as anti-OX40, SV.IL12, and SV.IL12+anti-OX40. Proteins of interest were stained and are indicated by color in each image: Ki-67 (red), CD3 (green), CD8 (magenta), and DAPI nuclear staining appears in blue. Bars represent means ± SEM (C), and each symbol represents an individual mouse (A and D). Statistical significance was determined with the Kruskal-Wallis test followed by the Dunn’s test. Results are representatives of at least two independent experiments.
Figure 7.
SV.IL12 Triggers Innate Immune Response and Induces iNOS Expression in MyC-CaP.Fluc Tumors
MyC-CaP.Fluc (A) or CT26.Fluc (B) tumors were harvested after 2 weeks of treatment from control and treated mice. Tumors were stained by multiplex immunofluorescence. Representative images are shown for untreated (control), anti-OX40, SV.IL12, and SV.IL12+anti-OX40 treated mice. Proteins of interest were stained and are indicated by color in each image: iNOS (cyan), arginase 1 (green), and CD11b (magenta). DAPI nuclear staining appears in blue.
In summary, our findings clearly show that even in absence of direct SV tumor targeting, SV.IL12 in combination with anti-OX40 alters the tumor microenvironment in two distinct solid tumors through an indirect and immunity driven mechanism that enhances T cell infiltration and intratumoral T cell immunity.
Discussion
We have developed a practical strategy for cancer immunotherapy using an OV and anti-OX40. This strategy takes advantage of the preexisting T cell immune repertoire in vivo, removing the need to know about present tumor antigens. We have shown in this study that the combination of replication-deficient SV.IL12 and anti-OX40 amplifies these antitumor T cells and induces their action throughout the body against two distinct solid tumors, reversing effectively local tumor-mediated immune suppression. This effect was specific for combination therapy and was not observed during SV.IL12 or anti-OX40 treatment alone.
Interestingly, both tumor models required different treatment frequencies to achieve the observed therapeutic efficacy of combination therapy. The reason for this discrepancy is not fully understood, but we suggest that it is multifactorial. First, both tumor models are being tested in two different mouse strains, which have different immune responses against infection/inflammation and cancer formation. For example, FVB mice have a Th2 bias in cellular immunity as compared to a strain such as BALB/c.52 The phenotype bias may, therefore, greatly impact the immunologic surveillance as well as the ability to respond to immune-based cancer therapies, as previously reported.53, 54, 55, 56, 57, 58 Furthermore, several studies comparing the sensitivity of FVB and BALB/c mice to a chemical or viral induction of cancer reported that FVB mice are more susceptible to tumor formation than are BALB/c mice.59,60 Taken together, these results suggest that BALB/c mice are naturally more resistant to cancer compared to FVB mice and, therefore, might require less frequent injections of SV.IL12 and anti-OX40. Second, the tumor microenvironment is very different in both tumor models, with differences in the cell type and composition. It has been published that the tumor mutational load correlates with better response rates to immunotherapies.61,62 Prostate cancer models have been described to have low tumor mutational burden and, therefore, are less sensitive to immunotherapies.63, 64, 65 In contrast, CT26 colon cancer cells are relatively more immunogenic with a higher tumor mutational burden and show better response rates to immunotherapies.66,67
The synergistic therapeutic effect between peritoneally delivered SV.IL12 and anti-OX40 antibody can be explained by the fact that SV.IL12 induced the expression of the OX40 target on CD4 T cells in vivo. The mechanism by which SV.IL12 enhances OX40 expression on CD4 T cells is unclear, but viruses are known to stimulate OX40 expression on anti-viral CD 4 T cells in the presence of their cognate antigen.68 Cytokines IL-12 and IFNγ are likely to be important, as previously reported by Sagiv-Barfi et al.69 They prolong survival and allow maintenance of these cells. The study of Sagiv-Barfi et al.69 demonstrated that stimulation with either the Toll-like receptor 9 ligand CpG or the Toll-like receptor 7/8 ligand single-stranded RNA (ssRNA) enhances OX40 expression on CD4 T cells through myeloid-derived cytokine production of IL-12, IFNγ, and TNF⍺. Due to the fact that SV.IL12 carries a ssRNA, infects myeloid cells, and induces IL-12 expression in infected cells as well as induces IFNγ production in T cells, the same mechanism is likely to apply.
Using SV.IL12 in combination therapy has many advantages compared to other immunostimulatory agents, such as CpG. OVs can promote tumor killing via various mechanisms, including direct tumor lysis, stimulation of enhanced local and systemic immune responses, modulation of the tumor microenvironment, and providing virally encoded immunomodulatory molecules. First, we show that Sindbis replication-deficient OV vector expressing IL-12 overcomes the need to rely on adequate tumor immune infiltration prior to treatment start. Pre-clinical and clinical data have shown that replication-competent OV vectors can markedly increase immune cell infiltration, including macrophages, CD4 T cells, and cytotoxic CD8 T cells into the local tumor microenvironment.8,33,70, 71, 72 This “priming” by viral infection can change a “cold” tumor microenvironment into a “hot” one with the influx of a multitude of immune cells and cytokines. Interestingly, immunohistochemistry of CT26.Fluc and MyC-CaP.Fluc tumor sections clearly showed that non-spreading SV.IL12 enhances T cell infiltration in both tumor models. However, the intensity of T cell infiltration was correlated with the ability of SV to transduce tumor cells directly, with much more infiltration in the SV-susceptible tumor model MyC-CaP.Fluc. Second, SV therapy does not require intratumoral injection, which is a potential drawback for many immunotherapies. SV overcomes this issue by being a blood-borne viral vector. It can travel to the tumor microenvironment and bind to overexpressed laminin receptors on cancer cells.73 In addition to its oncolytic potential, SV enters peripheral lymphoid organs, such as mediastinal lymph nodes, where it infects macrophages and activates T cells.32,33 Thus, even in absence of oncolytic activity, SV induces a systemic immune response, which has been shown to be required for effective immunotherapy.74 The addition of IL-12 into SV vector adds an additional support to the immune system, as IL-12 is a potent pro-inflammatory cytokine that promotes CD4 T cell differentiation into Th1 cells as well as directly activates CD8 T cells and enhances their cytotoxic potential.26,75
Our findings reveal that SV.IL12 in combination with anti-OX40 induces a dramatic expansion of ICOS+T-bet+ CD4 T cells, which was not observed in other treatment groups. The expansion of ICOS+ CD4 T cells is of interest, as previous studies also reported expansion of these cells during successful anti-CTLA4 therapy.3,36,76,77 In murine models, ICOS expression during anti-CTLA4 or anti-OX40 therapy was required for inducing optimal anti-tumor responses and tumor rejection, suggesting a crucial role of ICOS+ CD4 T cells on disease outcome.78,79 The increase in the frequency of ICOS+ CD4 T cells was also correlated with clinical benefit in a small cohort of patients with melanoma who received anti-CTLA4 therapy.37 Although many reports indicate a crucial role of ICOS+ CD4 T cells during anti-CTLA4 therapy, the precise function of these cells remains an open question. It is tempting to speculate that expansion of ICOS+ CD4 T cells improves the anti-tumor responses by enhancing T cell infiltration, cytolytic CD8 activity, and T cell memory formation. Addressing these possibilities is of great importance given the fundamental understanding that CD4 help is critical for the development of robust T cell responses, as well as recent findings that peripheral CD4 T cells are critical for effective immunotherapy.74
The high metabolic activity of cancer cells together with the poor vasculature blood supply in the tumor microenvironment can induce nutrient deprivation.14,15,38 These conditions can impair TCR signaling, glycolytic and mitochondrial metabolism, as well as decrease mitochondrial mass, all hallmarks of T effector cells, resulting in impaired anti-tumor effector functions of tumor-specific T cells.39, 40, 41, 42 Previous studies have reported the importance of metabolic reprogramming of T cells to rescue mitochondrial function, which resulted in increased anti-tumor responses in the tumor.39,80 Our data in two distinct models of cancer immunotherapy support the notion that SV.IL12 in combination with OX40 signaling provides the necessary metabolic support to T cells to generate an efficient anti-tumor response. This metabolic support is characterized most prominently by elevated mitochondrial function and mass in CD8 T cells as well as a switch to aerobic glycolysis in CD4 T cells. Previous reports demonstrated that aerobic glycolysis is crucial for T cell expansion and proliferation during the effector phase as well as promotes Th1 differentiation.19,81,82 These studies are in line with our results, showing enhanced Th1 differentiation and increased proliferation of T cells in mice treated with combined therapy. Furthermore, Myc has been shown to act as a key regulator of aerobic glycolysis in T cells.83 In accordance with that study, T cells from mice treated with SV.IL12 in combination with anti-OX40 demonstrated enhanced protein expression of c-Myc compared with all other groups. Thus, our study clearly shows that T cells are metabolically reprogrammed in the periphery during combination therapy. However, further experiments have to be performed to investigate whether the observed metabolic changes in peripheral T cells are also maintained in tumor-infiltrating T cells during SV treatment.
In conclusion, our data strongly show that the therapeutic efficacy of SV.IL12 with anti-OX40 is driven by T cell modulation and reprogramming of its metabolic state, in order to enhance the anti-tumor response in the periphery and in the tumor microenvironment. Furthermore, the use of SV allows these metabolically reprogrammed T cells to better infiltrate the tumor microenvironment, which is crucial for an adequate immunotherapy. Anti-OX40 antibody is currently being studied in phase 1 and 2 clinical trials. SV will be tested as a single agent in its first clinical trial in the third quarter of 2020. The results from our current preclinical studies provide a strong rationale for combining SV.IL12 with agonistic anti-OX40 antibodies in a therapeutic format in patients with solid tumors.
Materials and Methods
Cell Lines
Baby Hamster kidney (BHK), BALB/c colon carcinoma (CT26.WT [ATCC CRL-2638]), and FVB prostate carcinoma (MyC-CaP [ATCC CRL-3255]) cell lines were obtained from the American Type Culture Collection. Fluc-expressing CT26 and MyC-CaP cells (CT26.Fluc and MyC-CaP.Fluc) were generated by stable transfection of pGL4.20_Fluc plasmid.
BHK cells were maintained in α-modified minimum essential medium (α-MEM) (Corning Cellgro) with 5% fetal calf serum (FCS, Gibco) and 100 mg/mL penicillin-streptomycin (Corning Cellgro). CT26.Fluc and MyC-CaP.Fluc cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) containing 4.5 g/L glucose (Corning Cellgro) supplemented with 10% FCS, 100 mg/mL penicillin-streptomycin, 7.5 μg/mL puromycin, or 400 μg/mL gentamycin, respectively. All cell lines were cultured at 37°C and 5% CO2.
SV Production
All SV viral vectors used in these studies are replication defective. Vectors were produced as previously described. SV.IL12 plasmid used in this study was generated as published in 2002.84 SV empty is the same plasmid without an additional gene of interest (e.g., IL-12). SV.Luc was generated as described.22 SV.GFP was generated as published in 2012.85 Briefly, plasmids carrying the replicon (e.g., SinRep-IL12) or DHBB helper RNAs were linearized with XhoI. In vitro transcription was performed using the mMESSAGE mMACHINE RNA transcription kit (Ambion). Helper and replicon RNAs were then electroporated into BHK cells and incubated at 37°C in α-MEM supplemented with 10% FCS. After 12 h, the media were replaced with Opti-MEM (Gibco-BRL) supplemented with CaCl2 (100 mg/L) and cells were incubated at 37°C. After 24 h, the supernatant was collected, centrifuged to remove cellular debris, and frozen at −80°C. Vectors were titrated as previously described.84
In Vivo Experiments and Tumor Models
All experiments were performed in accordance with the Institutional Animal Care and Use Committee of New York University Health. Six- to 12-week-old female BALB/c mice were purchased from Taconic (Germantown, NY, USA) and age-matched male FVB/NJ mice were purchased from Jackson Laboratory.
Tumor Inoculation and Animal Studies
Treatment started on day 4 after i.p. inoculation of 7 × 104 CT26.Fluc cells or 105 cells of MyC-CaP.Fluc in 500 μL of Opti-MEM. For treatments, mice were randomized and SV (107 transduction units [TU]/mL), in a total volume of 500 μL, was injected i.p. into the left side of the animal once for CT26.Fluc and 4 days a week (days 1, 2, 3, and 4) for a total of 4 weeks for MyC-CaP.Fluc-inoculated mice. The immune checkpoint inhibitor anti-OX40 (clone OX-86, Bio X Cell) was injected i.p. into the left side of the animal at a dose of 250 μg per injection (once per week for the CT26.Fluc and three times per week for the MyC-CaP.Fluc tumor-bearing mice). Therapeutic efficacy of the treatment was monitored in two ways: tumor luminescence and survival. Noninvasive bioluminescent imaging was performed using the IVIS (in vivo imaging system) Spectrum imaging system (Caliper Life Sciences) at the indicated time points, and tumor growth was quantified using the Living Image 3.0 software (Caliper Life Sciences) as previously described.86 Relative tumor growth for each mouse was calculated by dividing total body counts of a given day by total body counts of the first IVIS image. Survival was monitored and recorded daily.
Flow Cytometry
For flow cytometry analysis, spleens and tumors were harvested from mice and processed as previously described.33 The extracted tumors were chopped into small pieces and incubated with a digestive mix containing RPMI 1640 with collagenase IV (50 μg/mL) and DNase I (20 U/mL) for 1 h at 37°C. Tumor samples had additional hyaluronidase V (50 μg/mL) in the digestive mix.
Spleens and digested tumors were mashed through a 70-μm strainer before red blood cells were lysed using ammonium-chloride-potassium (ACK) lysis buffer (Gibco). Cells were washed with PBS containing 1% FCS, and surface receptors were stained using various antibodies. Fluorochrome-conjugated antibodies against mouse CD3, CD4, CD44, ICOS, OX40, CD69, Foxp3, granzyme B, and T-bet were purchased from BioLegend. Fluorochrome-conjugated antibodies against mouse CD8a were purchased from BD Biosciences. MitoTracker Deep Red FM, MitoTracker Green, and fluorchrome-conjugated antibodies against CXCR3 and Ki-67 were purchased from Thermo Fisher Scientific. Stained cells were fixed with PBS containing 4% formaldehyde. For intracellular staining, the forkhead box P3 (FOXP3) staining buffer set was used (eBioscience). Flow cytometry analysis was performed on an LSR II machine (BD Biosciences) and data were analyzed using FlowJo (Tree Star).
T Cell Isolation
Total T cells were freshly isolated with the EasySep mouse T cell isolation kit. Freshly isolated lymphocytes were depleted of either CD4- or CD8-specific T cells using EasySep mouse CD4+ and CD8+ selection kits II. Isolation of T cells and depletions were performed according to the manufacturer’s protocols (STEMCELL Technologies).
ELISPOT
ELISPOT was performed as previously described.33 Splenocytes and T cells were prepared as described for flow cytometry. Mouse IFNγ ELISPOT was performed according to the manufacturer’s protocol (BD Biosciences). Lymphocytes (4 × 105 cells) and isolated T cells (8 × 104 cells) were plated per well overnight in RPMI 1640 supplemented with 10% FCS. No additional stimulus was used in the ELISPOT. As positive control, cells were stimulated with 5 ng/mL phorbol myristate acetate (PMA) + 1 μg/mL ionomycin.
Ex Vivo Cytotoxic Assay
T cells were isolated on day 7 and day 14 during treatment. T cells (8 × 105/mL) were co-cultured with CT26.Fluc cells (2 × 104/mL) or MyC-CaP.Fluc cells (2 × 104/mL) in a 24-well plate for 2 days in 1 mL of RPMI 1640 supplemented with 10% FCS. Cells were washed with PBS and lysed with 100 μL of M-PER mammalian protein extraction reagent (Promega) per well. Cytotoxicity was assessed based on the viability of CT26 cells, which was determined by measuring the luciferase activity in each well. Luciferase activity was measured by adding 100 μL of Steady-Glo reagent (Promega) to each cell lysate and measuring the luminescence using a GloMax portable luminometer (Promega).
CD8+ and CD4+ T Cell Depletion In Vivo
CD8+ T cells were depleted using anti-CD8 antibody (clone 2.43) (Bio X Cell, Lebanon, NH, USA). 0.1 mg of antibody in 0.2 mL of PBS was injected into each mouse, starting 1 day before the first SV treatment, and then every 4 days for 2 weeks. CD4+ T cells were depleted using anti-CD4 antibody (clone GK 1.5) (Bio X Cell, Lebanon, NH, USA). 0.4 mg was injected into each mouse, starting day 1 before the first treatment. Control mice were injected with PBS and isotype controls.
Quantitative Real-Time PCR
RNA was extracted from tumor samples using an RNeasy kit (QIAGEN), followed by cDNA synthesis with the iScript II kit (Bio-Rad). qRT-PCR was performed using iQ SYBR Green Supermix (Bio-Rad) and a StepOne real-time PCR detection system (Applied Biosystems). PCR conditions were as follows: 95°C for 10 min, followed by 40 cycles (94°C for 30 s, 58°C for 30 s) of amplification. For quantitation, CT values were normalized to GAPDH and expression was analyzed using the 2−ΔΔCT method. Primers for CXCL9, CXCL10, and GAPDH were used, which included the following: CXCL9 (forward, 5′-GAAGTCCGCTGTTCTTTTCC-3′, reverse, 5′-TTGACTTCCGTTCTTCAGTG-3′) and CXCL10 (forward, 5′-GCTGCAACTGCATCCATATC-3′, reverse, 5′-AGGAGCCCTTTTAGACCTTT-3′).
Transcriptome Analysis of T Cells
Total RNA was extracted from freshly isolated T cells on day 7 of treatment from spleens using an RNeasy kit (QIAGEN). For each group, three BALB/C mice or three FVB/J mice were used for biological repeats. RNA sequencing was done by the NYU Langone Genome Technology Center. RNA quality and quantity were analyzed. RNA sequencing libraries were prepared and loaded on the automated HiSeq 4000 sequencing system (Illumina) and run as single 50-nt reads.
Alignment and Differential Expression Analysis
Sequences were aligned to the mm10 mouse genome using Bowtie software (version 1.0.0)87with two mismatches allowed. Uniquely mapped reads were further processed by removing PCR duplicates with Picard (“Picard Tools,” Broad Institute, GitHub repository: http://broadinstitute.github.io/picard/). MarkDuplicates and transcripts were counted using HTSeq88 and differential gene expression was performed between all groups using DESeq.89 Differences in gene expression were considered significant with an adjusted p value <0.05.
GSEA and Enrichment Map Analysis
The network-based method enrichment map90 was used for gene-set enrichment visualization and interpretation of data. As a follow-up analysis of GSEA2,91 it reduces redundancy and helps in the interpretation of large gene sets and helps to quickly identify major enriched functional themes in the gene expression data. To perform this analysis, we first assigned a unique row identifier for each transcript and obtained DEGs through DESeq.89 These genes were then ranked and GSEA was performed in the GenePattern92 server using the GSEA pre-ranked module. We then obtained the gene identifiers corresponding to the gene names using the Bioconductor package org.Mm.eg.db, and the resulting positively and negatively regulated gene identifiers were used to generate enrichment maps in Cytoscape.93 The expression heatmap as drawn by Morpheus (https://software.broadinstitute.org/morpheus/). Highest and lowest expressions for each gene (row minimum and row maximum) were displayed as red or blue, respectively.
Measurement of OCRs and ECARs of T Cells
T cell metabolic output was measured by Seahorse technology as previously described.94 Purified T cells were plated at 6 × 105 cells/well in an Agilent Seahorse XF24 cell culture microplate. OCR and ECAR were measured using an Agilent Seahorse XFe24 metabolic analyzer following the procedure recommended by the manufacturer (Agilent Technologies). For the mitochondrial stress test, (1) oligomycin (1 μM), (2) carbonyl cyanide 4-(trifluoromethoxy)phenylhydrazone (FCCP) (1.5 μM), and (3) rotenone (100 nM) and antimycin A (1 μM) were injected sequentially through ports A, B, and C.
Immunoblot Analysis
Cells were lysed in M-PER mammalian protein extraction reagent according to the manufacturer’s protocol. Lysates were separated by SDS-PAGE on 4%–12% Bio-Rad gels, transferred to polyvinylidene fluoride (PVDF) membranes, blocked in 5% milk in Tris-buffered saline (TBS) buffer with 0.1% Tween 20 (TBST). Primary antibodies to c-Myc (Santa Cruz Biotechnology) and GAPDH (Thermo Fisher Scientific) were added at room temperature or overnight at 4°C. Secondary fluorescent antibodies (IRDye, LI-COR Biosciences) were added in 5% milk in TBST for 1 h at room temperature. An Odyssey Classic infrared imaging system was used for visualization.
Histochemistry and Multiplex Immunofluorescence (MIF)
Tumors of mice were collected, fixed in 4% paraformaldehyde (PFA) for 2 days and embedded in paraffin, sectioned, and H&E stained.
For MIF staining and imaging, 5-μm paraffin sections were stained with an Akoya Biosciences Opal multiplex automation kit on a Leica BondRX autostainer, according to the manufacturers’ instructions. Prior to incubation with the first primary antibody, sections underwent heat retrieval with Bond Epitope Retrieval Buffer 2 (Leica ER2, AR9640) and blocking. Primary antibodies in panel 1 were against CD3 (1:200, Bio-Rad, MCA1477T), CD8 (1:2,000, Cell Signaling Technology, 98941S), and Ki-67 (1:200, Abcam, AB16667). Primary antibodies in panel 2 recognized iNOS (1:1,000, GeneTex, GTX130246), Arg1 (1:750, GeneTex, GTX109242), granzyme B (1:1,000, Abcam, AB4059), CD11b (1:10,000, Abcam, AB133357), and F480 (1:250, Cell Signaling Technology, 70076S). Each primary antibody was followed by a cocktail of horseradish peroxidase-conjugated secondary antibodies against mouse and rabbit immunoglobulin G (IgG) (RTU, Akoya/PerkinElmer, catalog no. ARH1001) and then tyramide-mediated signal amplification (TSA) with covalent linkage of the individual Opal fluorophore (each 1:250, Opal 520 [FP1496001KT], 540 [FP1487001KT], 570 [FP1494001KT], 620 [FP1488001KT], 650 [FP1495001KT], or 690 [FP1497001KT], Akoya/PerkinElmer [catalog nos.]) to the tissue antigen. Antibodies were subsequently stripped using either ER1 (Leica, AR9961) or ER2 (Leica, AR9640) heat retrieval buffer, and the next round of immunostaining was initiated. After completion of the sequential incubations and stripping, slides were counterstained with spectral DAPI (Akoya/PerkinElmer, FP1490). Monoplex controls were used to confirm appropriate staining for antibodies integrated into the multiplex panels. Multispectral imaging was performed on a Vectra3 imaging system (Akoya/PerkinElmer) at ×20. The fluorophore emission signatures were captured by a multispectral camera and then unmixed with INFORM software (Akoya/PerkinElmer). Autofluorescence, obtained from an unstained slide, was removed from the composites and pseudo-colored images were exported as tif files.
Statistical Analysis
Statistical analysis was performed using GraphPad Prism 7.0 as described in the figure legends. All data are shown as mean ± SEM. Figures were prepared using GraphPad Prism 7, Adobe Photoshop, and ImageJ software. Treated groups were compared, with a one-way analysis using GraphPad Prism 7, to naive mice. Differences with a p value of <0.05 were considered significant: ∗p < 0.05, ∗∗p < 0.005, ∗∗∗p < 0.0001.
Author Contributions
I.S., S.O., and D.M. conceived the study. I.S., S.O., C.L., and K.K. designed experiments. I.S., S.O., C.L., K.K., A.M.H., C.P., and M.Y. performed mouse experiments and/or data analysis. I.S., O.S., and D.M. wrote the manuscript. All authors reviewed and edited the manuscript.
Conflicts of Interest
All authors are employed by NYU Langone School of Medicine and have no employment relationship or consultancy agreement with Cynvec. I.S., A.M.H., C.P., and D.M. are inventors on one or several issued patents and/or patent applications held by NYU that cover Sindbis treatment of neoplasia. As part of the Research and Licensing Agreement, authors who are inventors on patents are entitled to a portion of NYU Langone’s royalties received from Cynvec, should Sindbis vectors be approved by the US Food and Drug Administration (FDA) for the treatment of cancer. As part of the Licensing Agreement, NYU Langone has apportioned stock from its equity stake in Cynvec. The remaining authors declare no competing interests.
Acknowledgments
This work was supported by NIH Grant 5R44CA206606 and through a Research and Licensing Agreement between Cynvec and NYU Langone, which licenses the Sindbis technology to Cynvec. This work was also supported, in part, by the Experimental Pathology Research Laboratory at NYU Langone, which is partially supported by the Cancer Center Support Grant P30CA016087. The Vectra3 multispectral imaging system was purchased through Shared Instrumentation Grant S10 OD021747. We would like to thank the NYU High Throughput Biology Laboratory for Seahorse usage, training, and assistance. We are grateful to Shanshan Feng for support in viral vector preparation and titration.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omto.2020.04.012.
Supplemental Information
References
- 1.Pardoll D.M. The blockade of immune checkpoints in cancer immunotherapy. Nat. Rev. Cancer. 2012;12:252–264. doi: 10.1038/nrc3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sharma P., Allison J.P. The future of immune checkpoint therapy. Science. 2015;348:56–61. doi: 10.1126/science.aaa8172. [DOI] [PubMed] [Google Scholar]
- 3.Wei S.C., Levine J.H., Cogdill A.P., Zhao Y., Anang N.A., Andrews M.C., Sharma P., Wang J., Wargo J.A., Pe’er D., Allison J.P. Distinct cellular mechanisms underlie anti-CTLA-4 and anti-PD-1 checkpoint blockade. Cell. 2017;170:1120–1133.e17. doi: 10.1016/j.cell.2017.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li X., Wenes M., Romero P., Huang S.C., Fendt S.M., Ho P.C. Navigating metabolic pathways to enhance antitumour immunity and immunotherapy. Nat. Rev. Clin. Oncol. 2019;16:425–441. doi: 10.1038/s41571-019-0203-7. [DOI] [PubMed] [Google Scholar]
- 5.Galon J., Bruni D. Approaches to treat immune hot, altered and cold tumours with combination immunotherapies. Nat. Rev. Drug Discov. 2019;18:197–218. doi: 10.1038/s41573-018-0007-y. [DOI] [PubMed] [Google Scholar]
- 6.Ribas A., Dummer R., Puzanov I., VanderWalde A., Andtbacka R.H.I., Michielin O., Olszanski A.J., Malvehy J., Cebon J., Fernandez E. Oncolytic virotherapy promotes intratumoral T cell infiltration and improves anti-PD-1 immunotherapy. Cell. 2018;174:1031–1032. doi: 10.1016/j.cell.2018.07.035. [DOI] [PubMed] [Google Scholar]
- 7.Sun L., Funchain P., Song J.M., Rayman P., Tannenbaum C., Ko J., Mcnamara M., Marcela Diaz-Montero C., Gastman B. Talimogene laherparepvec combined with anti-PD-1 based immunotherapy for unresectable stage III-IV melanoma: a case series. J. Immunother. Cancer. 2018;6:36. doi: 10.1186/s40425-018-0337-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.LaRocca C.J., Warner S.G. Oncolytic viruses and checkpoint inhibitors: combination therapy in clinical trials. Clin. Transl. Med. 2018;7:35. doi: 10.1186/s40169-018-0214-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Senior M. Checkpoint inhibitors go viral. Nat. Biotechnol. 2019;37:12–17. doi: 10.1038/nbt.4327. [DOI] [PubMed] [Google Scholar]
- 10.Andtbacka R.H., Kaufman H.L., Collichio F., Amatruda T., Senzer N., Chesney J., Delman K.A., Spitler L.E., Puzanov I., Agarwala S.S. Talimogene laherparepvec improves durable response rate in patients with advanced melanoma. J. Clin. Oncol. 2015;33:2780–2788. doi: 10.1200/JCO.2014.58.3377. [DOI] [PubMed] [Google Scholar]
- 11.Zamarin D., Holmgaard R.B., Subudhi S.K., Park J.S., Mansour M., Palese P., Merghoub T., Wolchok J.D., Allison J.P. Localized oncolytic virotherapy overcomes systemic tumor resistance to immune checkpoint blockade immunotherapy. Sci. Transl. Med. 2014;6:226ra32. doi: 10.1126/scitranslmed.3008095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dias J.D., Hemminki O., Diaconu I., Hirvinen M., Bonetti A., Guse K., Escutenaire S., Kanerva A., Pesonen S., Löskog A. Targeted cancer immunotherapy with oncolytic adenovirus coding for a fully human monoclonal antibody specific for CTLA-4. Gene Ther. 2012;19:988–998. doi: 10.1038/gt.2011.176. [DOI] [PubMed] [Google Scholar]
- 13.Jiang H., Rivera-Molina Y., Gomez-Manzano C., Clise-Dwyer K., Bover L., Vence L.M., Yuan Y., Lang F.F., Toniatti C., Hossain M.B., Fueyo J. Oncolytic adenovirus and tumor-targeting immune modulatory therapy improve autologous cancer vaccination. Cancer Res. 2017;77:3894–3907. doi: 10.1158/0008-5472.CAN-17-0468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Delgoffe G.M. Filling the tank: keeping antitumor T cells metabolically fit for the long haul. Cancer Immunol. Res. 2016;4:1001–1006. doi: 10.1158/2326-6066.CIR-16-0244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Scharping N.E., Delgoffe G.M. Tumor microenvironment metabolism: a new checkpoint for anti-tumor immunity. Vaccines (Basel) 2016;4:46. doi: 10.3390/vaccines4040046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Allison K.E., Coomber B.L., Bridle B.W. Metabolic reprogramming in the tumour microenvironment: a hallmark shared by cancer cells and T lymphocytes. Immunology. 2017;152:175–184. doi: 10.1111/imm.12777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gupta S., Roy A., Dwarakanath B.S. Metabolic cooperation and competition in the tumor microenvironment: implications for therapy. Front. Oncol. 2017;7:68. doi: 10.3389/fonc.2017.00068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ocaña M.C., Martínez-Poveda B., Quesada A.R., Medina M.A. Metabolism within the tumor microenvironment and its implication on cancer progression: an ongoing therapeutic target. Med. Res. Rev. 2019;39:70–113. doi: 10.1002/med.21511. [DOI] [PubMed] [Google Scholar]
- 19.Pearce E.L., Poffenberger M.C., Chang C.H., Jones R.G. Fueling immunity: insights into metabolism and lymphocyte function. Science. 2013;342:1242454. [Google Scholar]
- 20.Aspeslagh S., Postel-Vinay S., Rusakiewicz S., Soria J.C., Zitvogel L., Marabelle A. Rationale for anti-OX40 cancer immunotherapy. Eur. J. Cancer. 2016;52:50–66. doi: 10.1016/j.ejca.2015.08.021. [DOI] [PubMed] [Google Scholar]
- 21.Zhang X., Xiao X., Lan P., Li J., Dou Y., Chen W., Ishii N., Chen S., Xia B., Chen K. OX40 costimulation inhibits Foxp3 expression and Treg induction via BATF3-dependent and independent mechanisms. Cell Rep. 2018;24:607–618. doi: 10.1016/j.celrep.2018.06.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tseng J.C., Levin B., Hurtado A., Yee H., Perez de Castro I., Jimenez M., Shamamian P., Jin R., Novick R.P., Pellicer A., Meruelo D. Systemic tumor targeting and killing by Sindbis viral vectors. Nat. Biotechnol. 2004;22:70–77. doi: 10.1038/nbt917. [DOI] [PubMed] [Google Scholar]
- 23.Bredenbeek P.J., Frolov I., Rice C.M., Schlesinger S. Sindbis virus expression vectors: packaging of RNA replicons by using defective helper RNAs. J. Virol. 1993;67:6439–6446. doi: 10.1128/jvi.67.11.6439-6446.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Menk A.V., Scharping N.E., Rivadeneira D.B., Calderon M.J., Watson M.J., Dunstane D., Watkins S.C., Delgoffe G.M. 4-1BB costimulation induces T cell mitochondrial function and biogenesis enabling cancer immunotherapeutic responses. J. Exp. Med. 2018;215:1091–1100. doi: 10.1084/jem.20171068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Granot T., Venticinque L., Tseng J.C., Meruelo D. Activation of cytotoxic and regulatory functions of NK cells by Sindbis viral vectors. PLoS ONE. 2011;6:e20598. doi: 10.1371/journal.pone.0020598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu J., Cao S., Kim S., Chung E.Y., Homma Y., Guan X., Jimenez V., Ma X. Interleukin-12: an update on its immunological activities, signaling and regulation of gene expression. Curr. Immunol. Rev. 2005;1:119–137. doi: 10.2174/1573395054065115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Henry C.J., Ornelles D.A., Mitchell L.M., Brzoza-Lewis K.L., Hiltbold E.M. IL-12 produced by dendritic cells augments CD8+ T cell activation through the production of the chemokines CCL1 and CCL17. J. Immunol. 2008;181:8576–8584. doi: 10.4049/jimmunol.181.12.8576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wilson D.C., Matthews S., Yap G.S. IL-12 signaling drives CD8+ T cell IFN-γ production and differentiation of KLRG1+ effector subpopulations during Toxoplasma gondii infection. J. Immunol. 2008;180:5935–5945. doi: 10.4049/jimmunol.180.9.5935. [DOI] [PubMed] [Google Scholar]
- 29.Ylikoski E., Lund R., Kyläniemi M., Filén S., Kilpeläinen M., Savolainen J., Lahesmaa R. IL-12 up-regulates T-bet independently of IFN-γ in human CD4+ T cells. Eur. J. Immunol. 2005;35:3297–3306. doi: 10.1002/eji.200526101. [DOI] [PubMed] [Google Scholar]
- 30.Szabo S.J., Kim S.T., Costa G.L., Zhang X., Fathman C.G., Glimcher L.H. A novel transcription factor, T-bet, directs Th1 lineage commitment. Cell. 2000;100:655–669. doi: 10.1016/s0092-8674(00)80702-3. [DOI] [PubMed] [Google Scholar]
- 31.Fountzilas C., Patel S., Mahalingam D. Review: oncolytic virotherapy, updates and future directions. Oncotarget. 2017;8:102617–102639. doi: 10.18632/oncotarget.18309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Granot T., Yamanashi Y., Meruelo D. Sindbis viral vectors transiently deliver tumor-associated antigens to lymph nodes and elicit diversified antitumor CD8+ T-cell immunity. Mol. Ther. 2014;22:112–122. doi: 10.1038/mt.2013.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Scherwitzl I., Hurtado A., Pierce C.M., Vogt S., Pampeno C., Meruelo D. Systemically administered Sindbis virus in combination with immune checkpoint blockade induces curative anti-tumor immunity. Mol. Ther. Oncolytics. 2018;9:51–63. doi: 10.1016/j.omto.2018.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang P.Y., Guo J.H., Hwang L.H. Oncolytic Sindbis virus targets tumors defective in the interferon response and induces significant bystander antitumor immunity in vivo. Mol. Ther. 2012;20:298–305. doi: 10.1038/mt.2011.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Joshi N.S., Cui W., Chandele A., Lee H.K., Urso D.R., Hagman J., Gapin L., Kaech S.M. Inflammation directs memory precursor and short-lived effector CD8+ T cell fates via the graded expression of T-bet transcription factor. Immunity. 2007;27:281–295. doi: 10.1016/j.immuni.2007.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ng Tang D., Shen Y., Sun J., Wen S., Wolchok J.D., Yuan J., Allison J.P., Sharma P. Increased frequency of ICOS+ CD4 T cells as a pharmacodynamic biomarker for anti-CTLA-4 therapy. Cancer Immunol. Res. 2013;1:229–234. doi: 10.1158/2326-6066.CIR-13-0020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Carthon B.C., Wolchok J.D., Yuan J., Kamat A., Ng Tang D.S., Sun J., Ku G., Troncoso P., Logothetis C.J., Allison J.P., Sharma P. Preoperative CTLA-4 blockade: tolerability and immune monitoring in the setting of a presurgical clinical trial. Clin. Cancer Res. 2010;16:2861–2871. doi: 10.1158/1078-0432.CCR-10-0569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chang C.H., Qiu J., O’Sullivan D., Buck M.D., Noguchi T., Curtis J.D., Chen Q., Gindin M., Gubin M.M., van der Windt G.J. Metabolic competition in the tumor microenvironment is a driver of cancer progression. Cell. 2015;162:1229–1241. doi: 10.1016/j.cell.2015.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Scharping N.E., Menk A.V., Moreci R.S., Whetstone R.D., Dadey R.E., Watkins S.C., Ferris R.L., Delgoffe G.M. The tumor microenvironment represses T cell mitochondrial biogenesis to drive intratumoral T cell metabolic insufficiency and dysfunction. Immunity. 2016;45:701–703. doi: 10.1016/j.immuni.2016.08.009. [DOI] [PubMed] [Google Scholar]
- 40.Ho P.C., Bihuniak J.D., Macintyre A.N., Staron M., Liu X., Amezquita R., Tsui Y.C., Cui G., Micevic G., Perales J.C. Phosphoenolpyruvate is a metabolic checkpoint of anti-tumor T cell responses. Cell. 2015;162:1217–1228. doi: 10.1016/j.cell.2015.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Siska P.J., Rathmell J.C. T cell metabolic fitness in antitumor immunity. Trends Immunol. 2015;36:257–264. doi: 10.1016/j.it.2015.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhao E., Maj T., Kryczek I., Li W., Wu K., Zhao L., Wei S., Crespo J., Wan S., Vatan L. Cancer mediates effector T cell dysfunction by targeting microRNAs and EZH2 via glycolysis restriction. Nat. Immunol. 2016;17:95–103. doi: 10.1038/ni.3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Buck M.D., O’Sullivan D., Pearce E.L. T cell metabolism drives immunity. J. Exp. Med. 2015;212:1345–1360. doi: 10.1084/jem.20151159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brown D.M. Cytolytic CD4 cells: direct mediators in infectious disease and malignancy. Cell. Immunol. 2010;262:89–95. doi: 10.1016/j.cellimm.2010.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mucida D., Husain M.M., Muroi S., van Wijk F., Shinnakasu R., Naoe Y., Reis B.S., Huang Y., Lambolez F., Docherty M. Transcriptional reprogramming of mature CD4+ helper T cells generates distinct MHC class II-restricted cytotoxic T lymphocytes. Nat. Immunol. 2013;14:281–289. doi: 10.1038/ni.2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reis B.S., Rogoz A., Costa-Pinto F.A., Taniuchi I., Mucida D. Mutual expression of the transcription factors Runx3 and ThPOK regulates intestinal CD4+ T cell immunity. Nat. Immunol. 2013;14:271–280. doi: 10.1038/ni.2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rao R.R., Li Q., Gubbels Bupp M.R., Shrikant P.A. Transcription factor Foxo1 represses T-bet-mediated effector functions and promotes memory CD8+ T cell differentiation. Immunity. 2012;36:374–387. doi: 10.1016/j.immuni.2012.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Verneris M.R., Karimi M., Baker J., Jayaswal A., Negrin R.S. Role of NKG2D signaling in the cytotoxicity of activated and expanded CD8+ T cells. Blood. 2004;103:3065–3072. doi: 10.1182/blood-2003-06-2125. [DOI] [PubMed] [Google Scholar]
- 49.Galon J., Costes A., Sanchez-Cabo F., Kirilovsky A., Mlecnik B., Lagorce-Pagès C., Tosolini M., Camus M., Berger A., Wind P. Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science. 2006;313:1960–1964. doi: 10.1126/science.1129139. [DOI] [PubMed] [Google Scholar]
- 50.Hwang W.T., Adams S.F., Tahirovic E., Hagemann I.S., Coukos G. Prognostic significance of tumor-infiltrating T cells in ovarian cancer: a meta-analysis. Gynecol. Oncol. 2012;124:192–198. doi: 10.1016/j.ygyno.2011.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Groom J.R., Luster A.D. CXCR3 in T cell function. Exp. Cell Res. 2011;317:620–631. doi: 10.1016/j.yexcr.2010.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kim E.M., Bae Y.M., Choi M.H., Hong S.T. Cyst formation, increased anti-inflammatory cytokines and expression of chemokines support for Clonorchis sinensis infection in FVB mice. Parasitol. Int. 2012;61:124–129. doi: 10.1016/j.parint.2011.07.001. [DOI] [PubMed] [Google Scholar]
- 53.Haabeth O.A., Lorvik K.B., Hammarström C., Donaldson I.M., Haraldsen G., Bogen B., Corthay A. Inflammation driven by tumour-specific Th1 cells protects against B-cell cancer. Nat. Commun. 2011;2:240. doi: 10.1038/ncomms1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xu X., Wang R., Su Q., Huang H., Zhou P., Luan J., Liu J., Wang J., Chen X. Expression of Th1- Th2- and Th17-associated cytokines in laryngeal carcinoma. Oncol. Lett. 2016;12:1941–1948. doi: 10.3892/ol.2016.4854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.De Monte L., Reni M., Tassi E., Clavenna D., Papa I., Recalde H., Braga M., Di Carlo V., Doglioni C., Protti M.P. Intratumor T helper type 2 cell infiltrate correlates with cancer-associated fibroblast thymic stromal lymphopoietin production and reduced survival in pancreatic cancer. J. Exp. Med. 2011;208:469–478. doi: 10.1084/jem.20101876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ochi A., Nguyen A.H., Bedrosian A.S., Mushlin H.M., Zarbakhsh S., Barilla R., Zambirinis C.P., Fallon N.C., Rehman A., Pylayeva-Gupta Y. MyD88 inhibition amplifies dendritic cell capacity to promote pancreatic carcinogenesis via Th2 cells. J. Exp. Med. 2012;209:1671–1687. doi: 10.1084/jem.20111706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gabitass R.F., Annels N.E., Stocken D.D., Pandha H.A., Middleton G.W. Elevated myeloid-derived suppressor cells in pancreatic, esophageal and gastric cancer are an independent prognostic factor and are associated with significant elevation of the Th2 cytokine interleukin-13. Cancer Immunol. Immunother. 2011;60:1419–1430. doi: 10.1007/s00262-011-1028-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Enninga E.A., Nevala W.K., Holtan S.G., Leontovich A.A., Markovic S.N. Galectin-9 modulates immunity by promoting Th2/M2 differentiation and impacts survival in patients with metastatic melanoma. Melanoma Res. 2016;26:429–441. doi: 10.1097/CMR.0000000000000281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hennings H., Glick A.B., Lowry D.T., Krsmanovic L.S., Sly L.M., Yuspa S.H. FVB/N mice: an inbred strain sensitive to the chemical induction of squamous cell carcinomas in the skin. Carcinogenesis. 1993;14:2353–2358. doi: 10.1093/carcin/14.11.2353. [DOI] [PubMed] [Google Scholar]
- 60.Woodworth C.D., Michael E., Smith L., Vijayachandra K., Glick A., Hennings H., Yuspa S.H. Strain-dependent differences in malignant conversion of mouse skin tumors is an inherent property of the epidermal keratinocyte. Carcinogenesis. 2004;25:1771–1778. doi: 10.1093/carcin/bgh170. [DOI] [PubMed] [Google Scholar]
- 61.Samstein R.M., Lee C.H., Shoushtari A.N., Hellmann M.D., Shen R., Janjigian Y.Y., Barron D.A., Zehir A., Jordan E.J., Omuro A. Tumor mutational load predicts survival after immunotherapy across multiple cancer types. Nat. Genet. 2019;51:202–206. doi: 10.1038/s41588-018-0312-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Maleki Vareki S. High and low mutational burden tumors versus immunologically hot and cold tumors and response to immune checkpoint inhibitors. J. Immunother. Cancer. 2018;6:157. doi: 10.1186/s40425-018-0479-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Olivier M., Hollstein M., Hainaut P. TP53 mutations in human cancers: origins, consequences, and clinical use. Cold Spring Harb. Perspect. Biol. 2010;2:a001008. doi: 10.1101/cshperspect.a001008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Alexandrov L.B., Nik-Zainal S., Wedge D.C., Campbell P.J., Stratton M.R. Deciphering signatures of mutational processes operative in human cancer. Cell Rep. 2013;3:246–259. doi: 10.1016/j.celrep.2012.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nik-Zainal S., Alexandrov L.B., Wedge D.C., Van Loo P., Greenman C.D., Raine K., Jones D., Hinton J., Marshall J., Stebbings L.A., Breast Cancer Working Group of the International Cancer Genome Consortium Mutational processes molding the genomes of 21 breast cancers. Cell. 2012;149:979–993. doi: 10.1016/j.cell.2012.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lechner M.G., Karimi S.S., Barry-Holson K., Angell T.E., Murphy K.A., Church C.H., Ohlfest J.R., Hu P., Epstein A.L. Immunogenicity of murine solid tumor models as a defining feature of in vivo behavior and response to immunotherapy. J. Immunother. 2013;36:477–489. doi: 10.1097/01.cji.0000436722.46675.4a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yu J.W., Bhattacharya S., Yanamandra N., Kilian D., Shi H., Yadavilli S., Katlinskaya Y., Kaczynski H., Conner M., Benson W. Tumor-immune profiling of murine syngeneic tumor models as a framework to guide mechanistic studies and predict therapy response in distinct tumor microenvironments. PLoS ONE. 2018;13:e0206223. doi: 10.1371/journal.pone.0206223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Boettler T., Moeckel F., Cheng Y., Heeg M., Salek-Ardakani S., Crotty S., Croft M., von Herrath M.G. OX40 facilitates control of a persistent virus infection. PLoS Pathog. 2012;8:e1002913. doi: 10.1371/journal.ppat.1002913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sagiv-Barfi I., Czerwinski D.K., Levy S., Alam I.S., Mayer A.T., Gambhir S.S., Levy R. Eradication of spontaneous malignancy by local immunotherapy. Sci. Transl. Med. 2018;10:eaan4488. doi: 10.1126/scitranslmed.aan4488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nakao A., Kasuya H., Sahin T.T., Nomura N., Kanzaki A., Misawa M., Shirota T., Yamada S., Fujii T., Sugimoto H. A phase I dose-escalation clinical trial of intraoperative direct intratumoral injection of HF10 oncolytic virus in non-resectable patients with advanced pancreatic cancer. Cancer Gene Ther. 2011;18:167–175. doi: 10.1038/cgt.2010.65. [DOI] [PubMed] [Google Scholar]
- 71.Nakao A., Kimata H., Imai T., Kikumori T., Teshigahara O., Nagasaka T., Goshima F., Nishiyama Y. Intratumoral injection of herpes simplex virus HF10 in recurrent breast cancer. Ann. Oncol. 2004;15:988–989. doi: 10.1093/annonc/mdh225. [DOI] [PubMed] [Google Scholar]
- 72.Kimata H., Imai T., Kikumori T., Teshigahara O., Nagasaka T., Goshima F., Nishiyama Y., Nakao A. Pilot study of oncolytic viral therapy using mutant herpes simplex virus (HF10) against recurrent metastatic breast cancer. Ann. Surg. Oncol. 2006;13:1078–1084. doi: 10.1245/ASO.2006.08.035. [DOI] [PubMed] [Google Scholar]
- 73.Wang K.S., Kuhn R.J., Strauss E.G., Ou S., Strauss J.H. High-affinity laminin receptor is a receptor for Sindbis virus in mammalian cells. J. Virol. 1992;66:4992–5001. doi: 10.1128/jvi.66.8.4992-5001.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Spitzer M.H., Carmi Y., Reticker-Flynn N.E., Kwek S.S., Madhireddy D., Martins M.M., Gherardini P.F., Prestwood T.R., Chabon J., Bendall S.C. Systemic immunity is required for effective cancer immunotherapy. Cell. 2017;168:487–502.e15. doi: 10.1016/j.cell.2016.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hsieh C.S., Macatonia S.E., Tripp C.S., Wolf S.F., O’Garra A., Murphy K.M. Development of TH1 CD4+ T cells through IL-12 produced by Listeria-induced macrophages. Science. 1993;260:547–549. doi: 10.1126/science.8097338. [DOI] [PubMed] [Google Scholar]
- 76.Vonderheide R.H., LoRusso P.M., Khalil M., Gartner E.M., Khaira D., Soulieres D., Dorazio P., Trosko J.A., Rüter J., Mariani G.L. Tremelimumab in combination with exemestane in patients with advanced breast cancer and treatment-associated modulation of inducible costimulator expression on patient T cells. Clin. Cancer Res. 2010;16:3485–3494. doi: 10.1158/1078-0432.CCR-10-0505. [DOI] [PubMed] [Google Scholar]
- 77.Chen H., Liakou C.I., Kamat A., Pettaway C., Ward J.F., Tang D.N., Sun J., Jungbluth A.A., Troncoso P., Logothetis C., Sharma P. Anti-CTLA-4 therapy results in higher CD4+ICOShi T cell frequency and IFN-γ levels in both nonmalignant and malignant prostate tissues. Proc. Natl. Acad. Sci. USA. 2009;106:2729–2734. doi: 10.1073/pnas.0813175106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Fu T., He Q., Sharma P. The ICOS/ICOSL pathway is required for optimal antitumor responses mediated by anti-CTLA-4 therapy. Cancer Res. 2011;71:5445–5454. doi: 10.1158/0008-5472.CAN-11-1138. [DOI] [PubMed] [Google Scholar]
- 79.Metzger T.C., Long H., Potluri S., Pertel T., Bailey-Bucktrout S.L., Lin J.C., Fu T., Sharma P., Allison J.P., Feldman R.M. ICOS promotes the function of CD4+ effector T cells during anti-OX40-mediated tumor rejection. Cancer Res. 2016;76:3684–3689. doi: 10.1158/0008-5472.CAN-15-3412. [DOI] [PubMed] [Google Scholar]
- 80.Balmer M.L., Hess C. Feeling worn out? PGC1α to the rescue for dysfunctional mitochondria in t cell exhaustion. Immunity. 2016;45:233–235. doi: 10.1016/j.immuni.2016.07.024. [DOI] [PubMed] [Google Scholar]
- 81.Peng M., Yin N., Chhangawala S., Xu K., Leslie C.S., Li M.O. Aerobic glycolysis promotes T helper 1 cell differentiation through an epigenetic mechanism. Science. 2016;354:481–484. doi: 10.1126/science.aaf6284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Roos D., Loos J.A. Changes in the carbohydrate metabolism of mitogenically stimulated human peripheral lymphocytes. I. Stimulation by phytohaemagglutinin. Biochim. Biophys. Acta. 1970;222:565–582. doi: 10.1016/0304-4165(70)90182-0. [DOI] [PubMed] [Google Scholar]
- 83.Wang R., Dillon C.P., Shi L.Z., Milasta S., Carter R., Finkelstein D., McCormick L.L., Fitzgerald P., Chi H., Munger J., Green D.R. The transcription factor Myc controls metabolic reprogramming upon T lymphocyte activation. Immunity. 2011;35:871–882. doi: 10.1016/j.immuni.2011.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tseng J.C., Levin B., Hirano T., Yee H., Pampeno C., Meruelo D. In vivo antitumor activity of Sindbis viral vectors. J. Natl. Cancer Inst. 2002;94:1790–1802. doi: 10.1093/jnci/94.23.1790. [DOI] [PubMed] [Google Scholar]
- 85.Suzme R., Tseng J.C., Levin B., Ibrahim S., Meruelo D., Pellicer A. Sindbis viral vectors target hematopoietic malignant cells. Cancer Gene Ther. 2012;19:757–766. doi: 10.1038/cgt.2012.56. [DOI] [PubMed] [Google Scholar]
- 86.Tseng J.C., Hurtado A., Yee H., Levin B., Boivin C., Benet M., Blank S.V., Pellicer A., Meruelo D. Using Sindbis viral vectors for specific detection and suppression of advanced ovarian cancer in animal models. Cancer Res. 2004;64:6684–6692. doi: 10.1158/0008-5472.CAN-04-1924. [DOI] [PubMed] [Google Scholar]
- 87.Langmead B., Trapnell C., Pop M., Salzberg S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Anders S., Pyl P.T., Huber W. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Anders S., Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Merico D., Isserlin R., Stueker O., Emili A., Bader G.D. Enrichment map: a network-based method for gene-set enrichment visualization and interpretation. PLoS ONE. 2010;5:e13984. doi: 10.1371/journal.pone.0013984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Mootha V.K., Lindgren C.M., Eriksson K.F., Subramanian A., Sihag S., Lehar J., Puigserver P., Carlsson E., Ridderstråle M., Laurila E. PGC-1α-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 92.Reich M., Liefeld T., Gould J., Lerner J., Tamayo P., Mesirov J.P. GenePattern 2.0. Nat. Genet. 2006;38:500–501. doi: 10.1038/ng0506-500. [DOI] [PubMed] [Google Scholar]
- 93.Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Scharping N.E., Menk A.V., Whetstone R.D., Zeng X., Delgoffe G.M. Efficacy of PD-1 blockade is potentiated by metformin-induced reduction of tumor hypoxia. Cancer Immunol. Res. 2017;5:9–16. doi: 10.1158/2326-6066.CIR-16-0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.