Abstract
Keratins are fibrous structural proteins that serve essential roles in forming the stratum corneum and protect the cells in this layer of skin from damage. Keratin 17 (KRT17) is a key member of the keratins, and dysregulated expression of KRT17 has been reported in various types of cancer, such as lung and gastric cancer. The present study aimed to identify the role of KRT17 in osteosarcoma and the underlying molecular mechanism. The expression of KRT17 in osteosarcoma tissues and cell lines was detected using reverse transcription-quantitative PCR (RT-qPCR) and western blotting. The effects of KRT17 on osteosarcoma cell proliferation and the Warburg effect in vitro were detected using CCK-8 and colony formation assays, cell cycle distribution analysis and metabolic measures. The effects of KRT17 on osteosarcoma cell proliferation in vivo were detected using a subcutaneous tumorigenesis model. The association between KRT17 and the AKT/mTOR/hypoxia-inducible factor 1α (HIF1α) pathway was detected using RT-qPCR and western blotting. The results demonstrated that KRT17 was highly expressed in osteosarcoma tissues and cell lines. Knockdown of KRT17 decreased osteosarcoma cell proliferation and colony formation, induced G1 phase arrest and inhibited glycolysis in vitro. Similarly, the suppression of KRT17 decreased osteosarcoma tumor growth in vivo. Knockdown of KRT17 decreased the expression of phosphorylated (p)-AKT, p-mTOR, HIF1α and the target gene of HIF1α glucose transporter 1. Restoring the expression of p-AKT, p-mTOR or HIF1α reversed the effect of KRT17 inhibition on cell proliferation and glycolysis. These results indicated that knockdown of KRT17 may be an effective method for treating osteosarcoma through inhibiting osteosarcoma cell proliferation and the Warburg effect by suppressing the AKT/mTOR/HIF1α pathway.
Keywords: keratin 17, osteosarcoma, Warburg effect, proliferation, AKT/mTOR/hypoxia-inducible factor 1α
Introduction
Osteosarcoma is one of the most common types of primary malignant cancer in bone tissues, primarily affecting adolescents and young adults (1). The majority of osteosarcomas occur in the metaphysis of the long bones of the extremities, whereas the involvement of the axial or the craniofacial bone can also be observed in the adult population (2). Despite the progress that has been made in treatment strategies for osteosarcoma, including surgical resection, chemotherapy and radiotherapy, the clinical prognosis remains poor, and the 5-year survival rate is still <20% due to the high rates of metastasis and recurrence (3,4). Although substantial attention has focused on the molecular mechanism behind osteosarcoma, the factors behind its occurrence and development remain unclear. Therefore, there is an urgent need to reveal the molecular mechanism of the development of osteosarcoma, to identify new molecular markers for early diagnosis, and to investigate new methods for treating this disease.
Compared with normal cells, tumor cells tend to undergo glycolysis despite the environment having sufficient oxygen; this phenomenon was first reported by Otto Warburg and is termed the Warburg effect (5). Through this metabolic rearrangement, tumor cells obtain enough energy to meet the requirements for growth and distant metastasis (6,7). Several oncogenes that regulate the Warburg effect and promote the development of osteosarcoma have been reported. For example, sphingosine-1-phosphate has been demonstrated to regulate the glycolytic enzyme phosphoglycerate mutase 1 to promote osteosarcoma proliferation (8). In addition, slit guidance ligand 2 has been reported to enhance the expression of 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 2 and to increase osteosarcoma cell proliferation and migration (9). Therefore, targeting the glycolytic pathway may be an appropriate strategy for the treatment of osteosarcoma.
Keratins, one of the largest gene families in humans, are a family of epithelial-specific intermediate filaments forming an indispensable part of the cytoskeleton (10). Keratins can be classified into two types, including 28 type I and 26 type II keratins (11). Keratins not only provide structural support for the cytoskeleton and regulate cellular metabolic processes, but also activate epithelial cell growth signaling pathways (12). Their expression is highly cell-type-specific, and mutation or loss of a keratin gene can cause or lead to a predisposition to several human diseases, induce the dysregulation of innate immunity, and produce increased sensitivity to apoptosis (13). Extensive research has been performed on the role of the keratin family in human cancer, which has attracted broad attention in the last two decades. For example, keratin 13 serves a role in reprogramming bone and brain metastases of human prostate cancer cells (14). In addition, keratin 5 and keratin 20 may be associated with urothelial bladder cancer (15). Keratin 17 (KRT17), a type I keratin, has also been identified to be expressed in several types of normal human epithelium, including the respiratory epithelium and urothelium (16). KRT17 was first reported to serve a positive role in epithelial cell proliferation and skin tumor growth (17). Furthermore, KRT17 has been reported to be associated with the development of various types of cancer, such as colorectal and breast cancer (18). However, its role in the progression of osteosarcoma remains unclear.
The present study aimed to investigate the role of KRT17 and its mechanism in osteosarcoma, and to determine whether KRT17 may be a novel target for the diagnosis and treatment of osteosarcoma.
Materials and methods
Patient samples
A total of 25 patients were enrolled in the present study. All patients were pathologically diagnosed with osteosarcoma and had received surgical resection at Guizhou Orthopedics Hospital (Guizhou, China) between June 2017 and June 2019. The patients included 17 males and 8 females, with a mean age of 19.15±13.17 years (range, 8–41 years). The inclusion criteria were as follows: i) The tissues were obtained during surgery and osteosarcoma was diagnosed by two pathologists; ii) the patients were diagnosed and treated for the first time; and iii) the patients were willing to participate. The exclusion criteria were as follows: i) Patients with other malignancies; ii) patients with other systemic diseases; iii) patients who had received treatment prior to admission; and iv) patients (and/or their families) who refused to participate. All 25 patients provided osteosarcoma tissue samples, of whom 10 also provided adjacent normal tissues. All specimens were obtained from patients following written informed consent form the patients or their parents/legal guardians. The study was approved by Guizhou Provincial Orthopedics Hospital Ethics Committee and was performed in accordance with the principles embodied in the Declaration of Helsinki.
Cell lines and cell culture
Human osteosarcoma cell lines (MG63, Saos2, U20S and 143B) and normal human osteoblast hFOB cells were obtained from the Cell Bank of the Chinese Academy of Sciences. The cells were cultured in Dulbecco's modified Eagle's medium (Gibco; Thermo Fisher Scientific, Inc.) containing 10% fetal bovine serum (Gibco; Thermo Fisher Scientific, Inc.), 0.1% penicillin and 0.1% streptomycin. MG63 (fibroblast morphology), Saos2 (epithelial morphology), U20S (epithelial morphology) and 143B (mixed morphology) cells were incubated at 37°C in a humidified atmosphere with 5% CO2, whereas hFOB cells were incubated at 35°C under the same atmosphere conditions.
Reverse transcription-quantitative PCR (RT-qPCR)
TRIzol reagent [Yeasen Biotechnology (Shanghai) Co., Ltd.] was used to isolate total RNA from osteosarcoma tissues, adjacent tissues, normal human osteoblast cells and osteosarcoma cells. PrimeScript RT Reagent kit reagent (Takara Bio, Inc.) was used to perform complementary DNA synthesis according to the manufacturer's instructions. Hieff® qPCR SYBR® Green Master Mix [Yeasen Biotechnology (Shanghai) Co., Ltd.] was used for the fluorescence quantification. The following primers were used: KRT17 forward, 5′-GGTGGGTGGTGAGATCAATGT-3′ and reverse, 5′-CGCGGTTCAGTTCCTCTGTC-3′; VEGF forward, 5′-AGGGCAGAATCATCACGAAGT-3′ and reverse, 5′-AGGGTCTCGATTGGATGGCA-3′; glucose transporter 1 (GLUT1) forward, 5′-GGCCAAGAGTGTGCTAAAGAA-3′ and reverse, 5′-ACAGCGTTGATGCCAGACAG-3′; MCL1 apoptosis regulator, Bcl2 family member (MCL1) forward, 5′-TGCTTCGGAAACTGGACATCA-3′ and reverse, 5′-TAGCCACAAAGGCACCAAAAG-3′; HIF1α forward, 5′-GAACGTCGAAAAGAAAAGTCTCG-3′ and reverse, 5′-CCTTATCAAGATGCGAACTCACA-3′; β-actin forward, 5′-CATGTACGTTGCTATCCAGGC-3′ and reverse, 5′-CTCCTTAATGTCACGCACGAT-3′. The reaction conditions were as follows: 95°C for 10 min, followed by 40 cycles of 95°C for 1 sec and 60°C for 60 sec. β-actin was used for normalization to determine the relative mRNA expression, which was analyzed using the 2−ΔΔCq method (19).
Western blotting
RIPA lysis buffer (Boster Biological Technology) containing a mixture of protease inhibitors was used to extract the proteins. The lysates were centrifuged at 8,000 × g at 4°C for 10 min to separate the proteins from the cell debris. The BCA assay kit (Boster Biological Technology) was used to measure the total protein concentration. The proteins (30 µg/lane) were separated by 10% SDS-PAGE and then transferred to PVDF membranes (EMD Millipore). Subsequently, 5% non-fat milk was used for blocking at room temperature for 2 h, after which the membranes were incubated overnight at 4°C with the following primary antibodies: Anti-KRT17 (1:1,000, cat no. 17516-1-AP), anti-cyclin D1 (1:1,000, cat no. 26939-1-AP), anti-CDK2 (1:1,000, cat no. 10122-1-AP), anti-CDK4 (1:1,000, cat no. 11026-1-AP, ProteinTech Group, Inc.), anti-CDK6 (1:1,000, cat no. 14052-1-AP, ProteinTech Group, Inc.), anti-AKT (1:1,000, cat no. 10176-2-AP), anti-phosphorylated (p)-AKT (1:1,000, cat no. 66444-1-Ig), anti-mTOR (1:1,000, cat no. 66888-1-Ig), anti-HIF1α (1:1,000, cat no. 20960-1-AP), anti-VEGF (1:1,000, cat no. 19003-1-AP), anti-GLUT1 (1:1,000, cat no. 21829-1-AP), anti-MCL1 (1:1,000, cat no. 16225-1-AP), anti-β-actin (1:1,000, cat no. 60008-1-Ig) (all from ProteinTech Group, Inc.) and anti-p-mTOR (1:500, cat no. 5536, Cell Signaling Technology, Inc.). After three washes with TBS containing 0.1% Tween-20, the membranes were incubated with the secondary horseradish peroxidase (HRP)-conjugated anti-mouse (1:3,000; cat no. BA1051) and anti-rabbit (1:3,000; cat no. BA1050; both from Boster Biological Technology) antibodies for 2 h at room temperature. Finally, protein bands were visualized using ChemiDoc™ XRS+ with Image Lab™ software (version 5.2.1; Bio-Rad Laboratories, Inc.). The expression levels of KRT17, cyclin D1, CDK2, CDK4, CDK6, HIF1α, VEGF, MCL1 and GLUT1 were relative to β-actin, whereas those of p-AKT and p-mTOR were analyzed as the ratios of p-AKT/AKT and p-mTOR/mTOR.
Retroviral infection and transfection
Human short hairpin RNA (shRNA) targeting KRT17 and negative control shRNA oligonucleotide sequences were cloned into pSuper-retro-puro viral vectors (Guangzhou RiboBio Co., Ltd.) to generate pSuper-retro-KRT17-RNAi(s) and pSuper-retro-scramble-RNAi(s). The shRNA sequences for KRT17 were as follows: shRNA1, 5′-GCGTGACCAGTATGAGAAGAT-3′; shRNA2, 5′-TGGTGCAGAGTGGCAAGAGTGAGAT-3′; and shRNA3, 5′-CTGACTCAGTACAAGAAAGAA-3′. The scrambled shRNA sequence 5′-wasTTCTCCGAACGTGTCACGT-3′ (all synthesized by Guangzhou RiboBio Co., Ltd). To obtain stable knockdown of KRT17 osteosarcoma cell lines, the cells were cultured in an environment containing 0.5 µg/ml puromycin for 2 weeks after infection. Osteosarcoma cells with the optimal stable knockdown of KRT17 and normal control (sh-scramble) were used for further study.
Cell Counting Kit-8 (CCK-8) assay
MG63 and Saos-2 cells were seeded in a 96-well plate at a density of 4×103 cells/well in a volume of 200 µl. On days 0, 1, 2, 3 and 4, the cells were incubated with CCK-8 (Boster Biological Technology) for 2 h at 37°C, and absorbance was measured at 450 nm using a microplate reader. For the rescue experiment, the AKT activator SC79 (5 µg/ml; MedChemExpress), mTOR activator MYH1485 (10 µM; MedChemExpress) and HIF activator ML228 (5 µM; MedChemExpress) were added after cell adherence, and the absorbance was measured at 450 nm after culturing for the time stated above.
Colony formation
MG63 and Saos-2 cells were seeded in a 6-well plate at a density of 1,000 cells/well. After culturing for 7 days at 37°C in a humidified incubator with 5% CO2, 4% paraformaldehyde was used to fix the cells for 15 min at room temperature, which were then stained with 0.1% crystal violet for 30 min at room temperature. Images of stained colonies were captured, and the number of colonies (≥10 mm2) was counted under the naked eye.
Cell cycle analysis
MG63 and Saos-2 cells were washed with PBS three times and fixed with 75% ethanol overnight at −20°C. The cells were then treated with 50 µg/ml propidium iodide solution (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's instructions and analyzed using a FACSCalibur flow cytometer (BD Biosciences). The results of the cell cycle distribution were analyzed using FlowJo software version 7.4.1 (FlowJo LLC).
Assays for measuring the extracellular acidification rate (ECAR) and cellular oxygen consumption rate (OCR)
The XFe Extracellular Flux Analyzer (Agilent Technologies, Inc.) was used to detect the ECAR and OCR in the present study. The ECAR was detected using an XF Glycolysis Stress Test kit (Agilent Technologies. Inc.), and the OCR was detected using an XF Cell Mito Stress Test kit. A total of 1×104 MG63 and Saos-2 cells were seeded in matched 96-well microplates. Following baseline measurements, 30 mM glucose, 2 µM oligomycin (an inhibitor of oxidative phosphorylation; Selleck Chemicals) and 10 mM 2-DG (an inhibitor of glycolysis; Selleck Chemicals) were added to each well at the specified time points to detect the ECAR. Similarly, 2 µM oligomycin, 5 µM FCCP (a reversible inhibitor of oxidative phosphorylation; Selleck Chemicals) and 1 µM rotenone (an inhibitor of mitochondrial complex I; Selleck Chemicals) plus 1 µM antimycin A (an inhibitor of mitochondrial complex III; Selleck Chemicals) were added in the appropriate order to detect the OCR and ECAR according to the operating steps described in a previous study (20). The data were collected and analyzed using XF-96 Wave software version 2.1 (Agilent Technologies. Inc.). The data were normalized based on the protein concentration.
Detection of glucose uptake, lactate production and ATP synthesis
To detect glucose uptake in each group, the Glucose Uptake Colorimetric assay kit (Invitrogen; Thermo Fisher Scientific, Inc.) was used according to the manufacturer's instruction. A total of 2,000 MG63 and Saos-2 cells were seeded in a 96-well plate and treated with the Krebs-Ringer-Phosphate-HEPES buffer containing 2% BSA (Wuhan Boster Biological Technology, Ltd.) for 40 min to starve the cells. Subsequently, 10 ml 2-DG (10 mM) was added to each well. Following culture for 20 min, the rate of glucose uptake was determined. To determine the lactate production and ATP synthesis of each group, Lactate assay kit II (Invitrogen; Thermo Fisher Scientific, Inc.) and ATP assay kit (Invitrogen; Thermo Fisher Scientific, Inc.), respectively, were used according to the manufacturer's instructions. A total of 2×106 MG63 and Saos-2 cells were lysed in 100 µl of the corresponding assay buffer and homogenized. Following centrifugation, the soluble fractions of lactate and ATP were assayed using 530 and 636 nm wavelengths in an ultraviolet spectrophotometer, respectively.
Animal experiments
All animal procedures were approved by the Ethics Committee of Guizhou Provincial Orthopedics Hospital. Female BLBA/c nude mice (6-week-old; n=10) were purchased from Beijing Huafukang Biotechnology Co., Ltd. and housed under specific pathogen-free conditions at 25°C with a 12-h light/dark cycle and free access to food and water. A total of 1×106 MG63 cells transfected with sh-KRT17 or sh-scramble were subcutaneously injected into the flanks of nude mice. Each group contained five mice. The animal health and behavior were monitored daily, and the volume of tumor tissue was measured weekly. The volume of tumor tissues were calculated using the following formula: Volume (mm3)=(length × width2)/2. When the maximum length of any tumor reached 15 mm or the volume of any tumor reached 800 mm3, the experiment was terminated. The mice were sacrificed in week 5 using cervical dislocation; death was verified by the absence of a heartbeat and the onset of rigor mortis, and the size of each tumor was determined.
Immunohistochemical staining
The tumor tissues isolated from nude mice were dehydrated and embedded in paraffin (Wuhan Boster Biological Technology, Ltd.) at room temperature. After cutting into 4-µm slices, tissue sections were deparaffinized using xylene and rehydrated in a descending alcohol series (100, 95, 90 and 80%) at room temperature. Following restoration with sodium citrate, the samples were treated with 3% H2O2 to block endogenous peroxidase activity, and then blocked with 5% BSA (Wuhan Servicebio Technology Co., Ltd.) for 30 min at room temperature. The specimens were subsequently incubated with a primary anti-KI67 (1:400; cat. no. 27309-1-AP; ProteinTech Group, Inc.) and anti-PCNA (1:400; cat. no. 10205-2-AP; ProteinTech Group, Inc.) antibodies for 12 h at 4°C, followed by incubation with HRP-conjugated secondary antibodies anti rabbit IgG (1:200; cat. no. G1210-2-A-100; Wuhan Servicebio Technology Co., Ltd.) for 2 h at room temperature. Following subsequent development using the Cell and Tissue Staining HRP-DAB kit (Beyotime Institute of Biotechnology) according to the manufacturer's protocol, images were obtained under a light microscope (magnification, ×400).
Statistical analysis
SPSS 20.0 (IBM Corp.) was used to perform the statistical analysis in the current study. Data are representative of at least three independent experiments and presented as the mean ± standard deviation. Unpaired Student's t-test was used to analyze the expression difference of KRT17 in osteosarcoma and corresponding adjacent tissues. The co-expression relationship between KRT17 and HIF1α in osteosarcoma tissues was analyzed using Pearson's correlation analysis. Two-tailed unpaired Student's t-test was used to analyze the statistical differences between two different groups of continuous variables. One-way analysis of variance (ANOVA) followed by Dunnett's post hoc test was performed to test the differences between control group and multiple experimental groups, while ANOVA followed by Bonferroni's post hoc test was used to test the differences among multiple groups. P<0.05 was considered to indicate a statistically significant difference.
Results
KRT17 is expressed in osteosarcoma tissues and cell lines
First, RT-qPCR was performed to analyze the expression of KRT17 in osteosarcoma tissues. The results demonstrated that the expression of KRT17 was significantly higher in osteosarcoma tissues (n=25) compared with in adjacent normal osteoblast tissues (n=10) (P<0.01; Fig. 1A). Similarly, the mRNA and protein levels of KRT17 were markedly increased in osteosarcoma lines, including MG63, Saos2, U2OS and 143B, compared with those in normal bone hFOB cells (P<0.01; Fig. 1B-D). The mRNA and protein levels of KRT17 were 2–3-fold higher in the MG63 and Saos2 cell lines compared with those in hFOB cells; therefore, these two cell lines were used for further study. 143B cells were excluded due to their mixed morphology resulting in large experimental error.
Figure 1.
KRT17 is highly expressed in osteosarcoma tissues and cell lines. (A) RT-qPCR was used to detect the expression in adjacent normal osteoblast (n=10) and osteosarcoma (n=25) tissues. (B) RT-qPCR was used to detect the expression of KRT17 in the normal osteoblast cell line hFOB and the osteosarcoma cell lines MG63, Saos2, U2OS and 143B. (C and D) Western blotting was used to detect the expression of KRT17 in the normal osteoblast cell line hFOB and the osteosarcoma cell lines MG63, Saos2, U2OS, and 143B. β-actin was used as a loading control. **P<0.01 vs. hFOB or as indicated. KRT17, keratin 17; RT-qPCR, reverse transcription-quantitative PCR.
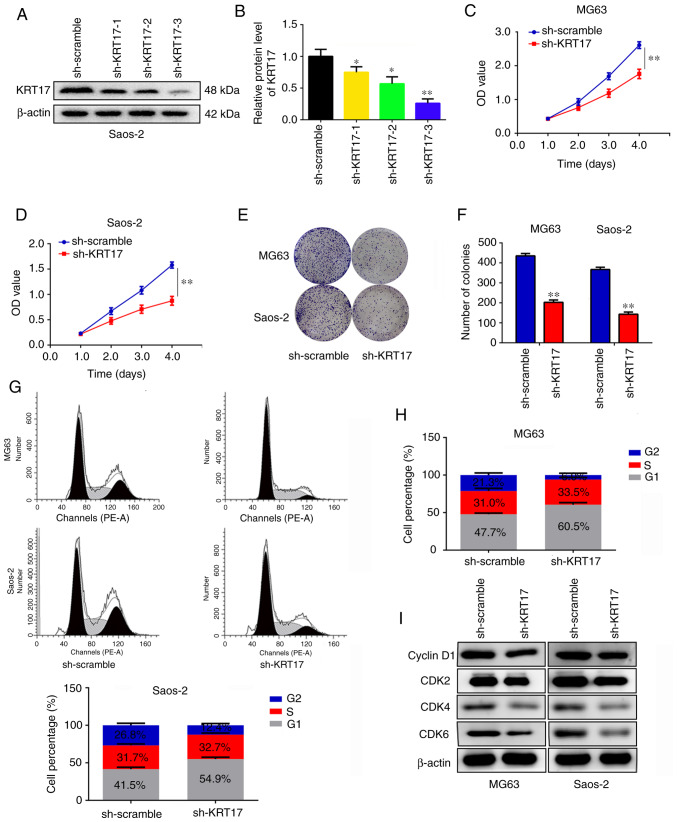
Knockdown of KRT17 decreases osteosarcoma cell proliferation and induces G1 phase arrest in vitro
To determine the role of KRT17 in osteosarcoma proliferation in vitro, its expression was disrupted using targeted KRT17 shRNA. Three potential KRT17-targeting shRNAs were used, and the results demonstrated that sh-KRT17-3 exhibited the strongest inhibition of KRT17 expression compared with the sh-scramble group (P<0.01; Fig. 2A and B). Therefore, sh-KRT17-3 was used to construct osteosarcoma cells with stable KRT17 knockdown for further experiments. CCK-8 assays were performed, and the results revealed that KRT17 knockdown significantly decreased the proliferation rate of MG63 and Saos2 cells compared with those transfected with sh-scramble (P<0.01; Fig. 2C and D). Similarly, it was identified that knockdown of KRT17 significantly decreased the numbers of MG63 and Saos2 cell colonies compared with those in the corresponding sh-scramble groups (P<0.01; Fig. 2E and F). Cell cycle distribution analysis was performed to detect the differences between the sh-scramble and sh-KRT17 groups; the results demonstrated that the proportion of cells in the G1 phase significantly increased in the sh-KRT17 groups of both MG63 and Saos2 cells compared with the respective sh-scramble groups, and a significant decrease of cells in the G2 phase was observed in the sh-KRT17 groups (P<0.05; Fig. 2G and H). In addition, the expression levels of cyclin D1, CDK2, CDK4 and CDK6 were examined in the sh-KRT17 and sh-scramble groups; it was observed that inhibition of KRT17 decreased the expression of cyclin D1, CDK2, CDK4 and CDK6 (Fig. 2I). These results indicated that knockdown of KRT17 decreased osteosarcoma cell proliferation and induced G1 phase arrest in vitro.
Figure 2.
Knockdown of KRT17 decreases osteosarcoma cell proliferation and induces G1 phase arrest in vitro. (A and B) The efficiency of three KRT17-targeting shRNAs was detected using western blotting. (C and D) Cell Counting Kit-8 assays were performed to detect the effects of KRT17 knockdown on osteosarcoma cell proliferation. (E and F) Colony formation assays were performed to detect the effects of KRT17 knockdown on osteosarcoma cell colony formation. (G and H) Cell cycle distribution was analyzed in the sh-scramble and sh-KRT17 groups of MG63 and Saos2 cells. (I) Western blotting was used to detect the expression of cyclin D1, CDK2, CDK4 and CDK6 in the sh-scramble and sh-KRT17 groups. *P<0.05, **P<0.01. KRT17, keratin 17; shRNA, short hairpin RNA; OD, optical density.
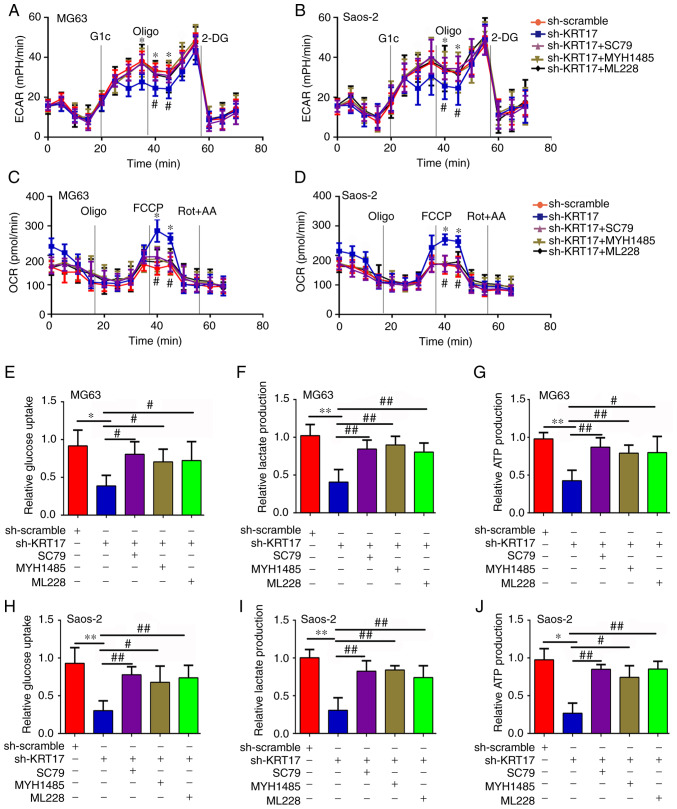
Knockdown of KRT17 decreases the Warburg effect of osteosarcoma cells
Previous studies have demonstrated that the Warburg effect serves a key role in the proliferation of osteosarcoma cells (21,22). Therefore, the present study investigated the effect of KRT17 on the Warburg effect of osteosarcoma cells. OCR, ECAR, glucose uptake, lactate production and ATP synthesis were analyzed in the sh-scramble and sh-KRT17 groups. The ECAR was significantly decreased in the sh-KRT17 group in both MG63 (at 30, 35 and 40 min) and Saos-2 cells (at 35 and 40 min) (P<0.05; Fig. 3A and B), whereas the basal and maximal OCR were significantly increased in the sh-KRT17 group compared with the sh-scramble group (P<0.05; Fig. 3C and D). In addition, the glucose uptake rate, lactate production and ATP synthesis were significantly decreased in MG63 and Saos-2 cells following KRT17 knockdown compared with the sh-scramble-transfected cells (P<0.05; Fig. 3E-J). Taken together, these results indicated that knockdown of KRT17 decreased the Warburg effect in osteosarcoma cells.
Figure 3.
Knockdown of KRT17 decreases the Warburg effect of osteosarcoma cells. (A and B) Using a Seahorse Bioscience XFp analyzer, the ECAR of MG63 and Saos2 cells in the sh-KRT17 and sh-scramble groups was measured. (C and D) Using a Seahorse Bioscience XFp analyzer, the OCR of MG63 and Saos2 cells with stable knockdown of KRT17 and sh-scramble cells was measured. (E-G) Glucose uptake, lactate production and ATP synthesis were measured in MG63 cells with stable knockdown of KRT17 and those transfected with sh-scramble. (H-J) Glucose uptake, lactate production and ATP synthesis were measured in Saos2 sh-KRT17 and sh-scramble cells. *P<0.05, **P<0.01 vs. sh-scramble. Glc, glucose; Oligo, oligomycin; 2-DG, 2-deoxy-D-glucose; FCCP, carbonyl cyanide 4-(trifluoromethoxy) phenylhydrazone; Rot, rotenone; AA, antimycin A; KRT17, keratin 17; ECAR, extracellular acidification rate; OCR, cellular oxygen consumption rate.
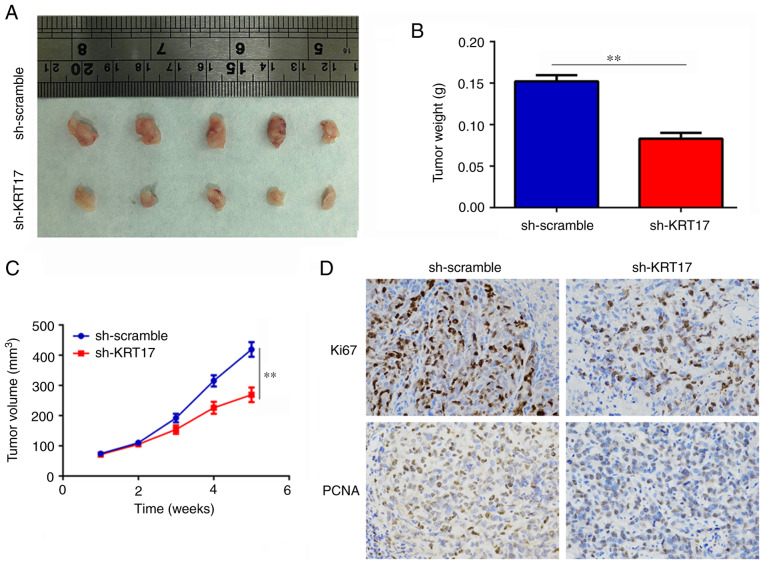
Knockdown of KRT17 decreases tumor growth in vivo
The effect of KRT17 on tumor growth in vivo was determined using a subcutaneous tumorigenesis model in nude mice. The results revealed that the rates of tumor growth were slower and the weights of the tumor were lower in the sh-KRT17 group compared with those in the sh-scramble group (P<0.01; Fig. 4A-C). In addition, the expression levels of KI67 and PCNA in tumor tissues from the sh-KRT17 group were significantly decreased compared with those in tissues from the sh-scramble group. (P<0.05; Fig. 4D). In summary, these results suggested that knockdown of KRT17 decreased osteosarcoma tumor growth in vivo.
Figure 4.
Knockdown of KRT17 decreases tumor growth in vivo. (A and B) The size and weight of the tumor tissues in the sh-scramble and sh-KRT17 groups. (C) The growth rate of osteosarcoma tumors in the sh-scramble and sh-KRT17 groups. (D) The expression of Ki67 and PCNA in tumor tissues from the sh-scramble and sh-KRT17 groups. **P<0.01. KRT17, keratin 17; sh, short hairpin.
KRT17 regulates the AKT/mTOR/HIF1α pathway
A previous study has demonstrated that KRT17 binds to stratifin and recruits it to the cytoplasm, where this complex activates the AKT/mTOR pathway (13). To determine whether KRT17 also regulates the AKT/mTOR pathway in osteosarcoma, western blotting was performed. The results demonstrated that total AKT and total mTOR levels changed insignificantly (P>0.05), whereas p-AKT and p-mTOR levels were significantly decreased in the sh-KRT17 group compared with those in the sh-scramble group (Fig. 5A-C). HIF1α is a key downstream protein of mTOR, which has been reported to serve a major role in cell proliferation and metabolic rearrangement, such as by increasing glycolysis (23). Therefore, the present study determined the expression of HIF1α following KRT17 inhibition and revealed that its level was also significantly decreased in the sh-KRT17 group compared with that in the sh-scramble group (P<0.05; Fig. 5A-C). Of note, correlation analysis revealed that KRT17 was co-expressed with HIF1α in osteosarcoma tissues (r=0.61, P<0.01; Fig. 5D). Furthermore, the expression levels of HIF1α target genes, such as VEGF, GLUT1 and MCL1, were determined, which demonstrated that the mRNA and protein levels of these genes were significantly decreased following KRT17 inhibition compared with those in the sh-scramble group (P<0.05; Fig. 5E-I). Among these target genes of HIF1α, GlUT1, which serves an important role in glycolysis, exhibited the strongest decrease.
Figure 5.
Inhibition of KRT17 suppresses the AKT/mTOR/HIF1α pathway. (A-C) The expression of AKT, p-AKT, mTOR, p-mTOR and HIF1α in the sh-scramble and sh-KRT17 groups was detected using western blotting. β-actin was used as a loading control. (D) The co-expression relationship between KRT17 and HIF1α in osteosarcoma tissues was determined using RT-qPCR and Pearson's correlation analysis. (E and F) The mRNA levels of VEGF, GLUT1 and MCL1 were determined using RT-qPCR in the sh-scramble and sh-KRT17 groups. (G-I) The protein levels of VEGF, GLUT1 and MCL1 were detected using western blotting in the sh-scramble and sh-KRT17 groups. *P<0.05, **P<0.01. KRT17, keratin 17; sh, short hairpin; p-, phosphorylated; HIF1α, hypoxia-inducible factor 1α; MCL1, MCL1 apoptosis regulator, Bcl2 family member; GLUT1, glucose transporter 1.
Restoration of the AKT/mTOR/HIF1α pathway decreases the inhibitory effect of KRT17 knockdown on osteosarcoma cell proliferation
To detect whether the AKT/mTOR/HIF1α pathway was involved in the KRT17 knockdown-induced inhibition of proliferation, activators of AKT (SC79), mTOR (MYH1485) and HIF1α (ML228) were employed. The results demonstrated that the expression of GLUT1 was increased when cells in the sh-KRT17 group were treated with SC79, MYH1485 or ML228 compared with the untreated sh-KRT17 group (Fig. 6A and B). The results of the CCK-8 assay revealed that the inhibitory effect of KRT17 knockdown on cell proliferation was reversed by SC79, MYH1485 or ML228 in MG63 and Saos2 cells (P<0.05; Fig. 6C and D). Similarly, the number of colonies was significantly increased when sh-KRT17 cells were treated with SC79, MYH1485 or ML228 compared with that in untreated sh-KRT17 cells (Fig. 6E and F). These results indicated that restoration of the AKT/mTOR/HIF1α pathway decreased the inhibitory effect of KRT17 knockdown on osteosarcoma cell proliferation.
Figure 6.
Restoration of the AKT/mTOR/HIF1α pathway reverses the effects of KRT17 knockdown on osteosarcoma cell proliferation and colony formation. (A and B) Western blotting was used to detect the expression of KRT17, AKT, p-AKT, mTOR, p-mTOR, HIF1α and GLUT1 in cells treated with sh-scramble or sh-KRT17 and SC79, MYH1485 and ML228. (C and D) Cell Counting Kit-8 assay was used to detect the cell proliferation in each group. (E and F) Colony formation assays were used to detect the colony formation of osteosarcoma cells in each group. **P<0.01 vs. sh-scramble; #P<0.05, ##P<0.01 vs. sh-KRT17. KRT17, keratin 17; sh, short hairpin; p-, phosphorylated; HIF1α, hypoxia-inducible factor 1α; GLUT1, glucose transporter 1.
Restoration of the AKT/mTOR/HIF1α pathway decreases the inhibitory effect of KRT17 knockdown on osteosarcoma cell glycolysis
The association between KRT17, the AKT/mTOR/HIF1α pathway and glycolysis was also investigated in the present study. In the ECAR analysis, the ECAR at 40 and 45 min was increased in sh-KRT17 cells upon treatment with activators of the AKT/mTOR/HIF1α pathway compared with untreated sh-KRT17 cells (P<0.05; Fig. 7A and B). In addition, the maximal OCR at 40 and 45 min was decreased in sh-KRT17 cells upon treatment with activators of the AKT/mTOR/HIF1α pathway compared with untreated sh-KRT17 cells (P<0.05; Fig. 7C and D). The rates of glucose uptake, lactate production and ATP synthesis were significantly increased when sh-KRT17 cells were treated with AKT, mTOR or HIF1α activators compared with those in untreated sh-KRT17 cells (P<0.05; Fig. 7E-J). These results indicated that restoration of the AKT/mTOR/HIF1α pathway decreased the inhibitory effect of KRT17 knockdown on osteosarcoma cell glycolysis.
Figure 7.
Restoration of the AKT/mTOR/HIF1α pathway decreases the inhibitory effect of KRT17 knockdown on osteosarcoma cell glycolysis. (A and B) The ECAR was detected in MG63 and Saos2 cells treated with sh-scramble or sh-KRT17 and SD79, MYH1485 and ML228. (C and D) The OCR was detected in each group of MG63 and Saos2 cells. (E-G) Glucose uptake, lactate production and ATP synthesis were measured in each group of MG63 cells. (H-J) Glucose uptake, lactate production and ATP synthesis were measured in each group of Saos2 cells. *P<0.05, **P<0.01 vs. sh-scramble; #P<0.05, ##P<0.01 vs. sh-KRT17. Glc, glucose; Oligo, oligomycin; 2-DG, 2-deoxy-D-glucose; FCCP, carbonyl cyanide 4-(trifluoromethoxy) phenylhydrazone; Rot, rotenone; AA, antimycin A; KRT-17, keratin 17; sh, short hairpin; HIF1α, hypoxia-inducible factor 1α; ECAR, extracellular acidification rate; OCR, cellular oxygen consumption rate.
Discussion
The present study identified the role of KRT17 in osteosarcoma and the underlying molecular mechanism. KRT17 is a member of the keratin family that serves a key role in protecting cells from damage or physical stress (24). Dysregulated expression of KRT17 has been reported in numerous types of cancer. Hu et al (25) have suggested that KRT17 is highly expressed in gastric cancer and is associated with poor outcome in those affected by this disease. In addition, Liu et al (26) have demonstrated that KRT17 has the potential to promote the proliferation, migration and invasion of lung adenocarcinoma cells. Khanom et al (13) have reported that the inhibition of KRT17 decreases the proliferation of oral cancer cells. Furthermore, Li et al (27) have demonstrated that KRT17 serves a key role in the resistance to paclitaxel in cervical cancer cells. Consistent with these previous studies, the present study demonstrated that KRT17 is increased in osteosarcoma cell tissues and cell lines. Knockdown of KRT17 significantly decreased the proliferation of osteosarcoma cells in vitro and in vivo. These results indicate that KRT17 may act as an oncogene in osteosarcoma.
Glycolysis is a common hallmark for cancer tissues as cancer cells utilize energy via glycolysis rather than by the tricarboxylic acid cycle (21). Based on glycolysis, cancer cells have enough energy for proliferation, migration and metastasis (28). The results of the present study demonstrated that inhibition of KRT17 significantly increased the OCR and decreased the ECAR, ATP production, lactate production and glucose uptake of osteosarcoma cells compared with those in the control group.
Previous studies have reported that the AKT/mTOR pathway is activated in various types of cancer, including osteosarcoma (29,30). Activated mTOR promotes cell proliferation by promoting the phosphorylation of downstream proteins (31). A previous study has demonstrated that KRT17 can bind with stratifin and increase the phosphorylation level of AKT (13). In Ewing's sarcoma, KRT17 has also been reported to have the capacity to activate the AKT pathway (32). Therefore, the present study determined the expression of proteins in the AKT pathway, with the results revealing that the levels of p-AKT and p-mTOR were decreased in KRT17-knockdown cells compared with those in the normal control group.
HIF1α is one of the downstream proteins of mTOR (33). Previous studies have demonstrated that activated mTOR can maintain the stability of HIF1α (34,35). Increased HIF1α translocates into the nucleus and binds to the promoters of its target genes, such as VEGF, GLUT1 and MCL1 (36–38). Through the regulation of its target genes, HIF1α serves roles in cancer cell proliferation, angiopoiesis and glycolysis (39). Based on the significant effects of KRT17 on osteosarcoma glycolysis, the present study considered whether HIF1α was regulated by KRT17 via the AKT/mTOR pathway; consistent with this speculation, it was identified that the expression of HIF1α was significantly decreased in sh-KRT17 osteosarcoma cells, as was that of its target genes, such as VEGF, MCL1 and GLUT1. Among these, GLUT1, which serves a key role in cell glycolysis, was decreased the most significantly. In addition, the results of the correlation analysis demonstrated that KRT17 was co-expressed with HIF1α. In summary, these results indicate that there may be a regulatory relationship between KRT17 and HIF1α via the AKT/mTOR pathway. To confirm these conclusions, AKT, mTOR and HIF1α activators were used, and the results demonstrated that the restoration of the AKT/mTOR/HIF1α pathway reversed the effects of KRT17 knockdown on osteosarcoma cell function, including proliferation and glycolysis.
In conclusion, the results of the present study demonstrated that KRT17 was highly expressed in osteosarcoma tissues and osteosarcoma cell lines. Knockdown of KRT17 decreased osteosarcoma cell proliferation and glycolysis by inhibiting the AKT/mTOR/HIF1α pathway. Therefore, KRT17 may be a novel biomarker for osteosarcoma diagnosis, as well as an effective target for treatment.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
XY, CY and WH performed the experiments and were responsible for data collection, analysis and interpretation of the results. XY, TC, QW, BQ and FP provided clinical samples for the experiments and validated the data. XY and BT were responsible for the experimental design, analysis and interpretation of data. All authors have read and approved the final version of the manuscript.
Ethics approval and consent to participate
The study of clinical samples was approved by the Ethics Committee of Guizhou Provincial Orthopedics Hospital and was performed in accordance with the principles embodied in the Declaration of Helsinki. All patients provided written informed consent to participate in the study. All animal procedures were approved by the Ethics Committee of Guizhou Provincial Orthopedics Hospital.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Melim C, Jarak I, Veiga F, Figueiras A. The potential of micelleplexes as a therapeutic strategy for osteosarcoma disease. 3 Biotech. 2020;10:147. doi: 10.1007/s13205-020-2142-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moore DD, Luu HH. Osteosarcoma. Cancer Treat Res. 2014;162:65–92. doi: 10.1007/978-3-319-07323-1_4. [DOI] [PubMed] [Google Scholar]
- 3.Bishop MW, Janeway KA, Gorlick R. Future directions in the treatment of osteosarcoma. Curr Opin Pediatr. 2016;28:26–33. doi: 10.1097/MOP.0000000000000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fan TM, Roberts RD, Lizardo MM. understanding and modeling metastasis biology to improve therapeutic strategies for combating osteosarcoma progression. Front Oncol. 2020;10:13. doi: 10.3389/fonc.2020.00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baltazar F, Afonso J, Costa M, Granja S. Lactate beyond a waste metabolite: Metabolic affairs and signaling in malignancy. Front Oncol. 2020;10:231. doi: 10.3389/fonc.2020.00231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liberti MV, Locasale JW. Correction to: ‘The warburg effect: How does it benefit cancer cells?’: [Trends in Biochemical Sciences, 41 (2016) 211] Trends Biochem Sci. 2016;41:287. doi: 10.1016/j.tibs.2016.01.004. [DOI] [PubMed] [Google Scholar]
- 7.Schwartz L, Supuran CT, Alfarouk KO. The warburg effect and the hallmarks of cancer. Anticancer Agents Med Chem. 2017;17:164–170. doi: 10.2174/1871520616666161031143301. [DOI] [PubMed] [Google Scholar]
- 8.Shen Y, Zhao S, Wang S, Pan X, Zhang Y, Xu J, Jiang Y, Li H, Zhang Q, Gao J, et al. S1P/S1PR3 axis promotes aerobic glycolysis by YAP/c-MYC/PGAM1 axis in osteosarcoma. Ebiomedicine. 2019;40:210–223. doi: 10.1016/j.ebiom.2018.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhao SJ, Shen YF, Li Q, He YJ, Zhang YK, Hu LP, Jiang YQ, Xu NW, Wang YJ, Li J, et al. SLIT2/ROBO1 axis contributes to the Warburg effect in osteosarcoma through activation of SRC/ERK/c-MYC/PFKFB2 pathway. Cell Death Dis. 2018;9:390. doi: 10.1038/s41419-018-0419-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rajabi M, Ali A, McConnell M, Cabral J. Keratinous materials: Structures and functions in biomedical applications. Mater Sci Eng C, Mater Biol Appl. 2020;110:110612. doi: 10.1016/j.msec.2019.110612. [DOI] [PubMed] [Google Scholar]
- 11.Donato RK, Mija A. Keratin associations with synthetic, biosynthetic and natural polymers: An extensive review. Polymers (Basel) 2019;12(pii):E32. doi: 10.3390/polym12010032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yi H, Yoon HN, Kim S, Ku NO. The role of keratins in the digestive system: Lessons from transgenic mouse models. Histochem Cell Biol. 2018;150:351–359. doi: 10.1007/s00418-018-1695-4. [DOI] [PubMed] [Google Scholar]
- 13.Khanom R, Nguyen CT, Kayamori K, Zhao X, Morita K, Miki Y, Katsube K, Yamaguchi A, Sakamoto K. Keratin 17 Is induced in oral cancer and facilitates tumor growth. PLoS One. 2016;11:e161163. doi: 10.1371/journal.pone.0161163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li Q, Yin L, Jones LW, Chu GC, Wu JB, Huang JM, Li Q, You S, Kim J, Lu YT, et al. Keratin 13 expression reprograms bone and brain metastases of human prostate cancer cells. Oncotarget. 2016;7:84645–84657. doi: 10.18632/oncotarget.13175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eckstein M, Wirtz RM, Gross-Weege M, Breyer J, Otto W, Stoehr R, Sikic D, Keck B, Eidt S, Burger M, et al. mRNA-Expression of KRT5 and KRT20 defines distinct prognostic subgroups of muscle-invasive urothelial bladder cancer correlating with histological variants. Int J Mol Sci. 2018;19(pii):E3396. doi: 10.3390/ijms19113396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li D, Ni XF, Tang H, Zhang J, Zheng C, Lin J, Wang C, Sun L, Chen B. KRT17 Functions as a tumor promoter and regulates proliferation, migration and invasion in pancreatic cancer via mTOR/S6k1 pathway. Cancer Manag Res. 2020;12:2087–2095. doi: 10.2147/CMAR.S243129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Depianto D, Kerns ML, Dlugosz AA, Coulombe PA. Keratin 17 promotes epithelial proliferation and tumor growth by polarizing the immune response in skin. Nat Genet. 2010;42:910–914. doi: 10.1038/ng.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang L, Zhang S, Wang G. Keratin 17 in disease pathogenesis: From cancer to dermatoses. J Pathol. 2019;247:158–165. doi: 10.1002/path.5178. [DOI] [PubMed] [Google Scholar]
- 19.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 20.Pan C, Liu Q, Wu X. HIF1α/miR-520a-3p/AKT1/mTOR feedback promotes The proliferation and glycolysis of gastric cancer cells. Cancer Manag Res. 2019;11:10145–10156. doi: 10.2147/CMAR.S223473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kobliakov VA. The mechanisms of regulation of aerobic glycolysis (warburg effect) by oncoproteins in carcinogenesis. Biochemistry (Mosc) 2019;84:1117–1128. doi: 10.1134/S0006297919100018. [DOI] [PubMed] [Google Scholar]
- 22.Abbaszadeh Z, Cesmeli S, Biray Avci C. Crucial players in glycolysis: Cancer progress. Gene. 2020;726:144158. doi: 10.1016/j.gene.2019.144158. [DOI] [PubMed] [Google Scholar]
- 23.Liao Z, She C, Ma L, Sun Z, Li P, Zhang X, Wang P, Li W. KDELR2 Promotes glioblastoma tumorigenesis targeted by HIF1a via mTOR signaling pathway. Cell Mol Neurobiol. 2019;39:1207–1215. doi: 10.1007/s10571-019-00715-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen P, Shen Z, Fang X, Wang G, Wang X, Wang J, Xi S. Silencing of keratin 17 by lentivirus-mediated short hairpin RNA inhibits the proliferation of PANC-1 human pancreatic cancer cells. Oncol Lett. 2020;19:3531–3541. doi: 10.3892/ol.2020.11469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hu H, Xu DH, Huang XX, Zhu CC, Xu J, Zhang ZZ, Zhao G. Keratin 17 promotes tumor growth and is associated with poor prognosis in gastric Cancer. J Cancer. 2018;9:346–357. doi: 10.7150/jca.19838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu J, Liu L, Cao L, Wen Q. Keratin 17 promotes lung adenocarcinoma progression by enhancing cell proliferation and invasion. Med Sci Monit. 2018;24:4782–4790. doi: 10.12659/MSM.909350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li J, Chen Q, Deng Z, Chen X, Liu H, Tao Y, Wang X, Lin S, Liu N. KRT17 confers paclitaxel-induced resistance and migration to cervical cancer cells. Life Sci. 2019;224:255–262. doi: 10.1016/j.lfs.2019.03.065. [DOI] [PubMed] [Google Scholar]
- 28.Liu L, Chai L, Ran J, Yang Y, Zhang L. BAI1 acts as a tumor suppressor in lung cancer A549 cells by inducing metabolic reprogramming via the SCD1/HMGCR module. Carcinogenesis. 2020 Apr 7; doi: 10.1093/carcin/bgaa036. ([Epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 29.Ma H, Su R, Feng H, Guo Y, Su G. Long noncoding RNA UCA1 promotes osteosarcoma metastasis through CREB1-mediated epithelial-mesenchymal transition and activating PI3K/AKT/mTOR pathway. J Bone Oncol. 2019;16:100228. doi: 10.1016/j.jbo.2019.100228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mei L, Sang W, Cui K, Zhang Y, Chen F, Li X. Norcantharidin inhibits proliferation and promotes apoptosis via c-Met/Akt/mTOR pathway in human osteosarcoma cells. Cancer Sci. 2019;110:582–595. doi: 10.1111/cas.13900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu F, Na L, Li Y, Chen L. Roles of the PI3K/AKT/mTOR signalling pathways in neurodegenerative diseases and tumours. Cell Biosci. 2020;10:54. doi: 10.1186/s13578-020-00416-0. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 32.Sankar S, Tanner JM, Bell R, Chaturvedi A, Randall RL, Beckerle MC, Lessnick SL. A novel role for keratin 17 in coordinating oncogenic transformation and cellular adhesion in Ewing sarcoma. Mol Cell Biol. 2013;33:4448–4460. doi: 10.1128/MCB.00241-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Spirina LV, Kondakova IV, Yurmazov ZA, Usynin EA, Slonimskaya EM, Lushnikova NA, Podnebesnova DV. VHL expression in kidney cancer: Relation to metastasis development, transcription and growth factors and component of Akt/m-TOR signaling pathway. Bull Exp Biol Med. 2019;167:671–675. doi: 10.1007/s10517-019-04596-9. [DOI] [PubMed] [Google Scholar]
- 34.Lyu X, Wang J, Guo X, Wu G, Jiao Y, Faleti OD, Liu P, Liu T, Long Y, Chong T, et al. EBV-miR-BART1-5P activates AMPK/mTOR/HIF1 pathway via a PTEN independent manner to promote glycolysis and angiogenesis in nasopharyngeal carcinoma. PLoS Pathog. 2018;14:e1007484. doi: 10.1371/journal.ppat.1007484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cheng SC, Quintin J, Cramer RA, Shepardson KM, Saeed S, Kumar V, Giamarellos-Bourboulis EJ, Martens JH, Rao NA, Aghajanirefah A, et al. mTOR- and HIF-1alpha-mediated aerobic glycolysis as metabolic basis for trained immunity. Science. 2014;345:1250684. doi: 10.1126/science.1250684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang Y, Huang Y, Liu H, Su D, Luo F, Zhou F. Long noncoding RNA CDKN2B-AS1 interacts with miR-411-3p to regulate ovarian cancer in vitro and in vivo through HIF-1a/VEGF/P38 pathway. Biochem Biophys Res Commun. 2019;514:44–50. doi: 10.1016/j.bbrc.2019.03.141. [DOI] [PubMed] [Google Scholar]
- 37.Shi L, He C, Li Z, Wang Z, Zhang Q. FBP1 modulates cell metabolism of breast cancer cells by inhibiting the expression of HIF-1α. Neoplasma. 2017;64:535–542. doi: 10.4149/neo_2017_407. [DOI] [PubMed] [Google Scholar]
- 38.Lou S, Wang Y, Yu Z, Guan K, Kan Q. Curcumin induces apoptosis and inhibits proliferation in infantile hemangioma endothelial cells via downregulation of MCL-1 and HIF-1α. Medicine (Baltimore) 2018;97:e9562. doi: 10.1097/MD.0000000000009562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang H, Li ZY, Xu ZH, Chen YL, Lu ZY, Shen DY, Lu JY, Zheng QM, Wang LY, Xu LW, et al. The prognostic value of miRNA-18a-5p in clear cell renal cell carcinoma and its function via the miRNA-18a-5p/HIF1A/PVT1 pathway. J Cancer. 2020;11:2737–2748. doi: 10.7150/jca.36822. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.