Abstract
Backgrounds
Ovarian cancer is one of the most common gynecological malignancies and mortality ranks the highest in cancer-associated death in females’ worldwide. Here, we attempted to evaluate the effect of DANCR on the biological behavior of transforming growth factor-β (TGF-β) stimulated ovarian cancer cells.
Material/Methods
The expression of DANCR in ovarian cancer cells (A2780 and SKOV3) treated with TGF-β were detected by quantitative real-time polymerase chain reaction (qRT-PCR). DANCR silencing was constructed using lentiviral transfection in ovarian cancer cells. The Cell Counting Kit-8 (CCK-8), flow cytometry and Transwell assays were performed to measure some cytology index. Western blot was utilized to explore the effect of DANCR on Krüppel-like factor 5 (KLF5) expression.
Results
The expression of DANCR in cancer cells (A2780 and SKOV3) treated with TGF-β was significantly higher. DANCR silencing suppressed cell viability, migration and invasion, and induced cell apoptosis of TGF-β treated ovarian cancer cells. Bioinformatics analysis showed that DANCR served as a sponge for miR-214, and also showed that KLF5 was targeted by miR-214. In addition, DANCR could inhibit the expression of KLF5.
Conclusions
We are the first to report that knockdown of DANCR could affect the biological process of ovarian cancer cells treated with TGF-β by sponging miR-214, which may provide new therapeutic ideas of ovarian cancer.
MeSH Keywords: MicroRNAs; Ovarian Neoplasms; RNA, Long Noncoding
Background
Ovarian cancer is the most lethal malignancy of the female reproductive system and mortality ranks the highest in cancer-associated death in females’ worldwide [1]. Despite great advances in ovarian cancer diagnostic and therapeutic strategies, the clinical prognosis of patients remains unfavorable, with a 5-year survival rate of 10–30% [2]. Hence, it is of important clinical significance to explore the molecular mechanisms involved in ovarian carcinogenesis and diagnosis.
As a set of non-coding RNA, long noncoding RNAs (lncRNAs) are characterized by no or limited protein-coding potential [3]. LncRNAs have been verified to function in diverse cellular processes including cell cycle, autophage, and apoptosis [4]. Accumulated evidence has shown that aberrant expression of lncRNAs is associated with the progression of ovarian cancer. For instance, DQ786243 aggravates ovarian cancer development [5]. Meanwhile, FEZF1-AS1 has been shown to exert an oncogenic role in ovarian cancer [6]; and lncRNA HOTTIP aggravates the progression of ovarian cancer [7]. DANCR (differentiation antagonizing non-protein coding RNA) has been reported to interfere with the progress of cell differentiation [8].
Transforming growth factor (TGF) is a complex multi-functional cytokine, which is widely involved in various pathophysiological processes in mammals [9]. In recent years, many studies have reported that TGF-β plays an extremely complex role in cancer initiation and development. In early carcinogenesis, TGF-β may repress cell proliferation, but promoted tumor invasion and metastasis through various mechanisms in the progressive stage [10–12]. Previous research has demonstrated the facilitation effect of TGF-β on the malignant biological behavior of ovarian cancer cells [13–15]. Additionally, DANCR level was reported to positively correlated with TGF-β receptor expression in cervical cancer cell [16]. However, whether DANCR could interfere with the promotion effect of TGF-β in ovarian cancer cell remains still unclear. In the current study, we attempted to investigate the effect of DANCR on the biological behavior of TGF-β stimulated ovarian cancer cells, and the underlying mechanism was further elucidated.
Material and Methods
Cell culture and transfection
Two ovarian cancer cell lines (A2780 and SKOV3) were attained from the Cell Bank of the Chinese Academy (Shanghai, China). All cells were maintained in DMEM (Hyclone, South Logan, UT, USA) containing 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin under an atmosphere of a humidified air and 5% CO2 at 37°C. For TGF-β treatment, the cells were serum-starved for 12 hours and then treated with 10 ng/mL TGF-β for 48 hours as previously described [14]. DANCR small interfering RNA (si-DANCR), miR-214 inhibitor, as well as negative controls were all designed by GenePharma (Shanghai, China). After cultured to a confluency of 50–60%, cells were transfected with si-DANCR and miR-214 inhibitor alone or in combination with Lipofectamine 2000 (Invitrogen, MA, USA) in accordance with manufacturer’s protocol.
Cell viability
A total of 2×103 cells of A2780 and SKOV3 cell line were planted in 96-well plate and cultured in a humid atmosphere with 5% CO2 at 37°C for 72 hours. Next, 10 μL Cell Counting Kit-8 (CCK-8) solution (Beyotime, Shanghai, China) was added into each well for 3 hours incubation. The absorbance of each well at 490 nm was measured at 24, 48, and 72 hours using FLx800 Fluorescence Microplate Reader (Biotek, USA).
Cell migration and invasion
For cell migration assays, 1×105 A2780 and SKOV3 cells were resuspended in serum-free medium (Gibco; Thermo Fisher Scientific, Inc.) and then plated into the upper chambers. Then 20% FBS (Gibco; Thermo Fisher Scientific, Inc.) was added to the lower chamber. For the invasion assay, Transwell inserts (Fisher Scientific, Waltham, MA, USA) were coated with Matrigel (BD, Franklin Lakes, NJ, USA). Stained cells were counted, and images were captured with an Olympus BX51 light microscope (magnification, 200×; Olympus Corporation).
Cell apoptosis
After stained with FITC/Annexin V and propidium iodide (PI), cells apoptosis detection was immediately performed using a FACScan (Beckman Coulter, Fullerton, CA, USA) and analyzed using CellQuest software.
Quantitative real time polymerase chain reaction (qRT-PCR) analysis
Total RNA was isolated from A2780 and SKOV3 cells using the TRIzol kit (Invitrogen, Carlsbad, CA, USA). Real-time polymerase chain reaction (RT-PCR) was performed using the Applied Biosystems 7500 Real-time PCR System (Applied Biosystem). The primers were as follows:
DANCR forward 5′‐GCGCCACTATGTAGCGGGTT‐3′ and reverse 5′‐TCAATGGCTTGTGCCTGTAGTT‐3′;
miR-214 forward 5′‐CTCAACTGGTGTCGTGGAGTCGGCAATTCA GTTGAGACTGCCTG‐3′ and reverse 5′‐ACACTCCAGCTGGGACAGCAGGCACA‐3′;
Krüppel-like factor 5 (KLF5) forward 5′-ATCGAGATGTTCGCTCGTGC-3′ and reverse 5′‐TTTAAAGGCAGACACTGAGTCAG‐3′;
U6 forward 5′‐GCTCGCTTCGGCAGCACA‐3′ and reverse 5′‐GAGGTA TTCGCA CCAGAG GA‐3′; and GAPDH forward 5′‐ACCACA GTCCATGCCATCCAC‐3′ and reverse 5′‐TCCACCACCCTGTTGCTGTA‐3′. The relative expression was analysis by using 2−ΔΔCT method and normalized to GAPDH or U6.
Western blotting analysis
Equal quantities of protein were separated via sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS‐PAGE) on a 10% gel and transferred to polyvinylidene fluoride (PVDF) membranes. After blocking with 5% skimmed milk, the membrane was incubated with the primary antibodies anti-KLF5 (ab230861) at 4°C overnight, followed by incubating with anti-rabbit horseradish peroxidase-conjugated secondary antibody and the reaction detected using enhanced chemiluminescence detection system.
Luciferase reporter assay
DANCR fragments containing wild type or mutant miR-214 binding sites was constructed by Genomeditech (Shanghai, China) and inserted into pGL3 Basic vector. Cells were co-transfected with the luciferase reporter constructs, miR-630 mimics, and Renilla luciferase construct (Promega). After 12 hours cultivation, the relative luciferase activities were measured using the Dual-Luciferase Reporter System (Promega).
Statistical analysis
Differences were calculated with Student’s t-test or one-way ANOVA. Pearson’s correlation analysis was used to analyze the expression correlation. All measurement data were expressed as the mean±standard deviation. All statistical analyses were utilized the SPSS 17.0 software and GraphPad Prism 6. A statistically significant difference was defined as P<0.05.
Results
DANCR silencing suppressed cell viability, migration and invasion, induced cell apoptosis of TGF-β treated ovarian cancer cells
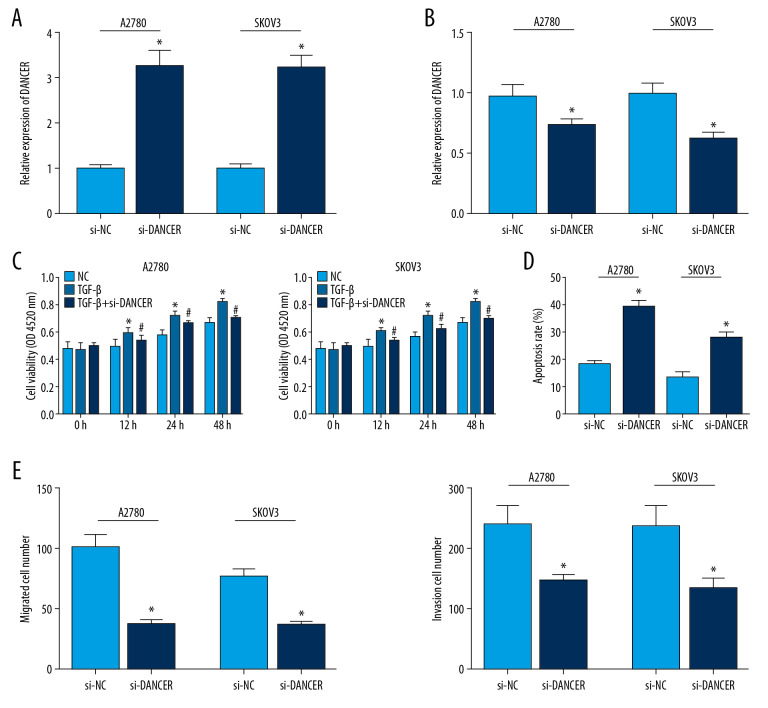
Initially, 2 ovarian cancer cell lines (A2780 and SKOV3) were stimulated with 10 ng/mL TGF-β for 48 hours. After treating with TGF-β, the DANCR level in cancer cells was obviously higher than that in untreated control cells (Figure 1A). And then we constructed stable DANCR knockdown in A2780 and SKOV3 cells via siRNA transfection (Figure 1B). The CCK-8 assay showed knockdown of DANCR reduced the proliferation of TGF-β treated ovarian cancer cells obviously (Figure 1C). In contrast to si-NC control group, the migration and invasion ability of cells was limited after transfection of DANCR knockdown (Figure 1D). Furthermore, flow cytometry analysis demonstrated DANCR silencing promoted a higher cell apoptosis percentage (Figure 1E).
Figure 1.
(A–E) DANCR silencing suppressed cell viability, migration and invasion, and induced cell apoptosis of TGF-β treated ovarian cancer cells. (A) The expression of DANCR in cancer cells (A2780 and SKOV3) treated with TGF-β was obviously higher than that in untreated control cells. (B) DANCR silencing was established in A2780 and SKOV3 cells treated with TGF-β. (C) The CCK-8 assay showed knockdown of DANCR reduced the proliferation of TGF-β treated ovarian cancer cells obviously. (D) Flow cytometry analysis demonstrated DANCR silencing promoted a higher cell apoptosis percentage than that in the control group. * P<0.05 compared to control group; # P<0.05 compared to TGF-β group. TGF-β – transforming growth factor-β; CCK-8 – Cell Counting Kit-8.
DANCR served as a sponge for miR-214
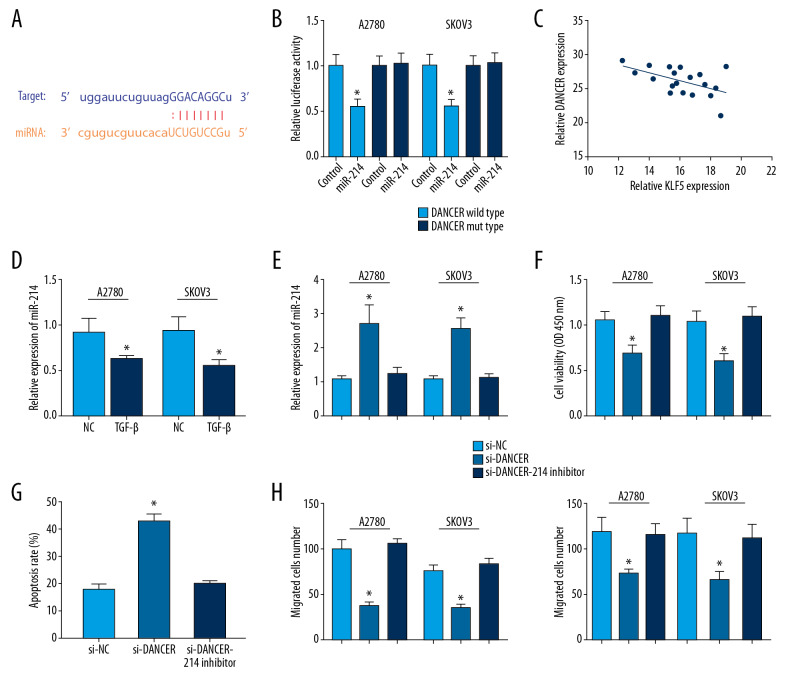
LncRNAs could serve as molecular sponges for miRNAs in some diseases [4]. We observed miR-214 had a potential binding site with DANCR using starBase v2.0 (Figure 2A). In DANCR wild type, miR-214 mimic could repress the fluorescence (Figure 2B). MiR-214 expression was negatively correlated with DANCR expression in ovarian cancer cells (Figure 2C). Besides, miR-214 expression level was notably downregulated in TGF-β treated ovarian cancer cells (Figure 2D). Under TGF-β-stimulated condition, DANCR silencing could elevate miR-214 expression level significantly in A2780 and SKOV3 cells, which was reversed by miR-214 inhibitor (Figure 2E). We found that MiR-214 inhibitor could abolish the suppression by si-DANCR in 2 cell lines (Figure 2F). In addition, miR-214 inhibitor could increase the apoptosis rate of TGF-β treatment in 2 cell lines transfected with DANCR silencing (Figure 2G). MiR-214 inhibitor could abolish the repression by si-DANCR in the migration and invasion number in 2 cell lines (Figure 2H).
Figure 2.
DANCR act as a sponge for miR-214. (A) Bioinformatics analysis showed that miR-214 has a potential binding site with DANCR. (B) MiR-214 mimic repressed the luciferase activity of only DANCR wild type in A2780 and SKOV3 cells. (C) MiR-214 expression was negatively correlated with DANCR expression in ovarian cancer cells. (D) MiR-214 expression level was notably downregulated in TGF-β treated ovarian cancer cells. (E) DANCR silencing could elevate miR-214 expression level significantly in A2780 and SKOV3 cells treated with TGF-β, which was reversed by miR-214 inhibitor. (F) MiR-214 inhibitor could abolish the suppression by si-DANCR. (G) MiR-214 inhibitor could increase the apoptosis rate of TGF-β treated 2 cell lines transfected with DANCR silencing. (H) MiR-214 inhibitor could rescue the suppression by si-DANCR in the migration and invasion number of 2 cells. * P<0.05 compared to control group. TGF-β – transforming growth factor-β.
KLF5 was a direct target of miR-214
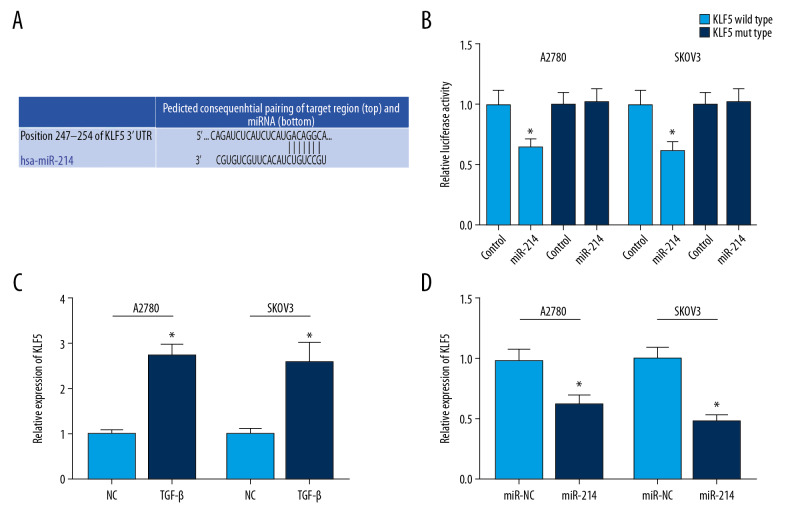
A highly conserved putative binding site was identified at KLF5 3′-UTR using TargetScan (Figure 3A). In KLF5 wild type, miR-214 mimic could repress the fluorescence (Figure 3B). Besides, we observed that KLF5 mRNA level was remarkably elevated in ovarian cancer cells treated with TGF-β (Figure 3C). In addition, miR-214 mimic obviously suppressed KLF5 expression (Figure 3D).
Figure 3.
(A) KLF5 is targeted by miR-214 using TargetScan. (B) A highly conserved putative binding site was identified at KLF5 3′-UTR using TargetScan. (C) KLF5 mRNA expression was significantly increased in ovarian cancer cells treated with TGF-β. (D) MiR-214 mimic obviously suppressed KLF6 expression. * P<0.05 compared to control group. TGF-β – transforming growth factor-β.
DANCR could suppress KLF5 expression
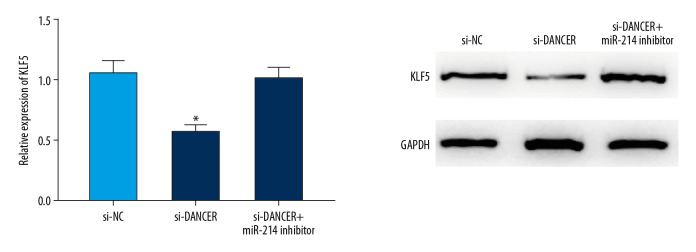
Furthermore, we explored the role of DANCR in the expression of KLF5 in ovarian cancer cells treated with TGF-β using qRT-PCR and western blot. We observed that DANCR knockdown could suppress the expression level of KLF5 in TGF-β stimulated cancer cells, which was reversed by miR-214 inhibitor (Figure 4).
Figure 4.
DANCR could suppress KLF5 expression. DANCR knockdown could suppress the expression level of KLF5 in TGF-β stimulated cancer cells, which was rescued by miR-214 inhibitor. * P<0.05 compared to control group. TGF-β – transforming growth factor-β.
Discussion
As a novel lncRNA, almost all research has focused on the role of DANCR in malignant tumors. Some studies recently have reported DANCR could accelerate ovarian cancer progression [17–19]. In this study, we further explored the underlying mechanism of DANCR in ovarian cancer in vitro. Initially, we observed the proliferation of ovarian cancer cells was significantly stronger under TGF-β stimulation. Subsequently, we observed the induced effect of DANCR on the TGF-β treated ovarian cancer cells. Besides, DANCR silencing promoted cell apoptosis under TGF-β stimulation.
Various studies have suggested that lncRNAs may execute its regulatory function in gene expression by sponging miRNAs [20]. MicroRNAs (miRNAs) are small endogenous non-coding RNA molecules, consisting of approximately 21–25 nucleotides. Past research has confirmed abnormal miRNAs influence on the progression of ovarian cancer [21,22]. In this study, we predicted miR-214 had a potential binding site with DANCR using bioinformatics analysis. Some research has revealed that miR-214 may act as an anti-oncogene, suppressing ovarian cancer development [23,24]. Similarly, we observed miR-214 expression level was notably downregulated in TGF-β treated ovarian cancer cells. In addition, miR-214 inhibitor could abolish the repression by si-DANCR in the biological behavior of ovarian cancer cells treated with TGF-β.
MiRNAs could mediate gene expression by sponging mRNAs, leading to post-transcriptional inhibition or mRNA degradation [25]. Our study demonstrated KLF5 is a direct target of miR-214. Krüppel-like factor 5 (KLF5) is a zinc finger-containing transcription factor that belongs to the KLF family [26]. Recently, KLF5 has been identified as a promoter in the development of some human cancers like gastric cancer [27], hepatocellular carcinoma [28], and lung cancer [29]. Similarly, KLF5 level was elevated in ovarian cancer cells. In addition, we observed knockdown of DANCR could inhibit the expression of KLF5. And the recovery experiment showed miR-214 mimic could reverse the suppression of DANCR silencing in TGF-β stimulated cancer cells. These data implied DANCR may promote the ovarian cancer progression by regulating miR-214/KLF5 expression.
Conclusions
This is the first report of DANCR expression upregulated in ovarian cancer cells under TGF-β stimulation. We that knockdown of DANCR could affect the biological process of ovarian cancer cells treated with TGF-β by sponging miR-214, which may provide new therapeutic ideas of ovarian cancer. We did not determine how much the substance-circumstance-phenomenon accounts for ovarian cancer progression; however, considering the importance these substances in pivotal roles in other cancers, we believe that this study data can contribute to further elucidate ovarian cancer progression and pathogenesis.
Acknowledgement
This work was supported by Cangzhou Central Hospital.
Footnotes
Conflict of interest
None.
Source of support: Departmental sources
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. Cancer J Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 2.Nick AM, Coleman RL, Ramirez PT, Sood AK. A framework for a personalized surgical approach to ovarian cancer. Nat Rev Clin Oncol. 2015;12:239–45. doi: 10.1038/nrclinonc.2015.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mattick JS, Rinn JL. Discovery and annotation of long noncoding RNAs. Nat Struct Mol Biol. 2015;22:5–7. doi: 10.1038/nsmb.2942. [DOI] [PubMed] [Google Scholar]
- 4.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: Insights into functions. Nat Rev Genet. 2009;10:155–59. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 5.Yan H, Silva MA, Li H, et al. Long noncoding RNA DQ786243 interacts with miR-506 and promotes progression of ovarian cancer through targeting cAMP responsive element binding protein 1. J Cell Biochem. 2018;119:9764–80. doi: 10.1002/jcb.27295. [DOI] [PubMed] [Google Scholar]
- 6.Zhao X, Cheng Z, Wang J. Long noncoding RNA FEZF1-AS1 promotes proliferation and inhibits apoptosis in ovarian cancer by activation of JAK-STAT3 pathway. Med Sci Monit. 2018;24:8088–95. doi: 10.12659/MSM.911194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zou T, Wang PL, Gao Y, Liang WT. Long noncoding RNA HOTTIP is a significant indicator of ovarian cancer prognosis and enhances cell proliferation and invasion. Cancer Biomark. 2019;25:133–39. doi: 10.3233/CBM-181727. [DOI] [PubMed] [Google Scholar]
- 8.Kretz M, Webster DE, Flockhart RJ, et al. Suppression of progenitor differentiation requires the long noncoding RNA ANCR. Genes Dev. 2012;26:338–43. doi: 10.1101/gad.182121.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bierie B, Moses HL. TGF-beta and cancer. Cytokine Growth Factor Rev. 2006;17:29–40. doi: 10.1016/j.cytogfr.2005.09.006. [DOI] [PubMed] [Google Scholar]
- 10.Dumont N, Arteaga CL. Targeting the TGF beta signaling network in human neoplasia. Cancer Cell. 2003;3:531–36. doi: 10.1016/s1535-6108(03)00135-1. [DOI] [PubMed] [Google Scholar]
- 11.Padua D, Massague J. Roles of TGFbeta in metastasis. Cell Res. 2009;19:89–102. doi: 10.1038/cr.2008.316. [DOI] [PubMed] [Google Scholar]
- 12.Roberts AB, Wakefield LM. The two faces of transforming growth factor beta in carcinogenesis. Proc Natl Acad Sci USA. 2003;100:8621–23. doi: 10.1073/pnas.1633291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cardenas H, Vieth E, Lee J, et al. TGF-beta induces global changes in DNA methylation during the epithelial-to-mesenchymal transition in ovarian cancer cells. Epigenetics. 2014;9:1461–72. doi: 10.4161/15592294.2014.971608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qiu X, Cheng JC, Zhao J, et al. Transforming growth factor-beta stimulates human ovarian cancer cell migration by up-regulating connexin43 expression via Smad2/3 signaling. Cell Signal. 2015;27:1956–62. doi: 10.1016/j.cellsig.2015.07.010. [DOI] [PubMed] [Google Scholar]
- 15.Yeung TL, Leung CS, Wong KK, et al. TGF-beta modulates ovarian cancer invasion by upregulating CAF-derived versican in the tumor microenvironment. Cancer Res. 2013;73:5016–28. doi: 10.1158/0008-5472.CAN-13-0023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cao L, Jin H, Zheng Y, et al. DANCR-mediated microRNA-665 regulates proliferation and metastasis of cervical cancer through the ERK/SMAD pathway. Cancer Sci. 2019;110:913–25. doi: 10.1111/cas.13921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gao YQ, Cheng HY, Liu KF. Long non-coding RNA DANCR upregulates IGF2 expression and promotes ovarian cancer progression. Eur Rev Med Pharmacol Sci. 2019;23:3621–26. doi: 10.26355/eurrev_201905_17785. [DOI] [PubMed] [Google Scholar]
- 18.Lin X, Yang F, Qi X, et al. LncRNA DANCR promotes tumor growth and angiogenesis in ovarian cancer through direct targeting of miR-145. Mol Carcinog. 2019;58:2286–96. doi: 10.1002/mc.23117. [DOI] [PubMed] [Google Scholar]
- 19.Pei CL, Fei KL, Yuan XY, Gong XJ. LncRNA DANCR aggravates the progression of ovarian cancer by downregulating UPF1. Eur Rev Med Pharmacol Sci. 2019;23:10657–63. doi: 10.26355/eurrev_201912_19763. [DOI] [PubMed] [Google Scholar]
- 20.Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505:344–52. doi: 10.1038/nature12986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kafshdooz L, Pourfathi H, Akbarzadeh A, et al. The role of microRNAs and nanoparticles in ovarian cancer: A review. Artif Cells Nanomed Biotechnol. 2018;46:241–47. doi: 10.1080/21691401.2018.1454931. [DOI] [PubMed] [Google Scholar]
- 22.Liu W, Wang S, Zhou S, et al. A systems biology approach to identify microRNAs contributing to cisplatin resistance in human ovarian cancer cells. Mol Biosyst. 2017;13:2268–76. doi: 10.1039/c7mb00362e. [DOI] [PubMed] [Google Scholar]
- 23.Chen Y, Du H, Bao L, Liu W. LncRNA PVT1 promotes ovarian cancer progression by silencing miR-214. Cancer Biol Med. 2018;15:238–50. doi: 10.20892/j.issn.2095-3941.2017.0174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu Y, Lin J, Zhai S, et al. MicroRNA-214 suppresses ovarian cancer by targeting beta-catenin. Cell Physiol Biochem. 2018;45:1654–62. doi: 10.1159/000487733. [DOI] [PubMed] [Google Scholar]
- 25.He L, Hannon GJ. MicroRNAs: Small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–31. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 26.Bieker JJ. Kruppel-like factors: Three fingers in many pies. J Biol Chem. 2001;276:34355–58. doi: 10.1074/jbc.R100043200. [DOI] [PubMed] [Google Scholar]
- 27.Yang T, Chen M, Yang X, et al. Down-regulation of KLF5 in cancer-associated fibroblasts inhibit gastric cancer cells progression by CCL5/CCR5 axis. Cancer Biol Ther. 2017;18:806–15. doi: 10.1080/15384047.2017.1373219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang J, Chu Y, Xu M, et al. miR-21 promotes cell migration and invasion of hepatocellular carcinoma by targeting KLF5. Oncol Lett. 2019;17:2221–27. doi: 10.3892/ol.2018.9843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang H, Shao F, Guo W, et al. Knockdown of KLF5 promotes cisplatin-induced cell apoptosis via regulating DNA damage checkpoint proteins in non-small cell lung cancer. Thorac Cancer. 2019;10:1069–77. doi: 10.1111/1759-7714.13046. [DOI] [PMC free article] [PubMed] [Google Scholar]