Abstract
Pancreatic cancer has extremely poor prognosis, warranting the discovery of novel therapeutic and prognostic markers. The expression of polymeric immunoglobulin receptor (pIgR), a key component of the mucosal immune system, is increased in several cancers. However, its clinical relevance in pancreatic cancer remains unclear. In the present study, the prognostic value of pIgR in pancreatic cancer patients after surgical resection was assessed and it was determined that the expression of pIgR was correlated with poor prognosis. Ten pancreatic cancer patient-derived xenograft (PDX) lines were established, followed by next-generation sequencing of tumor tissues from these lines after standard chemotherapy. Immunohistochemical analysis of chemoresistance-related molecules using 77 pancreatic cancer tissues was also performed. The expression of pIgR mRNA in the PDX group treated with anticancer drugs was higher than in the untreated group. High pIgR expression in tissue specimens from 77 pancreatic cancer patients was significantly associated with poor prognosis and was revealed to be an independent prognostic factor, predicting poor outcomes. High pIgR mRNA and protein levels were independent prognostic factors, indicating that pIgR could be a novel predictor for poor prognosis of pancreatic cancer patients.
Keywords: biomarker, chemoresistance, poor prognosis, polymeric immunoglobulin receptor, patient-derived xenograft, pancreatic cancer
Introduction
Pancreatic cancer is an aggressive disease, frequently diagnosed (in ~80% of cases) at an advanced stage (1) when the tumor is unresectable due to infiltration of local arteries or distant metastasis (2). Unfortunately, pancreatic resection is possible in few (5-10%) patients (3). Pancreatic adenocarcinoma remains one of the deadliest cancers, with a five-year survival rate of 6–8% (4,5) and a median duration of survival of less than two years even in surgically-treated patients (5).
Pancreatic cancer is the fourth-leading cause of cancer-related deaths in the USA (4) and is predicted to become the second-leading cause among individuals older than 65 years by 2030 (6). Globally, this trend has remained unchanged, with more than 200000 deaths attributed to pancreatic cancer annually (7).
The overall survival (OS) of pancreatic cancer patients has not improved significantly in the past 30 years (1). The high mortality rate is probably due to diagnosis at an advanced stage when the currently available therapies have limited effects. The addition of nab-paclitaxel (nab-PTX) to gemcitabine (GEM) increased the OS from 6.6 to 8.7 months (8,9). For patients in good physical condition, treatment with FOLFIRINOX (5-fluorouracil, oxaliplatin, and irinotecan) increased the survival to 11.1 months compared to the OS for GEM alone, but had potentially severe side effects (10).
Chemoresistance is a reason for high mortality in pancreatic cancer (7,11). Gemcitabine and nab-PTX are the standard of care for treating advanced pancreatic cancer (8,9), exhibiting benefits over GEM. However, systemic chemotherapy has a limited effect on OS due to low response rates and chemoresistance, ascribed to the poorly understood mechanism of action. Early diagnosis using novel biomarkers is an important goal for pancreatic cancer researchers.
The polymeric immunoglobulin receptor (pIgR) responsible for transcytosis of polymeric Igs (dimeric IgA and pentameric IgM) across mucosal surfaces (12) facilitates the secretion of IgA and IgM, the first-line antibodies against infection (13,14). The extracellular portion of pIgR is then cleaved off as a secretory component (SC) bound to polymeric IgA, protecting it from proteolytic degradation (14) and ensuring effective mucosal secretion (15). pIgR is expressed on epithelial cells and is upregulated by proinflammatory cytokines in response to viral and bacterial infections, thus linking innate and adaptive immunity (13,14,16,17). The extracellular component of pIgR can be cleaved to produce the SC that is not bound to IgA, which then acts as a scavenger on the mucosal lining (14).
pIgR is highly expressed in several cancers; its upregulation was detected in colon (18), breast (19,20), endometrial (21), bladder (22), hepatocellular (23,24), epithelial ovarian (25), and esophageal and gastric cancers (26). High levels of the SC were also detected in the sera of patients with lung (27,28), pancreatic (29), and colon cancer with liver metastasis (30). However, the clinical significance and prognostic value of pIgR remain unclear.
Herein, pIgR, as a novel protein associated with chemoresistance, was investigated using pancreatic cancer patient-derived xenograft (PDX) lines. To demonstrate the association between pIgR expression and clinicopathological features, the expression and prognostic ability of pIgR were evaluated using immunohistochemistry (IHC) of 77 human pancreatic cancer tissues following surgical resection.
Materials and methods
Establishment of pancreatic cancer PDX lines
NSG mice, obtained from The Jackson Laboratory, were housed aseptically in plastic cages at 22±1°C under 45±10% relative humidity and a 12-h light/12-h dark cycle. All mice were fed a standard diet and were allowed free access to food and water. All experiments involving animals were performed in accordance with the care and use guidelines of the Kanagawa Cancer Center Research Institute, Japan. The study was approved by the Research Ethics Committee of Kanagawa Cancer Center Research Institute (approval no. 176).
Surgically resected tumor tissues from ten pancreatic cancer patients (age range, 51–79 years; mean age, 67.2 years; 4 males and 6 females; Table SI) were subcutaneously transplanted into 6–12-week-old mice using transplantation needles (Fig. S1A) as previously described (31,32). These patients provided informed consent and tumor tissues were resected at the Kanagawa Cancer Center Hospital (Kanagawa, Japan) from June, 2008 to March, 2015. Briefly, after each patient underwent surgery, fresh tumor tissues were cut into small pieces of approximately 0.8–5.0 mm3 using scissors or minced under sterile conditions. A small incision was made on the lower back of the mouse near the base on the hindlimb, and a transplant needle was inserted until the tip reached the dorsal subcutaneous area of the upper part of the back, followed by closure. This method was used to prevent outflow of the engraftment. These PDX lines were designated as Generation 1 (G1). When the tumor volumes in G1 mice reached 1000 mm3, the tumor tissues were removed and re-implanted into other NSG mice. Ten PDX lines up to G7 were generated after repeated passaging (Fig. S1B).
Antitumor effects of chemotherapy in the pancreatic cancer PDX model
Anticancer drugs [GEM or GEM + nab-PTX] or saline (used as a control) were administered intraperitoneally to PDX mice with 200–400 mm3 tumors. Tumor volumes (mm3) were determined weekly using the formula [length (mm)2 × width (mm)]/2. The chemotherapy was administered via intra-abdominal injection on days 1, 3, and 6.
Identification of chemoresistance-related molecules
Tumor tissues from treated and control PDX lines were harvested when the tumor volume in the control group exceeded 1500 mm3. After the tumor-bearing mice were sacrificed, the tumors were cut into 4-µm thick pieces that were immediately frozen in liquid nitrogen. The frozen tissue was crushed using a Cryo-Press (Microtec Co., Ltd.). Total RNA was isolated and purified with ZR-Duet DNA/RNA MiniPrep (Zymo Research Corp.). RNA quality was assessed using the Agilent 2100 Bioanalyzer (Agilent Technologies, Inc.), followed by next-generation sequencing (NGS) on an Illumina HiSeq 4000 system at BGI with paired-end sequencing. The read lengths were 2×100 bp for GEM monotherapy and 2×150 bp for GEM + nab-PTX combination therapy.
Transcriptome analysis
Paired-end reads were mapped to all the RefSeq human (hg38 coordinates) and mouse (mm10 coordinates) transcripts using Bowtie 1.1.2 (33), allowing up to one mismatch, and reads mapped to both the species or to multiple genes were discarded. To avoid bias due to read length differences, only the first 100 bp of each read for samples with read lengths of 150 bp were used for mapping. Reads mapping to noncoding transcripts were removed, and the remaining were used for gene expression profiling of human cancer and mouse stromal cells (34). The expression values were normalized for cancer and stromal cells independently; the sum of expression values below the 95th percentile being 300000.
Bioinformatics analysis using The Cancer Genome Atlas (TCGA; http://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga) database
The Human Protein Atlas (http://www.proteinatlas.org) was used as a tool for antibody-based biomarker discovery (35). TCGA RNASeq data for pancreatic cancer were used to generate Kaplan-Meier curves on the basis of pIgR expression. Patients were classified into two groups, and the association between survival and gene expression was examined. The expression cut-off based on the fragments per kilobase of exon model per million reads mapped (FPKM) value that yielded the maximal difference between the two groups with regard to survival at the lowest log-rank P-value was selected.
Immunohistological analysis of samples from pancreatic cancer patients
A consecutive cohort of 77 histopathologically confirmed pancreatic cancer patients who underwent surgical resection at Showa University Hospital in Tokyo, Japan, from January 1, 2008 to December 31, 2017 was included (Table I). Patients with no history of chemotherapy (GEM or GEM + nab-PTX) were included. Archived formalin-fixed, paraffin-embedded (FFPE) tumor tissues from resected specimens from the cohort of patients were used. Histological classifications were based on the World Health Organization system. Tumor staging was performed per the criteria described in the UICC TNM classification (7th edition, 2009). The included patients had stage IIA or stage IIB disease, per the 7th edition of UICC. The study protocol was approved by the Ethics Committee of the Showa University School of Medicine (Tokyo, Japan; Approval no. 2611) and adhered to the principles of the Declaration of Helsinki.
Table I.
Correlation between pIgR expression and clinicopathological features in 77 cases of pancreatic cancer.
| pIgR expression | |||
|---|---|---|---|
| Characteristics | Low n=47 | High n=30 | P-value |
| Age (years) (mean ± SD) | 72.4 | 69.8 | 0.339a |
| Sex | 0.0608b | ||
| Male | 23 | 14 | |
| Female | 24 | 16 | |
| Tumor location | 0.6588b | ||
| Pancreatic head | 29 | 20 | |
| Pancreatic body/tail | 18 | 10 | |
| Histological type | 0.1008b | ||
| Adenocarcinoma | 43 | 30 | |
| Others | 4 | 0 | |
| TNM (UICC 7th) | 0.3395b | ||
| IIA | 14 | 6 | |
| IIB | 33 | 24 | |
| Histological differentiation | 0.1638b | ||
| G1 | 16 | 15 | |
| G2-4 | 31 | 15 | |
| Lymphatic invasion | 0.8813b | ||
| ly0, ly1 | 18 | 12 | |
| ly2, ly3 | 29 | 18 | |
| Venous invasion | 0.0697b | ||
| v0, v1 | 3 | 6 | |
| v2, v3 | 44 | 24 | |
| Perineural invasion | 0.4773b | ||
| ne01 | 11 | 5 | |
| ne23 | 36 | 25 | |
| Resection margin | 0.0618b | ||
| R0 | 32 | 19 | |
| R1-2 | 13 | 10 | |
| Adjuvant chemotherapy | 0.8645b | ||
| Absent | 21 | 14 | |
| Present | 26 | 16 | |
Student's t-test.
Pearson Chi-square test. pIgR, polymeric immunoglobulin receptor.
Immunohistological analysis of pIgR expression in human pancreatic cancer tissues was performed using standard protocols employing a Leica Bond system (Leica Microsystems GmbH). Briefly, heat-mediated antigen retrieval in FFPE tissue sections (3-µm thick) was performed in sodium citrate buffer (pH 9.0) for 20 min at 100°ç. The sections were incubated with an anti-pIgR antibody (1:500 dilution; product code ab96196; Abcam) for 15 min at room temperature, and a signal was detected using a horseradish peroxidase-conjugated compact polymer system [HRP-polymer secondary antibody; Goat Anti-Rabbit IgG H&L (HRP polymer); product code ab214880; Abcam]. DAB was applied as the chromogen and incubated for 5 min at room temperature. The sections were counterstained with hematoxylin for 5 min at room temperature and viewed under a bright-field microscope at an ×100 magnification.
The immunostained sections were reviewed and scored separately by two pancreas-specialist pathologists who were blinded to the clinical parameters. pIgR expression in the tumor tissue was assessed by comparing the staining intensity with the percentage of immunoreactive cells. The staining intensity was graded based on the following criteria: 0, negative; 1, weak; 2, moderate; 3, strong (uncolored, light yellow, yellowish brown, and brown, respectively). The staining percentages were graded based on the proportion of positively stained tumor cells as 0, 1, 2, and 3 for 0%, 1–25%, 26–50%, and ≥51% positive tumor cells. The pIgR staining was scored using the following formula: Overall score=positive percentage score × staining intensity score. The pIgR expression was evaluated based on the staining index (scored as 0, 1, 2, 3, 4, 6 or 9). A score of 0 was designated as 0, >0 and ≤2 as 1, >2 and ≤6 as 2, and >6 and ≤9 as 3. Tumor samples graded as level 0 or 1 had low pIgR expression, whereas those graded as level 2 or 3 had high pIgR expression. In addition, the percentage of pIgR-stained area was quantified using the Hybrid Cell Count BZ-X800 software (Keyence) (36).
For analysis of clinicopathological factors, an invasive factor was evaluated based on the pancreatic cancer classification for further stratification, as recommended by the Japan Pancreas Society. Lymphatic invasion was graded depending on the degree of invasion: ly0, negative; ly1, weak; ly2, moderate; ly3, strong. Similarly, venous invasion was graded as v0, v1, v2 or v3, and perineural invasion was graded as ne0, ne1, ne2 or ne3. Evaluation of histological differentiation and resection margin was performed according to the 7th edition of UICC. Histological differentiation was graded per the following criteria: G1, well differentiated; G2, moderately differentiated; G3, poorly differentiated; G4, undifferentiated. For the resection margin, the grading was performed as follows: R0, negative resection margins; R1, microscopic tumor infiltration; R2, macroscopic residual tumor.
Statistical analysis
Statistical significance was analyzed using JMP Pro 14.0 (SAS Institute Inc.). Associations between the IHC status of pIgR expression and various clinicopathological characteristics were evaluated using Student's t-tests or Pearson chi-square tests. Classification and regression tree analysis were used to assess the optimal prognostic cut-off for pIgR expression in OS studies. OS was defined as the interval, in months, between the initial pancreatic resection surgery and either death or the last observation. Kaplan-Meier analysis and the log-rank test were applied for estimating the differences in OS depending on high or low pIgR expression. Univariate and multivariate analyses were based on the Cox proportional hazards regression model. All the tests were two-sided, and results with P-values <0.05 were considered to indicate a statistically significant difference.
Results
Identification of a chemoresistance-related molecule
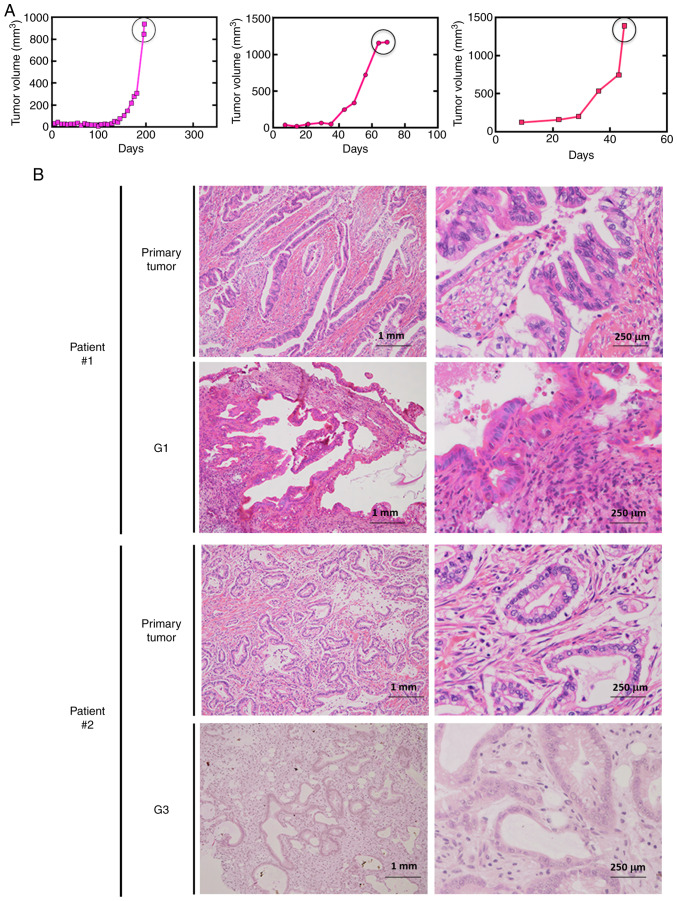
Ten pancreatic cancer PDX lines were established (Table SI). The tumor growth curve for one representative line is presented in Fig. 1A. The tumor tissues of the PDX lines retained the pathological (Fig. 1B) and genetic (data not shown) features of the original pancreatic cancer tissues, even after repeated passages.
Figure 1.
Establishment of pancreatic cancer PDX lines. (A) Tumor growth curves for a representative pancreatic PDX line. When the tumor volume reached ~1,000 mm3, the mouse was sacrificed, and the tumor was isolated and transplanted into another mouse. (B) Preserved morphological characteristics in xenograft tumors in NSG mice. H&E staining of the primary tumors and subsequent generations of PDXs for two patients. The pathological diagnosis of the primary tumors was tubular adenocarcinoma. PDX, patient-derived xenograft; H&E, hematoxylin and eosin.
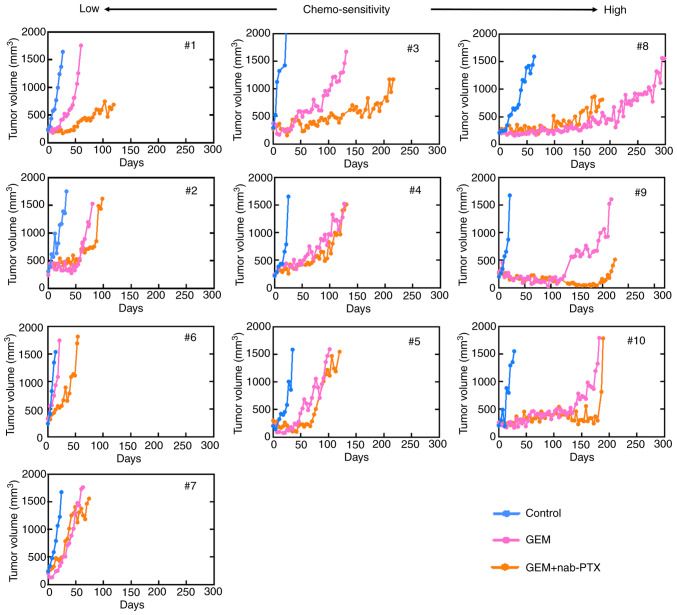
To identify the chemoresistance-related molecules, the antitumor effects of the mono- or combination therapy in the PDX models were characterized in terms of tumor growth after the treatments (Fig. 2). Although antitumor effects were transiently observed in some cases in the treatment group, tumor growth was finally observed in all the lines.
Figure 2.
Tumor growth curves revealing the comparison of control and chemotherapy groups. PDX mice in the chemotherapy group were treated with GEM or GEM + nab-PTX until the tumor volume was 1,500 mm3 or more. Although there were PDX lines with high and low chemo-sensitivity, all became chemo-resistant eventually, and an increase in the tumor volume was also observed in the chemotherapy group. PDX, patient-derived xenograft; GEM, gemcitabine; nab-PTX, nab-paclitaxel.
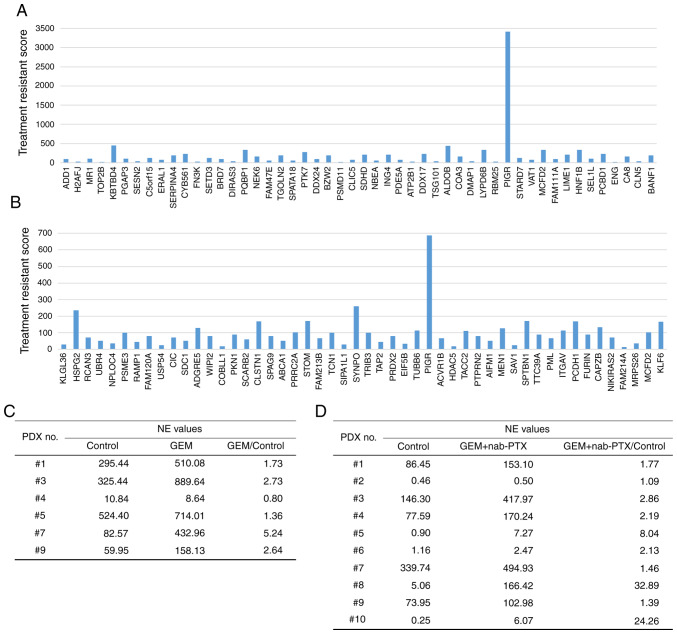
The mRNA expression in tumor tissues was analyzed using NGS. We performed NGS of tumor tissues from PDXs before and after treatment with standard chemotherapy. The number of genes analyzed by RNA sequencing using NGS was 26732. Among them, there were 193 genes with FPKM values of 10 or more and expression fold-change ratios between the treatment group and control group of 3 or more (Table SII). Using the normalized expression (NE) values, the ratio of the control to treated PDX lines was calculated. For the mono- and combination therapies, genes with NE values greater than 10 and NE ratios (treated group to control group) greater than 2 were selected (Fig. 3A and B). The ratio of NE values for pIgR mRNA expression (Fig. 3C and D) between the treated and control groups was greater than 1 for most of the lines treated with GEM or GEM + nab-PTX. This analysis revealed that the expression of pIgR mRNA was more increased in the PDX group that received chemotherapy than in untreated PDXs. These findings indicated that pIgR mRNA was expressed before the administration of anticancer drugs and may have been induced by chemotherapy. Moreover, the expression level of pIgR mRNA was higher in the treated group than in the control group, which may indicate that increased expression of pIgR mRNA was involved in chemoresistance.
Figure 3.
Identification of chemotherapy resistance-related molecules. (A and B) NGS analysis for the (A) GEM administration and (B) GEM + nab-PTX administration groups. Treatment resistance score was defined as the NE value ratio (treated group/control group) × NE value difference (treated group-control group). (C and D) The ratio of NE values for pIgR expression between the treated and control groups tended to be greater than 1.0 for most of the PDXs treated with (C) GEM or (D) GEM + nab-paclitaxel. NGS, next-generation sequencing; GEM, gemcitabine; nab-PTX, nab-paclitaxel; NE, normalized expression; pIgR, polymeric immunoglobulin receptor; PDX, patient-derived xenografts.
Prognostic analysis from TCGA RNA database for pancreatic cancer
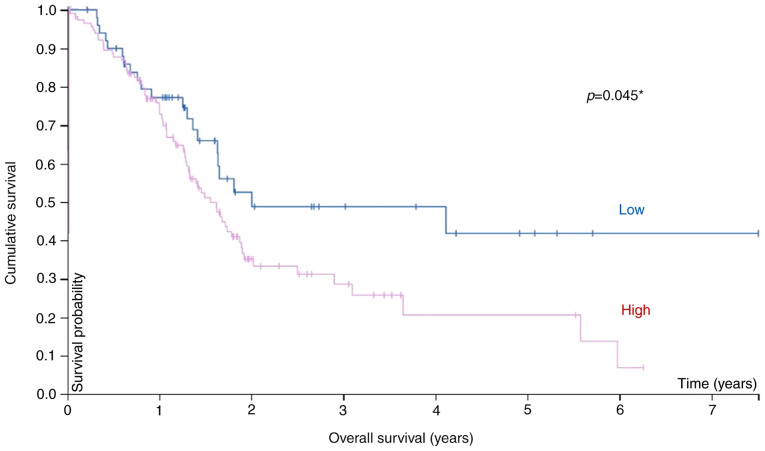
Additional analysis of pIgR expression and OS in 171 pancreatic cancer samples from TCGA database revealed an association between survival and the mRNA level (Fig. 4). Pancreatic cancer patients (stages I–IV) were divided into high and low pIgR mRNA groups using 41 FPKM as the cut-off value, and the prognosis of each group was examined. Patients in the high-pIgR mRNA group exhibited significantly poorer survival rates than those in the low pIgR mRNA group (P=0.045). The five-year survival rate was 42 and 21% for the low- and high-pIgR mRNA groups. Thus, it was inferred that pIgR may be a putative prognostic biomarker of pancreatic cancer.
Figure 4.
Kaplan-Meier plot summarizing the results from analysis of the associations between pIgR mRNA expression and patient survival in TCGA pancreatic cancer database (n=171). Red line, high expression (n=119); blue line, low expression (n=52). pIgR, polymeric immunoglobulin receptor; TCGA, The Cancer Genome Atlas. *Statistically significant.
Immunohistological and prognostic analysis in pancreatic cancer patients
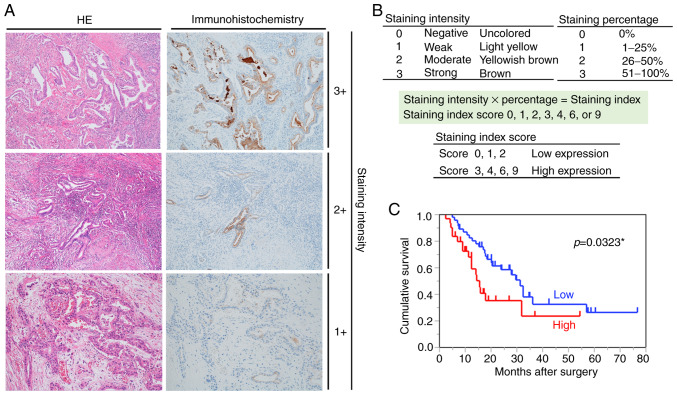
We corroborated our findings in the PDX model with the expression of pIgR in clinical specimens, the clinicopathological features of which are summarized in Table I. Immunohistochemistry revealed that pIgR was strongly expressed on the tumor cells (Fig. 5A). The samples were divided into the high [39.0% (30/77)] and low [61.0% (47/77)] pIgR groups (Table I), as described in the Materials and methods section (Fig. 5B).
Figure 5.
Relationship between pIgR expression and prognosis. (A) Immunohistochemical staining for pIgR in pancreatic cancer tissues (magnification, ×100). The left and right images show the same sample tissue blocks and correspond to the staining intensity. Left, H&E staining; right, immunohistochemical staining for pIgR. (B) Criteria for determination of pIgR expression levels. The pIgR expression levels in immunostaining were determined based on the intensity of staining and percentage of stained cells. The staining intensity and staining percentage criteria are presented. (C) Kaplan-Meier survival analysis in patients with pancreatic cancer (n=77, revealing OS based on the expression of pIgR protein. Red line, high-expression group (n=47), blue line, low-expression group (n=30). pIgR, polymeric immunoglobulin receptor; H&E, hematoxylin and eosin; OS, overall survival. *Statistically significant.
Furthermore, whether pIgR was useful for prognostic evaluation of pancreatic cancer was analyzed. Notably, there was no statistical difference in patient or pathological features, including age (P=0.34), sex (P=0.06), tumor location (head or body/tail, P=0.66), histological type (adenocarcinoma or others, P=0.10), TNM stage (IIA or IIB, P=0.34), histological differentiation (P=0.16), lymphatic invasion (P=0.88), venous invasion (P=0.07), perineural invasion (P=0.48), resection margin (P=0.06), and adjuvant chemotherapy (P=0.86), due to the separation of patients into the two groups of high and low pIgR expression (Table I). Next, the association between the pIgR expression levels and patient survival was assessed using Kaplan-Meier analysis with log-rank tests. Among 77 pancreatic cancer patients, those with high pIgR expression had poorer survival rates than those with low pIgR expression (P=0.0323) (Fig. 5C). Furthermore, the percentage of pIgR-stained area was analyzed using the Hybrid Cell Count software with respect to survival time and the expression of pIgR determined by two pathologists. The percentage of pIgR-stained area was significantly associated with the expression of pIgR as determined by the pathologists (P<0.001) (Fig. S2A). In addition, higher percentages of pIgR-stained area determined by the image analysis software were associated with shorter OS (r=−0.3801, P=0.00065) (Fig. S2B).
Univariate analysis of OS revealed four prognostic parameters: Histological differentiation (P=0.0012), resection margin (P=0.0004), adjuvant chemotherapy (P<0.0001), and pIgR expression (P=0.0404). Multivariate analysis using Cox proportional hazards revealed that histological differentiation (P=0.0004), resection margin (P=0.0004), adjuvant chemotherapy (P=0.0013), and pIgR expression (P=0.0045) were independent prognostic factors for poor outcome (Table II).
Table II.
Univariate and multivariate analyses of prognostic factor for overall survival in 77 pancreatic cancer patients.
| Univariate analysis | Multivariate analysis | |||||
|---|---|---|---|---|---|---|
| Clinicopathological factors | HR | 95% CI | P-valuea | HR | 95% CI | P-valuea |
| Age(years) | ||||||
| (≦71 vs. >71) | 1.03 | 0.54–1.92 | 0.9214 | – | – | – |
| Sex | ||||||
| (male vs. female) | 0.57 | 0.30–1.06 | 0.076 | – | – | – |
| TNM stage UICC 7th | ||||||
| (IIA vs. IIB) | 0.59 | 0.26–1.21 | 0.1575 | – | – | – |
| Tumor location | ||||||
| (body/tail vs. head) | 0.65 | 0.33–1.22 | 0.1861 | – | – | – |
| Histological differentiation | ||||||
| (G1 vs. G2, G3, G4) | 0.34 | 0.16–0.66 | 0.0012b | 0.26 | 0.11–0.56 | 0.0004b |
| Lymphatic invasion | ||||||
| (ly0, ly1 vs. ly2, ly3) | 0.71 | 0.37–1.31 | 0.2754 | – | – | – |
| Venous invasion | ||||||
| (v0, v1 vs. v2, v3) | 0.94 | 0.35–2.09 | 0.8849 | – | – | – |
| Perineural invasion | ||||||
| (ne0, ne1 vs. ne2, ne3) | 0.62 | 0.27–1.29 | 0.2108 | – | – | – |
| Resection margin | ||||||
| (R0 vs. R1, R2) | 0.36 | 0.22–0.63 | 0.0004b | 0.28 | 0.14–0.56 | 0.0004b |
| Adjuvant chemotherapy | ||||||
| (present vs. absent) | 0.26 | 0.13–0.50 | <0.0001b | 0.34 | 0.17–0.71 | 0.0013b |
| pIgR | ||||||
| (low vs. high) | 0.56 | 0.27–0.97 | 0.0404b | 0.35 | 0.17–0.71 | 0.0045b |
Cox proportional hazard model.
Statistically significant. 95% CI, 95% confidence interval; pIgR, polymeric immunoglobulin receptor.
Discussion
Pancreatic cancer is characterized by late diagnosis, early metastasis, limited response to chemotherapy, and extremely poor prognosis (7,37,38), highlighting the need for novel prognostic biomarkers and therapeutic targets. Using PDX lines, it was determined that pIgR mRNA was upregulated after treatment with anticancer drugs, indicating that pIgR may be associated with chemoresistance. It was also revealed that high pIgR expression was an independent prognostic factor for poor survival in pancreatic cancer patients. Analysis of TCGA data supported our findings at the RNA level in pancreatic cancer patients.
Various mouse tumor models and tumor cell lines have been used for predicting the efficacy and possible toxicities of anticancer drugs in cancer patients (31,39). However, the results of these studies do not necessarily reflect human clinical data (40), primarily due to the lack of a tumor microenvironment in such cell and tumor models. Moreover, studies in mice are also not always translatable to human patients (41,42). Therefore, better models, reflecting human clinical pathology, are required.
PDX lines of immunodeficient mice have emerged as relevant in vivo models for human tumors (43); they not only recapitulate the interactions with the host microenvironment but also reflect tumor heterogeneity, including cancer stem cells. In the present study the PDX lines were confirmed to retain most of the histological and genetic characteristics of their donor tumors and remained stable after repeated passaging. In pancreatic cancer, it is often challenging to perform surgical resection or biopsy using endoscopic ultrasound/fine needle aspiration at diagnosis and to collect sufficient pancreatic tissue samples (44,45). Therefore, PDX models are optimal for research in pancreatic cancer pathology (46).
It was determined that an increased pIgR mRNA level was associated with chemoresistance. However, variations in mRNA expression do not always correspond to changes in protein expression owing to various post-transcriptional protein modifications (47–49). Thus, mRNA abundance can be a poor predictor of the protein levels. Analysis of colon and rectal tumor proteomics data in TCGA database showed that mRNA abundance did not reliably predict the differences in protein abundance between tumors (50). To confirm the correlation between the mRNA and protein levels of pIgR and the relationship between pIgR protein and clinicopathological factors, pIgR protein expression was evaluated in tissues from 77 pancreatic cancer patients. It was revealed that high pIgR expression was an independent prognostic factor for survival in pancreatic cancer patients.
Few studies have investigated the role of pIgR in cancers, especially in pancreatic cancer. Improved survival of patients with low pIgR expression and low in vitro and ex vivo stromal activity was observed using data from 88 pancreatic ductal adenocarcinoma patient samples; however, pIgR expression alone had no statistically significant effect on the survival (51). The present data revealed statistical significance for pIgR expression. Low pIgR expression in pancreatic adenocarcinoma was significantly associated with progressive disease, a shorter time to recurrence, and death within five years (52); however, our results stand in contrast with these, possibly because the target patient population was different, with the patient cohort in the previous study having intestinal as well as pancreatobiliary type adenocarcinoma. Moreover, differences in surgical procedures and different types of anticancer drugs may have led to contrasting results. Further investigations are required to determine the relationship between pIgR expression and its role in pancreatic cancer patients.
A limitation of the present study was the subjective assessment of IHC staining. Because a single analytical method for IHC has not been established, expression analysis of pIgR in the tumor tissues was performed using previously reported methods (53–55). To decrease subjectivity, the use of image analysis software is an option (56). However, the specificity and sensitivity of automated software remain unclear (57), and therefore, a semiquantitative assessment strategy was deemed more appropriate. In addition, none of the patients had received neoadjuvant chemotherapy before surgical resection. The inclusion of specimens obtained after neoadjuvant chemotherapy would have been ideal to compare tissues with and without chemotherapy. However, only chemo-naïve patients were included in this study because neoadjuvant chemotherapy is not yet permitted in Japan. Third, for survival analysis using TCGA RNA database, the optimal expression cut-off that yielded the maximal difference between the two groups with regard to survival at the lowest log-rank P-value was selected; nonetheless, this cut-off may not be suitable for other patient groups.
In summary, in the present study it was revealed that the pIgR mRNA level was associated with chemoresistance, and that high expression of pIgR protein was significantly associated with poor prognosis of pancreatic cancer patients. Thus, pIgR may be a novel predictor of poor prognosis of pancreatic cancer patients after surgical resection and a promising candidate for targeted therapy of pancreatic cancer.
Supplementary Material
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used during the present study are available from the corresponding author upon reasonable request. All data generated or analyzed during this study are included in this published article. The results published in the present study are in part based upon data generated by the TCGA Research Network: https://www.cancer.gov/tcga.
Authors' contributions
RO, TT, SK and SW conceived and designed this study. RO, YK, KH, AH, TI, YH, HA, JT, KY, KA, MW, MS, RO, TA, MM, TT and SW collected samples and recorded the general data and the observation indicators of the patients. EY, TS and SW performed all the experiments using PDX models. SI and DK analyzed the data for NGS analysis. MT, NO and TN performed the pathological diagnosis and analyzed the immunostained samples. RO analyzed all the data and wrote the first draft of the manuscript. JT, KY, TT, SK and SW critically reviewed and corrected the manuscript. All authors reviewed and approved the final version of the manuscript.
Ethics approval and consent to participate
The study protocol was approved by the Ethics Committee of the Showa University School of Medicine (Tokyo, Japan; Approval no. 2611) and adhered to the principles of the Declaration of Helsinki. All experiments involving animals were performed in accordance with the care and use guidelines of the Kanagawa Cancer Center Research Institute, Japan. This study was approved by the Research Ethics Committee of Kanagawa Cancer Center Research Institute (approval no. 176).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Hidalgo M, Cascinu S, Kleeff J, Labianca R, Löhr JM, Neoptolemos J, Real FX, Van Laethem JL, Heinemann V. Addressing the challenges of pancreatic cancer: Future directions for improving outcomes. Pancreatology. 2015;15:8–18. doi: 10.1016/j.pan.2014.10.001. [DOI] [PubMed] [Google Scholar]
- 2.Lefebvre AC, Maurel J, Boutreux S, Bouvier V, Reimund JM, Launoy G, Arsene D. Pancreatic cancer: Incidence, treatment and survival trends-1175 cases in Calvados (France) from 1978 to 2002. Gastroenterol Clin Biol. 2009;33:1045–1051. doi: 10.1016/j.gcb.2009.04.011. [DOI] [PubMed] [Google Scholar]
- 3.Kocher HM, Alrawashdeh W. Pancreatic cancer. BMJ Clin Evid. 2010;2010(pii):0409. [PMC free article] [PubMed] [Google Scholar]
- 4.Siegel RL, Miller KD, Jemal A. Cancer statistics in USA, 2018. CA Cancer J Clin. 2018;68:7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 5.Li HB, Zhou J, Zhao FQ. A prognostic nomogram for disease-specific survival in patients with pancreatic ductal adenocarcinoma of the head of the pancreas following pancreaticoduodenectomy. Med Sci Monit. 2018;24:6313–6321. doi: 10.12659/MSM.909649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rahib L, Smith BD, Aizenberg R, Rosenzweig AB, Fleshman JM, Matrisian LM. Projecting cancer incidence and deaths to 2030: The unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res. 2014;74:2913–2921. doi: 10.1158/0008-5472.CAN-14-0155. [DOI] [PubMed] [Google Scholar]
- 7.Kamisawa T, Wood LD, Itoi T, Takaori K. Pancreatic cancer. Lancet. 2016;388:73–85. doi: 10.1016/S0140-6736(16)00141-0. [DOI] [PubMed] [Google Scholar]
- 8.Von Hoff DD, Ervin T, Arena FP, Chiorean EG, Infante J, Moore M, Seay T, Tjulandin SA, Ma WW, Saleh MN, et al. Increased survival in pancreatic cancer with nab-paclitaxel plus gemcitabine. N Engl J Med. 2013;369:1691–1703. doi: 10.1056/NEJMoa1304369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goldstein D, El-Maraghi RH, Hammel P, Heinemann V, Kunzmann V, Sastre J, Scheithauer W, Siena S, Tabernero J, Teixeira L, et al. Nab-Paclitaxel plus gemcitabine for metastatic pancreatic cancer: Long-term survival from a phase III trial. J Natl Cancer Inst. 2015;107:1–10. doi: 10.1093/jnci/dju413. [DOI] [PubMed] [Google Scholar]
- 10.Conroy T, Desseigne F, Ychou M, Bouché O, Guimbaud R, Bécouarn Y, Adenis A, Raoul JL, Gourgou-Bourgade S, de la Fouchardière C, et al. FOLFIRINOX versus gemcitabine for metastatic pancreatic cancer. N Engl J Med. 2011;364:1817–1825. doi: 10.1056/NEJMoa1011923. [DOI] [PubMed] [Google Scholar]
- 11.Cecconi D, Palmieri M, Donadelli M. Proteomics in pancreatic cancer research. Proteomics. 2011;11:816–828. doi: 10.1002/pmic.201000401. [DOI] [PubMed] [Google Scholar]
- 12.Mostov KE, Kraehenbuhl JP, Blobel G. Receptor-mediated transcellular transport of immunoglobulin: Synthesis of secretory component as multiple and larger transmembrane forms. Proc Natl Acad Sci USA. 1980;77:7257–7261. doi: 10.1073/pnas.77.12.7257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rojas R, Apodaca G. Immunoglobulin transport across polarized epithelial cells. Nat Rev Mol Cell Biol. 2002;3:944–955. doi: 10.1038/nrm972. [DOI] [PubMed] [Google Scholar]
- 14.Kaetzel CS. The polymeric immunoglobulin receptor: Bridging innate and adaptive immune responses at mucosal surfaces. Immunol Rev. 2005;206:83–99. doi: 10.1111/j.0105-2896.2005.00278.x. [DOI] [PubMed] [Google Scholar]
- 15.Phalipon A, Corthésy B. Novel functions of the polymeric Ig receptor: Well beyond transport of immunoglobulins. Trends Immunol. 2003;24:55–58. doi: 10.1016/S1471-4906(02)00031-5. [DOI] [PubMed] [Google Scholar]
- 16.Denning GM. IL-4 and IFN-gamma synergistically increase total polymeric IgA receptor levels in human intestinal epithelial cells. Role of protein tyrosine kinases. J Immunol. 1996;156:4807–4814. [PubMed] [Google Scholar]
- 17.Kvale D, Løvhaug D, Sollid LM, Brandtzaeg P. Tumor necrosis factor-alpha up-regulates expression of secretory component, the epithelial receptor for polymeric Ig. J Immunol. 1988;140:3086–3089. [PubMed] [Google Scholar]
- 18.Poger ME, Hirsch BR, Lamm ME. Synthesis of secretory component by colonic neoplasms. Am J Pathol. 1976;82:327–338. [PMC free article] [PubMed] [Google Scholar]
- 19.Harris JP, Caleb MH, South MA. Secretory component in human mammary carcinoma. Cancer Res. 1975;35:1861–1864. [PubMed] [Google Scholar]
- 20.Harris JP, South MA. Secretory component: A glandular epithelial cell marker. Am J Pathol. 1981;105:47–53. [PMC free article] [PubMed] [Google Scholar]
- 21.DeSouza LV, Krakovska O, Darfler MM, Krizman DB, Romaschin AD, Colgan TJ, Siu KW. mTRAQ-based quantification of potential endometrial carcinoma biomarkers from archived formalin-fixed paraffin-embedded tissues. Proteomics. 2010;10:3108–3116. doi: 10.1002/pmic.201000082. [DOI] [PubMed] [Google Scholar]
- 22.Rossel M, Billerey C, Bittard H, Ksiazek P, Alber D, Revillard JP, Vuitton DA. Alterations in polymeric immunoglobulin receptor expression and secretory component levels in bladder carcinoma. Urol Res. 1991;19:361–366. doi: 10.1007/BF00310151. [DOI] [PubMed] [Google Scholar]
- 23.Rossel M, Seilles E, Voigt JJ, Vuitton D, Legait N, Revillard JP. Polymeric Ig receptor expression in hepatocellular carcinoma. Eur J Cancer. 1992;28:1120–1124. doi: 10.1016/0959-8049(92)90469-I. [DOI] [PubMed] [Google Scholar]
- 24.Ai J, Tang Q, Wu Y, Xu Y, Feng T, Zhou R, Chen Y, Gao X, Zhu Q, Yue X, et al. The role of polymeric immunoglobulin receptor in inflammation-induced tumor metastasis of human hepatocellular carcinoma. J Natl Cancer Inst. 2011;103:1696–1712. doi: 10.1093/jnci/djr360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Berntsson J, Lundgren S, Nodin B, Uhlén M, Gaber A, Jirström K. Expression and prognostic significance of the polymeric immunoglobulin receptor in epithelial ovarian cancer. J Ovarian Res. 2014;7:26. doi: 10.1186/1757-2215-7-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fristedt R, Gaber A, Hedner C, Nodin B, Uhlén M, Eberhard J, Jirström K. Expression and prognostic significance of the polymeric immunoglobulin receptor in esophageal and gastric adenocarcinoma. J Transl Med. 2014;12:83. doi: 10.1186/1479-5876-12-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xiao T, Ying W, Li L, Hu Z, Ma Y, Jiao L, Ma J, Cai Y, Lin D, Guo S, et al. An approach to studying lung cancer-related proteins in human blood. Mol Cell Proteomics. 2005;4:1480–1486. doi: 10.1074/mcp.M500055-MCP200. [DOI] [PubMed] [Google Scholar]
- 28.Rossel M, Brambilla E, Billaud M, Vuitton DA, Blanc-Jouvan F, Biichle S, Revillard JP. Nonspecific increased serum levels of secretory component in lung tumors: Relationship to the gene expression of the transmembrane receptor form. Am J Respir Cell Mol Biol. 1993;9:341–346. doi: 10.1165/ajrcmb/9.3.341. [DOI] [PubMed] [Google Scholar]
- 29.Makawita S, Smith C, Batruch I, Zheng Y, Rückert F, Grützmann R, Pilarsky C, Gallinger S, Diamandis EP. Integrated proteomic profiling of cell line conditioned media and pancreatic juice for the identification of pancreatic cancer biomarkers. Mol Cell Proteomics. 2011;10 doi: 10.1074/mcp.M111.008599. M111.008599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kvale D, Norstein J, Meling GI, Børmer OP, Brandtzaeg P, Langmark F, Rognum TO. Circulating secretory component in relation to early diagnosis and treatment of liver metastasis from colorectal carcinomas. J Clin Pathol. 1992;45:568–571. doi: 10.1136/jcp.45.7.568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morton CL, Houghton PJ. Establishment of human tumor xenografts in immunodeficient mice. Nat Protoc. 2007;2:247–250. doi: 10.1038/nprot.2007.25. [DOI] [PubMed] [Google Scholar]
- 32.Chijiwa T, Kawai K, Noguchi A, Sato H, Hayashi A, Cho H, Shiozawa M, Kishida T, Morinaga S, Yokose T, et al. Establishment of patient-derived cancer xenografts in immunodeficient NOG mice. Int J Oncol. 2015;47:61–70. doi: 10.3892/ijo.2015.2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Komura D, Isagawa T, Kishi K, Suzuki R, Sato R, Tanaka M, Katoh H, Yamamoto S, Tatsuno K, Fukayama M, et al. CASTIN: A system for comprehensive analysis of cancer-stromal interactome. BMC Genomics. 2016;17:899. doi: 10.1186/s12864-016-3207-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pontén F, Jirström K, Uhlen M. The human protein atlas-A tool for pathology. J Pathol. 2008;216:387–393. doi: 10.1002/path.2440. [DOI] [PubMed] [Google Scholar]
- 36.Iida Y, Tanaka H, Sano H, Suzuki Y, Shimizu H, Urano T. Ectopic expression of PCSK9 by smooth muscle cells contributes to aortic dissection. Ann Vasc Surg. 2018;48:195–203. doi: 10.1016/j.avsg.2017.10.005. [DOI] [PubMed] [Google Scholar]
- 37.Zijlstra M, Bernards N, de Hingh IH, van de Wouw AJ, Goey SH, Jacobs EM, Lemmens VE, Creemers GJ. Does long-term survival exist in pancreatic adenocarcinoma? Acta Oncol. 2016;55:259–264. doi: 10.3109/0284186X.2015.1096020. [DOI] [PubMed] [Google Scholar]
- 38.Ying H, Dey P, Yao W, Kimmelman AC, Draetta GF, Maitra A, DePinho RA. Genetics and biology of pancreatic ductal adenocarcinoma. Genes Dev. 2016;30:355–385. doi: 10.1101/gad.275776.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jin K, Li G, Cui B, Zhang J, Lan H, Han N, Xie B, Cao F, He K, Wang H, et al. Assessment of a novel VEGF targeted agent using patient-derived tumor tissue xenograft models of colon carcinoma with lymphatic and hepatic metastases. PLoS One. 2011;6:e28384. doi: 10.1371/journal.pone.0028384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yada E, Wada S, Yoshida S, Sasada T. Use of patient-derived xenograft mouse models in cancer research and treatment. Futur Sci OA. 2018;4:FSO271. doi: 10.4155/fsoa-2017-0136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hidalgo M, Amant F, Biankin AV, Budinská E, Byrne AT, Caldas C, Clarke RB, de Jong S, Jonkers J, Mælandsmo GM, et al. Patient-derived xenograft models: An emerging platform for translational cancer research. Cancer Discov. 2014;4:998–1013. doi: 10.1158/2159-8290.CD-14-0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cho SY, Kang W, Han JY, Min S, Kang J, Lee A, Kwon JY, Lee C, Park H. An integrative approach to precision cancer medicine using patient-derived xenografts. Mol Cells. 2016;39:77–86. doi: 10.14348/molcells.2016.2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fujii E, Suzuki M, Matsubara K, Watanabe M, Chen YJ, Adachi K, Ohnishi Y, Tanigawa M, Tsuchiya M, Tamaoki N. Establishment and characterization of in vivo human tumor models in the NOD/SCID/γcnull mouse. Pathol Int. 2008;58:559–567. doi: 10.1111/j.1440-1827.2008.02271.x. [DOI] [PubMed] [Google Scholar]
- 44.Erickson RA, Sayage-Rabie L, Beissner RS. Factors predicting the number of EUS-guided fineneedle passes for diagnosis of pancreatic malignancies. Gastrointest Endosc. 2000;51:184–190. doi: 10.1016/S0016-5107(00)70416-0. [DOI] [PubMed] [Google Scholar]
- 45.LeBlanc JK, Ciaccia D, Al-Assi MT, McGrath K, Imperiale T, Tao LC, Vallery S, DeWitt J, Sherman S, Collins E. Optimal number of EUS-guided fineneedle passes needed to obtain acorrect diagnosis. Gastrointest Endosc. 2004;59:475–481. doi: 10.1016/S0016-5107(03)02863-3. [DOI] [PubMed] [Google Scholar]
- 46.Ohkuma R, Yada E, Ishikawa S, Komura D, Ishizaki H, Tamada K, Kubota Y, Hamada K, Ishida H, Hirasawa Y, et al. High expression of olfactomedin-4 is correlated with chemoresistance and poor prognosis in pancreatic cancer. PLoS One. 2020;15:e0226707. doi: 10.1371/journal.pone.0226707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.De Sousa Abreu R, Penalva LO, Marcotte EM, Vogel C. Global signatures of protein and mRNA expression levels. Mol Biosyst. 2009;5:1512–1526. doi: 10.1039/b908315d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schwanhüusser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W, Selbach M. Global quantification of mammalian gene expression control. Nature. 2011;473:337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- 49.Wu L, Candille SI, Choi Y, Xie D, Jiang L, Li-Pook-Than J, Tang H, Snyder M. Variation and genetic control of protein abundance in humans. Nature. 2013;499:79–82. doi: 10.1038/nature12223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang B, Wang J, Wang X, Zhu J, Liu Q, Shi Z, Chambers MC, Zimmerman LJ, Shaddox KF, Kim S, et al. Proteogenomic characterization of human colon and rectal cancer. Nature. 2014;513:382–387. doi: 10.1038/nature13438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Arumugam P, Bhattacharya S, Chin-Aleong J, Capaso M, Kocher HM. Expression of polymeric immunoglobulin receptor and stromal activity in pancreatic ductal adenocarcinoma. Pancreatology. 2017;17:295–302. doi: 10.1016/j.pan.2017.01.013. [DOI] [PubMed] [Google Scholar]
- 52.Fristedt R, Elebro J, Gaber A, Jonsson L, Heby M, Yudina Y, Nodin B, Uhlén M, Eberhard J, Jirström K. Reduced expression of the polymeric immunoglobulin receptor in pancreatic and periampullary adenocarcinoma signifies tumour progression and poor prognosis. PLoS One. 2014;9:e112728. doi: 10.1371/journal.pone.0112728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu F, Ye P, Bi T, Teng L, Xiang C, Wang H, Li Y, Jin K, Mou X. COLORECTAL polymeric immunoglobulin receptor expression is correlated with hepatic metastasis and poor prognosis in colon carcinoma patients with hepatic metastasis. Hepatogastroenterology. 2014;61:652–659. [PubMed] [Google Scholar]
- 54.Wang X, Du J, Gu P, Jin R, Lin X. Polymeric immunoglobulin receptor expression is correlated with poor prognosis in patients with osteosarcoma. Mol Med Rep. 2014;9:2105–2110. doi: 10.3892/mmr.2014.2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Niu H, Wang K, Wang Y. Polymeric immunoglobulin receptor expression is predictive of poor prognosis in glioma patients. Int J Clin Exp Med. 2014;7:2185. [PMC free article] [PubMed] [Google Scholar]
- 56.Aguilar-Mahecha A, Hassan S, Ferrario C, Basik M. Microarrays as validation strategies in clinical samples: Tissue and protein microarrays. OMICS. 2006;10:311–326. doi: 10.1089/omi.2006.10.311. [DOI] [PubMed] [Google Scholar]
- 57.Simon R, Sauter G. Tissue microarrays for miniaturized high-throughput molecular profiling of tumors. Exp Hematol. 2002;30:1365–1372. doi: 10.1016/S0301-472X(02)00965-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used during the present study are available from the corresponding author upon reasonable request. All data generated or analyzed during this study are included in this published article. The results published in the present study are in part based upon data generated by the TCGA Research Network: https://www.cancer.gov/tcga.