Abstract
Sorafenib is the first-line treatment for advanced hepatocellular carcinoma (HCC). Since many HCC patients experience drug resistance, there is an urgent need to discover more effective therapeutic strategies to overcome drug resistance. Long non-coding RNAs (lncRNAs) play an important role in tumor drug resistance. However, research on the role of lncRNA H19 in sorafenib resistance in HCC is quite limited. In the present study, CCK-8 assay, RT-qPCR, EdU staining, immunofluorescence staining, and western blot analysis were used to detect the effect of lncRNA H19 on sorafenib resistance of HCC cells. H19 expression was found to be negatively related to sorafenib sensitivity in HCC cells. Knockdown of lncRNA H19 elevated sorafenib sensitivity by suppressing epithelial-mesenchymal transition (EMT) in HCC cells. H19 upregulated miR-675 expression. miR-675 inhibitor decreased the cell viability in sorafenib-treated HCC cells, while miR-675 overexpression had the opposite effect on the treated cells. When the cells were pretreated with miR-675 mimic, H19 siRNA did not alter the effect of miR-675 on sorafenib sensitivity. In conclusion, our study provides new clues for further clinical treatment of sorafenib-resistant liver cancer patients.
Keywords: long non-coding RNA, H19, miR-675, chemosensitivity, epithelial-mesenchymal transition, hepatocellular carcinoma
Introduction
Hepatocellular carcinoma (HCC) is the most common primary malignant tumor of the liver. It is the fifth most common cancer in the word and the third most common cause of cancer-related death (1,2). HCC is commonly diagnosed at the advanced stage due to the asymptomatic features of early HCC, and patients with advanced HCC have a poor life expectancy with an average survival time of 7 months (3). Sorafenib, a multi-kinase inhibitor, is the only treatment option for advanced liver cancer (4). Although the use of sorafenib has improved the survival rate of patients with liver cancer, treatment resistance associated with sorafenib has become a major obstacle to treatment efficacy (2). Therefore, it is important to determine the molecular mechanisms underlying sorafenib resistance.
Long non-coding RNAs (lncRNAs) are non-protein-coding RNAs with more than 200 nucleotides. Many studies have shown that lncRNAs contribute to the drug resistance in many types of cancer (5,6). For example, lncRNA H19 contributes to 5-fluorouracil (5-FU) resistance in colorectal cancer by promoting autophagy (7). Downregulation of lncRNA HOTAIR was found to increase the sensitivity of cisplatin to ovarian cancer by inhibiting cisplatin-induced autophagy (8). H19 was demonstrated to mediate the chemosensitivity of breast cancer cells via the Wnt pathway and epithelial-mesenchymal transition (EMT) (9). lncRNA UCA1 confers tamoxifen resistance in endocrine therapy of breast cancer by regulating the EZH2/p21 axis and the PI3K/AKT signaling pathway (10). lncRNA HULC was found to attenuate the chemosensitivity of HCC cells by stabilizing Sirt1 to trigger autophagy (11). Among the lncRNAs, H19 was first described as an oncofetal transcript more than 30 years ago (12). Since its discovery, emerging evidence shows that H19 expression is upregulated in many types of cancer (7). However, research on lncRNA H19 in sorafenib resistance in HCC is quite limited.
The aim of the present study was to determine the mechanism of sorafenib resistance and to lay a foundation for achieving greater treatment efficacy. We focused on the mechanism by which lncRNA H19 regulates sorafenib resistance in liver cancer. Our findings provide new clues for further clinical treatment of patients with sorafenib-resistant liver cancer.
Materials and methods
Cell lines and transfection
Human HCC cell lines (Huh7, Hep3B, SNU-449, SNU-387) were purchased from the American Type Culture Collection (ATCC; Manassas, VA, USA). Huh7 was cultured in Dulbecco's modified Eagle's medium (DMEM, Gibco; Thermo Fisher Scientific, Inc.). Hep3B cells were cultured in Mimimum Essential Medium (MEM) (Gibco; Thermo Fisher Scientific, Inc.) SNU-387 and SNU-449 cells were cultured in RPMI-1640 medium (Gibco; Thermo Fisher Scientific, Inc.). All cells were placed in a humidified incubator containing 5% carbon dioxide (CO2) at 37°C with 10% fetal bovine serum (FBS, Gibco; Thermo Fisher Scientific, Inc.).
For transfection, H19 siRNA or the negative control were purchased from GenePharma (Shanghai, China). We used the mix of three sequences of siRNAs for knockdown. The sequences of the H19 siRNAs are as follows: H19-Homo-1552: Sense, 5′-CCCACAACAUGAAAGAAAUUU-3′ and antisense, 5′-AUUUCUUUCAUGUUGUGGGUU-3′; H19-Homo-1617: Sense, 5′-CCUCUAGCUUGGAAAUGAAUU-3′ and antisense, 5′-UUCAUUUCCAAGCUAGAGGUU-3′; and H19-Homo-1712: Sense, 5′-UCAUCAGCCCAACAUCAAAUU-3′ and antisense, 5′-UUUGAUGUUGGGCUGAUGAUU-3′; negative control (NC): Sense: 5′-UUCUCCGAACGUGUCACGUUU-3′ and antisense: 5′-ACGUGACACGUUCGGAGAAUU-3′; miR-675 mimic (5′-UGGUGCGGAGAGGGCCCACAGUG-3′ and 5′-CUGUGGGCCCUCUCCGCACCAUU-3′); inhibitor (5′-CACUGUGGGCCCUCUCCGCACCA-3′), and negative control were purchased by RIBO (Guangzhou, China). Transfections were achieved using Lipofectamine 2000 reagent (Life Technologies; Thermo Fisher Scientific, Inc.) according to the manufacturer's instructions. The transfected HCC cells were collected after 48 h of transfection for further experiments.
RT-qPCR
Total RNA was extracted from the cultured cells using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.), and RNA concentration was determined by spectrophotometry. Single-stranded cDNA was synthesized using a cDNA synthesis kit (Takara) according to the manufacturer's instructions. Reverse transcription-polymerase chain reaction assays were performed using Applied Biosystems SYBR Green Mix kits (Applied Biosystems). The amplification conditions were: 40 cycles of 5 sec at 95°C and 30 sec at 60°C. The 2−ΔΔCq method was used for quantitative gene expression (13). The following primers were used: H19-homo-F, 5′-TCCCAGAACCCACAACATGA-3′ and H19-homo-R, 5′-TTCACCTTCCAGAGCCGATT-3′; ACTB-homo-F, 5′-TGGCACCCAGCACAATGAA-3′ and ACTB-homo-R, 5′-CTAAGTCATAGTCCGCCTAGAAGCA-3′; Hsa-miR-675: 5′-TGGTGCGGAGAGGGCCCACAGTG-3′.
EdU assay
For the EdU staining assay, cells (1×105/well) were permeabilized and stained with Click-iT EdU Imaging Kit (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's instructions. Briefly, HCC cells under different conditions were incubated for 24 h. After removing the culture medium, the cells were fixed with 1 ml of 3.7% formaldehyde in phosphate-buffered saline (PBS) for 15 min and incubated with 0.5% Triton X-100 in PBS for 20 min. Then, the cells were incubated with 0.5 ml of Click-iT reaction cocktail at room temperature for 30 min in the dark. After removal of the reaction cocktail, cells were washed with 3% BSA in PBS followed by PBS. After addition of DAPI in PBS, cells were incubated for 20 min at room temperature in the dark. The cells were then washed with PBS, and images were obtained under an inverted fluorescence microscope (magnification ×200; Olympus Corp.).
Western blot analysis
Total proteins were extracted using RIPA buffer (Beyotimes Biotechnology, Guangzhou) and quantified using the bicinchoninic acid method. Proteins (40 µg/lane) were separated on a 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and then transferred to PVDF membranes (Millipore). The blots were incubated overnight with the following primary antibodies: E-cadherin (cat. no. 3195, dilution 1:1,000; Cell Signaling Technology, Inc.), vimentin (cat. no. 5741, dilution 1:1,000; Cell Signaling Technology, Inc.), and GAPDH (cat. no. 2118, dilution 1:1,000; Cell Signaling Technology, Inc.). After three washes, the membranes were incubated with a horseradish peroxidase-conjugated secondary antibody (anti-rabbit-HPR, cat. no. 7074, dilution 1:2,000; Cell Signaling Technology, Inc.) and visualized by electrochemiluminescence (ECL).
CCK-8 assay
Cell viability was detected using the Cell Counting Kit-8 (CCK-8) assay (Dojindo). Briefly, cells were seeded at 5,000 cells per well in 96-well plates and cultured for 12 h at 37°C. The cells were then subjected to different concentrations of sorafenib for 48 h. Then CCK-8 solution (10 µl/ well) was added to each well and the cells were incubated for additional 2–4 h at 37°C. The values of absorbance at 490 nm were measured using a microplate reader (Bio-Tek Instruments, Inc.).
Statistical analysis
Data are expressed as the mean ± SD. All statistical analyses were performed using GraphPad 6.0 statistical software (GraphPad Software, Inc.). Differences between two groups were analyzed using the Student's t-test. One-way analysis of variance (ANOVA) with Tukey's post hoc test was used to analyze differences among multiple groups. P<0.05 was assigned to indicate a statistical significance.
Results
Expression of H19 correlates with sorafenib sensitivity in HCC cells
We collected samples from 18 patients with HCC and examined the expression of H19 by RT-qPCR. We found significant upregulation of H19 in HCC tissue samples, compared with that noted in the matched adjacent normal tissues (Fig. S1A). In order to investigate the role of lncRNA H19 in drug resistance, HCC cells were treated with various concentrations of sorafenib for 48 h, and the CCK-8 assay was then performed. IC50 values of sorafenib in SNU-387 and SNU-449 cells were higher than these values in the Huh7 and Hep3B cells (Fig. 1A and B). Meanwhile, we detected the level of H19 in HCC cell lines. The results revealed that the expression of H19 in SNU-387 and SNU-449 cells was higher than that in the Huh7 and Hep3B cells (Fig. 1C). Compared with the untreated control, the EdU-positive cell ratio in all four HCC cell lines exposed to sorafenib was significantly lower (Fig. 1D). The EdU-positive cell ratios in the SNU-387 and SNU-449 cells with higher H19 expression were less than that in the Huh7 and Hep3B cells with lower H19 expression. These results show that H19 expression is negatively related to sorafenib sensitivity in HCC cells, suggesting that H19 is an oncogene.
Figure 1.
H19 expression is negatively related to sorafenib sensitivity in HCC cell linws (Huh7, Hep3B, SNU-449, SNU-387). (A) Cell viability was examined by CCK-8 assay in four HCC cell lines subjected to sorafenib treatment. (B) Statistical analysis of the IC50 in four HCC cell lines subjected to sorafenib treatment. (C) Relative mRNA levels of H19 as measured by RT-qPCR and normalized to β-actin in four HCC cell lines. (D) Cell proliferation was examined by EdU assay in four HCC cell lines subjected to sorafenib treatment. **P<0.01, ***P<0.001, compared to the control. HCC, hepatocellular carcinoma; IC50, half maximal inhibitory concentration.
H19 knockdown sensitizes HCC cells to sorafenib in vitro
To further investigate the effect of H19 on the chemoresistance of HCC cells against sorafenib, four HCC cell lines were transfected with H19 siRNA. CCK-8 and EdU assays were utilized to evaluate cell proliferative capacity. As shown in Fig. 2A, there was no significant difference in sensitivity to sorafenib between the control group and the NC siRNA group for all cell lines. The H19 siRNA groups showed significantly higher sensitivity to sorafenib than the control group and the NC siRNA group for all four HCC cell lines. Consistent with the CCK-8 results, H19 downregulation attenuated the EdU-positive cell ratio. The EdU-positive cell ratio in the H19 siRNA group was lower than that in the control group and NC siRNA group in all cell lines (Fig. 2B). H19 knockdown efficiency was evaluated by RT-qPCR (Fig. 2C). These results suggest that H19 knockdown sensitizes HCC cells to sorafenib in vitro.
Figure 2.
H19 knockdown sensitizes HCC cells to sorafenib in vitro. (A) Sorafenib sensitivity of HCC cells transfected with NC siRNA or H19 siRNA. (B) Proliferative capacity of HCC cells transfected with NC siRNA or H19 siRNA and subjected to sorafenib treatment. **P<0.01, ***P<0.001, compared with the sorafenib group. (C) H19 expression in HCC cells transfected with NC siRNA or H19 siRNA and subjected to sorafenib treatment. *P<0.05, **P<0.01, ***P<0.001, compared to the control. HCC, hepatocellular carcinoma; NC, negative control.
H19 knockdown suppresses epithelial-mesenchymal transition (ETM)
To investigate the mechanism by which H19 knockdown sensitizes HCC cells to sorafenib, immunofluorescence staining and western blot analysis were performed. HCC cells were transfected with NC siRNA or H19 siRNA. We found that siRNA H19-transfected cells showed stronger E-cadherin immunofluorescence compared with the untreated control and NC siRNA-transfected cells, while vimentin immunofluorescence was less than that noted in the untreated control and NC siRNA-transfected cells (Fig. 3A). Consistent with immunofluorescence results, western blot analysis showed that silencing of H19 by siRNA transfection resulted in a significant increase in E-cadherin and a decrease in vimentin in all four HCC cell lines examined (Fig. 3B). Moreover, the results of the migration and invasion assays revealed that H19 siRNA exerted a distinct inhibitory effect on the migration distance and cell numbers that had migrated in the four HCC cell lines (Fig. S2). These findings indicate that knockdown of lncRNA H19 may enhance sorafenib sensitivity in HCC cells by inhibiting EMT.
Figure 3.
H19 knockdown inhibits EMT and H19 positively regulates miR-675. (A) Immunofluorescence staining showed that E-cadherin (green) and vimentin (green) levels were altered after transfection of NC siRNA or H19 siRNA in the SNU-449 and SNU-387 cells. (B) E-cadherin and vimentin as assessed by western blot analysis. GAPDH was used as loading control. (C) Relative mRNA levels of miR-675 were measured by RT-qPCR and normalized to U6 after transfection of NC siRNA or H19 siRNA in four HCC cell lines. *P<0.05, **P<0.01, ***P<0.001, compared to the control. HCC, hepatocellular carcinoma; NC, negative control; EMT, epithelial-mesenchymal transition.
As previously reported, H19 was found to play a role in breast cancer by upregulating the expression of miR-675 (14). To study the effect of miR-675 on HCC, we first detected miR-675 expression in the collected samples. Notably, the expression of miR-675 in HCC tissues was significantly higher than that in normal tissues (Fig. S1B). Our results showed that H19 positively regulates miR-675. We next evaluated the level of miR-675 in HCC cells after transfection with H19 siRNA. The level of miR-675 in the siRNA H19-transfected group was significantly lower than that noted in the control group and NC group, indicating that miR-675 may be a target gene of lncH19 (Fig. 3C).
miR-675 regulates the sensitivity of HCC cells to sorafenib
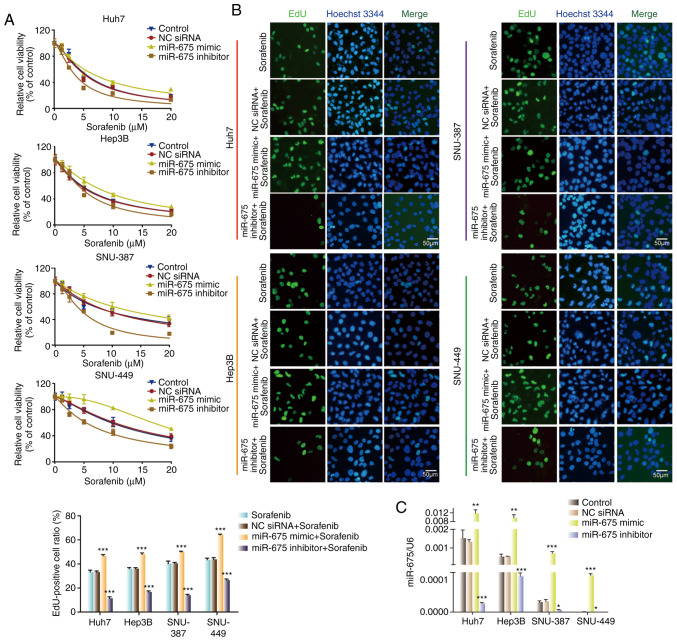
To investigate the function of miR-675 on sorafenib sensitivity of HCC cells, we transfected the four HCC cell lines with miR-675 mimic, inhibitor, or negative control. miR-675 mimic upregulated miR-675 expression while the inhibitor downregulated its expression (Fig. 4C). CCK-8 and EdU assays were used to assess the impact of miR-675 on cell proliferation. As shown in Fig. 4A and B, miR-675 mimic increased the cell viability and the number of EdU-positive cells compared with the control and NC groups. In contrast, miR-675 inhibitor reduced the cell viability and the number of EdU-positive cells compared with the control and NC groups. These results showed that miR-675 is involved in the regulation of cell sensitivity to sorafenib in HCC cells.
Figure 4.
miR-675 regulates sensitivity of HCC cells to sorafenib. (A) Sorafenib sensitivity of HCC cells transfected with NC siRNA, miR-675 mimic, or miR-675 inhibitor. (B) Proliferation of HCC cells transfected with NC siRNA, miR-675 mimic, or miR-675 inhibitor in the presence of sorafenib. ***P<0.001, compared with the sorafenib group. (C) Expression of miR-675 in HCC cells transfected with NC siRNA, miR-675 mimic, or miR-675 inhibitor in the presence of sorafenib. *P<0.05, **P<0.01, ***P<0.001. HCC, hepatocellular carcinoma; NC, negative control.
miR-675 influences epithelial-mesenchymal transition (ETM)
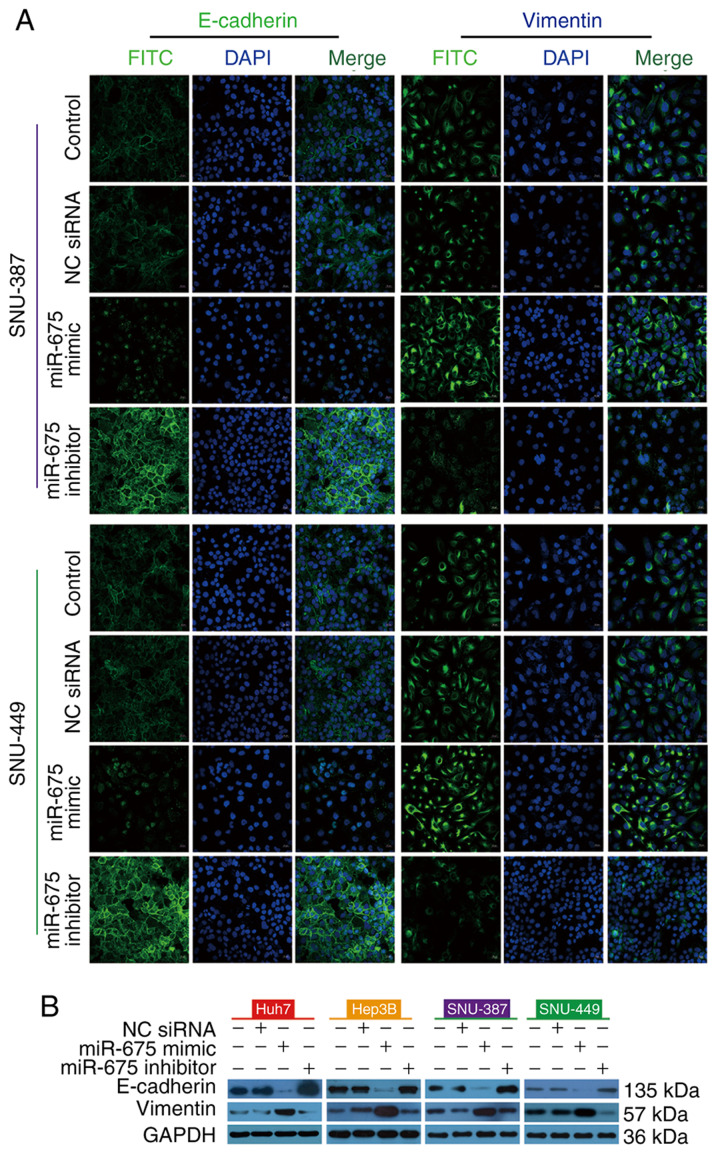
To investigate the effect of miR-675 on EMT, we transfected HCC cells with miR-675 mimic, inhibitor, or negative control. Immunofluorescence staining showed that miR-675 mimic reduced E-cadherin immunofluorescence and enhanced vimentin immunofluorescence. In contrast, miR-675 inhibitor increased E-cadherin immunofluorescence and decreased vimentin immunofluorescence (Fig. 5A). Consistent with the results of immunofluorescence staining, miR-675 mimic reduced the expression of E-cadherin protein and enhanced the expression of vimentin protein in the four HCC lines while a miR-675 inhibitor had the opposite effect (Fig. 5B). Taken together, these findings suggest that overexpression of miR-675 promotes EMT while inhibition of miR-675 can suppress EMT.
Figure 5.
miR-675 alters EMT. (A) Immunofluorescence showed that the staining of E-cadherin (green) and vimentin (green) was altered after transfection with the miR-675 mimic, inhibitor, or NC siRNA in SNU-449 and SNU-387 cells. (B) E-cadherin and vimentin levels in HCC cells transfected with NC siRNA, miR-675 mimic, or miR-675 inhibitor in the presence of sorafenib. HCC, hepatocellular carcinoma; NC, negative control; EMT, epithelial-mesenchymal transition.
miR-675 mediates the regulatory effect of lncRNA H19 on sorafenib sensitivity
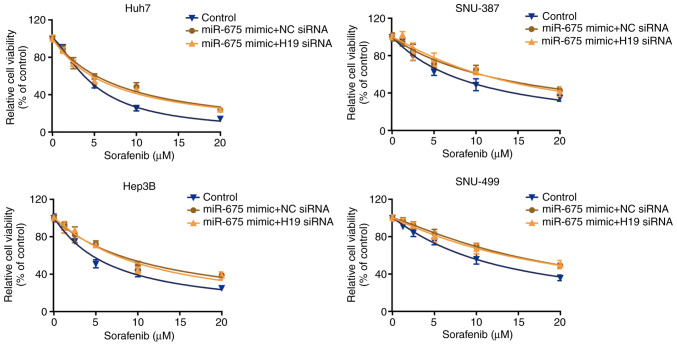
To examine whether miR-675 participates in the regulatory effect of lncRNA H19 on sensitivity to sorafenib, we transfected the four cell lines with miR-675 mimic, and then treated them with H19 siRNA. We found that H19 siRNA did not change the effect of miR-675 on sorafenib sensitivity, which was confirmed by CCK-8 assay (Fig. 6). These data indicate that miR-675 mediates the regulatory effect of lncRNA H19 on sorafenib sensitivity.
Figure 6.
miR-675 mediates the regulatory effect of lncRNA H19 on sorafenib sensitivity. Sorafenib sensitivity of HCC cells transfected with NC siRNA or H19 siRNA was evaluated by CCK-8 assay in the presence of the miR-675 mimic for 48 h. HCC, hepatocellular carcinoma; NC, negative control.
Discussion
Our findings revealed that H19 expression is negatively related to sorafenib sensitivity in hepatocellular carcinoma (HCC) cells, and that knockdown of H19 could sensitize HCC cells to sorafenib. Mechanistically, the results demonstrated that long non-coding RNA (lncRNA) H19 knockdown can suppress epithelial-mesenchymal transition (EMT) to enhance the sensitivity of HCC cells to sorafenib. Moreover, we found that knockdown of H19 was associated with an obvious reduction in the expression level of miR-675 in HCC cells. miR-675 mimic transfection promoted sorafenib resistance in the HCC cells by enhancing EMT, while the miR-675 inhibitor had the opposite effect. H19 knockdown did not influence the effect of miR-675 on sorafenib sensitivity in HCC cells.
lncRNA H19 is located on chromosome 11 in humans and has been characterized as an oncogenic lncRNA in a few types of cancer due to its promotion of cell proliferation and chemoresistance (15–17). In the present study, we first found that H19 expression is negatively related to sorafenib sensitivity in HCC cells based on CCK-8 and EdU assays. After transfection of H19 siRNA, we observed that the sensitivity of the liver cancer cells to sorafenib was significantly increased. These data suggest that H19 is an oncogene.
Epithelial-mesenchymal transition (EMT) is a biologic process whereby polarized epithelial cells lose epithelial characteristics and develop a mesenchymal phenotype. In EMT, cell adhesion molecules (such as E-cadherin) are lost while mesenchymal markers (such as vimentin) are induced, leading to the loss of polarity and enhancement of tumor cell migration and invasion. As a result, tumor cells become more insensitive to antitumor drugs (2,18,19). Many reports have shown that tumor drug resistance is frequently accompanied by EMT in different types of cancer, including lung cancer (20), pancreatic cancer (21), gastric cancer (22), and breast cancer (23). In HCC, long-term exposure of liver cancer cells to sorafenib can induce epithelial-mesenchymal transition to develop resistance, increase the risk of invasive and rebound growth (24). PDCD2 sensitizes HepG2 cells to sorafenib by inhibiting mesenchymal transformation of epithelial cells (25). Therefore, we hypothesized that knockdown of H19 would sensitize HCC cells to sorafenib via EMT. To test this hypothesis, we detected the levels of EMT-related proteins after silencing of H19. The results showed that E-cadherin expression was increased and vimentin expression was decreased by both immunofluorescence and western blot analysis, indicating that EMT was suppressed.
MicroRNAs (miRNAs) are a class of single-stranded non-coding RNAs of 18–22 nucleotides that regulate gene expression at the post-transcriptional level by targeting and degrading specific mRNAs (26,27). In many cancers, miRNAs can serve as oncogenes or tumor-suppressor genes and participate in chemosensitivity. For example, microRNA-31-5p regulates chemosensitivity by blocking the nuclear location of PARP1 in hepatocellular carcinoma (27). miR-20a-5p regulates gemcitabine chemosensitivity by targeting RRM2 in pancreatic cancer cells and serves as a predictor for gemcitabine-based chemotherapy (28). miR-494 was found to increase the chemosensitivity to doxorubicin of gastric cancer cells by targeting phosphodiesterase 4D (29).
Accumulating evidence suggests that H19 may function as a reservoir of miR-675. Li et al found that miR-675 is embedded in the first exon of the H19 transcript (30). Smits et al demonstrated that H19 is the primary miRNA precursor of miR-675 in both humans and mice (31). Similarly, it was also reported that transfection with H19 complementary DNA containing the pri-miR-675 hairpin increased the expression of mature miR-675 in human kidney 293T cells (32). Therefore, we performed RT-qPCR analysis to detect the level of miR-675 after H19 silencing, and found that H19 knockdown decreased the expression of miR-675. We then examined whether miR-675 regulates the sensitivity of HCC cells to sorafenib. Transfection of miR-675 mimic made HCC cells insensitive to sorafenib, as shown by CCK-8 and EdU assays. In contrast, miR-675 inhibitor sensitized HCC cells to sorafenib. Mechanistically, miR-675 alters chemosensitivity via EMT. Finally, to confirm that H19 regulates sorafenib resistance through miR-675, we transfected four HCC cell lines with miR-675 mimic and then treated them with H19 siRNA. The results showed that there was no significant difference between the miR-675 group and the H19+miR-675 group.
There are some limitations in the present study. Firstly, H19 overexpression experiments were previously attempted but the plasmid was not constructed successfully. Secondly, in vivo experiments were attempted but it was not possible to successfully establish the model. We think the limitation of the technical condition may be the main reason. In addition, we doubt that the cell viability could be insufficient to initiate tumorigenesis in vivo. Third, considering there were many studies to report that H19 is the precursor of miR-675, we simply verified these findings in HCC but did not investigate a direct regulatory relationship between H19 and miR-675. Further investigations may resolve these issues.
In conclusion, this study showed that knockdown of H19 sensitized HCC cells to sorafenib by downregulating miR-675, thereby preventing EMT. Thus, these findings provide evidence to define H19 as a potential therapeutic target for hepatocellular carcinoma.
Supplementary Material
Acknowledgements
Not applicable.
Funding
The present study was supported by grants from the Natural Science Foundation of Henan Province (nos. 162300410274 and 182300410298), and the National Natural Science Foundation of China (no. 81273260).
Availability of data and material
All data generated or analyzed during this study are included in this published article.
Authors' contributions
Conceptualization of the research study was carried out by FX and WC. The research methodology was designed by JS and JM. Software analysis of the data was carried out by XL. Validation of the results was carried out by LY. Data curation was conducted by ZL and HL. Writing of the original draft preparation was carried out by YX and writing, review and editing of the manuscript was done by YL. All authors read and approved the manuscript and agree to be accountable for all aspects of the research in ensuring that the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Ethics approval and consent to participate
The present study was approved by the Ethics Committee of Henan Provincial People's Hospital and written informed consent was obtained from all patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.El-Serag HB, Rudolph KL. Hepatocellular carcinoma: Epidemiology and molecular carcinogenesis. Gastroenterology. 2007;132:2557–2576. doi: 10.1053/j.gastro.2007.04.061. [DOI] [PubMed] [Google Scholar]
- 2.Zhu YJ, Zheng B, Wang HY, Chen L. New knowledge of the mechanisms of sorafenib resistance in liver cancer. Acta Pharmacol Sin. 2017;38:614–622. doi: 10.1038/aps.2017.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Giannini EG, Farinati F, Ciccarese F, Pecorelli A, Rapaccini GL, Di Marco M, Benvegnù L, Caturelli E, Zoli M, Borzio F, et al. Prognosis of untreated hepatocellular carcinoma. Hepatology. 2015;61:184–190. doi: 10.1002/hep.27443. [DOI] [PubMed] [Google Scholar]
- 4.Lai HH, Li CW, Hong CC, Sun HY, Chiu CF, Ou DL, Chen PS. TARBP2-mediated destabilization of Nanog overcomes sorafenib resistance in hepatocellular carcinoma. Mol Oncol. 2019;13:928–945. doi: 10.1002/1878-0261.12449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ma H, Yuan L, Li W, Xu K, Yang L. The LncRNA H19/miR-193a-3p axis modifies the radio-resistance and chemotherapeutic tolerance of hepatocellular carcinoma cells by targeting PSEN1. J Cell Biochem. 2018;119:8325–8335. doi: 10.1002/jcb.26883. [DOI] [PubMed] [Google Scholar]
- 6.Wang J, Lv B, Su Y, Wang X, Bu J, Yao L. Exosome-mediated transfer of lncRNA HOTTIP promotes cisplatin resistance in gastric cancer cells by regulating HMGA1/miR-218 axis. Onco Targets Ther. 2019;12:11325–11338. doi: 10.2147/OTT.S231846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang M, Han D, Yuan Z, Hu H, Zhao Z, Yang R, Jin Y, Zou C, Chen Y, Wang G, et al. Long non-coding RNA H19 confers 5-Fu resistance in colorectal cancer by promoting SIRT1-mediated autophagy. Cell Death Dis. 2018;9:1149. doi: 10.1038/s41419-018-1187-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yu Y, Zhang X, Tian H, Zhang Z, Tian Y. Knockdown of long non-coding RNA HOTAIR increases cisplatin sensitivity in ovarian cancer by inhibiting cisplatin-induced autophagy. J BUON. 2018;23:1396–1401. [PubMed] [Google Scholar]
- 9.Gao H, Hao G, Sun Y, Li L, Wang Y. Long noncoding RNA H19 mediated the chemosensitivity of breast cancer cells via Wnt pathway and EMT process. Onco Targets Ther. 2018;11:8001–8012. doi: 10.2147/OTT.S172379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li Z, Yu D, Li H, Lv Y, Li S. Long non-coding RNA UCA1 confers tamoxifen resistance in breast cancer endocrinotherapy through regulation of the EZH2/p21 axis and the PI3K/AKT signaling pathway. Int J Oncol. 2019;54:1033–1042. doi: 10.3892/ijo.2019.4679. [DOI] [PubMed] [Google Scholar]
- 11.Xiong H, Ni Z, He J, Jiang S, Li X, He J, Gong W, Zheng L, Chen S, Li B, et al. LncRNA HULC triggers autophagy via stabilizing Sirt1 and attenuates the chemosensitivity of HCC cells. Oncogene. 2017;36:3528–3540. doi: 10.1038/onc.2016.521. [DOI] [PubMed] [Google Scholar]
- 12.Pachnis V, Belayew A, Tilghman SM. Locus unlinked to alpha-fetoprotein under the control of the murine raf and Rif genes. Proc Natl Acad Sci USA. 1984;81:5523–5527. doi: 10.1073/pnas.81.17.5523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 14.Müller V, Oliveira-Ferrer L, Steinbach B, Pantel K, Schwarzenbach H. Interplay of lncRNA H19/miR-675 and lncRNA NEAT1/miR-204 in breast cancer. Mol Oncol. 2019;13:1137–1149. doi: 10.1002/1878-0261.12472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ding D, Li C, Zhao T, Li D, Yang L, Zhang B. LncRNA H19/miR-29b-3p/PGRN axis promoted epithelial-mesenchymal transition of colorectal cancer cells by acting on Wnt signaling. Mol Cells. 2018;41:423–435. doi: 10.14348/molcells.2018.2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lei Y, Guo W, Chen B, Chen L, Gong J, Li W. Tumor-released lncRNA H19 promotes gefitinib resistance via packaging into exosomes in non-small cell lung cancer. Oncol Rep. 2018;40:3438–3446. doi: 10.3892/or.2018.6762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Duan S, Li M, Wang Z, Wang L, Liu Y. H19 induced by oxidative stress confers temozolomide resistance in human glioma cells via activating NF-κB signaling. OncoTargets Ther. 2018;11:6395–6404. doi: 10.2147/OTT.S173244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maheswaran T, Rushbrook SM. Epithelial-mesenchymal transition and the liver: Role in hepatocellular carcinoma and liver fibrosis. J Gastroenterol Hepatol. 2012;27:418–420. doi: 10.1111/j.1440-1746.2012.07060.x. [DOI] [PubMed] [Google Scholar]
- 19.Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest. 2009;119:1420–1428. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fukuda K, Takeuchi S, Arai S, Katayama R, Nanjo S, Tanimoto A, Nishiyama A, Nakagawa T, Taniguchi H, Suzuki T, et al. Epithelial-to-mesenchymal transition is a mechanism of ALK inhibitor resistance in lung cancer independent of ALK mutation status. Cancer Res. 2019;79:1658–1670. doi: 10.1158/0008-5472.CAN-18-2052. [DOI] [PubMed] [Google Scholar]
- 21.Gaianigo N, Melisi D, Carbone C. EMT and treatment resistance in pancreatic cancer. Cancers (Basel) 2017;9:E122. doi: 10.3390/cancers9090122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sun J, Xu Z, Lv H, Wang Y, Wang L, Ni Y, Wang X, Hu C, Chen S, Teng F, et al. eIF5A2 regulates the resistance of gastric cancer cells to cisplatin via induction of EMT. Am J Transl Res. 2018;10:4269–4279. [PMC free article] [PubMed] [Google Scholar]
- 23.Dong H, Hu J, Zou K, Ye M, Chen Y, Wu C, Chen X, Han M. Activation of LncRNA TINCR by H3K27 acetylation promotes trastuzumab resistance and epithelial-mesenchymal transition by targeting MicroRNA-125b in breast cancer. Mol Cancer. 2019;18:3. doi: 10.1186/s12943-018-0931-9. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 24.van Malenstein H, Dekervel J, Verslype C, Van Cutsem E, Windmolders P, Nevens F, van Pelt J. Long-term exposure to sorafenib of liver cancer cells induces resistance with epithelial-to-mesenchymal transition, increased invasion and risk of rebound growth. Cancer Lett. 2013;329:74–83. doi: 10.1016/j.canlet.2012.10.021. [DOI] [PubMed] [Google Scholar]
- 25.Liu H, Wang M, Liang N, Guan L. PDCD2 sensitizes HepG2 cells to sorafenib by suppressing epithelial mesenchymal transition. Mol Med Rep. 2019;19:2173–2179. doi: 10.3892/mmr.2019.9860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang XH, Wang Q, Chen JS, Fu XH, Chen XL, Chen LZ, Li W, Bi J, Zhang LJ, Fu Q, et al. Bead-based microarray analysis of microRNA expression in hepatocellular carcinoma: MiR-338 is downregulated. Hepatol Res. 2009;39:786–794. doi: 10.1111/j.1872-034X.2009.00502.x. [DOI] [PubMed] [Google Scholar]
- 27.Que KT, Zhou Y, You Y, Zhang Z, Zhao XP, Gong JP, Liu ZJ. MicroRNA-31-5p regulates chemosensitivity by preventing the nuclear location of PARP1 in hepatocellular carcinoma. J Exp Clin Cancer Res. 2018;37:268. doi: 10.1186/s13046-018-0930-0. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 28.Lu H, Lu S, Yang D, Zhang L, Ye J, Li M, Hu W. MiR-20a-5p regulates gemcitabine chemosensitivity by targeting RRM2 in pancreatic cancer cells and serves as a predictor for gemcitabine-based chemotherapy. Biosci Rep. 2019;39:BSR20181374. doi: 10.1042/BSR20181374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peng QP, Du DB, Ming Q, Hu F, Wu ZB, Qiu S. MicroRNA 494 increases chemosensitivity to doxorubicin in gastric cancer cells by targeting phosphodiesterases 4D. Cell Mol Biol (Noisy-le-grand) 2018;64:62–66. doi: 10.14715/cmb/2017.64.15.10. [DOI] [PubMed] [Google Scholar]
- 30.Li DY, Busch A, Jin H, Chernogubova E, Pelisek J, Karlsson J, Sennblad B, Liu S, Lao S, Hofmann P, et al. H19 induces abdominal aortic aneurysm development and progression. Circulation. 2018;138:1551–1568. doi: 10.1161/CIRCULATIONAHA.117.032184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smits G, Mungall AJ, Griffiths-Jones S, Smith P, Beury D, Matthews L, Rogers J, Pask AJ, Shaw G, VandeBerg JL, et al. Conservation of the H19 noncoding RNA and H19-IGF2 imprinting mechanism in therians. Nat Genet. 2008;40:971–976. doi: 10.1038/ng.168. [DOI] [PubMed] [Google Scholar]
- 32.Cai X, Cullen BR. The imprinted H19 noncoding RNA is a primary microRNA precursor. RNA. 2007;13:313–316. doi: 10.1261/rna.351707. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article.