Summary
Histone variant H2A.Z-containing nucleosomes are incorporated at most eukaryotic promoters. This incorporation is mediated by the conserved SWR1 complex, which replaces histone H2A in canonical nucleosomes with H2A.Z in an ATP-dependent manner. Here, we show that promoter-proximal nucleosomes are highly heterogeneous for H2A.Z in Saccharomyces cerevisiae, with substantial representation of nucleosomes containing one, two, or no H2A.Z molecules. SWR1-catalyzed H2A.Z replacement in vitro occurs in a stepwise and unidirectional fashion, one H2A.Z-H2B dimer at a time, producing heterotypic nucleosomes as intermediates and homotypic H2A.Z nucleosomes as end products. The ATPase activity of SWR1 is specifically stimulated by H2A-containing nucleosomes without ensuing histone H2A eviction. Remarkably, further addition of free H2A.Z-H2B dimer leads to hyperstimulation of ATPase activity, eviction of nucleosomal H2A-H2B and deposition of H2A.Z-H2B. These results suggest that the combination of H2A-containing nucleosome and free H2A.Z-H2B dimer acting as both effector and substrate for SWR1 governs the specificity and outcome of the replacement reaction.
Graphical Abstract
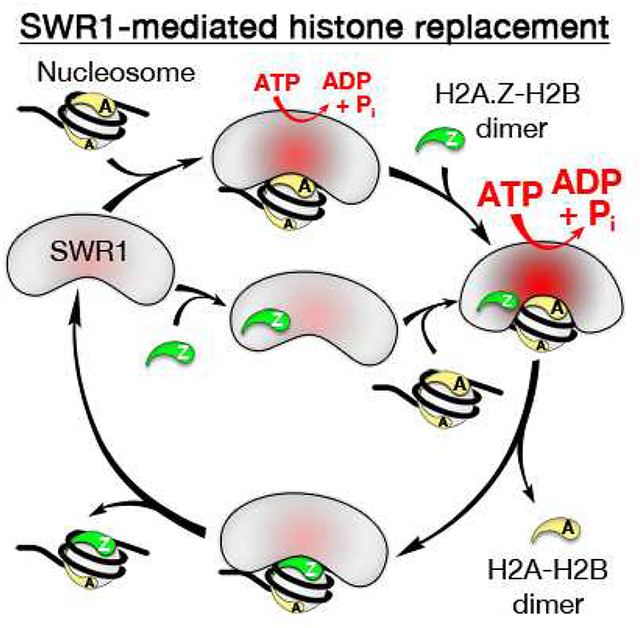
Introduction
The eukaryotic genome is packaged into chromatin within the cell nucleus. The fundamental packaging unit of chromatin is the nucleosome, which consists of an octameric histone core around which 147 base pairs (bp) of DNA are wrapped in ~1.7 left-handed superhelical turns, plus linker DNA of variable length between adjacent nucleosome core particles (Kornberg and Lorch, 1999). The canonical nucleosome, containing two each of the four main histones H2A, H2B, H3 and H4, is representative of the bulk of chromatin in the cell nucleus. However, a minor fraction of the nucleosome population is assembled from nonallelic histone variants, which have an important role in major chromosome activities of the cell, including transcription, DNA replication, and repair (Ausio, 2006).
The widely conserved histone H2A.Z variant shares 60% sequence identity with the canonical H2A histone and plays essential, non-redundant roles in higher eukaryotes (Guillemette and Gaudreau, 2006). In yeasts, H2A.Z is not essential, but cells exhibit slow growth (Carr et al., 1994; Santisteban et al., 2000), chromosome instability (Carr et al., 1994; Krogan et al., 2004), gene silencing defects (Meneghini et al., 2003), and sensitivity to genotoxic and environmental stress (Jackson and Gorovsky, 2000; Kobor et al., 2004; Mizuguchi et al., 2004). Crystallographic studies have shown that H2A.Z-containing nucleosomes are in general structurally similar to canonical nucleosomes but possess distinct internal and surface features (Suto et al., 2000). Biophysical studies also reported differences in nucleosome stability, positioning, and higher order interactions (Zlatanova and Thakar, 2008). Interestingly, it has been recently demonstrated that purified nucleosomes containing both histone H2A.Z and the histone H3.3 variant are the least stable among native nucleosomes to salt induced dissociation (Jin and Felsenfeld, 2007; Zhang et al., 2005).
Genome-wide mapping of nucleosome distribution indicates that the vast majority of budding yeast promoters have a stereotypical chromatin architecture, characterized by two well-positioned nucleosomes (+1 and −1) flanking an 80- to 230-base pair region, which is relatively depleted for histones and is commonly referred to as the ‘nucleosome-free region’ (NFR) (Cairns, 2009; Jiang and Pugh, 2009b; Weiner et al., 2010). With its NFR-proximal edge covering the transcription start site (TSS), the +1 nucleosome acts as a barrier that occludes the TSS and helps position downstream nucleosomes in the coding region (Jiang and Pugh, 2009b). Formaldehyde crosslinking and chromatin immunoprecipitation (ChIP) experiments conducted on the budding yeast Saccharomyces cerevisiae first demonstrated that histone H2A.Z (called Htz1) is enriched at intergenic regions upstream of PHO5 and GAL1 even under repressed conditions (Santisteban et al., 2000). It was subsequently shown in genome-wide studies that H2A.Z is dramatically enriched at the promoter-proximal +1 and −1 nucleosomes (Albert et al., 2007; Raisner et al., 2005), with enrichment diminishing progressively away from the promoter (Albert et al., 2007). The presence of H2A.Z nucleosomes surrounding most yeast promoters in the absence of transcription has led to the proposal that H2A.Z-containing nucleosomes help poise genes for transcription (Jin and Felsenfeld, 2007; Li et al., 2005; Santisteban et al., 2000; Zhang et al., 2005). In metazoans, H2A.Z is localized to nucleosomes proximal to promoters of active genes (Rando and Chang, 2009). More recently, H2A.Z has also been implicated in DNA repair (Morrison and Shen, 2009) and in suppression of spurious non-coding transcription (Zofall et al., 2009). The molecular function of H2A.Z in transcription and DNA repair remains obscure.
Previous studies have shown that the 14-subunit S. cerevisiae SWI/SNF-related SWR1 complex (SWR1) is required for the incorporation of H2A.Z (Kobor et al., 2004; Krogan et al., 2003; Mizuguchi et al., 2004). Human counterparts of SWR1, named SRCAP and p400, have also been identified (Gevry et al., 2007; Ruhl et al., 2006). SWR1 is itself enriched at promoters, coincident with the maxima of H2A.Z distribution (Venters and Pugh, 2009). The recruitment of SWR1 to promoters is attributed in part to the bromodomain-containing Bdf1 subunit of SWR1 and its interaction with acetylated histone H3 and H4 tails (Altaf et al., 2010; Koerber et al., 2009; Raisner et al., 2005)
How SWR1 carries out the ATP-dependent replacement of nucleosomal H2A with H2A.Z is not well understood. Studies from our laboratory have shown that the histone replacement reaction can be sufficiently reconstituted in vitro with purified components (Luk et al., 2007; Mizuguchi et al., 2004). This basic reaction has also been demonstrated with purified components from mammalian and insect cells, and can be enhanced by acetylation of the nucleosomal substrate (Altaf et al., 2010; Kusch et al., 2004; Ruhl et al., 2006). In the replacement reaction, the H2A.Z-H2B dimer is delivered as a unit to SWR1 (Mizuguchi et al., 2004; Ruhl et al., 2006) specifically to its Swc2 subunit. Delivery is assisted by an H2A.Z-specific chaperone Chz1, which is displaced upon H2A.Z-H2B binding (Luk et al., 2007). A second binding site for H2A.Z-H2B was recently localized to the N-terminal domain of the Swr1 ATPase subunit (Wu et al., 2009). The binding of H2A.Z-H2B to SWR1 is independent of ATP (Wu et al., 2005).
Other important steps of the histone replacement reaction involve the ATP-dependent eviction of nucleosomal H2A-H2B and insertion of H2A.Z-H2B. However, the mechanisms by which these steps are carried out are obscure. It is also unclear whether SWR1 replaces one or both histone H2A-H2B dimers in a canonical nucleosome with H2A.Z-H2B, producing heterotypic (AZ) or homotypic (ZZ) H2A.Z-containing nucleosomes. In vitro reconstitution by salt dialysis shows that the two species can be reconstituted from purified histones and DNA (Chakravarthy et al., 2004; Suto et al., 2000). Therefore, it is of particular interest to determine whether promoter-proximal H2A.Z nucleosomes are organized in the AZ or ZZ state because they are indistinguishable by standard ChIP procedures (Albert et al., 2007; Raisner et al., 2005). Mutation of the ATP-binding pocket of the Swr1 ATPase subunit, and studies with nonhydrolyzable ATP analogs documented that ATP hydrolysis is an absolute requirement for the histone replacement reaction (Mizuguchi et al., 2004). How the ATPase activity of the SWR1 complex is transduced to the eviction of H2A-H2B and insertion of H2A.Z-H2B, and whether the ATPase activity is regulated in the course of the reaction are unknown.
In this study, we investigated whether the homotypic and heterotypic states of H2A.Z-containing nucleosomes are present at the promoters of budding yeast in vivo. We found that promoter-proximal nucleosomes are highly heterogeneous in histone variant composition, with substantial representation of nucleosomes containing one, two or no H2A.Z molecules. To further understand this phenomenon, we developed an in vitro assay to distinguish between compositional states, and found that the histone replacement reaction is stepwise and unidirectional, i.e. progressing from AA (canonical) to AZ to ZZ nucleosomes. Further investigation of the underlying mechanism showed that ATP hydrolysis by the SWR1 complex is specifically activated by H2A-containing nucleosome and additionally by H2A.Z-H2B dimer, leading to histone replacement. These results lead to a model in which specific activation of SWR1 by the two in vivo histone substrates drives the stepwise, unidirectional pathway of histone H2A.Z replacement.
Results
Both AZ and ZZ nucleosomes are present in Saccharomyces cerevisiae
To investigate whether budding yeast nucleosomes contain one or two copies of the H2A.Z variant, we performed co-immunoprecipitation (co-IP) analysis with the use of a diploid strain in which one allele of HTZ1, the gene encoding S. cerevisiae H2A.Z, is left untagged and the second HTZ1 allele bears a Flag epitope tag (HTZ1FLAG) to facilitate purification. Cells in asynchronous culture were fixed with formaldehyde to preserve nucleosome integrity, and mononucleosomes were generated by MNase digestion (Figure S1A). We immunoprecipitated Htz1Flag-containing nucleosomes with anti-Flag antibodies and analyzed their composition by reversal of crosslinking and Western blotting. Probing with anti-Htz1 antibodies showed that untagged Htz1 co-purifies with Htz1Flag, indicating the presence of homotypic H2A.Z (ZZ) nucleosomes in yeast cells (Figure 1A). Moreover, reprobing the same blot with anti-H2A antibodies shows co-purification of H2A with Htz1Flag, demonstrating the existence of heterotypic H2A.Z (AZ) nucleosomes as well (Figure 1B). Based on the experimentally determined ratios (Z:ZF = 0.29, Figure 1A; A:ZF = 0.49, Figures 1C and 1D), we calculated the relative distribution of ZZ and AZ nucleosomes to be ~35% and ~65%, respectively (Figure S1B).
Figure 1. Isolation of homotypic ZZ and heterotypic AZ nucleosomes.

(A-B) Histone co-IP analysis of mononucleosomes prepared from fixed diploid HTZ1FLAG/HTZ1 cells (yEL021). A shows the SDS-PAGE and anti-Htz1 (α-Htz1) Western analyses of MNase-treated nuclear extract (Input), anti-Flag (α-Flag) IP flow-through (FT), and α-Flag immunoprecipitates eluted with Flag peptides (Flag eluate). 20, 10, 5, and 2 μL of the Flag eluate were loaded in lane 3, 4, 5, and 6, respectively. Lane 3 was imaged from a separate Western blot. The ratio of untagged Htz1 to Htz1Flag for the Flag eluate is 0.29 ± 0.08 (average and range of two Western analyses). The membrane was stripped and re-probed with anti-H2A (α-H2A) antibodies in B.
(C-D) The Flag eluate of A was quantified by α-Htz1 and α-H2A Western analyses using recombinant Htz1 and H2A standards. The estimated molar ratio of H2A and Htz1Flag in the Flag eluate is 0.49.
(E) Co-IP and Western analyses of the Flag eluate from G1-arrested, asynchronous haploid cells (yJL036). Numbers indicate quantification of the Htz1Flag Western signal relative to H2A.
(F) FACS analysis of asynchronous and G1-arrested cells.
(G-H) Quantification of total H2A and Htz1 polypeptides in the nuclear extract (Input) of G1-arrested cells. Asterisk (*) indicates a cross-reactive band. The two panels in H are imaged from the same Western blot.
In principle, AZ nucleosomes could be generated from AA nucleosomes by stepwise replacement with H2A.Z-H2B dimers. This replacement could occur in a replication-independent manner in all phases of the cell cycle, including the G1 phase (S. Sen, C. Wu unpublished observations). In addition, AZ nucleosomes could arise as a consequence of disruption of ZZ nucleosomes and reassembly with a mixed histone dimer pool during DNA replication in S phase. The latter contribution can be minimized in our analysis by the use of yeast cells arrested in G1 phase by alpha-mating factor (Figure 1F). Under these conditions, a haploid yeast strain carrying Htz1Flag as the sole copy still exhibits substantial co-purification of H2A with Htz1Flag (~75% compared to asynchronous cells) (Figure 1E).
We measured the relative proportion of H2A.Z to H2A bound to chromatin in G1-arrested cells by quantitative Western blotting using purified bacterially expressed Htz1 and H2A as protein standards (Figures 1G and 1H). Htz1 constitutes ~9% of total H2A-like histones in chromatin, comparable to previous results obtained for mammalian cells (4%) (West and Bonner, 1980).
ZZ and AZ nucleosomes are enriched at promoters
The presence of homotypic and heterotypic H2A.Z nucleosomes in G1-arrested cells prompted us to map their genomic locations. Both AZ and ZZ nucleosomes could be enriched at promoters genome-wide, or they could be differentially distributed amongst distinct sets of genes. To distinguish between these possibilities, we used sequential IP to fractionate the heterogeneous nucleosome population into three subpopulations representing ZZ, AZ and AA nucleosomes. We first immunopurified Htz1Flag-containing nucleosomes from haploid yeast cells (expressing Htz1Flag as sole source) with the use of anti-Flag antibodies to isolate ZZ and AZ nucleosomes in the bound fraction, followed by secondary IP of the eluate with anti-H2A antibodies to separate ZZ from AZ nucleosomes (Figure S2A). Western blotting of bound and flow-through fractions confirmed that IP was highly efficient (Figure S2B). In addition, the flow-through fraction from the first anti-Flag IP, which is quantitatively depleted for AZ and ZZ nucleosomes, was subjected to additional IP with anti-H2A to give a bound fraction highly enriched for AA nucleosomes.
We mapped the locations of each distinct nucleosomal population by hybridization of amplified, fluorescently labeled DNA to oligonucleotide tiling microarrays covering two yeast chromosomes (chromosome 3 and 6, plus other selected regions), at 10-bp resolution, for both DNA strands. The results are presented as normalized ratios of nucleosomal to genomic DNA fluorescence (Figures 2A and S3). Consistent with previous studies (Albert et al., 2007; Raisner et al., 2005), we confirmed that Htz1-containing nucleosomes (Z-total) map predominantly to the promoter-proximal +1 and −1 nucleosomes, with enrichment tapering off away from the promoter (Figure 2). Interestingly, we found that the subpopulations of AZ and ZZ nucleosomes are similarly enriched at most promoters (Figure 2A). This is especially evident in the normalized average profiles for 466 nucleosomes in the +1 position (Figure 2B). The relative abundances of AZ and ZZ nucleosomes at the +1 location are highly correlated (R = 0.89), arguing against differential enrichment of AZ or ZZ nucleosomes for a specific subset of promoters (Figure S2C). The average AZ and ZZ nucleosome profiles surrounding the promoter region also show differences. ZZ nucleosome enrichment is more restricted to the −1 and +1 positions, while AZ enrichment is comparatively lower and declines more gradually away from the promoter (Figure 2B).
Figure 2. Genomic distribution of the AA, AZ, and ZZ nucleosomes.

(A) Tiling microarray data of a representative region in chromosome 3 showing the genomic distribution of the Z-total (orange), ZZ (green), AZ (purple), AA (red), and Total (blue) nucleosomes. The data are presented as the normalized ratio of nucleosomal and genomic DNA signal. Grey bars indicate coding regions. (B) Normalized average nucleosome distribution in and around the +1 nucleosome center of 466 genes (Jiang and Pugh, 2009a). Circles illustrate the estimated positions of the −1, +1, +2, +3, and +4 nucleosomes.
Substantial presence of AA nucleosomes at promoters
Previous studies of H2A.Z enrichment at promoters genome-wide did not include canonical (AA) nucleosomes, which are commonly assumed to be depleted at promoters. To test this assumption, we determined the genomic distribution of the purified AA nucleosome subpopulation on tiling microarrays (Figure S2A). As anticipated, the normalized AA nucleosome distribution is similar to that observed for the total nucleosome pool (Total) (Figures 2A, 2B, and S3). However, the abundance of AA nucleosomes at promoters is surprisingly substantial, despite enrichment of the ZZ and AZ variants. This is especially evident at the −1 and +1 nucleosome positions, where H2A.Z is thought to be predominant but in fact exhibits a similar abundance to canonical H2A (Figure 2B). We conclude that steady-state histone variation at promoter-proximal nucleosomes is quite heterogeneous in a population of budding yeast, showing significant levels of both variant and canonical nucleosomes. Clustering analysis of H2A.Z nucleosome distributions for the TATA-containing and TATA-less promoters shows that histone heterogeneity appears to be a common feature of most yeast promoters, irrespective of gene category (Figure S2D).
SWR1 generates nucleosomal AZ intermediate and ZZ end product in vitro
The steady-state level of H2A.Z at promoter-proximal nucleosomes is a product of opposing H2A.Z assembly and disassembly pathways in vivo. Incorporation of H2A.Z in nucleosomes is catalyzed by the SWR1 chromatin remodeling complex, which could convert AA nucleosomes to the ZZ state by replacing both nucleosomal H2A-H2B dimers with Htz1-H2B in a concerted reaction. Alternatively, SWR1 could replace the H2A-H2B dimers in a stepwise manner involving AZ nucleosomes as a reaction intermediate. To distinguish these models, we developed a new histone replacement assay.
In this assay, immobilized arrays of canonical nucleosomes are incubated with SWR1 purified from an htz1Δ strain, Htz1Flag-H2B dimer and ATP as previously described (Figure 3A). The chromatin product is then subjected to MNase digestion to liberate mononucleosomes. Because Htz1 bears a 3xFlag epitope tag, replacement of one nucleosomal H2A-H2B dimer with Htz1Flag-H2B retards the native electrophoretic mobility of the nucleosome, and replacement of two dimers retards the mobility further. Thus, in an ATP-dependent, limited replacement reaction, three nucleosomal species with discrete mobility can be resolved by non-denaturing PAGE (Figures 3A and 3B). We examined the identities of each nucleosome species by Western blotting, and confirmed that the top, middle and bottom gel bands correspond to ZZ nucleosomes with two Flags (ZFZF), AZ nucleosomes with one Flag (AZF), and AA nucleosomes, respectively (Figure S4A).
Figure 3. In vitro assay showing the stepwise assembly of AZ and ZZ nucleosomes.

(A,B) Overview and experiment for the in vitro histone replacement assay. Bead-bound canonical nucleosome arrays (depicted with three nucleosomes for simplicity) were incubated with Htz1Flag-H2B dimer (chaperoned by Chz1, not depicted), SWR1, and ATP for 1 hr (Step 1). After washing, the chromatin was digested with MNase to liberate mononucleosomes (Step 2), which were subsequently analyzed by non-denaturing PAGE (Step 3). AA (top), AZF (middle), and ZFZF (bottom) nucleosomes were detected by SYBR Green staining.
(C) In vitro histone replacement time course. SWR1-mediated histone replacement reactions were stopped at various times by bead-pulldown and washing. Nucleosomal products were analyzed as described in A. Middle panel: densitometric measurement of the indicated gel region. Right panel: peak height versus time.
The detection of AZ nucleosomes in a partial replacement reaction suggests that the heterotypic H2A.Z nucleosome may be a reaction intermediate. To investigate this possibility, we monitored the progression of the SWR1-catalyzed replacement reaction in vitro. Consistent with the hypothesis, we found that the AZ species briefly accumulates upon the addition of ATP, reaching a maximum at 30 minutes, followed by a gradual decrease over time (Figures 3C and S4B). By contrast, the ZZ species continues to accumulate past 30 minutes, reaching a plateau where ZZ nucleosomes represent the bulk of the nucleosome population, while AA nucleosomes are correspondingly diminished to a minor fraction (Figures 3C and S4B). Thus, reaction kinetics suggests that SWR1 converts AA nucleosomes to the AZ and ZZ species in a stepwise manner.
Data of the above experiment does not inform whether a fully replaced ZZ nucleosome is the reaction end product, or a substrate for additional rounds of H2A.Z replacement (i.e. H2A.Z replacing H2A.Z). We addressed this question by first generating a mixed population of immobilized AA, AZ, and ZZ nucleosomes by a partial replacement reaction in which Htz1-H2B dimers provided to SWR1 bear a fluorescent Alexa633 label on Htz1 and a Flag tag on H2B (Htz1Alexa-H2BFlag dimers) (Figure 4A). [Analysis of an aliquot by MNase digestion confirms that mononucleosome products exhibit retarded electrophoretic mobility and Alexa633 fluorescence depending on the extent of replacement — the bottom band corresponding to unreplaced nucleosomes, and the middle and top bands to nucleosomes containing one and two Htz1Alexa-H2BFlag dimers, respectively (Figures 4A and 4B, lane I).] A second round of SWR1-mediated histone replacement using untagged, unlabeled Htz1-H2B dimers enabled us to evaluate if the two Htz1Alexa-H2BFlag dimers in the ZZ nucleosome were replaceable, as shown by a loss of Alexa633 fluorescence, SYBR green staining, and Htz1 content in the top nucleosome band (Figure 4A). However, all three indicators remained essentially unchanged after the second SWR1 reaction, indicating that SWR1 does not catalyze replacement of ZZ nucleosomes with new H2A.Z-H2B dimer (Figure 4B).
Figure 4. AA or AZ nucleosomes together with Htz1-H2B dimer are the specific substrates for SWR1.
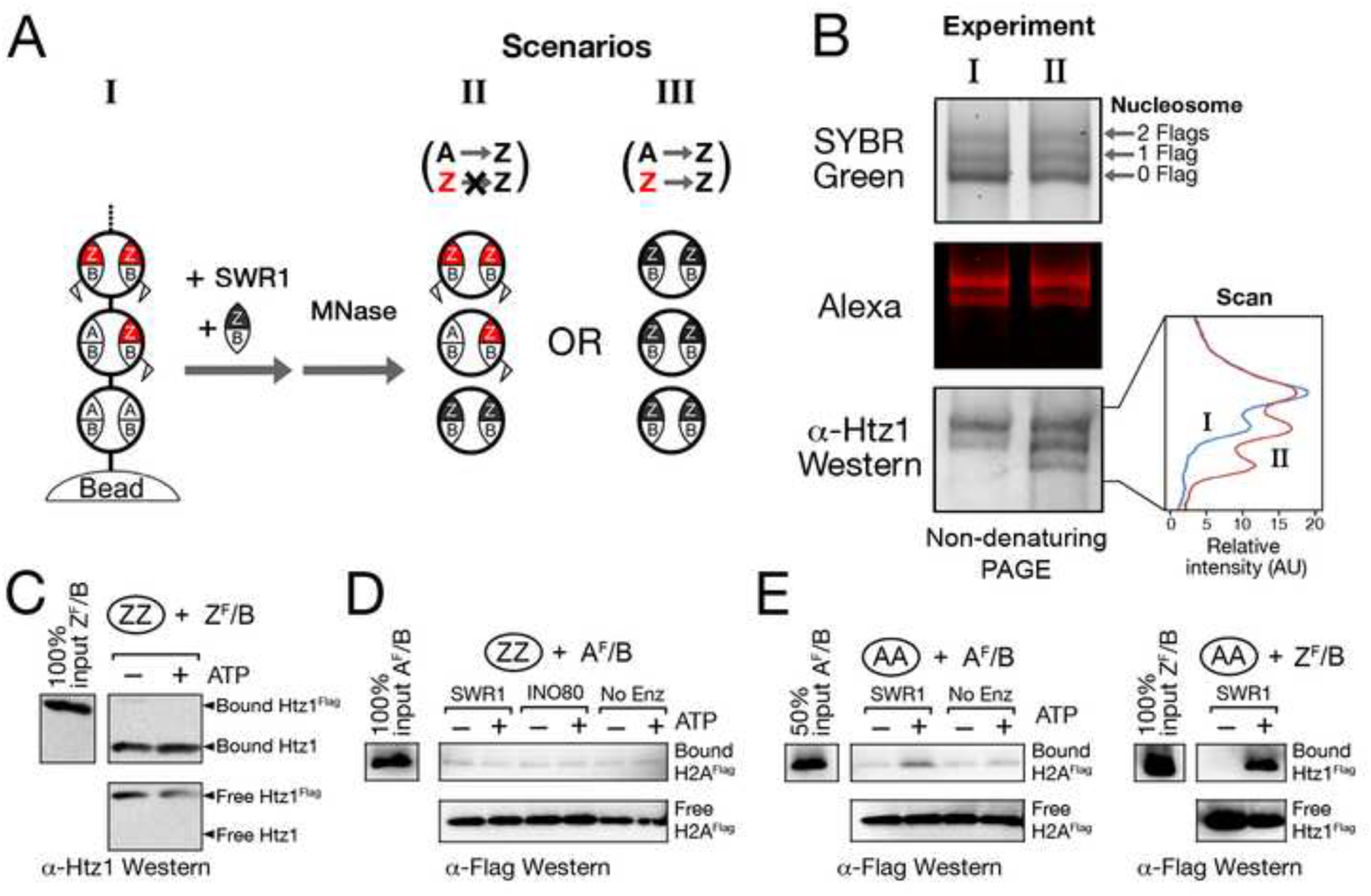
(A,B) Overview and experiment for the in vitro histone replacement assay. Nucleosomal arrays bearing a mixed population of AA, AZ, and ZZ nucleosomes were marked with Htz1Alexa-H2BFlag dimers. After incubating with unlabeled, untagged Htz1-H2B, SWR1, and ATP, two potential scenarios depicted in II and III could occur. B is the experiment. Red: Htz1Alexa; Flag: H2BFLAG. Scan: densitometric analysis of the α-Htz1 Western blot.
(C-E) Standard histone replacement assay (Mizuguchi et al., 2004). Immobilized AA or ZZ nucleosomal arrays were incubated with SWR1 (or INO80), native Flag-epitope tagged histone dimers, and ATP where indicated. 60 nM of dimers and approximately 15 nM nucleosome equivalents were used. The arrays were washed with 0.4 M KCl before SDS elution and Western analysis. Top panel: SDS eluted fraction of the chromatin bound histones. Bottom panel: free histones in the supernatant fraction. AA and ZZ ovals indicate the type of nucleosomal arrays used. ZF/B: Htz1Flag-H2B dimer, AF/B: H2AFlag-H2B dimer.
This experiment also permitted us to confirm directly that the heterotypic AZ nucleosome (middle band) is a substrate for SWR1-catalyzed histone replacement, by virtue of a potential increase in Htz1 content without a change in electrophoretic mobility (Figures 4A and 4B, lane II). We found that the middle band indeed shows a major increase in the Htz1 Western blotting signal, demonstrating that the AZ nucleosome, like the AA nucleosome (bottom band) is a substrate for SWR1 activity (Figure 4B, bottom panel, lane II). Taken together, these results provide compelling evidence that the AZ and ZZ nucleosomes are bona fide intermediate and end product, respectively, of the SWR1-mediated histone replacement reaction.
No reverse replacement of ZZ nucleosomes with H2A-H2B dimers
We confirmed that SWR1 does not replace ZZ nucleosomes with H2A.Z-H2B dimers using immobilized ZZ nucleosome arrays reconstituted from bacterially expressed histones. Incubation of these arrays with Flag-tagged histone dimer, SWR1, and ATP showed that SWR1 failed to replace ZZ nucleosome with Htz1Flag-H2B dimers even when dimers were in excess relative to nucleosomes (Figure 4C).
Next, we examined if AZ and AA nucleosomes could be produced from the ZZ species though a reverse reaction, by incubation of immobilized ZZ nucleosome arrays with excess H2AFlag-H2B dimers, SWR1, and ATP. This reaction also failed to produce detectable incorporation of H2AFlag above background in the bead-bound chromatin fraction (Figure 4D). We also tested whether the related INO80 remodeling complex could mediate a reverse replacement reaction, and found no detectable ATP-driven exchange of H2AFlag into ZZ nucleosomal arrays under reaction conditions (Figure 4D). Thus, other mechanisms may be responsible for the displacement of H2A.Z and re-assembly of the canonical nucleosome. By contrast, incubation of AA nucleosome arrays with saturating H2A-H2B dimers (60 nM) gave a small but reproducible level of ATP-dependent incorporation of H2AFlag into chromatin (Figure 4E), consistent with previous findings (Mizuguchi et al., 2004). We conclude that the histone replacement pathway mediated by SWR1 is unidirectional, with strong substrate specificity for H2A-containing nucleosomes and the Htz1-H2B dimer.
Canonical nucleosomes specifically stimulate SWR1 ATPase
Histone variant replacement by the multi-component SWR1 complex involves interaction with at least three essential substrates: ATP, the H2A-containing nucleosome, and the Htz1-H2B dimer. The differential utilization of H2A-containing nucleosomes suggests that SWR1 recognizes H2A- over H2A.Z-containing nucleosomes. Specific recognition could be a consequence of differential nucleosome binding and/or activation of the ATPase activity of SWR1. We examined whether AA and ZZ nucleosomes differentially stimulate the ATPase activity of SWR1, with the use of a real-time fluorescence assay that monitors production of inorganic phosphate from ATP hydrolysis (Brune et al., 1994).
Previously, we reported that the SWR1 complex exhibits nucleosome-stimulated ATPase activity as shown by hydrolysis of P32-ATP (Mizuguchi et al., 2004). This was confirmed by the fluorescence assay, which shows strong stimulation of ATP hydrolysis by conventional nucleosomes, and not by naked DNA (Figures 5A and S5A). Analysis of initial rates indicate a ~2.5-fold increase of ATP hydrolysis at saturating nucleosome and ATP levels. Strikingly, similar concentrations of ZZ nucleosomes failed to stimulate the ATPase activity of the SWR1 complex (Figures 5A, S5A). This demonstrates for the first time that SWR1 can functionally discriminate between conventional and variant nucleosomes. By contrast, INO80 and SWI/SNF exhibit no differential stimulation of ATPase activity by saturating levels of AA and ZZ nucleosomes (Figures 5B, 5C, and S5). Interestingly, both free H2A-H2B and Htz1-H2B dimers failed to stimulate the ATPase activity of SWR1 in the absence of nucleosomes, suggesting H2A-specific recognition must be in the context of nucleosome architecture (Figure 5D).
Figure 5. AA but not ZZ nucleosomes stimulate SWR1 ATPase.
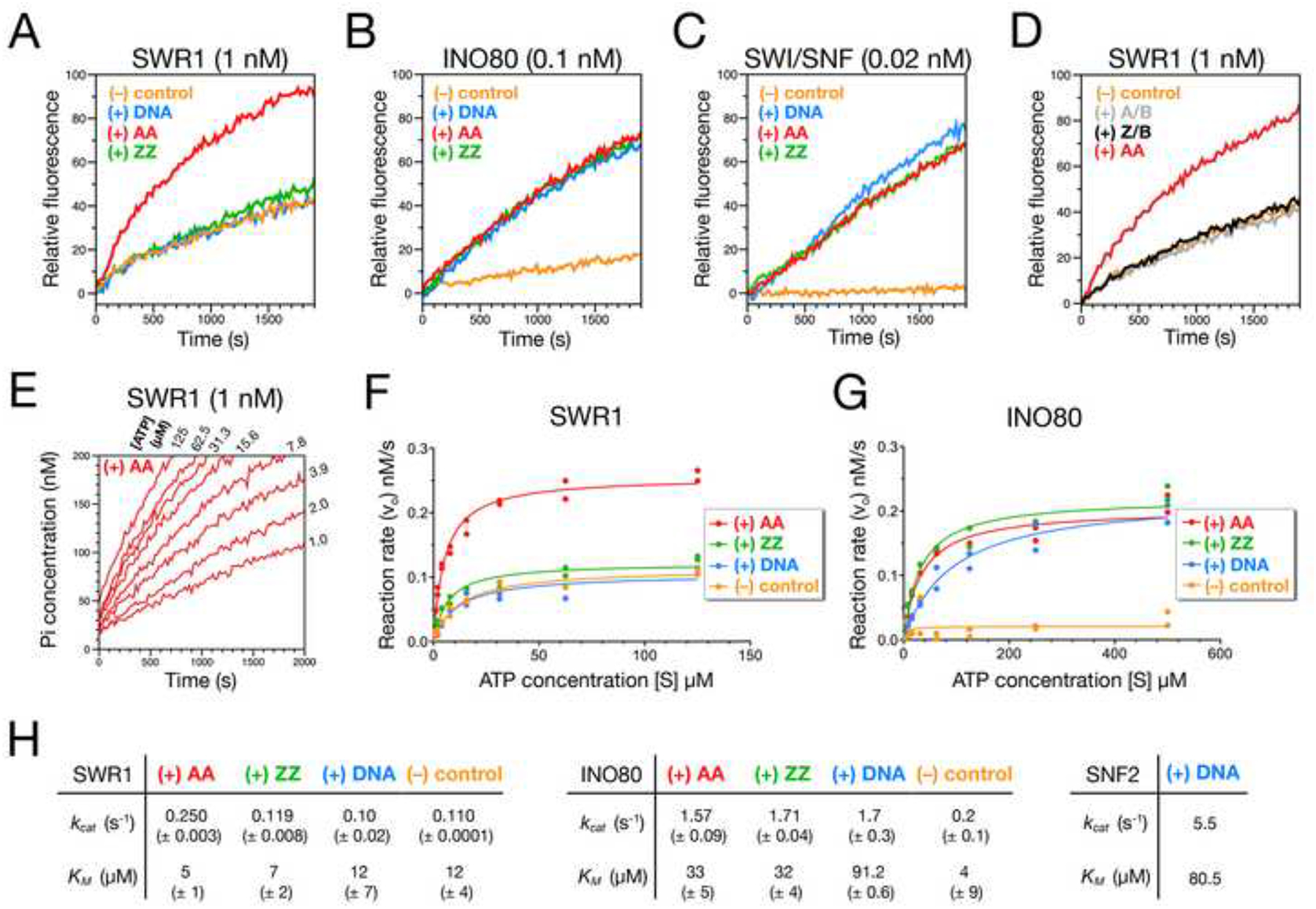
(A-C) ATPase assay for chromatin remodelers. Inorganic phosphate (Pi) produced during ATP hydrolysis was monitored in real-time by MDCC-PBP, which increases in fluorescence upon phosphate binding (Brune et al., 1994). Reactions were performed at 23°C in the absence (orange) or presence of 15 nM AA nucleosomes (red), ZZ nucleosomes (green), or free DNA (blue). ATP was added ~20 sec before the first measurement (zero time) to final concentrations of 62.5 μM for SWR1 and 500 μM for INO80 and SWI/SNF. Relative fluorescence was set as zero at zero time for all reactions.
(D) ATPase assay for SWR1 in the absence (orange) or presence of 15 nM recombinant Htz1-H2B dimer (Z/B, black), H2A-H2B dimer (A/B, grey), or AA nucleosomes (red).
(E) ATPase assay for SWR1 in the presence of AA nucleosomes and various ATP concentrations. Phosphate concentrations (calculated based on a linear phosphate standard curve) were plotted against time. Initial rate (vo) was determined by the slope of the linear part of each curve (between 0–300 sec).
(F,G) Plots of initial rate versus substrate (ATP) concentrations for 1 nM SWR1 and 0.1 nM INO80 in the presence or absence of 15 nM AA nucleosomes, ZZ nucleosomes, or DNA. The kinetic parameters VMAX and KM were determined by non-linear fitting of the Michaelis-Menten curve over plotted values.
(H) Turnover number kcat (obtained from dividing Vmax by total enzyme concentration), and KM for the ATPase of SWR1, INO80, and SWI/SNF in the presence or absence of 15 nM AA, ZZ nucleosomes, or DNA. Errors represent the range of two measurements.
We determined kinetic parameters for ATP hydrolysis by the SWR1 complex (Figures 5E, 5F, and 5H). SWR1 has an enzyme turnover rate (kcat) of 0.1 s−1 in the presence or absence of DNA. The kcat remains essentially the same when SWR1 is incubated with ZZ nucleosomes, but increases to 0.25 s−1 (2.5-fold) with saturating AA nucleosomes (Figures 5F and 5H). Hence, binding of H2A-containing nucleosomes to SWR1 stimulates ATPase activity by increasing the catalytic efficiency of the enzyme. The Michaelis constant (KM), which represents the ATP concentration at half maximal velocity (½ Vmax) shows little change for canonical and variant nucleosomes (5 μM and 7 μM, respectively). In comparison, the stimulated SWI/SNF has a kcat of 5.5 s−1 and KM of 80.5 μM, values consistent with previous determinations (Smith and Peterson, 2005) (Figure 5H).
Nucleosome stimulation of SWR1 ATPase not sufficient for H2A-H2B eviction
The stimulation of SWR1 ATPase by incubation with conventional nucleosomes raised the question of whether such ATP hydrolysis would be sufficient for eviction of the nucleosomal H2A-H2B dimer to facilitate Htz1-H2B deposition. To test this hypothesis, we incubated SWR1 with immobilized arrays of conventional nucleosomes carrying epitope-tagged histone H2AHA and monitored H2AHA-H2B eviction in the supernatant fraction by Western blotting. While the SWR1-catalyzed Htz1 replacement reaction in the presence of Htz1-H2B dimer (and histone chaperone) occurs robustly with quantitative eviction of H2AHA, we did not detect any eviction of histone H2AHA in the absence of Htz1Flag-H2B dimer (Figure 6A). Thus, the stimulation of ATP hydrolysis provided solely by canonical nucleosome effector is inadequate for eviction of nucleosomal H2A-H2B. Moreover, eviction of H2A-H2B and insertion of Htz1-H2B appear to be coupled processes.
Figure 6. Further binding of H2A.Z-H2B dimer hyperactivates SWR1 ATPase and evicts nucleosomal H2A-H2B.

(A) Standard histone replacement assay (Mizuguchi et al., 2004). Immobilized AA nucleosomal arrays (reconstituted with H2AHA histone) were incubated with SWR1, Htz1Flag-H2B (Z/B), histone chaperones and ATP where indicated. Top panel: Western analyses of chromatin bound histones eluted by SDS. Bottom panel: Western analyses of unincorporated histones. 22 nM of Chz1 or FACT was added to facilitate possible histone eviction.
(B) ATPase assay for SWR1 in the presence of 15 nM AA nucleosomes and 15 nM Htz1-H2B dimers (purple). Red is the AA only control.
(C) Kinetic parameters of SWR1 ATPase in the presence of 15 nM AA nucleosomes and 15 nM Htz1-H2B dimers (purple). For comparison, the curve and parameters for AA alone (red) are reproduced from Figures 5H and 5F, respectively. Errors represent the range of two measurements.
(D) Specificity of SWR1 ATPase hyperstimulation. ATPase assay was performed as described in Figure 5A except with different combinations of nucleosomes (Nuc) and histone dimers. Z/B: Htz1-H2B dimers, A/B: H2A-H2B dimers, AA: AA nucleosomes, ZZ: ZZ nucleosomes, (−) control: no dimer or nucleosome.
Htz1-H2B dimer and canonical nucleosome specifically activate SWR1
The requirement for free Htz1-H2B dimers for SWR1-mediated H2A.Z replacement prompted us to investigate whether addition of Htz1-H2B to SWR1 further increases ATP hydrolysis. Indeed, we observed a clear hyperstimulation of the ATPase activity of SWR1 when saturating Htz1-H2B dimers (15 nM) and canonical nucleosomes are both provided to SWR1 in the reaction (Figures 6B and 6D). The hyperstimulated ATPase activity exhibits a kcat of 0.45 s−1, which represents an additional 1.8-fold increase in the kcat relative to the stimulation by nucleosomes only (a 4.1-fold increase in total), with little change of KM (Figure 6C). We observed less hyperstimulation when H2A-H2B dimers were substituted for Htz1-H2B at the same molar concentration (Figure 6D, left panel). Importantly, a 4-fold increase of H2A-H2B dimer (~60 nM) hyperstimulated ATPase activity to nearly maximal level (Figure 6D, right panel), while hyperstimulation of the ATPase activity upon addition of Htz1-H2B or H2A-H2B dimers (at either concentration) to ZZ nucleosomes is much lower (Figure 6D). Given that incorporation of new H2A in canonical nucleosomal arrays is low (Figure 4E) under conditions where ATPase activity is high, these findings indicate that high ATPase activity per se is not sufficient for histone replacement. It is the presence of the correct, in vivo substrates that ensures efficient coupling of the high ATPase activity to histone replacement.
Discussion
The steady-state level of H2A.Z at promoter-proximal nucleosomes is a consequence of the opposing pathways of H2A.Z incorporation and H2A.Z eviction. Our observation of three distinct variant states of promoter nucleosomes in a cell population is complementary to previous mapping studies of H2A.Z in budding yeast (Albert et al., 2007; Raisner et al., 2005; Santisteban et al., 2000). The comparable representation of AA and ZZ states suggests that the AA-to-ZZ and ZZ-to-AA pathways are balanced for many genes, without one pathway dominating. However, this balance can be shifted, for example, at highly transcribed promoters (top 10% RNA Pol II occupancy), where the ZZ and AZ states are under-represented relative to the AA state for the +1 nucleosome position (Figure S2E), suggesting that H2A.Z eviction is occurring at a faster rate than incorporation. The greater restriction of the ZZ than AZ state to +1 and −1 nucleosome positions is interesting, and may be a consequence of the stepwise nature of the histone replacement reaction and the local concentration of SWR1 recruited to gene promoters (Venters and Pugh, 2009; Yoshida et al., 2010).
Our in vitro studies show that SWR1 is capable of stepwise deposition of H2A.Z-H2B into canonical nucleosomes, coupled with H2A-H2B eviction, to give a fully replaced variant nucleosome. However, once incorporated, H2A.Z cannot be evicted by SWR1, even in excess of either H2A.Z-H2B or H2A-H2B dimers under otherwise identical reaction conditions. Therefore, the SWR1-mediated pathway of H2A.Z replacement is unidirectional, terminating with ZZ nucleosomes. It is possible that a reverse reaction from the ZZ to AA nucleosome state requires different conditions, co-factors, or modifications of the SWR1 enzyme or histone substrates. Alternatively, return to the AA state may occur through separate pathways. For example, other SWI/SNF family members might possess the capability for specific replacement of nucleosomal H2A.Z-H2B with H2A-H2B, but we have not observed such activity for INO80, a chromatin remodeling complex paralogous to SWR1, under conditions where INO80 displays robust nucleosome- or DNA-stimulated ATPase and histone octamer sliding activities (Figures 5, S5 and unpublished observations).
More likely, the well-documented high histone H3 turnover rate at promoters implies promoter-specific nucleosome disassembly, i.e. eviction of H2A.Z-H2B and H3-H4, and subsequent nucleosome reassembly with new histones (Dion et al., 2007; Rufiange et al., 2007), thereby completing the dynamic cycling of H2A and H2A.Z at gene promoters (Figure 7A). These processes are likely to be mediated by a combination of SWI/SNF family enzymes (Barbaric et al., 2007; Gutierrez et al., 2007; Lorch et al., 2006), RNA polymerase (Weiner et al., 2010), and core histone chaperones (Corpet and Almouzni, 2009; Das et al., 2010). In addition, the in vivo lability of H2A.Z-containing nucleosomes as reflected in salt-sensitivity should also contribute (Henikoff et al., 2009; Jin and Felsenfeld, 2007; Zhang et al., 2005). Indeed, histone modifications at promoters correlate with the signatures of newly deposited histones, such as H3K56Ac and H4K16deAc (Rando and Chang, 2009).
Figure 7. A model for SWR1-mediated histone replacement.
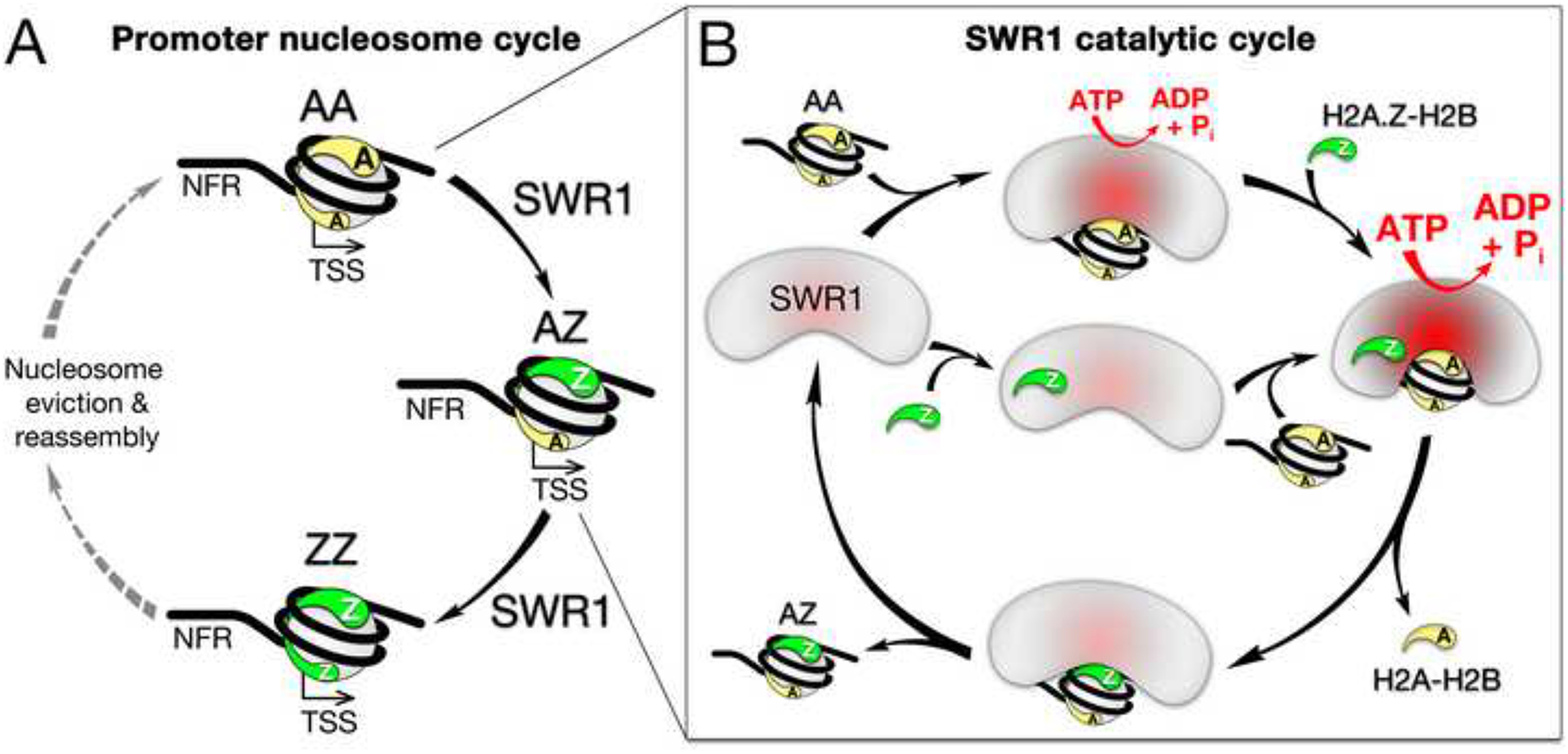
(A) Promoter nucleosome cycle: An AA nucleosome at the +1 promoter-proximal position is converted to the AZ and ZZ states by SWR1 via a stepwise, unidirectional pathway. The ZZ nucleosome is subsequently converted back to the AA state though pathways most likely involving nucleosome eviction and reassembly with an AA nucleosome (dotted grey arrows).
(B) SWR1 catalytic cycle: SWR1 stochastically binds to one face of an AA nucleosome and the H2A.Z-H2B dimer leading to hyperstimulation of ATPase activity (deep red) and a conformational change in SWR1 (shown) required for histone replacement. The newly incorporated Z face of the AZ nucleosome deactivates the ATPase and stops further histone replacement activity. The AZ nucleosome dissociates and rebinds stochastically on the A face for a second replacement reaction.
The directional nature of the H2A.Z replacement pathway implies that SWR1 must functionally differentiate between ZZ and AA (or AZ) nucleosomes. We have traced this differentiation, at least in part, to a specific, 2.5-fold increase of the ATPase activity (kcat) of SWR1 induced by AA, but not ZZ nucleosomes. However, this level of stimulation is insufficient for the eviction of H2A-H2B from nucleosomes. Only after further addition of free H2A.Z-H2B dimers is the ATPase activity of SWR1 hyperstimulated (4-fold increase of kcat), concurrent with H2A-H2B eviction and H2A.Z-H2B deposition. However, a hyperstimulated SWR1 ATPase is only necessary, but not sufficient to mediate robust histone replacement as saturating free H2A-H2B dimers can hyperstimulate SWR1 ATPase to nearly maximal level but with substantially reduced histone replacement (Figures 4E and 6D). This finding implies that unique features of H2A.Z-H2B dimer, in addition to stimulating ATP hydrolysis, enhance histone replacement by allosterically coupling the ATPase motor to histone transactions. This additional molecular specificity seems biologically necessary, given that H2A-H2B dimers should be in excess over H2A.Z-H2B dimers in vivo.
Overall, our data suggest a model in which SWR1 binding to and recognition of its two in vivo histone substrates (one face of the AA nucleosome and the H2A.Z-H2B dimer) leads to hyperstimulation of ATPase activity as well as a conformational change in SWR1 required for displacement of H2A-H2B and insertion of H2A.Z-H2B (Figure 7B). The order of SWR1 binding to nucleosomes and H2A.Z-H2B dimers should be stochastic. The newly incorporated Z face of the AZ nucleosome deactivates the ATPase and stops further histone replacement. The AZ nucleosome subsequently dissociates from and reassociates with SWR1 in a stochastic fashion (Figures 7B and 7C). In the second round, recognition by SWR1 of the A face of the AZ nucleosome and new H2A.Z-H2B dimer binding restimulates SWR1 activity to catalyze replacement of the remaining nucleosomal H2A-H2B with H2A.Z-H2B. Functional recognition of the A face of an AA or AZ nucleosome and the requirement for free H2A.Z-H2B dimer ensures that only these effectors, which are also substrates for SWR1, are productively utilized. This provides a way of controlling the specificity and outcome of the replacement reaction, which terminates with the ZZ nucleosome.
The SWR1 complex contains multiple ATP-binding subunits, including Swr1, actin, actin-related proteins Arp4 and Arp6, and the Rvb1-Rvb2 dodecamer, members of the AAA+ family of ATPases (Jha and Dutta, 2009; Mizuguchi et al., 2004). We have previously found that a mutation (K727G substitution) in the ATP-binding motif of the Swr1 subunit is sufficient to abrogate Htz1 replacement in vivo and in vitro without affecting assembly of the SWR1 complex (Mizuguchi et al., 2004). The ATPase activity of the purified mutant enzyme is neither stimulated by AA nucleosomes nor hyperstimulated by further addition of Htz1-H2B dimer (Figure S6). These findings indicate that the Swr1 ATPase is the key subunit whose activity is governed, directly and/or indirectly, by the histone effectors.
It will be interesting to define the molecular determinants within the canonical nucleosome and the H2A.Z-H2B dimer that are specifically recognized by the SWR1 complex, to identify the SWR1 components interacting with the nucleosome, and to follow the fate of the evicted H2A-H2B dimer. Other questions are the importance of the two Htz1-binding modules in SWR1 (Swc2, and the N-terminus of the Swr1 subunit itself), the relationship between ATPase activity, DNA translocase activity, and unwrapping of nucleosomal DNA, the timing and coupling of H2A eviction and H2A.Z insertion, and the structural transformations of SWR1 that accompany these events. Our present findings and the biochemical assays we have developed should facilitate future investigations on the mechanism of histone H2A.Z replacement.
Experimental Procedures
Immunopurification of AA, AZ, and ZZ nucleosomes
Crude chromatin was isolated from formaldehyde-fixed yeast cells as described in Liang and Stillman (1997) and digested with MNase to mononucleosomal level. Sequential IP was performed with the use of anti-Flag M2 agarose (Sigma) and anti-H2A antibodies (Active Motif) bound to nProtein A-sepharose (GE Healthcare).
Amplification and labeling of nucleosomal DNA for microarray analysis
Nucleosomal DNA and MNase-treated genomic DNA control were amplified by ligation-mediated PCR according to the protocol described in Johnson et al. (2008). Labeling was performed using the BioPrime Plus labeling kit (Invitrogen) according to the manufacturer’s protocol.
Microarrays
Custom tiling microarrays were designed based on the Agilent 4 × 180K platform. Each microarray contained approximately 150,000 biological probes spanning selected genomic regions. The tiling probes were spaced, on average, 10 bp apart and covered both the sense and anti-sense DNA strands.
Normalization of microarray data for different nucleosomal species
Given that Htz1 is the only H2A variant in budding yeast, normalization of microarray data was performed based on the assumptions that, to a first approximation, the sum of Z-total and AA nucleosomes is equal to the Total nucleosome signal, and that the sum of AZ and ZZ nucleosomes is equal to the Z-total nucleosome signal. Details are provided in supplemental Figure S3 and legend.
In vitro histone replacement assay
The SWR1 histone replacement assay was performed according to Mizuguchi et al. (2004), except the immobilized nucleosomal arrays (80 ng DNA equivalents) were digested with 0.16 U/μL MNase (+ 2 mM CaCl2) to liberate the nucleosomes. The reactions were stopped with 10 mM EDTA before analyzed by non-denaturing PAGE.
In vitro histone replacement assay using fluorescently labeled Htz1-H2B substrate
To generate the mixed AA, AZ, and ZZ nucleosomal substrate for the experiment in Figure 4B, AA nucleosomal arrays were incubated with SWR1 pre-charged with Htz1Alexa-H2BFlag and with Htz1-H2BFlag. The resulting chromatin had comparable levels of AA, AZ, and ZZ nucleosomes, which also exhibited comparable Alexa633 fluorescence for the AZ and ZZ nucleosomal species. In the chase step, the labeled nucleosomes were incubated with SWR1 pre-charged with the unlabeled, untagged Htz1-H2B. After washing, the nucleosomal products were released by MNase digestion and analyzed by non-denaturing PAGE as described above.
ATPase assay
ATPase assay was performed based on the procedure described in Brune et al. (1994). In this assay, inorganic phosphate (Pi) produced during ATP hydrolysis is monitored by the fluorophore-modified phosphate binding protein MDCC-PBP (Phosphate Sensor, Invitrogen), which increases in fluorescence upon Pi binding. Measurements were performed at 23°C on a Wallac Victor plate reader using a 405 nm excitation, 460 nm emission filter set.
Supplementary Material
Acknowledgements
We thank W.H. Wu for the yeast strain SWR1-FL htz1Δ, J. Landry for the yeast strain yJL036. We also thank H. Cam and C. Rubin for advice on microarray techniques. F. Pugh and members of the Wu lab for critical reading of the manuscript and anonymous reviewers for helpful suggestions. This work was supported by the intramural research program of the National Cancer Institute (C.W.) and the Leukemia and Lymphoma Society (E.L. and A.R.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Albert I, Mavrich TN, Tomsho LP, Qi J, Zanton SJ, Schuster SC, and Pugh BF (2007). Translational and rotational settings of H2A.Z nucleosomes across the Saccharomyces cerevisiae genome. Nature 446, 572–576. [DOI] [PubMed] [Google Scholar]
- Altaf M, Auger A, Monnet-Saksouk J, Brodeur J, Piquet S, Cramet M, Bouchard N, Lacoste N, Utley RT, Gaudreau L, et al. (2010). NuA4-dependent acetylation of nucleosomal histones H4 and H2A directly stimulates incorporation of H2A.Z by the SWR1 complex. J Biol Chem 285, 15966–15977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausio J (2006). Histone variants--the structure behind the function. Brief Funct Genomic Proteomic 5, 228–243. [DOI] [PubMed] [Google Scholar]
- Barbaric S, Luckenbach T, Schmid A, Blaschke D, Horz W, and Korber P (2007). Redundancy of chromatin remodeling pathways for the induction of the yeast PHO5 promoter in vivo. J Biol Chem 282, 27610–27621. [DOI] [PubMed] [Google Scholar]
- Brune M, Hunter JL, Corrie JE, and Webb MR (1994). Direct, real-time measurement of rapid inorganic phosphate release using a novel fluorescent probe and its application to actomyosin subfragment 1 ATPase. Biochemistry 33, 8262–8271. [DOI] [PubMed] [Google Scholar]
- Cairns BR (2009). The logic of chromatin architecture and remodelling at promoters. Nature 461, 193–198. [DOI] [PubMed] [Google Scholar]
- Carr AM, Dorrington SM, Hindley J, Phear GA, Aves SJ, and Nurse P (1994). Analysis of a histone H2A variant from fission yeast: evidence for a role in chromosome stability. Mol Gen Genet 245, 628–635. [DOI] [PubMed] [Google Scholar]
- Chakravarthy S, Bao Y, Roberts VA, Tremethick D, and Luger K (2004). Structural characterization of histone H2A variants. Cold Spring Harb Symp Quant Biol 69, 227–234. [DOI] [PubMed] [Google Scholar]
- Corpet A, and Almouzni G (2009). Making copies of chromatin: the challenge of nucleosomal organization and epigenetic information. Trends Cell Biol 19, 29–41. [DOI] [PubMed] [Google Scholar]
- Das C, Tyler J, and Churchill M (2010). The histone shuffle: histone chaperones in an energetic dance. Trends in Biochemical Sciences in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dion MF, Kaplan T, Kim M, Buratowski S, Friedman N, and Rando OJ (2007). Dynamics of replication-independent histone turnover in budding yeast. Science 315, 1405–1408. [DOI] [PubMed] [Google Scholar]
- Gevry N, Chan HM, Laflamme L, Livingston DM, and Gaudreau L (2007). p21 transcription is regulated by differential localization of histone H2A.Z. Genes Dev 21, 1869–1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillemette B, and Gaudreau L (2006). Reuniting the contrasting functions of H2A.Z. Biochem Cell Biol 84, 528–535. [DOI] [PubMed] [Google Scholar]
- Gutierrez JL, Chandy M, Carrozza MJ, and Workman JL (2007). Activation domains drive nucleosome eviction by SWI/SNF. EMBO J 26, 730–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S, Henikoff JG, Sakai A, Loeb GB, and Ahmad K (2009). Genome-wide profiling of salt fractions maps physical properties of chromatin. Genome Res 19, 460–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson JD, and Gorovsky MA (2000). Histone H2A.Z has a conserved function that is distinct from that of the major H2A sequence variants. Nucleic Acids Res 28, 3811–3816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jha S, and Dutta A (2009). RVB1/RVB2: running rings around molecular biology. Mol Cell 34, 521–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang C, and Pugh BF (2009a). A compiled and systematic reference map of nucleosome positions across the Saccharomyces cerevisiae genome. Genome Biol 10, R109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang C, and Pugh BF (2009b). Nucleosome positioning and gene regulation: advances through genomics. Nat Rev Genet 10, 161–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin C, and Felsenfeld G (2007). Nucleosome stability mediated by histone variants H3.3 and H2A.Z. Genes Dev 21, 1519–1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson DS, Li W, Gordon DB, Bhattacharjee A, Curry B, Ghosh J, Brizuela L, Carroll JS, Brown M, Flicek P, et al. (2008). Systematic evaluation of variability in ChIP-chip experiments using predefined DNA targets. Genome Res 18, 393–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobor MS, Venkatasubrahmanyam S, Meneghini MD, Gin JW, Jennings JL, Link AJ, Madhani HD, and Rine J (2004). A protein complex containing the conserved Swi2/Snf2-related ATPase Swr1p deposits histone variant H2A.Z into euchromatin. PLoS Biol 2, E131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koerber RT, Rhee HS, Jiang C, and Pugh BF (2009). Interaction of transcriptional regulators with specific nucleosomes across the Saccharomyces genome. Mol Cell 35, 889–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornberg RD, and Lorch Y (1999). Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell 98, 285–294. [DOI] [PubMed] [Google Scholar]
- Krogan NJ, Baetz K, Keogh MC, Datta N, Sawa C, Kwok TC, Thompson NJ, Davey MG, Pootoolal J, Hughes TR, et al. (2004). Regulation of chromosome stability by the histone H2A variant Htz1, the Swr1 chromatin remodeling complex, and the histone acetyltransferase NuA4. Proc Natl Acad Sci U S A 101, 13513–13518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan NJ, Keogh MC, Datta N, Sawa C, Ryan OW, Ding H, Haw RA, Pootoolal J, Tong A, Canadien V, et al. (2003). A Snf2 family ATPase complex required for recruitment of the histone H2A variant Htz1. Mol Cell 12, 1565–1576. [DOI] [PubMed] [Google Scholar]
- Kusch T, Florens L, Macdonald WH, Swanson SK, Glaser RL, Yates JR 3rd, Abmayr SM, Washburn MP, and Workman JL (2004). Acetylation by Tip60 is required for selective histone variant exchange at DNA lesions. Science 306, 2084–2087. [DOI] [PubMed] [Google Scholar]
- Li B, Pattenden SG, Lee D, Gutierrez J, Chen J, Seidel C, Gerton J, and Workman JL (2005). Preferential occupancy of histone variant H2AZ at inactive promoters influences local histone modifications and chromatin remodeling. Proc Natl Acad Sci U S A 102, 18385–18390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang C, and Stillman B (1997). Persistent initiation of DNA replication and chromatin-bound MCM proteins during the cell cycle in cdc6 mutants. Genes Dev 11, 3375–3386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorch Y, Maier-Davis B, and Kornberg RD (2006). Chromatin remodeling by nucleosome disassembly in vitro. Proc Natl Acad Sci U S A 103, 3090–3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luk E, Vu ND, Patteson K, Mizuguchi G, Wu WH, Ranjan A, Backus J, Sen S, Lewis M, Bai Y, et al. (2007). Chz1, a nuclear chaperone for histone H2AZ. Mol Cell 25, 357–368. [DOI] [PubMed] [Google Scholar]
- Meneghini MD, Wu M, and Madhani HD (2003). Conserved histone variant H2A.Z protects euchromatin from the ectopic spread of silent heterochromatin. Cell 112, 725–736. [DOI] [PubMed] [Google Scholar]
- Mizuguchi G, Shen X, Landry J, Wu WH, Sen S, and Wu C (2004). ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science 303, 343–348. [DOI] [PubMed] [Google Scholar]
- Morrison AJ, and Shen X (2009). Chromatin remodelling beyond transcription: the INO80 and SWR1 complexes. Nat Rev Mol Cell Biol 10, 373–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raisner RM, Hartley PD, Meneghini MD, Bao MZ, Liu CL, Schreiber SL, Rando OJ, and Madhani HD (2005). Histone variant H2A.Z marks the 5’ ends of both active and inactive genes in euchromatin. Cell 123, 233–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rando OJ, and Chang HY (2009). Genome-wide views of chromatin structure. Annu Rev Biochem 78, 245–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rufiange A, Jacques PE, Bhat W, Robert F, and Nourani A (2007). Genome-wide replication-independent histone H3 exchange occurs predominantly at promoters and implicates H3 K56 acetylation and Asf1. Mol Cell 27, 393–405. [DOI] [PubMed] [Google Scholar]
- Ruhl DD, Jin J, Cai Y, Swanson S, Florens L, Washburn MP, Conaway RC, Conaway JW, and Chrivia JC (2006). Purification of a human SRCAP complex that remodels chromatin by incorporating the histone variant H2A.Z into nucleosomes. Biochemistry 45, 5671–5677. [DOI] [PubMed] [Google Scholar]
- Santisteban MS, Kalashnikova T, and Smith MM (2000). Histone H2A.Z regulats transcription and is partially redundant with nucleosome remodeling complexes. Cell 103, 411–422. [DOI] [PubMed] [Google Scholar]
- Smith CL, and Peterson CL (2005). A conserved Swi2/Snf2 ATPase motif couples ATP hydrolysis to chromatin remodeling. Mol Cell Biol 25, 5880–5892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suto RK, Clarkson MJ, Tremethick DJ, and Luger K (2000). Crystal structure of a nucleosome core particle containing the variant histone H2A.Z. Nat Struct Biol 7, 1121–1124. [DOI] [PubMed] [Google Scholar]
- Venters BJ, and Pugh BF (2009). A canonical promoter organization of the transcription machinery and its regulators in the Saccharomyces genome. Genome Res 19, 360–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner A, Hughes A, Yassour M, Rando OJ, and Friedman N (2010). High-resolution nucleosome mapping reveals transcription-dependent promoter packaging. Genome Res 20, 90–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West MH, and Bonner WM (1980). Histone 2A, a heteromorphous family of eight protein species. Biochemistry 19, 3238–3245. [DOI] [PubMed] [Google Scholar]
- Wu WH, Alami S, Luk E, Wu CH, Sen S, Mizuguchi G, Wei D, and Wu C (2005). Swc2 is a widely conserved H2AZ-binding module essential for ATP-dependent histone exchange. Nat Struct Mol Biol 12, 1064–1071. [DOI] [PubMed] [Google Scholar]
- Wu WH, Wu CH, Ladurner A, Mizuguchi G, Wei D, Xiao H, Luk E, Ranjan A, and Wu C (2009). N terminus of Swr1 binds to histone H2AZ and provides a platform for subunit assembly in the chromatin remodeling complex. J Biol Chem 284, 6200–6207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida T, Shimada K, Oma Y, Kalck V, Akimura K, Taddei A, Iwahashi H, Kugou K, Ohta K, Gasser SM, et al. (2010). Actin-related protein Arp6 influences H2A.Z-dependent and -independent gene expression and links ribosomal protein genes to nuclear pores. PLoS Genet 6, e1000910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Roberts DN, and Cairns BR (2005). Genome-wide dynamics of Htz1, a histone H2A variant that poises repressed/basal promoters for activation through histone loss. Cell 123, 219–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zlatanova J, and Thakar A (2008). H2A.Z: view from the top. Structure 16, 166–179. [DOI] [PubMed] [Google Scholar]
- Zofall M, Fischer T, Zhang K, Zhou M, Cui B, Veenstra TD, and Grewal SI (2009). Histone H2A.Z cooperates with RNAi and heterochromatin factors to suppress antisense RNAs. Nature 461, 419–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


