Abstract
Background
Rett syndrome (RTT) is a progressive neurodevelopmental disease that is characterized by abnormalities in cognitive, social, and motor skills. RTT is often caused by mutations in the X-linked gene encoding methyl-CpG binding protein 2 (MeCP2). The mechanism by which impaired MeCP2 induces the pathological abnormalities in the brain is not understood. Both patients and mouse models have shown abnormalities at molecular and cellular level before typical RTT-associated symptoms appear. This implies that underlying mechanisms are already affected during neurodevelopmental stages.
Methods
To understand the molecular mechanisms involved in disease onset, we used an RTT patient induced pluripotent stem cell (iPSC)-based model with isogenic controls and performed time-series of proteomic analysis using in-depth high-resolution quantitative mass spectrometry during early stages of neuronal development.
Results
We provide mass spectrometry-based quantitative proteomic data, depth of about 7000 proteins, at neuronal progenitor developmental stages of RTT patient cells and isogenic controls. Our data gives evidence of proteomic alteration at early neurodevelopmental stages, suggesting alterations long before the phase that symptoms of RTT syndrome become apparent. Significant changes are associated with the GO enrichment analysis in biological processes cell-cell adhesion, actin cytoskeleton organization, neuronal stem cell population maintenance, and pituitary gland development, next to protein changes previously associated with RTT, i.e., dendrite morphology and synaptic deficits. Differential expression increased from early to late neural stem cell phases, although proteins involved in immunity, metabolic processes, and calcium signaling were affected throughout all stages analyzed.
Limitations
The limitation of our study is the number of RTT patients analyzed. As the aim of our study was to investigate a large number of proteins, only one patient was considered, of which 3 different RTT iPSC clones and 3 isogenic control iPSC clones were included. Even though this approach allowed the study of mutation-induced alterations due to the usage of isogenic controls, results should be validated on different RTT patients to suggest common disease mechanisms.
Conclusions
During early neuronal differentiation, there are consistent and time-point specific proteomic alterations in RTT patient cells carrying exons 3–4 deletion in MECP2. We found changes in proteins involved in pathway associated with RTT phenotypes, including dendrite morphology and synaptogenesis. Our results provide a valuable resource of proteins and pathways for follow-up studies, investigating common mechanisms involved during early disease stages of RTT syndrome.
Keywords: Rett syndrome, iPSC, Neuron differentiation, Quantitative mass spectrometry, TMT-10plex
Background
Rett syndrome (RTT) is a severe neurodevelopmental disorder that mainly affects females with a frequency of ~ 1:10,000 [1]. Clinical features of RTT start to present around 6–18 months of age and include deceleration of head growth, abnormalities in cognitive, social and motor skill development, and seizures [2, 3]. Postmortem studies showed increased density of neurons in combination with reduced soma sizes in RTT patient compared to healthy control brains [4, 5]. RTT neurons show a decrease in dendritic branching and a reduced number of dendritic spines and synapses [6, 7]. While studies suggest affected neurodevelopment starting at early stages, the molecular mechanisms underlying neuropathology in RTT is not understood.
In 90–95% of the RTT cases, the disease is caused by dominant loss-of-function mutations in the X-linked gene encoding methyl-CpG binding protein 2 (MeCP2) [8]. Random X chromosome inactivation in females results in somatic mosaics with normal and mutant MECP2 [9]. Males carrying a MECP2 mutation are not viable or suffer from severe symptoms and die early in life [10]. MeCP2 is described as a nuclear protein modulating gene expression, via binding to methylated DNA and hundreds of target genes. These modulations take place through direct repression or activation of genes or by means of DNA modulation and secondary gene regulation. Consequently, mutations in MECP2 lead to miss-regulation of hundreds of genes, including those influencing brain development and neuronal maturation [11–14]. So far, research in RTT focused on genomic and transcriptomic studies [15–17] and less so on proteome changes [18, 19]; although as molecular effectors of cellular processes, these are better predictors of pathological states. Recent advances in mass spectrometry-based proteomics now facilitate the study of global protein expression and quantification [20]. Considering the broad and complex regulating functions of MeCP2, modulating multiple cellular processes, we need insight into the final molecular effectors reflected by perturbation at the protein level to understand pathological states.
Here, we used an iPSC-based RTT model and performed proteome analysis on iPSC-derived neuronal stem cells (NES cells) carrying an MECP2 exons 3–4 mutation [21]. Earlier studies proved that iPSCs from RTT patients reflect disease-specific characteristics, including changes in neuronal differentiation at early stages of development [22, 23]. However, we lack knowledge on the precise molecular mechanisms at the onset of disease. To study early alterations in the proteome of RTT cells compared to isogenic controls (iCTR), we performed a high-resolution mass spectrometry-based quantitative proteomics at different time points during neuronal stem cell development (Fig. 1). We show that the difference between RTT and iCTR, in terms of the number of differentially expressed proteins, begins at early stages and increases at later progenitor stages. Interestingly, a large group of these proteins are involved in cellular processes, implicated in classical features of typical RTT phenotypes, such as dendrite formation and axonal growth. Proteins involved in immunity and metabolic processes are consistently changed between RTT and iCTR at all time points studied. Here, we provide a resource of target proteins and pathways for further studies into molecular mechanisms involved in early RTT disease stages.
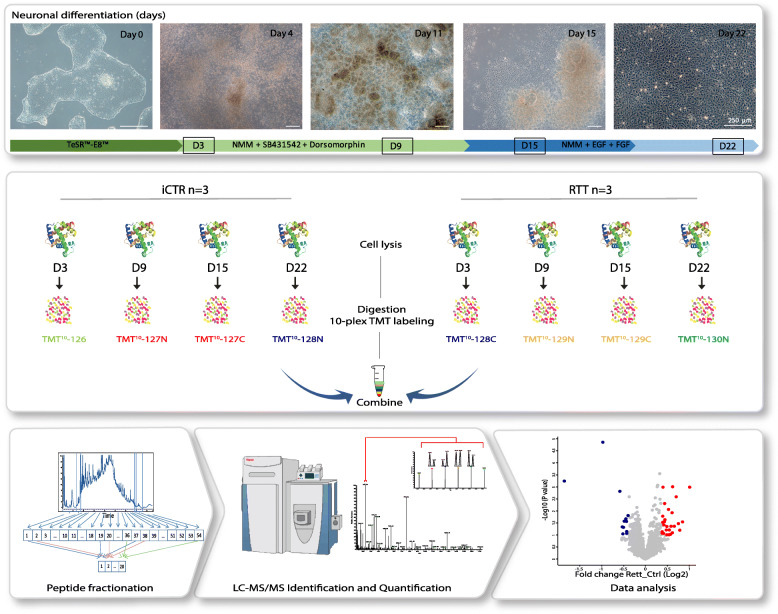
Fig. 1.
Overview of experimental workflow. iPSC differentiation towards neuronal stem cells. Different colors in arrows indicate change of medium. Squares mark days of sample collection. Samples at indicated time points (four time points) for (three clones) iCTR and (three clones) RTT were processed for proteomic analysis. In total, 24 samples were subjected for tryptic digestion, TMT-based isotope labeling, high-pH fractionation, and LC MS/MS analysis. Different bioinformatic approaches were then used to analyze the data
Methods
Cell culture and isogenic controls
RTT patient fibroblasts were derived from the Cell lines and DNA bank of Rett syndrome, X-linked mental retardation, and other genetic diseases at the University Siena in Italy via the Network of Genetic Biobanks Telethon. We used fibroblast lines carrying MECP2 mutation, showing a deletion in exons 3 and 4 of the MECP2 gene (RTT Ex3-4), (RTT#2282C2). Fibroblasts were derived frozen, thawed, and expanded in fibroblast medium (DMEM-F12, 20% FBS, 1% NEAA, 1% Pen/Strep, 50 μM β-Mercaptoethanol). To generate pure RTT, i.e., cells expressing affected X chromosome, and isogenic control, i.e., cells expressing the healthy X chromosome, fibroblasts were detached from cell culture plate, and single fibroblasts were seeded in a 96-well plate. Cells were further expanded and characterized for their MeCP2 state by immunocytochemistry and PCR [24]. All of our experiments were exempt from the approval of the institutional review board.
Reprogramming
Reprogramming of fibroblasts was performed as described before [24]. In brief, fibroblasts were detached from cell culture plate and washed with PBS. 4 × 105 cells were resuspended in 400 μl Gene Pulser® Electroporation Buffer Reagent (BioRad) with 23.4 μg of each episomal plasmid (Addgene, Plasdmid #27078, #27080, #27076), containing the reprogramming genes OCT4, SOX2, KLF4, and C-MYC. Cell solution was carefully mixed and electroporated with three pulses of 1.6 kV, capacitance of 3 μF and a resistance of 400 Ω (Gene Pulser II (BioRad)). Fibroblasts were left for recovery in Fibroblast medium without antibiotics, containing 10 μM Rock inhibitor (Y-27632). After cells reached a confluence of 60–70%, medium was changed to TeSR™-E7™ (STEMCELL). Colonies appeared after 21–28 days. These were picked manually and maintained in TeSR™-E8™ (STEMCELL). iPSC lines were characterized for pluripotency [24]. Six iPSC lines derived from one individual were selected and used in the present study, three iCTR clones and three RTT Ex3-4 clones.
Differentiation of neuronal stem cells
The 6 iPSC lines were differentiated towards neuronal stem cells as described before [21, 25]. As described in the paper by Shi et al., this protocol of cortical neurogenesis follows the same temporal order as occurs in vivo. iPSCs were plated in high density on Geltrex®-coated wells of a 12-well plate in TeSR™-E8™ with 10 μM Rock inhibitor. Medium was changed daily for 2 days. Afterwards, half of the medium was changed daily with Neuro-Maintenance-Medium (NMM) (1:1 DMEM/F12+GlutaMAX:Neurobasal Medium, 1x B27, 1xN2, 2.5 μg/ml insulin, 1.5 mM L-glutamin, 100 μM NEAA, 50 μM 2-mercaptoethanol, 1% penicillin/streptomycin) containing 1 μM dorsomorphin and 10 μM SB431542 up to day 12. At days 10–12, rosette structures appeared, which were manually picked and further cultured on Poly-L-Ornithin (0.01%)/laminin (20 μg/ml) coated cell culture plates in NMM medium containing EGF (20 ng/ml) and FGF-2 (20 ng/ml). Half of medium was changed daily and cells were cultured up to day 22.
Immunocytochemistry
To perform immunocytochemistry, cells were fixated with 4% paraformaldehyde and blocked with blocking buffer containing 5% normal goat serum (Gibco®), 0.1% bovine serum albumin (SigmaAldrich) and 0.3% Triton X-100 (SigmaAldrich). Primary antibody incubation for MeCP2 (D4F3, CellSignaling, 1:200, rabbit), OCT3/4 (C-10, Santa Cruz, 1:1000, mouse), SSEA4 (Developmental Studies Hybridoma Bank, 1:50, mouse), TRA1-60 (Santa Cruz, 1:200, mouse), TRA1-81 (Millipore, 1:250, mouse) and SOX2 (Millipore, 1:1000, rabbit) was performed in blocking buffer over night at 4 °C. Next day, cells were washed, and secondary antibody Alexa Fluor® 488 (ThermoFisher, 1:1000, mouse or rabbit) and Alexa Fluor® 594 (ThermoFisher, 1:1000, mouse or rabbit) were applied in blocking buffer for 1 h at room temperature. To identify cell nuclei, DAPI was used for 5 min before cells were mounted with Fluoromount™ (Sigma-Aldrich).
RNA collection, sequencing, and PCR analysis
To isolate RNA samples, standard TRIzol®-Chloroform isolation was done. RNA was stored at – 80 °C until further processing. For PCR analysis, RT-PCR was performed. cDNA was synthesized by using SuperScriptIV-Kit (ThermoFisher) following manufacturer’s recommendations and could be stored until further processing at – 20 °C. To perform PCR, different primer sets were used (Table 1) and PCR was executed with Phire Hot Start II DNA Polymerase (ThermoFisher).
Table 1.
Primers used for iPSC characterization
| OCT3/4 | Fwd: | GAC AGG GGG AGG GGA GGA GCT AGG |
| Rev: | CTT CCC TCC AAC CAG TTG CCC CAA AC | |
| SOX2 | Fwd: | GGG AAA TGG GAG GGG TGC AAA AGA GG |
| Rev: | TTG CGT GAG TGT GGA TGG GAT TGG TG | |
| NANOG | Fwd: | CAG CCC CGA TTC TTC CAC CAG TCC C |
| Rev: | CGG AAG ATT CCC AGT CGG GTT CAC C | |
| C-MYC | Fwd: | GCG TCC TGG GAA GGG AGA TCC GGA GC |
| Rev: | TTG AGG GGC ATC GTC GCG GGA GGC TG | |
| TDGF1 | Fwd: | TGC TGC TCA CAG GGC CCG ATA CTT C |
| Rev: | TCC TTT CGA GCT CAG TGC ACC ACA AAA C | |
| UTF1 | Fwd: | CAG ATC CTA AAC AGC TCG CAG AAT |
| Rev: | GCG TAC GCA AAT TAA AGT CCA GA | |
| DNMT3B | Fwd: | CAG GAG ACC TAC CCT CCA CA |
| Rev: | TGT CTG AAT TCC CGT TCT CC | |
| MECP2 (Set 1) | Fwd: | GGA GAA AAG TCC TGG AAG C |
| Rev: | CTT CAC GGC TTT CTT TTT GG | |
| MECP2 (Set 2) | Fwd: | CACGGAAGCTTAAGCAAAGG |
| Rev: | CTGGAGCTTTGGGAGATTTG | |
| EIF4G2 | Rev: | CTT CAC GGC TTT CTT TTT GG |
| Rev: | AGT TGT TTG CTG CGG AGT TGT CAT CTC GTC |
Western blotting
Frozen cell pellets were lysed by adding WB-Lysate buffer (50 mM Hepes ph 7.5, 150 mM NaCl, 1 mM EDTA, 2.5 mM EGTA, 0.1% TritonX-100, 10% glycerol, 1 mM DTT). To determine protein concentration, Bradford-Test was performed, and 30 μg of sample was used. For SDS-PAGE, pre-casted gels were used (Biorad) and ran in 10× Tris/glycine buffer for Western Blots and Native Gels (Biorad #1610734). Gels were blotted in tank-blotter (Biorad) on PVDF membranes (Biorad) according to manufactures protocol. After protein transfer, blots were blocked in 5% BSA/TBS for 1 h and stained for SOX2 (1:100, Millipore AB5603), SOX9 (1:250; cell signaling 82630) and ß-actin (1:1000; Chemicon, C4 MAB 1501) in 5% BSA/TBS over night at 4 °C. Next day, blots were washed and stained with secondary antibodies in 5% BSA/TBS for 1 h at room temperature. After another 3 TBS washes, blots were stained with SuperSignal™ West Femto Maximum Sensitivity Substrate (ThermoFisher) and analyzed with LiCor analyser.
Sample collection
Samples were collected at different days throughout the differentiation. First samples were taken at day 3 (D3) of protocol, 1 day after medium change towards NMM with dorsomorphin and SB431542. Second samples were taken at day 9 (D9), before rosette structures were cut, reminiscent to the early stage of secondary neurulation [26] followed by third sample collection at day 15 (D15), after rosettes were manually picked, comparable to complete neural tube formation state. Finally, fourth samples were taken at day 22 (D22), after first passage was performed, and cells were recovered. To collect, cells were washed once with PBS and then scraped off the cell culture plate. Solution was collected in an Eppendorf Microtube and centrifuged at maximum speed for 5 min. Supernatant was discarded, and pellet was frozen at – 80 °C until further processing for mass spectrometry.
Cell lysis and protein digestion
Samples were lysed, reduced, and alkylated in lysis buffer (1% sodium deoxycholate (SDC), 10 mM tris(2-carboxyethyl)phosphine hydrochloride (TCEP), 40 mM chloroacetamide (CAA), and 100 mM TRIS, pH 8.0 supplemented with phosphatase inhibitor (PhosSTOP, Roche)) and protease inhibitor (Complete mini EDTA-free, Roche). After sonication, samples were centrifugated at 20,000×g for 20 min. Protein concentration was estimated by a BCA protein assay. Reduction was done with 5 mM ammonium bicarbonate and dithiothreitol (DTT) at 55 °C for 30 min followed by alkylation with 10 mM iodoacetamide for 30 min in dark. Proteins were then digested into peptides by LysC (protein-enzyme ratio 1:50) at 37 °C for 4 h and trypsin (protein-enzyme ratio 1:50) at 37 °C for 16 h. Peptides were then desalted using C18 solid phase extraction cartridges (Waters).
Tandem mass tag (TMT) 10 plex labeling
Aliquots of ~ 100 μg of each sample were chemically labeled with TMT reagents (Thermo Fisher) according to Fig. 1. In total, three TMT mixtures were created for each biological replicate. Peptides were resuspended in 80 μl resuspension buffer containing 50 mM HEPES buffer and 12.5% acetonitrile (ACN, pH 8.5). TMT reagents (0.8 mg) were dissolved in 80 μl anhydrous ACN of which 20 μl was added to the peptides. Following incubation at room temperature for 1 h, the reaction was then quenched using 5% hydroxylamine in HEPES buffer for 15 min at room temperature. The TMT-labeled samples were pooled at 1:1 ratios followed by vacuum centrifuge to near dryness and desalting using Sep-Pak C18 cartridges.
Off-line basic pH fractionation
Before the mass spectrometry analysis, the TMT mixture was fractionated and pooled using basic pH Reverse Phase HPLC. Samples were solubilized in buffer A (5% ACN, 10 mM ammonium bicarbonate, pH 8.0) and subjected to a 50 min linear gradient from 18 to 45% ACN in 10 mM ammonium bicarbonate pH 8 at flow rate of 0.8 ml/min. We used an Agilent 1100 pump equipped with a degasser and a photodiode array (PDA) detector and Agilent 300 Extend C18 column (5 μm particles, 4.6 mm i.d., and 20 cm in length). The peptide mixture was fractionated into 54 fractions and consolidated into 20. Samples were acidified with 10% formic acid and vacuum-dried followed by re-dissolving in 5% formic acid/5% ACN for LC-MS/MS processing.
Mass spectrometry analysis
We used nanoflow LC-MS/MS using Orbitrap Lumos (Thermo Fisher Scientific) coupled to an Agilent 1290 HPLC system (Agilent Technologies). Trap column of 20 mm × 100 μm inner diameter (ReproSil C18, Dr Maisch GmbH, Ammerbuch, Germany) was used followed by a 40 cm × 50 μm inner diameter analytical column (ReproSil Pur C18-AQ (Dr Maisch GmbH, Ammerbuch, Germany)). Both columns were packed in-house. Trapping was done at 5 μl/min in 0.1 M acetic acid in H2O for 10 min, and the analytical separation was done at 300 nl/min for 2 h by increasing the concentration of 0.1 M acetic acid in 80% acetonitrile (v/v). The mass spectrometer was operated in a data-dependent mode, automatically switching between MS and MS/MS. Full-scan MS spectra were acquired in the Orbitrap from m/z 350–1500 with a resolution of 60,000 FHMW, automatic gain control (AGC) target of 200,000 and maximum injection time of 50 ms. Ten most intense precursors at a threshold above 5000 were selected with an isolation window of 1.2 Da after accumulation to a target value of 30,000 (maximum injection time was 115 ms). Fragmentation was carried out using higher-energy collisional dissociation (HCD) with collision energy of 38% and activation time of 0.1 ms. Fragment ion analysis was performed on Orbitrap with resolution of 60,000 FHMW and a low mass cut-off setting of 120 m/z.
Data processing
Mass spectra were processed using Proteome Discover (version 2.1, Thermo Scientific). Peak list was searched using the Swissprot database (version 2014_08) with the search engine Sequest HT. The following parameters were used. Trypsin was specified as enzyme, and up to two missed cleavages were allowed. Taxonomy was set for Homo sapiens, and precursor mass tolerance was set to 50 ppm with 0.05 Da fragment ion tolerance. TMT tags on lysine residues and peptide N termini and oxidation of methionine residues were set as dynamic modifications, and carbamidomethylation on cysteine residues was set as static modification. For the reporter ion quantification, integration tolerance was set to 20 ppm with the most confident centroid method. Results were filtered to a false discovery rate (FDR) below 1%. Finally, peptides lower than 6 amino-acid residues were discarded. Within each TMT experiment, reporter intensity values were normalized by summing the values across all peptides in each channel and then corrected for each channel by having the same summed value. After that, the normalized S/N values were summed for all peptides. Finally, proteins were log2 transformed and normalized by median subtraction. Proteins were included that are identified in 2 out of 3 clones.
Data visualization
The software Perseus was used for data analysis and to generate the plots. Volcano plots for each time point were generated, and up- or downregulated proteins were considered significant with a fold change cut-off = 1.3. Functional analysis to enrich to GO terms was done on David Database, and pathway enrichment analysis was done on Reactome Functional Interaction (http://www.reactome.org/). Furthermore, protein interaction network was performed using Cytoscape, Gnenmania plugin.
Statistics
Volcano plots were generated for each time point and because of the tight ratios typically observed in TMT quantification [27], we selected a cut-off for proteins being up- or downregulated with a p value ≤ 0.1 and ≥ 1.3-fold change difference in RTT compared to iCTR. These cutoffs are based on the observed distribution in the volcano plot and are affected by the ratio compression observed in isobaric quantification strategies, caused by impure MS1 precursor isolation [28]. To study protein subsets consistently dysregulated at all time points, we grouped all RTT at all time points together and all iCTR at all time points together. A volcano plot was generated, and proteins were considered significantly up- or downregulated with a p value ≤ 0.1 and ≥ 1.3-fold change difference in RTT compared to iCTR. To measure protein abundance changes in SOX2 and SOX9 by western blot analysis, we calculated the relative intensity by normalizing the values to the loading control ACTIN using the ImageJ software. Considering normal distribution, the mean values of the three RTT samples and three iCTR samples were compared by using a standard unpaired t test and considered significantly different when p < 0.05.
Results
Generation of iPSC-derived neuronal progenitors from RTT and isogenic controls
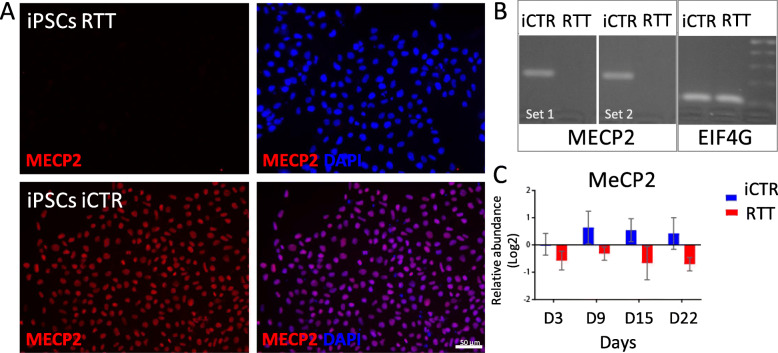
RTT patient and iCTR fibroblasts were reprogrammed into iPSCs via electroporation of reprogramming plasmids [24]. Pluripotency was confirmed using classic assays, including immunocytochemistry (Additional file 1: Figure S1a) and RNA expression (Additional file 1: Figure S1b). Expression of mutated and healthy MeCP2 in the RTT and iCRT lines was confirmed by immunocytochemistry and PCR for MECP2 (Fig. 2a, b). Additionally, the protein expression level of MeCP2 detected by mass spectrometry showed a higher expression of MeCP2 in iCTR relative to RTT samples (Fig. 2c, Additional file 1: Figure S1c). A detectable low expression of mutant alleles, here MeCP2 expression in mutant RTT lines, could be caused by the so-called co-isolation issue in TMT experiments [27]. Generation of NES cells was performed as described before [21]. Neuronal induction into neuronal rosette structures was monitored by visual inspection and appeared after approximately 12 days of neuronal development initiation (Fig. 1). Time points for sample collection were chosen along the differentiation towards neuronal progenitor cells.
Fig. 2.
iCTR and RTT cell line validation. a Representative immunohistochemical staining for MeCP2 in RTT iPSC line (upper) and iCTR iPSC line (lower). b PCR results of iCTR and RTT iPSC lines for two different primer sets spanning deletion Del_Ex3-4 and Exon2 as positive control. c Relative abundance level of MeCP2 in RTT NES and iCTR NES lines at different time points of sample collection
MS-based quantitative proteomics during neuronal development
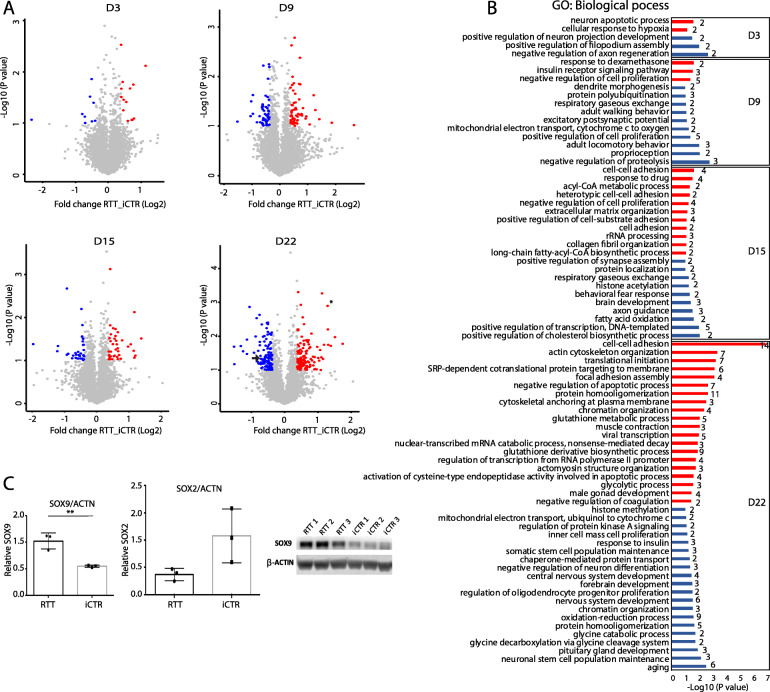
To study proteomic changes between RTT and iCTR during neuronal development, cell lysates at indicated time points were subjected to tryptic digestion, high-pH fractionation followed by high-resolution tandem mass spectrometry (LC-MS/MS) analysis and TMT-10plex quantification (Fig. 1). In total, we identified up to 7702 proteins, of which 3658 proteins were identified in all samples (Additional file 1: Figure S2, Table S1). Next, to determine protein expression changes over the time points, we compared RTT versus iCTR and considered proteins with a p value ≤ 0.1 and a fold change ≥ 1.3 in 2 out of 3 clones as significantly regulated (Fig. 3a, Table S2). This resulted in 23 up- or downregulated proteins at D3, 111 at D9, 72 at D15, and 243 at D22, between RTT and iCTR. We then compared the overlap of these regulated proteins across the different time points, which we presented in a Venn diagram (Additional file 1: Figure S3). We noticed that the majority of the regulated proteins only have 0–2% overlap between the different time points. To better understand what biological processes are involved at each time point, we performed Gene Ontology (GO) enrichment analysis with respect to biological functions (Fig. 3b). GO analysis on the significantly upregulated proteins in RTT versus iCTR revealed the GO terms neuron apoptotic process and cellular response to hypoxia at D3. GO terms related to downregulated proteins in RTT at D3 involved negative regulation of axon regeneration, positive regulation of filopodium assembly, and positive regulation of neuron projection development. At D9, insulin receptor signaling pathway was upregulated whereas excitatory postsynaptic potential was downregulated in RTT. At D15, cell-cell adhesion and acyl-CoA metabolic process were upregulated, and terms such as axon guidance, brain development, and histone acetylation were downregulated. Furthermore, at D22, GO terms related to cell-cell adhesion and actin cytoskeleton organization were upregulated, and nervous system development and forebrain development were downregulated in RTT. Terms such as brain development were downregulated in D15 as well as D22. Gene set enrichment analysis (GSEA) of all up- and downregulated proteins further revealed that RTT-associated proteins were strongly enriched in gene sets such as apoptosis, DNA repair, and in metabolism (Additional file 1: Figure S4, Table S3). To further verify the results of the mass spectrometry, we performed western blot analysis for proteins SOX2 and SOX9, transcription factors with pivotal role in development and differentiation [29, 30], which showed significant differences in expression levels between iCTR and RTT lines at D22 (Fig. 3a, c). In line with mass spectrometry data, western blot analysis showed a significant increase in SOX9 expression levels in RTT lines when compared to iCTR (p = 0.0057, unpaired t test), and a decrease in SOX2 expression in RTT lines at D22, although this did not reach statistical significance (p = 0.07, unpaired t test). Together, both approaches demonstrate that SOX2 and SOX9 were differentially expressed between RTT and iCTR, thereby validating our findings that RTT samples show proteome changes at early neurodevelopmental changes
Fig. 3.
Volcano plots and GO analysis at different time points. a Volcano plot demonstrating proteins differentially regulated in RTT compared to iCTR at each time point of neuronal development. Each data point represents a single quantified protein. The x-axis represents the log2-fold change in abundance (RTT/iCTR) and y-axis the − log10 (p value). Threshold for significant proteins is chosen for p value cut-off (0.1) and fold change ≥ 1.3. Proteins in blue indicate for downregulation and in red indicate for upregulation in RTT. Arrow in day 22 points for SOX2 expression, and asterisk points for SOX9 expression. b Gene Ontology analysis of the significant proteins on their biological function at each time point of neuronal differentiation. Red indicates the upregulated and blue the downregulated biological processes. The numbers indicate the genes enriched for each term. c Western blot analysis showing SOX9 and SOX2 expression in iCTR NES lines at D22. Significant increase in SOX9 expression in RTT samples, and SOX2 shows a trend towards decrease in RTT samples
A previous study identified perturbed astrocyte differentiation of RTT-iPSCs, suggesting skewed differentiation of neural progenitor cells into neuronal cell lineage [31]. Here, we studied whether we could find similar changes, i.e., increased neuronal marker MAP2 and decreased glia marker (e.g., ATP1A2, CLU, and SLC1A3) expression. While these astrocyte and neuronal markers are expressed in our samples, we found no significant differences between iCTR and RTT samples (Additional file 1: Figure S3). Furthermore, while the authors showed higher expression of LIN28 in RTT samples, we identified two isoforms (LIN28A and B) with no differences between RTT and iCTR samples. In order to evaluate whether the altered proteomes are present in the human brain, we compared our proteomic altered data to the Allen Brain atlas. This is showing individual gene expression in the different brain areas (Supplementary Table S4). This revealed high variability of expression between transcriptomics and proteomics. The majority of the proteins have measurable expression levels in human brain in vivo. Overall, we show that proteins associated with neuronal development are differentially expressed in RTT at early stages of neuronal differentiation.
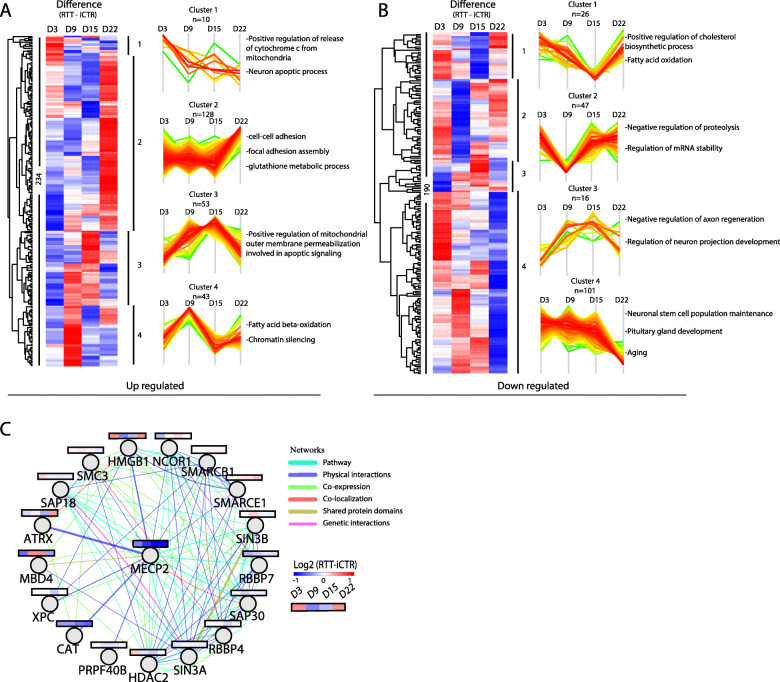
Coordinated proteome alteration during neuronal development in RTT syndrome
To gain insight into how the differentially expressed proteins in RTT behave across time points, we further analyzed all the significantly up- or downregulated proteins at D3, D9, D15, and D22. This resulted in 234 significantly up and 190 downregulated proteins. The average log2 values of RTT were extracted with iCTR for each time point, and the difference between RTT and iCTR is shown in a heat map (Fig. 4a). To obtain an unbiased view of the differentially expressed proteins during neuronal differentiation, we performed cluster analysis on the significantly up- or downregulated proteins. This resulted in four clusters for both the up- or downregulated proteins with distinct expression profiles. Cluster 1 contains proteins strongly upregulated in D3 that are involved in neuron apoptotic processes and cytochrome c release from mitochondria-related GO terms. Cluster 2 represents upregulated proteins at D22 that are involved in adhesion assembly and glutathione metabolic processes. Cluster 3 contains proteins upregulated at D9 and D15 having a role in apoptotic signaling, and cluster 4 represents proteins strongly upregulated at D9 having a role in oxidation and chromatin silencing. In the downregulated proteins, cluster 1 represents proteins that showed a strong downregulation at D15 mainly involved in cholesterol biosynthesis and fatty acid oxidation (Fig. 4b). Cluster 2 represents proteins strongly downregulated at D9, which are associated with regulation of proteolysis and mRNA stability. Cluster 3 of the downregulated proteins in RTT shows a decrease expression profile in D3 which are involved in axon regeneration and neuron projection development, and cluster 4 covers many proteins strongly downregulated in D22 involved in processes such as neuronal stem cell maintenance, pituitary gland development, and aging. Collectively, our data reveals a changing, stage-specific pattern of the differentially expressed proteins during neuronal development in RTT.
Fig. 4.
Up- and downregulated proteins and MeCP2-binding partners. a Heat map of all significant up- and b downregulated proteins with a p value ≤ 0.1 and ≥ 1.3-fold change between RTT and iCTR. The Z score of the difference between RTT and iCTR is given for each day with the corresponding cluster analysis and the GO terms for biological processes. c Network analysis of MeCP2-binding proteins identified in our data. The average log2 ratio RTT-iCTR over time is color coded for each protein over the time course of neuronal development. Edges are color coded according to the network as indicated
MeCP2 network analysis
To further investigate the proteins that are targets by the MeCP2 protein, we drew a protein interaction network (Cytoscape, Genemania plugin) using MeCP2 protein as input (Fig. 4c). The data covered 20 MeCP2-interacting proteins of which 18 proteins were identified in our data. To further investigate how the MeCP2- interacting proteins change over time in RTT, we extracted the average log2 values of iCTR by RTT samples to represents the difference at each time point. As expected, MeCP2 is downregulated along the course of neuronal differentiation in RTT samples compared to iCTR. The proteins are tightly interconnected around HDAC2, SIN3A, RBBP4, SAP30, SMARCE1, SMARCB1, and MeCP2 but to a lesser extend around CAT, XPC, PRPF40B, and MBD4. The network revealed several RNA/DNA binding proteins of which MeCP2 and CAT are one of the most downregulated proteins in RTT. While some proteins in the network, such as SMARCB1 and SIN3A, stayed constant over time, others showed changing levels, such as MBD4 and HMGB1. Interestingly, MBD4, next to MeCP2, is a member of the methyl-CpG-binding domain (MBD) family. Overall, we searched for MeCP2-binding partners and showed how these proteins change along the course of neuronal differentiation in RTT and iCTR.
Protein subsets consistently dysregulated at all time points
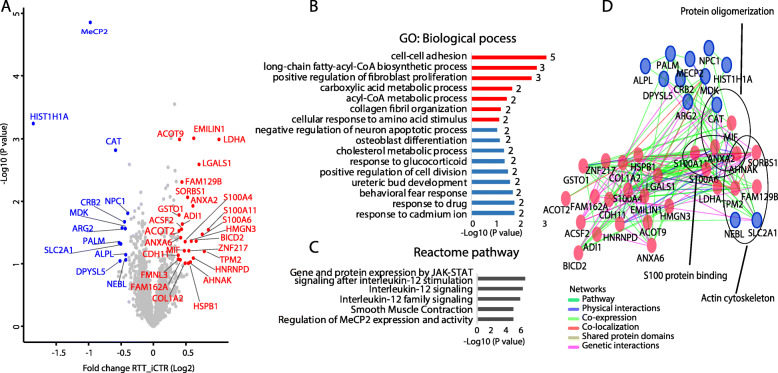
To study proteins differentially expressed between RTT and iCTR regardless the time point of differentiation, we grouped all RTT and all iCTR samples from all time points together. Due to the tight ratios typically observed in TMT quantification [27], we selected a cut-off for proteins being up- or downregulated with a p value ≤ 0.1 and ≥ 1.3-fold change difference in RTT compared to iCTR based on the observed distribution in the volcano plot (Fig. 5a). We identified 27 proteins being up and 12 proteins being downregulated in RTT compared to iCTR. As expected, MeCP2 was one of the most strongly downregulated proteins in RTT. GO analysis revealed biological processes such as cell-cell adhesion and acyl-CoA metabolic processes to be upregulated (Fig. 5b), which are also upregulated at individual time points D15 and D22 (Fig. 4a). In contrast, several processes such as response to cadmium ion, response to drug, and behavioral fear response were downregulated in RTT (Fig. 5b). Analysis of the differentially regulated proteins using Reactome pathway analysis revealed among others, JAK/STAT signaling after Interleukin-12 stimulation and regulation of MeCP2 expression and activity to be differentially expressed in RTT versus iCTR (Fig. 5c). To further visualize the connectivity among these significant proteins, we analyzed their protein networks in the Cytoscape tool (Genemania plugin). A high degree of connectivity, such as being co-expressed and having shared genetic interactions, around these proteins was identified. Interestingly, the majority of the proteins are involved in immunity, actin cytoskeleton organization, and calcium binding (Fig. 5d). Together, we show that proteins associated with immunity and metabolic processes are differentially expressed in RTT in a time-point independent manner during differentiation towards neuronal progenitors.
Fig. 5.
Volcano plot all days and network analysis. a Volcano plot demonstrating proteins differentially expressed in RTT versus iCTR after pooling all time points of neuronal development. The x-axis represents the log2-fold change in abundance (RTT/iCTR) and y-axis the − log10 (p value). Threshold for significant proteins is chosen for p value cutt-off (0.1) and fold change ≤ 1.3. Upregulated proteins in RTT are shown in red, and downregulated are shown in blue. b GO analysis on the biological process of the significant proteins. X-axis represents the − log10 (p value), and red and blue colors indicate for up- and downregulated proteins in RTT, respectively. The numbers represent the genes enriched for each term. c Reactome pathway analysis of the significant proteins. d Network analysis of the significant proteins by Cytoscape plugin GeneMania. Red indicates for upregulated, and blue indicates for downregulated proteins in RTT
Discussion
To gain insight into mechanisms involved in early neurodevelopmental changes in RTT patients with MeCP2 deficiency, we performed mass spectrometry-based quantitative proteomic analysis on samples derived from RTT patient and isogenic control iPSCs at days 3, 9, 15, and 22 of neuronal induction. We identified altered proteins associated to multiple pathways, of which cell-cell adhesion and actin cytoskeleton organization show most significant upregulations, and pituitary gland development and neuronal stem cell population maintenance show most significant downregulations at D22. At D3, D9, and D15, other and less dysregulated pathways were found, including neuron apoptotic process and negative regulation of axon regeneration (D3), response to dexamethasone and negative regulation of proteolysis (D9), and cell-cell adhesion and positive regulation of cholesterol biosynthetic process (D15) (Fig. 3). The number of altered proteins, as well as an increase in the fold change of protein alteration in RTT samples, gradually increased from day 3 to day 22. However, at all time points, altered pathways associated to neurodevelopment or neurogenesis with at least a 1.3-fold up- or downregulation, could be identified in RTT samples. This report shows that exons 3–4 deletion in MECP2 results in protein expression changes at neuronal progenitor stages and provides a resource of proteins and pathways for further exploration to identify common disease mechanisms underlying RTT and related phenotypes.
RTT samples present protein expression changes that become more apparent from early to late neuronal progenitor stages
The altered proteins in the RTT cultures gradually increased from D3 to D22 samples. As RTT patients start to show disease-related clinical symptoms at 6–18 months of age, it is likely molecular changes already start at prenatal stages and progress over time [8]. Based on mouse studies, expression of MeCP2 is already present during early embryogenesis [32] and gradually increases at later stages, suggesting its progressive importance in brain development [14, 33]. In line with this, it is expected that lack of MeCP2 will increasingly affect gene regulation and so the number of altered proteins, which converge and lead to clinical manifestation. While not confirmed on protein levels, transcriptional alterations in RTT samples during early neuronal developmental stages have been shown [34, 35]. Changes in the transcriptome were already observed at the pluripotent stem cell stage when iPSCs of RTT patients were compared to human embryonic stem cells [36]. Here, we identified alterations on protein levels occurring only 3 days after neuronal induction of pluripotent stem cells. This time point is comparable to a brain developmental stage of 3 months of gestational age [37]. So, our study supports the hypothesis of an early post gestational onset of molecular changes, leading to clinical phenotype at early postnatal stages.
Protein expression changes in neuronal progenitor cells from RTT patients
The most significant upregulated proteins are associated with GO terms cell-cell adhesion and actin cytoskeleton organization. In previous studies, dysregulation of the actin cytoskeletal organization was observed on transcriptional level and associated with phenotypic alterations as abnormal dendrite formation and dendritic growth [38–41]. However, these studies reported a transcriptional downregulation of the mentioned pathways, which is in contrast to our results. Due to the poor correlation of transcriptome and proteome [42] as well as the early developmental time points in our study, different effects on the named pathways can be assumed. As specifically actin play a very dynamic role in neurodevelopment, it is important to compare samples with regard to their developmental stage [43, 44]. On the other hand, the downregulation of proteins in pathway neuronal stem cell population maintenance we found was observed in mouse cells before, showing a reduced proliferation rate of neuronal stem cells as well as alterations in neuronal development [45]. Also, we found downregulated pathways indicative of affected nervous system development and forebrain development, which is associated with the typical neuronal phenotype in RTT. Reduced dendritic outgrowth and decreased spine density has been shown in RTT mouse models [46–49] as well as neuronal morphologies typical for juvenile brains in RTT postmortem tissue [50–52]. Interestingly, already at day 3 and day 9 of neuronal induction we identified downregulation of the pathways dendrite morphogenesis and axon guidance, which are associated with specific morphological alterations observed in postmortem tissue from RTT patients [47, 50, 51]. The second most downregulated pathway in our study pituitary gland development was also associated with RTT before [53]. In patients with RTT syndrome, pathologist identified a reduced size of the pituitary gland [53] and dysregulation of thyroid hormone release [54]. Interestingly, one of the significantly upregulated protein at D22 was AUTS2 (Supplementary Table S2), which was found before in a MeCP2-deficient mouse model at postnatal day 60 on transcriptomic and proteomic level. This is an autism susceptibility gene implicated in various neurological disorders such as autism spectrum disorder, which has similar disease symptoms as RTT syndrome. Interestingly, all these alterations found at early neurodevelopmental stages are associated to RTT phenotypes observed at later stages.
Worth mentioning are proteins, which levels, are also dysregulated in other neurodevelopmental disorders, which patients show an overlap in neurological dysfunctions of RTT patients. We found changed levels in DOCK6 (at D3) [55, 56], NPC1 (at D9) [57, 58], CDK5 (at D9) [59, 60], HINT1 (at D9) [61, 62], CSTB (at D9) [63], ACSL4 (at D22) [64, 65], KIF5C (at D22) [66, 67], and TUBB2A (at D22) [68], which are respectively dysregulated in epilepsy, Niemann-Pick disease, mental retardation, cortical dysplasia, lissencephaly, neurotonia, and axonal neuropathy. Even though these candidate proteins were not associated with RTT before, these could be key in affected neuronal development in RTT and other neurological disorders, and therefore could be of special interest in identifying new disease mechanisms. Overall, our findings suggest that dysregulation of MeCP2 affects protein expression associated with neurodevelopmental functions at early stages of neuronal differentiation. Reported protein changes provide starting points for further research, especially those that have been identified in other neurodevelopmental disorders that present similar phenotypes.
Expression changes of MeCP2-interacting proteins in RTT and iCTR
Network analysis using GeneMANIA [69] revealed part of these proteins to be interacting with MeCP2. Interestingly, of these interacting proteins, MBD4 is a member of the MBD family of proteins together with MeCP2. Mutations in these functional important domains tend to cause RTT-associated phenotypes [70–72]. Furthermore, HMGB1, which was downregulated in D9 and D15 in our data, was previously shown to be lower expressed in hippocampal granule neurons of Mecp2 KO mice [7].
Protein profile changes consistently dysregulated at all time points of neuronal differentiation
When all RTT versus iCTR samples from different time points were pooled, data revealed differentially expressed proteins in RTT involved in immunity, calcium binding, and metabolism. This analysis allowed us to compensate for limited amount of biological replicates in such high-throughput technology. Several proteins associated with metabolic processes were differentially expressed in RTT, including ACSF2, ACOT2, ACOT9, LDHA, MIF, NPC1, CAT, and GSTO1. Recent evidence indicated there is perturbed lipid metabolism in RTT based on findings in brain and peripheral patient tissue, as well as metabolic dysfunctions, based on mouse studies, supporting a role for these pathways in RTT phenotypes [40, 73–76]. A previous study involving brain extracts of mice heterogeneous for MeCP2 demonstrated that disturbances in the metabolism produces changes in the morphological and biochemical development of the brains during early brain formation [74]. Another study further observed that models with synaptic defects during development fail to couple to metabolic pathways by using human independent databases [75]. Also, others earlier reported on upregulated GSTO1 transcript levels in RTT patients’ lymphocytes together with several other mitochondrial-related genes [76]. These observations point towards involvement of the metabolic pathway in affected neuronal development.
Furthermore, our results indicate dysregulated proteins associated with immunological processes as interleuking-12 signaling. Interestingly, several publications have indicated a potential association of the immune system with MeCP2 dysregulation [77–79]. For instance, children with MECP2 duplication show immunological abnormalities and suppressed IFN-ϒ [77]. Others showed that MeCP2 deficient patients, as well as CDKL5-related RTT patients, show dysregulated cytokine release suggestive of defective regulation of inflammatory responses [78]. Further, microglia have been proposed as key cell type in affected neurodevelopment in RTT [79]. Together, the identification of altered interleuking-12 signaling in our study supports involvement of the immune system in RTT phenotypes.
Lastly, our findings suggest changes in the calcium homeostasis in RTT, as several proteins associated with calcium signaling were altered in RTT samples. Disturbances in calcium homeostasis during early postnatal development were reported before in Mecp2 knockout model and were suggested to play a major role in disturbed neuronal signaling in RTT [34, 80]. Based on iPSC research, the authors identified alterations in calcium signaling in RTT neurons indicative of an immature phenotype [40]. Also, others suggested involvement of calcium homeostatis in affected neuronal maturation in RTT [81]. Therefore, our findings confirm calcium signaling involvement in RTT phenotypes that already manifest at neuronal progenitor cell stages.
Altogether, our findings show pathway involvement associated to immunity, calcium signaling, and metabolism, in line with earlier studies, but now confirmed at proteome level and at much earlier developmental stages than indicated before.
Limitations
A limitation of our study is the investigation of cells from only one donor. Although three different iPSC RTT and three isogenic control clones were included, findings should be validated in other RTT patient lines to suggest common disease mechanisms. At present, our in-depth comprehensive approach was only possible with the current number of replicates. A follow-up study with more patients might be more feasible by focusing on a few target proteins.
Conclusion
Already at neuronal progenitor cell stages, RTT patient cells show altered protein expression levels. While both RTT patients and mouse models of RTT already show abnormalities before typical RTT-associated symptoms appear [82, 83], others never reported on neuronal phenotype specific protein changes at such early neurodevelopmental cell stages. Much of our understanding of how MeCP2 deficiency contributes to RTT disease is derived from genomic and transcriptomic studies. So far, only a few proteomic studies have been performed involving RTT human derived tissue [19, 84, 85]. The current study provides mass spectrometry-based quantitative proteomic data, depth of 7702 proteins, using an earlier developed iPSC-based models involving RTT patient and isogenic control cells [24]. We showed most significant changes in pathways associated with cell-cell adhesion and actin cytoskeleton organization as well as neuronal stem cell population maintenance and pituitary gland development and identified changes in protein expression associated with dendrite morphology or synaptic defects, previously associated with RTT [22, 35]. All these alterations were identified only shortly after neuronal induction, much earlier than actual clinical phenotypes appear. Proteins involved in immunity and metabolism, also in line with previous studies on RTT pathology [49, 73], are consistently differentially expressed. Insight into the set of altered signaling pathways or proteins, as found in this study, could support identification of underlying disease mechanisms to guide better understanding of disease phenotypes. Presented results highlight the early pre-natal onset of RTT and deserve further study.
Supplementary information
Additional file 1: Figure S1. iPSC characterisation. a. Exemplary characterisation of iPSC lines. Immunocytochemistry for pluripotency marker (OCT3/4, SOX2, SSEA4, TRA1-60, TRA1-81). b. PCR-analysis for pluripotency marker. c. MeCP2 expression of the three iCTR and RTT samples at each time point.
Additional file 2: Figure S2. Number of proteins identified. A bar chart showing the number of proteins identified in each biological replicate and time point.
Additional file 3: Figure S3. Venn diagram. a. Number of proteins decreased expressed in RTT at different time points. b. Number of proteins increased expressed in RTT at different time points. c. Overview of the number of proteins altered in RTT.
Additional file 4: Figure S4. Top gene sets enriched in RTT-iPSCs. Proteins below p=0.1 are ranked by GSEA based on their differential expression level. Black vertical lines indicate the position where members of a pathway appear in the ranked gene list.
Acknowledgements
Specimens were provided by the Cell lines and DNA bank of Rett Syndrome, X-linked mental retardation and other genetic disease, member of the Telethon Network of Genetic Biobanks (project no. GTB12001), funded by Telethon Italy, and of the EuroBioBank network.
Abbreviations
- iPSC
Induced pluripotent stem cell
- MeCP2
Methyl-CpG binding protein 2
- RTT
Rett syndrome
- iCTR
Isogenic controls
- GO
Gene Ontology
- TMT
Tandem mass tag
Authors’ contributions
S.V.M., L.H., M.A., and V.M.H. contributed to conception and design of the study. S.V.M. processed all mass spectrometry data and analysis. L.H. prepared all samples. D.H. helped with the cell lysis and TMT labeling. D.H., M.A., and V.M.H. contributed to acquisition. M.A. and V.M.H. supervised drafting final manuscript. The author(s) read and approved the final manuscript.
Funding
This work was funded by the EC under FP7-PEOPLE-2013 (607508) and The Netherlands Organization for Scientific Research (NWO VICI 453-14-005). VMH is supported by ZonMw (VIDI 917-12-343).
Availability of data and materials
All mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD013327
Username: reviewer46083@ebi.ac.uk
Password: ABIw2h3I
Ethics approval and consent to participate
All experiments were exempt from approval of Medical Ethical Toetsingscommissie (METC), Institutional Review Board of the VU medical centre.
Consent for publication
Not applicable
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Suzy Varderidou-Minasian, Lisa Hinz, Maarten Altelaar and Vivi M. Heine contributed equally to this work.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s13229-020-00344-3.
References
- 1.Chahrour M, Zoghbi HY. The story of Rett syndrome: from clinic to neurobiology. Neuron. 2007;56(3):422–437. doi: 10.1016/j.neuron.2007.10.001. [DOI] [PubMed] [Google Scholar]
- 2.Neul JL, Kaufmann WE, Glaze DG, Christodoulou J, Clarke AJ, Bahi-Buisson N, et al. Rett syndrome: revised diagnostic criteria and nomenclature. Annals of neurology. 2010;68(6):944–950. doi: 10.1002/ana.22124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Percy AK, Neul JL, Glaze DG, Motil KJ, Skinner SA, Khwaja O, et al. Rett syndrome diagnostic criteria: lessons from the Natural History Study. Annals of neurology. 2010;68(6):951–955. doi: 10.1002/ana.22154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Akbarian S. The neurobiology of Rett syndrome. The Neuroscientist : a review journal bringing neurobiology, neurology and psychiatry. 2003;9(1):57–63. doi: 10.1177/1073858402239591. [DOI] [PubMed] [Google Scholar]
- 5.Marchetto MC, Winner B, Gage FH. Pluripotent stem cells in neurodegenerative and neurodevelopmental diseases. Human molecular genetics. 2010;19(R1):R71–R76. doi: 10.1093/hmg/ddq159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Qiu Z, Sylwestrak EL, Lieberman DN, Zhang Y, Liu XY, Ghosh A. The Rett syndrome protein MeCP2 regulates synaptic scaling. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2012;32(3):989–994. doi: 10.1523/JNEUROSCI.0175-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smrt RD, Eaves-Egenes J, Barkho BZ, Santistevan NJ, Zhao C, Aimone JB, et al. Mecp2 deficiency leads to delayed maturation and altered gene expression in hippocampal neurons. Neurobiol Dis. 2007;27(1):77–89. doi: 10.1016/j.nbd.2007.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Amir RE, Van den Veyver IB, Wan M, Tran CQ, Francke U, Zoghbi HY. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nature genetics. 1999;23(2):185–188. doi: 10.1038/13810. [DOI] [PubMed] [Google Scholar]
- 9.Trappe R, Laccone F, Cobilanschi J, Meins M, Huppke P, Hanefeld F, et al. MECP2 mutations in sporadic cases of Rett syndrome are almost exclusively of paternal origin. American journal of human genetics. 2001;68(5):1093–1101. doi: 10.1086/320109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schanen C, Francke U. A severely affected male born into a Rett syndrome kindred supports X-linked inheritance and allows extension of the exclusion map. American journal of human genetics. 1998;63(1):267–269. doi: 10.1086/301932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lewis JD, Meehan RR, Henzel WJ, Maurer-Fogy I, Jeppesen P, Klein F, et al. Purification, sequence, and cellular localization of a novel chromosomal protein that binds to methylated DNA. Cell. 1992;69(6):905–914. doi: 10.1016/0092-8674(92)90610-o. [DOI] [PubMed] [Google Scholar]
- 12.Nan X, Ng HH, Johnson CA, Laherty CD, Turner BM, Eisenman RN, et al. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature. 1998;393(6683):386–389. doi: 10.1038/30764. [DOI] [PubMed] [Google Scholar]
- 13.Chahrour M., Jung S. Y., Shaw C., Zhou X., Wong S. T. C., Qin J., Zoghbi H. Y. MeCP2, a Key Contributor to Neurological Disease, Activates and Represses Transcription. Science. 2008;320(5880):1224–1229. doi: 10.1126/science.1153252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gabel HW, Kinde B, Stroud H, Gilbert CS, Harmin DA, Kastan NR, et al. Disruption of DNA-methylation-dependent long gene repression in Rett syndrome. Nature. 2015;522(7554):89–93. doi: 10.1038/nature14319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rodrigues DC, Kim DS, Yang G, Zaslavsky K, Ha KC, Mok RS, et al. MECP2 is post-transcriptionally regulated during human neurodevelopment by combinatorial action of RNA-binding proteins and miRNAs. Cell Rep. 2016;17(3):720–734. doi: 10.1016/j.celrep.2016.09.049. [DOI] [PubMed] [Google Scholar]
- 16.Shovlin S, Tropea D. Transcriptome level analysis in Rett syndrome using human samples from different tissues. Orphanet journal of rare diseases. 2018;13(1):113. doi: 10.1186/s13023-018-0857-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Colak D, Al-Dhalaan H, Nester M, Albakheet A, Al-Younes B, Al-Hassnan Z, et al. Genomic and transcriptomic analyses distinguish classic Rett and Rett-like syndrome and reveals shared altered pathways. Genomics. 2011;97(1):19–28. doi: 10.1016/j.ygeno.2010.09.004. [DOI] [PubMed] [Google Scholar]
- 18.Matarazzo V, Ronnett GV. Temporal and regional differences in the olfactory proteome as a consequence of MeCP2 deficiency. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(20):7763–7768. doi: 10.1073/pnas.0307083101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cortelazzo A, Guerranti R, De Felice C, Signorini C, Leoncini S, Pecorelli A, et al. A plasma proteomic approach in Rett syndrome: classical versus preserved speech variant. Mediators of inflammation. 2013;2013:438653. doi: 10.1155/2013/438653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Altelaar AFM, Munoz J, Heck AJR. Next-generation proteomics: towards an integrative view of proteome dynamics. Nature Reviews Genetics. 2012;14:35. doi: 10.1038/nrg3356. [DOI] [PubMed] [Google Scholar]
- 21.Nadadhur AG, Emperador Melero J, Meijer M, Schut D, Jacobs G, Li KW, et al. Multi-level characterization of balanced inhibitory-excitatory cortical neuron network derived from human pluripotent stem cells. PloS one. 2017;12(6):e0178533. doi: 10.1371/journal.pone.0178533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Andoh-Noda T, Akamatsu W, Miyake K, Matsumoto T, Yamaguchi R, Sanosaka T, et al. Differentiation of multipotent neural stem cells derived from Rett syndrome patients is biased toward the astrocytic lineage. Molecular brain. 2015;8:31. doi: 10.1186/s13041-015-0121-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim KC, Choi CS, Kim JW, Han SH, Cheong JH, Ryu JH, et al. MeCP2 modulates sex differences in the postsynaptic development of the valproate animal model of autism. Molecular neurobiology. 2016;53(1):40–56. doi: 10.1007/s12035-014-8987-z. [DOI] [PubMed] [Google Scholar]
- 24.Hinz L, Hoekstra SD, Watanabe K, Posthuma D, Heine VM. Generation of isogenic controls for in vitro disease modelling of X-chromosomal disorders. Stem Cell Reviews and Reports. 2018. [DOI] [PMC free article] [PubMed]
- 25.Shi Y, Kirwan P, Livesey FJ. Directed differentiation of human pluripotent stem cells to cerebral cortex neurons and neural networks. Nature protocols. 2012;7:1836. doi: 10.1038/nprot.2012.116. [DOI] [PubMed] [Google Scholar]
- 26.Fedorova V, Vanova T, Elrefae L, Pospisil J, Petrasova M, Kolajova V, et al. Differentiation of neural rosettes from human pluripotent stem cells in vitro is sequentially regulated on a molecular level and accomplished by the mechanism reminiscent of secondary neurulation. Stem cell research. 2019;40:101563. doi: 10.1016/j.scr.2019.101563. [DOI] [PubMed] [Google Scholar]
- 27.Altelaar AFM, Frese CK, Preisinger C, Hennrich ML, Schram AW, Timmers HTM, et al. Benchmarking stable isotope labeling based quantitative proteomics. Journal of Proteomics. 2013;88:14–26. doi: 10.1016/j.jprot.2012.10.009. [DOI] [PubMed] [Google Scholar]
- 28.Karp NA, Huber W, Sadowski PG, Charles PD, Hester SV, Lilley KS. Addressing accuracy and precision issues in iTRAQ quantitation. Molecular & cellular proteomics : MCP. 2010;9(9):1885–1897. doi: 10.1074/mcp.M900628-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.De Santa BP, Bonneaud N, Boizet B, Desclozeaux M, Moniot B, Sudbeck P, et al. Direct interaction of SRY-related protein SOX9 and steroidogenic factor 1 regulates transcription of the human anti-Mullerian hormone gene. Molecular and cellular biology. 1998;18(11):6653–6665. doi: 10.1128/mcb.18.11.6653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rizzino A. Sox2 and Oct-3/4: a versatile pair of master regulators that orchestrate the self-renewal and pluripotency of embryonic stem cells. Wiley interdisciplinary reviews Systems biology and medicine. 2009;1(2):228–236. doi: 10.1002/wsbm.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim JJ, Savas JN, Miller MT, Hu X, Carromeu C, Lavallee-Adam M, et al. Proteomic analyses reveal misregulation of LIN28 expression and delayed timing of glial differentiation in human iPS cells with MECP2 loss-of-function. PloS one. 2019;14(2):e0212553. doi: 10.1371/journal.pone.0212553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shahbazian MD, Antalffy B, Armstrong DL, Zoghbi HY. Insight into Rett syndrome: MeCP2 levels display tissue- and cell-specific differences and correlate with neuronal maturation. Human molecular genetics. 2002;11(2):115–124. doi: 10.1093/hmg/11.2.115. [DOI] [PubMed] [Google Scholar]
- 33.Cohen DR, Matarazzo V, Palmer AM, Tu Y, Jeon OH, Pevsner J, et al. Expression of MeCP2 in olfactory receptor neurons is developmentally regulated and occurs before synaptogenesis. Mol Cell Neurosci. 2003;22(4):417–429. doi: 10.1016/s1044-7431(03)00026-5. [DOI] [PubMed] [Google Scholar]
- 34.Marchetto MC, Carromeu C, Acab A, Yu D, Yeo GW, Mu Y, et al. A model for neural development and treatment of Rett syndrome using human induced pluripotent stem cells. Cell. 2010;143(4):527–539. doi: 10.1016/j.cell.2010.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim KY, Hysolli E, Park IH. Neuronal maturation defect in induced pluripotent stem cells from patients with Rett syndrome. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(34):14169–14174. doi: 10.1073/pnas.1018979108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tanaka Y, Kim KY, Zhong M, Pan X, Weissman SM, Park IH. Transcriptional regulation in pluripotent stem cells by methyl CpG-binding protein 2 (MeCP2) Human molecular genetics. 2014;23(4):1045–1055. doi: 10.1093/hmg/ddt500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Keeney JG, Davis JM, Siegenthaler J, Post MD, Nielsen BS, Hopkins WD, et al. DUF1220 protein domains drive proliferation in human neural stem cells and are associated with increased cortical volume in anthropoid primates. Brain Struct Funct. 2015;220(5):3053–3060. doi: 10.1007/s00429-014-0814-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bhattacherjee A, Mu Y, Winter MK, Knapp JR, Eggimann LS, Gunewardena SS, et al. Neuronal cytoskeletal gene dysregulation and mechanical hypersensitivity in a rat model of Rett syndrome. Proceedings of the National Academy of Sciences of the United States of America. 2017;114(33):E6952–E6e61. doi: 10.1073/pnas.1618210114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Griesi-Oliveira K, Suzuki AM, Alves AY, Mafra A, Yamamoto GL, Ezquina S, et al. Actin cytoskeleton dynamics in stem cells from autistic individuals. Scientific reports. 2018;8(1):11138. doi: 10.1038/s41598-018-29309-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pacheco NL, Heaven MR, Holt LM, Crossman DK, Boggio KJ, Shaffer SA, et al. RNA sequencing and proteomics approaches reveal novel deficits in the cortex of Mecp2-deficient mice, a model for Rett syndrome. Molecular autism. 2017;8:56. doi: 10.1186/s13229-017-0174-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ehrhart F, Coort SL, Eijssen L, Cirillo E, Smeets EE, Bahram Sangani N, et al. Integrated analysis of human transcriptome data for Rett syndrome finds a network of involved genes. The World Journal of Biological Psychiatry. 2019:1–14. [DOI] [PubMed]
- 42.Hack CJ. Integrated transcriptome and proteome data: the challenges ahead. Brief Funct Genomic Proteomic. 2004;3(3):212–219. doi: 10.1093/bfgp/3.3.212. [DOI] [PubMed] [Google Scholar]
- 43.Blue ME, Kaufmann WE, Bressler J, Eyring C, O'Driscoll C, Naidu S, et al. Temporal and regional alterations in NMDA receptor expression in Mecp2-null mice. Anat Rec (Hoboken). 2011;294(10):1624–1634. doi: 10.1002/ar.21380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pacary E, Azzarelli R, Guillemot F. Rnd3 coordinates early steps of cortical neurogenesis through actin-dependent and -independent mechanisms. Nat Commun. 2013;4:1635. doi: 10.1038/ncomms2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Alessio N, Riccitiello F, Squillaro T, Capasso S, Del Gaudio S, Di Bernardo G, et al. Neural stem cells from a mouse model of Rett syndrome are prone to senescence, show reduced capacity to cope with genotoxic stress, and are impaired in the differentiation process. Exp Mol Med. 2018;50(3):1. doi: 10.1038/s12276-017-0005-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee LJ, Tsytsarev V, Erzurumlu RS. Structural and functional differences in the barrel cortex of Mecp2 null mice. The Journal of comparative neurology. 2017;525(18):3951–3961. doi: 10.1002/cne.24315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Armstrong DD. Neuropathology of Rett syndrome. Mental retardation and developmental disabilities research reviews. 2002;8(2):72–76. doi: 10.1002/mrdd.10027. [DOI] [PubMed] [Google Scholar]
- 48.Johnston MV, Jeon OH, Pevsner J, Blue ME, Naidu S. Neurobiology of Rett syndrome: a genetic disorder of synapse development. Brain & development. 2001;23(Suppl 1):S206–S213. doi: 10.1016/s0387-7604(01)00351-5. [DOI] [PubMed] [Google Scholar]
- 49.Kaufmann WE, Johnston MV, Blue ME. MeCP2 expression and function during brain development: implications for Rett syndrome’s pathogenesis and clinical evolution. Brain & development. 2005;27(Suppl 1):S77–s87. doi: 10.1016/j.braindev.2004.10.008. [DOI] [PubMed] [Google Scholar]
- 50.Armstrong DD. Rett syndrome neuropathology review 2000. Brain & development. 2001;23(Suppl 1):S72–S76. doi: 10.1016/s0387-7604(01)00332-1. [DOI] [PubMed] [Google Scholar]
- 51.Jellinger K, Seitelberger F. Neuropathology of Rett syndrome. American journal of medical genetics Supplement. 1986;1:259–288. doi: 10.1002/ajmg.1320250528. [DOI] [PubMed] [Google Scholar]
- 52.Boggio EM, Lonetti G, Pizzorusso T, Giustetto M. Synaptic determinants of rett syndrome. Frontiers in synaptic neuroscience. 2010;2:28-. [DOI] [PMC free article] [PubMed]
- 53.Murakami JW, Courchesne E, Haas RH, Press GA, Yeung-Courchesne R. Cerebellar and cerebral abnormalities in Rett syndrome: a quantitative MR analysis. AJR Am J Roentgenol. 1992;159(1):177–183. doi: 10.2214/ajr.159.1.1609693. [DOI] [PubMed] [Google Scholar]
- 54.Stagi S, Cavalli L, Ricci S, Mola M, Marchi C, Seminara S, et al. Parathyroid hormone levels in healthy children and adolescents. Hormone Research in Paediatrics. 2015;84(2):124–129. doi: 10.1159/000432399. [DOI] [PubMed] [Google Scholar]
- 55.Sukalo M, Tilsen F, Kayserili H, Muller D, Tuysuz B, Ruddy DM, et al. DOCK6 mutations are responsible for a distinct autosomal-recessive variant of Adams-Oliver syndrome associated with brain and eye anomalies. Hum Mutat. 2015;36(11):1112. doi: 10.1002/humu.22830. [DOI] [PubMed] [Google Scholar]
- 56.Pisciotta L, Capra V, Accogli A, Giacomini T, Prato G, Tavares P, et al. Epileptic encephalopathy in Adams-Oliver syndrome associated to a New DOCK6 mutation: a peculiar behavioral phenotype. Neuropediatrics. 2018;49(3):217–221. doi: 10.1055/s-0038-1639372. [DOI] [PubMed] [Google Scholar]
- 57.Li Rong, Pradhan Manisha, Xu Miao, Roeder Amanda, Beers Jeanette, Zou Jizhong, Liu Chengyu, Porter Forbes D., Zheng Wei. An induced pluripotent stem cell line (TRNDi001-D) from a Niemann-Pick disease type C1 (NPC1) patient carrying a homozygous p. I1061T (c. 3182T>C) mutation in the NPC1 gene. Stem Cell Research. 2020;44:101737. doi: 10.1016/j.scr.2020.101737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wheeler S, Sillence DJ. Niemann-Pick type C disease: cellular pathology and pharmacotherapy. Journal of neurochemistry. 2019. [DOI] [PubMed]
- 59.Ohshima T, Hirasawa M, Tabata H, Mutoh T, Adachi T, Suzuki H, et al. Cdk5 is required for multipolar-to-bipolar transition during radial neuronal migration and proper dendrite development of pyramidal neurons in the cerebral cortex. Development (Cambridge, England) 2007;134(12):2273–2282. doi: 10.1242/dev.02854. [DOI] [PubMed] [Google Scholar]
- 60.Magen D, Ofir A, Berger L, Goldsher D, Eran A, Katib N, et al. Autosomal recessive lissencephaly with cerebellar hypoplasia is associated with a loss-of-function mutation in CDK5. Hum Genet. 2015;134(3):305–314. doi: 10.1007/s00439-014-1522-5. [DOI] [PubMed] [Google Scholar]
- 61.Scarpini G, Spagnoli C, Salerno GG, Rizzi S, Frattini D, Fusco C. Autosomal recessive axonal neuropathy caused by HINT1 mutation: new association of a psychiatric disorder to the neurologic phenotype. Neuromuscular disorders : NMD. 2019;29(12):979. doi: 10.1016/j.nmd.2019.05.001. [DOI] [PubMed] [Google Scholar]
- 62.Wang Z, Lin J, Qiao K, Cai S, Zhang VW, Zhao C, et al. Novel mutations in HINT1 gene cause the autosomal recessive axonal neuropathy with neuromyotonia. Eur J Med Genet. 2019;62(3):190–194. doi: 10.1016/j.ejmg.2018.07.009. [DOI] [PubMed] [Google Scholar]
- 63.Mancini GM, Schot R, de Wit MC, de Coo RF, Oostenbrink R, Bindels-de Heus K, et al. CSTB null mutation associated with microcephaly, early developmental delay, and severe dyskinesia. Neurology. 2016;86(9):877–878. doi: 10.1212/WNL.0000000000002422. [DOI] [PubMed] [Google Scholar]
- 64.Bhat SS, Schmidt KR, Ladd S, Kim KC, Schwartz CE, Simensen RJ, et al. Disruption of DMD and deletion of ACSL4 causing developmental delay, hypotonia, and multiple congenital anomalies. Cytogenet Genome Res. 2006;112(1-2):170–175. doi: 10.1159/000087531. [DOI] [PubMed] [Google Scholar]
- 65.Gazou A, Riess A, Grasshoff U, Schaferhoff K, Bonin M, Jauch A, et al. Xq22.3-q23 deletion including ACSL4 in a patient with intellectual disability. Am J Med Genet A. 2013;161a(4):860–864. doi: 10.1002/ajmg.a.35778. [DOI] [PubMed] [Google Scholar]
- 66.Poirier K, Lebrun N, Broix L, Tian G, Saillour Y, Boscheron C, et al. Mutations in TUBG1, DYNC1H1, KIF5C and KIF2A cause malformations of cortical development and microcephaly. Nature genetics. 2013;45(6):639–647. doi: 10.1038/ng.2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Willemsen MH, Ba W, Wissink-Lindhout WM, de Brouwer AP, Haas SA, Bienek M, et al. Involvement of the kinesin family members KIF4A and KIF5C in intellectual disability and synaptic function. Journal of medical genetics. 2014;51(7):487–494. doi: 10.1136/jmedgenet-2013-102182. [DOI] [PubMed] [Google Scholar]
- 68.Cushion TD, Paciorkowski AR, Pilz DT, Mullins JG, Seltzer LE, Marion RW, et al. De novo mutations in the beta-tubulin gene TUBB2A cause simplified gyral patterning and infantile-onset epilepsy. American journal of human genetics. 2014;94(4):634–641. doi: 10.1016/j.ajhg.2014.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Montojo J, Zuberi K, Rodriguez H, Kazi F, Wright G, Donaldson SL, et al. GeneMANIA Cytoscape plugin: fast gene function predictions on the desktop. Bioinformatics (Oxford, England) 2010;26(22):2927–2928. doi: 10.1093/bioinformatics/btq562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Moretti P, Zoghbi HY. MeCP2 dysfunction in Rett syndrome and related disorders. Current opinion in genetics & development. 2006;16(3):276–281. doi: 10.1016/j.gde.2006.04.009. [DOI] [PubMed] [Google Scholar]
- 71.Shahbazian MD, Zoghbi HY. Rett syndrome and MeCP2: linking epigenetics and neuronal function. American journal of human genetics. 2002;71(6):1259–1272. doi: 10.1086/345360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cukier HN, Rabionet R, Konidari I, Rayner-Evans MY, Baltos ML, Wright HH, et al. Novel variants identified in methyl-CpG-binding domain genes in autistic individuals. Neurogenetics. 2010;11(3):291–303. doi: 10.1007/s10048-009-0228-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kyle Stephanie M., Vashi Neeti, Justice Monica J. Rett syndrome: a neurological disorder with metabolic components. Open Biology. 2018;8(2):170216. doi: 10.1098/rsob.170216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Forrest CM, Kennedy PG, Rodgers J, Dalton RN, Turner C, Darlington LG, et al. Kynurenine pathway metabolism following prenatal KMO inhibition and in Mecp2(+/-) mice, using liquid chromatography-tandem mass spectrometry. Neurochemistry international. 2016;100:110–119. doi: 10.1016/j.neuint.2016.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sullivan CR, Mielnik CA, O'Donovan SM, Funk AJ, Bentea E, DePasquale EA, et al. Connectivity analyses of bioenergetic changes in schizophrenia: identification of novel treatments. Molecular neurobiology. 2019;56(6):4492–4517. doi: 10.1007/s12035-018-1390-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pecorelli A, Leoni G, Cervellati F, Canali R, Signorini C, Leoncini S, et al. Genes related to mitochondrial functions, protein degradation, and chromatin folding are differentially expressed in lymphomonocytes of Rett syndrome patients. Mediators of inflammation. 2013;2013:137629. doi: 10.1155/2013/137629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yang T, Ramocki MB, Neul JL, Lu W, Roberts L, Knight J, et al. Overexpression of methyl-CpG binding protein 2 impairs T(H)1 responses. Science translational medicine. 2012;4(163):163ra58. doi: 10.1126/scitranslmed.3004430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Leoncini S, De Felice C, Signorini C, Zollo G, Cortelazzo A, Durand T, et al. Cytokine dysregulation in MECP2- and CDKL5-related Rett syndrome: relationships with aberrant redox homeostasis, inflammation, and omega-3 PUFAs. Oxidative medicine and cellular longevity. 2015;2015:421624. doi: 10.1155/2015/421624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ferreri Andrés J. M., Illerhaus Gerald, Zucca Emanuele, Cavalli Franco. Flows and flaws in primary central nervous system lymphoma. Nature Reviews Clinical Oncology. 2010;7(8):1–2. doi: 10.1038/nrclinonc.2010.9-c1. [DOI] [PubMed] [Google Scholar]
- 80.Mironov SL, Skorova E, Hartelt N, Mironova LA, Hasan MT, Kugler S. Remodelling of the respiratory network in a mouse model of Rett syndrome depends on brain-derived neurotrophic factor regulated slow calcium buffering. The Journal of physiology. 2009;587(Pt 11):2473–2485. doi: 10.1113/jphysiol.2009.169805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Dong Q, Liu Q, Li R, Wang A, Bu Q, Wang KH, et al. Mechanism and consequence of abnormal calcium homeostasis in Rett syndrome astrocytes. eLife. 2018;7. [DOI] [PMC free article] [PubMed]
- 82.Charman T, Cass H, Owen L, Wigram T, Slonims V, Weeks L, et al. Regression in individuals with Rett syndrome. Brain & development. 2002;24(5):281–283. doi: 10.1016/s0387-7604(02)00058-x. [DOI] [PubMed] [Google Scholar]
- 83.De Filippis B, Ricceri L, Laviola G. Early postnatal behavioral changes in the Mecp2-308 truncation mouse model of Rett syndrome. Genes, brain, and behavior. 2010;9(2):213–223. doi: 10.1111/j.1601-183X.2009.00551.x. [DOI] [PubMed] [Google Scholar]
- 84.Cheng T-L, Chen J, Wan H, Tang B, Tian W, Liao L, et al. Regulation of mRNA splicing by MeCP2 via epigenetic modifications in the brain. Scientific reports. 2017;7:42790. doi: 10.1038/srep42790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Pecorelli A, Cervellati C, Cortelazzo A, Cervellati F, Sticozzi C, Mirasole C, et al. Proteomic analysis of 4-hydroxynonenal and nitrotyrosine modified proteins in RTT fibroblasts. The international journal of biochemistry & cell biology. 2016;81(Pt B):236–245. doi: 10.1016/j.biocel.2016.08.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Figure S1. iPSC characterisation. a. Exemplary characterisation of iPSC lines. Immunocytochemistry for pluripotency marker (OCT3/4, SOX2, SSEA4, TRA1-60, TRA1-81). b. PCR-analysis for pluripotency marker. c. MeCP2 expression of the three iCTR and RTT samples at each time point.
Additional file 2: Figure S2. Number of proteins identified. A bar chart showing the number of proteins identified in each biological replicate and time point.
Additional file 3: Figure S3. Venn diagram. a. Number of proteins decreased expressed in RTT at different time points. b. Number of proteins increased expressed in RTT at different time points. c. Overview of the number of proteins altered in RTT.
Additional file 4: Figure S4. Top gene sets enriched in RTT-iPSCs. Proteins below p=0.1 are ranked by GSEA based on their differential expression level. Black vertical lines indicate the position where members of a pathway appear in the ranked gene list.
Data Availability Statement
All mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD013327
Username: reviewer46083@ebi.ac.uk
Password: ABIw2h3I