Abstract
We have demonstrated in vitro transcription (IVT) of cDNA sequences from purified Jurkat T-cell mRNA immobilized on microfluidic packed beds down to single-cell quantities. The microfluidically amplified antisense-RNA (aRNA) was nearly identical in length and quantity compared with benchtop reactions using the same starting sample quantities. Microarrays were used to characterize the number and population of genes in each sample, allowing comparison of the microfluidic and benchtop processes. For both benchtop and microfluidic assays, we measured the expression of approximately 4000 to 9000 genes for sample amounts ranging from 20 pg to 10 ng (2 to 1000 cell equivalents), corresponding to 41 to 93% of the absolute number of genes detected from a 100 ng total RNA control sample. Concordance of genes detected between methods (benchtop vs. microfluidic) and repeats (microfluidic vs. microfluidic) typically exceeded 90%. Validation of microarray by Real-time PCR of a panel of five genes suggests transcription of genes present is approximately six times more efficient with the microfluidic IVT compared with benchtop processing. Microfluidic IVT introduces no bias to the gene expression profile of the sample and provides more efficient transcription of mRNA sequences present at the single-cell level.
Introduction
Many biological samples are limited in quantity and cannot be reacquired, making it critical that the analyses performed on such precious samples generate as much data as possible. Thus, technologies and assays are needed that allow for manipulation of samples down to single-cell concentrations.
Global RNA expression analysis can be performed using the microarray gene expression platform. Microarrays allow simultaneous measurements of more than 24 000 genes, making the platform useful for comparison between the genomes of control versus diseased samples. One problem is that microarrays require microgram quantities of RNA be applied to the platform. Typical single-cell mRNA amounts range from 0.1 to 0.5 pg, requiring 106 to 107 fold amplification of the original material before adequate concentrations are generated for application to most microarray platforms.1
Several mRNA amplification methods exist,2-4 many of which have been applied to single-cell amplification on the benchtop with varying levels of success.5-7 Some methods have also been commercialized, and are available as reagent kits.‡. Each has advantages, such as high amplification efficiencies or preservation of gene expression profiles; along with shortcomings, such as requiring more sample RNA (10 to 100 ng) than can be recovered using small needle biopsy or laser capture microdissection, several processing steps, and/or careful control of reaction times. Common to all methods, mRNA is reverse transcribed (RT) to cDNA, producing a mixture of DNA fragments ranging from 200 to 8000 bases reflecting the different genes transcribed within a cell. RT is challenging because most reverse transcriptase enzymes work poorly on dilute mRNA due to low processivity and transcription rates,8 making it difficult to generate an accurate cDNA representation of the mRNA in the cell(s). Volume reduction through microfluidics has shown to be effective at increasing the local recovery and concentration of mRNA allowing rapid and accurate RT down to single-cell levels.9-14
Linear amplification using T7 RNA polymerase (in vitro transcription, or IVT) is the most commonly used RNA amplification method for microarrays because it can be used to continuously and isothermally transcribe double-stranded cDNA (ds cDNA) into antisense-RNA (aRNA) without significant bias to the gene expression levels as all fragments are amplified at an equal rate.2 Though IVT is relatively slow (≈ 14 h per reaction) compared with PCR, gives relatively low yields when used for benchtop single-cell analysis, and limited to approximately 103 to 104 fold amplification per reaction, it is currently the method of choice for global expression studies because it reliably delivers high-quality amplified aRNA for use in downstream applications.
In this work we examined the IVT reaction in a microfluidic system apart from the other steps of the linear amplification (Eberwine) process. Though it is possible to generate cDNA sequences using reverse transcription on-chip, synthesis of a relatively large amount of cDNA on the benchtop eliminated sample-to-sample cDNA synthesis variations. Any differences in aRNA profiles were therefore linked specifically with the IVT reactions performed, which allowed us to draw conclusions about the benefits of miniaturizing this step.
We opted to study IVT by making ds cDNA on beads on the benchtop, then used aliquots of those beads on the device. We generated a stock of ds cDNA on beads, packed aliquots of those beads into columns in a microfluidic device, and used those cDNA templates to generate aRNA for subsequent analyses. The cDNA microspheres were packed against a sieve valve12 to form bead columns approximately 0.4 to 1 nL in volume, which could withstand a continuous flow of reagents and enzymes. The advantages of working with cDNA sequences on beads packed into a nL-scale column include (1) a simple flow-through configuration eliminating further bead manipulation, reducing transfer losses to/from the channel, and facilitating the integration towards multistep sample preparation; and (2) allowing multiple amplifications to be rerun on the same template. We performed benchtop IVT reactions in parallel with the microfluidic experiments using approximately equal amounts of cDNA-functionalized beads in each. We show using off-chip Real-time PCR that the efficiency of first round transcription from cDNA to aRNA using the microfluidic device was about six times higher than benchtop methods. Our results also show approximately 4000 to 9000 genes detected using microarray quantification for both microfluidic- and benchtop-processed samples. This represents between approximately 41 to 93%, respectively, of the absolute total number of genes detected using a second round benchtop amplification from 100 ng total Jurkat T-cell RNA as a control. Concordance of gene expression between microfluidic and benchtop amplification from similar sample sizes were typically about 90%. Thus, it is feasible to perform microscale IVT amplifications on ds cDNA reproducibly while maintaining the expression profile of the sample compared with the benchtop method. Furthermore, this method has the potential for facile integration with other existing mRNA capture and reverse transcription techniques, enabling potentially higher-resolution and broader gene expression profiling using low concentration RNA samples than existing state-of-the-art single-cell analysis techniques.
Materials and methods§
Device
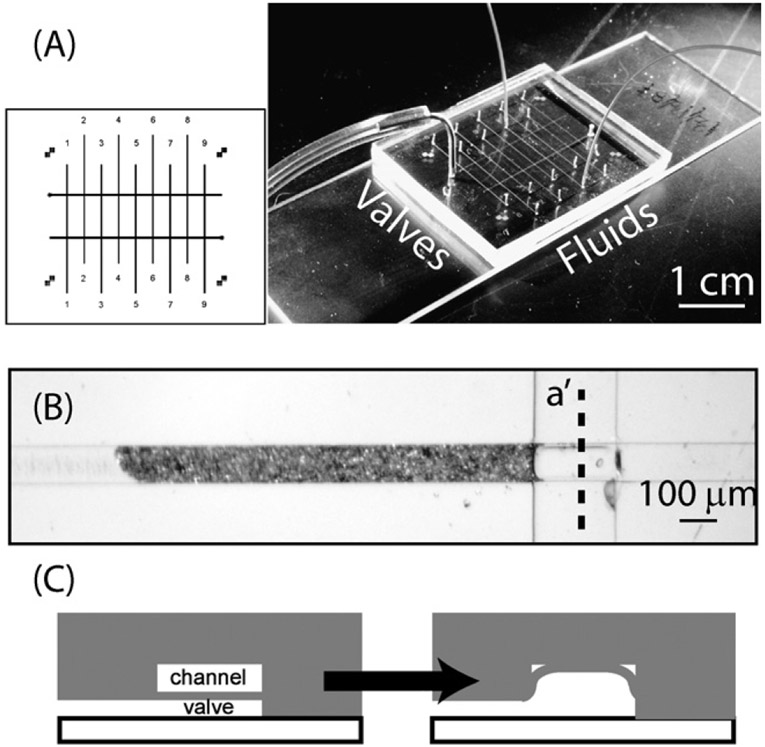
The device was made using polydimethylsiloxane (PDMS) multilayer soft-lithography techniques described elsewhere.15 Here, we employed “push up” style valves across a rectangular cross-section to create a sieve. These allow fluid to flow through but retain particles smaller than the gap left by a partially-sealed channel.12 Flow channels measuring 100 μm wide × 10 μm tall × 15 μm long, with two valve channels measuring 200 pm wide x 10 mm high x 20 mm long (Fig. 1). The thin membrane (active valving) layer was spin-coated at 500 rpm for 6 s, ramped up to 3500 rpm for 10 s, then held at that speed for 60 s. This produced films that were between 15 and 18 μm thick. We used nine-channel devices in the experiments, which allowed for parallelization and increased the number of experiments that could be performed on a single chip. Fluid flow holes were punched using a TiN coated round punch (CR0180115N26R4, Technical Innovations). Valve access holes were punched using a 0.75 mm Unicore punch (Harris). These yielded holes that sealed by PDMS compression to 360 μm and 790 μm tubing, respectively.
Fig. 1.
Microfluidic device description. (A) The device measures 25 mm long × 35 mm wide × 3 mm high and is arranged with parallel flow channels (vertical lines) controlled by sieve valves (horizontal lines). The photo shows a packaged device with flow and pneumatic lines attached. (B) Typical columns of 6 μm beads (horizontal) pack against the valve (vertical) and fill about 1 nL. (C) Viewed through cross-section a′ (see B), a closed sieve valve leaves a small gap that traps beads but allows liquid to flow through.
After multilayer bonding, the device was sealed against a 25 mm × 75 mm × 1 mm glass slide using a homebuilt microwave oxygen plasma system.16,17 The device and glass slide were first cleaned with adhesive tape (Scotch brand, 3M) to remove dust. Non-bonding surfaces were left protected with tape, and the device and slide were placed into a glass vacuum desiccator (140 mm, Wheaton USA) with aluminium foil covering the bottom (pressure reduced to approximately 1 Torr). The desiccator was placed into a microwave oven (Panasonic NN-S949BA) and microwaved for 6 s at 360 W (30% power). The glass desiccator was removed from the microwave and the vacuum was slowly released. The device was then placed on the glass slide to bond, using gentle pressure to contact all areas without blocking the channels. The finished device was then placed in an oven at 80 °C for at least 2 h to further enhance the PDMS–glass bond.
Pneumatic valves (HA010E1-6VDC, Humphrey Valves) were used with a four-channel manifold at 240 kPa (35 psi). Nylon single-barbed tube fittings (Part No. 5463K125, McMaster-Carr) were used to interface to Microbore Tygon tubing (Autoanalysis Tubing 1 mm (0.025′) ID, Cole Parmer). Stainless steel tubing (0.79 mm OD, Upchurch) was inserted into the end of the Tygon tubing, with the other end of the stainless steel tubing inserted into the device to interface the valve layer.
A thermoelectric heater/cooler stage was made to regulate the temperature between 10 and 65 ° C. The stage consisted of a bottom aluminium layer (75 mm wide × 125 mm long × 25 mm tall) with two holes drilled through the side and tapped for hose barb fittings for 6.35 mm tubing (1/4′). Tygon tubing (6.35 mm ID) was attached to the hose barbs and water was pumped through the aluminium layer using a PT submersible fountain pump (PT-08MIX). The top layer was a piece of copper (75 mm wide × 125 mm long × 3 mm tall) with screw holes tapped at the corners. A nine-pin D-sub connector, used to power the thermoelectric element (TE-127-1.4-1.15, TE Tech) and incorporate a thermocouple, is attached to the top plate with a custom-machined piece of stainless steel. The temperature was controlled using a TE Tech TC-24/25 RS-232 and PS-12-8.4 fixed voltage power supply and custom-written Labview™ software.
Fluids were delivered into the device using a two-syringe infusion/withdraw pump with RS-232 communication (Pump 11, Harvard Apparatus). The pump was controlled using either the front panel or a custom-written Labview code that interfaces through the RS-232 port. This allowed the pump to be programmed for long runs with pulse flow (typically 16 h at 10.4 nL min−1 average flow, scheduled as 10 s flow at 104 nL min−1 and 90 s stopped flow). The syringes were 50 μL GasTight with 26s blunt end needles (Hamilton). Tubing was 360 μm OD × 100 μm ID PEEK (Upchurch), cut to approximately 20 cm lengths. Tubing–syringe interconnects were formed using PDMS cast around 30 gauge copper wire (Arcor) cured at 80 °C overnight. The wire was removed and the pieces were cut into roughly 10 mm sections. Tubing and syringes were inserted without lubricants and sealed to pressures above 350 kPa. The other end of the tubing was inserted directly into the PDMS device, and also sealed to pressures above 350 kPa. Outlet tubing fed to 1.5 mL centrifuge tubes, where samples were collected on ice in tubes covered with parafilm (Pechiney Plastic Packaging Co.).
Device channels, tubing, syringes, and work surfaces were cleaned using RNase Away (Invitrogen) to deactivate any RNase, and then rinsed with Bind and Wash buffer [BW; 0.1 mol L−1 Tris-HCl (Quality Biological, Gaithersburg MD), 15 mmol L−1 NaCl (Quality Biological), a mass fraction of 0.02% Tween 20 (Sigma), and DEPC-treated water (Quality Biological)]. It was critical to avoid RNase contamination during the experiments in order to prevent degradation of the RNA. All other reagents and equipment purchased for use in this study were designated as either clean, molecular biology-grade or RNase-free.
Preparation of cDNA sequences
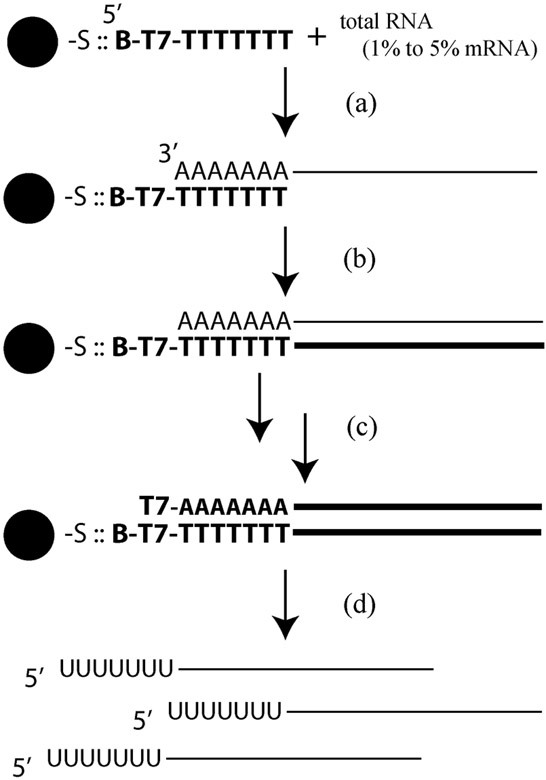
The experimental workflow is shown in Fig. 2. All experiments up to generation of the ds cDNA were performed as benchtop assays [Fig. 2 steps (a) to (c)]. First, 5 μL of streptavidin-coated 6 μm polystyrene microspheres (Polysciences) were washed three times in 100 μL BW buffer and centrifuged to collect (6000 rpm, 2 min; Eppendorf 5415D) following the manufactures recommendations, then suspended in 5 μL BW. One μL (20 ng) of diluted custom primers [5′/52-Bio/GGCCAGTGAATTGTAATACGACTCACTATAGGGAGGCGG(T24)VN-3′; Integrated DNA Technologies (IDT)] were added to the beads and allowed to bind at room temperature for 30 min. The beads were again washed three times in 100 μL BW buffer to remove unincorporated primers. Next, 1 μL of Jurkat total RNA (Stratagene, Cat# 540107-41; used at 1 mg mL−1, 0.1 mg mL−1, and 0.01 mg mL−1, for 1 μg, 0.1 μg, and 0.01 μg total RNA, respectively) was added and allowed to hybridize for 60 min at room temperature with occasional agitation every 15 min. Under these conditions, mRNA (representing 1 to 5% of total RNA and most with poly(A) tails) can then be isolated from total RNA. Zero μg μL−1 control RNA preparations were processed under the same conditions.
Fig. 2.
Description of Eberwine protocol used for generating first and second strand cDNA, and aRNA. Functionalized microbeads are mixed with total RNA samples to hybridize the poly(A) tail of the mRNA to an oligo(dT) strand (a). The RNA is then reverse transcribed (b), followed by RNA digestion and second strand synthesis (i.e., ds cDNA) (c). The IVT reaction is then performed on the ds cDNA to generate amplified aRNA (d), which is the reaction under investigation.
The reverse transcription mix (MessageAmp II, Cat# 1751, Ambion/Applied Biosystems) was added directly to this mixture along with DEPC-treated water to a total volume of 20 μL and incubated at 42 °C for 2 h (flicking the tube every 20 min). This generated the first strand of cDNA, which was attached to the bead.
Next, the second strand reaction mix (MessageAmp II) containing RNase H and DNA polymerase was added to the RT-bead mixture at 16 °C following the manufacturer’s instructions, with the reaction time reduced from 2 to 1 h (flicking the tube every 20 min). After the reaction was completed, the beads were washed three times with BW buffer to remove the reagents, leaving only the ds cDNA immobilized on the beads and resuspended to approximately 20 μL.
Packing the columns onto the microdevice
Columns of 6 μm latex beads with cDNA attached were formed by syringe pumping approximately 0.5 μL of cDNA-function-alized bead slurry (from a 1 : 3 dilution of 5 μL stock bead slurry in BW buffer) into the device with the sieve valve closed. The columns ranged from 0.3 to 1 nL in volume, and were typically 0.5 nL. The total amount of cDNA on each column was determined by dilution-calculation and estimated based on the length of the column, with 1% of the total bead library equaling about 1 nL of bead column. Each bead column was rinsed with approximately 2 μL BW buffer. For benchtop comparison, an equivalent amount of cDNA beads were added to a 1.5 mL centrifuge tube.
IVT reaction
The IVT reaction was performed both on-chip and on benchtop using MessageAmp II kit reagents, modified with a mass fraction of 0.02% Tween 20 to help wet the beads and PDMS channels. Typically, 10 μL of reaction solution (1 μL each of 10 × IVT reaction buffer, CTP, GTP, ATP, UTP, 0.2% Tween 20 in DEPC-treated water, and T7 RNA polymerase, plus 3 μL of DEPC-treated water) was used in both benchtop and on-chip experiments. RNase-free 1.5 mL centrifuge tubes were used for benchtop reactions. On-chip, the device and outlet tubing was pre-filled with BW buffer, and the syringes and inlet tubing were primed with the IVT reaction mix. A custom-built temperature-controlled plate was used to thermostat the device at 37 °C. The reaction product was collected on ice in 1.5 mL centrifuge tubes covered with parafilm. Typical reactions were 16 h (overnight), and the flow rate across the microspheres adjusted to yield approximately 9 μL of aRNA solution in the centrifuge collection tube. The aRNA product was purified using the MessageAmp kit as suggested by the manufacturer. Product was either stored at −80 °C for up to 1 week, or processed in a second round of benchtop amplification following the manufacturer’s recommendation.
Second round IVT amplification was necessary for samples analyzed on the Illumina microarray platform. The aRNA generated from either the microfluidic device or benchtop was used for second round amplification up to the IVT reaction, as suggested for the MessageAmp II aRNA Amplification protocol. The IVT portion of the reaction was performed using reagents from the MessageAmp-Biotin Enhanced kit (Ambion, Cat# AM1791) as suggested by the manufacturer. Biotin nucleotide incorporation was necessary for later gene expression signal detection on the microarray. Biotin-labeled aRNA was purified and RNA profiles examined using the BioAnalyzer’s Nano6000 Assay (Agilent). RNA concentrations were determined using the Nanodrop (Nanodrop) and RiboGreen (Invitrogen).
Microarray assays
The Human RefSeq_8 microarray platform (Illumina) was used for gene expression analyses. The Human RefSeq_8 microarrays contain eight arrays per slide, each with probe-sets corresponding to over 24 000 well-characterized targets, allowing for hybridization of eight different samples. Except for the zero aRNA control sample, 850 ng of biotin-labeled aRNA was hybridized to an array as recommended by Illumina. Expression microarrays were washed, stained and scanned to determine the target signal intensities, as recommended in the Illumina expression manual.
Data analysis was performed using the Gene Expression Module of the BeadStudio™ Data Analysis software (Illumina), with 95% confidence for signal intensity above baseline.
cDNA generation for Real-time Q-PCR validation
The aRNA generated on either the microfluidic device or benchtop was purified, and first strand cDNA generated according the second round procedure of the MessageAmp II Kit. First strand cDNA product for a 1 μg Jurkat T-cell total RNA control sample was processed in the same manner. Samples were stored at −20 ° C until ready for use. All primers except for beta-actin (ACTB) were synthesized by IDT. Beta-actin primers were a generous gift from BioTrove Inc. (Woburn, MA) and Dr. Heidi Erickson. The primer sequences are listed in Table 1.
Table 1.
Gene panel primer pairs. The primer pairs used for each gene are listed, with the forward listed above the reverse
| Gene symbol | Primer pair sequences |
|---|---|
| CHEK2 | 5′-ATCCAAAGGCACGTTTTACG-3′ 5′-ACAACACAGCAGCACACACA-3′ |
| CEBPZ | 5′-GACTTTGCTGGCTCATTTCA-3′ 5′-CGTTCAGCCTCCCATCTAAG-3′ |
| ACTB | 5′-AACCGCGAGAAGATGACCC-3′ 5′-ATCACGATGCCAGTGGTACG-3′ |
| NUP54 | 5′-TGAACTCATCTGTGGGAGGA-3′ 5′-CAGACCACAGTCATGGTTGC-3′ |
| PHB | 5′-GGCTGAGCAACAGAAAAAGG-3′ 5′-GCTGGCAGGTAGGTGATGTT-3′ |
Prior to use, the cDNA samples and primer sequences were mixed as instructed with the Sybr-green Q-PCR master mix (PE Applied Biosystems). Real-time quantitative PCR was performed using the default cycling conditions of the Applied Biosystems 7900 instrument. Relative transcript comparison levels were determined based on the delta comparative threshold (DCt) as suggested by PE Applied Biosystems.
Results and discussion
Gene expression profiles
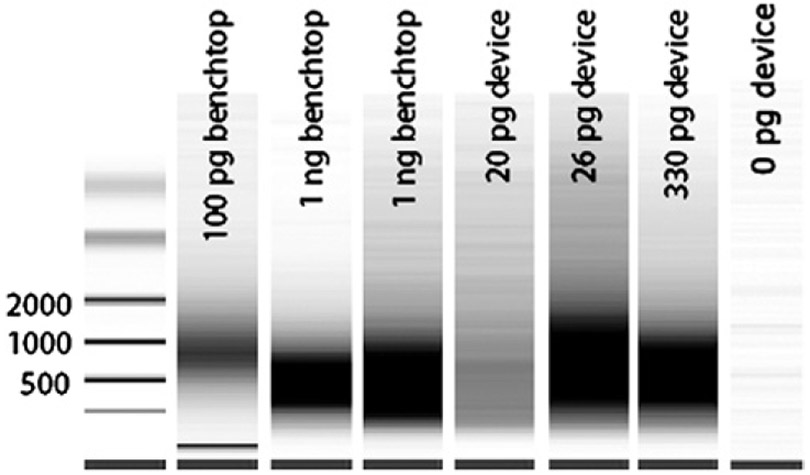
The first challenge of microfluidic IVT was to ensure that accurate, full length reproductions of the transcriptome were generated. Different species of mRNA have different lengths distributed between approximately 200 to 8000 bases long and maximal concentration at approximately 1000 bases. Pseudogels (electropherograms) of product aRNA typically produce a “smear” distribution, with the greatest intensity taken to be the average transcript length. If multiple amplifications are performed to increase the amount of aRNA, the average transcript length is reduced for each subsequent round due to the random priming of the aRNA for reverse transcription. Both benchtop and device methods gave smears with average transcript lengths of approximately 500 bases after two rounds of amplification (Fig. 3). This indicates that the aRNA is amplifying full length for each round and that the RNA polymerase has achieved equivalent processivity for both methods. It is also possible to roughly quantify the amount of aRNA produced from the amplification reaction(s) by analyzing the electropherograms and by Nanodrop quantification. In all cases, we achieved microgram quantities of material after two rounds of amplification (Table 2). For the lowest concentration samples containing 20 pg total RNA (0.2 to 1 pg of mRNA), this represents a 106 to 107 fold amplification for the two-round process, or 1000 to 3000 fold per round. The fact that both microfluidic and benchtop samples amplified to approximately the same amount suggests that the RNA polymerization reaction may be the limiting step in the process and improvements in mass transfer in the microfluidic device do not drive the reaction faster. One possible explanation for this is that reaction kits employ high concentrations of nucleotides and enzyme to overcome the quiescent conditions in microcentrifuge tubes where convective mass transport is small. Another explanation could be that substantial amount of nonspecific “junk” RNA is produced with the same length as the template. Molecular crowding does not appear to be an issue on the bead surface because high concentration samples gave as much or more amplified material as low concentration samples with the total amount of beads kept nearly constant. To assess the true levels of genetic material, both microarrays and Real-time PCR must be performed.
Fig. 3.
Pseudogels demonstrating the aRNA profiles from benchtop- and microfluidic-processed samples. Samples were amplified a second round to increase the aRNA concentration and incorporate biotinylated nucleotides for signal detection on microarrays. The peak intensity of the aRNA range from 500–1000 bases for all samples which indicates optimal RT and full length IVT.
Table 2.
Amplification of mRNA. The approximate concentrations of samples after two rounds of amplification show approximately 106–107 fold amplification, assuming 2% of each total RNA sample is mRNA. Sample sizes are listed as a range of the total RNA of the sample input, grouped by the same bead set and method, with the number of samples given by n. The average amount of aRNA after two rounds of amplification was the same for both on-chip and benchtop methods
| Sample size/pg |
Method | Average/μg | Stdev/μg | n | Amplification factor |
|---|---|---|---|---|---|
| 20–60 | Chip | 4.3 | 1.9 | 4 | 5 × 106 |
| 50–100 | Benchtop | 3.6 | 1.5 | 3 | 2 × 106 |
| 300–500 | Chip | 13 | 9.8 | 3 | 2 × 106 |
| 500–1000 | Benchtop | 5.7 | 4.5 | 4 | 4 × 105 |
Microarray characterization of microfluidic IVT compared with benchtop IVT
Microarrays provide a screening platform on which more than 104 potential genes can be analyzed simultaneously for potential disease markers. Though the gene expression values are less quantitative than Real-time PCR methods, more information can be obtained about the global expression profiles relative to control samples. Microarrays are primarily a discovery tool and are most useful when comparing moderately- to highly-expressed genes. Low-copy-number genes are significantly more difficult to detect with any certainty except in high concentration samples containing μg quantities of RNA, and are best left to PCR analyses.
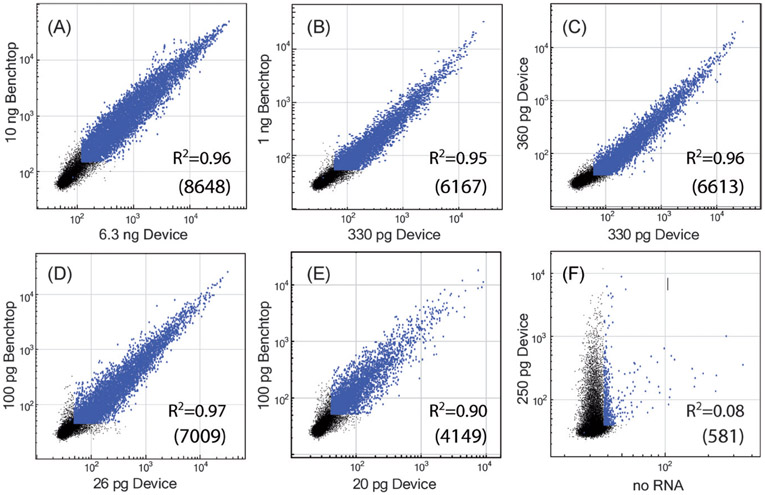
We successfully detected 4000 to 9000 genes using twice-amplified samples, when input total RNA concentrations ranged from 20 pg to 10 ng of total RNA (Fig. 4). The expression levels of the samples showed that the microfluidic method produced no statistically significant amplification bias relative to the benchtop amplification using similar starting amounts of material. Concordances between the number and level of genes detected using the benchtop and microfluidic amplification methods were typically in excess of 90%. To test reproducibility from run-to-run in the microfluidic device, we compared amplified samples prepared on consecutive days. The numbers of genes detected using approximately 330 and 360 pg samples were nearly equivalent (see Fig. 4, 7348 vs. 7391), and concordance between the two samples was 6613 genes. For lower sample amounts (26 vs. 52 pg), the numbers of genes detected were significantly more variable (5908 vs. 8513), with the number of concordant genes at 5512, corresponding to approximately 57% of the control sample. The variability of gene expression observed for the low concentrations is likely associated with the technical difficulties associated with handling such limited amounts, where loss of even small amounts can lead to substantial variability.
Fig. 4.
Microarray data show excellent agreement between the expression profiles obtained using benchtop (two rounds) and microfluidic + benchtop (two rounds) amplification. The number of concordant genes between the two samples are listed on each chart in parentheses. (A) For the highest concentration samples processed, 94% of detected genes were concordant, amounting to 8648. Reducing the starting sample amount reduced the ultimate number of genes detected, though concordance was still high (B). Device amplification was also reproducible, as nearly equivalent expression profiles were obtained from different runs on the same cDNA sequences (C). At low starting amounts of material (two to three cell equivalents; D and E), the number of genes detected was still very good, with detected genes concordant with benchtop amplification ranging from 4149 to 7009, or 43 to 72% of the control sample. When no sample was present (F), concordance with an aRNA-positive sample was very poor. For reference, one cell contains approximately 10 pg of RNA. Points in blue have p < 0.05, and black points fall below the confidence limit (intensities less than ≈200).
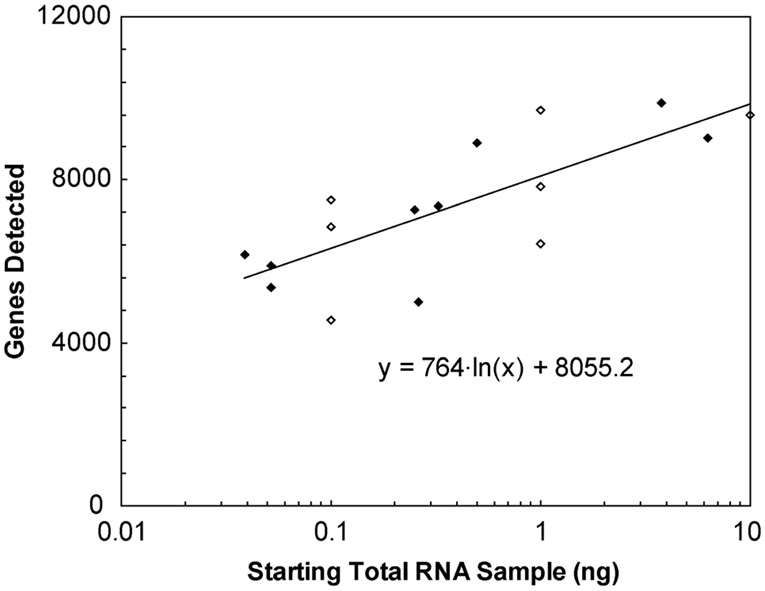
We attempted to correlate gene call rates with the starting sample size. The gene expression was plotted vs. the starting sample size, combining microfluidic and benchtop amplified sample data sets. We observed a semi-log correlation (Fig. 5). For every decade reduction in starting template, there appears to be an average drop in detection of about 1700 genes, which extrapolates down to about 4700 genes for 10 pg (single-cell) samples. It is possible that benchtop RT processing is inefficient and can lead to sampling errors, limiting the amount of moderate- to low-expression genes in the aliquots of cDNA. Putting more aRNA onto each microarray may improve detection slightly, but background noise (“junk” RNA) would increase and likely negate any gains. Another way to increase sensitivity could be to incorporate the capture and RT with the IVT on a single microfluidic device. Others have described mRNA capture and RT being approximately four times more efficient on a microfluidic device than using benchtop protocols.11 Under those conditions, incorporating these functions onto the device and performing single-cell assays would yield an increase in the number of genes detected by approximately 2400, assuming a logarithmic sample-detection relationship. Such improvements could lead to detection of additional genes with biological and biomedical benefits.
Fig. 5.
Correlation between gene call rates and sample size. Microarray gene call rates (after two rounds of amplification) were plotted vs. the starting sample size from both microfluidic (♦) and benchtop amplification (◊) experiments. Detection appears to increase logarithmically with increasing starting sample size. Though the raw data are noisy, regression analysis of the effect of starting sample size on gene detection shows a positive correlation with 95% confidence.
Real-time PCR analysis of microarray results
To assess the quality of the amplified aRNA, single round IVT samples were subjected first to traditional PCR to test for the presence/absence of genes previously determined with high or moderate expression levels (i.e., GAPDH and ITGB1, respectively). These tested positive, giving confidence that the amplified material was genomic (data not shown). For validation and more quantitative assessment of transcripts detected using the microarrays, we then performed Real-time PCR on once-amplified samples using the benchtop and microfluidic protocols from approximately 50 pg of total RNA (approximately five cell equivalents). The panel of high- and moderate expression genes included CHEK2, CEBPZ, ACTB, NUP54, and PHB (Table 3).
Table 3.
Validation of microarray using Real-time PCR. Real-time PCR was performed measuring critical threshold (Ct) values for five genes of varying expression levels on a total RNA control, benchtop amplified, and device amplified samples. In all cases, the genes tested present (except for no template), with significantly lower Ct for the device amplified samples. This indicates higher amplification on-chip in this concentration range compared with bead-based benchtop amplification.a
| Sample | CHEK2 | CEPBZ | ACTB | NUP54 | PHB |
|---|---|---|---|---|---|
| 1 μg control | 24.5 ± 0.1 | 25.1 ± 0.4 | 16.3 ± 0.2 | 27.9 ± 0.0 | 20.5 ± 0.2 |
| Benchtop | 30.9 ± 0.8 | 31.5 ± 0.5 | 26.2 ± 0.9 | 37.0 ± 0.2 | 26.2 ± 1.2 |
| Device | 28.8 ± 3.6 | 28.0 ± 0.4 | 24.0 ± 0.3 | 35.3 ± 1.0 | 23.2 ± 0.9 |
| No template | -ND- | -ND- | -ND- | -ND- | 35.2 ± 1.2 |
(All Ct values = avg ± stdev based on four samples, except the No template which is two samples; ND = not determined—below level of detection).
Using the ΔCt method, we compared Ct values from a 1 μg Jurkat total RNA control sample with the Ct values for the benchtop amplification and found an average increase of 7.5 ± 0.8, or about 181 fold more dilute. On-chip Ct values were 5.0 ± 1.2 cycles greater than the 1 μg sample, or 32 times lower. This indicates approximately six fold more aRNA of each of these genes is transcribed on the device. We estimate an amplification factor of 400 to 900 on-chip from only 0.5 to 2 pg of starting mRNA. Furthermore, the relative levels obtained from Real-time PCR compare well with those observed on the microarray. Hence, microfluidic IVT appears to give a more efficient amplification of the present cDNA than the benchtop protocol, especially for samples of 100 pg or less.
Discussion of integrating IVT with capture and RT
Performing a complete amplification process on a chip would require the ability to pass different reagents over the beads at different temperatures and wash the columns between the steps. As already noted, previous devices used to capture and RT mRNA11,12 strongly resemble the device design employed here. One could envision using the same packed column to capture, RT, generate a second cDNA strand, and perform the IVT by simply flowing different reaction mixtures over the beads at different temperatures for each particular enzymatic reaction step.
To achieve sufficient material for microarray characterization from single-cell samples, a second round of amplification would likely still be necessary. However, it is not clear that integrating the second round onto a microfluidic device would provide any additional benefit because the amount of aRNA generated from the first round could be easily amplified using benchtop techniques. Hence, the second round amplification process is not limited by the same factors as the first round, and is therefore not likely to see the same kinds of improvements.
Conclusions
Microfluidic IVT yields aRNA that is an unbiased representation of gene expression in the original sample, with amplification factors around 600 for low concentration of input total RNA. We successfully processed samples with as little as 20 pg of starting sample, amounting to about two cell equivalents, and up to approximately 10 ng.
Sample profiles were analyzed using electrophoresis to characterize average transcript length, and were shown to be high-quality and full length compared with the established benchtop methods. Microarrays were used to explore the global gene expression profiles of Jurkat T-cells with microfluidic IVT and compared with the existing benchtop protocols. We observed no significant difference in gene expression between the two methods. Repeatability of the microfluidic IVT method was also examined and found to be excellent. A basic analysis of microarray gene detection vs. starting sample size was performed to help estimate the impact of improved processing on gene detection.
Finally, Real-time PCR was used to validate the microarray data, quantify gene expression levels from a panel of five genes, and allow estimation of overall amplification yields. We determined that microfluidic IVT produced about six fold more efficient transcription of the present cDNA compared with the benchtop IVT for sample sizes of approximately 50 pg. We anticipate that full integration of the Eberwine process, incorporating capture through the IVT, would increase moderate- and low-copy gene detection on microarrays by approximately 2400 at the single-cell level.
The advantages of this process are the increased efficiency of IVT compared with the equivalent benchtop method; the IVT process is amenable to parallelization and integration with other bead-based RNA processes such as purification and reverse transcription; and little or no sample manipulation is required once the cDNA is synthesized on the beads, thereby reducing sample losses. Though a radical departure from conventional tube-based sample processing, microfluidic amplification methods provide a means to improve characterization of gene expression from low concentration and single-cell samples.
Acknowledgements
This research was supported in part by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research. The authors thank Dr. Yonghong Wang for depositing the microarray data into NCBI Gene Expression Omnibus (GEO) repository.
Footnotes
Commercially available kits, such as the MessageAmp™ aRNA Amplification Kit from Ambion Inc. (Austin, TX), and SMART™ mRNA Amplification kit from Clontech (Mountain View, CA) are now available.
Certain commercial equipment, instruments, or materials are identified in this paper to foster understanding. Such identification does not imply recommendation or endorsement by the National Institute of Standards and Technology, nor does it imply that the materials or equipment identified are necessarily the best available for the purpose.
References
- 1.Livesey F, Brief. Funct. Genomic. Proteomic, 2003, 2, 31–36. [DOI] [PubMed] [Google Scholar]
- 2.van Gelder R, von Zastrow M, Yool A, Dement W, Barchas J and Eberwine J, Proc. Natl. Acad. Sci. U. S. A, 1990, 87, 1663–1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Iscove N, Barbara M, Gu M, Gibson M, Modi C and Winegarden N, Nat. Biotechnol, 2002, 20, 940–943. [DOI] [PubMed] [Google Scholar]
- 4.Petalidis L, Bhattacharyya S, Morris G, Collins V, Freeman T and Lyons P, Nucleic Acids Res, 2003, 31, e142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Viale A, Li J, Tiesman J, Hester S, Massimi A, Griffin C, Grills G, Khitrov G, Lilley K, Knudtson K, Ward B, Kornacker K, Chu C, Auer H and Brooks A, J. Biomol. Techniques, 2007, 18, 150–161. [PMC free article] [PubMed] [Google Scholar]
- 6.Tietjen I, Rihel J, Cao Y, Koentges G, Zakhary L and Dulac C, Neuron, 2003, 38, 161–175. [DOI] [PubMed] [Google Scholar]
- 7.Nygaard V and Hovig E, Nucleic Acids Res, 2006, 34, 996–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coffin J, Hughes S and Varmus H, Retroviruses, Cold Spring Harbor Laboratory Press, Plainview, New York, 1997. [PubMed] [Google Scholar]
- 9.Jiang G and Harrison D, Analyst, 2000, 125, 2176–2179. [DOI] [PubMed] [Google Scholar]
- 10.Bontoux N, Dauphinot L, Vitalis T, Studer V, Chen Y, Rossier J and Potier M-C, Lab Chip, 2008, 8, 443–450. [DOI] [PubMed] [Google Scholar]
- 11.Zhong J, Chen Y, Marcus J, Scherer A, Quake S, Taylor C and Weiner L, Lab Chip, 2008, 8, 68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marcus JS, Anderson WF and Quake SR, Anal. Chem, 2006, 78, 3084–3089. [DOI] [PubMed] [Google Scholar]
- 13.Satterfield B, Stern S, Caplan M, Hukari K and West J, Anal. Chem, 2007, 79, 6230–6235. [DOI] [PubMed] [Google Scholar]
- 14.Hong JW, Studer V, Hang G, Anderson WF and Quake SR, Nat. Biotechnol, 2004, 22, 435–439. [DOI] [PubMed] [Google Scholar]
- 15.Unger M, Chou H, Thorsen T, Scherer A and Quake S, Science, 2001, 288, 113–116. [DOI] [PubMed] [Google Scholar]
- 16.Ginn B and Steinbock O, Langmuir, 2003, 19, 8117–8118. [Google Scholar]
- 17.Hui A, Wang G, Lin B and Chan W-T, Lab Chip, 2005, 5, 1173–1177. [DOI] [PubMed] [Google Scholar]
- 18.Edgar R, Domrachev M and Lash A, Nucleic Acids Res., 2002, 30, 207–210. [DOI] [PMC free article] [PubMed] [Google Scholar]