Abstract
Gastric cancer is a deadly disease and remains the third leading cause of cancer-related death worldwide. The 5-year overall survival rate of patients with early-stage localized gastric cancer is more than 60%, whereas that of patients with distant metastasis is less than 5%. Surgical resection is the best option for early-stage gastric cancer, while chemotherapy is mainly used in the middle and advanced stages of this disease, despite the frequently reported treatment failure due to chemotherapy resistance. Therefore, there is an unmet medical need for identifying new biomarkers for the early diagnosis and proper management of patients, to achieve the best response to treatment. Long non-coding RNAs (lncRNAs) in body fluids have attracted widespread attention as biomarkers for early screening, diagnosis, treatment, prognosis, and responses to drugs due to the high specificity and sensitivity. In the present review, we focus on the clinical potential of lncRNAs as biomarkers in liquid biopsies in the diagnosis and prognosis of gastric cancer. We also comprehensively discuss the roles of lncRNAs and their molecular mechanisms in gastric cancer chemoresistance as well as their potential as therapeutic targets for gastric cancer precision medicine.
Keywords: LncRNA, Gastric cancer, Precision medicine, Early diagnosis, Cancer treatment, Chemoresistance
Background
Gastric cancer is one of the most common malignancies worldwide, with more than one million new cases every year, and remains the third leading cause of cancer-related deaths [1, 2]. The clinical stage at the time of diagnosis directly determines the prognosis of patients with this disease. The patients with localized, early-stage gastric cancer usually have a high 5-year overall survival (OS) rate (> 60%), whereas the 5-year OS rates for gastric cancer patients with local and distant metastasis dramatically decrease to 30 and 5%, respectively [2]. Unfortunately, due to the occult and atypical nature of early clinical symptoms of gastric cancer, more than 60% of patients have local or distant metastases at the time of diagnosis [2]. For patients with early gastric cancer, surgical resection is the best treatment option; for patients who cannot undergo surgical resection or patients with advanced metastases, chemotherapy is the most important treatment [3, 4]. However, poor or even no response to chemotherapy is often observed in gastric cancer patients because of the intrinsic or acquired resistance, which becomes the most common cause of treatment failure [5]. Therefore, the low rate of early diagnosis and chemotherapy resistance constitute the main contributions to the poor prognosis of gastric cancer.
To date, the biomarkers commonly used in early screening for gastric cancer include carcinoembryonic antigen (CEA), alpha-fetoprotein (AFP), carbohydrate antigen 19–9 (CA19–9), CA72–4, CA125, etc. [6, 7]. However, the sensitivities and positive rates of these biomarkers are poor; their sensitivities in the diagnosis of gastric cancer are from 4.7 to 33.3%, and the positive rates of CEA, CA199, and CA724 only range from 21.1 to 30% [7–9]. The diagnosis of gastric cancer still depends on upper gastrointestinal endoscopy, but its clinical application is limited because of the invasiveness and high cost [10]. Therefore, there is an urgent need for minimal-invasive or non-invasive detection approaches, as well as highly specific biomarkers, to improve gastric cancer early diagnosis and survival outcomes.
Long non-coding RNAs (lncRNAs) have attracted increasing attention as cancer biomarkers for early screening, diagnosis, prognosis, and responses to drug treatment [11–13]. A recent study has shown that the expression of lncRNA MNX1-AS1 (MNX1 antisense RNA 1) is significantly increased in gastric cancer tissues and associated with the poor prognosis of gastric cancer patients [14]. LncRNA SNHG11 (small nucleolar RNA host gene 11) has been reported as a potential biomarker for early detection of colon cancer and a new therapeutic target of this disease [15]. A stroma-related lncRNA panel has been found to predict recurrence and adjuvant chemotherapy benefit in patients with early-stage colon cancer [16]. LncRNAs are involved in the acquired resistance to chemotherapy [17, 18], and targeting lncRNA can reverse drug resistance and enhance the sensitivity of cancer cells to chemotherapy [19]. Given the importance of lncRNAs in cancer, a better understanding of their roles in the early diagnosis, treatment, prognosis, and drug resistance of gastric cancer may provide new insights for precise treatment and individualized management of patients with this disease.
The regulation of lncRNA expression and the roles of lncRNAs in gastric cancer progression and metastasis have been extensively discussed in several recent reviews [20–24]. In the present review, we focus on the clinical evidence of lncRNAs as biomarkers in liquid biopsies in the early diagnosis and prognosis of gastric cancer. We also comprehensively discuss the roles of lncRNAs and their molecular mechanisms in gastric cancer chemoresistance, as well as their potential as therapeutic targets for gastric cancer precise medicine.
An overview of lncRNAs
The Encyclopedia of DNA Elements (ENCODE) project has revealed that only about 1.2% of human transcripts (RNAs) encode proteins and more than 98% of human transcripts are non-protein-coding RNAs (ncRNAs), such as lncRNAs, circular RNAs (circRNAs), microRNAs (miRNAs), and small nucleolar RNAs (snoRNAs) [25]. LncRNAs are the transcripts of more than 200 nucleotides, accounting for 80 to 90% of all ncRNAs and are characterized by low expression levels, poor interspecies conservation, and high expression coefficient of variance [26, 27].
According to their genomic localization and evolutionary lineage, lncRNAs can be divided into intergenic lncRNAs, intronic lncRNAs, exonic lncRNAs, sense lncRNAs, and antisense lncRNAs. Intergenic lncRNAs (also called lincRNAs) are transcribed from genomic regions between coding genes, while intronic lncRNAs overlap entirely with introns of protein-coding genes and exonic lncRNAs overlap entirely or partially with exons of protein-coding genes [28, 29]. The transcriptional orientation of lncRNAs can be in sense or antisense when compared with the transcriptional orientation of the protein-coding genes [30]. Besides, lncRNAs can be classified into nuclear lncRNAs and cytoplasmic lncRNAs based on the subcellular localization, which is critical for their functions. Most lncRNAs are located in the nucleus and only about 15% are in the cytoplasm [31]. Nuclear lncRNAs mainly regulate the transcription or mRNA processing, e.g. lncRNA XIST (X inactive specific transcript), MALAT1 (metastasis associated lung adenocarcinoma transcript 1), and NEAT1 (nuclear paraspeckle assembly transcript 1) functioning as transcription regulators [32–34]. Cytoplasmic lncRNAs are more often involved in post-transcriptional regulation, such as playing the role of miRNA sponges. Du et al. have demonstrated that cytoplasmic localization is an important factor in determining the sponge efficacy of lncRNA TUG1 (taurine up-regulated 1) [35]. Cytoplasmic lncRNA PVT1 (plasmacytoma variant translocation 1) has been found to act as a competitive endogenous RNA (ceRNA) against miR-214-3p and promote the progression of colon cancer [36].
LncRNAs were initially considered as “junk” or “genomic dark matter” without function. With the deepening of research in recent years, lncRNAs have been found to participate widely in various physiological and pathological processes of organisms. In the human body, lncRNAs not only regulate the physiological processes such as cell proliferation, differentiation, and apoptosis but also participate in regulating various pathological processes of the body, such as cancer, cardiovascular diseases, autoimmune diseases, diabetes, and more [37–40]. The specific function of lncRNAs is to regulate gene expression at the pre-transcriptional, transcriptional, and post-transcriptional levels. At the pre-transcriptional level, lncRNA regulates gene expression by gene modification, histone modification, and chromatin remodeling, without changing the DNA sequences of the organisms [41, 42]. During transcription, lncRNA interacts with transcription factors to regulate gene transcription [43]. At the post-transcriptional level, lncRNA acts as a precursor of some miRNAs to regulate gene expression, or as a ceRNA to regulate the translation of the corresponding mRNA [44]. However, due to the large number of lncRNAs, the functions of most lncRNAs are still unclear and require further comprehensive research.
LncRNAs as liquid biopsy biomarkers of gastric cancer
The development of liquid biopsies has opened a new era for precision medical treatment of human cancer. Because of their minimal-invasive or non-invasive characteristics and high public acceptance, liquid biopsies can be conducted more frequently for early screening, diagnosis, and prognosis of cancer. Besides, liquid biopsies can be collected at specific time intervals to monitor responses to treatment, drug resistance, recurrence, and metastasis of cancer. Added benefits are that, unlike tissue biopsies obtained from only one tumor area, liquid biopsies may better reflect the genetic characteristics of all tumor subclones in patients [45]. LncRNAs are widely distributed in peripheral plasma/serum, saliva, gastric juice, urine, semen, and other liquids and play important roles in various aspects of human physiological and pathological processes [46–50]. Based on the aforementioned benefits, a comprehensive understanding of the current research status of lncRNAs is critical for the further development of them as cancer biomarkers in liquid biopsies.
Accumulating evidence suggests the usefulness of lncRNAs as liquid biopsy biomarkers for human cancer. LncRNAs in peripheral blood plasma/serum have been demonstrated as biomarkers for various types of human cancer, such as lung, breast, and colon cancer [51–53]. LncRNAs in saliva have been mainly used as biomarkers for head and neck cancer, such as oral, pharyngeal, and laryngeal cancer [54, 55]. LncRNAs in gastric juice and urine have also been reported as biomarkers of gastric cancer and urinary system cancer, respectively [49]. Of note, the urinary level of lncRNA PCA3 (prostate cancer associated 3) has been used as a biomarker for the diagnosis of prostate cancer in clinical applications [56, 57]. Although there is no report on lncRNAs in semen, recent studies have shown that miRNAs in semen may be used as biomarkers for prostate cancer [58]. To date, almost all attention has been paid to the lncRNAs in plasma/serum and gastric juice but not in other liquid biopsies as biomarkers of gastric cancer, which has been comprehensively discussed in this section.
LncRNAs in plasma/serum as diagnostic and prognostic biomarkers of gastric cancer
The development of a disease often leads to changes in the plasma/serum composition, which can be detected to reflect the status of the disease [59]. LncRNAs, which are freely circulating in the plasma/serum or packaged in exosomes, have all of the characteristics of ideal biomarkers because they are stable over long periods at room temperature, during repeated freeze-thaw cycles, or at different pH values [60]. More importantly, the plasma/serum levels of lncRNAs are mostly the same as those in the primary tumor tissues, thus precisely reflecting the characteristics of the tumors [61, 62]. In addition, the collection of plasma/serum samples at different time points is relatively convenient for monitoring the progress of the disease [63–66].
LncRNAs in plasma/serum as diagnostic biomarkers of gastric cancer
A large number of circulating lncRNAs have been reported as biomarkers for the diagnosis of gastric cancer (as summarized in Table 1), which have obvious advantages over the diagnostic biomarkers in clinical applications. Xian et al. have found that lncRNA HULC (hepatocellular carcinoma upregulated long noncoding RNA) and ZNFX1-AS1 (ZNFX1 antisense RNA 1) can distinguish gastric cancer patients from healthy controls and have proposed them as biomarkers for diagnosing gastric cancer [77]. The receiver operator characteristic curve (ROC) analysis has shown that the area under curve (AUC) values for HULC and ZNFX1-AS1 are 0.65 and 0.85, respectively, which are higher than those of traditional serum biomarkers, including CEA (0.62), CA19–9 (0.56), CY211 (0.59), and neuron-specific enolase (NSE, 0.56) [77]. Jin et al. have further confirmed that HULC is more sensitive and specific than CEA and CA724 as a diagnostic marker of gastric cancer [82]. Yang et al. have found that the AUC values of lncRNA PANDAR (promoter of CDKN1A antisense DNA damage activated RNA), FOXD2-AS1 (FOXD2 adjacent opposite strand RNA 1), and SMARCC2 (SWI/SNF related, matrix associated, actin dependent regulator of chromatin subfamily c member 2) as diagnostic biomarkers of gastric cancer are 0.77, 0.7, and 0.75, respectively, which are similar to the AUC value of combined CEA, AFP, CA125, CA153, and CA199 [97]. Feng et al. have also demonstrated that lncRNA B3GALT5-AS1 (B3GALT5 antisense RNA 1) is better than CEA and CA19–9 as a diagnostic biomarker of gastric cancer [87]. Zhou et al. have recently reported that lncRNA C5orf66-AS1 (C5orf66 antisense RNA 1) can be utilized for the diagnosis of gastric cancer with the AUC value of 0.688 [67]. More importantly, lncRNA C5orf66-AS1 has further been shown to predict early gastric cancer with the AUC value of 0.789 [67].
Table 1.
LncRNAs in plasma/serum as diagnostic and prognostic biomarkers of gastric cancer
| LncRNA | Biomarker type | Expression | Cases | Sensitivity | Specificity | AUC | Sample | Refs |
|---|---|---|---|---|---|---|---|---|
| C5orf66-AS1 | Diagnostic | Down | 200 patients with GC and 278 non-GC | 77.5% | 53.6% | 0.688 | Serum | [67] |
| PTCSC3 | Diagnostic/Prognostic | Down | 68 patients with GC and 60 healthy controls | N/A | N/A | 0.92 | Plasma | [68] |
| ARHGAP27P1 | Diagnostic/Prognostic | Down | 53 patients with GC and 53 healthy controls | 75.5% | 60.4% | 0.732 | Plasma | [61] |
| TUBA4B | Diagnostic/Prognostic | Down | 37 patients with GC and 37 healthy controls | N/A | N/A | 0.8075 | Plasma | [69] |
| LINC00086 | Diagnostic/Prognostic | Down | 168 patients with GC and 74 healthy controls | 72.6% | 83.8% | 0.860 | Plasma | [70] |
| DGCR5 | Diagnostic/Prognostic | Down | 34 patients with GC and 34 healthy controls | N/A | N/A | 0.722 | Plasma | [71] |
| SNHG17 | Diagnostic/Prognostic | Up | 67 patients with GC and 67 healthy controls | N/A | N/A | 0.748 | Plasma | [72] |
| MEF2C-AS1 | Diagnostic | Down | 46 patients with GC and 21 healthy controls | N/A | N/A | 0.733 | Plasma | [73] |
| MT1JP | Diagnostic/Prognostic | Down | 34 patients with GC and 34 healthy controls | N/A | N/A | 0.649 | Plasma | [64] |
| GACAT2 | Diagnostic/Prognostic | Up | 117 patients with GC and 80 healthy controls | 87.2% | 28.2% | 0.622 | Plasma | [74] |
| RMRP | Diagnostic | Down | 83 patients with GC and 90 healthy controls | 59.1% | 67.8% | 0.693 | Plasma | [49] |
| UCA1 | Diagnostic | Up | 20 patients with GC and 20 healthy controls | 89.2% | 80.3% | 0.928 | Plasma | [75] |
| LINC00152 | Diagnostic | Up | 79 patients with GC and 81 healthy controls | 48.1% | 85.2% | 0.657 | Plasma | [76] |
| ZNFX1-AS1 | Diagnostic | Up | 50 patients with GC and 50 healthy controls | 84% | 68% | 0.85 | Plasma | [77] |
| GASL1 | Diagnostic | Down | 112 patients with GC and 56 healthy controls | N/A | N/A | N/A | Plasma | [78] |
| GASL1 | Diagnostic/Prognostic | Down | 88 patients with GC and 72 healthy controls | N/A | N/A | 0.8945 | Serum | [79] |
| MALAT1 | Diagnostic/Prognostic | Up | 64 patients with GC and 64 healthy controls | N/A | N/A | 0.8984 | Plasma | [80] |
| MALAT1 | Prognostic | Up | 36 GC/NDM and 36 GC/DM | N/A | N/A | N/A | Plasma | [81] |
| HULC | Diagnostic | Up | 50 patients with GC and 50 healthy controls | 58% | 80% | 0.65 | Plasma | [77] |
| HULC | Diagnostic | Up | 100 patients with GC and110healthy controls | 82% | 83.6% | 0.888 | Serum | [82] |
| H19 | Diagnostic | Up | 43 patients with GC and 34 healthy controls | 74% | 58% | 0.64 | Plasma | [83] |
| H19 | Diagnostic | Up | 70 patients with GC and 70 healthy controls | 82.9% | 72.9% | 0.838 | Plasma | [84] |
| H19 | Diagnostic | Up | 35 patients with GC and 25 healthy controls | 90.9% | 100% | 0.982 | Plasma | [85] |
| H19 | Diagnostic | Up | 40 patients with GC and 42 healthy controls | 87.2% | 37.2% | 0.643 | Plasma | [86] |
| B3GALT5-AS1 | Diagnostic | Up | 107 patients with GC and 87 healthy controls | 87.4% | 74.7% | 0.816 | Serum | [87] |
| HOXA11-AS | Diagnostic/Prognostic | Up | 94 patients with GC and 40 healthy controls | 78.7% | 97.8% | 0.924 | Serum | [88] |
| SNHG6 | Diagnostic/Prognostic | Up | 114 patients with GC and 99 healthy controls | N/A | N/A | N/A | Serum | [89] |
| DANCR | Diagnostic/Prognostic | Up | 55 patients with GC and 39 healthy controls | 72.7% | 79.5% | 0.816 | Serum | [90] |
| LINC00978 | Diagnostic | Up | 38 patients with GC and 31 healthy controls | 80% | 70% | 0.831 | Serum | [91] |
| ZFAS1 | Diagnostic/Prognostic | Up | 77 patients with GC and 60 healthy controls | 76.6% | 63.9% | 0.727 | Plasma | [92] |
| Exosomal ZFAS1 | Diagnostic/Prognostic | Up | 60 patients with GC and 37 healthy controls | 71.7% | 75.7% | 0.792 | Serum | [93] |
| Exosomal lncUEGC1 | Diagnostic | Up | 51 patients with GC and 60 healthy controls | N/A | N/A | 0.8760 | Plasma | [65] |
| Exosomal lncUEGC2 | Diagnostic | Up | 51 patients with GC and 60 healthy controls | N/A | N/A | 0.7582 | Plasma | [65] |
| Exosomal PCSK2–2:1 | Diagnostic/Prognostic | Down | 63 patients with GC and 29 healthy controls | 84% | 86.5% | 0.896 | Serum | [94] |
| Exosomal GNAQ-6:1 | Diagnostic | Down | 43 patients with GC and 27 healthy controls | 83.7% | 55.6% | 0.732 | Serum | [95] |
| Exosomal MIAT | Diagnostic/Prognostic | Up | 109 patients with GC and 50 healthy controls | N/A | N/A | 0.892 | Serum | [96] |
| PANDAR | Diagnostic | Up | 109 patients with GC and 106 healthy controls | N/A | N/A | 0.767 | Plasma | [97] |
| FOXD2-AS1 | Up | N/A | N/A | 0.700 | ||||
| SMARCC2 | Up | N/A | N/A | 0.748 | ||||
| Combined | N/A | N/A | N/A | 0.839 | ||||
| H19 | Diagnostic | Up | 62 patients with GC and 40 healthy controls | 74.19% | 90.0% | 0.854 | Plasma | [98] |
| MEG3 | Down | 95.16% | 42.50% | 0.638 | ||||
| miR-675-5p | Up | 77.42% | 52.50% | 0.661 | ||||
| Combined | N/A | 88.87% | 85% | 0.927 | ||||
| CTC-501O10.1 | Diagnostic | Up | 100 patients with GC and 100 healthy controls | 90% | 51% | 0.74 | Plasma | [99] |
| AC100830.4 | Up | 84% | 58% | 0.73 | ||||
| RP11-210 K20.5 | Up | 89% | 55% | 0.737 | ||||
| Combined | N/A | 99% | 49% | 0.764 | ||||
| INHBA-AS1 | Diagnostic | Up | 51 patients with GC and 53 healthy controls | N/A | N/A | 0.855 | Plasma | [100] |
| MIR4435–2HG | Up | N/A | N/A | 0.882 | ||||
| CEBPA-AS1 | Up | N/A | N/A | 0.785 | ||||
| AK001058 | Up | N/A | N/A | 0.852 | ||||
| Combined | N/A | N/A | N/A | 0.921 | ||||
| TINCR | Diagnostic | Up | 80 patients with GC and 80 healthy controls | 69% | 56% | 0.66 | Plasma | [66] |
| CCAT2 | Up | 85% | 51% | 0.63 | ||||
| AOC4P | Up | 86% | 41% | 0.67 | ||||
| BANCR | Up | 75% | 78% | 0.81 | ||||
| LINC00857 | Up | 93% | 26% | 0.61 | ||||
| Combined | N/A | 82% | 87% | 0.91 | ||||
| FAM49B-AS | Diagnostic | Up | 223 patients with GC and 223 healthy controls | N/A | N/A | 0.609 | Plasma | [101] |
| GUSBP11 | Up | N/A | N/A | 0.635 | ||||
| CTDHUT | Up | N/A | N/A | 0.762 | ||||
| Combined | N/A | 77.5% | 73.9% | 0.818 | ||||
| United CA242, CA724 | N/A | 93.2% | 86.6% | 0.952 | ||||
| H19 | Diagnostic | Up | 32 patients with GC and 30 healthy controls | 68.75% | 56.67% | 0.724 | Plasma | [102] |
| United CEA | N/A | N/A | N/A | 0.804 | ||||
| CTC-497E21.4 | Diagnostic | Up | 110 patients with GC and 84 healthy controls | 81.82% | 75.00% | 0.848 | Serum | [103] |
| United CEA, CA199 | N/A | 96.36% | 42.86% | 0.896 | ||||
| Exosomal HOTTIP | Diagnostic | Up | 126 patients with GC and 120 healthy controls | 69.8% | 85.0% | 0.827 | Serum | [104] |
| United CEA, CA199, CA724 | N/A | N/A | N/A | 0.870 |
N/A Not available; AUC Area under curve; GC Gastric cancer
Circulating lncRNAs have better biomarker values when combined, e.g. combining lncRNA PANDAR, FOXD2-AS1, and SMARCC2 increases the AUC value to 0.84 [97]. The combination of lncRNA CTC-501O10.1, AC100830.4, and RP11-210 K20.5 has been found to improve the sensitivity of the diagnosis to 99% [99]. The combination of lncRNA INHBA-AS1 (INHBA antisense RNA 1), MIR4435–2HG (MIR4435–2 host gene), CEBPA-AS1 (CEBPA divergent transcript), and AK001058 has increased the AUC value to 0.92 [100]. Also, the combination of lncRNA TINCR (terminal differentiation-induced ncRNA), CCAT2 (colon cancer associated transcript 2), AOC4P (amine oxidase copper containing 4, pseudogene), BANCR (BRAF-activated non-protein coding RNA), and LINC00857 has increased the AUC value to 0.91, the sensitivity to 82%, and the specificity to 87% [66]. Meanwhile, combining lncRNAs and miRNAs have also improved their diagnostic efficiency, e.g. the AUC value of lncRNA H19 (H19 imprinted maternally expressed transcript) and MEG3 (maternally expressed 3) combined with miR-675-5p is 0.93 while the specificity and sensitivity are 88.9 and 85%, respectively [98]. However, the sensitivities of H19, MEG3, and miR-675-5p are 74.19, 95.16, and 77.42%, respectively, their respective specificities are 90.0, 42.50, and 52.50%, and their AUC values range from 0.638 to 0.854 [98]. Moreover, lncRNAs combined with traditional serum tumor markers have improved the diagnostic efficiency, e.g. lncRNA CTC-497E21.4 combined with CEA and CA199 has increased the AUC value to 0.9 [103]. Using the lncRNA FAM49B-AS (FAM49B antisense RNA), GUSBP11 (GUSB pseudogene 11), and CTDHUT (CTD highly upregulated transcript) combined with A242 and CA724, the AUC value, sensitivity, and specificity have been increased to 0.95, 93.2, and 86.6%, respectively [101].
LncRNAs in plasma/serum as prognostic biomarkers of gastric cancer
Tumor size, stage, depth of invasion, lymph node metastasis, distant metastasis, and pathological type are the relevant factors for the prognosis of cancer patients [105]. Circulating lncRNAs have been associated with these prognosis-related factors and have been demonstrated as prognostic biomarkers of gastric cancer (as summarized in Table 1). It has been found that the expression levels of lncRNA GASL1 (growth arrest associated lncRNA 1), PTCSC3 (papillary thyroid carcinoma susceptibility candidate 3), and MALAT1 are significantly correlated with tumor size, TNM (tumor, node, metastasis) stage, and distant metastasis of gastric cancer, respectively [68, 79, 80]. The expression levels of lncRNA SNHG6, ARHGAP27P1 (Rho GTPase activating protein 27 pseudogene 1), DANCR (differentiation antagonizing non-protein coding RNA), DGCR5 (DiGeorge syndrome critical region gene 5), MT1JP (metallothionein 1 J, pseudogene), SNHG17, and ZFAS1 (ZNFX1 antisense RNA 1) are closely related to the TNM stage, tumor invasion depth, and lymph node metastasis of gastric cancer [61, 64, 71, 72, 89, 90, 92]. It has also been reported that lncRNA HOXA11-AS (HOXA11 antisense RNA) and TUBA4B (tubulin alpha 4b) are tightly correlated with the tumor size, TNM stage, and lymph node metastasis of gastric cancer [69, 88]. Importantly, the Kaplan-Meier survival curve analysis has indicated that the patients with low expression of HOXA11-AS have a better survival rate, whereas the patients with low expression of TUBA4B have a shorter survival time [69, 88]. Tan et al. have demonstrated a significant correlation between the expression level of lncRNA GACAT2 (gastric cancer associated transcript 2) and the lymph node metastasis, distant metastasis, and perineural invasion of gastric cancer [74]. In addition, Ji et al. have shown that LINC00086 expression level is significantly associated with tumor size, lymph node metastasis, TNM stage, and the levels of CEA and CA19–9, while the gastric cancer patients with low expression of LINC00086 have low survival rates [70].
Exosomal lncRNAs in plasma/serum as diagnostic and prognostic biomarkers of gastric cancer
In the blood, long RNAs may be packaged into extracellular vesicles, which makes them more stable in plasma/serum. According to their diameters, the extracellular vesicles are classified into apoptotic bodies (50–5000 nm), microvesicles (50–1000 nm), and exosomes (30–100 nm) [106]. Apoptotic bodies are produced by cells undergoing programmed cell death, microvesicles are vesicles directly released from cell membranes, and exosomes are intracellular in origin [107, 108]. Among these types of vesicles, exosomes are the most abundant reservoir of lncRNAs [106]. Due to their intracellular origin and high quantities of long RNAs, circulating exosomal lncRNAs have been proposed as promising biomarkers for gastric cancer [109].
Compared with traditional diagnostic biomarkers (CEA, CA724, and CA199), circulating exosomal lncRNA PCSK2–2:1 (proprotein convertase subtilisin/kexin type 2–2:1) and GNAQ-6:1 (G protein subunit alpha q-6:1) have been reported as better biomarkers for distinguishing gastric cancer patients from healthy people. The AUCs (0.9 and 0.74, respectively), sensitivities (84 and 83.7%, respectively), and specificities (86.5 and 55.6%, respectively) of PCSK2–2:1 and GNAQ-6:1 are significantly better than the best traditional diagnostic biomarker CA724, which only has an AUC value of 0.57, a sensitivity of 56%, and a specificity of 65.5% [94, 95]. It has also been shown that the exosomal PCSK2–2:1 level is significantly related to the tumor size, TNM stage, and venous infiltration and may be developed as a prognostic biomarker of gastric cancer [94]. Lin et al. have found that the expression levels of exosomal lncRNA UEGC1 (ENST00000568893) and UEGC2 (ENST00000378432.1) are increased in patients with gastric cancer [65]. The stability tests have shown that almost all plasma UEGC1 is encapsulated by exosomes and has a higher AUC value while UEGC2 is only partially encapsulated by exosomes, suggesting that UEGC1 is more suitable to be developed as a diagnostic biomarker for early gastric cancer [65]. Xu et al. have shown that the serum level of exosomal lncRNA MIAT (myocardial infarction associated transcript) is significantly increased in gastric cancer patients, which is associated with worse clinical variables and shorter survival [96]. Moreover, it has been found that the serum exosomal MIAT is down-regulated in patients after treatment but markedly up-regulated in patients suffering recurrence [96]. Furthermore, exosomal lncRNAs combined with serum tumor markers have shown improved diagnostic accuracy, e.g., exosomal lncRNA HOTTIP (HOXA distal transcript antisense RNA) combined with CEA, CA199, and CA724 have been found to increase the AUC value from 0.83 to 0.87 [104].
Taken together, lncRNAs in plasma/serum have shown great potential as biomarkers for the diagnosis and prognosis of gastric cancer. Importantly, the combinations, including but not limited to multiple lncRNAs combinations, lncRNA and miRNA combinations, and lncRNA and serum tumor marker combinations usually have better values as diagnostic biomarkers compared to an individual lncRNA. The exosomal lncRNAs in plasma/serum have also shown an advantage as biomarkers due to their high stability; however, further verification studies are needed. Moreover, controversial results have been obtained for the same lncRNA in gastric cancer. LncRNA H19 has been demonstrated as a diagnostic biomarker of gastric cancer with a large range of AUC values (0.6–0.98) in recent studies, which may be correlated with the individual differences [83–86]. Further investigations with larger sample size are warranted for improving accuracy and precision. The specific source and molecular mechanisms of lncRNAs in plasma/serum are yet to be determined.
LncRNAs in gastric juice as diagnostic and prognostic biomarkers of gastric cancer
Gastric juice is directly secreted by the gastric mucosa and can sensitively reflect the pathological state of the stomach, making it an ideal sample for studying gastric cancer [110]. Recent studies have shown that lncRNAs in gastric juice are specific and their expression levels may be inconsistent with those in tissue and plasma. Fei et al. have found that the expression level of LINC00982 is significantly decreased in tumor tissues but increased in gastric juice from patients with gastric cancer [111]. Similar results have been obtained for lncRNA RMRP (RNA component of mitochondrial RNA processing endoribonuclease) and AA174084 by Shao et al. [49, 112]. It has been speculated that some lncRNAs may be secreted actively by gastric cancer cells during the disease process or partly by exosomes or other pathways [112].
To date, several lncRNAs, including RMRP, AA174084, PVT1, H19, LINC00982, ABHD11-AS1 (ABHD11 antisense RNA 1), UCA1 (urothelial cancer associated 1), and LINC00152 have been identified from gastric juice and demonstrated as biomarkers for gastric cancer. The sensitivities, specificities, and AUC values of these newly characterized diagnostic biomarkers of gastric cancer range from 41 to 56.4%, 75.4 to 93.4%, and 0.65 to 0.85, respectively. Furthermore, the expression level of AA174084 in gastric juice has been correlated with tumor size, tumor stage, Lauren type, and CEA level in the gastric juice, and a higher AA174084 level in gastric juice indicates a poorer prognosis of gastric cancer patients [112]. The expression level of ABHD11-AS1 in gastric juice has also been associated with the tumor size, tumor stage, and CEA level in the blood, while the high level of ABHD11-AS1 suggests an increased risk of gastric cancer recurrence [113]. Therefore, AA174084 and ABHD11-AS1 can be used for both the diagnosis and prognosis of gastric cancer. In addition, the combination of gastric juice ABHD11-AS1, serum CEA, and gastric juice CEA can improve the diagnostic accuracy of early gastric cancer [113].
In summary, due to the high specificity and reliability, gastric juice lncRNAs can be used as biomarkers for the diagnosis and prognosis of gastric cancer. However, an individual gastric juice lncRNA always has high specificity but insufficient sensitivity as a biomarker. More combination studies, such as the combination of multiple gastric juice lncRNAs, the combination of gastric juice lncRNAs with plasma lncRNAs, and the combination of gastric juice lncRNAs with serum tumor markers may be carried out to increase the sensitivity. Also, further investigations are needed to explore the specific source and molecular mechanisms of gastric juice lncRNAs.
LncRNA-mediated regulation of chemoresistance in gastric cancer
Chemotherapy is the main treatment option for patients with advanced gastric cancer, while drug resistance is the major cause of gastric cancer treatment failure. The mechanisms of cancer chemoresistance include, but not limited to, drug degradation, amplification and overexpression of oncogenes, anti-apoptosis, immune escape, epithelial-mesenchymal transition (EMT), cancer stemness, autophagy, epigenetic modifications, and up-regulation of multidrug resistance (MDR)-related genes [114–119]. Recent studies have shown that lncRNAs are widely involved in regulating various mechanisms of cancer chemoresistance [120]. LncRNAs have been found to regulate drug resistance by acting as a ceRNA or directly binding to mRNAs or proteins and modulating their expression and/or functions. In this section, we provide a summary of the molecular mechanisms for lncRNAs-mediated gastric cancer chemoresistance (as summarized in Table 2).
Table 2.
Mechanisms of chemotherapy resistance mediated by lncRNAs
| LncRNA | Effection | Drugs | Pathway/target | Mechanism | Refs |
|---|---|---|---|---|---|
| MALAT1 | Inducing | 5-FU, DDP, VCR | miR-23B-3P, ATG12, miR-30b, ATG5, SOX2, nanog | CeRNA, Inducing autophagy, Increasing cancer stemness | [121–123] |
| AK022798 | Inducing | DDP | caspase8, caspase3, MRP1 | Inhibiting cell apoptosis, Regulating MDR-related genes | [124] |
| CRAL | Reversing | DDP | miR-505, CYLD, PI3K/AKT | CeRNA, Promoting DNA damage and apoptosis | [125] |
| ROR | Inducing | ADR, VCR | MRP1 | Inhibiting cell apoptosis, Regulating MDR-related genes | [126] |
| XLOC_006753 | Inducing | 5-FU, DDP | PI3K/AKT/mTOR, caspase9, Wnt/β-catenin, Vimentin, Snail | Inhibiting cell apoptosis, Promoting EMT | [127] |
| MACC1-AS1 | Inducing | 5-FU, OXA | miR-145-5p, CD133, OCT4, SOX2, LIN28 | CeRNA, Increasing cancer stemness | [128] |
| D63785 | Inducing | DOX | miR-422a, MEF2D | CeRNA, Inhibiting cell apoptosis | [129] |
| LINC01433 | Inducing | DOX, DDP | YAP, USP9X | Inhibiting cell apoptosis | [130] |
| HOXD-AS1 | Inducing | DDP |
EZH2, PDCD4, H3K27me3 |
Epigenetically silencing PDCD4 via recruiting EZH2 | [131] |
| HULC | Reversing | DDP, ADM, 5-FU | FOXM1 | Suppressing autophagy, Promoting cell apoptosis | [132, 133] |
| PCAT-1 | Inducing | DDP | miR-128, ZEB1, EZH2, PTEN, H3K27me3 | CeRNA, Promoting EMT, Epigenetically silencing PTEN via recruiting EZH2 | [134, 135] |
| CASC2 | Reversing | DDP | miR-19a | CeRNA, Promoting cell apoptosis | [136] |
| HOTAIR | Inducing | DDP, 5-FU, ADM, MMC, PTX |
miR-17-5p, PTEN, miR-217, miR-34a, PI3K/AKT, Wnt/β-catenin miR-126 |
CeRNA, Promoting EMT, Regulating MDR-related genes, Inhibiting cell apoptosis, Promoting cell proliferation | [137–140] |
| THOR | Inducing | DDP | SOX9 | Increasing cancer stemness | [141] |
| BLACAT1 | Inducing | OXA | miR-361, ABCB1 | CeRNA, Inhibiting apoptosis and promoting invasion, Regulating MDR-related genes | [142] |
| GHET1 | Inducing | DDP | BAK, BCL-2, MDR1, MRP1 | Inhibiting cell apoptosis, Regulating MDR-related genes | [143] |
| ANRIL | Inducing | 5-FU, DDP | MDR1, MRP1 | Regulating MDR-related genes | [144] |
| UCA1 | Inducing | ADM, DDP, 5-FU | PARP, BCL-2, miR-27b, caspase-3 | CeRNA, Inhibiting cell apoptosis | [145, 146] |
| PVT1 | Inducing | PTX, 5-FU, DDP | BCL-2, MDR1, MRP1, mTOR, HIF-1α | Inhibiting cell apoptosis | [147, 148] |
| MRUL | Inducing | ADM, VCR | ABCB1 | Inhibiting cell apoptosis | [149] |
| SNHG5 | Inducing | DDP | BCL-2, BAX, MDR1, MRP1 | Inhibiting cell apoptosis, Regulating MDR-related genes | [150] |
| DANCR | Inducing | DDP | MDR1, MRP1 | Inhibiting cell apoptosis, Regulating MDR-related genes | [151] |
| FAM84B-AS | Inducing | DDP | FAM84B, caspase3, caspase7, caspase9, BCL2, BCL-xL | Inhibiting cell apoptosis | [152] |
| BCAR4 | Inducing | DDP | Wnt/β-catenin, Nanog, OCT3/4, SOX2, c-Myc, KLF4 | Increasing cancer stemness | [153] |
| NEAT1 | Inducing | ADM | N/A | Inhibiting cell apoptosis, Promoting invasion | [154] |
| LEIGC | Reversing | 5-FU | CDH1, E-cad, Vimentin, Twist, Slug, ZEB1, Snail | Inhibiting EMT | [155] |
| CASC9 | Inducing | ADM, PTX | MDR1 | Inhibiting cell apoptosis, Promoting cell proliferation | [156] |
| HOTTIP | Inducing | DDP, ADM, 5-FU | E-cad, ZO1, N-cad, Vimentin, ZEB1, Twist | Promoting EMT | [157] |
| HCP5 | Inducing | OXA, 5-FU | MiR-3619-5p, SOX2, OCT4, LIN28, CD1331 | CeRNA, Increasing cancer stemness | [158] |
N/A Not available; AUC Area under curve
LncRNA-mediated cell apoptosis
Many anticancer drugs have been found to induce apoptosis and apoptosis-related signaling networks [159, 160]. However, the dysregulation of apoptosis often leads to drug resistance and treatment failure [161]. There are two major pathways of apoptosis, i.e. the extrinsic and intrinsic pathways (mitochondrial pathway) [162, 163]. The extrinsic pathway is initiated by the attachment of death receptors with their death initiating ligands, such as Fas cell surface death receptor (FAS) binding to FAS ligand (FASL), tumor necrosis factor receptor 1 (TNFR1) binding to tumor necrosis factor alpha (TNFα), and TRAIL cell surface receptors 1 and 2 (TRAILR1/2) binding to TNF-related apoptosis-inducing ligand (TRAIL) [163]. Consequently, an adaptor molecule, FAS-associated death domain protein (FADD) couples the death receptors, which leads to the activation of caspase-8 and caspase-10 [163]. Either activated caspase-8 or caspase-10 can directly cleave and activate caspase-3, caspase-6, or caspase-7, thereby promoting apoptosis. Alternatively, irreparable genetic damage, hypoxia, and other internal stimulation can activate apoptosis through the internal mitochondrial pathway. Subsequently, BH3-only protein members, BAX (BCL-2 associated X, apoptosis regulator) and BAK (BCL-2 antagonist/killer), which belong to the B-cell lymphoma-2 (BCL-2) family, can neutralize the anti-apoptotic proteins BCL-2 and BCL-xL (B-cell lymphoma-extra large) [162, 163]. Simultaneously, activation of BAX/BAK can increase the permeability of the mitochondrial outer membrane (MOM) and release different apoptosis mediators, such as cytochrome c, which can activate caspase-9. In turn, caspase-9 cleaves and activates caspase-3, caspase-6, and caspase-7, thus triggering apoptosis [161, 164]. Moreover, PI3K (phosphatidylinositol 3-kinase)/AKT (serine/threonine protein kinase B), Hippo, Wnt/β-catenin, and HIF-1α (hypoxia-inducible factor-1α) signaling pathways are involved in regulating apoptosis. Recent studies have shown that lncRNAs can regulate gastric cancer chemoresistance by modulating these apoptosis-related signaling pathways (Fig. 1).
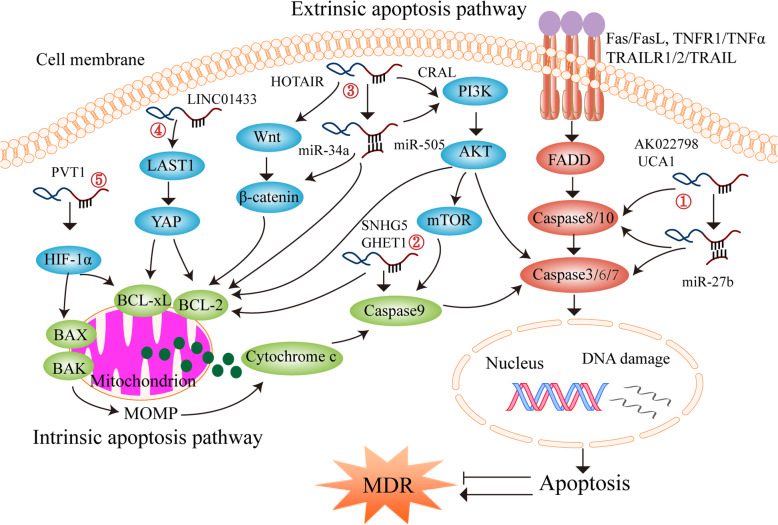
Fig. 1.
LncRNAs regulate chemoresistance through apoptosis. There are two apoptosis pathways, i.e. extrinsic pathway and intrinsic pathway (mitochondrial pathway). ①-② LncRNAs act as a ceRNA, directly bind to mRNAs or proteins, and regulate multidrug resistance (MDR) through extrinsic and intrinsic pathways of apoptosis. ③-⑤ LncRNAs also regulate apoptosis-mediated MDR through PI3K/AKT, Wnt/β-catenin, Hippo, and HIF-1α signaling pathways
Extrinsic apoptosis pathway
The abnormal expression of caspase-8 and caspase-3 leads to the inhibition of apoptosis and chemotherapy resistance [165]. Hang et al. have reported that the overexpression of lncRNA AK022798 down-regulates the expression of caspase-8 and caspase-3 and inhibits the extrinsic apoptosis pathway, leading to cisplatin (DDP) resistance in gastric cancer cells, while interference with AK022798 increases the expression levels of caspase-8 and caspase-3 and promotes apoptosis, reversing chemotherapy resistance in vitro [124]. Fang et al. have revealed that lncRNA UCA1 functions as a sponge of miR-27b to down-regulate caspase-3 expression and inhibit extrinsic apoptosis pathway, thereby inducing the resistance of gastric cancer cells to DDP, adriamycin (ADR), and 5-fluorouracil (5-FU). It has further been shown that silencing UCA1 increases the expression level of caspase-3, thus promoting apoptosis and reversing MDR in gastric cancer cells in vitro [145, 146].
Intrinsic apoptosis pathway (mitochondrial pathway)
The pro-apoptotic proteins (BAX, BAK) and anti-apoptotic proteins (BCL-2, BCL-xL) maintain a dynamic balance in regulating the mitochondrial apoptosis pathway, while the broken balance often causes cancer progression and chemoresistance [166, 167]. Li et al. have shown that lncRNA SNHG5 expression is remarkably higher in DDP-resistant gastric cancer patients and cells [150]. Further mechanism study has revealed that SNHG5 down-regulates BAX expression and up-regulates BCL-2 expression, thereby inhibiting apoptosis and promoting DDP resistance of gastric cancer cells. Similar results have been obtained for lncRNA GHET1 (gastric carcinoma proliferation enhancing transcript 1) by Zhang et al [143]. Moreover, interfering with GHET1 expression causes an increase in BAX level and a decrease in BCL-2 level, thus enhancing the sensitivity of BGC823 and SGC7901 cells to chemotherapy [143]. Du et al. have reported that lncRNA PVT1 inhibits apoptosis and enhances 5-FU resistance of gastric cancer by activating BCL-2 [147]. A Kaplan-Meier analysis has shown that therapy without 5-FU significantly improves the first progression survival and OS of gastric cancer patients with high PVT1 expression, while these patients do not experience survival-related benefits from 5-FU-based chemotherapy [147]. Zhang et al have shown that lncRNA FAM84B-AS (FAM84B antisense RNA) increases the expression levels of BCL-2 and BCL-xL and decreases the expression levels of caspase-9, caspase-3, and caspase-7, consequently inhibiting apoptosis and causing gastric cancer cell resistance to DDP; however, silencing FAM84B-AS enhances gastric cancer cell sensitivity to DDP in vitro and in vivo [152].
PI3K/AKT signaling pathway
The PI3K/AKT signaling pathway plays an important role in regulating apoptosis and drug resistance. The activation of PI3K/AKT pathway inhibits apoptosis, leading to tumor progression, drug resistance, and treatment failure, while inhibition of PI3K/AKT signaling reverses drug resistance by inducing apoptosis [168, 169]. In gastric cancer, Wang et al. have reported that CRAL (cisplatin resistance-associated lncRNA) functions as a ceRNA to reverse gastric cancer DDP resistance via the miR-505/CYLD (cylindromatosis)/AKT axis [125]. It has been found that CRAL is mainly located in the cytoplasm and sponges the endogenous miR-505, consequently increasing CYLD expression, suppressing AKT activation, and enhancing the sensitivity of gastric cancer cells to DDP in vitro and in vivo [125]. Zeng et al. have reported that the knockdown of XLOC_006753 can reduce the expression levels of PI3K, p-AKT (Thr308/Ser473), and p-mTOR (phosphorylation mechanistic target of rapamycin kinase), thus activating caspase-9 to promote apoptosis and reverse DDP and 5-FU resistance in gastric cancer cells in vitro [127]. Cheng et al. have demonstrated that lncRNA HOTAIR (HOX transcript antisense RNA) is significantly up-regulated in gastric cancer patients and DDP-resistant cells [137]. HOTAIR has further been found to target miR-34a and activate the PI3K/AKT pathway, consequently decreasing the expression of caspase-3 and BAX, increasing the expression of BCL-2, inhibiting apoptosis, and inducing DDP resistance in gastric cancer cells in vitro and in vivo [137].
Hippo signaling pathway
The Hippo signaling pathway is closely associated with apoptosis and MDR by regulating its downstream effectors, Yes-associated protein (YAP) and large tumor suppressor kinase 1 (LATS1) [170]. Recent studies have shown that activation of the Hippo signaling pathway inhibits apoptosis by decreasing the BAX/BCL-2 ratio [171], whereas the downregulation of YAP expression can promote apoptosis [172]. Zhang et al. have shown that LINC01433 decreases the phosphorylation of YAP by disrupting the YAP-LATS1 association. Meanwhile, YAP directly binds to the LINC01433 promoter region and activates its transcription [130]. The formation of the LINC01433-YAP feedback loop suppresses apoptosis and induces resistance to doxorubicin (DOX) and DDP. It has also been found that LINC01433 knockdown significantly increases the sensitivity of gastric cancer cells to DOX and DDP [130].
Wnt/β-catenin signaling pathway
Wnt/β-catenin signaling pathway has been demonstrated as an important regulator of cell proliferation, differentiation, and apoptosis, and its abnormal activation is related to MDR in cancer [173, 174]. Targeting the Wnt/β-catenin signaling pathway is a new hope for reversing cancer drug resistance [175, 176]. Cheng et al. have reported that lncRNA HOTAIR directly binds to miR-34a, reduces its expression level, and increases the expression of Wnt and β-catenin [137]. The interference with HOTAIR can decrease the expression of Wnt and β-catenin, thereby increasing the BAX/BCL-2 ratio, activating caspase-3, promoting apoptosis, and reversing DDP resistance in gastric cancer cells in vitro and in vivo [137].
HIF-1α signaling pathway
Activation of the HIF-1α signaling pathway is critical for cancer cells adapting to the hypoxic environment, which can mediate apoptosis through the mitochondrial pathway [177]. Recent studies have shown that HIF-1α regulates the mitochondrial apoptosis pathway and MDR by breaking the dynamic balance between the pro-apoptotic proteins (BAX, BAK) and anti-apoptotic proteins (BCL-2, BCL-xL) [178, 179]. Zhang et al. have reported that lncRNA PVT1 is highly expressed in DDP resistant gastric cancer cells and tumor tissues from DDP resistant gastric cancer patients, up-regulates the expression of HIF-1α, inhibits apoptosis, and induces DDP resistance [148]. It has further been shown that silencing PVT1 can reduce the expression of HIF-1α and enhance the sensitivity of gastric cancer cells to DDP [148].
LncRNA-mediated EMT
EMT is a biological process in which epithelial cells lose their polarity and transform into mesenchymal cells with the ability to move freely [115]. The expression and/or function of epithelial genes such as E-cadherin (E-cad), Claudin, cytokeratins (CKs), and zona occludens 1 (ZO1) are lost during the transition, whereas the expression levels of genes that define the mesenchymal phenotype, such as Vimentin, fibronectin, N-cadherin (N-cad), and matrix metalloproteinases (MMPs) are elevated [180]. The process of EMT is mainly regulated by transcription factors, including zinc-finger-binding transcription factors Snail1 and Snail2, the basic helix-loop-helix (bHLH) factors Twist1 and Twist2, and the zinc-finger E-box-binding homeobox factors ZEB1 and ZEB2 (Fig. 2) [181]. EMT leads to the degradation of adhesion structures between tumor cells, increasing invasiveness and causing chemoresistance and treatment failure [182].
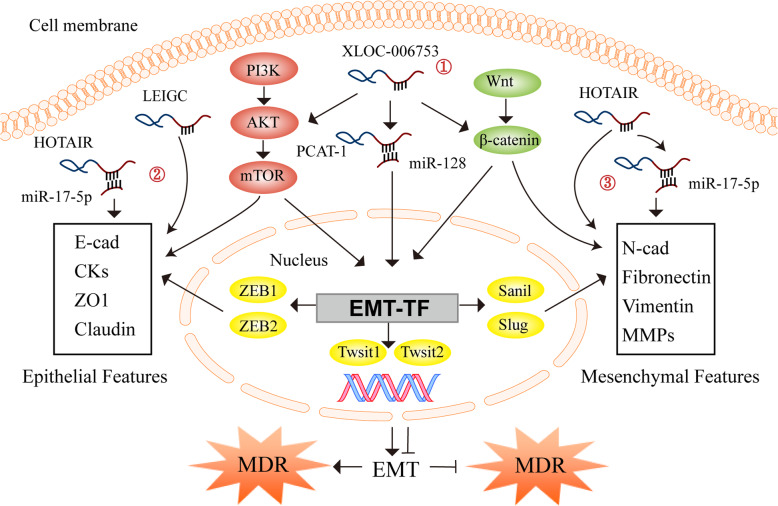
Fig. 2.
LncRNAs regulate EMT-mediated chemoresistance. ① LncRNAs act as a ceRNA, directly bind to mRNAs or proteins, and regulate EMT-mediated multidrug resistance (MDR) by modulating PI3K/AKT and Wnt/β-catenin signaling pathways. ②-③ LncRNAs also regulate EMT-mediated MDR by targeting EMT markers or transcription factors
LncRNAs have recently been found to play an important role in the process of drug resistance caused by EMT [183, 184]. LncRNAs regulate EMT-mediated resistance in gastric cancer by regulating EMT markers or transcription factors (Fig. 2). Mao et al. have demonstrated that the expression of lncRNA HOTTIP is up-regulated in MDR gastric cancer cells, which decreases the expression of E-cad and ZO1, increases the expression of N-cad, Vimentin, ZEB1, and Twist, and induces EMT [157]. Conversely, silencing HOTTIP can reverse EMT and enhance the sensitivity of MDR gastric cancer cells to DDP, ADR, and 5-FU in vitro [157]. Han et al. have found that lncRNA LEIGC expression is significantly down-regulated in tumor tissues from human gastric cancer patients, which causes the decreased expression of E-cad and the increased expression of Vimentin, Twist, Slug, ZEB1, and Snail, as well as EMT and resistance of gastric cancer cells to 5-FU [155]. Jia et al. have shown that lncRNA HOTAIR directly targets miR-17-5p to down-regulate E-cad expression and up-regulate the expression of N-cad and Vimentin, thereby inducing EMT and the resistance of gastric cancer cells to DDP, ADR, mitomycin (MMC), and 5-FU [138]. Guo et al. have reported that lncRNA PCAT-1 (prostate cancer associated transcript 1) is highly expressed in DDP-resistant gastric cancer tissues and cells [134]. Mechanistically, PCAT-1 competitively binds to miR-128, upregulates ZEB1 expression, and induces EMT and DDP resistance [134]. Zeng et al. have reported that lncRNA XLOC_006753 expression is up-regulated in gastric cancer tissues and MDR gastric cancer cell lines, and the knockdown of XLOC_006753 can reduce the expression levels of PI3K, p-AKT (Thr308/Ser473), p-mTOR, β-catenin, Vimentin, and Snail, thus reversing EMT and enhancing the sensitivity of gastric cancer cells to DDP and 5-FU in vitro [127].
LncRNA-mediated cancer cell stemness
Cancer stem cells (CSCs) are a subset of cancer cells with the ability to self-renew and differentiate, which can lead to tumor growth, metastasis, and drug resistance [185]. CSCs play a pivotal role in drug resistance and cancer treatment failure because they have channel proteins to efflux anticancer drugs, which leads to the decreased concentration of drugs in the cells and then induces MDR [185]. The stemness markers of CSCs mainly include cluster of differentiation 24 (CD24), CD29, CD44, CD133, nanog, SRY-box transcription factor 2 (SOX2), SOX9, LIN28, OCT1/2/4, c-Myc, kruppel like factor 4 (KLF4), aldehyde dehydrogenase 1 (ALDH1), and essential specific antigen (ESA) (Fig. 3) [186, 187]. The gain or loss of cancer cell stemness is regulated by the stemness-related pathways and stemness markers [188]. Therefore, targeting the cancer cell stemness-related pathways or markers is an important strategy to reverse drug resistance and enhance drug sensitivity.
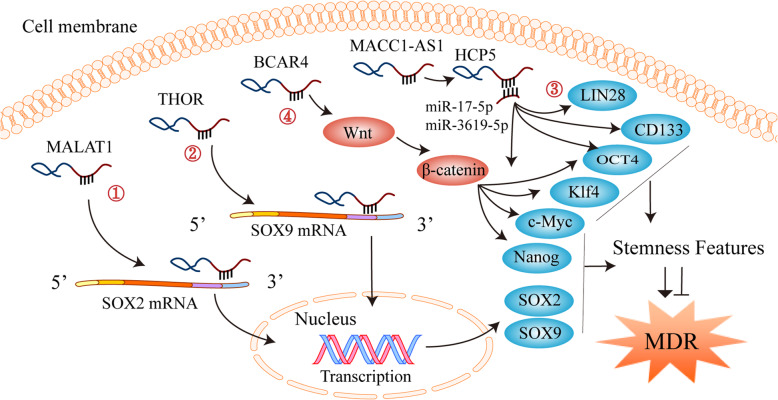
Fig. 3.
LncRNAs regulate chemoresistance through cancer cell stemness. ①-③ LncRNAs act as a ceRNA, directly bind to mRNAs or proteins, and regulate cancer cell stemness and multidrug resistance (MDR) by modulating stemness-related markers. ④ LncRNAs also regulate cancer cell stemness and MDR through modulating Wnt/β-catenin signaling pathway
LncRNAs have been reported to regulate gastric cancer cell stemness and MDR by modulating stemness-related pathways or markers (Fig. 3). Activation of the Wnt/β-catenin pathway has been found to promote the stemness of cancer cells [189, 190]. Wang et al. have reported that lncRNA BCAR4 (breast cancer anti-estrogen resistance 4) is highly expressed in DDP-resistant gastric cancer cells. Further studies have shown that BCAR4 activates the Wnt/β-catenin signaling pathway and up-regulates the expression of stemness markers nanog, OCT3/4, SOX2, c-Myc, and KLF4, which further enhance gastric cancer cell stemness and DDP resistance [153]. He et al. have found that lncRNA MACC1-AS1 (MACC1 antisense RNA 1) competitively antagonizes miR-145-5p, thereby up-regulating the levels of diacylglycerol cholinephosphotransferase (CPT1) and acetyl-CoA synthetase (ACS) to participate in fatty acid oxidation (FAO), increasing the expression of CD133, OCT4, SOX2, and LIN28, and inducing the resistance of gastric cancer cells to 5-FU and oxaliplatin (OXA) [128]. Unsurprisingly, the knockdown of MACC1-AS1 attenuates the stemness of gastric cancer cells and reverses MDR [128]. Song et al. have shown that THOR (testis-associated highly conserved oncogenic long non-coding RNA) is highly expressed in gastric cancer tissues and cells, whereas THOR knockdown decreases the expression of SOX9 through directly binding to its 3′UTR, thus inhibiting gastric cancer cell stemness and reversing the resistance of gastric cancer cells to DDP [141]. Xiao et al. have demonstrated that lncRNA MALAT1 directly binds to SOX2 mRNA, enhances its stability, and increases its expression, which further promotes the stemness of gastric cancer cells and induces DDP resistance [121]. Wu et al. have found that lncRNA HCP5 (histocompatibility leukocyte antigen complex P5) drives FAO by sponging miR-3619-5p and promoting stemness and the resistance of gastric cancer cells to 5-FU and OXA [158].
LncRNA-mediated autophagy
Autophagy is an evolutionarily conserved cellular process, through which damaged organelles and superfluous proteins are degraded, thereby maintaining the correct cellular balance [191]. The process of autophagy is divided into five distinct stages (Fig. 4): 1) initiation, 2) vesicle nucleation, 3) vesicle elongation, 4) vesicle fusion, and 5) cargo degradation [192]. Firstly, various stresses (deficiency of oxygen, ultraviolet rays, or exposure to toxic agents) trigger autophagy, and then the assembly of the Unc-51-like kinase 1 (ULK1) complex, comprising ULK1, autophagy-related genes 13 (ATG13), and ATG101 induces nucleation of the autophagy-isolation membrane. Following nucleation, the elongation of the isolation membrane is regulated by the ATG12-ATG5-ATG16 complex. Moreover, the isolation membrane collects cellular materials to degrade and form an autophagosome, which is regulated by ATG8/LC3 (microtubule-associated protein 1A/1B-light chain 3) complex. Subsequently, the autophagolysosome is formed through the fusion of autophagosome and lysosome, which is mediated by Ras-related protein 7 (Rab7) and FYVE and coiled-coil protein 1 (FYCO1) transport proteins. Finally, cellular components are degraded and recycled to supply energy to the cells due to the action of hydrolytic enzymes [193].
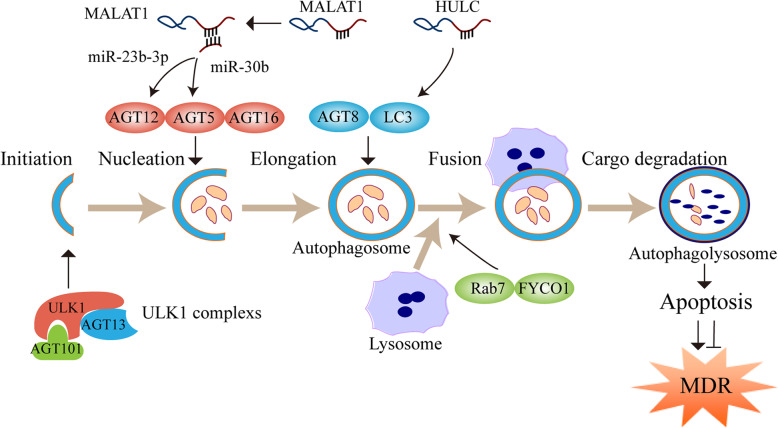
Fig. 4.
LncRNAs regulate autophagy-mediated chemoresistance. The process of autophagy is divided into five distinct stages: initiation, vesicle nucleation, vesicle elongation, vesicle fusion, and cargo degradation. LncRNAs act as a ceRNA, directly bind to mRNAs or proteins, and regulate autophagy-mediated multidrug resistance (MDR) by targeting ATGs or ATG-LC3 complex
Recent studies have shown a paradoxical role of autophagy in cancer [194]. Autophagy is a double-edged sword of cancer MDR; it not only participates in the development of MDR and protects cancer cells from chemotherapy but also promotes cell death and mediates chemosensitization in MDR cancer cells with insufficient apoptosis [195, 196]. In gastric cancer, lncRNAs are widely involved in regulating various stages of autophagy as well as autophagy-mediated MDR (Fig. 4). Hu et al. have demonstrated that lncRNA MALAT1 acts as a ceRNA for miR-23b-3p and attenuates the inhibitory effects of miR-23b-3p on ATG12 expression, thus inducing autophagy-mediated resistance of gastric cancer cells to DDP and vincristine (VCR) in vitro and in vivo [122]. It has been found that MALAT1 is highly expressed in DDP-resistant AGS and HGC-27 cells [122, 123]. MALAT1 also binds to miR-30b and increases ATG5 expression, whereas MALAT1 knockdown can suppress autophagy and enhance the chemosensitivity of gastric cancer cells [122, 123]. Xin et al have found that lncRNA HULC interacts with forkhead box M1 (FOXM1) and stabilizes this protein, thus increasing the ratio of LC3-II/LC3-I and inducing autophagy-mediated DDP resistance [132]. As expected, silencing HULC has been shown to inhibit autophagy and enhance chemotherapy sensitivity of gastric cancer cells in vitro and in vivo [132].
LncRNA regulates MDR-related genes
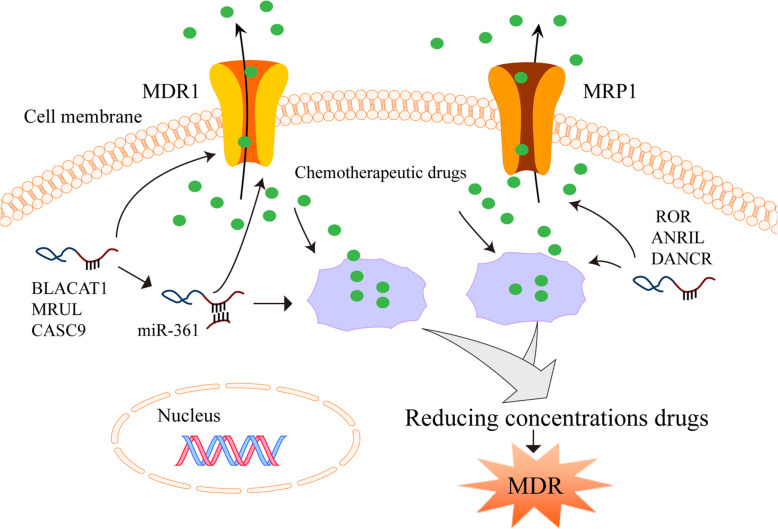
Ample evidence suggests that the expression of ATP-binding cassette (ABC) transporters, especially multidrug resistance protein 1 (MDR1, also known as P-glycoprotein or P-gp) and multidrug resistance-associated protein 1 (MRP1), which are encoded by the ABC subfamily B member 1 (ABCB1) and the ABC subfamily C member 1 (ABCC1), respectively, confers resistance to chemotherapy [197]. The ABC transporters export chemotherapeutic drugs out of the cells, resulting in resistance with reduced concentrations of the drugs intracellularly. The transporters sequestrate intracellular drugs into membrane vesicles in the cytoplasm, which also causes chemotherapy resistance (Fig. 5) [198].
Fig. 5.
LncRNAs regulate chemoresistance through modulating MDR-related genes. The ABC transporters export chemotherapy drugs out of the cells, leading to resistance with reduced concentrations of the drugs intracellularly. The transporters also sequestrate intracellular drugs into membrane vesicles in the cytoplasm, resulting in chemotherapy resistance. LncRNAs can act as a ceRNA, directly bind to mRNAs or proteins, and regulate MDR through up-regulating the expression of MDR-related genes (MDR1 and MRP1)
In gastric cancer, lncRNAs participate in the acquisition of chemotherapy resistance by regulating MDR-related genes (Fig. 5). Wang et al have shown that lncRNA ROR (regulator of reprogramming) expression is positively associated with MDR and poor prognosis of patients with gastric cancer [126]. It has also been reported that ROR depletion reduces MRP1 expression and reverses resistance to ADR and VCR [126]. Wu et al have reported that lncRNA BLACAT1 (bladder cancer associated transcript 1) accelerates the OXA-resistance acquisition of gastric cancer cells by targeting miR-361 and increasing MDR1 protein expression in vitro and in vivo [142]. Moreover, Wang et al have demonstrated that MRUL (MDR-related and upregulated lncRNA) exerts an enhancer-like role in up-regulating MDR1 expression, whereas MRUL knockdown reduces MDR1 expression and reverses resistance to ADR and VCR in vitro and in vivo [149]. Shang et al have found that lncRNA CASC9 (cancer susceptibility 9) is overexpressed in BGC823 and SGC7901 cells that are resistant to paclitaxel (PTX) or ADR [156]. Further studies have shown that CASC9 knockdown decreases MDR1 expression and restores the sensitivity of gastric cancer cells to PTX and ADR in vitro [156]. It has also been found that lncRNA ANRIL (antisense noncoding RNA in the INK4 locus) is highly expressed in DDP-resistant and 5-FU-resistant gastric cancer tissues and cells [144]. Importantly, ANRIL expression is positively correlated with the expression of MDR1 and MRP1 while ANRIL knockdown down-regulates the expression of MDR1 and MRP1 and reverses MDR [144]. Xu et al have shown that the overexpression of lncRNA DANCR up-regulates the expression of MDR1 and MRP1 and induces DDP resistance of gastric cancer cells in vitro [151].
LncRNA-mediated epigenetic modifications
Epigenetic modifications of histones can regulate resistance to anticancer drugs because cancer cells can develop drug resistance by reprogramming epigenetic networks to maintain their intrinsic homeostasis [199]. For example, the demethylation of H3K4 promotes DDP resistance of cancer cells while restoration of H3K4 methylation reverses such resistance [200]. Further, histone deacetylases regulate the functional equilibrium of histone acetylation and deacetylation, and its dysfunction leads to chemotherapy resistance [201].
In gastric cancer, lncRNAs also contribute to chemotherapy resistance by regulating histone methylation. Ye et al have found that lncRNA HOXD-AS1 (HOXD antisense RNA 1) is highly expressed in DDP-resistant gastric cancer tissues and cells [131]. Mechanism studies have shown that HOXD-AS1 epigenetically inhibits PDCD4 expression by binding to the histone methyltransferase enhancer of zeste homologue 2 (EZH2) on the promoter of PDCD4, thus increasing H3K27me3 level and inducing DDP resistance in gastric cancer cells [131]. Li et al have shown that lncRNA PCAT-1 epigenetically silences phosphatase and tensin homolog (PTEN) by binding to EZH2, which also increases H3K27me3 level and causes DDP resistance [135]. More importantly, the knockdown of either HOXD-AS1 or PCAT-1 enhances the sensitivity of DDP-resistant gastric cancer cells to DDP.
Perspectives and future directions
Overall, this review provides compelling evidence for lncRNAs as biomarkers for diagnosis, prognosis, and regulator of chemoresistance in gastric cancer. Because lncRNAs in the circulation (serum/plasma) or gastric juice are easy to obtain with non-invasive methods, they have great advantages as biomarkers for early screening, diagnosis, and prognosis of gastric cancer. Currently, lncRNA PCA3 in urine has been used as an early screening biomarker of prostate cancer [56, 57]. Therefore, it is of great clinical value to validate lncRNAs in serum/plasma or gastric juice as biomarkers for gastric cancer. Considering that most of the studies cited in this review are single-center trials with small samples, the results may be biased. Next, more in-depth studies are needed to accelerate the clinical applications of lncRNAs, such as increasing the sample size or conducting multi-center research to reduce the errors caused by individual differences.
LncRNAs are also involved in the regulation of chemotherapy resistance by modulating the signaling pathways related to apoptosis, EMT, cancer cell stemness, and autophagy, the expression of MDR-related genes, and epigenetic modifications. Therefore, targeting lncRNAs may be a promising strategy to enhance chemosensitivity and improve the efficacy of gastric cancer chemotherapy [202]. Previous studies have shown that treatment without 5-FU significantly improves the first progression survival and overall survival of gastric cancer patients with high PVT1 expression [147]. However, the patients harboring PVT1 overexpression do not obtain survival-related benefits from 5-FU-based chemotherapy [147]. Therefore, it is of great importance to further characterize lncRNAs in liquid biopsies as a guide to precision medicine for gastric cancer patients.
There is an increasing interest in targeting lncRNAs for gastric cancer therapy. However, concerns have also been raised about the therapeutic potential of targeting a single lncRNA and the current targeting strategies. First of all, despite the great progress in understanding the structures and functions of lncRNAs since their discovery, the study of lncRNAs is still a burgeoning research field and we have only touched on the tip of this iceberg. Furthermore, given the large number of lncRNAs and their up-regulation or down-regulation in gastric cancer, it is critically needed to determine the most clinically relevant lncRNAs in this disease. Of note, lncRNAs are poorly conserved among different species. Therefore, the lncRNA-targeting strategies that are developed by utilizing various animal models and cell culture systems cannot be easily extended to human applications. The latest advances in CRISPR (clustered regularly interspaced short palindromic repeats)/Cas9 gene knockout, knock-in, and point mutations may help to understand the biological role of lncRNAs. At the same time, the development of human primary cell models and patient-derived tumor xenograft (PDX) animal models may be helpful for investigating the role of lncRNAs and developing lncRNA-targeting strategies. In the near future, the development of lncRNA-targeted cancer therapy seems to be very promising.
In conclusion, accumulating evidence has shown the potential of lncRNAs as biomarkers in liquid biopsies throughout the entire management process of gastric cancer, including diagnosis, selection of chemotherapeutics, monitoring of curative effects, and prognosis.
Acknowledgements
We thank the current and former members of our laboratories and collaborators for their contributions to the publications cited in this review article. The research field in lncRNA is rapidly growing, and we apologize for not being able to cite all the recent publications, due to space limitation.
Abbreviations
- 5-FU
5-fluorouracil
- ABC
ATP-binding cassette
- ABCB1
ABC subfamily B member 1
- ABCC1
ABC subfamily C member 1
- ABHD11
Abhydrolase domain containing 11
- ABHD11-AS1
ABHD11 antisense RNA 1
- ACS
Acetyl-CoA synthetase
- ADR
Adriamycin
- AFP
Alpha fetoprotein
- AKT
serine/threonine protein kinase B
- ALDH1
Aldehyde dehydrogenase 1
- ANRIL
Antisense noncoding RNA in the INK4 locus
- AOC4P
Amine oxidase copper containing 4, pseudogene
- ARHGAP27P1
Rho GTPase activating protein 27 pseudogene 1
- ATG
Autophagy-related genes
- AUC
Area under curve
- B3GALT5
Beta-1,3-galactosyltransferase 5
- B3GALT5-AS1
B3GALT5 antisense RNA 1
- BAK
BCL-2 antagonist/killer
- BANCR
BRAF-activated non-protein coding RNA
- BAX
BCL-2 associated X, apoptosis regulator
- BCAR4
Breast cancer anti-estrogen resistance 4
- BCL-2
B-cell lymphoma-2
- BCL-xL
B-cell lymphoma-extra large
- BLACAT1
Bladder cancer associated transcript 1
- C5orf66-AS1
C5orf66 antisense RNA 1
- CA
Carbohydrate antigen
- CASC
Cancer susceptibility
- CCAT2
Colon cancer associated transcript 2
- CD
Cluster of differentiation
- CDH1
Cadherin 1
- CEA
Carcinoembryonic antigen
- CEBPA
CCAAT enhancer binding protein alpha
- CEBPA-AS1
CEBPA divergent transcript
- CeRNA
Competitive endogenous RNA
- circRNAs
Circular RNAs
- CKs
Cytokeratins
- c-Myc/MYC
MYC proto-oncogene, bHLH transcription factor
- CPT1
Diacylglycerol cholinephosphotransferase
- CRAL
Cisplatin resistance-associated lncRNA
- CRISPR
Clustered regularly interspaced short palindromic repeats
- CSCs
Cancer stem cells
- CTDHUT
CTD highly upregulated transcript
- CYLD
Cylindromatosis
- DANCR
Differentiation antagonizing non-protein coding RNA
- DDP
Cisplatin
- DGCR5
DiGeorge syndrome critical region gene 5
- DOX
Doxorubicin
- E-cad
E-cadherin
- EMT
Epithelial-mesenchymal transition
- ESA
Essential specific antigen
- EZH2
Enhancer of zeste homologue 2
- FADD
FAS-associated death domain protein
- FAM49B
Family with sequence similarity 49, member B
- FAM49B-AS
FAM49B antisense RNA
- FAM84B
Family with sequence similarity 84, member B
- FAM84B-AS
FAM84B antisense RNA
- FAO
Fatty acid oxidation
- FAS
Fas cell surface death receptor
- FASL
FAS ligand
- FOXD2
Forkhead box D2
- FOXD2-AS1
FOXD2 adjacent opposite strand RNA 1
- FOXFOXMM1
Forkhead box M1
- FYCO1
FYVE and coiled-coil protein 1
- GACAT2
Gastric cancer associated transcript 2
- GASL1
Growth arrest associated lncRNA 1
- GHET1
Gastric carcinoma proliferation enhancing transcript 1
- GNAQ-6:1
G protein subunit alpha q-6:1
- GUSBP11
GUSB pseudogene 11
- H19
H19 imprinted maternally expressed transcript
- HCP5
Histocompatibility leukocyte antigen complex P5
- HIF-1α
Hypoxia-inducible factor-1α
- HOTAIR
HOX transcript antisense RNA
- HOTTIP
HOXA distal transcript antisense RNA
- HOXA11
Homeobox A11
- HOXA11-AS
HOXA11 antisense RNA
- HOXD
Homeobox D cluster
- HOXD-AS1
HOXD antisense RNA
- HULC
Hepatocellular carcinoma upregulated long noncoding RNA
- INHBA
Inhibin subunit beta A
- INHBA-AS1
INHBA antisense RNA 1
- KLF4
Kruppel Like Factor 4
- LATS1
Large tumor suppressor kinase 1
- LC3
Microtubule associated protein 1 light chain 3 alpha
- LEIGC
-
LncRNA
chr2:118381039–118,383,698
- LincRNAs
Intergenic lncRNAs
- LncRNAs
Long non-coding RNAs
- MACC1
MET transcriptional regulator MACC1
- MACC1-AS1
MACC1 antisense RNA 1
- MALAT1
Metastasis associated lung adenocarcinoma transcript 1
- MDR
Multidrug resistance
- MDR1
Multidrug resistance protein 1
- MEF2C
Myocyte enhancer factor 2C
- MEF2C-AS1
MEF2C antisense RNA 1
- MEF2D
Myocyte enhancer factor 2D
- MEG3
Maternally expressed 3
- MIR4435–2HG
MIR4435–2 host gene
- MIAT
Myocardial infarction associated transcript
- miRNAs
microRNAs
- MMC
Mitomycin
- MMPs
Matrix metalloproteinases
- MNX1
Motor neuron and pancreas homeobox 1
- MNX1-AS1
MNX1 antisense RNA 1
- MOM
Mitochondrial outer membrane
- MOMP
Mitochondrial outer membrane permeability
- MRP1
Multidrug resistance-associated protein 1
- MRUL
MDR-related and upregulated lncRNA
- MT1JP
Metallothionein 1 J, pseudogene
- mTOR
Mechanistic target of rapamycin kinase
- N-cad
N-cadherin
- ncRNAs
Noncoding RNAs
- NEAT1
Nuclear paraspeckle assembly transcript 1
- NSE
Neuron-specific enolase
- OCT
Organic cation/carnitine transporter
- OS
Overall survival
- OXA
Oxaliplatin
- PANDAR
Promoter of CDKN1A antisense DNA damage activated RNA
- PARP
Poly-(ADP-ribose) polymerase
- PCA3
Prostate cancer associated 3
- PCAT1
Prostate cancer associated transcript 1
- PDCD4
Programmed cell death 4
- PDX
Patient-derived tumor xenograft
- P-gp
P-glycoprotein
- PI3K
Phosphatidylinositol 3-kinase
- PCSK2–2:1
Proprotein convertase subtilisin/kexin type 2–2:1
- PTCSC3
Papillary thyroid carcinoma susceptibility candidate 3
- PTEN
Phosphatase and tensin homolog
- PTX
Paclitaxel
- PVT1
Plasmacytoma variant translocation 1
- Rab7
Ras-related protein 7
- RMRP
RNA component of mitochondrial RNA processing endoribonuclease
- ROC
Receiver operator characteristic curve
- ROR
Regulator of reprogramming
- SMARCC2
SWI/SNF related, matrix associated, actin dependent regulator of chromatin subfamily c member 2
- SNHG
Small nucleolar RNA host gene
- snoRNAs
Small nucleolar RNAs
- SOX
SRY-box transcription factor
- THOR
Testis-associated highly conserved oncogenic long non-coding RNA
- TINCR
Terminal differentiation-induced ncRNA
- TNFR1
Tumor necrosis factor receptor 1
- TNFα
Tumor necrosis factor alpha
- TNM
Tumor, node, metastasis
- TRAIL
TNF-related apoptosis-inducing ligand
- TRAILR1/2
TRAIL cell surface receptors 1 and 2
- TUBA4B
Tubulin alpha 4b
- TUG1
Taurine up-regulated 1
- UCA1
Urothelial cancer associated 1
- UEGC1
ENST00000568893.1
- UEGC2
ENST00000378432.1
- ULK1
Unc-51-like kinase 1
- USP9X
Ubiquitin specific peptidase 9 X-linked
- UTR
Untranslated regions
- VCR
Vincristine
- XIST
X inactive specific transcript
- YAP
Yes-associated protein
- ZEB1
Zinc finger E-box binding homeobox 1
- ZFAS1
ZNFX1 antisense RNA 1
- ZNFX1
Zinc finger NFX1-type containing 1
- ZO1
Zona occludens 1
Authors’ contributions
JJQ and XDC conceptualized the manuscript. SMR and SWM collected the literature. LY and ZYX collected the literature, wrote the manuscript, and made the figures. JJQ and XDC edited and made significant revisions to the manuscript. All authors read and approved the final manuscript.
Funding
This study was supported by National Natural Science Foundation of China (81903842, 81573953, 81703753, 81973634), Zhejiang Chinese Medical University Startup Funding (111100E014), Zhejiang Provincial Science and Technology Projects (2018C37045), Natural Science Foundation of Zhejiang Province (LY18H290006), Zhejiang Provincial Medical and Healthy Science and Technology Projects (WKJ-ZJ-1728, 2016KYB220), and Program of Zhejiang Provincial TCM Sci-tech Plan (2016ZZ012, 2018ZY006, 2018ZB044, 2019ZZ010, 2020ZZ005).
Availability of data and materials
Not applicable.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Li Yuan and Zhi-Yuan Xu contributed equally to this work.
Contributor Information
Li Yuan, Email: 201611010611014@zcmu.edu.cn.
Zhi-Yuan Xu, Email: xuzy@zjcc.org.cn.
Shan-Ming Ruan, Email: shanmingruan@zcmu.edu.cn.
Shaowei Mo, Email: 201811112511317@zcmu.edu.cn.
Jiang-Jiang Qin, Email: jqin@zcmu.edu.cn, Email: zylysjtu@hotmail.com.
Xiang-Dong Cheng, Email: chengxd516@126.com.
References
- 1.Fitzmaurice C, Abate D, Abbasi N, Abbastabar H, Abd-Allah F, Abdel-Rahman O, Abdelalim A, Abdoli A, Abdollahpour I, Abdulle ASM, et al. Global, Regional, and National Cancer Incidence, Mortality, Years of Life Lost, Years Lived With Disability, and Disability-Adjusted Life-Years for 29 Cancer Groups, 1990 to 2017: A systematic analysis for the global burden of disease study. JAMA Oncol. 2019;5:1749. [DOI] [PMC free article] [PubMed]
- 2.Thrift Aaron P., El-Serag Hashem B. Burden of Gastric Cancer. Clinical Gastroenterology and Hepatology. 2020;18(3):534–542. doi: 10.1016/j.cgh.2019.07.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ham IH, Oh HJ, Jin H, Bae CA, Jeon SM, Choi KS, Son SY, Han SU, Brekken RA, Lee D, Hur H. Targeting interleukin-6 as a strategy to overcome stroma-induced resistance to chemotherapy in gastric cancer. Mol Cancer. 2019;18:68. doi: 10.1186/s12943-019-0972-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wagner AD, Syn NL, Moehler M, Grothe W, Yong WP, Tai BC, Ho J, Unverzagt S. Chemotherapy for advanced gastric cancer. Cochrane Database Syst Rev. 2017;8:Cd004064. doi: 10.1002/14651858.CD004064.pub4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Biagioni A, Skalamera I, Peri S, Schiavone N, Cianchi F, Giommoni E, Magnelli L, Papucci L. Update on gastric cancer treatments and gene therapies. Cancer Metastasis Rev. 2019;38:537–548. doi: 10.1007/s10555-019-09803-7. [DOI] [PubMed] [Google Scholar]
- 6.Yu J, Zheng W. An alternative method for screening gastric Cancer based on serum levels of CEA, CA19-9, and CA72-4. J Gastrointest Cancer. 2018;49:57–62. doi: 10.1007/s12029-016-9912-7. [DOI] [PubMed] [Google Scholar]
- 7.Shimada H, Noie T, Ohashi M, Oba K, Takahashi Y. Clinical significance of serum tumor markers for gastric cancer: a systematic review of literature by the task force of the Japanese gastric Cancer association. Gastric Cancer. 2014;17:26–33. doi: 10.1007/s10120-013-0259-5. [DOI] [PubMed] [Google Scholar]
- 8.He CZ, Zhang KH, Li Q, Liu XH, Hong Y, Lv NH. Combined use of AFP, CEA, CA125 and CAl9-9 improves the sensitivity for the diagnosis of gastric cancer. BMC Gastroenterol. 2013;13:87. doi: 10.1186/1471-230X-13-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hu Ping-Jen, Chen Ming-Yao, Wu Ming-Shun, Lin Ying-Chin, Shih Ping-Hsiao, Lai Chih-Ho, Lin Hwai-Jeng. Clinical Evaluation of CA72-4 for Screening Gastric Cancer in a Healthy Population: A Multicenter Retrospective Study. Cancers. 2019;11(5):733. doi: 10.3390/cancers11050733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Suzuki T, Kitagawa Y, Nankinzan R, Yamaguchi T. Early gastric cancer diagnostic ability of ultrathin endoscope loaded with laser light source. World J Gastroenterol. 2019;25:1378–1386. doi: 10.3748/wjg.v25.i11.1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Necula L, Matei L, Dragu D, Neagu AI, Mambet C, Nedeianu S, Bleotu C, Diaconu CC, Chivu-Economescu M. Recent advances in gastric cancer early diagnosis. World J Gastroenterol. 2019;25:2029–2044. doi: 10.3748/wjg.v25.i17.2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhuo W, Liu Y, Li S, Guo D, Sun Q, Jin J, Rao X, Li M, Sun M, Jiang M, et al. Long Noncoding RNA GMAN, Up-regulated in Gastric Cancer Tissues, Is Associated With Metastasis in Patients and Promotes Translation of Ephrin A1 by Competitively Binding GMAN-AS. Gastroenterology. 2019;156:676–691. doi: 10.1053/j.gastro.2018.10.054. [DOI] [PubMed] [Google Scholar]
- 13.Zhang E, He X, Zhang C, Su J, Lu X, Si X, Chen J, Yin D, Han L, De W. A novel long noncoding RNA HOXC-AS3 mediates tumorigenesis of gastric cancer by binding to YBX1. Genome Biol. 2018;19:154. doi: 10.1186/s13059-018-1523-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shuai Y, Ma Z, Liu W, Yu T, Yan C, Jiang H, Tian S, Xu T, Shu Y. TEAD4 modulated LncRNA MNX1-AS1 contributes to gastric cancer progression partly through suppressing BTG2 and activating BCL2. Mol Cancer. 2020;19:6. doi: 10.1186/s12943-019-1104-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu Wei, Zhou Gai, Wang Huizhi, Liu Yawen, Chen Baoding, Chen Wei, Lin Chen, Wu Shuhui, Gong Aihua, Xu Min. Circulating lncRNA SNHG11 as a novel biomarker for early diagnosis and prognosis of colorectal cancer. International Journal of Cancer. 2020;146(10):2901–2912. doi: 10.1002/ijc.32747. [DOI] [PubMed] [Google Scholar]
- 16.Zhou Rui, Sun Huiying, Zheng Siting, Zhang Jingwen, Zeng Dongqiang, Wu Jianhua, Huang Zhenhua, Rong Xiaoxiang, Bin Jianping, Liao Yulin, Shi Min, Liao Wangjun. A stroma‐related lncRNA panel for predicting recurrence and adjuvant chemotherapy benefit in patients with early‐stage colon cancer. Journal of Cellular and Molecular Medicine. 2020;24(5):3229–3241. doi: 10.1111/jcmm.14999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang Q, Li K, Huang X, Zhao C, Mei Y, Li X, Jiao L. Yang H: lncRNA SLC7A11-AS1 promotes Chemoresistance by blocking SCF (beta-TRCP)-mediated degradation of NRF2 in pancreatic Cancer. Mol Ther Nucleic Acids. 2020;19:974–985. doi: 10.1016/j.omtn.2019.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huan L, Guo T, Wu Y, Xu L, Huang S, Xu Y, Liang L, He X. Hypoxia induced LUCAT1/PTBP1 axis modulates cancer cell viability and chemotherapy response. Mol Cancer. 2020;19:11. doi: 10.1186/s12943-019-1122-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gu N, Wang X, Di Z, Xiong J, Ma Y, Yan Y, Qian Y, Zhang Q, Yu J. Silencing lncRNA FOXD2-AS1 inhibits proliferation, migration, invasion and drug resistance of drug-resistant glioma cells and promotes their apoptosis via microRNA-98-5p/CPEB4 axis. Aging (Albany NY) 2019;11:10266–10283. doi: 10.18632/aging.102455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ghafouri-Fard S, Taheri M. Long non-coding RNA signature in gastric cancer. Exp Mol Pathol. 2019;113:104365. doi: 10.1016/j.yexmp.2019.104365. [DOI] [PubMed] [Google Scholar]
- 21.Virgilio E, Giarnieri E, Giovagnoli MR, Montagnini M, Proietti A, D'Urso R, Mercantini P, Balducci G, Cavallini M. Long non-coding RNAs in the gastric juice of gastric cancer patients. Pathol Res Pract. 2018;214:1239–1246. doi: 10.1016/j.prp.2018.07.023. [DOI] [PubMed] [Google Scholar]
- 22.Zong W, Ju S, Jing R, Cui M. Long non-coding RNA-mediated regulation of signaling pathways in gastric cancer. Clin Chem Lab Med. 2018;56:1828–1837. doi: 10.1515/cclm-2017-1139. [DOI] [PubMed] [Google Scholar]
- 23.Nasrollahzadeh-Khakiani M, Emadi-Baygi M, Schulz WA, Nikpour P. Long noncoding RNAs in gastric cancer carcinogenesis and metastasis. Brief Funct Genomics. 2017;16:129–145. doi: 10.1093/bfgp/elw011. [DOI] [PubMed] [Google Scholar]
- 24.Sun W, Yang Y, Xu C, Xie Y, Guo J. Roles of long noncoding RNAs in gastric cancer and their clinical applications. J Cancer Res Clin Oncol. 2016;142:2231–2237. doi: 10.1007/s00432-016-2183-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.The ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. [DOI] [PMC free article] [PubMed]
- 26.Liao Q, Shen J, Liu J, Sun X, Zhao G, Chang Y, Xu L, Li X, Zhao Y, Zheng H, et al. Genome-wide identification and functional annotation of plasmodium falciparum long noncoding RNAs from RNA-seq data. Parasitol Res. 2014;113:1269–1281. doi: 10.1007/s00436-014-3765-4. [DOI] [PubMed] [Google Scholar]
- 27.Johnsson P, Lipovich L, Grander D, Morris KV. Evolutionary conservation of long non-coding RNAs; sequence, structure, function. Biochim Biophys Acta. 1840;2014:1063–1071. doi: 10.1016/j.bbagen.2013.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ma L, Bajic VB, Zhang Z. On the classification of long non-coding RNAs. RNA Biol. 2013;10:925–933. doi: 10.4161/rna.24604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alessio Enrico, Bonadio Raphael Severino, Buson Lisa, Chemello Francesco, Cagnin Stefano. A Single Cell but Many Different Transcripts: A Journey into the World of Long Non-Coding RNAs. International Journal of Molecular Sciences. 2020;21(1):302. doi: 10.3390/ijms21010302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mondal T, Juvvuna PK, Kirkeby A, Mitra S, Kosalai ST, Traxler L, Hertwig F, Wernig-Zorc S, Miranda C, Deland L, et al. Sense-Antisense lncRNA Pair Encoded by Locus 6p22.3 Determines Neuroblastoma Susceptibility via the USP36-CHD7-SOX9 Regulatory Axis. Cancer Cell. 2018;33:417–434. doi: 10.1016/j.ccell.2018.01.020. [DOI] [PubMed] [Google Scholar]
- 31.Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R, Willingham AT, Stadler PF, Hertel J, Hackermuller J, Hofacker IL, et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316:1484–1488. doi: 10.1126/science.1138341. [DOI] [PubMed] [Google Scholar]
- 32.Katsel P, Roussos P, Fam P, Khan S, Tan W, Hirose T, Nakagawa S, Pletnikov MV, Haroutunian V. The expression of long noncoding RNA NEAT1 is reduced in schizophrenia and modulates oligodendrocytes transcription. NPJ Schizophr. 2019;5:3. doi: 10.1038/s41537-019-0071-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang B, Arun G, Mao YS, Lazar Z, Hung G, Bhattacharjee G, Xiao X, Booth CJ, Wu J, Zhang C, Spector DL. The lncRNA Malat1 is dispensable for mouse development but its transcription plays a cis-regulatory role in the adult. Cell Rep. 2012;2:111–123. doi: 10.1016/j.celrep.2012.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McHugh CA, Chen CK, Chow A, Surka CF, Tran C, McDonel P, Pandya-Jones A, Blanco M, Burghard C, Moradian A, et al. The Xist lncRNA interacts directly with SHARP to silence transcription through HDAC3. Nature. 2015;521:232–236. doi: 10.1038/nature14443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Du Z, Sun T, Hacisuleyman E, Fei T, Wang X, Brown M, Rinn JL, Lee MG, Chen Y, Kantoff PW, Liu XS. Integrative analyses reveal a long noncoding RNA-mediated sponge regulatory network in prostate cancer. Nat Commun. 2016;7:10982. doi: 10.1038/ncomms10982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shang AQ, Wang WW, Yang YB, Gu CZ, Ji P, Chen C, Zeng BJ, Wu JL, Lu WY, Sun ZJ, Li D. Knockdown of long noncoding RNA PVT1 suppresses cell proliferation and invasion of colorectal cancer via upregulation of microRNA-214-3p. Am J Physiol Gastrointest Liver Physiol. 2019;317:G222–g232. doi: 10.1152/ajpgi.00357.2018. [DOI] [PubMed] [Google Scholar]
- 37.Bianchessi V, Badi I, Bertolotti M, Nigro P, D'Alessandra Y, Capogrossi MC, Zanobini M, Pompilio G, Raucci A, Lauri A. The mitochondrial lncRNA ASncmtRNA-2 is induced in aging and replicative senescence in endothelial cells. J Mol Cell Cardiol. 2015;81:62–70. doi: 10.1016/j.yjmcc.2015.01.012. [DOI] [PubMed] [Google Scholar]
- 38.Mathieu EL, Belhocine M, Dao LT, Puthier D. Spicuglia S: [functions of lncRNA in development and diseases] Med Sci (Paris) 2014;30:790–796. doi: 10.1051/medsci/20143008018. [DOI] [PubMed] [Google Scholar]
- 39.Dallner OS, Marinis JM, Lu YH, Birsoy K, Werner E, Fayzikhodjaeva G, Dill BD, Molina H, Moscati A, Kutalik Z, et al. Dysregulation of a long noncoding RNA reduces leptin leading to a leptin-responsive form of obesity. Nat Med. 2019;25:507–516. doi: 10.1038/s41591-019-0370-1. [DOI] [PubMed] [Google Scholar]
- 40.Westra HJ, Martinez-Bonet M, Onengut-Gumuscu S, Lee A, Luo Y, Teslovich N, Worthington J, Martin J, Huizinga T, Klareskog L, et al. Fine-mapping and functional studies highlight potential causal variants for rheumatoid arthritis and type 1 diabetes. Nat Genet. 2018;50:1366–1374. doi: 10.1038/s41588-018-0216-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang CY, Colognori D, Sunwoo H, Wang D, Lee JT. PRC1 collaborates with SMCHD1 to fold the X-chromosome and spread Xist RNA between chromosome compartments. Nat Commun. 2019;10:2950. doi: 10.1038/s41467-019-10755-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huang M, Wang H, Hu X. Cao X: lncRNA MALAT1 binds chromatin remodeling subunit BRG1 to epigenetically promote inflammation-related hepatocellular carcinoma progression. Oncoimmunology. 2019;8:e1518628. doi: 10.1080/2162402X.2018.1518628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xiu B, Chi Y, Liu L, Chi W, Zhang Q, Chen J, Guo R, Si J, Li L, Xue J, et al. LINC02273 drives breast cancer metastasis by epigenetically increasing AGR2 transcription. Mol Cancer. 2019;18:187. doi: 10.1186/s12943-019-1115-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang W, Hu W, Wang Y, An Y, Song L, Shang P, Yue Z. Long non-coding RNA UCA1 promotes malignant phenotypes of renal cancer cells by modulating the miR-182-5p/DLL4 axis as a ceRNA. Mol Cancer. 2020;19:18. doi: 10.1186/s12943-020-1132-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Barlebo Ahlborn L, Ostrup O. Toward liquid biopsies in cancer treatment: application of circulating tumor DNA. Apmis. 2019;127:329–336. doi: 10.1111/apm.12912. [DOI] [PubMed] [Google Scholar]
- 46.Zuo Z, Hu H, Xu Q, Luo X, Peng D, Zhu K, Zhao Q, Xie Y, Ren J. BBCancer: an expression atlas of blood-based biomarkers in the early diagnosis of cancers. Nucleic Acids Res. 2020;48:D789–d796. doi: 10.1093/nar/gkz942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhan Y, Du L, Wang L, Jiang X, Zhang S, Li J, Yan K, Duan W, Zhao Y, Wang L, et al. Expression signatures of exosomal long non-coding RNAs in urine serve as novel non-invasive biomarkers for diagnosis and recurrence prediction of bladder cancer. Mol Cancer. 2018;17:142. doi: 10.1186/s12943-018-0893-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xie Z, Zhou F, Yang Y, Li L, Lei Y, Lin X, Li H, Pan X, Chen J, Wang G, et al. Lnc-PCDH9-13:1 is a hypersensitive and specific biomarker for early hepatocellular carcinoma. EBioMedicine. 2018;33:57–67. doi: 10.1016/j.ebiom.2018.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shao Y, Ye M, Li Q, Sun W, Ye G, Zhang X, Yang Y, Xiao B, Guo J. LncRNA-RMRP promotes carcinogenesis by acting as a miR-206 sponge and is used as a novel biomarker for gastric cancer. Oncotarget. 2016;7:37812–37824. doi: 10.18632/oncotarget.9336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Darbandi M, Darbandi S, Agarwal A, Baskaran S, Dutta S, Sengupta P, Khorram Khorshid HR, Esteves S, Gilany K, Hedayati M, et al. Reactive oxygen species-induced alterations in H19-Igf2 methylation patterns, seminal plasma metabolites, and semen quality. J Assist Reprod Genet. 2019;36:241–253. doi: 10.1007/s10815-018-1350-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lin Y, Leng Q, Zhan M, Jiang F. A plasma long noncoding RNA signature for early detection of lung Cancer. Transl Oncol. 2018;11:1225–1231. doi: 10.1016/j.tranon.2018.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ozgur E, Ferhatoglu F, Sen F, Saip P, Gezer U. Circulating lncRNA H19 may be a useful marker of response to neoadjuvant chemotherapy in breast cancer. Cancer Biomark. 2020;27:11–17. doi: 10.3233/CBM-190085. [DOI] [PubMed] [Google Scholar]
- 53.Fang C, Zan J, Yue B, Liu C, He C, Yan D. Long non-coding ribonucleic acid zinc finger antisense 1 promotes the progression of colonic cancer by modulating ZEB1 expression. J Gastroenterol Hepatol. 2017;32:1204–1211. doi: 10.1111/jgh.13646. [DOI] [PubMed] [Google Scholar]
- 54.Arantes L, De Carvalho AC, Melendez ME, Lopes Carvalho A. Serum, plasma and saliva biomarkers for head and neck cancer. Expert Rev Mol Diagn. 2018;18:85–112. doi: 10.1080/14737159.2017.1404906. [DOI] [PubMed] [Google Scholar]
- 55.Gomes CC, de Sousa SF, Calin GA, Gomez RS. The emerging role of long noncoding RNAs in oral cancer. Oral Surg Oral Med Oral Pathol Oral Radiol. 2017;123:235–241. doi: 10.1016/j.oooo.2016.10.006. [DOI] [PubMed] [Google Scholar]
- 56.Groskopf J, Aubin SM, Deras IL, Blase A, Bodrug S, Clark C, Brentano S, Mathis J, Pham J, Meyer T, et al. APTIMA PCA3 molecular urine test: development of a method to aid in the diagnosis of prostate cancer. Clin Chem. 2006;52:1089–1095. doi: 10.1373/clinchem.2005.063289. [DOI] [PubMed] [Google Scholar]
- 57.Lemos AEG, Matos ADR, Ferreira LB, Gimba ERP. The long non-coding RNA PCA3: an update of its functions and clinical applications as a biomarker in prostate cancer. Oncotarget. 2019;10:6589–6603. doi: 10.18632/oncotarget.27284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Selth LA, Roberts MJ, Chow CW, Marshall VR, Doi SA, Vincent AD, Butler LM, Lavin MF, Tilley WD, Gardiner RA. Human seminal fluid as a source of prostate cancer-specific microRNA biomarkers. Endocr Relat Cancer. 2014;21:L17–L21. doi: 10.1530/ERC-14-0234. [DOI] [PubMed] [Google Scholar]
- 59.Henriksen K, O'Bryant SE, Hampel H, Trojanowski JQ, Montine TJ, Jeromin A, Blennow K, Lonneborg A, Wyss-Coray T, Soares H, et al. The future of blood-based biomarkers for Alzheimer's disease. Alzheimers Dement. 2014;10:115–131. doi: 10.1016/j.jalz.2013.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang J, Yang K, Yuan W, Gao Z. Determination of serum Exosomal H19 as a noninvasive biomarker for bladder Cancer diagnosis and prognosis. Med Sci Monit. 2018;24:9307–9316. doi: 10.12659/MSM.912018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang G, Xu Y, Zou C, Tang Y, Lu J, Gong Z, Ma G, Zhang W, Jiang P. Long noncoding RNA ARHGAP27P1 inhibits gastric cancer cell proliferation and cell cycle progression through epigenetically regulating p15 and p16. Aging (Albany NY) 2019;11:9090–9110. doi: 10.18632/aging.102377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tang X, Yu L, Bao J, Jiang P, Yan F. Function of long noncoding RNA UCA1 on gastric Cancer cells and its Clinicopathological significance in plasma. Clin Lab. 2019;65. [DOI] [PubMed]
- 63.Giraldez MD, Spengler RM, Etheridge A, Goicochea AJ, Tuck M, Choi SW, Galas DJ, Tewari M. Phospho-RNA-seq: a modified small RNA-seq method that reveals circulating mRNA and lncRNA fragments as potential biomarkers in human plasma. EMBO J. 2019;38:e101695. [DOI] [PMC free article] [PubMed]
- 64.Xu Y, Zhang G, Zou C, Zhang H, Gong Z, Wang W, Ma G, Jiang P, Zhang W. LncRNA MT1JP suppresses gastric Cancer cell proliferation and migration through MT1JP/MiR-214-3p/RUNX3 Axis. Cell Physiol Biochem. 2018;46:2445–2459. doi: 10.1159/000489651. [DOI] [PubMed] [Google Scholar]
- 65.Lin LY, Yang L, Zeng Q, Wang L, Chen ML, Zhao ZH, Ye GD, Luo QC, Lv PY, Guo QW, et al. Tumor-originated exosomal lncUEGC1 as a circulating biomarker for early-stage gastric cancer. Mol Cancer. 2018;17:84. doi: 10.1186/s12943-018-0834-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang K, Shi H, Xi H, Wu X, Cui J, Gao Y, Liang W, Hu C, Liu Y, Li J, et al. Genome-wide lncRNA microarray profiling identifies novel circulating lncRNAs for detection of gastric Cancer. Theranostics. 2017;7:213–227. doi: 10.7150/thno.16044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhou Q, Li H, Jing J, Yuan Y, Sun L. Evaluation of C5orf66-AS1 AS a potential biomarker for predicting early gastric Cancer and its role in gastric carcinogenesis. Onco Targets Ther. 2020;13:2795–2805. doi: 10.2147/OTT.S239965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhang G, Chi N, Lu Q, Zhu D, Zhuang Y. LncRNA PTCSC3 is a biomarker for the treatment and prognosis of gastric Cancer. Cancer Biother Radiopharm. 2020;35:77–81. doi: 10.1089/cbr.2019.2991. [DOI] [PubMed] [Google Scholar]
- 69.Guo J, Li Y, Duan H, Yuan L. LncRNA TUBA4B functions as a competitive endogenous RNA to inhibit gastric cancer progression by elevating PTEN via sponging miR-214 and miR-216a/b. Cancer Cell Int. 2019;19:156. doi: 10.1186/s12935-019-0879-x. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 70.Ji B, Huang Y, Gu T, Zhang L, Li G, Zhang C. Potential diagnostic and prognostic value of plasma long noncoding RNA LINC00086 and miR-214 expression in gastric cancer. Cancer Biomark. 2019;24:249–255. doi: 10.3233/CBM-181486. [DOI] [PubMed] [Google Scholar]
- 71.Xu Y, Zhang G, Zou C, Gong Z, Wang S, Liu J, Ma G, Liu X, Zhang W, Jiang P. Long noncoding RNA DGCR5 suppresses gastric cancer progression by acting as a competing endogenous RNA of PTEN and BTG1. J Cell Physiol. 2019;234:11999–12010. doi: 10.1002/jcp.27861. [DOI] [PubMed] [Google Scholar]
- 72.Zhang G, Xu Y, Wang S, Gong Z, Zou C, Zhang H, Ma G, Zhang W, Jiang P. LncRNA SNHG17 promotes gastric cancer progression by epigenetically silencing of p15 and p57. J Cell Physiol. 2019;234:5163–5174. doi: 10.1002/jcp.27320. [DOI] [PubMed] [Google Scholar]
- 73.Luo T, Zhao J, Lu Z, Bi J, Pang T, Cui H, Yang B, Li W, Wang Y, Wu S, Xue X. Characterization of long non-coding RNAs and MEF2C-AS1 identified as a novel biomarker in diffuse gastric cancer. Transl Oncol. 2018;11:1080–1089. doi: 10.1016/j.tranon.2018.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tan L, Yang Y, Shao Y, Zhang H, Guo J. Plasma lncRNA-GACAT2 is a valuable marker for the screening of gastric cancer. Oncol Lett. 2016;12:4845–4849. doi: 10.3892/ol.2016.5297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gao J, Cao R, Mu H. Long non-coding RNA UCA1 may be a novel diagnostic and predictive biomarker in plasma for early gastric cancer. Int J Clin Exp Pathol. 2015;8:12936–12942. [PMC free article] [PubMed] [Google Scholar]
- 76.Li Q, Shao Y, Zhang X, Zheng T, Miao M, Qin L, Wang B, Ye G, Xiao B, Guo J. Plasma long noncoding RNA protected by exosomes as a potential stable biomarker for gastric cancer. Tumour Biol. 2015;36:2007–2012. doi: 10.1007/s13277-014-2807-y. [DOI] [PubMed] [Google Scholar]
- 77.Xian HP, Zhuo ZL, Sun YJ, Liang B, Zhao XT. Circulating long non-coding RNAs HULC and ZNFX1-AS1 are potential biomarkers in patients with gastric cancer. Oncol Lett. 2018;16:4689–4698. doi: 10.3892/ol.2018.9199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wang Bing, Chen Huawei, Zhang Yang. Involvement of GASL1 in postoperative distant recurrence of gastric adenocarcinoma after gastrectomy distal resection and the possible mechanism. Journal of Cellular Biochemistry. 2019;120(7):11454–11461. doi: 10.1002/jcb.28423. [DOI] [PubMed] [Google Scholar]
- 79.Peng C, Li X, Yu Y, Chen J. LncRNA GASL1 inhibits tumor growth in gastric carcinoma by inactivating the Wnt/beta-catenin signaling pathway. Exp Ther Med. 2019;17:4039–4045. doi: 10.3892/etm.2019.7409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhu K, Ren Q. Zhao Y: lncRNA MALAT1 overexpression promotes proliferation, migration and invasion of gastric cancer by activating the PI3K/AKT pathway. Oncol Lett. 2019;17:5335–5342. doi: 10.3892/ol.2019.10253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Xia H, Chen Q, Chen Y, Ge X, Leng W, Tang Q, Ren M, Chen L, Yuan D, Zhang Y, et al. The lncRNA MALAT1 is a novel biomarker for gastric cancer metastasis. Oncotarget. 2016;7:56209–56218. doi: 10.18632/oncotarget.10941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Jin C, Shi W, Wang F, Shen X, Qi J, Cong H, Yuan J, Shi L, Zhu B, Luo X, et al. Long non-coding RNA HULC as a novel serum biomarker for diagnosis and prognosis prediction of gastric cancer. Oncotarget. 2016;7:51763–51772. doi: 10.18632/oncotarget.10107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Arita T, Ichikawa D, Konishi H, Komatsu S, Shiozaki A, Shoda K, Kawaguchi T, Hirajima S, Nagata H, Kubota T, et al. Circulating long non-coding RNAs in plasma of patients with gastric cancer. Anticancer Res. 2013;33:3185–3193. [PubMed] [Google Scholar]
- 84.Zhou X, Yin C, Dang Y, Ye F, Zhang G. Identification of the long non-coding RNA H19 in plasma as a novel biomarker for diagnosis of gastric cancer. Sci Rep. 2015;5:11516. doi: 10.1038/srep11516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mohamed WA, Schaalan MF, Ramadan B. The expression profiling of circulating miR-204, miR-182, and lncRNA H19 as novel potential biomarkers for the progression of peptic ulcer to gastric cancer. J Cell Biochem. 2019;120:13464–13477. doi: 10.1002/jcb.28620. [DOI] [PubMed] [Google Scholar]
- 86.Yoruker EE, Keskin M, Kulle CB, Holdenrieder S, Gezer U. Diagnostic and prognostic value of circulating lncRNA H19 in gastric cancer. Biomed Rep. 2018;9:181–186. doi: 10.3892/br.2018.1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Feng W, Zong W, Li Y, Shen X, Cui X, Ju S. Abnormally expressed long noncoding RNA B3GALT5-AS1 may serve AS a biomarker for the diagnostic and prognostic of gastric cancer. J Cell Biochem. 2020;121:557–565. doi: 10.1002/jcb.29296. [DOI] [PubMed] [Google Scholar]
- 88.Liu Y, Zhang YM, Ma FB, Pan SR, Liu BZ. Long noncoding RNA HOXA11-AS promotes gastric cancer cell proliferation and invasion via SRSF1 and functions as a biomarker in gastric cancer. World J Gastroenterol. 2019;25:2763–2775. doi: 10.3748/wjg.v25.i22.2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Li Y, Li D, Zhao M, Huang S, Zhang Q, Lin H, Wang W, Li K, Li Z, Huang W, et al. Long noncoding RNA SNHG6 regulates p21 expression via activation of the JNK pathway and regulation of EZH2 in gastric cancer cells. Life Sci. 2018;208:295–304. doi: 10.1016/j.lfs.2018.07.032. [DOI] [PubMed] [Google Scholar]
- 90.Pan L, Liang W, Gu J, Zang X, Huang Z, Shi H, Chen J, Fu M, Zhang P, Xiao X, et al. Long noncoding RNA DANCR is activated by SALL4 and promotes the proliferation and invasion of gastric cancer cells. Oncotarget. 2018;9:1915–1930. doi: 10.18632/oncotarget.23019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Fu Min, Huang Zhenhua, Zang Xueyan, Pan Lei, Liang Wei, Chen Jingyan, Qian Hui, Xu Wenrong, Jiang Pengcheng, Zhang Xu. Long noncoding RNA LINC00978 promotes cancer growth and acts as a diagnostic biomarker in gastric cancer. Cell Proliferation. 2017;51(1):e12425. doi: 10.1111/cpr.12425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zhou H, Wang F, Chen H, Tan Q, Qiu S, Chen S, Jing W, Yu M, Liang C, Ye S, Tu J. Increased expression of long-noncoding RNA ZFAS1 is associated with epithelial-mesenchymal transition of gastric cancer. Aging (Albany NY) 2016;8:2023–2038. doi: 10.18632/aging.101048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Pan L, Liang W, Fu M, Huang ZH, Li X, Zhang W, Zhang P, Qian H, Jiang PC, Xu WR, Zhang X. Exosomes-mediated transfer of long noncoding RNA ZFAS1 promotes gastric cancer progression. J Cancer Res Clin Oncol. 2017;143:991–1004. doi: 10.1007/s00432-017-2361-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Cai C, Zhang H, Zhu Y, Zheng P, Xu Y, Sun J, Zhang M, Lan T, Gu B, Li S, Ma P. Serum Exosomal long noncoding RNA pcsk2-2:1 as a potential novel diagnostic biomarker for gastric Cancer. Onco Targets Ther. 2019;12:10035–10041. doi: 10.2147/OTT.S229033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Li S, Zhang M, Zhang H, Hu K, Cai C, Wang J, Shi L, Ma P, Xu Y, Zheng P. Exosomal long noncoding RNA lnc-GNAQ-6:1 may serve as a diagnostic marker for gastric cancer. Clin Chim Acta. 2020;501:252–257. doi: 10.1016/j.cca.2019.10.047. [DOI] [PubMed] [Google Scholar]
- 96.Xu H, Zhou J, Tang J, Min X, Yi T, Zhao J, Ren Y. Identification of serum exosomal lncRNA MIAT as a novel diagnostic and prognostic biomarker for gastric cancer. J Clin Lab Anal. 2020:e23323.. [DOI] [PMC free article] [PubMed]
- 97.Yang Z, Sun Y, Liu R, Shi Y, Ding S. Plasma long noncoding RNAs PANDAR, FOXD2-AS1, and SMARCC2 AS potential novel diagnostic biomarkers for gastric cancer. Cancer Manag Res. 2019;11:6175–6184. doi: 10.2147/CMAR.S201935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ghaedi H, Mozaffari MAN, Salehi Z, Ghasemi H, Zadian SS, Alipoor S, Hadianpour S, Alipoor B. Co-expression profiling of plasma miRNAs and long noncoding RNAs in gastric cancer patients. Gene. 2019;687:135–142. doi: 10.1016/j.gene.2018.11.034. [DOI] [PubMed] [Google Scholar]
- 99.Liu J, Wang J, Song Y, Ma B, Luo J, Ni Z, Gao P, Sun J, Zhao J, Chen X, Wang Z. A panel consisting of three novel circulating lncRNAs, is it a predictive tool for gastric cancer? J Cell Mol Med. 2018;22:3605–3613. doi: 10.1111/jcmm.13640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ke D, Li H, Zhang Y, An Y, Fu H, Fang X, Zheng X. The combination of circulating long noncoding RNAs AK001058, INHBA-AS1, MIR4435-2HG, and CEBPA-AS1 fragments in plasma serve as diagnostic markers for gastric cancer. Oncotarget. 2017;8:21516–21525. doi: 10.18632/oncotarget.15628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zheng R, Liang J, Lu J, Li S, Zhang G, Wang X, Liu M, Wang W, Chu H, Tao G, et al. Genome-wide long non-coding RNAs identified a panel of novel plasma biomarkers for gastric cancer diagnosis. Gastric Cancer. 2019;22:731–741. doi: 10.1007/s10120-018-00915-7. [DOI] [PubMed] [Google Scholar]
- 102.Hashad D, Elbanna A, Ibrahim A, Khedr G. Evaluation of the role of circulating long non-coding RNA H19 as a promising novel biomarker in plasma of patients with gastric Cancer. J Clin Lab Anal. 2016;30:1100–1105. doi: 10.1002/jcla.21987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Zong W, Feng W, Jiang Y, Ju S, Cui M, Jing R. Evaluating the diagnostic and prognostic value of serum long non-coding RNA CTC-497E21.4 in gastric cancer. Clin Chem Lab Med. 2019;57:1063–1072. doi: 10.1515/cclm-2018-0929. [DOI] [PubMed] [Google Scholar]
- 104.Zhao R, Zhang Y, Zhang X, Yang Y, Zheng X, Li X, Liu Y, Zhang Y. Exosomal long noncoding RNA HOTTIP as potential novel diagnostic and prognostic biomarker test for gastric cancer. Mol Cancer. 2018;17:68. doi: 10.1186/s12943-018-0817-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Tirada N, Aujero M, Khorjekar G, Richards S, Chopra J, Dromi S, Ioffe O. Breast Cancer tissue markers, genomic profiling, and other prognostic factors: a primer for radiologists. Radiographics. 2018;38:1902–1920. doi: 10.1148/rg.2018180047. [DOI] [PubMed] [Google Scholar]
- 106.Dong L, Lin W, Qi P, Xu MD, Wu X, Ni S, Huang D, Weng WW, Tan C, Sheng W, et al. Circulating long RNAs in serum extracellular vesicles: their characterization and potential application as biomarkers for diagnosis of colorectal Cancer. Cancer Epidemiol Biomark Prev. 2016;25:1158–1166. doi: 10.1158/1055-9965.EPI-16-0006. [DOI] [PubMed] [Google Scholar]
- 107.Hauser P, Wang S, Didenko VV. Apoptotic bodies: selective detection in extracellular vesicles. Methods Mol Biol. 2017;1554:193–200. doi: 10.1007/978-1-4939-6759-9_12. [DOI] [PubMed] [Google Scholar]
- 108.van Niel G, D'Angelo G, Raposo G. Shedding light on the cell biology of extracellular vesicles. Nat Rev Mol Cell Biol. 2018;19:213–228. doi: 10.1038/nrm.2017.125. [DOI] [PubMed] [Google Scholar]
- 109.Fu M, Gu J, Jiang P, Qian H, Xu W, Zhang X. Exosomes in gastric cancer: roles, mechanisms, and applications. Mol Cancer. 2019;18:41. doi: 10.1186/s12943-019-1001-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Sung J, Kim N, Lee J, Hwang YJ, Kim HW, Chung JW, Kim JW, Lee DH. Associations among gastric juice pH, atrophic gastritis, intestinal metaplasia and helicobacter pylori infection. Gut Liver. 2018;12:158–164. doi: 10.5009/gnl17063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Fei ZH, Yu XJ, Zhou M, Su HF, Zheng Z, Xie CY. Upregulated expression of long non-coding RNA LINC00982 regulates cell proliferation and its clinical relevance in patients with gastric cancer. Tumour Biol. 2016;37:1983–1993. doi: 10.1007/s13277-015-3979-9. [DOI] [PubMed] [Google Scholar]
- 112.Shao Y, Ye M, Jiang X, Sun W, Ding X, Liu Z, Ye G, Zhang X, Xiao B, Guo J. Gastric juice long noncoding RNA used as a tumor marker for screening gastric cancer. Cancer. 2014;120:3320–3328. doi: 10.1002/cncr.28882. [DOI] [PubMed] [Google Scholar]
- 113.Yang Y, Shao Y, Zhu M, Li Q, Yang F, Lu X, Xu C, Xiao B, Sun Y, Guo J. Using gastric juice lncRNA-ABHD11-AS1 AS a novel type of biomarker in the screening of gastric cancer. Tumour Biol. 2016;37:1183–1188. doi: 10.1007/s13277-015-3903-3. [DOI] [PubMed] [Google Scholar]
- 114.Leary Meghan, Heerboth Sarah, Lapinska Karolina, Sarkar Sibaji. Sensitization of Drug Resistant Cancer Cells: A Matter of Combination Therapy. Cancers. 2018;10(12):483. doi: 10.3390/cancers10120483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Chakraborty S, Mir KB, Seligson ND, Nayak D, Kumar R, Goswami A. Integration of EMT and cellular survival instincts in reprogramming of programmed cell death to anastasis. Cancer Metastasis Rev. 2020. [DOI] [PubMed]
- 116.Du Bowen, Shim Joong. Targeting Epithelial–Mesenchymal Transition (EMT) to Overcome Drug Resistance in Cancer. Molecules. 2016;21(7):965. doi: 10.3390/molecules21070965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Dong J, Qin Z, Zhang WD, Cheng G, Yehuda AG, Ashby CR, Jr, Chen ZS, Cheng XD, Qin JJ. Medicinal chemistry strategies to discover P-glycoprotein inhibitors: An update. Drug Resist Updat. 2020;49:100681. doi: 10.1016/j.drup.2020.100681. [DOI] [PubMed] [Google Scholar]
- 118.Qin JJ, Yan L, Zhang J, Zhang WD. STAT3 as a potential therapeutic target in triple negative breast cancer: a systematic review. J Exp Clin Cancer Res. 2019;38:195. doi: 10.1186/s13046-019-1206-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Qin JJ, Cheng XD, Zhang J, Zhang WD. Dual roles and therapeutic potential of Keap1-Nrf2 pathway in pancreatic cancer: a systematic review. Cell Commun Signal. 2019;17:121. doi: 10.1186/s12964-019-0435-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Zhao W, Shan B, He D, Cheng Y, Li B, Zhang C, Duan C. Recent Progress in characterizing long noncoding RNAs in Cancer drug resistance. J Cancer. 2019;10:6693–6702. doi: 10.7150/jca.30877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Xiao Y, Pan J, Geng Q, Wang G. LncRNA MALAT1 increases the stemness of gastric cancer cells via enhancing SOX2 mRNA stability. FEBS Open Bio. 2019;9:1212–1222. doi: 10.1002/2211-5463.12649. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 122.YiRen H, YingCong Y, Sunwu Y, Keqin L, Xiaochun T, Senrui C, Ende C, XiZhou L, Yanfan C. Long noncoding RNA MALAT1 regulates autophagy associated chemoresistance via miR-23b-3p sequestration in gastric cancer. Mol Cancer. 2017;16:174. doi: 10.1186/s12943-017-0743-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Xi Z, Si J, Nan J. LncRNA MALAT1 potentiates autophagyassociated cisplatin resistance by regulating the microRNA30b/autophagyrelated gene 5 axis in gastric cancer. Int J Oncol. 2019;54:239–248. doi: 10.3892/ijo.2018.4609. [DOI] [PubMed] [Google Scholar]
- 124.Hang Q, Sun R, Jiang C, Li Y. Notch 1 promotes cisplatin-resistant gastric cancer formation by upregulating lncRNA AK022798 expression. Anti-Cancer Drugs. 2015;26:632–640. doi: 10.1097/CAD.0000000000000227. [DOI] [PubMed] [Google Scholar]
- 125.Wang Z, Wang Q, Xu G, Meng N, Huang X, Jiang Z, Chen C, Zhang Y, Chen J, Li A, et al. The long noncoding RNA CRAL reverses cisplatin resistance via the miR-505/CYLD/AKT axis in human gastric cancer cells. RNA Biol. 2020:1–14. [DOI] [PMC free article] [PubMed]
- 126.Wang S, Chen W, Yu H, Song Z, Li Q, Shen X, Wu Y, Zhu L, Ma Q. Xing D: lncRNA ROR promotes gastric Cancer drug resistance. Cancer Control. 2020;27:1073274820904694. doi: 10.1177/1073274820904694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Zeng L, Liao Q, Zou Z, Wen Y, Wang J, Liu C, He Q, Weng N, Zeng J, Tang H, et al. Long non-coding RNA XLOC_006753 promotes the development of multidrug resistance in gastric Cancer cells through the PI3K/AKT/mTOR signaling pathway. Cell Physiol Biochem. 2018;51:1221–1236. doi: 10.1159/000495499. [DOI] [PubMed] [Google Scholar]
- 128.He W, Liang B, Wang C, Li S, Zhao Y, Huang Q, Liu Z, Yao Z, Wu Q, Liao W, et al. MSC-regulated lncRNA MACC1-AS1 promotes stemness and chemoresistance through fatty acid oxidation in gastric cancer. Oncogene. 2019;38:4637–4654. doi: 10.1038/s41388-019-0747-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Zhou Z, Lin Z, He Y, Pang X, Wang Y, Ponnusamy M, Ao X, Shan P, Tariq MA, Li P, Wang J. The long noncoding RNA D63785 regulates chemotherapy sensitivity in human gastric Cancer by targeting miR-422a. Mol Ther Nucleic Acids. 2018;12:405–419. doi: 10.1016/j.omtn.2018.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Zhang C, Qian H, Liu K, Zhao W, Wang L. A feedback loop regulation of LINC01433 and YAP promotes malignant behavior in gastric Cancer cells. Onco Targets Ther. 2019;12:7949–7962. doi: 10.2147/OTT.S222903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Ye Y, Yang S, Han Y, Sun J, Xv L, Wu L, Ming L. HOXD-AS1 confers cisplatin resistance in gastric cancer through epigenetically silencing PDCD4 via recruiting EZH2. Open Biol. 2019;9:190068. doi: 10.1098/rsob.190068. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 132.Xin L, Zhou Q, Yuan YW, Zhou LQ, Liu L, Li SH, Liu C. METase/lncRNA HULC/FoxM1 reduced cisplatin resistance in gastric cancer by suppressing autophagy. J Cancer Res Clin Oncol. 2019;145:2507–2517. doi: 10.1007/s00432-019-03015-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Zhang Y, Song X, Wang X, Hu J, Jiang L. Silencing of LncRNA HULC enhances chemotherapy induced apoptosis in human gastric Cancer. J Med Biochem. 2016;35:137–143. doi: 10.1515/jomb-2015-0016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Guo Y, Yue P, Wang Y, Chen G, Li Y. PCAT-1 contributes to cisplatin resistance in gastric cancer through miR-128/ZEB1 axis. Biomed Pharmacother. 2019;118:109255. doi: 10.1016/j.biopha.2019.109255. [DOI] [PubMed] [Google Scholar]
- 135.Li H, Ma X, Yang D, Suo Z, Dai R, Liu C. PCAT-1 contributes to cisplatin resistance in gastric cancer through epigenetically silencing PTEN via recruiting EZH2. J Cell Biochem. 2020;121:1353–1361. doi: 10.1002/jcb.29370. [DOI] [PubMed] [Google Scholar]
- 136.Li Y, Lv S, Ning H, Li K, Zhou X, Xv H, Wen H. Down-regulation of CASC2 contributes to cisplatin resistance in gastric cancer by sponging miR-19a. Biomed Pharmacother. 2018;108:1775–1782. doi: 10.1016/j.biopha.2018.09.181. [DOI] [PubMed] [Google Scholar]
- 137.Cheng C, Qin Y, Zhi Q, Wang J, Qin C. Knockdown of long non-coding RNA HOTAIR inhibits cisplatin resistance of gastric cancer cells through inhibiting the PI3K/Akt and Wnt/beta-catenin signaling pathways by up-regulating miR-34a. Int J Biol Macromol. 2018;107:2620–2629. doi: 10.1016/j.ijbiomac.2017.10.154. [DOI] [PubMed] [Google Scholar]
- 138.Jia J, Zhan D, Li J, Li Z, Li H, Qian J. The contrary functions of lncRNA HOTAIR/miR-17-5p/PTEN axis and Shenqifuzheng injection on chemosensitivity of gastric cancer cells. J Cell Mol Med. 2019;23:656–669. doi: 10.1111/jcmm.13970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Wang H, Qin R, Guan A, Yao Y, Huang Y, Jia H, Huang W, Gao J. HOTAIR enhanced paclitaxel and doxorubicin resistance in gastric cancer cells partly through inhibiting miR-217 expression. J Cell Biochem. 2018;119:7226–7234. doi: 10.1002/jcb.26901. [DOI] [PubMed] [Google Scholar]
- 140.Yan Jin, Dang Yini, Liu Shiyu, Zhang Yifeng, Zhang Guoxin. LncRNA HOTAIR promotes cisplatin resistance in gastric cancer by targeting miR-126 to activate the PI3K/AKT/MRP1 genes. Tumor Biology. 2016;37(12):16345–16355. doi: 10.1007/s13277-016-5448-5. [DOI] [PubMed] [Google Scholar]
- 141.Song H, Xu Y, Shi L, Xu T, Fan R, Cao M, Xu W, Song J. LncRNA THOR increases the stemness of gastric cancer cells via enhancing SOX9 mRNA stability. Biomed Pharmacother. 2018;108:338–346. doi: 10.1016/j.biopha.2018.09.057. [DOI] [PubMed] [Google Scholar]
- 142.Wu X, Zheng Y, Han B, Dong X. Long noncoding RNA BLACAT1 modulates ABCB1 to promote oxaliplatin resistance of gastric cancer via sponging miR-361. Biomed Pharmacother. 2018;99:832–838. doi: 10.1016/j.biopha.2018.01.130. [DOI] [PubMed] [Google Scholar]
- 143.Zhang X, Bo P, Liu L, Zhang X, Li J. Overexpression of long non-coding RNA GHET1 promotes the development of multidrug resistance in gastric cancer cells. Biomed Pharmacother. 2017;92:580–585. doi: 10.1016/j.biopha.2017.04.111. [DOI] [PubMed] [Google Scholar]
- 144.Lan WG, Xu DH, Xu C, Ding CL, Ning FL, Zhou YL, Ma LB, Liu CM, Han X. Silencing of long non-coding RNA ANRIL inhibits the development of multidrug resistance in gastric cancer cells. Oncol Rep. 2016;36:263–270. doi: 10.3892/or.2016.4771. [DOI] [PubMed] [Google Scholar]
- 145.Shang C, Guo Y, Zhang J, Huang B. Silence of long noncoding RNA UCA1 inhibits malignant proliferation and chemotherapy resistance to adriamycin in gastric cancer. Cancer Chemother Pharmacol. 2016;77:1061–1067. doi: 10.1007/s00280-016-3029-3. [DOI] [PubMed] [Google Scholar]
- 146.Fang Q, Chen X, Zhi X. Long non-coding RNA (LncRNA) Urothelial carcinoma associated 1 (UCA1) increases multi-drug resistance of gastric Cancer via Downregulating miR-27b. Med Sci Monit. 2016;22:3506–3513. doi: 10.12659/MSM.900688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Du P, Hu C, Qin Y, Zhao J, Patel R, Fu Y, Zhu M, Zhang W, Huang G. LncRNA PVT1 mediates Antiapoptosis and 5-fluorouracil resistance via increasing Bcl2 expression in gastric Cancer. J Oncol. 2019;2019:9325407. doi: 10.1155/2019/9325407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Zhang XW, Bu P, Liu L, Zhang XZ, Li J. Overexpression of long non-coding RNA PVT1 in gastric cancer cells promotes the development of multidrug resistance. Biochem Biophys Res Commun. 2015;462:227–232. doi: 10.1016/j.bbrc.2015.04.121. [DOI] [PubMed] [Google Scholar]
- 149.Wang Y, Zhang D, Wu K, Zhao Q, Nie Y, Fan D. Long noncoding RNA MRUL promotes ABCB1 expression in multidrug-resistant gastric cancer cell sublines. Mol Cell Biol. 2014;34:3182–3193. doi: 10.1128/MCB.01580-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Li M, Zhang YY, Shang J, Xu YD. LncRNA SNHG5 promotes cisplatin resistance in gastric cancer via inhibiting cell apoptosis. Eur Rev Med Pharmacol Sci. 2019;23:4185–4191. doi: 10.26355/eurrev_201905_17921. [DOI] [PubMed] [Google Scholar]
- 151.Xu YD, Shang J, Li M, Zhang YY. LncRNA DANCR accelerates the development of multidrug resistance of gastric cancer. Eur Rev Med Pharmacol Sci. 2019;23:2794–2802. doi: 10.26355/eurrev_201904_17554. [DOI] [PubMed] [Google Scholar]
- 152.Zhang Y, Li Q, Yu S, Zhu C, Zhang Z, Cao H, Xu J. Long non-coding RNA FAM84B-AS promotes resistance of gastric cancer to platinum drugs through inhibition of FAM84B expression. Biochem Biophys Res Commun. 2019;509:753–762. doi: 10.1016/j.bbrc.2018.12.177. [DOI] [PubMed] [Google Scholar]
- 153.Wang L, Chunyan Q, Zhou Y, He Q, Ma Y, Ga Y, Wang X. BCAR4 increase cisplatin resistance and predicted poor survival in gastric cancer patients. Eur Rev Med Pharmacol Sci. 2017;21:4064–4070. [PubMed] [Google Scholar]
- 154.Zhang J, Zhao B, Chen X, Wang Z, Xu H, Huang B. Silence of long noncoding RNA NEAT1 inhibits malignant biological behaviors and chemotherapy resistance in gastric Cancer. Pathol Oncol Res. 2018;24:109–113. doi: 10.1007/s12253-017-0233-3. [DOI] [PubMed] [Google Scholar]
- 155.Han Y, Ye J, Wu D, Wu P, Chen Z, Chen J, Gao S, Huang J. LEIGC long non-coding RNA acts as a tumor suppressor in gastric carcinoma by inhibiting the epithelial-to-mesenchymal transition. BMC Cancer. 2014;14:932. doi: 10.1186/1471-2407-14-932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Shang C, Sun L, Zhang J, Zhao B, Chen X, Xu H, Huang B. Silence of cancer susceptibility candidate 9 inhibits gastric cancer and reverses chemoresistance. Oncotarget. 2017;8:15393–15398. doi: 10.18632/oncotarget.14871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Mao Z, Wu Y, Zhou J, Xing C. Salinomycin reduces epithelial-mesenchymal transition-mediated multidrug resistance by modifying long noncoding RNA HOTTIP expression in gastric cancer cells. Anti-Cancer Drugs. 2019;30:892–899. doi: 10.1097/CAD.0000000000000786. [DOI] [PubMed] [Google Scholar]
- 158.Wu H, Liu B, Chen Z, Li G, Zhang Z. MSC-induced lncRNA HCP5 drove fatty acid oxidation through miR-3619-5p/AMPK/PGC1alpha/CEBPB axis to promote stemness and chemo-resistance of gastric cancer. Cell Death Dis. 2020;11:233. doi: 10.1038/s41419-020-2426-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Wang W, Qin JJ, Voruganti S, Nijampatnam B, Velu SE, Ruan KH, Hu M, Zhou J, Zhang R. Discovery and characterization of dual inhibitors of MDM2 and NFAT1 for pancreatic Cancer therapy. Cancer Res. 2018;78:5656–5667. doi: 10.1158/0008-5472.CAN-17-3939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Wang W, Yang J, Liao YY, Cheng G, Chen J, Cheng XD, Qin JJ, Shao Z. Cytotoxic Nitrogenated Azaphilones from the Deep-Sea-derived fungus Chaetomium globosum MP4-S01-7. J Nat Prod. 2020;83:1157–66. [DOI] [PubMed]
- 161.Mohammad RM, Muqbil I, Lowe L, Yedjou C, Hsu HY, Lin LT, Siegelin MD, Fimognari C, Kumar NB, Dou QP, et al. Broad targeting of resistance to apoptosis in cancer. Semin Cancer Biol. 2015;35(Suppl):S78–s103. doi: 10.1016/j.semcancer.2015.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.D'Arcy MS. Cell death: a review of the major forms of apoptosis, necrosis and autophagy. Cell Biol Int. 2019;43:582–592. doi: 10.1002/cbin.11137. [DOI] [PubMed] [Google Scholar]
- 163.Pfeffer CM, Singh ATK. Apoptosis: a target for anticancer therapy. Int J Mol Sci. 2018;19:448. [DOI] [PMC free article] [PubMed]
- 164.Cavalcante Giovanna C., Schaan Ana Paula, Cabral Gleyce Fonseca, Santana-da-Silva Mayara Natália, Pinto Pablo, Vidal Amanda F., Ribeiro-dos-Santos Ândrea. A Cell’s Fate: An Overview of the Molecular Biology and Genetics of Apoptosis. International Journal of Molecular Sciences. 2019;20(17):4133. doi: 10.3390/ijms20174133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Fianco Giulia, Contadini Claudia, Ferri Alessandra, Cirotti Claudia, Stagni Venturina, Barilà Daniela. Caspase-8: A Novel Target to Overcome Resistance to Chemotherapy in Glioblastoma. International Journal of Molecular Sciences. 2018;19(12):3798. doi: 10.3390/ijms19123798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.García-Aranda Marilina, Pérez-Ruiz Elisabet, Redondo Maximino. Bcl-2 Inhibition to Overcome Resistance to Chemo- and Immunotherapy. International Journal of Molecular Sciences. 2018;19(12):3950. doi: 10.3390/ijms19123950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Binju Mudra, Amaya-Padilla Monica Angelica, Wan Graeme, Gunosewoyo Hendra, Suryo Rahmanto Yohan, Yu Yu. Therapeutic Inducers of Apoptosis in Ovarian Cancer. Cancers. 2019;11(11):1786. doi: 10.3390/cancers11111786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Verret B, Cortes J, Bachelot T, Andre F, Arnedos M. Efficacy of PI3K inhibitors in advanced breast cancer. Ann Oncol. 2019;30:x12–x20. doi: 10.1093/annonc/mdz381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Lin KN, Jiang YL, Zhang SG, Huang SY, Li H. Grape seed proanthocyanidin extract reverses multidrug resistance in HL-60/ADR cells via inhibition of the PI3K/Akt signaling pathway. Biomed Pharmacother. 2020;125:109885. doi: 10.1016/j.biopha.2020.109885. [DOI] [PubMed] [Google Scholar]
- 170.Nguyen CDK, Yi C. YAP/TAZ signaling and resistance to Cancer therapy. Trends Cancer. 2019;5:283–296. doi: 10.1016/j.trecan.2019.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Ye C, Wang W, Xia G, Yu C, Yi Y, Hua C, Tu F, Shen L, Chen C, Sun W, Zheng Z. A novel curcumin derivative CL-6 exerts antitumor effect in human gastric cancer cells by inducing apoptosis through hippo-YAP signaling pathway. Onco Targets Ther. 2019;12:2259–2269. doi: 10.2147/OTT.S196914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Yang XM, Cao XY, He P, Li J, Feng MX, Zhang YL, Zhang XL, Wang YH, Yang Q, Zhu L, et al. Overexpression of rac GTPase activating protein 1 contributes to proliferation of cancer cells by reducing hippo signaling to promote cytokinesis. Gastroenterology. 2018;155:1233–1249.e1222. doi: 10.1053/j.gastro.2018.07.010. [DOI] [PubMed] [Google Scholar]
- 173.Chiarini F, Paganelli F, Martelli AM, Evangelisti C. The role played by Wnt/beta-catenin signaling pathway in acute lymphoblastic leukemia. Int J Mol Sci. 2020;21:1098. [DOI] [PMC free article] [PubMed]
- 174.Roy S, Kar M, Roy S, Padhi S, Kumar A, Thakur S, Akhter Y, Gatto G, Banerjee B. Inhibition of CD44 sensitizes cisplatin-resistance and affects Wnt/beta-catenin signaling in HNSCC cells. Int J Biol Macromol. 2020;149:501–512. doi: 10.1016/j.ijbiomac.2020.01.131. [DOI] [PubMed] [Google Scholar]
- 175.Ram Makena M, Gatla H, Verlekar D, Sukhavasi S. M KP, K CP: Wnt/beta-catenin signaling: the culprit in pancreatic carcinogenesis and therapeutic resistance. Int J Mol Sci. 2019;20:4242. [DOI] [PMC free article] [PubMed]
- 176.Harb J, Lin PJ, Hao J. Recent development of Wnt signaling pathway inhibitors for Cancer therapeutics. Curr Oncol Rep. 2019;21:12. doi: 10.1007/s11912-019-0763-9. [DOI] [PubMed] [Google Scholar]
- 177.Yin J, Ni B, Liao WG, Gao YQ. Hypoxia-induced apoptosis of mouse spermatocytes is mediated by HIF-1alpha through a death receptor pathway and a mitochondrial pathway. J Cell Physiol. 2018;233:1146–1155. doi: 10.1002/jcp.25974. [DOI] [PubMed] [Google Scholar]
- 178.Méndez-Blanco Carolina, Fondevila Flavia, Fernández-Palanca Paula, García-Palomo Andrés, van Pelt Jos, Verslype Chris, González-Gallego Javier, Mauriz José L. Stabilization of Hypoxia-Inducible Factors and BNIP3 Promoter Methylation Contribute to Acquired Sorafenib Resistance in Human Hepatocarcinoma Cells. Cancers. 2019;11(12):1984. doi: 10.3390/cancers11121984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Ge X, Pan MH, Wang L, Li W, Jiang C, He J, Abouzid K, Liu LZ, Shi Z, Jiang BH. Hypoxia-mediated mitochondria apoptosis inhibition induces temozolomide treatment resistance through miR-26a/bad/Bax axis. Cell Death Dis. 2018;9:1128. doi: 10.1038/s41419-018-1176-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Lu W, Kang Y. Epithelial-Mesenchymal plasticity in Cancer progression and metastasis. Dev Cell. 2019;49:361–374. doi: 10.1016/j.devcel.2019.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Olteanu GE, Mihai IM, Bojin F, Gavriliuc O, Paunescu V. The natural adaptive evolution of cancer: the metastatic ability of cancer cells. Bosn J Basic Med Sci. 2020. [DOI] [PMC free article] [PubMed]
- 182.Teeuwssen, Fodde Wnt Signaling in Ovarian Cancer Stemness, EMT, and Therapy Resistance. Journal of Clinical Medicine. 2019;8(10):1658. doi: 10.3390/jcm8101658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Liu B, Wu S, Ma J, Yan S, Xiao Z, Wan L, Zhang F, Shang M. Mao a: lncRNA GAS5 reverses EMT and tumor stem cell-mediated gemcitabine resistance and metastasis by targeting miR-221/SOCS3 in pancreatic Cancer. Mol Ther Nucleic Acids. 2018;13:472–482. doi: 10.1016/j.omtn.2018.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Dong H, Hu J, Zou K, Ye M, Chen Y, Wu C, Chen X, Han M. Activation of LncRNA TINCR by H3K27 acetylation promotes Trastuzumab resistance and epithelial-mesenchymal transition by targeting MicroRNA-125b in breast Cancer. Mol Cancer. 2019;18:3. doi: 10.1186/s12943-018-0931-9. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 185.Yadav AK, Desai NS. Cancer stem cells: acquisition, characteristics, therapeutic implications, targeting strategies and future prospects. Stem Cell Rev Rep. 2019;15:331–355. doi: 10.1007/s12015-019-09887-2. [DOI] [PubMed] [Google Scholar]
- 186.Lathia J, Liu H, Matei D. The clinical impact of Cancer stem cells. Oncologist. 2020;25:123–131. doi: 10.1634/theoncologist.2019-0517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Najafi M, Farhood B, Mortezaee K. Cancer stem cells (CSCs) in cancer progression and therapy. J Cell Physiol. 2019;234:8381–8395. doi: 10.1002/jcp.27740. [DOI] [PubMed] [Google Scholar]
- 188.Prasad S, Ramachandran S, Gupta N, Kaushik I, Srivastava SK. Cancer cells stemness: a doorstep to targeted therapy. Biochim Biophys Acta Mol basis Dis. 1866;2020:165424. doi: 10.1016/j.bbadis.2019.02.019. [DOI] [PubMed] [Google Scholar]
- 189.Wang J, Quan Y, Lv J, Gong S, Dong D. BRD4 promotes glioma cell stemness via enhancing miR-142-5p-mediated activation of Wnt/beta-catenin signaling. Environ Toxicol. 2020;35:368–376. doi: 10.1002/tox.22873. [DOI] [PubMed] [Google Scholar]
- 190.Liu Q, Sun J, Luo Q, Ju Y, Song G. Salinomycin suppresses tumorigenicity of liver cancer stem cells and Wnt/beta-catenin signaling. Curr Stem Cell Res Ther. 2020. [DOI] [PubMed]
- 191.Allen Elizabeth A., Baehrecke Eric H. Autophagy in animal development. Cell Death & Differentiation. 2020;27(3):903–918. doi: 10.1038/s41418-020-0497-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Levy JMM, Towers CG, Thorburn A. Targeting autophagy in cancer. Nat Rev Cancer. 2017;17:528–542. doi: 10.1038/nrc.2017.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Li X, He S, Ma B. Autophagy and autophagy-related proteins in cancer. Mol Cancer. 2020;19:12. doi: 10.1186/s12943-020-1138-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Yang Ying, Klionsky Daniel J. Autophagy and disease: unanswered questions. Cell Death & Differentiation. 2020;27(3):858–871. doi: 10.1038/s41418-019-0480-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Ho Cally J., Gorski Sharon M. Molecular Mechanisms Underlying Autophagy-Mediated Treatment Resistance in Cancer. Cancers. 2019;11(11):1775. doi: 10.3390/cancers11111775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Li YJ, Lei YH, Yao N, Wang CR, Hu N, Ye WC, Zhang DM, Chen ZS. Autophagy and multidrug resistance in cancer. Chin J Cancer. 2017;36:52. doi: 10.1186/s40880-017-0219-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Robey RW, Pluchino KM, Hall MD, Fojo AT, Bates SE, Gottesman MM. Revisiting the role of ABC transporters in multidrug-resistant cancer. Nat Rev Cancer. 2018;18:452–464. doi: 10.1038/s41568-018-0005-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Amawi H, Sim HM, Tiwari AK, Ambudkar SV, Shukla S. ABC transporter-mediated multidrug-resistant Cancer. Adv Exp Med Biol. 2019;1141:549–580. doi: 10.1007/978-981-13-7647-4_12. [DOI] [PubMed] [Google Scholar]
- 199.Hu Q, Baeg GH. Role of epigenome in tumorigenesis and drug resistance. Food Chem Toxicol. 2017;109:663–668. doi: 10.1016/j.fct.2017.07.022. [DOI] [PubMed] [Google Scholar]
- 200.Xu W, Zhou B, Zhao X, Zhu L, Xu J, Jiang Z, Chen D, Wei Q, Han M, Feng L, et al. KDM5B demethylates H3K4 to recruit XRCC1 and promote chemoresistance. Int J Biol Sci. 2018;14:1122–1132. doi: 10.7150/ijbs.25881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Schizas D, Mastoraki A, Naar L, Tsilimigras DI, Katsaros I, Fragkiadaki V, Karachaliou GS, Arkadopoulos N, Liakakos T, Moris D. Histone Deacetylases (HDACs) in gastric cancer: An update of their emerging prognostic and therapeutic role. Curr Med Chem. 2019. [DOI] [PubMed]
- 202.Arun G, Diermeier SD, Spector DL. Therapeutic targeting of long non-coding RNAs in Cancer. Trends Mol Med. 2018;24:257–277. doi: 10.1016/j.molmed.2018.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.