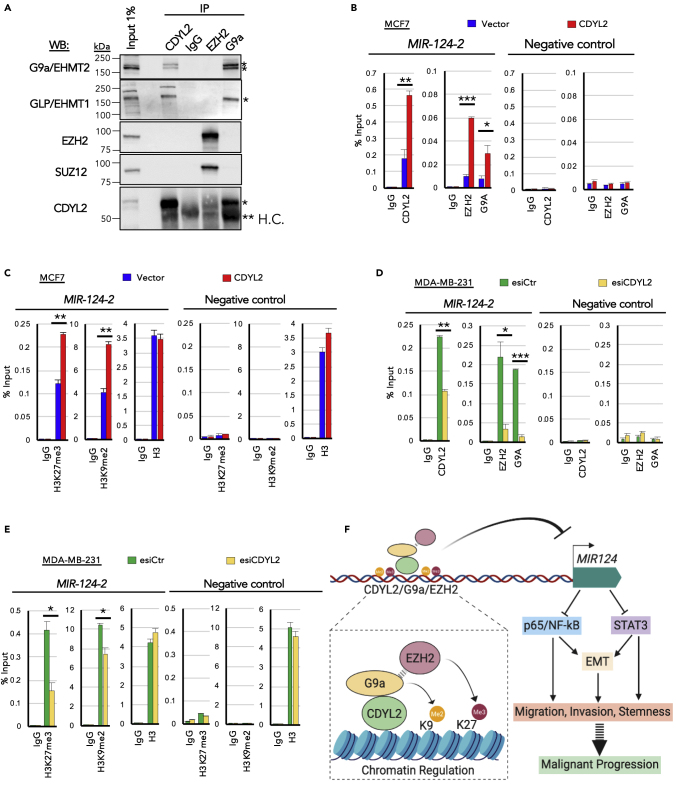
Figure 7.
CDYL2 Interaction with G9a, GLP, EZH2, and SUZ12 and Its Regulation of G9a, EZH2, H3K9me2, and H3K27me3 Levels Upstream of MIR124-2
(A) Immunoprecipitation (IP) of CDYL2, EZH2, and G9a was performed on MCF7 cell lysates and the presence of the indicated proteins in the resulting IP eluates determined by western blotting (WB). A non-specific IgG was used as negative control. Input lysate was used to assess the relative strength of each coIP signal. The experiment was repeated three times with similar results. (∗specific band; ∗∗H.C., IgG heavy chain).
(B) ChIP-qPCR analysis of the relative occupancy of CDYL2, EZH2, and G9a upstream of MIR124-2 in MCF7-CDYL2 compared with MCF7-Vector cells. IgG, negative control ChIP. qPCR analysis was also performed at an unrelated negative control sequence. Shown is the mean enrichment as a percentage of input of three independent experiments, ± SD. Significance was determined by t test (∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001).
(C) As in (B), except using antibodies specific to H3K9me2, H3K27me3, H3, and IgG. These ChIP analyses were conducted using the same lysates as in (B), so are paired analyses.
(D and E) Experiments were conducted as described in (B) and (C), except using chromatin lysates prepared from MDA-MB-231 cells treated with esiCDYL2 or esiLuc.
(F) Schematic model of the proposed contribution of CDYL2 to epigenetic regulation of MIR124, cell signaling, and malignancy-associated cellular processes.