Abstract
The regulation of cutaneous inflammatory processes is essential for the human skin to maintain homeostasis in the presence of the dense communities of resident microbes that normally populate this organ. Forming the hair follicle-associated sebaceous gland, sebocytes are specialized lipid-producing cells that can release inflammatory mediators. Cytokine and chemokine expression by pilosebaceous epithelial cells, i.e. sebocytes and follicular keratinocytes, has been proposed to contribute to the common human skin disease acne vulgaris. The underlying mechanisms that drive inflammatory gene expression in acne-involved pilosebaceous epithelial cells are still unknown as almost all sebaceous follicles contain dense concentrations of bacteria yet only some show an inflammatory reaction. In this study we hypothesized that metabolites from the abundant skin-resident microbe Propionibacterium acnes can influence cytokine expression from human sebocytes. We show that short-chain fatty acids (SCFAs) produced by Propionibacterium acnes under environmental conditions that favor fermentation will drive inflammatory gene expression from sebocytes. These molecules are shown to influence sebocyte behavior through two distinct mechanisms: the inhibition of histone deacetylase (HDAC) activity and the activation of fatty acid receptors. Depletion of HDAC8 and HDAC9 in human sebocytes resulted in an enhanced cytokine response to Toll-like receptor-2 activation that resembled the transcriptional profile of an acne lesion. These data provide a new insight into the regulation of inflammatory gene expression in the skin, further characterize the contribution of sebocytes to epidermal immunity, and demonstrate how changes in the metabolic state of the skin microbiome can promote inflammatory acne.
INTRODUCTION
Acne vulgaris is an inflammatory skin disorder involving hair follicles and associated sebaceous glands. Ranking third in global disease burden of chronic skin conditions, acne is a significant health care problem and a leading source of medical expense (1). It is widely thought that the skin-resident bacterium Cutibacterium acnes (formerly known as Propionibacterium acnes (2) and referred to throughout this work as P. acnes) is a driver of skin inflammation in this disease (3). Toll-like Receptor-2 (TLR2) ligands present on P. acnes have been shown to initiate inflammatory gene expression from cells such as keratinocytes and monocytes (4–7). However, P. acnes is an abundant member of the microbiome in healthy skin where inflammation is not activated by the presence of these TLR ligands (8, 9). Furthermore, despite the existence of this microbe in the majority of sebaceous follicles, only a select few at any time demonstrate the inflammation characteristic of acne. Thus, critical underlying events that regulate the inflammatory response to P. acnes are not completely understood.
Taking these observations into account, we hypothesized that a change in the local microenvironment of individual sebaceous follicles may lead to a change in the metabolic activity of P. acnes, and that this local shift in metabolites may be then responsible for promoting the inflammatory responses observed in acne. Supporting this hypothesis, our previous work demonstrated that short-chain fatty acids (SCFAs) produced by P. acnes in hypoxic, lipid-rich conditions—conditions that mimic an occluded, sebaceous follicle—have a robust proinflammatory effect on epidermal keratinocytes (10). Keratinocytes treated with SCFAs showed an enhanced cytokine response to TLR activation—a distinct and striking phenotype of the keratinocyte inflammatory response that is opposite to the well-documented anti-inflammatory effects of SCFAs on cells of myeloid origin (11–13). This proinflammatory response by keratinocytes could be linked to the inhibition of histone deacetylase (HDAC) activity by SCFAs, and the target of SCFAs was associated with unique HDACs (HDAC8 and HDAC9), as specific depletion of HDAC8 or HDAC9 led to an enhanced response to TLR activation in a manner similar to SCFA treatment. These observations suggested that these HDAC enzymes in particular are responsible for repressing excessive levels of proinflammatory gene expression in keratinocytes, and that when SCFAs that inhibit HDAC activity are produced by P. acnes then the epithelial surface breaks tolerance to these commensal microbes.
In addition to keratinocytes, sebocytes that make up the sebaceous glands associated with hair follicles have also been identified as a major contributor of proinflammatory cytokine and antimicrobial peptide expression in response to P. acnes (14–17). In fact, recent studies have identified immunomodulatory roles of human sebocytes through their production of cytokines, chemokines, and lipid mediators (18–20). Historically viewed primarily as lipid-producers, sebocytes are now recognized as another immunocompetent cell type that may participate in—and have regulatory effects on—inflammatory and immune processes in the skin (21–23). However, whether HDAC activity is involved in regulating the expression of inflammatory genes in sebocytes—and whether P. acnes may modulate this regulation through the production of SCFAs—is unknown.
In this study we set out to explore the effects of P. acnes metabolites and HDAC inhibition on the inflammatory response of sebocytes. Through this work, we demonstrate that human sebocytes, like keratinocytes, exhibit an increased response to TLR activation and P. acnes stimulation when HDAC activity is depleted. Furthermore, the same HDAC enzymes that were demonstrated to be important in keratinocytes—HDAC8 and HDAC9—play a role in regulating proinflammatory gene expression in sebocytes, as depletion of either of these proteins largely replicated the effects of SCFA treatment. Finally, we show that unlike keratinocytes, a portion of the proinflammatory effects of SCFAs in sebocytes is due to the activation of fatty acid receptors. Together, these observations show a role for lipids, hypoxia, and P. acnes in promoting the development of acne by both epigenetic and canonical inflammatory mechanisms, and define the sebocyte as a unique cell type that may participate in the inflammatory response of the skin.
MATERIALS AND METHODS
Reagents
Cell Culture
The sebocyte cell lines SEB-1 (24) and SZ95 (25) were maintained in Sebomed Basal Medium supplemented with 10% fetal bovine serium, 1X antibiotic-antimycotic, and recombinant human epidermal growth factor (rhEGF, 5 ng/mL) in incubators maintained at 37°C in 5% CO2. Cells were passaged before reaching 100% confluency in tissue culture flasks. For experiments, cells were switched to medium containing 1% FBS overnight before stimulation.
ELISA
BD OptEIA ELISA Kits were used to determine cytokine concentrations in cell supernatants (BD Biosciences). Kits were used according to manufacturer’s instructions and results were determined using a 96-well plate reader (Beckman-Coulter).
RNA Isolation and RT-qPCR
RNA from cultured sebocytes was isolated using the PureLink RNA Mini Kit (Thermo Fisher Scientific) according to manufacturer’s instructions. RNA concentration was determined using a NanoDrop 2000 Spectrophotometer (Thermo Fisher Scientific), and up to 1 ug of RNA was converted into cDNA using the iScript cDNA Synthesis Kit (Bio-RAD). 5 ng of cDNA per sample was used in each subsequent RT-qPCR reaction, performed in 20 uL reactions using Taqman Universal PCR Master Mix (Thermo Fisher Scientific) and Taqman Gene Expression Assays (Thermo Fisher Scientific, assay numbers described in Table S1). All RT-qPCR reactions were performed on a CFX96 Touch Real-Time PCR Detection System (Bio-RAD).
RNA Sequencing
Purified isolated RNA was submitted to the University of California, San Diego Institute for Genomic Medicine (UCSD IGM) core facility for quality analysis, library preparation, and high-throughput sequencing. RNA quality was assessed using the Agilent Tapestation instrument (Agilent). Libraries were constructed using TruSeq Stranded mRNA Library Prep Kits (Illumina) and analyzed on a HiSeq 4000 instrument (Illumina). Raw data was obtained from the UCSD IGM core and analyzed using Partek Flow software (Partek) to determine transcript abundance and identify differentially expressed genes. Data is deposited in the NCBI Sequence Read Archive (SRA) https://trace.ncbi.nlm.nih.gov/Traces/sra/?study=SRP151073 with the accession number SRP151073. Gene Ontology analysis was performed using the Metascape website (www.metascape.org) (26).
SDS-PAGE and Western Blotting
Total protein was isolated from cultured sebocytes by cell lysis on ice in 1X RIPA buffer (150 mM NaCl, 25 mM Tris-HCl pH 7.6, 0.1% SDS, 1% sodium deoxycholate, 1% NP-40) supplemented with 1X protease inhibitor cocktail (Roche) followed by incubation in a sonic water bath at 4°C for 30 minutes. Cellular debris was pelleted by centrifugation at 12,000 x g for 15 minutes at 4°C. Protein concentration in supernatants was then determined by BCA Assay (Pierce/ Thermo Fisher) and protein concentrations were normalized among samples. Equal amounts of protein were then denatured by heating at 100°C for 10 minutes in 1X Lamelli sample buffer (Bio-RAD), separated on 4–20% Mini-Protean TGX gels (Bio-RAD), and transferred to PVDF membranes using the Trans-blot Turbo system (Bio-RAD). Membranes were blocked with Odyssey Blocking Buffer (LI-COR) for 1 hour at room temperature, followed by overnight incubation at 4°C with primary antibodies. Membranes were washed 3 × 5 minutes in PBS + 0.05% Tween-20, then incubated for 1 hour, protected from light, at room temperature with IRDye-labeled species-specific secondary antibodies (LI-COR). Membranes subsequently washed with PBS+0.05% Tween-20 and PBS, after which membranes were visualized with the Odyssey Infrared Imager (LI-COR). Odyssey program software was used to perform densitometry analysis.
siRNA-mediated Gene Knockdown
Select genes were depleted from cultured sebocytes using Silencer Select siRNA oligonucleotides (Thermo Fisher Scientific, catalog numbers listed in Table S1). Briefly, sebocytes at 60% confluency in 6 cm tissue culture dishes were treated with siRNA oligonucleotides diluted to a final concentration of 10 nM in 5 mL of a 9:1 Sebomed Basal medium:OptiMEM Medium mixture containing 0.25% RNAimax Lipofectamine reagent (Thermo Fisher Scientific). Cells were treated for 18 hours with siRNA, after which media was changed and cells were allowed to recover for 24 hours. Cells were then collected and plated into appropriate 12-well, 24-well, or 96-well plates for subsequent analysis. Cell treatments occurred within 120 hours of original siRNA application, and RT-qPCR measurement of target transcript abundance was measured simultaneously with additional experiments.
Propionibacterium acnes culture and stimulation of sebocytes
Propionibacterium acnes strain ATCC6919 was maintained in Difco Reinforced Clostridial Medium (BD Biosciences). For stimulation of sebocytes, P. acnes was inoculated into Sebomed Basal Medium (with 10% FBS and 5 ng/mL rhEGF, without antibiotics) with or without the addition of 2% glycerol. Bacteria were cultured in anaerobic conditions using BD GasPak EX pouches (BD Biosciences) at 37°C for 7 days. Culture supernatants were then sterile filtered and added at 20% volume to Sebomed Basal Medium and used to stimulate sebocytes.
HDAC Activity Assays
Measurements of HDAC activity in sebocytes in the presence of short-chain fatty acids were performed using the Epigenase HDAC Activity/ Inhibition Direct Assay Kit (Epigentek) according to manufacturer’s instructions. Briefly, nuclear extracts from SEB-1 and SZ95 sebocyte cells were incubated in 8-well strips with an acetylated histone HDAC substrate in the presence or absence of ranging concentrations of propionate, butyrate, or trichostatin A at 37°C for 90 minutes. Test wells were subsequently incubated with capture antibody (room temperature, 60 minutes), detection antibody (room temperature, 30 min), and detection solution (room temperature, 10 minutes) with washing of wells between incubation steps. Stop solution was added to test wells after 10 minutes of incubation with detection solution, and the absorbance of wells in the test plate at 450 nm was measured in a plate reader (Beckman-Coulter). Inhibition of HDAC activity by propionate, butyrate or Trichostatin A was calculated using the following formula as directed by the manufacturer:
Inhibition%= (1- (Inhibitor sample OD- Blank OD)/(No inhibitor sample OD-Blank OD)) x 100.
RESULTS
Products of Cutibacterium (formerly Propionibacterium) acnes anaerobic fermentation increase cytokine expression and histone acetylation in sebocytes
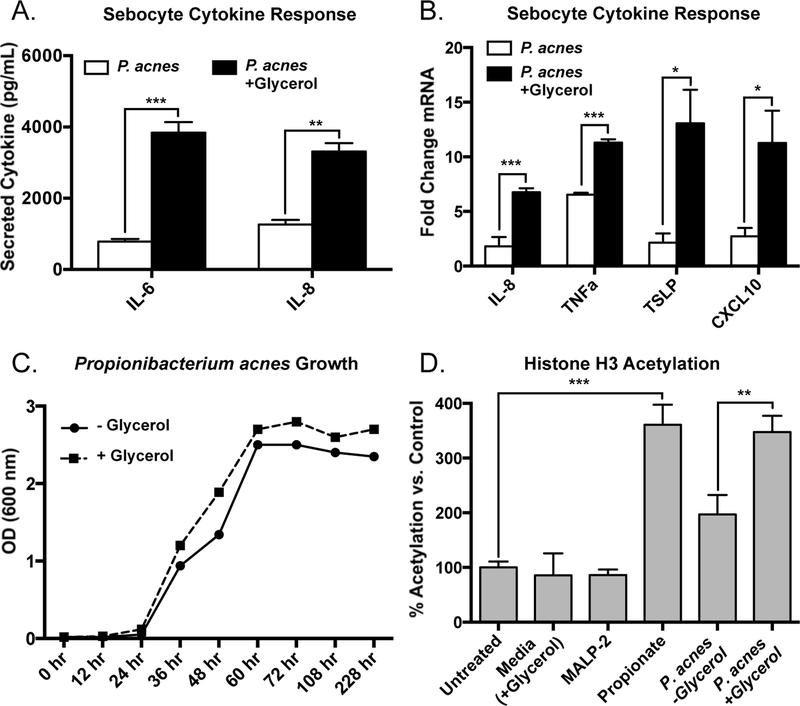
Sebaceous glands in the skin are exposed to resident microbes and their products and have been previously shown to produce inflammatory cytokines (14). To test the hypothesis that metabolic products of Cutibacterium (formerly Propionibacterium) acnes (P. acnes) could promote cytokine release from sebocytes, we first evaluated the release of IL-6 and IL-8 from a cultured sebocyte cell line (SEB-1) after exposure to supernatants of P. acnes (ATCC6919) that were grown under hypoxic conditions for 7 days in the presence or absence of glycerol. It has previously been demonstrated that glycerol, a naturally occurring metabolite on human skin that arises from the catalysis of sebaceous gland-derived triglycerides (27), is utilized by P. acnes under anaerobic conditions and results in the generation of short-chain fatty acids (28). The conditioned supernatant from these bacteria was then sterile-filtered and added to sebocytes. Exposure of sebocytes to supernatant from P. acnes grown without glycerol led to a modest release of IL-6 and IL-8, but when exposed to bacterial supernatants from P. acnes grown with glycerol a significant increase in these cytokines was seen (Figure 1a). Similarly, measurement of mRNA by qPCR for the inflammatory cytokines IL-8, TNFα, CXCL10 and TSLP demonstrated that sebocytes exposed to bacterial culture supernatant with glycerol had higher levels of gene expression than exposure to supernatant from cultures grown without glycerol (Figure 1b). These results did not appear to be a result of increased bacterial load in the glycerol-containing cultures as P. acnes growth and total abundance was similar in both culture conditions (Figure 1c).
Figure 1. Cutibacterium acnes fermentation products increase cytokine response and histone acetylation in human sebocytes.
A) Conditioned media from cultures of Cutibacterium (formerly Propionibacterium) acnes (P. acnes; strain ATCC6919) cultured with and without 2% glycerol was used to stimulate the SEB-1 human sebocyte cell line (20% volume of bacterial conditioned media in cell culture media). Levels of IL-6 and IL-8 were measured in cell culture supernatant by ELISA following 24 hours of stimulation. B) Proinflammatory cytokine induction as measured by RT-qPCR in SEB-1 sebocytes treated for 4 hours with P. acnes conditioned media as described in panel A; transcript abundances are normalized against the housekeeping gene GAPDH. C) Growth of P. acnes cultured with and without glycerol determined by measuring optical density at 600 nm on a spectrophotometer. D) Levels of global histone H3 acetylation in SEB-1 sebocytes treated with MALP-2 (200 ng/mL), propionate (2 mM), or bacterial conditioned media (20% volume) for 24 hours and measured using an AlphaLISA Assay kit. For graphs, data shown are mean +/− SEM of one experiment representative of at least three independent experiments with n=3 for each condition. *p<0.05; **p<0.01; ***p<0.001 as determined by student’s t-test.
P. acnes cultured with glycerol results in the generation of SCFAs, and the SCFAs propionate, butyrate, and valerate have been shown to inhibit histone deacetylase (HDAC) activity and increase histone acetylation (29–31). Increased histone acetylation can result in variable effects on cytokine expression depending on cell type (10, 32). We therefore next assessed if the crude bacterial supernatant resulted in a change in histone acetylation. Global histone H3 acetylation in sebocytes was increased by propionate, but was not affected by stimulation with the TLR2/6 ligand MALP-2 (Figure 1d). While the addition of supernatant from P. acnes grown without glycerol led to a modest increase in global H3 acetylation, supernatant from the glycerol-containing cultures led to a significantly higher increase in H3 acetylation, to levels comparable to treatment with propionate (Figure 1d).
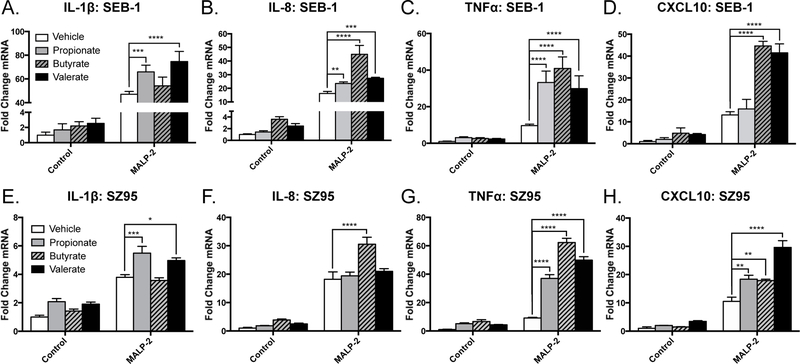
To determine if SCFAs alone could recapitulate the responses observed by sebocytes to the crude P. acnes supernatant, sebocytes were next exposed to a TLR2/6 ligand (MALP-2) in the presence or absence of 3 purified SCFAs: propionate, butyrate, and valerate. Consistent with our previous findings in human keratinocytes, we saw that SCFAs significantly enhanced the TLR-mediated transcriptional induction of various proinflammatory cytokines such as IL-1β, IL-8, TNFα, and CXCL10 from SEB-1 sebocytes (Figure 2a–d). This enhanced cytokine response in the presence of SCFAs was also seen with an independently derived sebocyte cell line (SZ95) (Figure 2e–h) (25). Taken together with our previous findings that P. acnes can generate SCFAs under anaerobic, lipid-rich conditions, these results suggest that production of SCFAs by P. acnes increases inflammatory cytokine expression induced by its TLR ligands from sebocytes.
Figure 2. Short-chain fatty acids increase proinflammatory gene expression from human sebocytes in response to TLR activation.
A-D) SEB-1 sebocytes or E-H) SZ95 sebocytes were stimulated with MALP-2 (200 ng/mL) for 4 hours with or without the addition of propionate, butyrate, or valerate (2 mM). RT-qPCR was used to measure the induction of the proinflammatory cytokines IL-1β (A,E), IL-8 (B, F), TNFα (C,G) and CXCL10 (D,H); transcript abundances are normalized against the housekeeping gene GAPDH. For graphs, data shown are mean +/− SEM of one experiment representative of at least three independent experiments with n=3 for each condition. *p<0.05; **p<0.01; ***p<0.001; ****p<0.0001 as determined by two-way ANOVA.
HDAC inhibition increases inflammatory gene expression from sebocytes
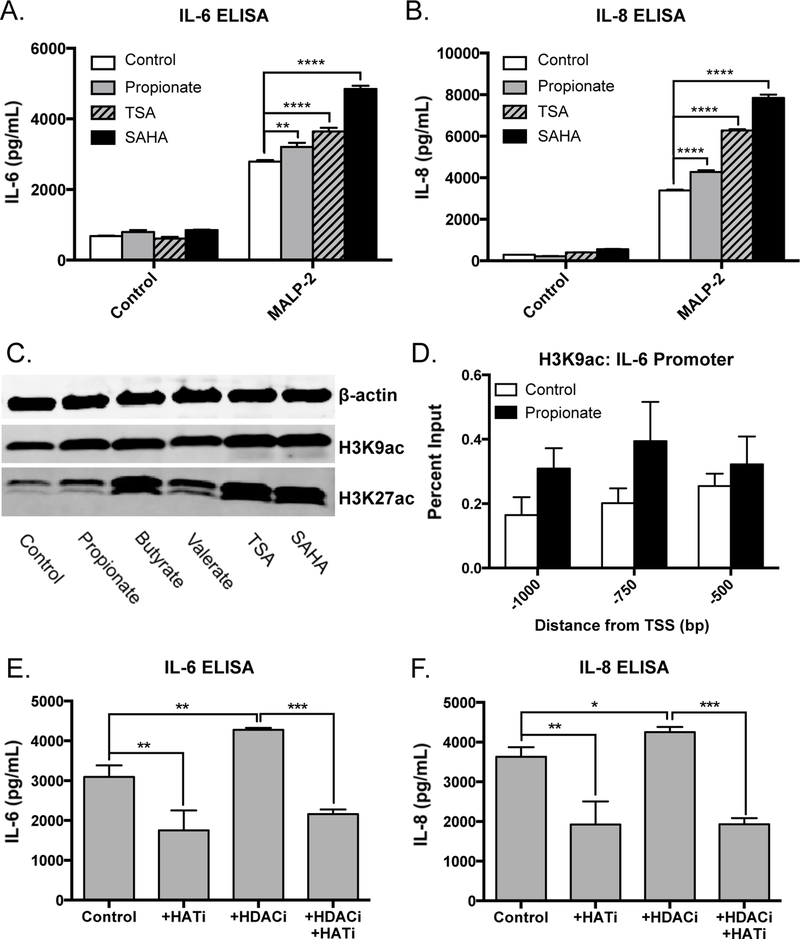
To further assess if SCFAs lead to increased inflammatory response by inhibition of HDACs, we compared responsiveness to propionate to two well-established chemical HDAC inhibitors (TSA and SAHA). We observed that both SCFAs and non-SCFA HDAC inhibitors all increased TLR-mediated production of IL-6 and IL-8 from SEB-1 sebocytes (Figure 3a–b). Consistent with their known ability to inhibit HDAC activity, these molecules led to increased global levels of histone acetylation in sebocytes as demonstrated by Western blot analysis of multiple lysine residues of histone H3 (H3K9ac, H3K27ac) (Figure 3c). Furthermore, we observed an increase in H3K9 acetylation throughout the promoter region of the IL-6 gene when sebocytes were treated with propionate (Figure 3d), suggesting that the increased TLR-mediated induction of this and other inflammatory genes by HDAC inhibitors may be due to changes in chromatin accessibility. In order to verify that the observed increase in histone acetylation in sebocytes treated with SCFAs was a direct result of inhibition of HDAC activity, HDAC activity was measured from nuclear extracts of SEB-1 and SZ95 cells in the presence of SCFAs. Both propionate and butyrate were seen to directly inhibit HDAC activity to a comparable degree as the well-established HDAC inhibitor Trichostatin A (Supplemental Figure 1).
Figure 3. Histone deacetylase inhibitors increase TLR-mediated cytokine production in sebocytes.
A-B) Levels of IL-6 and IL-8 secretion from SEB-1 sebocytes stimulated with MALP-2 (200 ng/mL) with or without the addition of the HDAC inhibitors propionate (2 mM), SAHA (100 nM), or TSA (100 nM) for 18 hours. C) Levels of acetylated histone residues (H3K9ac and H3K27ac) measured by Western blot in SEB-1 sebocytes treated with the indicated HDAC inhibitors for 8 hours. D) Levels of H3K9 acetylation in the IL-6 promoter region in SEB-1 sebocytes treated with or without propionate (2 mM) for 8 hours as measured by ChIP-qPCR. E-F) Levels of IL-6 and IL-8 secretion from SEB-1 sebocytes treated with MALP-2 (200 ng/mL) with or without the addition of the HDAC inhibitor butyrate (HDACi, 2 mM) or the histone acetyltransferase inhibitor anacardic acid (HATi, 4 uM) for 18 hours. For Western blots, one representative experiment out of three is shown. For graphs, data shown are mean +/− SEM of one experiment representative of at least three independent experiments with n=3 for each condition. *p<0.05; **p<0.01; ***p<0.001; ****p<0.0001 as determined by two-way ANOVA or student’s t-test.
While HDACs are responsible for the removal of acetylation marks in the genome, another class of enzymes, histone acetyltransferases (HATs), are responsible for the addition of these marks. Interestingly, when we stimulated sebocytes with anacardic acid—a chemical known to inhibit the action of HAT enzymes—we observed a decrease in TLR-mediated expression of IL-6 and IL-8, and a complete elimination of the proinflammatory effects of treatment with butyrate (Figure 3e–f). Together, these data demonstrate that sebocytes exhibit a proinflammatory phenotype in response to HDAC inhibition—a finding that contrasts to the anti-inflammatory effects of HDAC inhibitors on bone marrow derived immune cells like macrophages and dendritic cells.
Histone deacetylase 8 (HDAC8) and HDAC9 regulate inflammatory gene expression in sebocytes
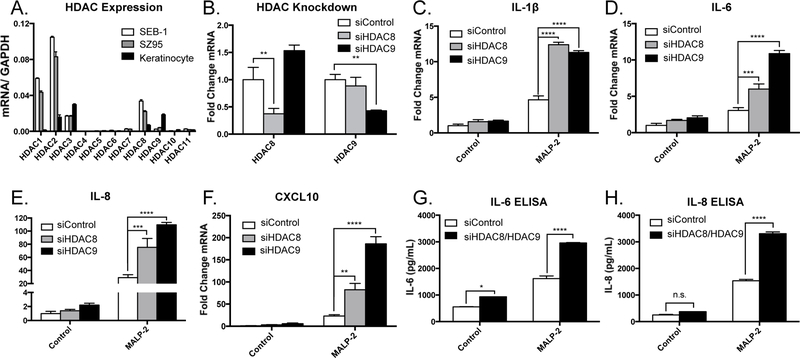
As short-chain fatty acids and other chemical HDAC inhibitors used in these experiments are broad-acting and inhibit the activity of numerous HDAC enzymes, we next sought to determine which individual HDACs may be important in the regulation of inflammatory gene expression in sebocytes. First, we characterized the expression of the 11 classical HDAC family members in sebocytes and compared them with expression levels from epidermal keratinocytes (Figure 4a). HDAC8 and HDAC9 were partially depleted using siRNA-mediated gene silencing (Figure 4b and Supplemental Figure 2a–d). This resulted in a significant amplification of MALP-2-mediated induction of IL-1β, IL-6, IL-8, and CXCL10 (Figure 4c–f). Expression of these cytokines was not increased in HDAC-depleted sebocytes without TLR activation. Furthermore, protein levels of IL-6 and IL-8 as measured by ELISA were also increased in sebocytes with partial silencing of HDAC8 and HDAC9 and treated with MALP-2 (Figure 4g–h). In order to validate that these findings were indeed specific to depletion of HDAC8 and HDAC9 and not an off-target effect of the silencing oligonucleotides used, we tested additional siRNA oligonucleotides specific for HDAC8 and HDAC9. Consistent with our initial results, partial depletion of HDAC8 and HDAC9 using alternate oligonucleotides significantly enhanced the TLR-mediated induction of IL-6 and IL-8 from SEB-1 sebocytes (Supplemental Figure 3a–c). These data indicate that HDAC8 and HDAC9 influence inflammatory cytokine expression in sebocytes, and suggest that the activity of SCFA on sebocytes is also mediated by inhibition of HDACs.
Figure 4. HDAC8 and HDAC9 control inflammatory gene expression in human sebocytes.
A) Expression pattern of the 11 classical HDAC enzymes in SEB-1 sebocytes and SZ95 sebocytes compared to normal human epidermal keratinocytes. B) Levels of HDAC8 and HDAC9 expression in SEB-1 sebocytes following siRNA-mediated gene knockdown; transcript abundances are normalized against the housekeeping gene GAPDH. C-F) Expression levels of the proinflammatory cytokines IL-1β, IL-6, IL-8, and CXCL10 (normalized against GAPDH) in SEB-1 sebocytes stimulated with MALP-2 (200 ng/mL) following siRNA-mediated depletion of HDAC8 or HDAC9. G-H) Levels of secreted IL-6 and IL-8 from SEB-1 sebocytes stimulated with MALP-2 (200 ng/mL) following siRNA-mediated depletion of HDAC8 and HDAC9. For graphs, data shown are mean +/− SEM of one experiment representative of at least three independent experiments with n=3 for each condition. *p<0.05; **p<0.01; ***p<0.001; ****p<0.0001 as determined by two-way ANOVA or student’s t-test.
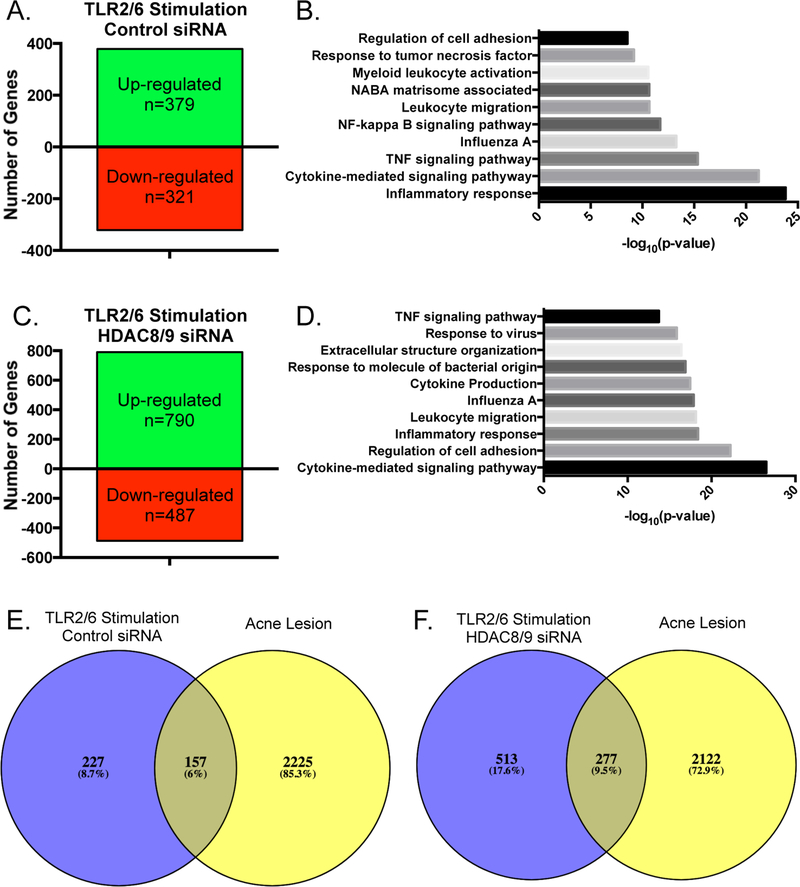
Having identified HDAC8 and HDAC9 as being important in restraining the sebocyte inflammatory response, we sought to use RNA-sequencing to evaluate the global transcriptomic effects of depletion of these enzymes. In sebocytes treated with scrambled control siRNA, stimulation with MALP-2 led to an up-regulation of 379 genes and a down-regulation of 321 genes (Figure 5a). Gene ontology analysis of the set of genes up-regulated by TLR2 activation in these cells identified enrichment of immune and inflammation-related pathways, such as inflammatory response, cytokine-mediated signaling pathway, and the TNF signaling pathway, among others (Figure 5b). Consistent with our hypothesis that HDAC8 and HDAC9 work to restrain inflammatory gene expression in sebocytes, cells depleted of HDAC8 and HDAC9 and stimulated by MALP-2 resulted in induction of over twice as many genes (790 vs. 379, Figure 5c). Furthermore, a more significant enrichment of pathways involved in inflammatory responses was observed, such as cytokine-medicated signaling pathway, regulation of cell adhesion, and leukocyte migration (Figure 5d).
Figure 5. Depletion of HDAC8 and HDAC9 increases global inflammatory response from human sebocytes.
SEB-1 sebocytes were treated with scrambled control siRNA or siRNA specific for HDAC8 and HDAC9, after which cells were treated with MALP-2 (200 ng/mL) for 4 hours. RNA was isolated and analyzed by RNA-sequencing. A-D) Numbers of genes up-regulated and down-regulated by 1.5 fold or greater by MALP-2 treatment in cells treated with control siRNA (A) or siRNA specific for HDAC8 and HDAC9 (C). Gene Ontology analysis of the sets of genes upregulated by MALP-2 treatment in cells treated with control siRNA (B) or siRNA specific for HDAC8 and HDAC9 (D). E-F) Genes induced by MALP-2 in SEB-1 sebocytes treated with control siRNA (E) or siRNA specific for HDAC8 and HDAC9 (F) were compared with the transcriptome of genes up-regulated by 2-fold or greater identified from an inflamed clinical acne lesion. RNA-sequencing was performed on n=1 representative sample for each condition.
Next, we compared the global inflammatory response of sebocytes with or without functional HDAC8 and HDAC9 to the inflammatory signature of an acne lesion. A total of 2382 genes were up-regulated in an inflamed acne lesion compared to healthy skin from the same donor. Comparison of this dataset with the list of genes induced by MALP-2 alone in sebocytes (treated with control siRNA), showed a relatively minor overlap of 157 genes (Figure 5e). However, when an acne lesion was compared with sebocytes lacking HDAC8 and HDAC9 and stimulated by MALP-2, almost twice as many genes overlapped, with 277 genes common between the samples (Figure 5f). These data demonstrate that when the activity of HDAC8 and HDAC9 are impaired in sebocytes, activation of Toll-like receptor 2 leads to the induction of many more inflammatory genes that are detected in clinical acne samples.
Free fatty acid receptor activation by SCFAs induces cytokine expression in sebocytes
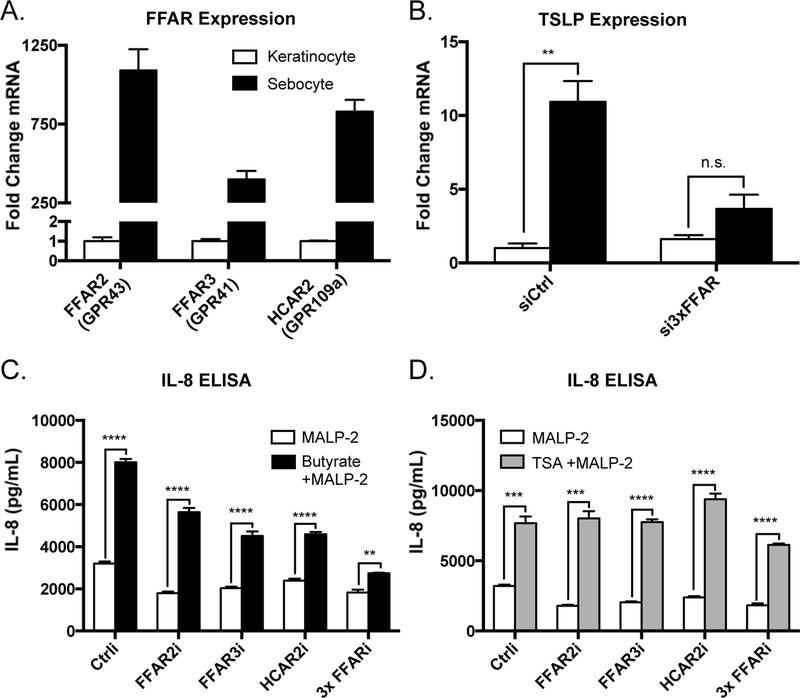
Finally, in addition to the capacity to inhibit HDAC activity, SCFAs also have the potential to interact with cell surface G protein-coupled receptors. In particular, short-chain fatty acids are known ligands of GPR41 (also known as free fatty acid receptor 3/ FFAR3), GPR43 (aka FFAR2), and GPR109a (aka hydroxycarboxylic acid receptor 2/ HCAR2) (33, 34). We next assessed if there was a direct response to SCFAs by sebocytes and sought to distinguish this response from the amplification of TLR signaling by the epigenetic mechanism of histone acetylation. Prior observations in keratinocytes showed no detectable response to SCFAs alone, which may be explained by the extremely low expression levels of the three relevant GPCRs by this cell type (10). In contrast, sebocytes express FFAR2, FFAR3, and HCAR2 at relatively high levels, several hundred-fold higher than keratinocytes (Figure 6a). Furthermore, SZ95 sebocytes treated with SCFAs in the absence of TLR2 ligands resulted in increased expression of the proinflammatory mediator thymic stromal lymphopoietin (TSLP), an effect that was lost when FFARs were depleted via siRNA-mediated knockdown (Figure 6b). When FFAR2, FFAR3, or HCAR2 were depleted individually from sebocytes, the SCFA-mediated increase in MALP-2-induced cytokine secretion was partially decreased (Figure 6c). Meanwhile, when cells were depleted of all three receptors in combination, the proinflammatory effect of SCFAs was almost completely abolished (Figure 6c). Importantly, this did not occur to the same extent when cells were activated by the non-SCFA HDAC inhibitor Trichostatin A, demonstrating that these cells still respond appropriately to inhibition of HDAC activity (Figure 6d). These data demonstrated that sebocytes, compared to other cell types in the skin, exhibit a unique response to SCFAs, which promote cytokine expression by both activation of G Protein-coupled receptors and inhibition of HDAC activity.
Figure 6. Free fatty acid receptor activation by short-chain fatty acids induces cytokine expression from sebocytes.
A) Expression patterns of the major free fatty acid receptors in SZ95 sebocytes compared to normal human epidermal keratinocytes; transcript abundances are normalized against GAPDH. B) Expression levels of the inflammatory mediator TSLP expressed in SZ95 sebocytes following stimulation with butyrate (2 mM) for 4 hours (transcript abundance normalized against GAPDH). C-D) IL-8 secretion from SZ95 sebocytes stimulated with MALP-2 (200 ng/mL) with or without butyrate (2 mM) or TSA (100 nM) for 18 hours following siRNA-mediated depletion of the indicated free fatty acid receptors. For graphs, data shown are mean +/− SEM of one experiment representative of at least two independent experiments with n=3 for each condition. *p<0.05; **p<0.01; ***p<0.001; ****p<0.0001 as determined by two-way ANOVA or student’s t-test.
DISCUSSION
These data show for the first time how metabolic products of skin-resident bacteria can induce inflammatory gene expression from sebocytes. While it has been known that sebocytes can respond to P. acnes through activation of Toll-like receptor 2, the question of why these bacteria only drive an inflammatory response in the skin in certain settings has remained unanswered. We have previously shown that when cultured in hypoxic conditions and supplied a lipid substrate—an environment reminiscent of a plugged, sebaceous follicle—P. acnes can generate high levels of short-chain fatty acids (SCFAs) through anaerobic fermentation. Interestingly, these molecules were shown to amplify Toll-like receptor-driven cytokine responses from epidermal keratinocytes through their inhibition of histone deacetylase function. In this study, we now demonstrate that cytokine expression in sebocytes is also regulated through this epigenetic mechanism, and also show that sebocytes have a unique capacity to enhance cytokine expression by direct activation of surface-bound free fatty acid receptors. Thus, sebocytes in the skin are uniquely sensitive to a change in metabolites by P. acnes and respond to short-chain fatty acids by two independent mechanisms.
The proinflammatory effects of HDAC inhibition in both sebocytes and keratinocytes, combined with the similar pattern of expression of the 11 classical HDAC family members observed in the two cell types, suggested that the same HDAC enzymes might control inflammatory gene expression in both keratinocytes and sebocytes. Indeed, specific depletion of HDAC8 or HDAC9—which we had previously demonstrated to control cytokine expression in keratinocytes—resulted in an amplified response to TLR activation in sebocytes. When examined at a global level by RNA-sequencing, the number of genes up-regulated in response to TLR2/6 activation was nearly doubled when HDAC8 and HDAC9 activity was inhibited. Furthermore, the set of genes induced in sebocytes depleted of HDAC8 and HDAC9 more closely resembled the transcriptome of whole tissue samples from acne, suggesting that inhibition of HDAC8 and HDAC9 in the sebaceous gland by P. acnes-derived SCFAs is modeling aspects of the pathophysiology of the disease. While the overlap with the sebocyte transcriptome still represents a relatively small portion of the genes identified from a clinical acne lesion, one must consider that the acne biopsy represents a heterogeneous mixture of various cell types from a physiological setting, while the sebocyte data represents a single cell type grown in culture. More precise measurements of the sebocyte contribution to an acne lesion—for example, through single cell sequencing of sebocytes from clinical specimens—could further elucidate the contribution of this cell type, and the importance of sebocyte HDAC activity, to acne inflammation in vivo.
Despite the commonalities in regard to HDAC8 and HDAC9, the impact of SCFAs on the inflammatory response of sebocytes and keratinocytes is not completely identical. As this work shows, SCFAs have a unique ability to induce cytokine expression from sebocytes through the activation of free fatty acid receptors, a finding that was not observed in prior work with keratinocytes. Some proinflammatory mediators, such as TSLP, were induced from sebocytes by treatment with SCFAs alone, even in the absence of TLR ligands. These effects may be due to the high expression of these FFARs in sebocytes, which were detected at several hundred-fold higher levels than in keratinocytes. These findings are interesting when taken together with previous studies demonstrating that free fatty acids present in sebum—such as lauric, palmitic, and oleic acids—can induce antimicrobial peptide expression from sebocytes (35). Together, these findings illustrate how the local follicular microenvironment, consisting of fatty acids from the host, resident microbes, and their metabolic products, can influence inflammatory and antimicrobial processes in the skin. Future studies to characterize the fatty acid profiles of healthy and diseased skin and investigate how different mixtures of fatty acids may further inform the relative role of these metabolites in skin inflammation. Additionally, evaluating the generation of SCFAs and other bioactive metabolites by P. acnes cultured with lipid sources that more closely resemble human sebum, as opposed to simply glycerol, may shed interesting light onto the impacts of microbial metabolism on skin inflammation.
The effects of P. acnes-derived short-chain fatty acids on inflammatory gene expression from sebocytes through both epigenetic and receptor-dependent mechanisms described in this study invite the question of whether distinct strains of P. acnes have a differential capacity to generate these molecules. While P. acnes proliferation and overgrowth in the follicle was once considered a driving factor behind the inflammation seen in acne, recent work has demonstrated that the overall abundance of microbes in healthy and affected follicles is actually quite comparable (8). Thus, the current view has shifted towards the idea that distinct strains of P. acnes are unique in their abilities to induce cytokine expression from sebocytes, keratinocytes, and cells of the innate immune system. Our results support speculation that SCFAs from P. acnes will drive cytokine expression prior to rupture of the follicle and may influence both the local pilosebacous unit as well as surrounding skin. Indeed, characterizations of the P. acnes populations isolated from the skin of acne patients and healthy controls have identified “acne-associated” and “healthy-associated” strains of P. acnes, which exert distinct effects on the immune and inflammatory responses of the skin (9, 36, 37). However, whether the ability to produce SCFAs—or the ratios of SCFAs produced by these different bacteria—contribute to the host response has yet to be investigated.
Overall, this study expands our knowledge into the regulation of inflammatory gene expression in the skin, an organ that must strictly control its capacity to respond to microbes. While this work was conducted using immortalized cell lines, the observation of the same trends in response to HDAC inhibition, specific depletion of HDAC8 and HDAC9, and depletion of free fatty acid receptors in two independently derived cell lines supports that these responses are not restricted to a cell line but more likely reflect the sebocyte itself. Replicating these experiments in primary human sebocytes isolated from skin samples would help support the notion that these phenomena may occur in vivo, but even these culture-based studies are not a completely accurate representation of the physiological setting of the pilosebaceous unit. In the future, genetic mouse models can be used to generate tissue-specific deletions of HDAC8 and HDAC9—from sebocytes, epidermal keratinocytes, or both—to gain a more in depth understanding of the role of these enzymes in regulating skin inflammation and immunity. However, the lack of appropriate animal models for inflammatory acne vulgaris makes it difficult to specifically link our findings to acne rather than skin inflammation in a broader sense.
In conclusion, this work helps to promote a model of skin inflammation where changes in the local microenvironment of the pilosebaceous unit drives metabolic shifts in resident skin microbes, resulting in the production of bioactive metabolites that promote the shift from the cutaneous immune system tolerating P. acnes colonization to one that is inflammatory. This shift may be the fundamental event in the pathophysiology of acne vulgaris.
Supplementary Material
Table 1.
| Reagent | Supplier | Catalog Number |
|---|---|---|
| Sebomed Basal Medium | Cedarlane/ Fisher | F8205 |
| Antibiotic-antimycotic | Life Technologies | 15240-062 |
| Recombinant human epidermal growth factor (rhEGF) | Sigma | E9644 |
| Fetal Bovine Serum (FBS) | Gemini | 900-108 |
| Sodium propionate | Sigma | P1880 |
| Sodium butyrate | Sigma | B5887 |
| Isovaleric acid | Sigma | 78651 |
| Trichostatin A | Sigma | T8552 |
| Suberoylanilide Hydroxamic Acid (SAHA) | Sigma | SML0061 |
| Macrophage-activating lipopeptide-2 (MALP-2) | Enzo/ VWR | 162-034-C500 |
| Taqman Universal PCR Master Mix | Life Technologies | 4318157 |
| Complete Protease Inhibitor Cocktail | Roche | 11697498001 |
| RNAimax Lipofectamine | Thermo Fisher | 56532 |
| HDAC Activity/ Inhibition Direct Assay | Epigentek | P-4034-96 |
| Silencer Select siRNA Oligonucleotides | See Table S1 | |
| Taqman gene expression assays | See Table S1 | |
| ELISA Kits | See Table S1 | |
Reagent list
Listed here are supplies and reagents, suppliers and catalog numbers.
ACKNOWLEDGEMENTS
We thank Dr. Diane Thiboutot from Pennsylvania State University Dermatology for providing the SEB-1 sebocyte cell line for use in these experiments. We also thank Dr. Yun “Larry” Tong from the UCSD Department of Dermatology for the acquisition of human samples used in immunohistochemistry and RNA-sequencing.
2. This work was supported by NIH R01AR074302, R01AR069653 and R01AI052453 to RLG.
Footnotes
3. Dr. Gallo is a consultant and has equity interest in MatriSys bioscience and Sente Inc.
4. Dr. Zouboulis owns an international patent on the SZ95 sebaceous gland cell line (WO2000046353).
REFERENCES
- 1.Moradi Tuchayi S, Makrantonaki E, Ganceviciene R, Dessinioti C, Feldman SR, and Zouboulis CC. 2015. Acne vulgaris. Nature reviews. Disease primers 1: 15029. [DOI] [PubMed] [Google Scholar]
- 2.Scholz CF, and Kilian M. 2016. The natural history of cutaneous propionibacteria, and reclassification of selected species within the genus Propionibacterium to the proposed novel genera Acidipropionibacterium gen. nov., Cutibacterium gen. nov. and Pseudopropionibacterium gen. nov. International journal of systematic and evolutionary microbiology 66: 4422–4432. [DOI] [PubMed] [Google Scholar]
- 3.Bojar RA, and Holland KT. 2004. Acne and Propionibacterium acnes. Clinics in dermatology 22: 375–379. [DOI] [PubMed] [Google Scholar]
- 4.Kim J, Ochoa MT, Krutzik SR, Takeuchi O, Uematsu S, Legaspi AJ, Brightbill HD, Holland D, Cunliffe WJ, Akira S, Sieling PA, Godowski PJ, and Modlin RL. 2002. Activation of Toll-Like Receptor 2 in Acne Triggers Inflammatory Cytokine Responses. The Journal of Immunology 169: 1535–1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jugeau S, Tenaud I, Knol AC, Jarrousse V, Quereux G, Khammari A, and Dreno B. 2005. Induction of toll-like receptors by Propionibacterium acnes. Br J Dermatol 153: 1105–1113. [DOI] [PubMed] [Google Scholar]
- 6.Qin M, Pirouz A, Kim MH, Krutzik SR, Garban HJ, and Kim J. 2014. Propionibacterium acnes Induces IL-1beta secretion via the NLRP3 inflammasome in human monocytes. J Invest Dermatol 134: 381–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vowels BR, Yang S, and Leyden JJ. 1995. Induction of proinflammatory cytokines by a soluble factor of Propionibacterium acnes: implications for chronic inflammatory acne. Infection and Immunity 63: 3158–3165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barnard E, Shi B, Kang D, Craft N, and Li H. 2016. The balance of metagenomic elements shapes the skin microbiome in acne and health. Scientific reports 6: 39491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fitz-Gibbon S, Tomida S, Chiu BH, Nguyen L, Du C, Liu M, Elashoff D, Erfe MC, Loncaric A, Kim J, Modlin RL, Miller JF, Sodergren E, Craft N, Weinstock GM, and Li H. 2013. Propionibacterium acnes Strain Populations in the Human Skin Microbiome Associated with Acne. J Invest Dermatol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sanford JA, Zhang L, Williams MR, Gangoiti JA, Huang CM, and Gallo RL. 2016. Inhibition of HDAC8 and HDAC9 by microbial short-chain fatty acids breaks immune tolerance of the epidermis to TLR ligands. Sci Immunol in press. [DOI] [PubMed] [Google Scholar]
- 11.Saemann MD, Bohmig GA, Osterreicher CH, Burtscher H, Parolini O, Diakos C, Stockl J, Horl WH, and Zlabinger GJ. 2000. Anti-inflammatory effects of sodium butyrate on human monocytes: potent inhibition of IL-12 and up-regulation of IL-10 production. FASEB journal : official publication of the Federation of American Societies for Experimental Biology 14: 2380–2382. [DOI] [PubMed] [Google Scholar]
- 12.Nastasi C, Candela M, Bonefeld CM, Geisler C, Hansen M, Krejsgaard T, Biagi E, Andersen MH, Brigidi P, Odum N, Litman T, and Woetmann A. 2015. The effect of short-chain fatty acids on human monocyte-derived dendritic cells. Scientific reports 5: 16148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chang PV, Hao L, Offermanns S, and Medzhitov R. 2014. The microbial metabolite butyrate regulates intestinal macrophage function via histone deacetylase inhibition. Proceedings of the National Academy of Sciences of the United States of America 111: 2247–2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nagy I, Pivarcsi A, Kis K, Koreck A, Bodai L, McDowell A, Seltmann H, Patrick S, Zouboulis CC, and Kemeny L. 2006. Propionibacterium acnes and lipopolysaccharide induce the expression of antimicrobial peptides and proinflammatory cytokines/chemokines in human sebocytes. Microbes and infection / Institut Pasteur 8: 2195–2205. [DOI] [PubMed] [Google Scholar]
- 15.Nakatsuji T, Liu YT, Huang CP, Zoubouis CC, Gallo RL, and Huang CM. 2008. Antibodies elicited by inactivated propionibacterium acnes-based vaccines exert protective immunity and attenuate the IL-8 production in human sebocytes: relevance to therapy for acne vulgaris. J Invest Dermatol 128: 2451–2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Selway JL, Kurczab T, Kealey T, and Langlands K. 2013. Toll-like receptor 2 activation and comedogenesis: implications for the pathogenesis of acne. BMC Dermatology 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li ZJ, Choi DK, Sohn KC, Seo MS, Lee HE, Lee Y, Seo YJ, Lee YH, Shi G, Zouboulis CC, Kim CD, Lee JH, and Im M. 2014. Propionibacterium acnes activates the NLRP3 inflammasome in human sebocytes. J Invest Dermatol 134: 2747–2756. [DOI] [PubMed] [Google Scholar]
- 18.Lovaszi M, Mattii M, Eyerich K, Gacsi A, Csanyi E, Kovacs D, Ruhl R, Szegedi A, Kemeny L, Stahle M, Zouboulis CC, Eyerich S, and Torocsik D. 2017. Sebum lipids influence macrophage polarization and activation. Br J Dermatol 177: 1671–1682. [DOI] [PubMed] [Google Scholar]
- 19.Mattii M, Lovaszi M, Garzorz N, Atenhan A, Quaranta M, Lauffer F, Konstantinow A, Kupper M, Zouboulis CC, Kemeny L, Eyerich K, Schmidt-Weber CB, Torocsik D, and Eyerich S. 2018. Sebocytes contribute to skin inflammation by promoting the differentiation of T helper 17 cells. Br J Dermatol 178: 722–730. [DOI] [PubMed] [Google Scholar]
- 20.Alestas T, Ganceviciene R, Fimmel S, Muller-Decker K, and Zouboulis CC. 2006. Enzymes involved in the biosynthesis of leukotriene B4 and prostaglandin E2 are active in sebaceous glands. Journal of molecular medicine 84: 75–87. [DOI] [PubMed] [Google Scholar]
- 21.Zoubouis CC 2001. Is acne vulgaris a genuine inflammatory disease? Dermatology 203: 277–279. [DOI] [PubMed] [Google Scholar]
- 22.Zouboulis CC, Jourdan E, and Picardo M. 2014. Acne is an inflammatory disease and alterations of sebum composition initiate acne lesions. Journal of the European Academy of Dermatology and Venereology : JEADV 28: 527–532. [DOI] [PubMed] [Google Scholar]
- 23.Lovaszi M, Szegedi A, Zouboulis CC, and Torocsik D. 2017. Sebaceous-immunobiology is orchestrated by sebum lipids. Dermato-endocrinology 9: e1375636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thiboutot D, Jabara S, McAllister JM, Sivarajah A, Gilliland K, Cong Z, and Clawson G. 2003. Human skin is a steroidogenic tissue: steroidogenic enzymes and cofactors are expressed in epidermis, normal sebocytes, and an immortalized sebocyte cell line (SEB-1). J Invest Dermatol 120: 905–914. [DOI] [PubMed] [Google Scholar]
- 25.Zouboulis CC, Seltmann H, Neitzel H, and Orfanos CE. 1999. Establishment and characterization of an immortalized human sebaceous gland cell line (SZ95). J Invest Dermatol 113: 1011–1020. [DOI] [PubMed] [Google Scholar]
- 26.Tripathi S, Pohl MO, Zhou Y, Rodriguez-Frandsen A, Wang G, Stein DA, Moulton HM, DeJesus P, Che J, Mulder LC, Yanguez E, Andenmatten D, Pache L, Manicassamy B, Albrecht RA, Gonzalez MG, Nguyen Q, Brass A, Elledge S, White M, Shapira S, Hacohen N, Karlas A, Meyer TF, Shales M, Gatorano A, Johnson JR, Jang G, Johnson T, Verschueren E, Sanders D, Krogan N, Shaw M, Konig R, Stertz S, Garcia-Sastre A, and Chanda SK. 2015. Meta- and Orthogonal Integration of Influenza “OMICs” Data Defines a Role for UBR4 in Virus Budding. Cell host & microbe 18: 723–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fluhr JW, Darlenski R, and Surber C. 2008. Glycerol and the skin: holistic approach to its origin and functions. Br J Dermatol 159: 23–34. [DOI] [PubMed] [Google Scholar]
- 28.Shu MWY, ; Yu J; Kuo S; Coda A; Jiang Y; Gallo RL; Huang CM 2013. Fermentation of Propionibacterium acnes, a commensal bacterium in the human skin microbiome, as skin probiotics against methicillin-resistant Staphylococcus aureus. PloS one 8: e55380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sealy L, and Chalkley R. 1978. The effect of sodium butyrate on histone modification. Cell 14: 115–121. [DOI] [PubMed] [Google Scholar]
- 30.Hinnebusch BF, Meng S, Wu JT, Archer SY, and Hodin RA. 2002. The effects of short-chain fatty acids on human colon cancer cell phenotype are associated with histone hyperacetylation. J Nutr 132: 1012–1017. [DOI] [PubMed] [Google Scholar]
- 31.Waldecker M, Kautenburger T, Daumann H, Busch C, and Schrenk D. 2008. Inhibition of histone-deacetylase activity by short-chain fatty acids and some polyphenol metabolites formed in the colon. The Journal of nutritional biochemistry 19: 587–593. [DOI] [PubMed] [Google Scholar]
- 32.Roger T, Lugrin J, Le Roy D, Goy G, Mombelli M, Koessler T, Ding XC, Chanson AL, Reymond MK, Miconnet I, Schrenzel J, Francois P, and Calandra T. 2011. Histone deacetylase inhibitors impair innate immune responses to Toll-like receptor agonists and to infection. Blood 117: 1205–1217. [DOI] [PubMed] [Google Scholar]
- 33.Brown AJ, Goldsworthy SM, Barnes AA, Eilert MM, Tcheang L, Daniels D, Muir AI, Wigglesworth MJ, Kinghorn I, Fraser NJ, Pike NB, Strum JC, Steplewski KM, Murdock PR, Holder JC, Marshall FH, Szekeres PG, Wilson S, Ignar DM, Foord SM, Wise A, and Dowell SJ. 2003. The Orphan G protein-coupled receptors GPR41 and GPR43 are activated by propionate and other short chain carboxylic acids. The Journal of biological chemistry 278: 11312–11319. [DOI] [PubMed] [Google Scholar]
- 34.Singh N, Gurav A, Sivaprakasam S, Brady E, Padia R, Shi H, Thangaraju M, Prasad PD, Manicassamy S, Munn DH, Lee JR, Offermanns S, and Ganapathy V. 2014. Activation of Gpr109a, receptor for niacin and the commensal metabolite butyrate, suppresses colonic inflammation and carcinogenesis. Immunity 40: 128–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nakatsuji T, Kao MC, Zhang L, Zouboulis CC, Gallo RL, and Huang CM. 2010. Sebum free fatty acids enhance the innate immune defense of human sebocytes by upregulating beta-defensin-2 expression. J Invest Dermatol 130: 985–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Agak GW, Kao S, Ouyang K, Qin M, Moon D, Butt A, and Kim J. 2018. Phenotype and Antimicrobial Activity of Th17 Cells Induced by Propionibacterium acnes Strains Associated with Healthy and Acne Skin. J Invest Dermatol 138: 316–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yu Y, Champer J, Agak GW, Kao S, Modlin RL, and Kim J. 2016. Different Propionibacterium acnes Phylotypes Induce Distinct Immune Responses and Express Unique Surface and Secreted Proteomes. J Invest Dermatol 136: 2221–2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.