Abstract
The single residue mutation of butyrylcholinesterase (BChEG117H) hydrolyzes a number of organophosphosphorus (OP) anticholinesterases. While other active site/proximal mutations have been investigated, none are sufficiently active to be prophylactically useful. In a fundamentally different, computer simulation-driven strategy, we identified a surface peptide loop (residues 278–285) exhibiting dynamic motions during catalysis and modified it via residue insertions. We evaluated these loop mutants using computer simulations, substrate kinetics, resistance to inhibition and enzyme reactivation assays using both the choline ester and OP substrates. A slight but significant increase in reactivation was noted with paraoxon with one of the mutants, and changes in KM and catalytic efficiency were noted in others. Simulations suggested weaker interactions between OP versus choline substrates and the active site of all enzymes. The results indicate that improvement of OP anticholinesterase hydrolysis through surface loop engineering may be a more effective strategy in an enzyme with higher intrinsic OP compound hydrolase activity.
Keywords: G117H, organophosphate hydrolysis, enzyme engineering, bioscavenging
Graphical Abstract
INTRODUCTION
Improved therapies to counteract the toxicity of organophosphorus (OP) anticholinesterases have been sought for decades. OP anticholinesterases are commonly used as insecticides. The OP nerve agents such as sarin (2-[fluoro(methyl)phosphoryl]oxypropane) also act as potent anticholinesterases and are some of the most toxic synthetic chemicals ever created. Sarin and or VX (N-[2-[ethoxy(methyl)phosphoryl]sulfanylethyl]-N-propan-2-ylpropan-2-amine) have been used in the Iraq-Iran civil war (1980s), the Aum Shinrikyo cult terrorist attacks in Japan (1994–95), the Syrian civil war (2013), and in the assassination of Kim Jong Nam (2017). More recently, one of a group of highly potent and lesser characterized chemicals referred to as Novichok agents was used in an attempted assassination of a former Russian spy, Sergei Skripal (2018)1–3.
OP insecticides and nerve agents elicit acute toxicity primarily through inhibition of acetylcholinesterase (AChE), leading to increased synaptic acetylcholine levels, overstimulation of the cholinergic receptors within the central and peripheral nervous systems and consequent signs of toxicity including increased secretions from parasympathetic end organs, tremors, seizures, respiratory depression and death4,5. Intoxicated individuals are generally treated with atropine (a muscarinic receptor antagonist), an oxime (e.g., pralidoxime, to reactivate inhibited AChE molecules), and a benzodiazepine (e.g., midazolam) to suppress seizures6. While current treatments can prevent lethality when given early after exposure, long-term neurological deficits have been demonstrated following severe OP anticholinesterase intoxication despite therapeutic intervention7,8. Thus, alternative and/or adjunctive treatments are being actively investigated.
Bioscavengers for OP anticholinesterases are proteins that bind to and inactivate the toxicants in the circulation before they can reach systemic target organs to elicit cholinergic toxicity. Bioscavengers were first tested for protection against OP compound toxicity by Wolfe and coworkers (1987)9. There are two types of bioscavengers: stoichiometric and catalytic. While stoichiometric scavengers bind OP molecules in a near 1:1 ratio, catalytic scavengers can reactivate following phosphylation by the OP, making them potentially much more efficient. Enzymes that catalyze OP anticholinesterases naturally (e.g. organophosphorus acid anhydrolase, paraoxonase, phosphotriesterase) have been purified and tested but often suffer from issues such as low catalytic rate, limited substrate selectivity and immunogenicity10,11. The stoichiometric bioscavenger enzyme butyrylcholinesterase (BChE) has essentially no effective catalytic rate with any OP substrates but exhibits a widespread binding affinity for them. Moreover, it has been demonstrated to be safe and effective with little immunogenicity at high doses12,13. Due to its stoichiometric interaction with an OP anticholinesterase, however, a very large dose of the enzyme is required, resulting in economic barriers preventing its widespread use as a prophylactic agent.
Kinetically, OP toxicants interact with BChE in a manner similar to choline esters14. A Michaelis complex is initially formed (described as the ratio of association vs dissociation, k1/k−1) which rapidly progresses to acylation (in the case of choline ester) or phosphylation (in the case of an OP inhibitor, k2). The subsequent kinetic step (k3) is markedly different between choline and OP substrates. In deacylation, a histidine-bound water molecule rapidly displaces the acyl group from the catalytic serine residue in the active site. In dephosphylation, the histidine-bound water molecule is sterically hindered from displacing the phosphorus atom, thus it remains covalently attached to the active site serine. While rapid deacylation with a choline substrate allows return of the enzyme to an effective catalyst, the slow dephosphylation step leaves the catalytic reaction blocked, allowing accumulation of acetylcholine, prolonged activation of cholinergic receptors and cholinergic toxicity.
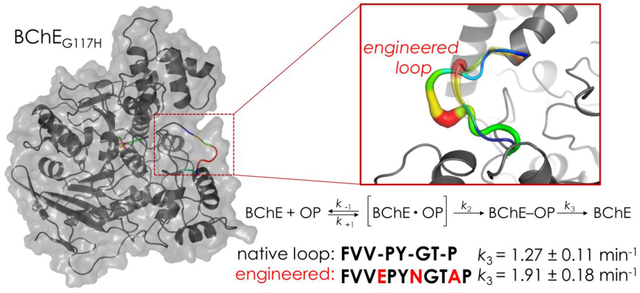
It was proposed over three decades ago that introduction of an alternative histidine near the active site serine may facilitate OP toxicant hydrolysis14. Subsequent evaluation of a series of single point mutations in the oxyanion hole of BChE to histidine found one mutant (BChEG117H) with a marked increase in reactivation with a number of OP anticholinesterases15,16. Despite showing increased catalysis compared to the native enzyme, the turnover rate of BChEG117H (kcat = 0.75 min−1 with paraoxon) remained much too low to be an effective catalytic scavenger in vivo17. For comparison, the turnover rate of human serum paraoxonase against paraoxon is 340–660 min−1, while the most active bacterial phosphotriesterases have a rate of 124,200 min−1 18,19. Since the characterization of BChEG117H, over 60 BChE mutants have tested the effects of alternating histidine positions, changing amino acids at the 117 position, adding positive charges in BChEG117H, and introducing negative charges to increase the nucleophilicity of histidine, among others20. To date, no other mutants have shown significant improvements compared to BChEG117H.
An emerging concept in enzyme function is that dynamic motions of peptide sequences on an enzyme’s surface, distant from the active site region, can influence the rate of a catalytic reaction21. It has been proposed that catalytic rate is affected by thermodynamical coupling of the hydration-shell, the bulk solvent, and the catalyzed reaction22. For a number of enzyme systems, networks of conserved residues have been discovered that span from the surface of the protein to the active site region, effectively coupling with the catalytic reaction mechanism23. In fact, enhancing energy flow through these networks may be a general approach for increasing enzyme-mediated catalysis. Conformational fluctuations in such peptide networks were shown to coincide with catalysis in the serine hydrolase lipase B from Candida antarctica, which utilizes the same catalytic triad as BChE and other serine hydrolases24. Importantly, insertion of a photosensitive azobenzene bridge joining the flexible surface loops on C. antarctica lipase B allowed enhanced photostimulation of catalysis. It is proposed that within limits, energy transfer from the surface loop to the catalytic site is influenced by the sequence, quantity and physicochemical nature of the peptides within the network. Thus, increasing the length of this loop and facilitating its interaction with the solvent is hypothesized to increase catalytic activity by increasing energy transfer. We hypothesized that applying these principles to BChEG117H would aid in the development of a rationally designed mutant with increased catalytic activity toward OP anticholinesterases.
Herein, we report computer simulations with BChEG117H that identified a catalysis-associated, dynamic surface peptide network at residues 278–285. We hypothesized that increasing the number of amino acids in this network with long side-chain, hydrophilic residues could lead to higher energy transfer to the catalytic site and increased catalytic activity. Five BChEG117H loop mutants were constructed and evaluated for kinetics of choline and OP substrate hydrolysis, resistance to inhibition and dephosphylation rate constants (k3) using three OP anticholinesterases (paraoxon, echothiophate [EthP] and diisopropylfluorophosphate [DFP]).
A number of substrate-specific differences were noted between BChEG117H and the five loop mutants tested, including a minimal but statistically significant increase in the dephosphylation rate constant with one loop mutant particularly when paraoxon was used as the substrate. The results provide initial evidence that modifying a surface peptide network on BChEG117H can indeed alter its kinetic interactions with OP substrates. Simulations suggested however that in contrast to the choline ester substrate, OP substrates had poor affinity for the active site region which is likely pivotal in the observed low catalytic rate of BChEG117H and the loop mutants studied.
MATERIAL AND METHODS
Chemicals and reagents:
Figure 1 shows the chemical structures of the three OP toxicants studied. Paraoxon (O,O’-diethyl-p-nitrophenyl phosphate; PO) (98.6% purity by HPLC) was purchased from ChemService (West Chester, PA). A 10 mM stock solution of paraoxon was prepared in 100% dry ethanol and kept desiccated under nitrogen at −80°C until use.
Figure 1.
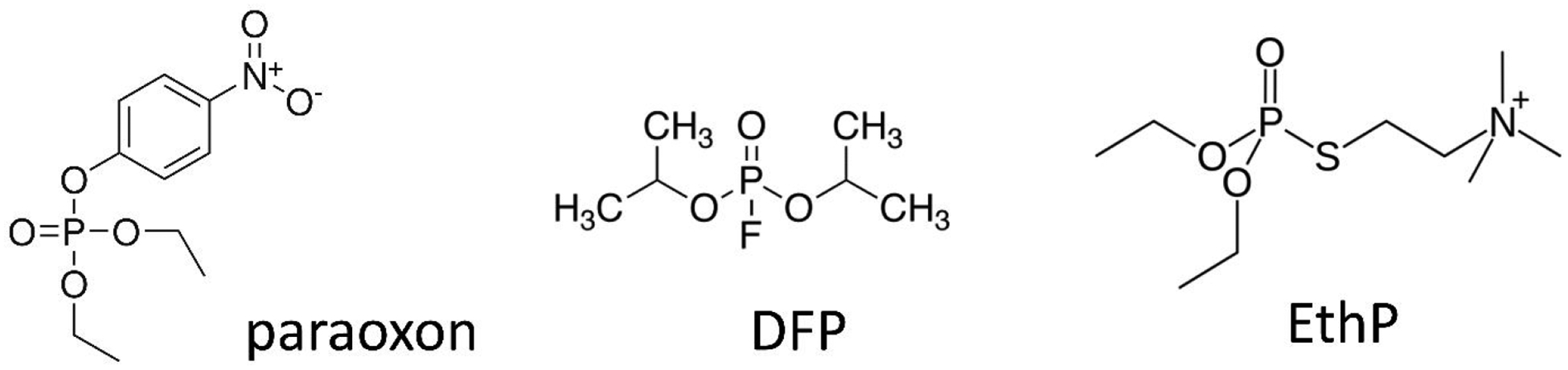
Structures of OP anticholinesterases (paraoxon, PO; diisopropylfluorophosphate, DFP; echothiophate, EthP) investigated in this study.
Diisopropylfluorophosphate (2-[fluoro(propan-2-yloxy)phosphoryl]oxypropane; DFP) (99% purity by NMR) was kindly provided by Derik Heiss at Battelle Memorial Institute (Columbus, OH) and stored as provided at −80°C25. Echothiophate iodide (2-diethoxyphosphorylsulfanylethyl(trimethyl)azanium; EthP) was originally obtained from Wyeth Ayerst and was a kind gift from Dr. Oksana Lockridge (University of Nebraska, Omaha, NE)26,27. Butyrylthiocholine iodide and all other chemicals and reagents were purchased from Sigma-Aldrich (St. Louis, MO).
Computer simulations and analysis:
Molecular dynamics (MD) simulations were performed to model native and engineered BChEG117H in complex with OP (PO, EthP and DFP) and choline (butyrylcholine, BCh) substrates in explicit water solvent. Note that the native enzyme considered for all modeling studies includes the G117H mutation, as it is required for catalytic activity. Model preparation and simulations were performed using the AMBER v14 suite of programs for biomolecular simulations28. AMBER’s ff14SB29 force-fields were used for all simulations. The parameters for the substrates were obtained using the protocol described in the AMBER manual. MD simulations were performed using NVIDIA graphical processing units (GPUs) and AMBER’s pmemd.cuda simulation engine using our lab protocols published previously30,31.
A total of twenty-four separate simulations were performed, based on the combination of native and engineered versions of BChEG117H in complex with PO, DFP and EthP, as well as with BCh. The enzyme was modeled based on the coordinates available in the protein data bank (PDB ID: 4BDS)32. The substrates were modeled based on the diester substrate coordinates for acetylcholine coordinates (PDB ID: 2ACE)33, the template for the diester bond was used and the remaining crystal structure and the atoms were added by AMBER’s leap program based on the substrate template developed using AMBER’s protocol. After processing the coordinates of the protein and substrate, all systems were neutralized by addition of counter-ions and the resulting system were solvated in a rectangular box of SPC/E water, with a 10 Å minimum distance between the protein and the edge of the periodic box. The prepared systems were equilibrated using a protocol described previously34. The equilibrated systems were then used to run 200 nanoseconds (ns) of production MD under constant energy conditions (NVE ensemble). The production simulations were performed at a temperature of 300 K and a time-step of 2 femtoseconds (with SHAKE applied to bonds and angles involving hydrogens). The production runs were performed under NVE conditions as this ensemble offers better computational stability and performance for longer MD simulations35. As NVE ensemble was used for production runs, these values correspond to initial temperature at start of simulations. Temperature adjusting thermostat was not used in simulations; over the course of 200 ns simulations the temperature fluctuated around 300 K with RMS fluctuations between 2–4 K, which is typical for well-equilibrated systems. A total of 10,000 conformational snapshots (stored every 20 ps) collected for each system was used for analysis.
Modeling the hydrolysis reaction catalyzed by BChE:
The enzyme mechanism of OP substrate hydrolysis by BChE has been previously investigated in detail by Zhan and coworkers based on quantum mechanical-molecular mechanical (QM/MM) modeling36. However, QM/MM studies of enzyme reactions are computationally expensive, particularly when the goal is to identify role of dynamics of distal residues. Therefore, to characterize the protein dynamics associated with the BChE enzyme mechanism, we performed with the faster classical (MD) simulations for a simple 4-state model of the reaction pathway. This pathway was described by the reactant state, two intermediate states and the product state for BChEG117H and used BCh as the model substrate, in order to identify intrinsic dynamics of BChE regions associated with the catalyzed reaction. The two intermediate states were modeled based on the previous acetylcholinesterase studies37. As classical MD simulations do not sample higher energy conformations adequately, we used the approach of applying restraints on important interatomic distances involved in the reaction mechanism during the MD simulations, which allows modeling of intermediate states (and transition states) with molecular mechanics and classical force-fields. This approach has been successfully used for investigating cocaine hydrolysis by BChE in the past38,39. Four separate MD simulations (based on the protocol described above) were performed for each of these states, each 100 ns in duration. See supporting information for further details.
Further, an additional MD simulation of 100 ns with paraoxon, as a prototype OP substrate, was also analyzed for loop regions of large protein flexibility. Paraoxon was selected for modeling of dynamics, as other detailed experimental studies (see resistance to inhibition described below) were performed with this substrate, and it is often used in comparative studies of OP anticholinesterase toxicity and cholinesterase biochemistry. The MD simulation with paraoxon as substrate was only performed in the reactant state, and the regions of large dynamical flexibility were found to be same as the ones when native substrate BCh was present. Therefore, results from 4-state models with OP substrate are expected to be similar to the ones with native BCh substrate.
RMSF10 calculations for protein dynamics:
The aggregated root-mean-square-fluctuations for top 10 quasi-harmonic modes (RMSF10) were used to calculate protein flexibility. It is well known that the slowest 10 modes contribute to the majority of fluctuations in proteins (>80%) and the use of RMSF10, instead of all modes, removes the faster stochastic motions of the protein, allowing focus on intrinsic dynamics of proteins40. Both these calculations were performed using AMBER’s ptraj analysis program. All trajectory conformations were first aligned to a common structure, to remove any translation and overall molecular rotation during the simulations.
Enzyme-substrate interactions:
The energy for the enzyme-substrate interactions was calculated as a sum of electrostatic and van der Waals energy between atom pairs. This protocol was previously developed to investigate other protein-substrate systems34,41.
| (1) |
Eel is the electrostatic contribution, Evdw is the van der Waals term and the summation runs over all atom pairs for the protein-substrate complex. The Eel and Evdw terms were computed as follows,
| (2) |
where qi are partial charges, and Aij, Bij are Lennard-Jones parameters. These parameters were obtained from the AMBER force field. A distance-dependent dielectric function was used:
| (3) |
B= εo−A; εo=78.4 for water; A= −8.5525; λ=0.003627 and k=7.7839.
All enzyme and substrate atom pairs were included in the calculations and resulting interaction energies were summed up per residue pair. The energies were calculated for 10,000 snapshots, every 20 ps, sampled during the full 200 ns simulation and were averaged over these 10,000 snapshots.
Expression and purification of BChEG117H and loop mutants:
We constructed a monomeric BChEG117H mutant (control with no insertions) and six monomeric BChEG117H loop mutants with three residue insertions into the residues 278–285 sequence with an identity of ENX, keeping glutamate and asparagine insertions constant but varying “X” with one of six different amino acids (A, G, I, P, R, and T). The constructs of all seven were cloned into the pMFH6 vector to express the mutants with a hexahistidine tag at the C-terminus. In vivo transposition of the expression cassette into a bacmid in DH10Bac cells, isolation of recombinant bacmid DNA, and transfection of the recombinant bacmid DNA into Spodoptera frugiperda Sf9 cells to generate recombinant baculovirus were performed according to the Bac-to-Bac Baculovirus Expression System (Invitrogen Life Technologies), The baculovirus stock was then amplified and tittered to infect Sf9 cells for large-scale expression of mutants. The expressed proteins, secreted in the medium, were purified using a Q Sepharose Fast Flow column followed by a Ni2+-NTA agarose column. Protein concentrations were determined on a NanoDrop Spectrophotometer (Thermo Scientific ND1000).
Substrate kinetics with BTCh:
Enzyme activity with the choline substrate was evaluated using a modified Ellman method42 using butyrylthiocholine (BTCh) as the substrate. The final concentration of BTCh was varied between 10 μM and 1 mM and change in absorbance was monitored at 412 nm for 5 min at 37°C using a Spectramax M2 microplate reader (Molecular Devices; Sunnyvale, CA). Enzyme activity was estimated based on the rate of appearance of the reaction product, 2-nitro-5-thiobenzoate (ε = 14,150 M−1 cm−1) and was uniformly corrected for non-enzymatic substrate hydrolysis using enzyme-free controls. Vmax and KM were determined using non-linear regression with the Michaelis-Menten equation. Turnover number, kcat, was calculated using the relationship Vmax = kcat[E], where [E] represents the concentration of BChE active sites in the reaction.
Resistance to inhibition assay:
Resistance of BChEG117H and the loop mutants to inhibition by the OP toxicants was evaluated similar to Wang and associates (2004)26. Working enzyme solutions were prepared prior to assay (2 μg/ml in 100 mM potassium phosphate buffer, pH 7.0). Twenty ng of BChEG117H or one of the loop mutants was added to either vehicle or paraoxon (50 μM), diisopropylfluorophosphate (5 μM) or echothiophate (100 μM) and allowed to pre-incubate for 10 min in a 96-well plate containing buffer (100 mM potassium phosphate, pH 7.0) and DTNB (0.5 mM). Following pre-incubation, 20 μl of BTCh (1 mM final concentration) was added to begin the reaction and enzyme activity was assayed as described above and plotted as percent of the respective vehicle control over time.
Reactivation rate (k3) determination:
Hydrolysis of OP compounds by BChE proceeds as shown in Figure 2.
Figure 2.

Schematic of the interaction of BChE and a model OP toxicant (paraoxon). First, an OP compound and BChE form a reversible Michaelis-complex (k1/k−1), however the bond quickly progresses through this step to phosphylation at a rate of k2. Spontaneous reactivation (k3) does not occur at an appreciable rate in the wild-type enzyme, but is detectable in the mutant BChEG117H16.
The relative rate of dephosphylation (k3), a measure of an enzyme’s ability to reactivate after inhibition, can be used as an indicator of the catalytic rate. The k3 values of BChEG117H and the loop mutants were determined as described by Lockridge and colleagues16. In brief, an equal volume of enzyme (20 μg/ml in 100 mM potassium phosphate buffer, pH 7.0) and an OP inhibitor (10 mM paraoxon or echothiophate) were pre-incubated for 1 min to achieve saturation of all enzyme active sites. Following incubation, a 5 μl sample was removed, rapidly diluted 400-fold into a cuvette containing the reaction components (0.5 mM DTNB and 1 mM BTCh in 100 mM potassium phosphate buffer, pH 7.0) and gently mixed. This large dilution reduces the final OP toxicant concentration to below its KM, allowing dissociation of any OP molecules from the reversible Michaelis complexes. Reactivation of the covalently bound enzyme was then followed at 412 nm at 37°C as described above. If the extent of reactivation was incomplete, the equation provided by Hovanec and colleagues was used as a correction for the observed k343. As this assay is highly time-sensitive, the timing between sample transfer and initiation of absorbance readings was kept constant at 5 sec.
Paraoxon hydrolysis:
Hydrolysis of the OP substrate paraoxon was determined using a direct photometric assay similar to that of Lockridge and colleagues16. BChEG117H and the loop mutants were diluted to 0.06 mg/ml using PBS. A stock solution of paraoxon (in dry ethanol) was diluted on the day of assay and added to the reaction in final concentrations between 10–500 μM (with 5% v/v ethanol included in all reactions). The final reaction volume of 100 μl contained 7.5 μl diluted enzyme and 72.5 μl 100 mM and 50 mM potassium phosphate buffer (pH 7.0), with the reaction being initiated by adding a 20 μl aliquot of paraoxon. The p-nitrophenol produced by hydrolysis of paraoxon was monitored at 405 nm for 1 h at 37°C. The extinction coefficient of p-nitrophenol (ε = 18,500 M−1 cm−1) was used to determine enzyme activity, corrected by subtracting non-enzymatic hydrolysis. Kinetic parameters were determined using non-linear regression with the Michaelis-Menten equation.
Statistical Analyses:
Differences in substrate kinetics parameters with both BTCh and paraoxon were evaluated using one-way ANOVA. For the resistance to inhibition and reactivation rate (k3) assays, statistical differences were determined using two-way ANOVA with enzyme and OP inhibitor as main effect variables. In all cases, a post hoc Dunnett’s multiple comparisons test was performed to determine differences among BChEG117H and the loop mutants. All statistical tests were conducted using GraphPad Prism software for Windows (La Jolla, CA), version 6.0.
RESULTS
Loop engineering:
For BChEG117H we identified two surface loops (residues 278–285 and 375–382) by computational modeling that showed large, dynamical motions coupled with the hydrolysis of substrate in the active site. The octapeptide comprising residues 278–285 (278FVVPYGTP285) in particular showed enhanced catalysis-associated dynamics. We therefore selected this loop as our initial target of engineering. We hypothesized that increasing the length of this loop through insertions of residues with long hydrophilic side chains to increase the surface area for thermodynamical coupling would improve the rate of catalysis of BChEG117H. To address the concern regarding change of secondary structure of the surface loop with insertions of the residues as well as to obtain ideas of suitable residues at these sites, we performed a BLASTP search, which identified a subject annotated as serine carboxypeptidase with the matched sequence of 78FVVEPYNGTIP88. Unfortunately, a structure of this protein was not available. A homology search with the identified serine carboxypeptidase, further identified wheat serine carboxypeptidase (PDB code 1BCR), whose structure indicates the corresponding region is on the surface as a partial loop. Based on this additional information, we postulated that inserting an additional three residues, a glutamate between V280 and P281, an asparagine between Y282 and G283, and a variable residue between T284 and P285 of BChEG117H would enhance dynamics and increase interaction with the solvent without significantly affecting the overall protein structure (Figure 3). Instead of using a fixed residue I in the third position, we incorporated VNK in the primer to encode a wild card residue (A, D, E, G, H, I, K, L, M, N, P, Q, R, S, T, or V).
Figure 3: A surface loop in BChE (residues 278–285) was selected for protein engineering.
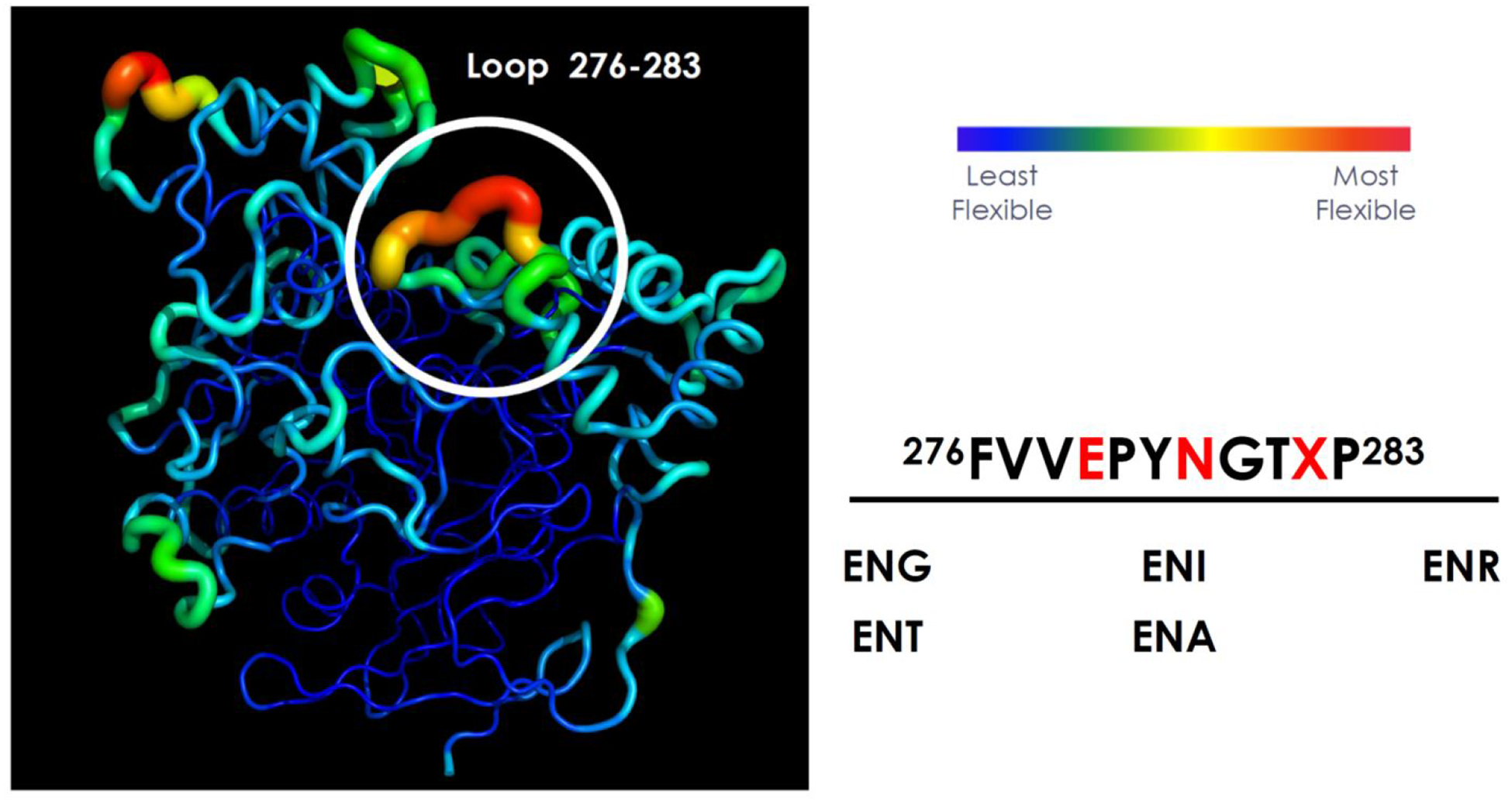
Dynamical motions associated with the enzyme were identified using RMSF10. The thin blue lines show rigid areas while the green to red areas with thicker tubes correspond to regions displaying large conformational fluctuations. The selected loop is in the highlighted circle. After design considerations, five mutants were selected for dynamical loop engineering. The red residues are inserted residues. In addition to E and N as the first two insertions, five different residues were considered at the third insertion site.
Number of residues inserted in engineered loop:
Figure 4 shows results of computational modeling to characterize the effect of increasing the length of the engineered loop on dynamics. Starting from the BChEG117H, one (E), two (E, N) and three (E, N, I) residues were inserted in the positions described above. Computational modeling based on MD simulations indicated that the dynamical flexibility increased markedly with insertion of three residues, while inserting only one led to a slight increase and inserting two showed a slight decrease. These results indicate that insertion of selected residues into the loop sequence may “fine tune” the structure enabling more interactions and increased loop dynamics, as predicted.
Figure 4:
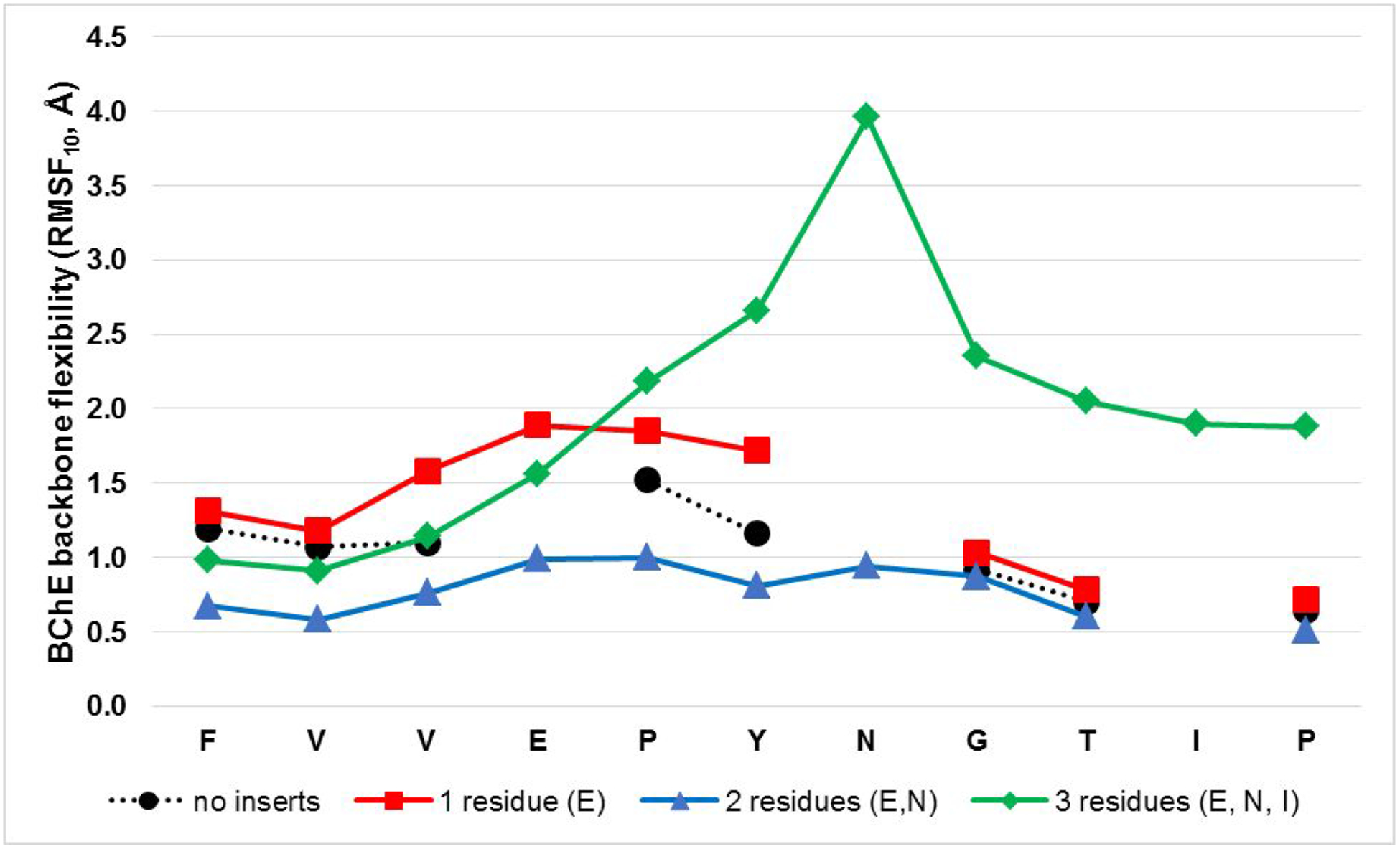
Increasing the number of residues inserted in engineered loop led to changes in dynamics. MD simulations indicated increased backbone flexibility as one or three residues were inserted, while inserting two residues showed a slight decrease (possibly due to an increase in structural interactions). Therefore, insertions of three residues was considered optimal for further evaluations.
BTCh substrate kinetics:
First, the activity of all six loop mutants with butyrylthiocholine (1 mM) was markedly lower than noted with BChEG117H expressed and purified in the same manner (mean U/mg protein ± SD: BChEG117H, 110.8 ± 5.4; ENT, 41.9 ± 4.8; ENG, 41.7 ± 3.2; ENA, 20.4 ± 1.6; ENI, 19.9 ± 2.2; ENR, 16.9 ± 0.9; ENP, 5.5 ± 0.8). The mutant ENP was computationally considered but its catalytic activity with butyrylthiocholine was so low that kinetic analyses proved difficult (the results were highly variable), and thus we did not evaluate ENP further.
Table 1 shows the kinetic parameters (kcat, KM, and kcat/KM) determined from the hydrolysis of butyrylthiocholine by BChEG117H and BChEG117H-derived loop mutants (n = 3 for each enzyme preparation). The turnover rate with BTCh was significantly reduced (40–84%) in all enzymes with loop insertions compared to BChEG117H (F5,156 = 5.25, p = 0.0002). A significant effect was also found in KM values (F5,156 = 58.1, p < 0.0001). Post hoc analysis indicated that ENG and ENR mutants had a significantly higher KM in comparison to BChEG117H. These parameters together as the ratio kcat/KM suggest that the catalytic efficiency of the loop mutants was reduced compared to BChEG117H. Note that the mutants were designed based on computational identification of increased dynamics of the loop region during modeling of paraoxon hydrolysis.
Table 1. Comparison of substrate kinetics parameters of BChEG117H and loop mutants using butyrylthiocholine as the substrate and analysis using the Michaelis-Menten equation.
Values are reported as mean ± SEM. An asterisk indicates a significant difference compared to BChEG117H.
|
kcat
a (min−1) |
KM (mM) |
kcat/KM (min−1 M−1) |
|
|---|---|---|---|
| G117H | 13884±374 | 1.21 ± 0.06 | 11.5 × 106 |
| +ENI | 2207 ± 76* | 1.7 ± 0.10 | 1.3 × 106 |
| +ENA | 4024 ± 204* | 1.68 ± 0.14 | 2.4 × 106 |
| +ENG | 8363 ± 1280* | 2.66 ± 0.60* | 3.1 × 106 |
| +ENR | 2568±359* | 3.68 ± 0.71* | 0.7 × 106 |
| +ENT | 5976±251* | 1.84 ± 0.13 | 3.2 × 106 |
kcat = Vmax/[enzyme active site]
Resistance to inhibition:
To evaluate enzyme-OP toxicant interactions, we first evaluated the enzyme’s ability to resist inhibition by a high concentration of an OP anticholinesterase following a 10-min incubation period, as shown in Figure 5. Preliminary assays allowed selection of OP toxicant concentrations that achieved near complete inhibition of BChEG117 (>99%; data not shown). Under these conditions, using paraoxon, BChEG117H was somewhat more resistant to inhibition compared to all five loop mutants studied, i.e., it retained 57% of its activity following paraoxon exposure while loop mutants retained only 31–44% of pre-exposure activity). In contrast, when DFP was used as the inhibitor, the loop mutants were generally more resistant to inhibition. Specifically, ENI, ENA, ENG, and ENR mutants retained nearly full activity (96–100%) following DFP exposure, which was higher than noted with BChEG117H (82%). A similar finding was noted following exposure to EthP, with three loop mutants (ENI, ENA, and ENR) being more resistant to inhibition than BChEG117H.
Figure 5:
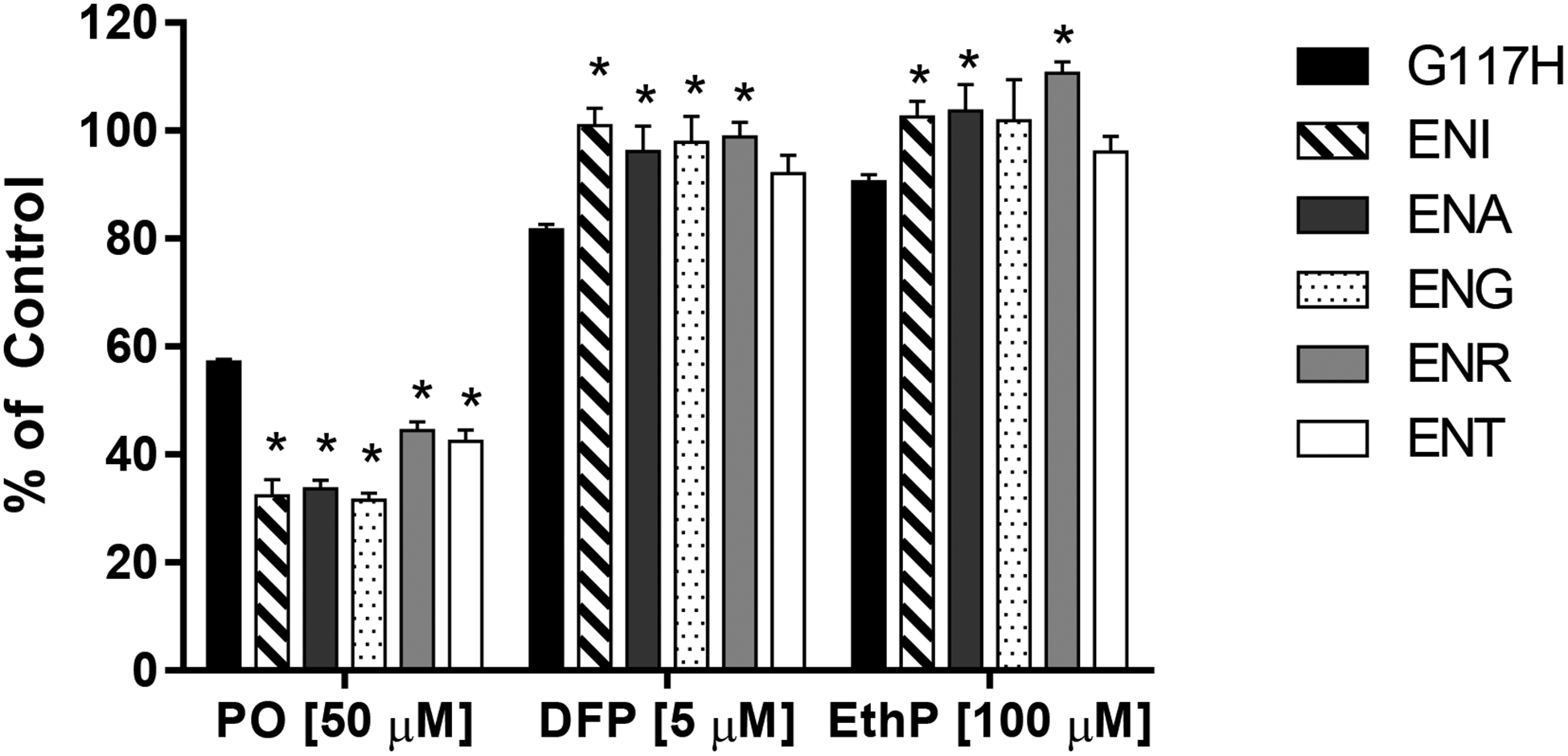
Comparative resistance to inhibition of BChEG117H and loop mutants following a 10-min exposure to paraoxon (50 μM final), DFP (5 μM final) or EthP (100 μM final). Data represent the mean ± SEM of three independent replicates. An asterisk indicates a significant difference compared to BChEG117H.
Reactivation Rate:
This assay starts with putatively complete occupation of all active sites of the BChEG117H molecules by pre-incubation in an excess of an OP inhibitor. Following a marked (400-fold) dilution, any OP inhibitor bound in a reversible Michaelis complex would dissociate, while OP molecules bound covalently to the active site serine would not. Thus, measuring the rate of recovery of catalytic activity under these conditions reflects the rate of dephosphylation. Baseline (100%) activity for each enzyme was estimated using enzyme without inhibitor. As shown in Figure 6A, when paraoxon or echothiophate was used, all enzymes showed an initial reduction in activity followed by recovery to 100% of baseline. With DFP as the inhibitor, complete reactivation was not evident, likely due to aging of the enzyme which results in irreversible inhibition44. The activity of DFP inhibited BChEG117H returned to 67 ± 2% of baseline while the various loop mutants showed recovery to 72–96% of baseline. The reactivation rate (k3) was calculated by plotting the difference between observed and estimated values versus time during the recovery period, as described previously by Lockridge and coworkers16. The k3 values of BChEG117H with echothiophate (1.28 min−1 ± 0.04) and paraoxon (1.27 min−1 ± 0.11) as substrate were relatively similar to those reported previously16. To our knowledge, the reactivation rate of BChEG117H with DFP has not been previously reported. The ENA loop mutant showed a slight but significant increase in k3 when pre-incubated with paraoxon, but no differences in k3 were noted with the ENA mutant using DFP or echothiophate. In contrast, Figure 6B shows the k3 of the ENI and ENR loop mutants with DFP was significantly decreased. Note that the reported k3 values in cases where recovery did not achieve 100% of baseline were corrected by the approach of Hovanec and colleagues43.
Figure 6:
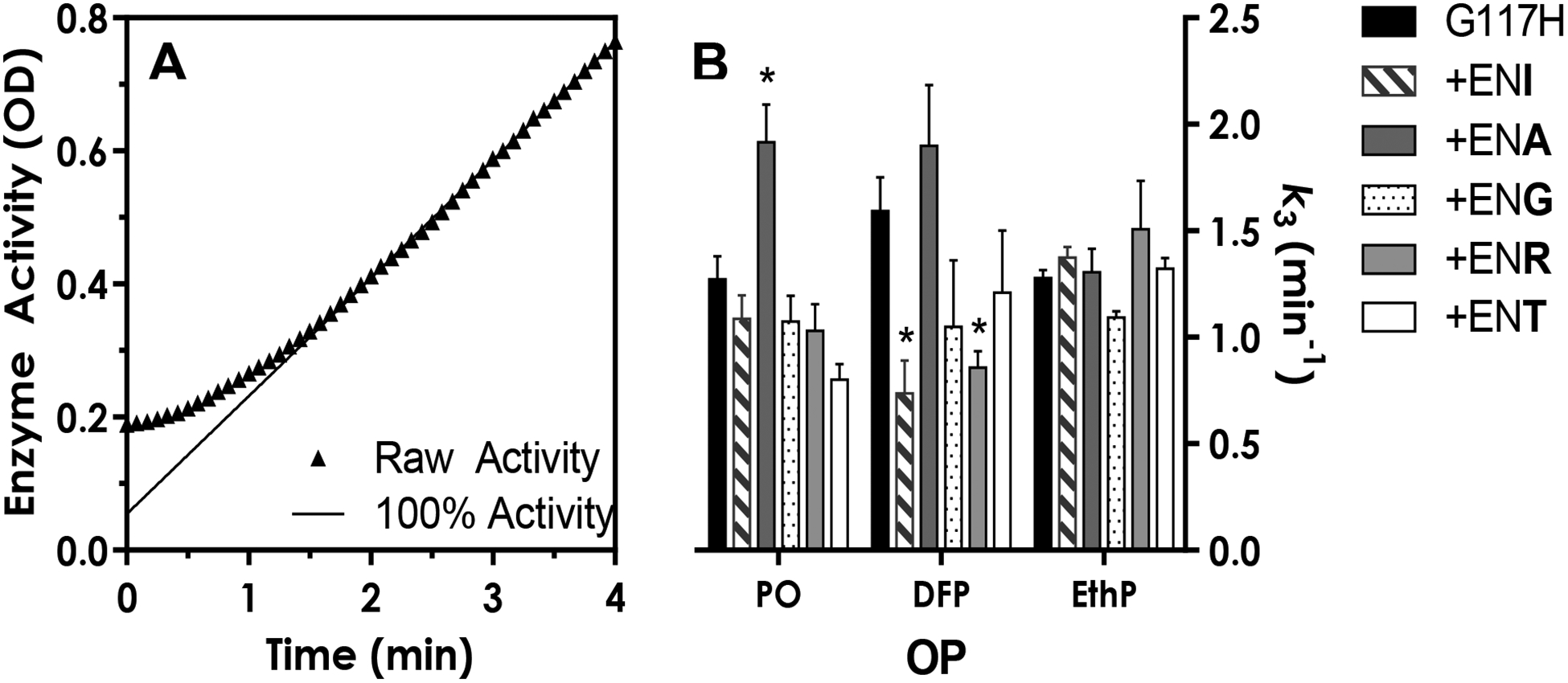
Recovery of activity in BChEG117H and loop mutants following exposure to 10 mM OP inhibitor (paraoxon, DFP or EthP). Enzyme was allowed to react with the OP toxicant for one minute and then diluted 400-fold into a solution containing substrate (BTChI, 1 mM). Figure 6A shows a representative plot of raw enzyme activity (ΔA412) following dilution including the reactivation and recovery phases. Figure 6B shows the mean ± SEM of k3 values. In the case of DFP, 100% reactivation was not achieved, thus reported k3 values have been corrected based on Hovanec et al43.
Paraoxon hydrolysis:
Substrate kinetics with paraoxon suggested that all loop mutants had reduced catalytic activity compared to BChEG117H. KM was significantly lower in the ENA mutant and there was a trend toward reduced KM with ENG and ENT (p = 0.062 and p = 0.052, respectively). Table 2 compares substrate kinetics among the different enzymes. The loop mutants ENA and ENG showed a trend toward higher catalytic efficiency (kcat/KM) compared to BChEG117H, but this appeared driven by the changes in KM and not catalytic activity. Changes in substrate binding affinity could be critical in the kinetics of OP toxicant hydrolysis. Therefore, we used computational modeling to compare binding of paraoxon, echothiophate and DFP to BChEG117H and the selected engineered loop mutants.
Table 2: Comparison of the substrate kinetics parameters of BChEG117H and loop mutants using paraoxon as the substrate.
Kinetic analysis was determined using non-linear regression with the classic Michaelis-Menten equation. Values are reported as mean ± SEM. An asterisk indicates a significant difference compared to BChEG117H.
|
kcat
a (min−1) |
KM (μM) |
kcat/KM (min−1 M−1) |
|
|---|---|---|---|
| G117H | 0.244 ± 0.03 | 107.6 ± 28.1 | 2.2 × 103 |
| ENI | 0.071 ± 0.003* | 75.82 ± 9.3 | 0.9 × 103 |
| ENA | 0.078 ± 0.004* | 19.72 ± 5.3* | 3.9 ×103 |
| ENG | 0.089 ± 0.005* | 24.23 ± 6.0* | 3.6 ×103 |
| ENR | 0.092 ± 0.005* | 77.62 ± 12.2 | 1.2 × 103 |
| ENT | 0.076 ± 0.005* | 23.59 ± 6.1* | 3.2 × 103 |
kcat = Vmax/[enzyme active site]
Inhibitor/substrate binding in the active site:
Simulations of enzyme-substrate interactions of BChEG117H and the five loop mutants indicated that OP substrates bound poorly to the active site region. Table 3 shows the interaction energies from conformational snapshots sampled during the MD simulations, and estimated using a sum of electrostatics and van der Waals interactions between the substrate and enzyme residues. In addition, the MD trajectories were also analyzed for enzyme-substrate interaction behavior.
Table 3: Enzyme-substrate interaction energy obtained from computations.
Averaged interaction energies (in kcal/mol) between substrate and full enzyme (all residues) and only the catalytic triad (residues H117, S198, and H438).
| BCh (174.262a) | paraoxon (275.195a) | DFP (184.146a) | echothiophate (383.228a) | |||||
|---|---|---|---|---|---|---|---|---|
| enzyme | triad | enzyme | triad | enzyme | triad | enzyme | triad | |
| G117H | −34.84 | −8.45 | −35.36 | −2.41 | −10.29 | −0.43 | −33.96 | −4.98 |
| ENI | −32.34 | −8.40 | −38.86 | −9.17b | −23.05 | −3.15 | −39.61 | −9.62b |
| ENG | −31.50 | −8.42 | −37.88 | −7.91b | −10.73 | −1.49 | −35.59 | −7.29b |
| ENA | −31.26 | −8.14 | −33.62 | −1.36 | −24.34 | −4.87 | −39.99 | −10.17b |
| ENT | −31.48 | −8.21 | −35.89 | −5.03 | −21.83 | −3.29 | −39.45 | −6.74b |
| ENR | −33.13 | −9.22 | −34.08 | −1.78 | −24.77 | −4.52 | −36.15 | −8.46b |
molecular weight,
indicates interaction ≥ in strength to native substrate.
Relatively similar interactions were noted between BCh as substrate and BChEG117H and the loop mutants, with the substrate remaining in the active site region for a relatively prolonged time (full 200 ns of MD trajectory). The movies of the MD simulations are available as supporting information. In contrast, when an OP substrate was modeled, it left the active site more rapidly (around 100 nanoseconds or less). These differences are reflected in Table 3, with lesser negative values in the case of the OP substrates. With paraoxon, (except with the ENI mutant) the substrate completely avoided the active site pocket after 50 ns or less, making only minimal contacts with the protein surface. With the ENI mutant, paraoxon remained in the active site with its aromatic group maintaining its original conformation while other moieties sampled alternate conformations. With DFP, the OP molecule quickly left the active site pocket as well. Moreover, DFP interactions with BChEG117H suggested the OP molecule completely dissociated from the protein and moved into the bulk-solvent indicating very weak interactions. Finally, with echothiophate, more stable interactions were noted with ENI and ENG mutants, but the OP molecule also rapidly left the active site in BChEG117H and the ENA loop-mutant. Interestingly, simulations with DFP suggested even lesser time-dependent interaction with the enzyme. Note that in cases where the active site region showed poor binding but the substrate still interacted with the protein, this was due to interactions with residues outside the active site. It should also be noted that OP substrates are larger than BCh and when interaction energy values are adjusted for molecular weight, the OP toxicant interaction with the enzymes appears even weaker.
DISCUSSION:
Enzymes are known to accelerate chemical reactions by several orders of magnitude, in some cases by as much as 1020 (that is 20 orders of magnitude). The essential contributions of primary, secondary, tertiary and quaternary structural characteristics of the protein in influencing catalytic activity are well known. More recently, protein dynamics has emerged as a contributing factor in enzyme-mediated catalysis21. It has been suggested that enzyme structure and dynamics of selective surface loop regions enable thermodynamical coupling between the enzyme surface and active site residues. This coupling provides energy to overcome the activation energy barrier. In this paradigm, increased motions of surface loops are critical for collecting energy from solvent and relaying it to the active site through a network of residues. The current study was designed to study the structure-dynamics-activity relationships in catalytic degradation of OP anticholinesterases by BChEG117H through rational engineering of a dynamic surface peptide network. The use of human butyrylcholinesterase (BChE) as a bioscavenger for organophosphate anticholinesterases is markedly limited by its stoichiometric interaction with OP toxicants.
Despite extensive research, little progress has been made in developing an effective catalytic bioscavenger based on BChE. Based on a growing body of evidence that protein dynamics influence enzyme catalysis, we applied these principles to a slow catalytic variant, BChEG117H, in an attempt to enhance its activity against OP toxicants and understand better the role of surface peptide networks in catalysis. Molecular dynamics simulations identified a dynamic surface loop on BChEG117H. The effects of peptide insertions into this peptide network on choline ester and OP substrate binding were modeled and enzyme activity evaluated in five recombinant loop mutants. The results confirm that surface peptide networks identified on this enzyme by molecular simulations can influence substrate-active site interactions and be potentially used to enhance the hydrolysis of catalytic bioscavengers.
To search for a more effective BChE catalyst, we followed a new paradigm where enzymes are viewed not only as biochemical but also as biophysical machines. The enzyme, in addition to providing appropriate active site residues for hydrolytic chemistry, also captures energy to overcome the energy barrier associated with substrate hydrolysis. This energy is collected from the solvent on the surface and transferred into the active site region. Therefore, we developed engineered mutants of the existing mutant BChEG117H to investigate potential changes in enzyme-solvent energy coupling. Using a computational approach, we identified a surface loop (residues 278–285) far from the active site region which showed increased dynamics compared to other regions of the enzyme during catalysis. Previously, we have shown that surface loop regions of an enzyme can exhibit increased dynamic motions during enzyme-solvent thermodynamical coupling24. The engineered surface loop mutants described herein were designed to increase the length and solvent interactions of this sequence by inserting three additional residues with long side-chains to putatively improve energy coupling with the solvent for enhanced energy transfer.
Interestingly, the engineered loop mutants studied all had a reduced catalytic efficiency (kcat/KM) with the choline substrate, BTCh. We did not expect any of our mutations to improve the catalytic rate using the choline substrate as it is already operating near the rate of diffusion (maximal speed). This is supported by the similar observation that BChEG117H also exhibits reduced activity with choline esters (e.g., butyrylthiocholine)16. Our data also suggested that binding of the substrate with all the tested loop mutants was reduced relative to binding to BChEG117H, suggesting the changes in the surface peptide network were indeed influencing physiochemical interactions in the active site region but in a negative manner relative to catalytic activity.
We conducted two assays to evaluate the enzymes’ interactions with three OP anticholinesterases. First, we tested the enzymes’ resistance to inhibition by a high concentration of OP. It should be noted that with this assay, the mechanism of resistance cannot be fully appreciated, but a change in resistance to inhibition may reflect a change in catalytic activity or substrate binding. We found that with DFP and echothiophate as substrates, the loop mutants ENI, ENA, and ENR were more resistant to inhibition than BChEG117H. Surprisingly, all loop mutants were less resistant to inhibition by paraoxon compared to BChEG117H. These conflicting results suggested that the selected peptide insertions differentially affected interaction with the different OP anticholinesterases. To evaluate further a mechanism of altered interactions between the enzyme and substrates, we measured enzyme reactivation, which is relatively unaffected by changes in substrate binding. Using paraoxon as the substrate, only the ENA mutant showed an improved catalytic efficiency. This increase, however, was not evident when paraoxon hydrolysis was evaluated separately, wherein we found a significant decrease in both kcat and KM. Taken together, these results suggested that the decreased KM with paraoxon had more impact on the enzyme’s apparent efficiency than any increase in reactivation. Reactivation data with DFP and EthP did not show significant increases in k3, suggesting that the noted increase in loop mutant resistance to inhibition is likely a reflection of altered binding. We do not however have direct hydrolysis data from DFP and echothiophate to confirm this possibility.
Computer simulations of BChEG117H and the engineered loop mutants with the choline ester substrate butyrylcholine and selected OP compounds provided insights into the observed experimental findings. BCh had strong interactions with all enzymes tested. However, the OP substrates showed mostly weak interactions, with the substrate showing unstable binding in most cases, rapidly leaving the active site pocket in less than 50 nanoseconds. This implied that the enzyme had limited time for effective hydrolysis. Interestingly the loop mutants showed slightly improved interactions with the OP substrates, particularly, paraoxon and echothiophate. This was supported by the increased kcat/KM for these mutants over BChEG117H.
In conclusion, surface loop insertions on BChEG117H resulted in relatively minimal changes in catalysis but appeared to decrease stability of the OP substrates within the active site region. Substrate/inhibitor-specific differences were noted between BChEG117H and the loop mutants with substrate kinetics and resistance to inhibition assays. We did observe a slight but significant increase in the dephosphylation rate constant with paraoxon with one (ENA) loop mutant. Our data suggest that increasing the interaction of identified hypermobile surface loops with the solvent may potentially yield improved catalytic interactions between OP toxicanthydrolyzing enzymes and their substrates. Further emphasis on substrate stability within the active site of mutants may be important in future studies. While other OP anticholinesterase hydrolases (e.g., paraoxonase, phosphotriesterase) have their own difficulties for prophylactic applications (e.g, immunogenicity, more limited substrate specificity)10,11, catalytic bioscavengers with higher intrinsic activity may be better candidates for evaluating the role of surface loop dynamics in enhancing OP toxicant hydrolysis. Understanding how surface loop networks and their dynamics contribute to catalysis may lead to further development of more effective catalytic bioscavengers for protection against OP toxicity.
Supplementary Material
ACKNOWLEDGEMENT:
These studies were supported by the Sitlington endowment from the Oklahoma State Board of Regents (CNP), an Interdisciplinary Toxicology Program Fellowship at Oklahoma State University (KH), an internal team-building grant from the Center for Veterinary Health Sciences (AE-1-50060, CNP, PKA, HJ), and a multi-PI grant from NIGMS/NIH to PKA (GM105978, PKA). Other sources of funding included HDTRA1-13-1-0042 (CNP), R01ES008739-20 (CNP), SBIR W911QY-18-C-0200 (CNP), R01GM58634 (HJ), R21 AI112662 (HJ) and Oklahoma Agricultural Experimental Station project OKL03054 (HJ).
Footnotes
SUPPORTING INFORMATION:
Sixteen video clips showing the 200 ns MD simulations between BChEG117H and loop-mutant enzymes against a choline substrate (BCh) and OP substrates (paraoxon, echothiophate, and DFP).
REFERENCES
- (1).Holstege CP, Kirk M, and Sidell FR (1997) Chemical warfare. Nerve agent poisoning. Crit. Care. Clin 13, 923–942. [DOI] [PubMed] [Google Scholar]
- (2).John H, van der Schans MJ, Koller M, Spruit HET, Worek F, Thiermann H, and Noort D (2018) Fatal sarin poisoning in Syria 2013: forensic verification within an international laboratory network. Forensic Toxicol 36, 61–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Paddock RC, and Sang-Hun C (2017, February 23) Kim Jong-nam Was Killed by VX Nerve Agent, Malaysians Say. nytimes.com. [Google Scholar]
- (4).Peter J, Sudarsan T, and Moran J (2014) Clinical features of organophosphate poisoning: A review of different classification systems and approaches. Indian J Crit Care Med 18, 805–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Ecobichon DJ (2001) Toxic Effects of Pesticides, in The C5 isozyme of serum cholinesterase and adult weight (Klassen CD, Ed.) 6 ed., pp 763–810. [Google Scholar]
- (6).Leikin JB, Thomas RG, Walter FG, Klein R, and Meislin HW (2002) A review of nerve agent exposure for the critical care physician. Crit. Care Med 30, 2346–2354. [DOI] [PubMed] [Google Scholar]
- (7).Chen Y (2012) Organophosphate-induced brain damage: mechanisms, neuropsychiatric and neurological consequences, and potential therapeutic strategies. Neurotoxicology 33, 391–400. [DOI] [PubMed] [Google Scholar]
- (8).Miyaki K, Nishiwaki Y, Maekawa K, Ogawa Y, Asukai N, Yoshimura K, Etoh N, Matsumoto Y, Kikuchi Y, Kumagai N, and Omae K (2005) Effects of sarin on the nervous system of subway workers seven years after the Tokyo subway sarin attack. J Occup Health 47, 299–304. [DOI] [PubMed] [Google Scholar]
- (9).Wolfe AD, Rush RS, Doctor BP, Koplovitz I, and Jones D (1987) Acetylcholinesterase prophylaxis against organophosphate toxicity. Fundam. Appl. Toxicol 9, 266–270. [DOI] [PubMed] [Google Scholar]
- (10).Nachon F, Brazzolotto X, Trovaslet M, and Masson P (2013) Progress in the development of enzyme-based nerve agent bioscavengers. Chem. Biol. Interact 206, 536–544. [DOI] [PubMed] [Google Scholar]
- (11).Wales ME, and Reeves TE (2012) Organophosphorus hydrolase as an in vivo catalytic nerve agent bioscavenger Drug Test Anal (John H, and Blum M-M, Eds.) 4, 271–281. [DOI] [PubMed] [Google Scholar]
- (12).Genovese RF, Sun W, Johnson CC, diTargiani RC, Doctor BP, and Saxena A (2010) Safety of administration of human butyrylcholinesterase and its conjugates with soman or VX in rats. Basic Clin. Pharmacol. Toxicol 106, 428–434. [DOI] [PubMed] [Google Scholar]
- (13).Reed BA, Sabourin CL, and Lenz DE (2017) Human butyrylcholinesterase efficacy against nerve agent exposure. J. Biochem. Mol. Toxicol 31. [DOI] [PubMed] [Google Scholar]
- (14).Järv J (1984) Stereochemical aspects of cholinesterase catalysis. Bioorganic Chemistry 12, 259–278. [Google Scholar]
- (15).Millard CB, Lockridge O, and Broomfield CA (1995) Design and expression of organophosphorus acid anhydride hydrolase activity in human butyrylcholinesterase. Biochemistry 34, 15925–15933. [DOI] [PubMed] [Google Scholar]
- (16).Lockridge O, Blong RM, Masson P, Froment MT, Millard CB, and Broomfield CA (1997) A single amino acid substitution, Gly117His, confers phosphotriesterase (organophosphorus acid anhydride hydrolase) activity on human butyrylcholinesterase. Biochemistry 36, 786–795. [DOI] [PubMed] [Google Scholar]
- (17).Geyer BC, Kannan L, Garnaud P-E, Broomfield CA, Cadieux CL, Cherni I, Hodgins SM, Kasten SA, Kelley K, Kilbourne J, Oliver ZP, Otto TC, Puffenberger I, Reeves TE, Robbins N, Woods RR, Soreq H, Lenz DE, Cerasoli DM, and Mor TS (2010) Plant-derived human butyrylcholinesterase, but not an organophosphorous-compound hydrolyzing variant thereof, protects rodents against nerve agents. Proc. Natl. Acad. Sci. U.S.A 107, 20251–20256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Smolen A, Eckerson HW, Gan KN, Hailat N, and La Du BN (1991) Characteristics of the genetically determined allozymic forms of human serum paraoxonase/arylesterase. Drug Metab. Dispos 19, 107–112. [PubMed] [Google Scholar]
- (19).Dumas DP, Durst HD, Landis WG, Raushel FM, and Wild JR (1990) Inactivation of organophosphorus nerve agents by the phosphotriesterase from Pseudomonas diminuta. Arch. Biochem. Biophys 277, 155–159. [DOI] [PubMed] [Google Scholar]
- (20).Lushchekina SV, Schopfer LM, Grigorenko BL, Nemukhin AV, Varfolomeev SD, Lockridge O, and Masson P (2018) Optimization of Cholinesterase-Based Catalytic Bioscavengers Against Organophosphorus Agents. Front Pharmacol 9, 211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Agarwal PK (2019) A Biophysical Perspective on Enzyme Catalysis. Biochemistry 58, 438–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Agarwal PK (2006) Enzymes: An integrated view of structure, dynamics and function. Microb. Cell Fact 5, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Ramanathan A, and Agarwal PK (2011) Evolutionarily conserved linkage between enzyme fold, flexibility, and catalysis. PLoS Biol 9, e1001193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Agarwal PK, Schultz C, Kalivretenos A, Ghosh B, and Broedel SEJ (2012) Engineering a Hyper-catalytic Enzyme by Photoactivated Conformation Modulation. Journal of Physical Chemistry Letters 3, 1142–1146. [Google Scholar]
- (25).Heiss DR, Zehnder DW, Jett DA, Platoff GE, Yeung DT, and Brewer BN (2016) Synthesis and Storage Stability of Diisopropylfluorophosphate. J. Chem 2016, 1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Wang Y, Schopfer LM, Duysen EG, Nachon F, Masson P, and Lockridge O (2004) Screening assays for cholinesterases resistant to inhibition by organophosphorus toxicants. Anal. Biochem 329, 131–138. [DOI] [PubMed] [Google Scholar]
- (27).Nachon F, Asojo OA, Borgstahl GEO, Masson P, and Lockridge O (2005) Role of water in aging of human butyrylcholinesterase inhibited by echothiophate: the crystal structure suggests two alternative mechanisms of aging. Biochemistry 44, 1154–1162. [DOI] [PubMed] [Google Scholar]
- (28).Case DA, Cheatham TE, Darden T, Gohlke H, Luo R, Merz KM, Onufriev A, Simmerling C, Wang B, and Woods RJ (2005) The Amber biomolecular simulation programs. J Comput Chem 26, 1668–1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Maier JA, Martinez C, Kasavajhala K, Wickstrom L, Hauser KE, and Simmerling C (2015) ff14SB: Improving the Accuracy of Protein Side Chain and Backbone Parameters from ff99SB. J. Chem. Theory Comput 11, 3696–3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Salomon-Ferrer R, Götz AW, Poole D, Le Grand S, and Walker RC (2013) Routine Microsecond Molecular Dynamics Simulations with AMBER on GPUs. 2. Explicit Solvent Particle Mesh Ewald. J. Chem. Theory Comput 9, 3878–3888. [DOI] [PubMed] [Google Scholar]
- (31).Narayanan C, Bernard DN, Bafna K, Gagné D, Chennubhotla CS, Doucet N, and Agarwal PK (2018) Conservation of Dynamics Associated with Biological Function in an Enzyme Superfamily. Structure 26, 426–436.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Nachon F, Carletti E, Ronco C, Trovaslet M, Nicolet Y, Jean L, and Renard P-Y (2013) Crystal structures of human cholinesterases in complex with huprine W and tacrine: elements of specificity for anti-Alzheimer’s drugs targeting acetyl- and butyryl-cholinesterase. Biochem. J 453, 393–399. [DOI] [PubMed] [Google Scholar]
- (33).Raves ML, Harel M, Pang YP, Silman I, Kozikowski AP, and Sussman JL (1997) Structure of acetylcholinesterase complexed with the nootropic alkaloid, (−)-huperzine A. Nat. Struct. Biol 4, 57–63. [DOI] [PubMed] [Google Scholar]
- (34).Agarwal PK (2004) Cis/trans isomerization in HIV-1 capsid protein catalyzed by cyclophilin A: insights from computational and theoretical studies. Proteins 56, 449–463. [DOI] [PubMed] [Google Scholar]
- (35).Beck DAC, and Daggett V (2004) Methods for molecular dynamics simulations of protein folding/unfolding in solution. Methods 34, 112–120. [DOI] [PubMed] [Google Scholar]
- (36).Yao Y, Liu J, and Zhan C-G (2012) Why does the G117H mutation considerably improve the activity of human butyrylcholinesterase against sarin? Insights from quantum mechanical/molecular mechanical free energy calculations. Biochemistry 51, 8980–8992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Cheng Y, Cheng X, Radic Z, and McCammon JA (2007) Acetylcholinesterase: mechanisms of covalent inhibition of wild-type and H447I mutant determined by computational analyses. J. Am. Chem. Soc 129, 6562–6570. [DOI] [PubMed] [Google Scholar]
- (38).Pan Y, Gao D, Yang W, Cho H, Yang G, Tai H-H, and Zhan C-G (2005) Computational redesign of human butyrylcholinesterase for anticocaine medication. Proc Natl Acad Sci USA 102, 16656–16661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Zheng F, Yang W, Ko M-C, Liu J, Cho H, Gao D, Tong M, Tai H-H, Woods JH, and Zhan C-G (2008) Most Efficient Cocaine Hydrolase Designed by Virtual Screening of Transition States. J. Am. Chem. Soc 130, 12148–12155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Ramanathan A, and Agarwal PK (2009) Computational identification of slow conformational fluctuations in proteins. J Phys Chem B 113, 16669–16680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Shukla S, Bafna K, Gullett C, Myles DAA, Agarwal PK, and Cuneo MJ (2018) Differential Substrate Recognition by Maltose Binding Proteins Influenced by Structure and Dynamics. Biochemistry 57, 5864–5876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Ellman GL, Courtney KD, Andres V, and Featherstone RM (1961) A New and Rapid Colorimetric Determination of Acetylcholinesterase Activity. Biochem. Pharmacol 7, 88–95. [DOI] [PubMed] [Google Scholar]
- (43).Hovanec JW, Broomfield CA, Steinberg GM, Lanks KW, and Lieske CN (1977) Spontaneous reactivation of acetylcholinesterase following organophosphate inhibition. I. An analysis of anomalous reactivation kinetics. Biochim. Biophys. Acta 483, 312–319. [DOI] [PubMed] [Google Scholar]
- (44).Masson P, Fortier PL, Albaret C, Froment MT, Bartels CF, and Lockridge O (1997) Aging of di-isopropyl-phosphorylated human butyrylcholinesterase. Biochem. J 327 (Pt 2), 601–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


