Abstract
Vaccinia TopIB (vTopIB), a 314-amino acid eukaryal type IB topoisomerase, recognizes and transesterifies at the DNA sequence 5’-(T/C)CCTT↓, leading to the formation of a covalent DNA-(3’-phosphotyrosyl274)-enzyme intermediate in the supercoil relaxation reaction. The C-terminal segment of vTopIB (amino acids 81-314), which engages the DNA minor groove at the scissile phosphodiester, comprises an autonomous catalytic domain that retains cleavage specificity, albeit with lower cleavage site affinity compared to the full-length enzyme. The N-terminal domain (amino acids 1-80) engages the major groove on the DNA face opposite the scissile phosphodiester. Whereas DNA contacts of the N-terminal domain have been implicated in DNA site affinity of vTopIB, it was not known whether the N-terminal domain per se could bind DNA. Here, using isothermal titration calorimetry, we demonstrate the ability of the isolated N-terminal domain to bind a CCCTT-containing 24-mer duplex with an apparent affinity that is ~2.2-fold higher than that for an otherwise identical duplex in which the pentapyrimidine sequence is changed to ACGTG. Analyses of the interactions of the isolated N-terminal domain with duplex DNA via solution NMR methods are consistent with its DNA contacts observed in DNA-bound crystal structures of full-length vTopIB. The chemical shift perturbations and changes in hydrodynamic properties triggered by CCCTT DNA versus non-CCCTT DNA suggest differences in DNA binding dynamics. The importance of key N-terminal domain contacts in the context of full-length vTopIB are underscored by assessing the effects of double-alanine mutations on DNA transesterification and its sensitivity to ionic strength.
INTRODUCTION
Type IB topoisomerases relax DNA supercoils by iteratively cleaving and rejoining one strand of the DNA duplex through a covalent DNA-(3’-phosphotyrosyl)-enzyme intermediate. Type IB topoisomerases are present in the proteomes of all eukarya, many bacterial phyla, a single archeal phylum (Thaumarcheaota), and several large eukaryal DNA viruses (e.g., vaccinia and mimivirus)1–5. TopIBs from bacteria and eukaryal viruses are small two-domain proteins that exemplify the minimal catalytic core of the type IB enzyme 3, 6–9; they comprise a phylogenetic cluster distinct from the eukaryal and archaeal TopIBs, which are larger proteins embellished by additional modules appended to the core structure 1, 10.
Vaccinia virus TopIB (vTopIB) is a 37 kDa monomeric protein (Figure S1) consisting of an N-terminal domain (N domain, amino acids 1-80) and a C-terminal domain (C domain, amino acids 81-314) connected by a flexible, protease-sensitive linker that becomes protease-resistant when vTopIB is bound to DNA6, 11. The C domain consists of ten α helices and a three-strand antiparallel β-sheet packed against one surface3. The smaller N domain comprises a five-strand antiparallel β-sheet with two short α helices (Figure S1)6. vTopIB displays stringent specificity for cleavage of double-stranded DNA (dsDNA) at the sequence 5′-(C/T)CCTTp↓N in the scissile strand 12, 13. vTopIB engages the DNA target site circumferentially as a C-shaped protein clamp in which the N domain interacts with the major groove on the face of the DNA helix opposite the cleavage site while the C domain interacts with the minor groove on the face of the helix that contains the scissile phosphodiester7, 14, 15 (Figure S1). The active site is located entirely within the C domain and consists of a set of four amino acids (R130, K167, R223, H265) that catalyzes the attack of the tyrosine nucleophile (Y274) on the scissile phosphodiester8, 16–21. The active site is not preassembled in the vTopIB apoenzyme; rather its assembly is triggered by recognition of specific nucleobases and backbone phosphates of the 5’-CCCTT-3’:3’-GGGAA-5’ target dsDNA sequence3, 8, 22–26. vTopIB relaxes DNA supercoils in a stepwise fashion in the following way: after nicking the DNA duplex by transesterification of vTopIB to the DNA 3’-phosphate end, the DNA rotates inside the protein clamp (with friction at the protein–DNA interface) and its 5’-OH end passes the tyrosine-3’-DNA adduct once every turn. The N domain of vTopIB (specifically the Y70 side chain) was identified as a contributor to the friction experienced during DNA rotation27.
The isolated C domain of vTopIB (amino acids 81-314) performs the same repertoire of reactions as the full-length enzyme: supercoil relaxation, site-specific DNA transesterification, and DNA strand transfer28. Subtraction of the N domain slowed the rate of single-turnover DNA cleavage by ~104-fold but had little effect on the rate of single-turnover re-ligation. Strand cleavage by the isolated C domain was inhibited by salt and by Mg2+, effects indicative of reduced affinity of non-covalent interactions with DNA. It was concluded that the while C domain is fully competent for transesterification chemistry, it is compromised with respect to a rate-limiting pre-cleavage conformational step contingent on DNA contacts made by the N domain28. Extensive mutagenesis of N domain amino acids in the context of full-length vTopIB implicated F59, R67, Q69, Y70, Y72, G73, and R80 as contributors to the noncovalent interaction of vTopIB with its DNA target site28–30. The mutational data are in accord with the DNA contacts made by the N-domain in the crystal structures determined by Van Duyne and colleagues7, 8 (Figure 1).
Figure 1.
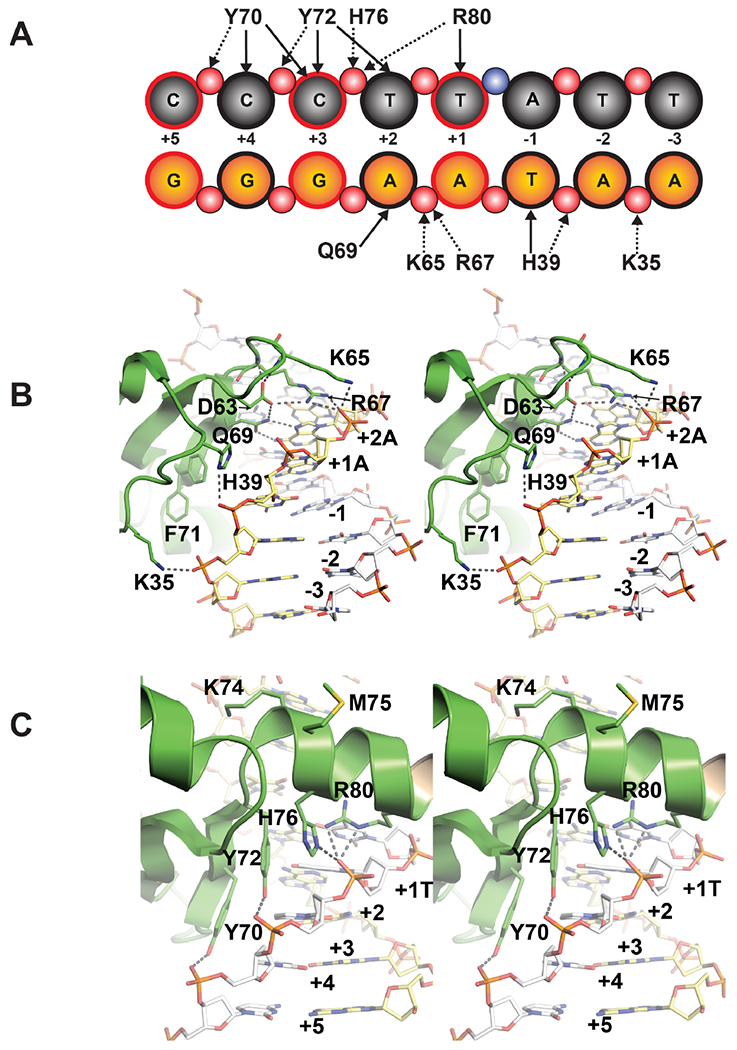
(A) Schematic representation of contacts between TopN and a segment of the 24-mer DNA duplex containing the 5’-CCCTT-3’ consensus sequence (spDNA). The grey circles represent the nucleosides of the scissile strand and orange circles represent those of the non-scissile strand; red circles represent backbone phosphate groups. Hydrophobic contacts and hydrogen bonds are represented by solid and dashed arrows, respectively. The scissile phosphate, represented by the blue circle, lies between the +1T and −1A nucleosides. The details of the contacts are summarized in Table S1. Bases that have been altered to generate the nsDNA duplex from the spDNA duplex are shown with a red (instead of a black) outline. Selected sidechain contacts made by TopN with the non-scissile strand (B) and the scissile strand (C) are shown as stereo images from PDB ID: 3IGC. Key residues and nucleobases are numbered.
Whereas the N domain of vTopIB clearly contributes to DNA binding and cleavage kinetics, it is not known whether it possesses an inherent DNA binding ability. Here we employ biophysical methods (isothermal calorimetry and NMR) to address three questions: (i) Does the N domain have an inherent ability to bind duplex DNA? (ii) Does the isolated N domain interact with duplex DNA in a mode that is consistent with that seen in the context of full-length vTopIB? (iii) Does the N domain display selectivity in its interaction with the CCCTT target sequence? Additionally, based on the findings here, and guided by crystal structures of poxvirus TopIB, we extend the mutational analysis of full-length vTopIB to a series of double-alanine mutations in the N domain at residues implicated in site recognition.
MATERIALS AND METHODS
Recombinant vTopIB N-domain
DNA encoding the N domain of vTopIB (amino acids 1-80, henceforth referred to as TopN) was PCR amplified from a pET-21b plasmid template encoding the full-length protein using an antisense primer that introduced a BamH1 site downstream of a stop codon at amino acid 81. The PCR product was inserted between the NdeI and BamH1 restriction sites of a pET-15b expression vector. The resulting plasmid encoding TopN fused to an N-terminal thrombin cleavable (His)6-tag was transformed into E. coli BL21 CodonPlus (DE3) RIL competent cells (Agilent Technologies). Cells from a single ampicillin-resistant transformant were introduced into M9 media containing ampicillin (100 mg/mL) and grown at 37 °C until the A600 reached 0.6. The M9 medium contained 1 g/L of 15NH4Cl (for uniform 15N-labeling) and 1.5 g/L 13C6-D-glucose (for uniform 15N, 13C-labeling) as the only sources of nitrogen and carbon, respectively. Expression was induced by addition of isopropyl β-D-1-thiogalactopyranoside (IPTG) to a final concentration of 400 μM. The temperature was reduced to 16 °C following induction and cells were grown under agitation for 24 hours. Bacteria were then harvested by centrifugation at 6750 g for 20 minutes at 4 °C and re-suspended in lysis buffer A (50 mM Tris-HCl, pH 7.5, 1 M NaCl, 10% glycerol) supplemented with 1 tablet of protease inhibitor (complete mini EDTA-free, Roche). Cells were lysed by sonication on ice and debris was removed by centrifugation at 24,400 g for 60 minutes. The soluble fraction was applied to a Talon cobalt metal affinity-column (Clontech Laboratories Inc.) pre-equilibrated with buffer B (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 10% glycerol) and incubated for 30 minutes. The (His)6-tagged protein was eluted with buffer B containing 400 mM imidazole and the eluate was dialyzed into buffer C (50 mM Tris-HCl, pH 8.5, 150 mM NaCl, 2.5 mM CaCl2) and cleaved with 1 unit of human thrombin (Enzyme Research) per 10 mg of fusion protein. The completion of the cleavage reaction was verified by SDS-PAGE. TopN was further purified by size exclusion chromatography using a Superdex 75 10/300 GL gel-filtration column (GE Biosciences) pre-equilibrated with either buffer D (50 mM sodium phosphate, pH 6.8, 150 mM NaCl, 1 mM DTT) or buffer E (25 mM 2-(N-morpholino) ethanesulfonic acid (MES), pH 6.4; henceforth referred to as the NMR buffer). Assignment of backbone resonances of TopN was carried out in buffer D and DNA titrations and relaxation measurements were carried out in NMR buffer. All NMR samples contained 10% D2O to facilitate field locking during NMR experiments. When partial deuteration was required, the H2O-based M9 media (described above) were replaced with D2O-based M9 media, and the expression and purification of TopN was carried out as described above.
Isothermal Titration Calorimetry
Isothermal titration calorimetry (ITC) measurements were performed using an iTC200 microcalorimeter (Malvern). Purified TopN was concentrated to 794 μM in NMR buffer by iterative centrifugal ultrafiltration using a filter with a cutoff size of 3 kDa (Millipore). 4 mM dsDNA stock solutions were used to prepare solutions of ~25 μM in NMR buffer. Reverse mode titrations were performed with DNA solutions or NMR buffer in the sample cell and TopN solution in the syringe. Titrations consisted of a preliminary 0.4 μL injection followed by 38 injections of 0.9 μL over 120 min. The results were fitted to a single-site binding model using the Origin 7.0 ITC data analysis module. More complex binding models were also attempted in the presence of the DNA duplex lacking the CCCTT target sequence (nsDNA, see below), but these did not produce fits with physically reasonable parameters. All measurements were performed in duplicate at 25 °C.
NMR Spectroscopy
NMR spectra were acquired using 15N-labeled or 15N, 13C-labeled or 15N, 2H-labeled or 15N, 13C, 2H-labeled TopN on Bruker Avance spectrometers operating at 500 MHz, 600 MHz, 700 MHz or 800 MHz or a Varian Inova spectrometer operating at 600 MHz. All spectrometers were equipped with cryogenic probes capable of applying pulsed field gradients along the z-axis. NMR experiments were performed at 25 °C. Spectral data were processed using NMRPipe31 and analyzed using the NMRViewJ32 software suite.
Backbone assignments were performed on TopN (~300 μM) using a standard backbone-directed triple-resonance strategy33. The following experiments were performed: HNCO (600 MHz; 512, 32 and 32 complex points with sweep-widths of 13.3, 33 and 13.0 ppm for the 1H, 15N and 13C dimensions, respectively), CBCANH, CBCA(CO)NH (600 MHz; 512, 32 and 52 complex points with sweep-widths of 13.3, 33 and 70 ppm for the 1H, 15N and 13C dimensions, respectively), and (H)C(CO)NH-TOCSY (600 MHz; 512, 32 and 64 complex points with sweep-widths of 13.3, 33 and 80 ppm for the 1H, 15N and 13C dimensions, respectively; mixing time = 18.2 ms). Representative spectra are shown in Figure S2. Whereas a significant number of resonance assignments were transferable to the various DNA-bound states of TopN from those in the apo state, additional experiments were necessary to complete the resonance assignments in the presence of DNA (see Figure S2 for representative spectra). For TopN in the presence of spDNA, assignments were obtained for a sample containing a protein:DNA ratio of 1:2 in NMR buffer. The following experiments were acquired: HNCO (700 MHz; 512, 32, 50 complex points and sweep-widths of 12, 35 and 13 ppm for the 1H, 15N and 13C dimensions, respectively), HNCA (700 MHz; 512, 32, and 60 complex points and sweep-widths of 12, 35 and 36 ppm for the 1H, 15N and 13C dimensions, respectively), and CBCA(CO)NH (700 MHz; 512, 32, 50 complex points and sweep-widths of 12, 35 and 65 ppm for the 1H, 15N and 13C dimensions, respectively). These spectra were acquired using non-uniform sampling and a 30% sampling schedule34. Time domain signals were reconstructed using the MDDGui software35 and processed as usual.
NMR-based Titrations
All titrations were carried out using 200 μM samples of TopN (15N-labeled for salt and DNA titrations at 500 or 600 MHz; 15N, 2H-labeled for DNA titrations at 800 MHz) in NMR buffer unless specified otherwise. The influence of salt on TopN chemical shifts was determined by acquiring 1H, 15N HSQC spectra (600 MHz; 512, 64 complex points and spectral-widths of 11.7 and 34 ppm for the 1H and 15N dimensions, respectively) at salt concentrations of 0, 50, 100 and 150 mM NaCl. Two additional sets of spectra were acquired for the sample containing 150 mM NaCl by addition of phosphate to final concentrations of 25 and 50 mM.
The influence of the dsDNA sequence on TopN was determined by utilizing 1H, 15N HSQC spectra at various concentrations of 24-mer spDNA and nsDNA duplexes. The spDNA sequence consisted of a duplex (Figure 1) previously used by Morham and Shuman36 containing the CCCTT sequence targeted by vTopIB. The nsDNA oligo was generated from spDNA by replacing the CCCTT sequence in the scissile stand (and its corresponding complementary bases in the non-scissile strand of the duplex) by ACGTG, while keeping the rest of the bases identical between the two constructs. DNA was purchased (IDT) as HPLC-purified single stranded oligomers and annealed in a PCR oven overnight. For spDNA (TopN concentration = 200 μM), 1H, 15N HSQC spectra (500 MHz; 512, 64 complex points and sweep-widths of 12.0 and 35 ppm for the 1H and 15N dimensions, respectively) were acquired for samples with final spDNA concentrations of 0, 10, 21.6, 25, 50, 59.6, 78.6, 100, 150, 200, 250, 300 and 400 μM. Additional 1H, 15N HSQC spectra (800 MHz; 512, 128 complex points and spectral-widths of 13 and 26 ppm for 1H and 15N dimensions, respectively) were acquired for samples with final spDNA concentrations of 400, 600 and 800 μM.
Corresponding titrations for nsDNA (TopN concentration = 200 μM) were performed using 1H, 15N-HSQC spectra acquired at 600 MHz (512, 128 complex points with spectral widths of 13.3 and 33 ppm for the 1H and 15N dimensions, respectively) or 800 MHz (512, 128 complex points and spectral widths of 13 and 26 ppm for the 1H and 15N dimensions, respectively) for samples with final DNA concentrations of 0, 20, 40, 60, 80, 100, 120, 140, 160, 200, 250, 300 and 400 μM at 600 MHz or 400, 600 and 800 μM at 800 MHz.
Titrations were also performed with a 21-mer DNA sequence that deviated significantly from both spDNA and nsDNA (nsDNA21, see top panel of Figure S4 for the sequence; TopN concentration = 60.7 μM unless otherwise stated) were performed using 1H, 15N-HSQC spectra acquired at 600 MHz (512, 128 complex points and spectral widths of 11.7 and 33.7 ppm for the 1H and 15N dimensions, respectively) for samples with final DNA concentrations of 0, 6 (TopN concentration = 74 μM), 30, 61 and 121 μM.
Chemical shift perturbations (CSPs, Δδ in ppm) for backbone amide resonances were calculated per residue using the following equation:
| (1) |
Where ΔδH and ΔδN represent the chemical shift differences in the 1H and 15N dimensions, respectively in the presence of a titrant. The chemical shifts of the apo state were used as reference unless otherwise stated.
NMR Spin-relaxation Measurements
Relaxation measurements were performed on 2H, 15N-labeled TopN samples in NMR buffer in either the apo state (TopN concentration = 400 μM) or in the presence of unlabeled 800 μM spDNA or 800 μM nsDNA (TopN concentrations in both cases = 200 μM). All relaxation experiments were performed at 800 MHz. Spin-lattice relaxation rates (R1) were measured using relaxation delays of 0.016, 0.064, 0.144, 0.256 (x2), 0.384, 0.544, 0.8 and 1.2 seconds for the apo state and the DNA complexes (512, 50 complex points and sweep widths of 13.9, 26 ppm for the 1H and 15N dimensions, respectively). Relaxation delays used for the measurement of spin-spin relaxation rates (R2) were 0, 0.016, 0.033, 0.049 (x2), 0.065, 0.082, 0.098 and 0.114 seconds for the apo state and in the presence of nsDNA (512, 50 complex points and sweep widths of 13.9, 26 ppm for the 1H and 15N dimensions, respectively) and 0, 0.016, 0.033, 0.049, 0.065, 0.115, 0.131 and 0.163 seconds in the presence of spDNA (512, 50 complex points and sweep widths of 13.9, 26 ppm for the 1H and 15N dimensions, respectively). {1H}-15N steady-state NOEs were acquired using the procedures described by Ferrage et. al37. Recycle delays of 6.5 seconds (512, 128 complex points and sweep widths of 13.9, 26 ppm for the 1H and 15N dimensions, respectively) with or without (for the reference experiment) 3 seconds of 1H saturation. Relaxation data were analyzed using the built-in functions available in the NMRViewJ suite32.
The rotational diffusion tensor of TopN alone and those in the presence of spDNA or nsDNA were determined employing methodology described before38 and using the R1, R2 and {1H}-15N steady-state NOEs from above. Residues that displayed {1H}-15N NOE values less than 0.6 or those determined to contain significant exchange contributions to their spin-spin relaxation rates (R2/R1 ratio larger than the mean by more than 1.5 times the standard deviation) were not used in the calculations. The co-ordinates for unit vectors of the backbone amides were obtained after addition of protons to the DNA-bound full-length structure of TopIB (PDB ID: 3IGC)8 using UCSF Chimera39. Relaxation data for F71 (and for Y72 that immediately follows it) that showed orientational heterogeneity in the crystal structure were not utilized in determining the rotational diffusion tensor. Three models, fully-asymmetric, axially-symmetric and isotropic diffusion, were tested and their statistical significance was determined by using the F-test. P values smaller than 0.05 were considered to be statistically significant.
Preparation of vTopIB Mutants
Double-alanine mutations were introduced into the vTopIB gene by two-stage PCR overlap extension. NdeI-BglII restriction fragments containing the mutated vTopIB genes were cloned into the T7-based expression vector pET16b. The inserts of wild-type and mutant vTopIB plasmids were sequenced completely to exclude the acquisition of unwanted changes during amplification and cloning. pET-vTopIB plasmids were transformed into E. coli BL21(DE3). Cultures (100 mL) derived from single transformants were grown at 37 °C in LB medium containing 0.1 mg/mL ampicillin until the A600 reached 0.6. The cultures were adjusted to 1 mM IPTG and incubation was continued at 37 °C for 3 hours with constant shaking. The cells were harvested by centrifugation, and the pellets were stored at −80 °C. All subsequent steps were performed at 4 °C. Thawed cells were re-suspended in 10 mL of buffer A’ (50 mM Tris-HCl, pH 7.5, 0.25 M NaCl, 10% sucrose). Lysozyme and Triton X-100 were added to final concentrations of 1 mg/mL and 0.1%, respectively. The lysates were sonicated to reduce viscosity, and insoluble material was removed by centrifugation for 50 minutes at 15,000 rpm in a Sorvall SS34 rotor. The supernatants were applied to 2 mL columns of Ni2+-nitrilotriacetic acid-agarose that had been equilibrated with buffer A’. The columns were washed with 10 mL of the same buffer and then eluted stepwise with 2 mL aliquots of 50, 100, 200, and 500 mM imidazole in buffer B’ (50 mM Tris-HCl, pH 8.0, 50 mM NaCl, 10% glycerol). The protein compositions of the column fractions were monitored by SDS-PAGE. Recombinant His10-tagged vTopIB was recovered predominantly in the 200 and 500 mM imidazole eluate fractions, which were combined and dialyzed against 50 mM NaCl in buffer C’ (50 mM Tris-HCl, pH 8.0, 2 mM EDTA, 5 mM dithiothreitol, 10% glycerol). The dialysates were applied to 1 mL columns of phosphocellulose that were pre-equilibrated with buffer C’. The columns were washed with 10 mL of buffer C’ and then eluted stepwise with 4 mL aliquots of 0.2, 0.5, and 1 M NaCl in 50 mM Tris-HCl (pH 7.5), 10% glycerol. vTopIB was recovered in the 1 M NaCl eluate fraction. The purified proteins were stored at −80°C.
Effect of Double-alanine Mutations on DNA Cleavage
Single-turnover DNA cleavage reaction mixtures containing (per 20 μL) 50 mM Tris-HCl (pH 7.5), 0.3 pmol (15 nM) 5′-32P-labeled 18-mer/30-mer DNA substrate, and 75 ng (~2 pmol; ~100 nM) wild-type (WT) or mutant vTopIB were incubated at 37 °C. The cleavage reactions were initiated by adding vTopIB to pre-warmed reaction mixtures. Aliquots (20 μL) were withdrawn at 5, 10, 20, 30, 60, 120 and 300 seconds and the reactions were quenched immediately by adding SDS to 1%. The samples were digested for 1 hour at 37 °C with 10 μg proteinase K, then adjusted to 47% formamide, heat-denatured, and analyzed by electrophoresis through a 17% polyacrylamide gel containing 7 M urea in TBE (90 mM Tris-borate, 2.5 mM EDTA). The reaction products (32P-labeled DNA-peptide adducts and residual uncleaved 18-mer DNA) were quantified by scanning the gel with a phosphorimager. The percent of input DNA cleaved at each time point (an average of two or three independent cleavage experiments ± SEM) was plotted as a function of time. All of the proteins attained endpoint values of 92-96% DNA cleavage. The apparent cleavage rates constants (kcl) were calculated by nonlinear regression fitting of the data to a one-phase association model in GraphPad Prism.
Effect of Salt on DNA Cleavage
Reaction mixtures (20 μL) containing 50 mM Tris-HCl (pH 7.5), 0.3 pmol 32P-labeled 18-mer/30-mer DNA, 75 ng vTopIB, and 0, 50, 100, 150, or 200 mM NaCl were incubated at 37 °C for either 10 seconds (WT, K34A-H39A, D63A-K65A), 30 seconds (H76A-R80A), or 3 minutes (R67A-Q69A). The reactions were quenched with SDS and the products were analyzed as described above. The extents of covalent complex formation were normalized to that of a control reaction without added salt (defined as 100%) and plotted as a function of NaCl concentration. Each datum is the average of two independent experiments.
RESULTS AND DISCUSSION
TopN Interacts with Duplex DNA
To test the ability of TopN to interact with dsDNA we utilized isothermal titration calorimetry (ITC). ITC measurements indicated that the 24-mer dsDNA carrying the consensus CCCTT sequence (spDNA) binds TopN with a slightly higher apparent affinity (KD = 17.0±3.2 μM; average of two independent measurements) compared to dsDNA that does not (nsDNA, KD = 37.7±1.0 μM) (see Figure 2 for representative examples). In comparison, using a nitrocellulose filter-binding assay, an apparent KD value for full-length vTopIB towards the consensus sequence was estimated to be ~4 nM under conditions similar to those used here13. While extracting accurate ΔH and ΔS data from thermograms such as the ones shown in Figure 2 is difficult, some interesting trends are noted. The binding is exothermic in both cases with ΔH=−20.8±1.3 Kcal•mol−1 and −23.5±2.7 Kcal•mol−1 for spDNA and nsDNA respectively. This is characteristic of a protein that engages the major groove of dsDNA40. The entropy change is also negative in both cases and slightly more favorable for spDNA than for nsDNA (ΔS= −48.0±4.6 and −58.5±8.9 cal•mol−1•deg−1 for spDNA and nsDNA respectively), suggesting a marginally larger electrostatic contribution to binding in the former case40. Having established the ability of TopN to engage dsDNA (albeit with ~4200-fold less avidity than full-length vTopIB), we proceeded to further investigate the nature of the interaction in residue-specific detail using solution NMR techniques.
Figure 2.
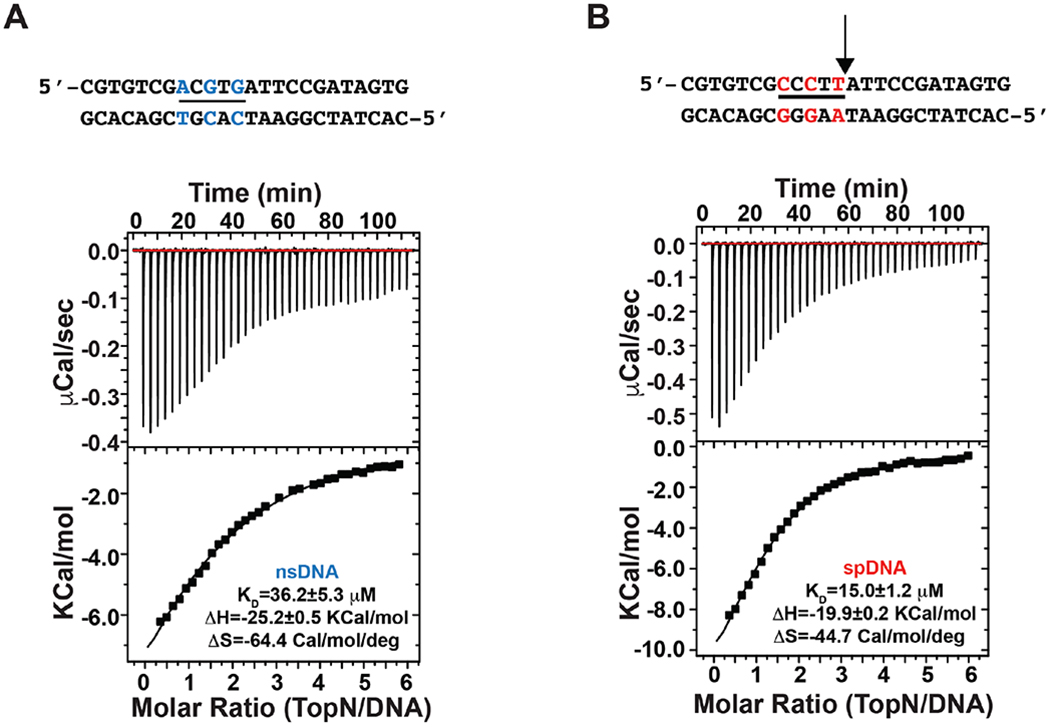
Measurement of the binding of TopN to nsDNA (A) and spDNA (B) duplexes via isothermal calorimetry (ITC). The data shown are representative of duplicate measurements. DNA duplexes used are shown at the top of the corresponding ITC traces. The bases of the specific CCCTT (underlined) sequence on the spDNA (red lettering) that have been altered to generate the nsDNA (blue lettering) are indicated. The arrow indicates the site of cleavage by vTopIB. The binding affinity (KD) and the corresponding ΔH and ΔS values obtained from the fits of the data represented are also indicated.
Resonance Assignment of TopN
The 1H, 15N HSQC spectrum of the isolated TopN shows excellent dispersion (Figure 3) and standard triple-resonance strategies33 could be utilized to obtain complete assignments of backbone 1H, 15N, 13Cα, 13C’ and sidechain 13Cβ resonances (see Figure S2 for representative data). 1H, 15N resonance assignments were also obtained for sidechain amides of N15, N16, N19, N25, Q32, Q69, Q78 and Q79 positions and the indole NH of W50. The secondary structure derived from chemical shifts using the PINE server41 indicates α-helices between N25-Q32 and between T49-L54; β-strands between A3-Y6, G9-F12, D42-V45, L57-G62, and R68-F71. These locations are largely consistent with that seen in the crystal structures of the isolated N domain6 and full-length poxvirus TopIB bound to dsDNA7, 8.
Figure 3.
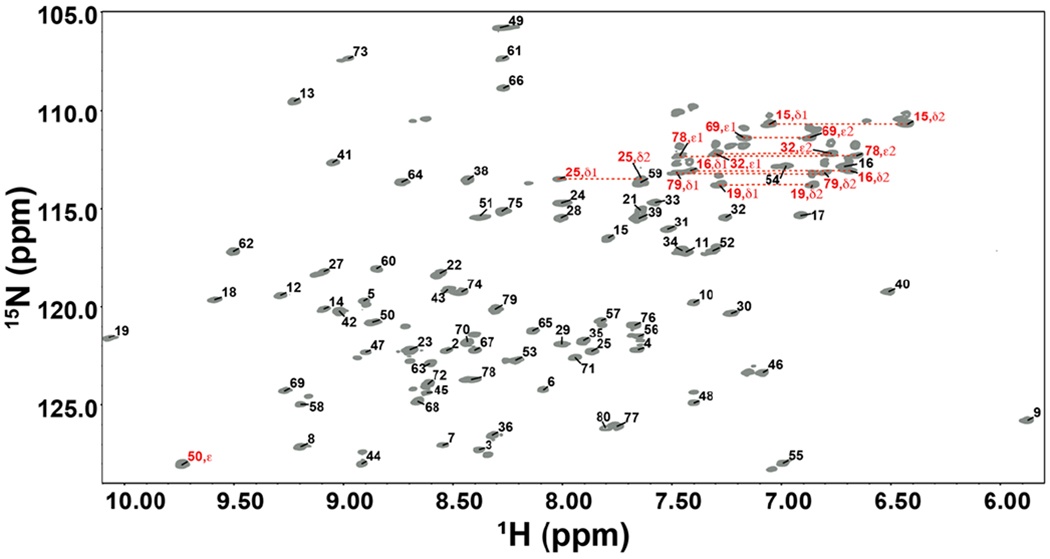
Assigned 15N, 1H HSQC (600 MHz) of TopN. Backbone assignments are labeled in black and sidechain assignments are labeled in red.
TopN Utilizes a Surface Similar to the N Domain in Full-length vTopIB to Engage DNA
An inspection of 15N, 1H HSQC spectra of TopN in the presence of a two-fold molar excess of spDNA (mean ± SD over the protein sequence, Δδ=0.10±0.10 ppm) or nsDNA (Δδ=0.11±0.11 ppm) reveals several chemical shift perturbations (CSPs, Figure 4) that are greater than those arising from the effects of salt (or phosphate) alone (Figure S3). The largest CSPs in the presence of spDNA or nsDNA are at the N-terminus, in the loop between α1 and β3, the loop between β4 and β5, and at the C-terminus. The relative magnitudes of the CSPs obtained in the presence of DNA (a polyelectrolyte) compared with those obtained in the presence of salt alone (see Figure S3) suggest that the CSPs are indicative of engagement with DNA.
Figure 4.

(A) Chemical shift perturbations (CSPs) in TopN in the presence of two molar equivalents of either spDNA (black) or nsDNA (red). Titrations were carried out under low salt conditions in NMR buffer (see text). Regions encompassing secondary structure elements are shaded and labeled. The C-terminal segment (shaded cyan, labeled α3) is helical only in the context of the full-length structure in complex with DNA. Amino acids whose sidechains make contact with DNA in the structure of the transition-state mimic of full-length vTopIB are indicated by (*). (B) For spatial representation, the CSPs in the presence of spDNA (top) and nsDNA (bottom) have been mapped onto the surface of the N-terminal domain in the crystal structure of vTopIB bound to the consensus sequence. Key residues that display significant perturbations are labeled. The C domain has been omitted for clarity.
Large CSPs (Δδ >0.18 ppm; Figure 4) seen at the C-terminus and in the α1-β3 and β4-β5 loops are largely consistent with the TopN-specific DNA contacts expected from the crystal structure of the complex of full-length TopIB with dsDNA containing the CCCTT sequence (Figure 1, Table S1). Significant CSPs in the α1-β3 loop in the presence of spDNA or nsDNA are noted for V34 (Δδ=0.24 ppm for spDNA, Δδ=0.30 ppm for nsDNA), K35 (0.25, 0.25), T38 (0.28, 0.38) and H39 (0.22, 0.44). K35 contacts the phosphate backbone of the non-scissile strand of the DNA duplex in the crystal structure, while H39 contacts both the sugar of the −1T position of the non-scissile strand and the phosphate backbone. Whereas D63 in the β4-β5 loop shows large perturbations for both spDNA and nsDNA (0.26, 0.20), S64 displays a large CSP value only for spDNA (0.23, 0.09). The nature of CSPs at the TopN C-terminus that contacts the scissile strand including several base-specific contacts (Y70 that contacts +3C and +4C; Y72 that contacts +2T and +3C; R80 that contacts +1T, see Figure 1, Table S1) are largely consistent between the two duplexes used in this study. Y70 (0.22, 0.19), K74 (0.26, 0.21), H76 (0.27, 0.30), V77 (0.44, 0.43), Q78 (0.19, 0.18) and N79 (0.29, 0.29) all show large perturbations for both spDNA and nsDNA. Similar overall patterns of CSPs are also seen in the presence of 21-mer DNA duplex (nsDNA21) that deviates significantly from both spDNA as well as nsDNA (see Figure S4) suggesting that the same regions of TopN are involved in engaging duplex DNA irrespective of the nucleotide sequence. Some large perturbations are also seen at the extreme N-terminus of TopN (specifically on M1, S2 and A3) in the presence of both spDNA and nsDNA (Figure 4). This region also shows a larger than average chemical shift response to the presence of salt alone (especially in the presence of phosphate, Figure S3). It is possible that these CSPs result from additional contacts of the flexible N-terminus with the phosphate backbone of the duplex DNA.
Differences in TopN Interactions with Specific and Non-specific DNA Duplexes
Changes in individual 1H and 15N resonance positions (including the sign of the shift i.e. upfield or downfield) induced on TopN by spDNA correlate extremely well (see Figure S5) with those induced by nsDNA, as indicated by the slopes (1H = 0.97, 15N = 0.98) and correlation coefficients (1H = 0.96, 15N = 0.97) both approaching 1, excluding a few significant outliers (1H: H39, L40, T49 and S64; 15N: T38, H39, R68, Q69, F71, R80). This suggests that the overall TopN binding modes of spDNA and nsDNA are similar, as indicated above. However several residues show significant differences in CSPs that exceed the mean by at least 3-times the standard deviations (>0.19 ppm, mean±SD = 0.04±0.05 ppm calculated over the sequence using the 1:2 TopN:spDNA state as reference, Figure 5). The largest differences involve H39 (Δδ=0.22 ppm) and L40 (Δδ=0.25 ppm). As mentioned above, H39 contacts the −1T and its sidechain is positioned between the +1 and −1 positions of the non-scissile strand. At ratios that exceed 1:2 TopN:spDNA (1:3, 1:4), no further changes in the CSP values are seen for either H39 or L40 (Figure S6). This contrasts with the gradually increasing perturbations seen for these residues beyond 2 molar equivalents of nsDNA (Figure S6). The non-linear trajectory of these resonances indicates that the association with nsDNA at these ratios deviates from a simple binding/unbinding behavior and suggests complex association dynamics at high ratios of nsDNA. A significant difference in the nature of perturbations induced by the two DNA duplexes is also seen for T49 (Δδ=0.19 ppm). In fact, this residue shows a high degree of perturbation in the presence of nsDNA but is largely unperturbed by spDNA. This effect is difficult to rationalize structurally given that in the TopIB•dsDNA structures it faces away from the bound DNA. It is likely that this is the result of a slight difference in the average orientation (and contacts with the DNA as mentioned above) of the extreme N-terminus (that lies in close proximity to T49) between the two cases. Note that since the N-terminus itself is flexible on the chemical shift timescale it does not itself show very large differences in perturbations. This is partially borne out by the enhanced fast dynamics (data not shown) for M1 in the presence of nsDNA compared to spDNA and an above average difference in CSPs (0.08 ppm) between the two cases.
Figure 5.
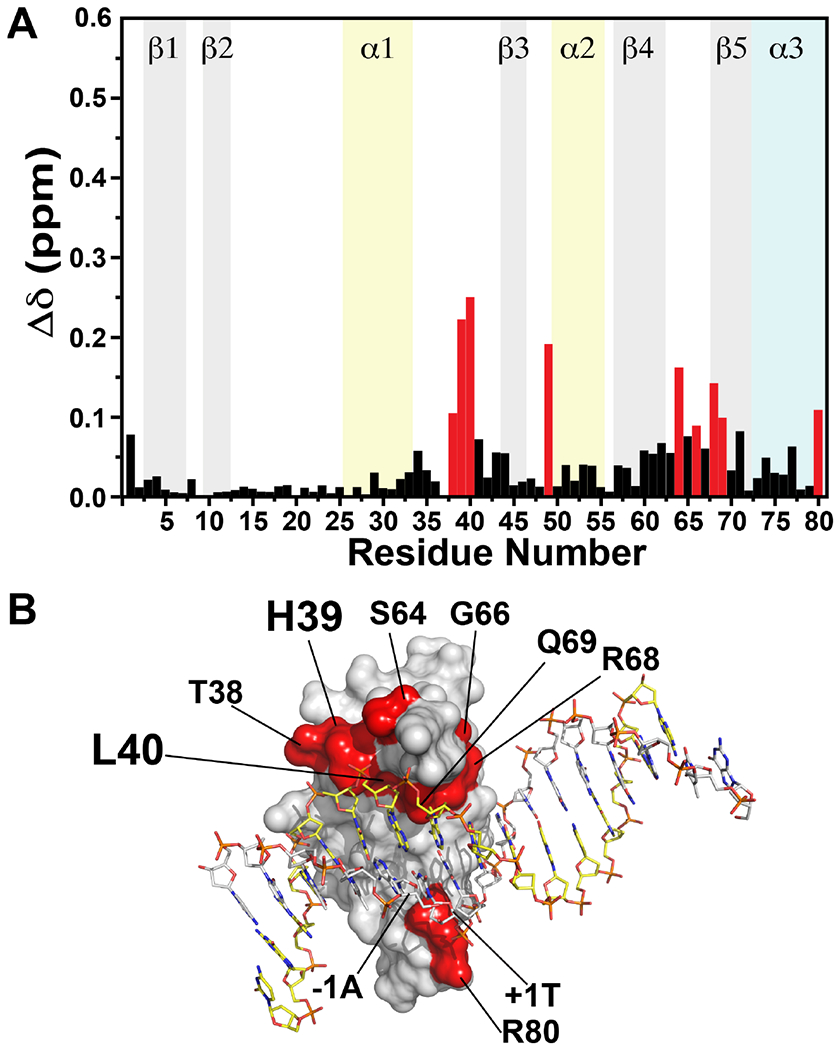
(A) Differences in chemical shifts for TopN in the presence of two molar equivalents of spDNA or nsDNA. Resonances in the presence of spDNA were used as reference to calculate the perturbations using Equation 1. Residues with the largest differences are shown in red. (B) Residues with significant differences in resonance positions in the presence of spDNA or nsDNA are mapped onto the surface of the N domain in the crystal structure of of vTopIB bound to the consensus sequence, and colored red. H39 and L40 that exhibit the most significant differences are labeled in larger font. The C domain has been omitted for clarity.
Additional smaller but significant differences (>0.09 ppm, mean + 1 SD) between the spDNA and nsDNA states are seen for T38, S64, G66, R68 and Q69, all of which lie on the surface that contacts the non-scissile strand. A significant difference (0.11 ppm) is also seen for R80 that makes a specific contact with the +1T position in spDNA (Figure 1, Table S1). Inspection of the sidechain resonances of Q69 (that makes a base-specific contact with the +2A of the non-scissile strand, Table S1) also reveals a significant difference in behavior in the presence of spDNA and nsDNA (Figure S7).
To explore electrostatic contributions to the interactions of spDNA and nsDNA with TopN, we studied the effect of increasing salt concentration on the stability of the complex as reflected by an alteration in the magnitude of the CSPs. Overall, the backbone resonances of TopN in the presence of spDNA appear to be more sensitive to salt content as expected from the ITC results discussed above, where the less negative ΔS value suggests a somewhat larger electrostatic contribution towards binding of spDNA compared to nsDNA40. At the highest salt content used in these studies (150 mM NaCl) most TopN resonances move very close to their positions in the apo state in NMR buffer containing the same level of salt suggesting a lack of interaction with DNA. A similar behavior is seen for the sidechain of Q69 for which the ε-resonances that are significantly broadened in the presence of spDNA (see Figure S7) are largely restored to their apo state in presence of 100 mM NaCl.
In order to best represent the salt sensitivity of the resonance positions of residues that are significantly perturbed in the presence of duplex DNA, we defined the following parameter (Δnorm,salt)
| (2) |
Δδapo,DNA represents the CSPs in the presence of either spDNA or nsDNA (at a 1:2 TopN:DNA ratio) referenced to the apo state in low-salt (0 mM NaCl) NMR buffer; Δδhigh,low represents the perturbations induced by either spDNA or nsDNA in high-salt (150 mM NaCl) NMR buffer referenced to low-salt NMR buffer. Δnorm,salt values vary from 0 to 1, with larger numbers representing residues that are significantly perturbed by the presence of DNA but show the least sensitivity to salt. As evident from Figure 6, the largest difference in Δnorm,salt values between spDNA and nsDNA is seen for H39. Additional residues, L40 and T49 also show large differences in Δnorm,salt values between spDNA and nsDNA. However, the Δnorm,salt values for these latter residues in the presence of spDNA are small due to small individual CSPs (Figure 4, also see below) leading to reduced values of the second factor in Equation 2.
Figure 6.
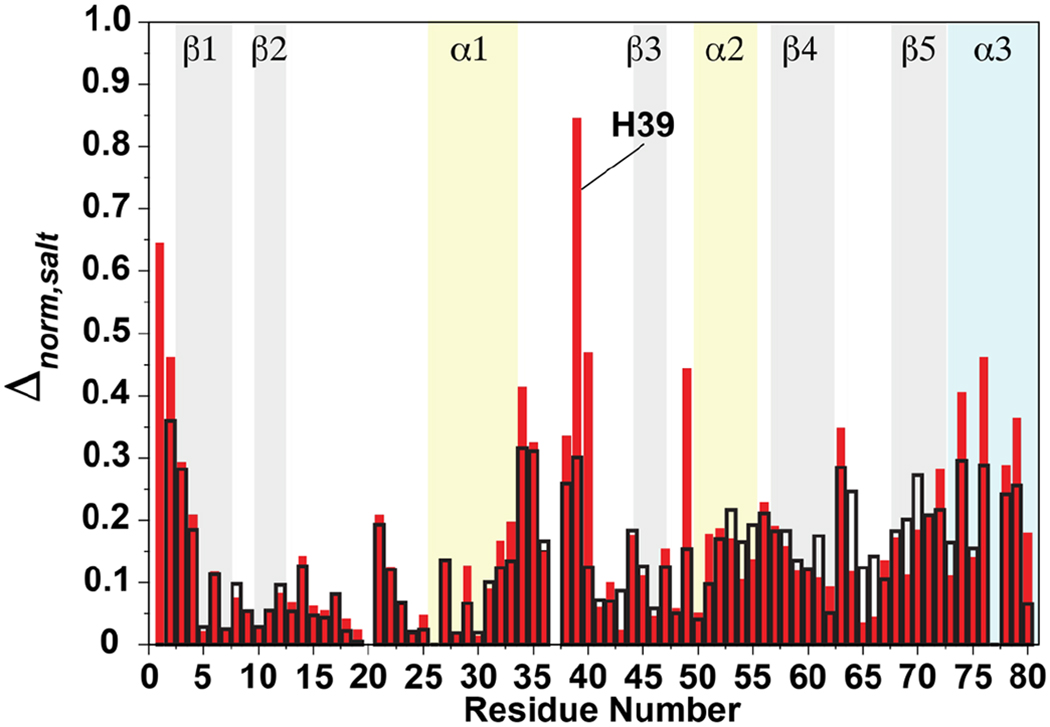
Δnorm,salt values calculated using Equation 2 for spDNA (open black bars) and nsDNA (closed red bars) plotted against residue number.
The crystal structures7, 8 of the vTopIB•dsDNA complex point to a polar interaction between the sidechain of H39 and the phosphate backbone of the non-scissile strand (Table S1). It appears that this mode of interaction is maintained for the association of TopN with spDNA. This is evident from the CSP of H39, that while substantial in the absence of salt, is significantly reduced with increasing salt concentration, suggesting a weakened interaction (Figure S3B). In contrast for nsDNA, H39 (that also has a large CSP in the low salt buffer) is less sensitive to salt compared to its behavior in the presence of spDNA (Figure S3C). A similar overall, though less drastic, trend is seen for T38 (see Figure S3; hence the smaller difference in Δnorm,salt values). L40 that has a significant CSP in the presence of nsDNA (but not for spDNA, hence the small Δnorm,salt value) has a minimal response to the presence of salt (see Figure S3C). Overall, these data suggest that electrostatics play a less significant role in the interactions involving at least part of the α1–β3 loop and nsDNA. For the other regions involved in binding dsDNA, some small differences in Δnorm,salt values are also seen at the C-terminus (e.g. for K74, H76, N79) suggesting some differences in electrostatic interactions between spDNA and nsDNA involving the C-terminus of TopN.
Hydrodynamics of TopoN in the Presence of Duplex DNA
To determine the change in the overall hydrodynamic properties of TopN upon interaction with duplex DNA we measured the 15N spin-lattice (R1) and spin-spin (R2) relaxation rates, together with {1H}-15N steady-state NOEs at 800 MHz in the apo state and in the presence of four molar equivalents of either spDNA or nsDNA (Figure S8). For apo TopN, analysis of the relaxation rates using protocols described before38, indicates an axially symmetric diffusion tensor with a modest departure from isotropic behavior (η=Dpara/Dperp=1.15±0.07) and a corresponding rotational correlation time, τc=1/(2Dpara+4Dperp)=6.1±0.1 ns. The τc value increases to 16.0±1.0 ns in the presence of spDNA providing further evidence of complex formation i.e. the generation of a significantly larger species. Additionally, there is a significant increase in motional anisotropy (η=1.42±0.23) as is expected upon association of a small, globular protein domain with a linear DNA duplex of significant size. This scenario is confirmed by a HYDRONMR42 calculation of a 1:1 complex TopN•spDNA complex based on the crystal structure (after removal of the C domain) that yields η=2.03 (τc=22.3 ns). For nsDNA, the isotropic diffusion tensor model that was deemed viable based on statistical analyses (see MATERIALS AND METHODS) showed a more significant increase in the τc=1/6Diso of 18.5±0.1 ns. The axially symmetric model (that did not meet the threshold of statistical significance) also predicts a larger τc=19.2±1.1 ns together with a substantial η=1.26±0.19. Analysis of relaxation data at a lower TopN:nsDNA molar ratio (1:2; data not shown) also yields a higher τc=17.4±0.3 ns and η=1.35±0.06. The larger τc for the nsDNA state compared to the spDNA state likely suggests a complex ensemble possibly containing species with multiple sites on the nsDNA being occupied by TopN molecules. Note that formation of small populations of minor species could also result in the increased global exchange contribution to the spin-spin relaxation rates (R2). Our procedure to eliminate relaxation rates with significant exchange contributions from the hydrodynamic calculations (described in the MATERIAL AND METHODS section) could be prone to inaccuracies when a significant proportion of R2 rates contain such contributions leading to errors in the rotational diffusion parameters extracted. However, the R1 rates for TopN in the presence of spDNA (mean±SD over the sequence, 0.70±0.20 sec−1) are systematically higher (on average by about 14% on a residue-by-residue basis) than those in the presence nsDNA (0.65±0.24 sec−1) in a 1:4 protein:DNA molar ratio. This suggests that the structural ensemble contains species that are, on average, larger in the case of nsDNA. This observation together with the fact that further changes in CSPs (and their nonlinear trajectories, Figure S6) are noted in the presence of nsDNA beyond 1:2 TopN to DNA molar ratios suggests complex binding events compared with what appears to be largely stoichiometric binding (and no changes in perturbations between the 1:2 and 1:4 molar ratios) seen for spDNA.
Double-alanine Mutagenesis of the N Domain
Comprehensive mutational analysis of vTopIB has provided deep insights into the structural basis for DNA binding and transesterification. Testing the effects of mutations at 171 individual amino acids has identified 10 residues that are critical or important for the transesterification reaction and 13 residues that contribute to DNA binding 12, 26, 30, 43. We deem a residue important for DNA cleavage when the single-turnover cleavage rate constant is suppressed by at least an order of magnitude by an alanine mutation. Residues involved in DNA cleavage fall into two classes. The first class participates directly with the Y274 nucleophile in transesterification chemistry; amino acids in this category are required for both cleavage and religation reactions, and they enhance the rates of both steps by 2 to 5 orders of magnitude (e.g. R130, K167, R223, H265). The second class is selectively required during the forward cleavage reaction and has little or lesser impact on the rate of religation; such residues (G132, Y136, S204, K220, N228) are implicated in the DNA-triggered conformational changes that assemble the full active site. For most of the amino acids implicated in DNA binding, an alanine mutation has little effect on the rate of single-turnover DNA cleavage (assayed under conditions of low ionic strength), but typically renders DNA cleavage sensitive to inhibition by salt29, 30, 43. Amino acids implicated in DNA binding include N domain residues F59, R67, Q69, Y70, Y72, G73, and R80.
Mutagenesis of vTopIB using the “one residue at a time” strategy has reached a point of diminishing returns. However, the crystal structures of DNA-bound poxvirus TopIB7, 44 empower further studies by illuminating potentially redundant atomic contacts at the protein-DNA interface, some of which have been probed functionally, and to good effect, by double-alanine mutagenesis15, 26. Here we applied the double alanine approach to 5 pairs of N domain residues: K35A-H39A, D63A-K65A, R67A-Q69A, F71A-K74A, and H76A-R80A. The His-tagged wild-type and mutant proteins were produced in E. coli and partially purified from soluble bacterial extracts by sequential Ni-agarose and phosphocellulose chromatography steps. The vTopIB polypeptide constituted the major species in the protein preparations, as determined by SDS-PAGE, and the extents of purification were essentially equivalent (Figure 7A).
Figure 7.
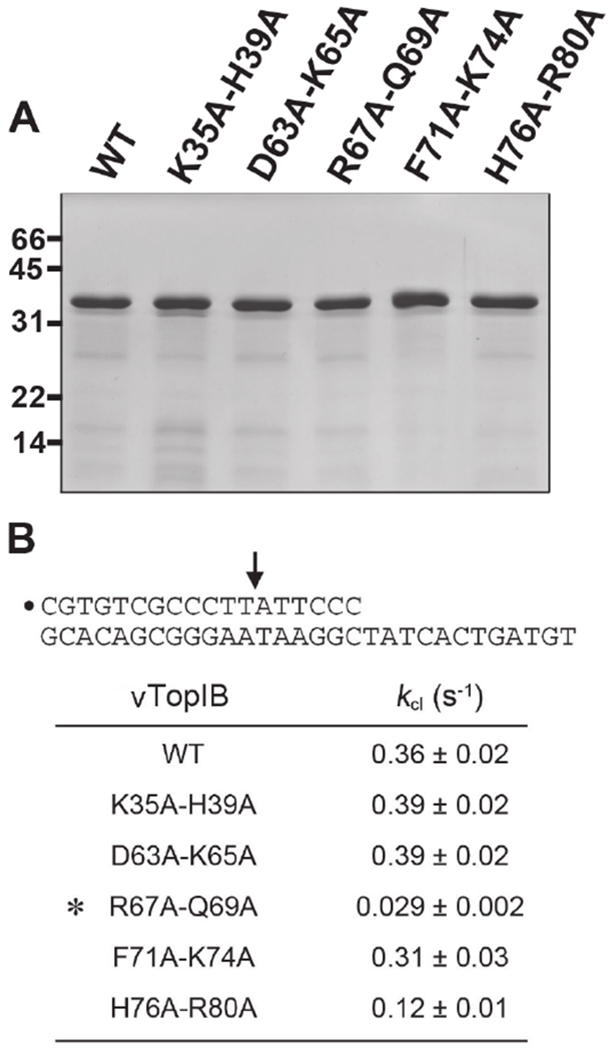
Effect of double-alanine mutations on DNA cleavage. (A) Aliquots (4 μg) of the phosphocellulose preparations of wild-type vTopIB and the indicated double-mutants were analyzed by SDS-PAGE. The Coomassie blue-stained gel is shown. The positions and sizes (kDa) of marker polypeptides are indicated on the left. (B) Single-turnover DNA cleavage reactions were performed as described under MATERIALS AND METHODS. The 5′-32P-labeled 18-mer/30-mer DNA substrate is depicted at the top with the 5′-32P-label denoted by • and the topoisomerase cleavage site indicated by an arrow. The apparent cleavage rate constants (kcl±SE) were calculated by nonlinear regression of the kinetic data to a one-phase association model. The asterisk denotes a mutation that elicits a >10-fold decrease in cleavage rate.
A suicide substrate containing a single CCCTT cleavage site was used to examine the cleavage transesterification reaction under single-turnover conditions in the absence of added salt. The substrate (shown in Figure 7B) consisted of a 5’-32P-labeled 18-mer scissile strand 5’-CGTGTCGCCCTTATTCCC annealed to an unlabeled 30-mer strand. Upon formation of the covalent protein-DNA adduct, the distal cleavage product 5’-ATTCCC is released and the topoisomerase becomes covalently trapped on the DNA. The apparent cleavage rate constant (kcl) for the wild-type enzyme was 0.36 s−1. The double mutants K35A-H39A (0.39 s−1) and D63A-K65A (0.39 s−1) were as active as wild-type in single-turnover DNA cleavage, signifying that simultaneous loss of contact of K35 and H39 with the –3 and –2 phosphates of the non-scissile strand was well tolerated, as was the simultaneous loss of the K35 contact with the scissile strand +1 phosphate and D63 coordination of R67 and Q69. The F71A-K74A mutant (0.39 s−1) was also benign. F71 and K74 are not themselves components of the DNA interface; rather they demarcate the structural transition between the N domain β4 strand into a long α helix of the catalytic domain that forms when vTopIB binds as a C-shaped clamp to the DNA duplex. The cleavage rate of the H76A-R80A mutant (0.12 s−1) was one third that of wild-type vTopIB, which does not meet our 10-fold criterion of significance for a mutational effect on cleavage. Thus, simultaneous loss of H76 and Arg80 contacts to the +3-scissile strand phosphate was fairly benign. By contrast, the cleavage rate constant of the R67A-Q69A double-mutant (0.029 s−1) was 12-fold slower than wild-type (Figure 7B) and the double mutation was more impactful than the single R67A (0.22 s−1) and Q69A (0.09 s−1) mutants characterized previously29, 30. We conclude that the Q69 contact with the –2A of the nonscissile strand and the R67 contact with the nonscissile strand –2 phosphate are collectively important for cleavage. We note that the specific contact of the Q69 sidechain with DNA in solution is underscored by the contrast in the chemical shift response of its ε position to spDNA and nsDNA as described above (Figure S7).
The effect of the R67A-Q69A double-mutation on the religation transesterification reaction was studied under single-turnover conditions by assaying the ability of preformed suicide intermediates to transfer the covalently held 5-32P-labeled 12-mer strand to a 5′ OH-terminated 18-mer strand to generate a 30-mer product. The religation reaction of wild-type TopIB was effectively complete by 5 seconds, the earliest time analyzed, in keeping with prior measurements of krel of 1.2 to 1.4 s−1 for vTopIB18, 45. The religation profile of the vTopIB-(R67A-Q69A)–DNA intermediate was indistinguishable from wild-type vTopIB, within the limits of the assay (i.e., krel was ≥0.4 s−1). Thus, the R67A-Q69A mutation specifically affects cleavage, but not religation, and hence not transesterification chemistry per se.
Effect of Salt on DNA Cleavage by N Domain Double-alanine Mutants
Many vTopIB mutants that cleave DNA with wild-type or near wild-type kinetics in the absence of salt are sensitized to inhibition of cleavage in the presence of salt, which is taken to indicate that such mutants have weakened non-covalent binding to the CCCTT target site. To address this issue for the N domain double-alanine mutants, we measured the amounts of covalent adduct formed in the presence of 50, 100, 150 and 200 mM NaCl and normalized the values to the extent of cleavage in reactions lacking salt. The reaction times for the wild-type and mutant enzymes were adjusted to attain similar extents of cleavage in the no salt controls and thus comparable sensitivity for the effects of salt. For example, cleavage was quenched after 10 seconds for WT, K34A-H39A, D63A-K65A, and F71A-K74A, 30 seconds for H76A-R80A, and 3 minutes for R67A-Q69A. We observed that the wild-type vTopIB was unaffected by up to 100 mM NaCl, but was inhibited by 20% at 150 mM NaCl and 46% at 200 mM NaCl (Fig. 8). In contrast, covalent adduct formation by each of the N domain double-alanine mutants was salt-sensitive. H76A-R80A was the most acutely sensitive, with cleavage being inhibited by 82% at 100 mM NaCl and by 95% at 150 mM NaCl, followed closely by R67A-Q69A, which was inhibited by 70% at 100 mM NaCl and by 93% at 150 mM NaCl (Fig. 8). The D63A-K65A and F71A-K74A mutants were inhibited by 38-45% at 100 mM NaCl and by 87-88% at 150 mM NaCl. The K35A-H39A protein was least sensitive, being inhibited by 21%, 69% and 91% at 100, 150, and 200 mM NaCl. We surmise that simultaneous loss of two N domain DNA contacts (or of two “structural” residues in the case of F71 and K74) compromises target site binding such that site recognition becomes sensitive to ionic strength. These results are largely consistent with NMR discussed earlier which demonstrate that K35, H39, D63, Q69, F71, K74 and H76 all show significant CSPs in the presence of spDNA and exhibit large responses to increasing concentration of salt in the buffer (Figure S3B).
Figure 8.
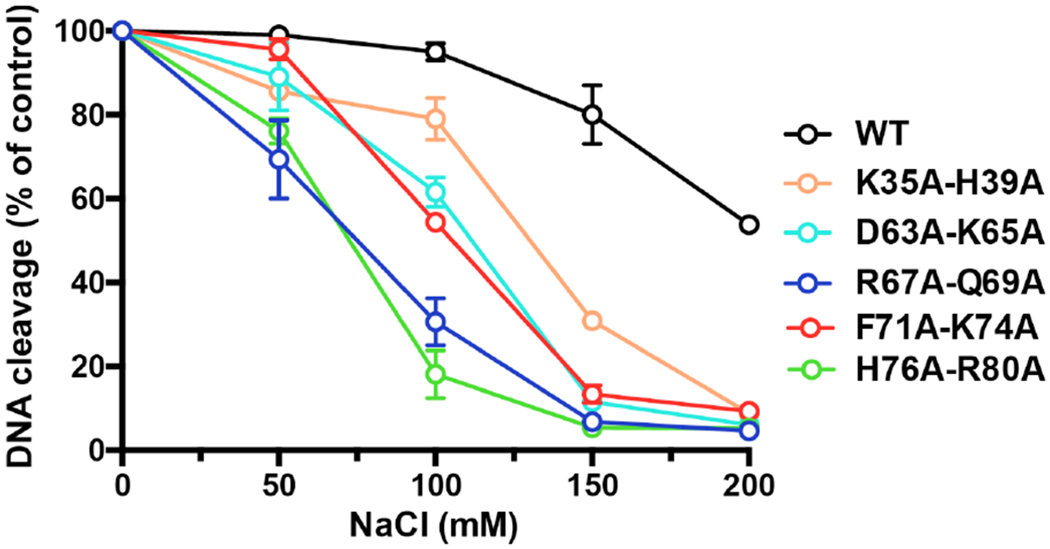
Salt-sensitivity of DNA cleavage. The extents of covalent complex formation normalized to that of a control reaction without added salt are plotted as a function of NaCl concentration for wild-type vTopIB and double-alanine mutants. Each datum is the average of two independent experiments.
CONCLUSIONS
Based on the ITC and NMR analysis described above, it is apparent that the isolated N domain (TopN) of vTopIB is capable of interacting with duplex DNA. Whereas TopN binds DNA duplexes that contain the CCCTT sequence and one that does not, with only a nominal ~2.2-fold difference in affinity, there are observable differences in the mode of association (this could in principle complicate an accurate estimation of affinity in the case of nsDNA). The largest deviation in CSPs between spDNA and nsDNA, both in the magnitude and in the response to salt, lies in the α1-β3 loop and includes residues H39 and L40. The formation of a complex of TopN with the specific sequence appears to be stoichiometric while additional interactions are seen in the case of a DNA duplex lacking this sequence possibly due to the formation of higher-order structures. In both cases, however, the regions of TopN that display the largest CSPs (the α1-β3 and β4-β5 loops and the extreme C-terminus) are largely consistent with the DNA contacts of the N domain seen in full-length vTopIB. Thus, our results suggest that the isolated N domain engages the duplex DNA using a surface that reflects the interaction in the context of full-length protein; further engagement of the C domain reinforces the specificity of the interaction and facilitates the formation of a high-affinity complex. Sekiguchi and Shuman had previously proposed that DNA backbone recognition and reaction chemistry are performed by the CTD of TopIB, whereas discrimination of the DNA sequence at the cleavage site is accomplished by a separate N-terminal domain15. While it has since been established that the C domain has an innate role in discrimination of the target sequence26, our data reinforces the importance of the N domain in DNA recognition. The fact that we find evidence of additional interactions of the isolated N domain in the presence of a DNA duplex missing the CCCTT sequence could imply that the absence of this sequence leads the N domain to sample additional stretches of DNA perhaps locking on once the specificity sequence is encountered.
Supplementary Material
Acknowledgments
Funding
This research was supported by National Science Foundation grant MCB 1412007 (RG) and National Institutes of Health grant GM46330 (SS). A grant G12 MD007603 is acknowledged for partial support of the NMR facilities at The City College of New York). RG is a member of the New York Structural Biology Center, a NYSTAR facility. This manuscript was written in part when RG was a Program Director at the NSF in the Molecular and Cellular Biosciences (MCB) division. His IR/D activities were supported by award MCB 1657192.
Footnotes
ASSOCIATED CONTENT
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.biochem.7b00042 8 Figures and 1 Table.
The authors declare no competing financial interests.
REFERENCES
- (1).Leppard JB, and Champoux JJ (2005) Human DNA topoisomerase I: relaxation, roles, and damage control, Chromosoma 114, 75–85. [DOI] [PubMed] [Google Scholar]
- (2).Dahmane N, Gadelle D, Delmas S, Criscuolo A, Eberhard S, Desnoues N, Collin S, Zhang H, Pommier Y, Forterre P, and Sezonov G (2016) topIb, a phylogenetic hallmark gene of Thaumarchaeota encodes a functional eukaryote-like topoisomerase IB, Nucleic Acids Res. 44, 2795–2805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Cheng C, Kussie P, Pavletich N, and Shuman S (1998) Conservation of structure and mechanism between eukaryotic topoisomerase I and site-specific recombinases, Cell 92, 841–850. [DOI] [PubMed] [Google Scholar]
- (4).Krogh BO, and Shuman S (2002) A poxvirus-like type IB topoisomerase family in bacteria, Proc. Natl. Acad. Sci. USA 99, 1853–1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Benarroch D, Claverie JM, Raoult D, and Shuman S (2006) Characterization of mimivirus DNA topoisomerase IB suggests horizontal gene transfer between eukaryal viruses and bacteria, J. Virol 80, 314–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Sharma A, Hanai R, and Mondragón A (1994) Crystal structure of the amino-terminal fragment of vaccinia virus DNA topoisomerase I at 1.6 A resolution, Structure 2, 767–777. [DOI] [PubMed] [Google Scholar]
- (7).Perry K, Hwang Y, Bushman FD, and Van Duyne GD (2006) Structural basis for specificity in the poxvirus topoisomerase, Mol. Cell 23, 343–354. [DOI] [PubMed] [Google Scholar]
- (8).Perry K, Hwang Y, Bushman FD, and Van Duyne GD (2010) Insights from the structure of a smallpox virus topoisomerase-DNA transition state mimic, Structure 18, 127–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Patel A, Shuman S, and Mondragón A (2006) Crystal structure of a bacterial type IB DNA topoisomerase reveals a preassembled active site in the absence of DNA, J. Biol. Chem 281, 6030–6037. [DOI] [PubMed] [Google Scholar]
- (10).Redinbo MR, Stewart L, Kuhn P, Champoux JJ, and Hol WG (1998) Crystal structures of human topoisomerase I in covalent and noncovalent complexes with DNA, Science 279, 1504–1513. [DOI] [PubMed] [Google Scholar]
- (11).Sekiguchi J, and Shuman S (1995) Proteolytic footprinting of vaccinia topoisomerase bound to DNA, J. Biol. Chem 270, 11636–11645. [DOI] [PubMed] [Google Scholar]
- (12).Shuman S (1998) Vaccinia virus DNA topoisomerase: a model eukaryotic type IB enzyme, Biochim. Biophys. Acta 1400, 321–337. [DOI] [PubMed] [Google Scholar]
- (13).Shuman S, and Prescott J (1990) Specific DNA cleavage and binding by vaccinia virus DNA topoisomerase I, J. Biol. Chem 265, 17826–17836. [PubMed] [Google Scholar]
- (14).Sekiguchi J, and Shuman S (1994) Vaccinia topoisomerase binds circumferentially to DNA, J. Biol. Chem 269, 31731–31734. [PubMed] [Google Scholar]
- (15).Sekiguchi J, and Shuman S (1996) Identification of contacts between topoisomerase I and its target DNA by site-specific photocrosslinking, EMBO J. 15, 3448–3457. [PMC free article] [PubMed] [Google Scholar]
- (16).Shuman S, Kane EM, and Morham SG (1989) Mapping the active-site tyrosine of vaccinia virus DNA topoisomerase I, Proc. Natl. Acad. Sci. USA 86, 9793–9797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Cheng C, Wang LK, Sekiguchi J, and Shuman S (1997) Mutational analysis of 39 residues of vaccinia DNA topoisomerase identifies Lys-220, Arg-223, and Asn-228 as important for covalent catalysis, J. Biol. Chem 272, 8263–8269. [DOI] [PubMed] [Google Scholar]
- (18).Wittschieben J, and Shuman S (1997) Mechanism of DNA transesterification by vaccinia topoisomerase: catalytic contributions of essential residues Arg-130, Gly-132, Tyr-136 and Lys-167, Nucleic Acids Res. 25, 3001–3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Petersen BO, and Shuman S (1997) Histidine 265 is important for covalent catalysis by vaccinia topoisomerase and is conserved in all eukaryotic type I enzymes, J. Biol. Chem 272, 3891–3896. [DOI] [PubMed] [Google Scholar]
- (20).Krogh BO, and Shuman S (2000) Catalytic mechanism of DNA topoisomerase IB, Mol. Cell 5, 1035–1041. [DOI] [PubMed] [Google Scholar]
- (21).Krogh BO, and Shuman S (2002) Proton relay mechanism of general acid catalysis by DNA topoisomerase IB, J. Biol. Chem 277, 5711–5714. [DOI] [PubMed] [Google Scholar]
- (22).Cheng C, and Shuman S (1999) Site-specific DNA transesterification by vaccinia topoisomerase: role of specific phosphates and nucleosides, Biochemistry 38, 16599–16612. [DOI] [PubMed] [Google Scholar]
- (23).Tian L, Sayer JM, Jerina DM, and Shuman S (2004) Individual nucleotide bases, not base pairs, are critical for triggering site-specific DNA cleavage by vaccinia topoisomerase, J. Biol. Chem 279, 39718–39726. [DOI] [PubMed] [Google Scholar]
- (24).Tian L, Claeboe CD, Hecht SM, and Shuman S (2004) Remote phosphate contacts trigger assembly of the active site of DNA topoisomerase IB, Structure 12, 31–40. [DOI] [PubMed] [Google Scholar]
- (25).Yakovleva L, Lai J, Kool ET, and Shuman S (2006) Nonpolar nucleobase analogs illuminate requirements for site-specific DNA cleavage by vaccinia topoisomerase, J. Biol. Chem 281, 35914–35921. [DOI] [PubMed] [Google Scholar]
- (26).Yakovleva L, Chen S, Hecht SM, and Shuman S (2008) Chemical and traditional mutagenesis of vaccinia DNA topoisomerase provides insights to cleavage site recognition and transesterification chemistry, J. Biol. Chem 283, 16093–16103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Koster DA, Croquette V, Dekker C, Shuman S, and Dekker NH (2005) Friction and torque govern the relaxation of DNA supercoils by eukaryotic topoisomerase IB, Nature 434, 671–674. [DOI] [PubMed] [Google Scholar]
- (28).Cheng C, and Shuman S (1998) A catalytic domain of eukaryotic DNA topoisomerase I, J. Biol. Chem 273, 11589–11595. [DOI] [PubMed] [Google Scholar]
- (29).Sekiguchi J, and Shuman S (1997) Mutational analysis of vaccinia virus topoisomerase identifies residues involved in DNA binding, Nucleic Acids Res. 25, 3649–3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Tian L, and Shuman S (2007) Vaccinia topoisomerase mutants illuminate roles for Phe59, Gly73, Gln69 and Phe215, Virology 359, 466–476. [DOI] [PubMed] [Google Scholar]
- (31).Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, and Bax A (1995) NMRPipe: a multidimensional spectral processing system based on UNIX pipes, J. Biomol. NMR 6, 277–293. [DOI] [PubMed] [Google Scholar]
- (32).Johnson BA (2004) Using NMRView to visualize and analyze the NMR spectra of macromolecules, Meth. Mol. Biol 278, 313–352. [DOI] [PubMed] [Google Scholar]
- (33).Sattler M, Schleucher J, and Griesinger C (1999) Heteronuclear multidimensi onal NMR experiments for the structure determination of proteins in solution employing pulsed field gradients, Prog. NMR Spectrosc 34, 93–158. [Google Scholar]
- (34).Lemak A, Gutmanas A, Chitayat S, Karra M, Fares C, Sunnerhagen M, and Arrowsmith CH (2011) A novel strategy for NMR resonance assignment and protein structure determination, J. Biomol. NMR 49, 27–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Orekhov VY, and Jaravine VA (2011) Analysis of non-uniformly sampled spectra with multi-dimensional decomposition, Prog. NMR Spectrosc 59, 271–292. [DOI] [PubMed] [Google Scholar]
- (36).Morham SG, and Shuman S (1992) Covalent and noncovalent DNA binding by mutants of vaccinia DNA topoisomerase I, J. Biol. Chem 267, 15984–15992. [PubMed] [Google Scholar]
- (37).Ferrage F, Cowburn D, and Ghose R (2009) Accurate sampling of high-frequency motions in proteins by steady-state 15N-{1H} nuclear Overhauser effect measurements in the presence of cross-correlated relaxation, J. Am. Chem. Soc 131, 6048–6049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Ghose R, Fushman D, and Cowburn D (2001) Determination of the rotational diffusion tensor of macromolecules in solution from NMR relaxation data with a combination of exact and approximate methods - Application to the determination of interdomain orientation in multidomain proteins, J. Magn. Reson 149, 204–217. [DOI] [PubMed] [Google Scholar]
- (39).Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, and Ferrin TE (2004) UCSF Chimera - a visualization system for exploratory research and analysis, J. Comput. Chem 25, 1605–1612. [DOI] [PubMed] [Google Scholar]
- (40).Privalov PL, Dragan AI, and Crane-Robinson C (2011) Interpreting protein/DNA interactions: distinguishing specific from non-specific and electrostatic from non-electrostatic components, Nucleic Acids Res. 39, 2483–2491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Eghbalnia HR, Wang L, Bahrami A, Assadi A, and Markley JL (2005) Protein energetic conformational analysis from NMR chemical shifts (PECAN) and its use in determining secondary structural elements, J. Biomol. NMR 32, 71–81. [DOI] [PubMed] [Google Scholar]
- (42).García de la Torre J, Huertas ML, and Carrasco B (2000) HYDRONMR: prediction of NMR relaxation of globular proteins from atomic-level structures and hydrodynamic calculations, J. Magn. Reson 147, 138–146. [DOI] [PubMed] [Google Scholar]
- (43).Krogh BO, and Shuman S (2001) Vaccinia topoisomerase mutants illuminate conformational changes during closure of the protein clamp and assembly of a functional active site, J. Biol. Chem 276, 36091–36099. [DOI] [PubMed] [Google Scholar]
- (44).Perry JJP, Cotner-Gohara E, Ellenberger T, and Tainer JA (2010) Structural dynamics in DNA damage signaling and repair, Curr. Opin. Struct. Biol 20, 283–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Nagarajan R, and Stivers JT (2006) Major groove interactions of vaccinia Topo I provide specificity by optimally positioning the covalent phosphotyrosine linkage, Biochemistry 45, 5775–5782. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


