Summary
There is pressing urgency to understand the pathogenesis of the severe acute respiratory syndrome coronavirus clade 2 (SARS-CoV-2), which causes the disease COVID-19. SARS-CoV-2 spike (S) protein binds angiotensin-converting enzyme 2 (ACE2), and in concert with host proteases, principally transmembrane serine protease 2 (TMPRSS2), promotes cellular entry. The cell subsets targeted by SARS-CoV-2 in host tissues and the factors that regulate ACE2 expression remain unknown. Here, we leverage human, non-human primate, and mouse single-cell RNA-sequencing (scRNA-seq) datasets across health and disease to uncover putative targets of SARS-CoV-2 among tissue-resident cell subsets. We identify ACE2 and TMPRSS2 co-expressing cells within lung type II pneumocytes, ileal absorptive enterocytes, and nasal goblet secretory cells. Strikingly, we discovered that ACE2 is a human interferon-stimulated gene (ISG) in vitro using airway epithelial cells and extend our findings to in vivo viral infections. Our data suggest that SARS-CoV-2 could exploit species-specific interferon-driven upregulation of ACE2, a tissue-protective mediator during lung injury, to enhance infection.
Keywords: scRNA-seq, interferon, ISG, ACE2, SARS-CoV-2, COVID-19, influenza, non-human primate, human, mouse
Graphical Abstract
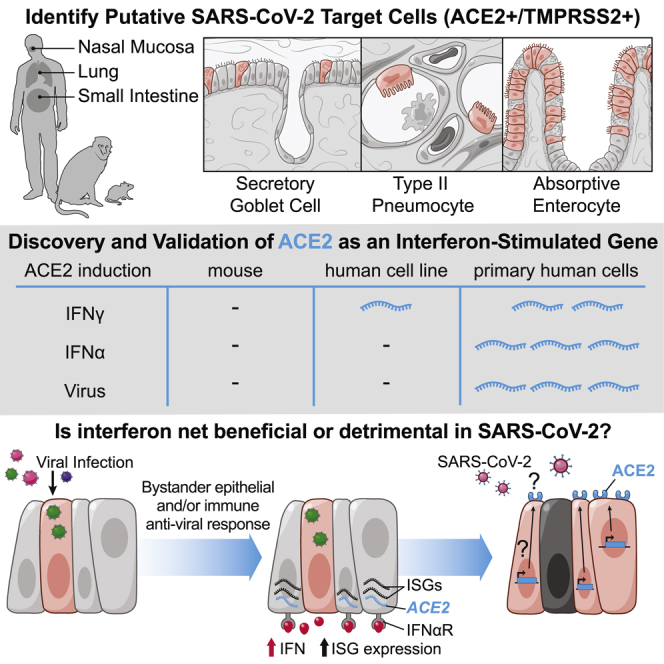
Highlights
-
•
Meta-analysis of human, non-human primate, and mouse single-cell RNA-seq datasets for putative SARS-CoV-2 targets
-
•
Type II pneumocytes, nasal secretory cells, and absorptive enterocytes are ACE2+TMPRSS2+
-
•
Interferon and influenza increase ACE2 in human nasal epithelia and lung tissue
-
•
Mouse Ace2 is not upregulated by interferon, raising implications for disease modeling
Analysis of single-cell RNA-seq datasets from human, non-human primate, and mouse barrier tissues identifies putative cellular targets of SARS-CoV-2 on the basis of ACE2 and TMPRSS2 expression. ACE2 represents a previously unappreciated interferon-stimulated gene in human, but not mouse, epithelial tissues, identifying anti-viral induction of a host tissue-protective mechanism, but also a potential means for viral exploitation of the host response.
Introduction
Human coronaviruses (CoVs) are single-stranded positive-sense RNA viruses that can cause mild to severe respiratory disease (Fung and Liu, 2019). Over the past two decades, zoonotic transmission events have led to the emergence of two highly pathogenic CoVs: severe acute respiratory syndrome (SARS)-CoV and Middle East respiratory syndrome (MERS)-CoV. SARS-CoV-2, which causes the disease known as COVID-19, was first reported in late 2019 (Coronaviridae Study Group of the International Committee on Taxonomy of V., 2020, Lu et al., 2020, Paules et al., 2020). COVID-19 is characterized by pneumonia, fever, cough, and occasional diarrhea (Guan et al., 2020, Holshue et al., 2020, Huang et al., 2020), and SARS-CoV-2 RNA has been reliably detected in nasopharyngeal swabs, sputum, and stool samples (Wang et al., 2020, Wölfel et al., 2020, Zou et al., 2020). As of April 19, 2020, SARS-CoV-2 continues to spread worldwide, and there are over 2,401,379 confirmed cases, 165,044 deaths, and 623,903 recovered individuals in 185 countries and regions (Dong et al., 2020a). Early models of COVID-19 transmission dynamics estimate one infectious individual infects slightly over two individuals; travel restrictions reduce that spread to one individual, although these figures might evolve as more accurate epidemiological data become available (Kucharski et al., 2020).
Work during the first SARS-CoV epidemic identified the human host factor angiotensin-converting enzyme 2 (ACE2) as the receptor for SARS-CoV (Li et al., 2003). SARS-CoV-2 spike (S) protein has been experimentally shown to bind ACE2 on host cells with significantly higher affinity than SARS-CoV-S (Hoffmann et al., 2020, Wrapp et al., 2020). The main host protease that mediates S protein activation on primary target cells and initial viral entry is the type II transmembrane serine protease TMPRSS2 (Glowacka et al., 2011, Hoffmann et al., 2020, Iwata-Yoshikawa et al., 2019, Matsuyama et al., 2010, Shulla et al., 2011, Walls et al., 2020). Other host proteases, such as furin, have also been suggested to promote the pathogenesis of this pandemic SARS-CoV-2 clade, but when and where they process S protein remains to be determined (Böttcher-Friebertshäuser et al., 2013, Bugge et al., 2009, Coutard et al., 2020, Walls et al., 2020). Binding of SARS-CoV-S to ACE2 results in receptor-mediated internalization (Grove and Marsh, 2011, Kuba et al., 2005). Importantly, ACE2 functions as a key tissue-protective component during severe acute lung injury (Imai et al., 2005, Kuba et al., 2005).
A tissue-level basis for understanding SARS-CoV tropism was proposed based on ACE2 histological staining and expression in human epithelia of the lung and small intestine (Hamming et al., 2004, Harmer et al., 2002, Jonsdottir and Dijkman, 2016). However, unlike the specific expression of CDHR3 (the rhinovirus-C receptor), which is resolved to ciliated epithelial cells of the upper airway (Griggs et al., 2017), the specific cell subsets within each tissue that express ACE2 remain unknown. Identifying the cell subsets targeted by SARS-CoV-2 (ACE2+) and those at greatest risk of direct infection (ACE2+TMPRSS2+) is critical for understanding and modulating host defense mechanisms and viral pathogenesis.
After cellular detection of viral entry into a host cell, interferon (IFN) induction of interferon-stimulated genes (ISGs) is essential for host antiviral defense in mice, non-human primates (NHPs), and humans (Bailey et al., 2014, Deeks et al., 2017, Dupuis et al., 2003, Everitt et al., 2012, Schneider et al., 2014, Utay and Douek, 2016). There are three distinct types of IFNs: type I IFNs (IFN-α and IFN-β), type II IFNs (IFN-γ), and type III IFNs (IFN-λ) (Broggi et al., 2020, Müller et al., 1994, Stetson and Medzhitov, 2006). Each appears to converge on almost indistinguishable responses, mediated through the binding of STAT1 homodimers or STAT1/STAT2 heterodimers to ISGs. However, mounting evidence suggests that each type of IFN might have a non-redundant role in host defense or immunopathology, particularly at epithelial barriers (Broggi et al., 2020, Iwasaki et al., 2017, Iwasaki and Pillai, 2014, Jewell et al., 2010).
Although the host response to SARS-CoV highlighted a role for IFNs, most studies assessed the effect of IFN restriction in cell lines that might not fully recapitulate the repertoire of ISGs present in primary human target cells (Bailey et al., 2014, de Lang et al., 2006, Sainz et al., 2004, Zheng et al., 2004). One study of SARS-CoV suggested the timing of the type I IFN response was critical in vivo (Channappanavar et al., 2016). Clinical therapy using approved IFNs has been attempted for SARS-CoV, MERS-CoV, and SARS-CoV-2 in the absence of a controlled trial to mixed effect, resulting in anecdotal evidence suggesting either rapid improvement or worsening of symptoms (Dong et al., 2020b, Lei et al., 2020, Li and De Clercq, 2020). Elucidating tissue- and cell-type-specific ISGs and their activity is essential for understanding the role of IFNs in host defense during human SARS-CoV-2 infection.
Massively parallel single-cell RNA-sequencing (scRNA-seq) is transforming our ability to comprehensively map the cell types, subsets, and states present during health and disease in barrier tissues (Ordovas-Montanes et al., 2020, Ordovas-Montanes et al., 2018, Smillie et al., 2019). This has been particularly evident in the elucidation of novel human epithelial and stromal cell subsets and states (Ordovas-Montanes et al., 2018, Regev et al., 2017, Ruiz García et al., 2019, Schiller et al., 2019, Smillie et al., 2019, Vieira Braga et al., 2019). Recently, scRNA-seq has been applied to better understand the cellular variation present during viral infection in vitro and in vivo (Russell et al., 2018, Steuerman et al., 2018). Global single-cell profiling efforts such as the Human Cell Atlas (HCA) initiative are ideally poised to rapidly share critical data and enhance our understanding of disease during emergent public health challenges (Sungnak et al., 2020).
Here, using published and unpublished datasets (all from non-SARS-CoV-2-infected samples), we analyze human, NHP, and mouse tissues that have been clinically identified to harbor virus in patients exhibiting COVID-19 symptoms. We provide a cautionary note on the interpretation of the scRNA-seq data presented below, given that many factors such as dissociation, profiling method, and sequencing depth can influence results (STAR Methods). Here, we focus our analysis and discussion on the specific subsets where ACE2 and TMPRSS2 are enriched and on relative comparisons within each dataset, rather than between datasets or equivalence to absolute numbers of total cells. Across several studies of human and NHP tissues, we found ISGs upregulated in ACE2-expressing cells.
Strikingly, by treating primary human upper airway basal cells with distinct types of inflammatory cytokines, we demonstrate that IFN-α drives ACE2 expression. Human influenza infection also induces broader expression of ACE2 in upper airway epithelial cells and is corroborated by publicly available databases. Overall, our data provide motivation to better understand the trade-offs of antiviral and/or IFN therapy in humans infected with SARS-CoV-2 in order to balance host restriction, tissue tolerance, and viral enhancement mechanisms (Davidson et al., 2015, Fung and Liu, 2019, Imai et al., 2005, Iwasaki et al., 2017, Kuba et al., 2005, Lei et al., 2020, Medzhitov et al., 2012, Zou et al., 2014). Importantly, although our findings identify similar cell subsets enriched for Ace2 in mice, neither in vitro nor in vivo IFN-stimulation nor in vivo viral challenge substantially alter Ace2 expression levels. The dynamic, species-specific and multifaceted role of IFN raises implications for pre-clinical COVID-19 disease modeling.
Results
Lung Epithelial Cell Expression of Host Factors Used by SARS-CoV-2 in Non-Human Primates and Humans
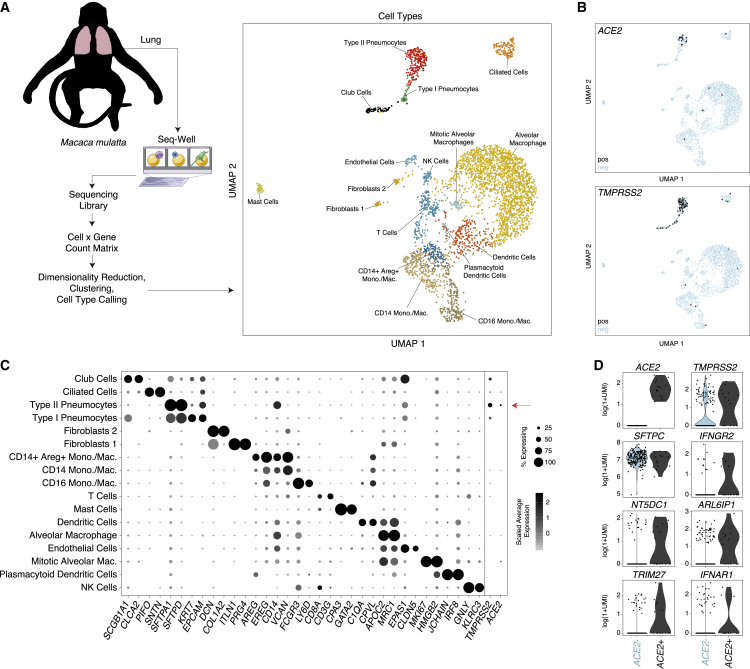
To investigate which cells within human and NHP tissues represent likely SARS-CoV-2 targets, we analyzed new and existing scRNA-seq datasets to assess which cell types express ACE2, alone or with TMPRSS2. In a previously unpublished dataset consisting of NHP (Macaca mulatta) lung tissue collected after necropsy of healthy adult animals and analyzed by using Seq-Well v1 (Gierahn et al., 2017), we recovered at least 17 distinct major cell types, including various lymphoid, myeloid, and stromal populations (Figures 1A–1C; Table S1; STAR Methods). ACE2 and TMPRSS2 were primarily expressed in epithelial cells, with 6.7% of type II pneumocytes expressing ACE2 and 3.8% co-expressing ACE2 and TMPRSS2 (Figures 1B and 1C). Notably, the only double-positive cells observed were classified within the type II pneumocyte population; however, we also identified TMPRSS2 expression within club cells, ciliated epithelial cells, and type I pneumocytes, albeit at diminished abundance and frequency compared with type II pneumocytes (Figure 1C; Table S1).
Figure 1.
Expression of ACE2 in Type II Pneumocytes in Healthy Lungs of Non-human Primates
(A) Schematic of protocol for isolation of lung tissue at necropsy from healthy non-human primates (M. mulatta, n = 3), creation of scRNA-seq libraries by using Seq-Well v1, and computational analysis to identify cell types by using unbiased methods. UMAP projection of 3,793 single cells, points colored by cell identity (see STAR Methods).
(B) Uniform manifold approximation and projection (UMAP) as in (A), points colored by detection of ACE2 (coronavirus receptor, top) or TMPRSS2 (coronavirus S protein priming for entry, bottom). Color coding is as follows: black, RNA positive; blue, RNA negative.
(C) Dot plot of 2 defining genes for each cell type (Table S1) (Bonferroni-adjusted p < 0.001) and ACE2 and TMPRSS2. Dot size represents fraction of cells within that type expressing a given gene, and color intensity represents binned count-based expression amount (log(scaled UMI+1)) among expressing cells. ACE2 is enriched in type II pneumocytes (6.7% expressing, Bonferroni-adjusted p = 8.62E−33), as is TMPRSS2 (29.5% expressing, Bonferroni-adjusted p = 8.73E−153). Of all type II pneumocytes, 3.8% co-express ACE2 and TMPRSS2 (Table S9). Red arrow indicates cell type with largest proportion of ACE2+TMPRSS2+ cells.
(D) Genes differentially expressed among ACE2+ and ACE2− type II pneumocytes. (SCDE package, FDR-adjusted p < 0.05 for IFNGR2, NT5DC1, ARL6IP1, and TRIM27; full results can be found in Table S1).
See also Table S1.
Next, we compared ACE2+ with ACE2− type II pneumocytes to explore broader gene programs that differentiate putative SARS-CoV-2 target cells from cells of a similar phenotype and ontogeny (Figure 1D; Table S1). Among genes significantly upregulated in ACE2+ type II pneumocytes, we observed IFNGR2 (false discovery rate [FDR]-adjusted p = 0.022), a receptor for type II IFNs. Notably, previous work has demonstrated limited anti-viral potency of IFN-γ for SARS-associated coronaviruses, compared with that of type I IFNs, at least in vitro (Sainz et al., 2004, Zheng et al., 2004). Other co-regulated genes of potential interest include TRIM27 (FDR-adjusted p = 0.025), as well as NT5DC1 (FDR-adjusted p = 0.003) and ARL6IP1 (FDR-adjusted p = 0.047), which were upregulated in the A549 adenocarcinoma alveolar basal epithelial cell line after exposure to IFN-α and IFN-γ for 6 h (Sanda et al., 2006). We found IFNAR1 consistently expressed among both ACE2+ type II pneumocytes and ACE2+TMPRSS2+ co-expressing type II pneumocytes, but its level of upregulation compared with all remaining pneumocytes did not meet statistical significance (FDR-adjusted p = 0.11). This analysis finds ACE2+ cells enriched within a rare fraction of secretory cells in NHPs and that ACE2 expression is co-regulated with genes involved in IFN responses.
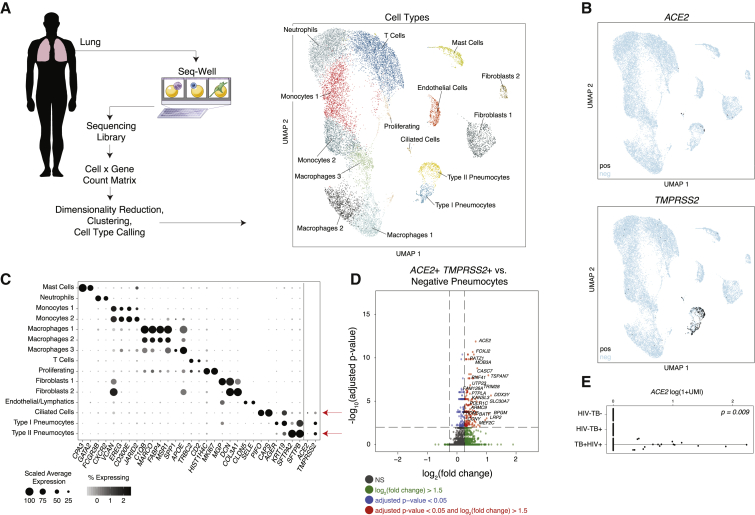
To assess whether the findings from NHP lung cells were similarly present in humans, we analyzed a previously unpublished scRNA-seq dataset derived from surgical resections of fibrotic lung tissue collected with Seq-Well S3 (Hughes et al., 2019). Unsupervised analysis identified multiple cell types and subtypes of immune cells (Figures 2A–2C; STAR Methods), as defined by the genes displayed in Figure 2C (full lists available in Table S2). Here, we found that ACE2 and TMPRSS2 were primarily expressed within type II pneumocytes and ciliated cells, in line with our analysis of the NHP-derived cells (Figures 1 and 2A, 2B). In type II pneumocytes (identified by unique expression of surfactant proteins SFTPC, SFTPB, and SFTPA1), we found 1.4% of cells expressing ACE2 (FDR-adjusted p = 1.35E−21), 34.2% expressing TMPRSS2 (FDR-adjusted p < 1E−300), and 0.8% co-expressing both. In ciliated cells, we found 7% were ACE2+ (FDR-adjusted p = 5E−64), 24.6% were TMPRSS2+ (FDR-adjusted p = 3.8E−30), and 5.3% co-expressed both.
Figure 2.
Select Lung Epithelial Cells from Control, HIV-1-Infected, and Mycobacterium-tuberculosis-Infected Human Donors Co-Express ACE2 and TMPRSS2
(A) Schematic of protocol for isolation of human lung tissue from surgical excess, creation of scRNA-seq libraries by using Seq-Well S3, and computational analysis to identify cell types by using unbiased methods. Shown on the right is a UMAP projection of 18,915 cells across 8 donors (n = 3 TB+HIV+; n = 3 TB+; n = 2 non-infected patients). Cells represented by points, colored according to cell type (see STAR Methods).
(B) UMAP projection as in (A), points colored by detection of ACE2 (top) or TMPRSS2 (bottom). Color coding is as follows: black, RNA positive; blue, RNA negative.
(C) Dot plot of 2 defining genes for each cell type (FDR-adjusted p < 0.001), and ACE2 and TMPRSS2; dot size represents fraction of cells within cell type expressing a given gene, and color intensity represents binned count-based expression amount (log(scaled UMI+1)) among expressing cells. All cluster-defining genes are provided in Table S2. Red arrow indicates cell types with largest proportion of ACE2+TMPRSS2+ cells.
(D) Volcano plot identifying significantly upregulated genes in ACE2+TMPRSS2+ pneumocytes compared with all remaining pneumocytes. Red points represent genes with a FDR-adjusted p < 0.05, and log2(fold change) >1.5. Text highlighting specific genes; the full list is available in Table S2.
(E) Expression of ACE2 across human donors by HIV and TB status (p = 0.009 by likelihood-ratio test).
See also Table S2.
As above, to assess for cellular pathways significantly co-expressed within putative target cells for SARS-CoV-2, we computed differentially expressed genes between ACE2+TMPRSS2+ type II pneumocytes and all other type II pneumocytes (Figures 2C and 2D; Table S2). We found significant enrichment of BATF among ACE2+TMPRSS2+ cells (FDR-adjusted p = 3.25E−7), which has been demonstrated previously to be upregulated by type I and type II IFNs (Murphy et al., 2013). Of note, we also observed TRIM28 co-expressed with ACE2 and TMPRSS2 among type II pneumocytes in this dataset (FDR-adjusted p = 2.34E−9), which might play a role in potentiating an IFN response in lung epithelial cells (Krischuns et al., 2018). Within this cohort of donors, 3 individuals were human immunodeficiency virus (HIV)+ and diagnosed with active tuberculosis, 3 donors had active tuberculosis and were HIV−, and 2 were negative for both pathogens. Surprisingly, we found that all of the ACE2+ cells across all cell types were derived from HIV+ Mycobacterium tuberculosis (Mtb)+ donors despite approximately equivalent recovery of epithelial cell types from all donors (likelihood-ratio test, p = 0.009) (Figure 2E). Given limited cell and patient numbers combined with potential sampling biases, we caution that this observation requires much broader cohorts to validate a potential role for co-infections; still, we note our observation is suggestive of a role for chronic IFNs in the induction of ACE2, given that HIV infection is associated with persistent upregulation of ISGs, and we observed elevated amounts of IFNAR2, IFI30, and IKBKB (Utay and Douek, 2016) (FDR-adjusted p = 1.1E−6, 8.8E−9, 1.57E−7, respectively; HIV+ versus HIV− epithelial cells).
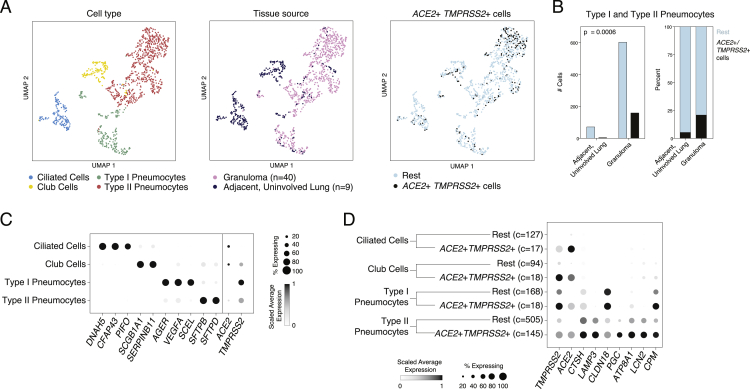
Next, using a previously unpublished scRNA-seq dataset consisting of granuloma and adjacent, uninvolved lung samples from Mtb-infected NHPs (Macaca fascicularis) collected with Seq-Well S3, we identified subsets of epithelial cells expressing ACE2 and TMPRSS2 (Figure S1; Table S3; STAR Methods). The majority of ACE2+TMPRSS2+ cells were, once again, type II pneumocytes (22%) and type I pneumocytes (9.7%) and were largely enriched within granulomatous regions compared with those in adjacent uninvolved lung (Figures S1B and S1C) (p = 0.006, Fisher Exact Test). ACE2+TMPRSS2+ type II pneumocytes expressed significantly higher amounts of antimicrobial effectors such as LCN2 compared with remaining type II pneumocytes (Figure S1D). Cells with club cell/secretory, type I pneumocyte, and ciliated cell types also contained some ACE2+TMPRSS2+ cells, but we did not have sufficient power to detect significantly differentially expressed genes between these cells and other cells within those clusters. Altogether, we identify ACE2+TMPRSS2+ cells in lower airways of humans and NHPs with consistent cellular phenotypes and evidence supporting a potential role for IFN-associated inflammation in upregulation of ACE2.
Figure S1.
NHP Tuberculosis Infected Lung and Granuloma, Related to Figures 1 and 2
(A). UMAP projection of epithelial cells (1,099 cells) colored by annotated cell type, tissue source, and gating as ACE2+TMPRSS2+ cells. ACE2+TMPRSS2+ cells comprise 11% of ciliated cells, 16% of club cells, 10% type I pneumocytes, and 22% type II pneumocytes. Data generated using Seq-Well S3 (Table S3).
(B). Number of cells (left) and % (right) ACE2+TMPRSS2+ cells by tissue source (granuloma versus uninvolved lung) and cell type. Ciliated cells and club cells were omitted from this analysis as we detected too few cells (< 7 total cells) belonging to these clusters in the granulomas. Statistical significance assessed by Fisher Exact Test (Table S3).
(C). Dot plot of top cluster defining genes for each epithelial cell type and ACE2 and TMPRSS2. Dot size represents fraction of cells expressing, and color intensity represents average log(normalized UMI + 1) among all cells in each group scaled between 0 and 1 by gene. ACE2 expression is enriched in club cells (Bimodal test, Bonferroni-corrected p < 0.001), ciliated cells (p < 0.005), and type I pneumocytes (p < 0.001). TMPRSS2 expression is enriched in type I pneumocytes (p < 0.001) and ciliated cells (p < 0.001) (Table S3).
(D). Dot plot of genes differentially expressed between ACE2+TMPRSS2+ epithelial cells versus rest (Bimodal test, Bonferroni-corrected p < 0.01, log fold change > 0.5). (Table S3, c = number of cells, n = number of animals).
Ileal Absorptive Enterocytes Express Host Factors Used by SARS-CoV-2
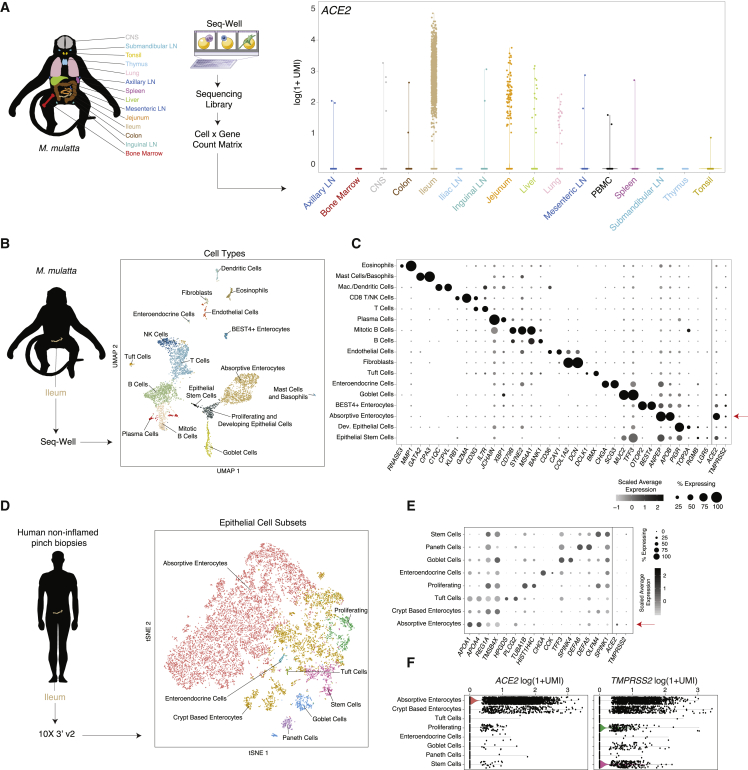
Next, we examined several other tissues for ACE2-expressing cells on the basis of the location of hallmark symptoms of COVID-19, focusing on the gastrointestinal tract due to reports of clinical symptoms and viral shedding (Xiao et al., 2020). Leveraging a previously unpublished scRNA-seq atlas of NHP (M. mulatta) tissues collected with Seq-Well v1, we observed that the majority of ACE2+ cells reside in the small intestine, principally within the ileum, jejunum, and, to a lesser extent, the liver and colon (Figure 3A; STAR Methods). Critically, we note that, in this experiment, the dissociation method used on each tissue was optimized to preserve immune cell recovery, and therefore under-sampled stromal and epithelial populations, as well as neurons from the brain. Within the ileum, we identified ACE2+ cells as absorptive enterocytes on the basis of specific expression of ACE2 within cells defined by APOA1, SI, FABP6, and ENPEP, among others, by a likelihood-ratio test (Figures 3B and 3C) (p < 1E−300, 62% of all absorptive enterocytes; see Table S4). All other epithelial subtypes expressed ACE2 to a lesser extent, and variably co-expressed ACE2 with TMPRSS2 (see Table S4 for full statistics).
Figure 3.
NHP and Human Ileal Absorptive Enterocytes Co-Express ACE2 and TMPRSS2
(A) Expression ACE2 across diverse tissues in healthy NHPs (n = 3 animals; 52,858 cells).
(B) Schematic of protocol for isolation of NHP ileum (n = 5) at necropsy for scRNA-seq using Seq-Well v1, and computational pipeline to identify cell types by using unbiased methods. Shown on the right is a UMAP projection of 4,515 cells colored by cell type.
(C) Dot plot of 2 defining genes for each cell type, with ACE2 and TMPRSS2. Dot size represents fraction of cells within cell type expressing a given gene, and color intensity represents binned count-based expression amounts (log(scaled UMI+1)) among expressing cells. All cluster defining genes are provided in Table S4. Red arrow indicates cell type with largest proportion of ACE2+TMPRSS2+ cells.
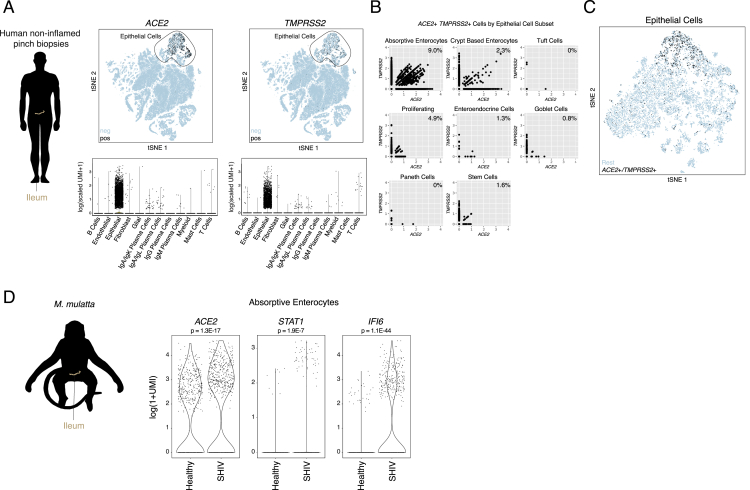
(D) Schematic of protocol for isolation of human ileal cells from endoscopic pinch biopsies in non-inflamed regions (n = 13). Shown on the right is a tSNE plot of 13,689 epithelial cells selected from original dataset generated by 10x 3′ v2 (see Figure S2), colored by cellular subsets.
(E). Dot plot of 2 defining genes for each cell type, with ACE2 and TMPRSS2. Dot size represents fraction of cells within cell type expressing a given gene, and color intensity represents binned count-based expression amounts (log(scaled UMI+1)) among expressing cells. All cluster defining genes are provided in Table S5. Red arrow indicates cell type with largest proportion of ACE2+TMPRSS2+ cells.
(F). Expression of ACE2 (left) and TMPRSS2 (right) among all epithelial subsets from human donors.
Persistent viral RNA in rectal swabs has been detected in pediatric infection, even after negative nasopharyngeal tests (Xu et al., 2020). In an additional dataset consisting of endoscopic biopsies from the terminal ileum of a human pediatric cohort (n = 13 donors, ranging in age from 10 to 18 years old), collected with 10X 3′ v2, we confirmed a large abundance of ACE2+ cells with selective expression within absorptive enterocytes (29.7% ACE2+, FDR-adjusted p = 2.46E−100) (Figures 3D and 3E; Table S5; STAR Methods). Furthermore, we identified a subset (888 cells, ∼6.5% of all epithelial cells) that co-express both genes (Figures S2A–S2C). We performed differential expression testing and GO-term enrichment using these cells relative to matched non-expressers to highlight putative biological functions enriched within them, such as metabolic processes and catalytic activity, and to identify shared phenotypes of ACE2+TMPRSS2+ ileal cells across both human and NHP cohorts (Table S5). We speculate that viral targeting of these cells, taken from patients without overt clinical viral infection, might help explain intestinal symptoms. Finally, we compared ileal absorptive enterocytes from healthy NHPs and NHPs infected with simian-human immunodeficiency virus (SHIV) and then treated for 6 months with anti-retroviral therapy (animal and infection characteristics published in Colonna et al., 2018) (STAR Methods). We found significant upregulation of ACE2, STAT1, and IFI6 within the absorptive enterocytes of SHIV-infected animals (which maintain chronically elevated amounts of IFNs and ISGs) compared with those of uninfected controls (FDR-adjusted p < 2E-7) (Figure S2D) (Deeks et al., 2017, Utay and Douek, 2016).
Figure S2.
Human and NHP Ileum, Related to Figure 3
(A). Top: tSNE projection of all cells from healthy pediatric human ileum within a previously-unpublished 10x 3′ v2 dataset (115,569 cells). Black: higher expression of ACE2 (left), TMPRSS2 (right). Bottom: Corresponding violin plots of expression values for ACE2 (left) and TMPRSS2 (right). Solid line: epithelial cells.
(B). Co-expression of ACE2 and TMPRSS2 by epithelial cell subset. Number indicates % of ACE2+TMPRSS2+ cells by cell subset.
(C). tSNE projection of 13,689 cells as in Figure 3D, cells colored by co-expression of ACE2 and TMPRSS2 (black).
(D). Expression of ACE2 and canonical interferon-responsive genes among absorptive enterocytes from Healthy (n = 2) and SHIV-infected, anti-retroviral treated animals (n = 3). Bonferroni-adjusted p-values by Wilcoxon test (healthy: 510 cells, SHIV-infected: 636 cells).
Upper Airway Expression of Host Factors Used by SARS-CoV-2
To identify potential viral target cells in nasal and sinus tissue, two regions that are frequently primary sites of exposure for coronaviruses, we analyzed existing scRNA-seq datasets from the human upper airway (inferior turbinate and ethmoid sinus mucosa) across a spectrum of healthy donors and individuals with allergic inflammation due to chronic rhinosinusitis (CRS) collected with Seq-Well v1 (Figure 4A; STAR Methods) (Ordovas-Montanes et al., 2018). We had previously noted a significantly enriched IFN-dominated gene signature in inferior turbinate secretory epithelial cells from both healthy and CRS donors compared with CRS samples from the ethmoid sinus, which were significantly enriched for interleukin-4 (IL-4)/IL-13 gene signatures (Giovannini-Chami et al., 2012, Ordovas-Montanes et al., 2018). We speculate that these cells, taken from clinically non-virally infected patients, yet constantly exposed to environmental viruses, might provide one of the earliest locations for coronaviruses to infect before spreading to other tissues. We observed significant enrichment of ACE2 expression in apical epithelial cells and, to a lesser extent, ciliated cells compared with all cell types recovered from surgically resected mucosa (1% of apical epithelial cells, FDR-adjusted p = 4.55E−6, n.s. in ciliated cells) (Figure 4B; Table S6).
Figure 4.
Healthy and Allergic Inflamed Human Nasal Mucosa Co-Express ACE2 and TMPRSS2 in a Subset of Goblet Secretory Cells
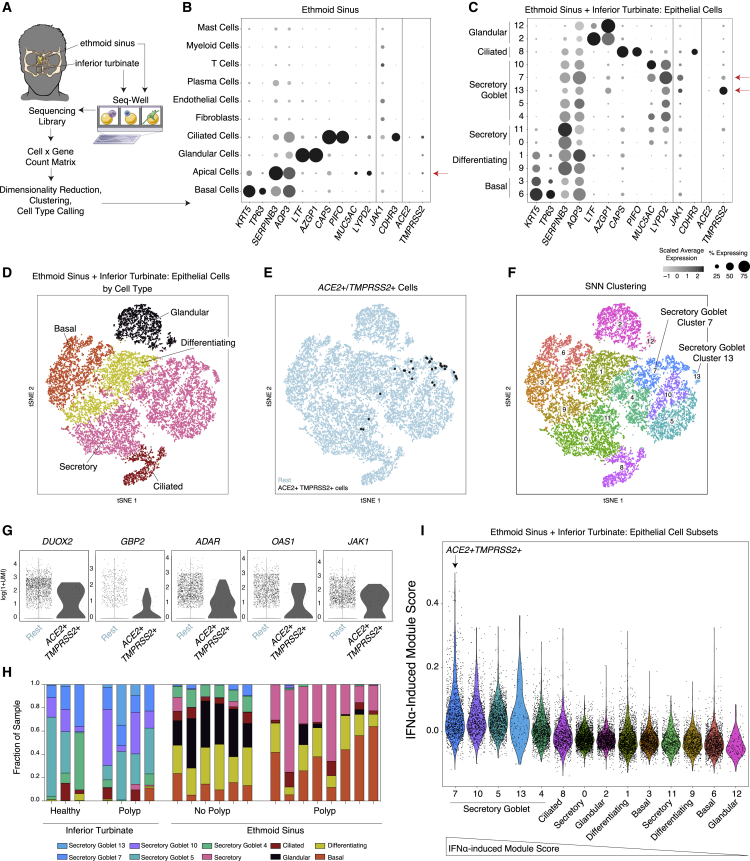
(A) Schematic for sampling of n = 12 ethmoid sinus surgical samples and n = 9 inferior turbinate nasal scrapings to generate scRNA-seq libraries by using Seq-Well v1. See Ordovas-Montanes et al., (2018).
(B) Dot plot of all cell types from ethmoid-sinus-derived cells (n = 6 non-polyp CRS samples, n = 6 polyp CRS samples). Two defining genes for each cell type, in addition to CDHR3 (rhinovirus receptor), ACE2, TMPRSS2, and JAK1. Dot size represents fraction of cells within that type expressing a given gene, and color intensity represents binned count-based expression amounts (log(scaled UMI+1)) among expressing cells (see Table S6 for statistics by subset). Red arrow indicates cell types with largest proportion of ACE2+TMPRSS2+ cells.
(C) Dot plot for 2 defining genes for each cell type identified from granular clustering of epithelial cells (18,325 single cells) derived from both ethmoid sinus and inferior turbinate sampling (healthy inferior turbinate [3,681 cells; n = 3 samples], polyp-bearing patient inferior turbinate [1,370 cells; n = 4 samples], non-polyp ethmoid sinus surgical samples [5,928 cells; n = 6 samples], and polyp surgical and scraping samples directly from polyp in ethmoid sinus [7,346 cells; n = 8 samples]). Red arrow indicates cell type with largest proportion of ACE2+TMPRSS2+ cells.
(D) tSNE of 18,325 single epithelial cells from inferior turbinate and ethmoid sinus (omitting immune cells). Colored by cell types 3,152 basal, 3,089 differentiating, 8,840 secretory, 1,105 ciliated, and 2,139 glandular cells.
(E) tSNE as in (D), identifying epithelial cells co-expressing ACE2 and TMPRSS2 (30 cells, black points).
(F) tSNE as in (D), colored by detailed cell types with higher granularity, as in (C).
(G) Individual differentially expressed genes between ACE2+TMPRSS2+ cells and all other secretory epithelial cells (see Table S6 for full gene list with statistics). Bonferroni-adjusted likelihood-ratio test p < 0.02 for all genes displayed.
(H) Stacked bar plot of each subset of epithelial cells among all epithelial cells by donor (each bar) and sampling location (noted below graph) (unpaired t test p < 0.00035 for Secretory Goblet 7 inferior turbinate versus ethmoid sinus; see Table S6 for raw values).
(I) Violin plot of cell clusters in respiratory epithelial cells (from Figures 4C and 4F) ordered by average expression of IFN-α-induced gene signatures, presented as a gene module score; non-normal distribution by Lilliefors test, Mann-Whitney U-test p = 2.2E−16, 1.21 effect size, IFN-α signature for Secretory Goblet Cluster 7 versus all epithelial cells. Arrow indicates cluster containing majority ACE2+TMPRSS2+ cells.
To better map putative SARS-CoV-2 targets among epithelial subsets, we employed a finer-grained clustering method applied to both ethmoid sinus surgical specimens and scrapings from the inferior turbinate and ethmoid sinus (Figures 4C–4F). Once again, we observed selective expression of ACE2 within a minority of cell types, with 1.3% of all secretory cells expressing ACE2 (Figure 4C) (FDR-adjusted p = 0.00023), specifically sub-clusters 7 and 13, which represent two varieties of secretory epithelial cell (Figures 4C, 4F, and 4G). Cluster 7 secretory cells are marked by S100P, LYPD2, PSCA, CEACAM5, and STEAP4; encompass some MUC5AC goblet cells; and contain the most significantly enriched ACE2 and TMPRSS2 expression (4% express ACE2, FDR-adjusted p = 7.32E−28; 28% express TMPRSS2, FDR-adjusted p = 2.15E−132; Table S6). We next explicitly gated cells by their TMPRSS2 and ACE2 expression, identifying a rare subset that co-expresses both, the majority of which fall within the “Secretory Cluster 7” cell type (Figures 4E and 4F) (30 cells, ∼0.3% of all upper airway secretory cells, 1.6% of goblet “Secretory Cluster 7”). These findings are aligned with concurrent work by the HCA Lung Biological Network on human nasal scRNA-seq data, which identified nasal secretory cells to be enriched for ACE2 and TMPRSS2 expression (Sungnak et al., 2020).
Although we identified co-expression of ACE2 and TMPRSS2 in few airway cells overall, we detected ACE2 and TMPRSS2 single- and double-positive cells in over 20 donors and thus posit that these genes are enriched in secretory cells and are not a product of individual-patient-driven variability (Figure S3A). Inferior turbinate scrapings collected on Seq-Well S3, which increases the resolution of lower-abundance transcripts compared with Seq-Well v1, revealed consistent and specific expression restricted to goblet secretory cells, but at a greater detection frequency in samples from the same donors (Figure S3B) (ACE2+ from 4.7% v1 to 9.8% S3; ACE2+TMPRSS2+ from 1.9% v1 to 4% S3) (Hughes et al., 2019). Using the gated ACE2+TMPRSS2+ cells, we tested for differentially expressed genes compared to the remaining secretory epithelial cells (full results provided in Table S6). Notably, we observed significant upregulation of ADAR, GBP2, OAS1, JAK1, and DUOX2 (FDR adjusted, all p < 0.02) within ACE2+TMPRSS2+ cells, potentially indicative of IFN signaling (Figure 4G). Almost all “Secretory Cluster 7” cells were from inferior turbinate scrapings of healthy and allergically inflamed individuals, few cells were from the ethmoid sinus tissue of patients with chronic rhinosinusitis without nasal polyps, and no cells were detected in polyp tissue (Figure 4H). Gene Ontology (GO) analysis of enriched genes in double-positive cells include processes related to intracellular cytoskeleton and macromolecular localization and catabolism, potentially involved in viral particle entry, packaging, and exocytosis (Fung and Liu, 2019).
Figure S3.
Nasal and Sinus Mucosa, Related to Figures 4 and 5
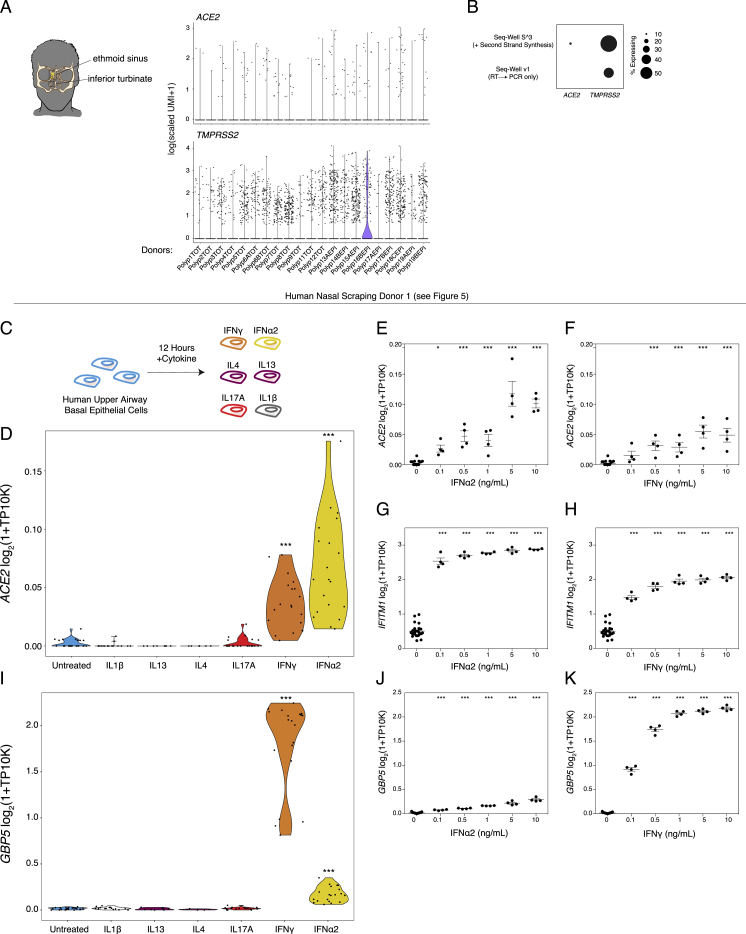
(A). Expression of ACE2 and TMPRSS2 across donors.
(B). Enhanced capture of ACE2 mRNA with second strand synthesis protocol employed in Seq-Well S3. Dot size represents fraction of cells expressing.
(C). Cultured human primary basal epithelial cells at confluence were treated with increasing doses (0.1 to 10ng/mL) of IFNα2, IFNγ, IL-4, IL-13, IL-17A, and IL-1B for 12 h and bulk RNA-seq analysis was performed (Replicate experiment using Human Donor 1 as in Figure 5)
(D). ACE2 expression by stimulation condition. Wilcoxon test between each cytokine (combined doses) versus rest: IFNα Bonferroni-adjusted p = 4.1E-07; IFNγ Bonferroni-adjusted p = 9.3E-03; all else n.s. ∗∗∗ p < 0.001.
(E). ACE2 expression by IFNα2 dose. Bonferroni-corrected t-test compared to 0 ng/mL condition: ∗∗∗ p < 0.001, ∗ p < 0.05.
(F). ACE2 expression by IFNγ dose. Bonferroni-corrected t-test compared to 0 ng/mL condition: ∗∗∗ p < 0.001, ∗ p < 0.05.
(G). IFITM1 expression by IFNα2 dose. Bonferroni-corrected t-test compared to 0 ng/mL condition: ∗∗∗ p < 0.001.
(H). IFITM1 expression by IFNγ dose. Bonferroni-corrected t-test compared to 0 ng/mL condition: ∗∗∗ p < 0.001.
(I). GBP5 expression among cultured human primary basal epithelial cells. Wilcoxon test: IFNα versus IFNγ Bonferroni-adjusted p = 2.94E-07; IFNγ Bonferroni-adjusted p = 9.3E-03. TP10K: transcripts per 10,000 reads. ∗∗∗ p < 0.001.
(J). GBP5 expression by IFNα2 dose. Bonferroni-corrected t-test compared to 0 ng/mL condition: ∗∗∗ p < 0.001.
(K). GBP5 expression by IFNγ dose. Bonferroni-corrected t-test compared to 0 ng/mL condition: ∗∗∗ p < 0.001.
We next utilized IFN-inducible gene sets of relevance to human airway epithelial cells, which we derived from a prior study by performing differential expression on a published dataset where air-liquid interface cultures from primary human nasal epithelial cells were treated with IFN-αA/D, IFN-β1a, IFN-γ, IL-4, or IL-13 (Giovannini-Chami et al., 2012, Ordovas-Montanes et al., 2018). Using these gene lists, we scored the human nasal epithelial cells analyzed by scRNA-seq described in Figures 4C and 4F and found significant concomitant upregulation of the IFN-α-stimulated gene set within ACE2+TMPRSS2+ secretory goblet cluster 7 (Figure 4I).
Type I Interferon IFN-α Drives ACE2 Expression in Primary Human Nasal Epithelial Cells
The meta-analysis described above consistently identified an association between ACE2 expression and canonical ISGs or components of the IFN-signaling pathway. This prompted us to investigate whether IFNs might play an active role in regulating ACE2 expression levels in specific target cell subsets, thus potentially allowing for a tissue-protective host response or increased viral binding of SARS-CoV-2 through ACE2. Our initial literature search indicated that IFN-γ and IL-4 downregulate the SARS-CoV receptor ACE2 in Vero E6 cells (African green monkey kidney epithelial cells [de Lang et al., 2006]), appearing to invalidate this hypothesis. Relatedly, in vitro stimulation of A549 cells, a commonly used cell line model for lung epithelia, with IFN-α, IFN-γ, and IFN-α+IFN-γ for 24 h did not identify ACE2 as an ISG (Russell et al., 2018). This is potentially explained by recent work that aimed to understand SARS-CoV-2 receptor usage by performing screening studies within cell line models and found that A549 cells did not express ACE2 and therefore represents a poor model to understand regulation of this gene (Letko et al., 2020). While conducting experiments to directly test the hypothesis that ACE2 is an ISG, we noted in our own gene lists used for scoring from Ordovas-Montanes et al., 2018 and in a supplementary extended table available from Giovannini-Chami et al., 2012 that ACE2 was in upregulated gene lists after exposure to Type I IFN.
We directly tested whether IFN-α induces ACE2 in primary human upper airway epithelial cells in greater detail. We cultured human primary basal (stem and progenitors) epithelial cells to confluence and treated them with increasing doses (0.1–10 ng/mL) of IFN-α2, IFN-γ, IL-4, IL-13, IL-17A, or IL-1β for 12 h and then performed bulk RNA-seq (Figure S3C). Only IFN-α2 and IFN-γ led to upregulation of ACE2 over the time period tested, and compared with all other cytokines, IFN-α2 lead to greater and more significant upregulation over all doses tested (Figure S3D,Wilcoxon test: IFN-α2 FDR-adjusted p = 4.1E−07; IFN-γ p = 9.3E-03,Figures S3E and S3F, all statistical tests compared with 0 ng/mL dose). We confirmed substantial and dose-dependent induction of canonical members of the interferon response after IFN-α2 and IFN-γ (Figures S3G and S3H). Conversely, we found that IFN-γ, relative to IFN-α2, induced potent upregulation of GBP5, a GTPase-like protein thought to act as a viral restriction factor through inhibiting furin-mediated protease activity, which could limit viral processing from infected cells, whereas IFN-α2 more robustly induced IFITM1 (Figure S3G–S3K) (Braun and Sauter, 2019).
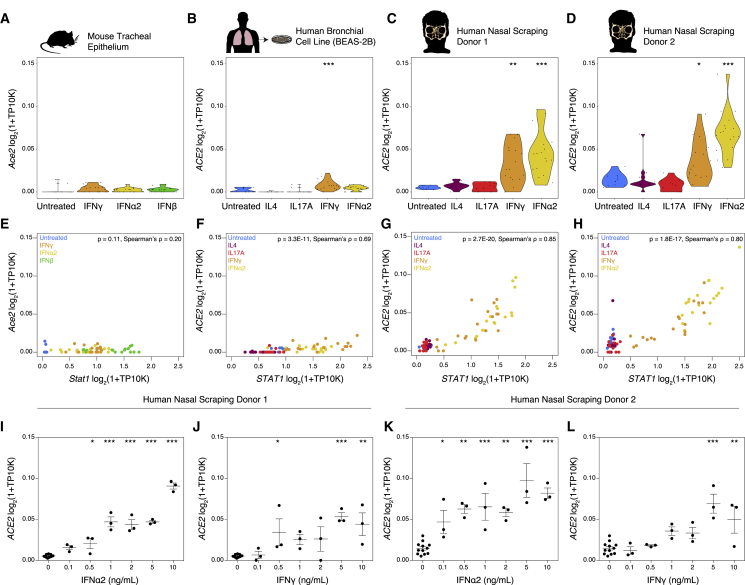
To further extend and substantiate these findings, as above, we stimulated primary mouse tracheal basal cells, the commonly used human bronchial cell line BEAS-2B, and upper airway basal cells from two human donors (Figure 5A-D). We confirmed appropriate induction of an IFN response in each cell type by performing differential expression testing between untreated cells and IFN-treated cells for each condition (Table S7). Within each cell type, stimulation with IFN-α2, IFN-γ, or IFN-β resulted in dose-dependent upregulation of canonical ISGs, including STAT1/Stat1, BST2/Bst2, XAF1/Xaf1, IFI35/Ifi35, MX1/Mx1, and GBP2/Gbp2. Notably, Ace2 expression was not robustly induced in basal cells derived from healthy mouse trachea under any interferon stimulation condition (Figure 5A). The magnitude of ACE2 upregulation was diminished in BEAS-2B cells compared to that in our original findings in primary human upper airway epithelial cells, but reached statistical significance compared with that of the untreated condition after IFN-γ exposure (Figure 5B). In primary basal cells derived from healthy nasal mucosa, we confirmed significant induction of ACE2 after IFN-α2 stimulation and, to a lesser extent, after stimulation with IFN-γ (IFN-α2-stimulated: both Bonferroni-adjusted p < 0.001; IFN-γ-stimulated: both Bonferroni-adjusted p < 0.05) (Figures 5C and 5D). Expression of ACE2 was significantly correlated with expression of STAT1 in all human cell types, with a larger effect size and correlation coefficient in primary human basal cells (Figure 5E-H). These experiments support a relationship between induction of the canonical IFN response, including key transcription factors and transcriptional regulation of the ACE2 locus. Finally, among primary human samples, we confirmed the dose-dependence of ACE2 upregulation after IFN-α2 or IFN-γ treatment and significant induction of ACE2 after IFN-α2 stimulation at concentrations as low as 0.1–0.5 ng/mL (Figure 5I-L).
Figure 5.
ACE2 is an Interferon-Stimulated Gene in Primary Human Barrier Tissue Epithelial Cells
(A–D) Basal epithelial cells from distinct sources were cultured to confluence and treated with increasing doses (0.1–10 ng/mL) of IFN-α2, IFN-γ, IL-4, IL-17A, and/or IFN-β for 12 h and bulk RNA-seq analysis was performed. Expression of ACE2 (human) or Ace2 (mouse) by cell type and stimulation condition. (A) Primary mouse basal cells from tracheal epithelium are shown. (B) BEAS-2B human bronchial cell line is shown. (C) Primary human basal cells from nasal scraping, Donor 1, is shown. (D) Primary human basal cells from nasal scraping, Donor 2. Abbreviation is as follows: TP10K, transcripts per 10,000 reads. ∗∗∗p < 0.001, ∗∗p < 0.01, ∗p < 0.05, Bonferroni-corrected t test compared with untreated condition.
(E–H) Co-expression of STAT1/Stat1 and ACE2/Ace2 by cell type. (E) Primary mouse basal cells from tracheal epithelium are shown. (F) BEAS-2B human bronchial cell line is shown. (G) Primary human basal cells from nasal scraping, Donor 1, are shown. (H) Primary human basal cells from nasal scraping, Donor 2 are shown. Abbreviation is as follows: TP10K, transcripts per 10,000 reads. Statistical significance assessed by Spearman’s rank correlation.
(I–L) Expression of ACE2 in primary human basal cells from nasal scrapings across a range of concentrations of IFN-γ or IFN-α2. (I) IFN-α2 dose response in Donor 1 (p < 0.001 by one-way ANOVA) is shown. (J) IFN-γ dose response in Donor 1 (p < 0.01 by one-way ANOVA) is shown. (K) IFN-α2 dose response in Donor 2 (p < 0.001 by one-way ANOVA) is shown. (L) IFN-γ dose response in Donor 2 (p < 0.001 by one-way ANOVA). Abbreviation is as follows: TP10K, transcripts per 10,000 reads. ∗∗∗p < 0.001, ∗∗p < 0.01, ∗p < 0.05, Bonferroni-corrected post hoc testing compared with 0 ng/mL condition.
See also Figures S3 and S4 and Table S7.
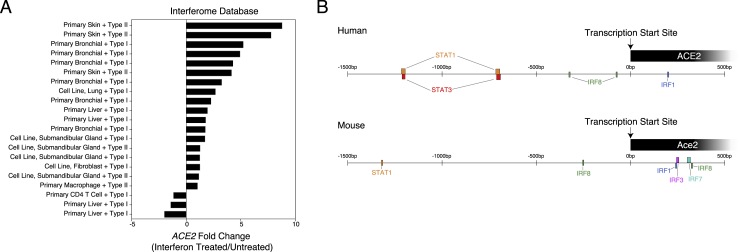
Next, using a publicly available resource (interferome.org) that hosts genomic and transcriptomic data from cells or tissues treated with IFN, we queried ACE2 expression within human and mouse cells, searching for datasets with a log2-fold-change of >1 or < −1 compared with untreated samples, including all IFN types (Rusinova et al., 2013). We recovered 21 datasets spanning 8 distinct primary tissues or cell lines with non-trivial changes in ACE2 expression after both type I and type II IFN treatment (Figure S4A). We observed substantial upregulation of ACE2 in primary skin and primary bronchial cells treated with either type I or type II IFN (> 5-fold upregulation compared with that in untreated cells), in strong support of our in vitro data (Figures 5C, 5D, 5G–5L, and S3D–S3F). Immune cell types, such as CD4 T cells and macrophages, were noticeably absent from datasets with a significant change in ACE2 expression after IFN stimulation or were even found to downregulate ACE2 (e.g., primary CD4 T cells + type I IFN) (Figure S4A, and in our analysis of scRNA-seq peripheral blood mononuclear cell data from Butler et al., (2018); data not shown).
Figure S4.
Published Studies of Epithelial Cells Following Interferon Treatment Related to Figure 5
(A). Fold change of ACE2 expression among human or mouse datasets following Type I or Type II interferon treatment compared to untreated control. Generated from publicly available microarray data curated at interferome.org. Includes all studies with abs(fold-change) > 1.
(B). Location of transcription factors binding regions spanning −1500 bp to +500 bp from the transcription start site of ACE2 (human, top) or Ace2 (mouse, bottom). Generated from TRANSFAC data using the interferome.org database (Matys et al., 2003, Rusinova et al., 2013).
Given that the majority of cells robustly upregulating ACE2 were epithelial, this observation potentially explains why previous analyses to define canonical ISGs within immune populations did not identify ACE2 as an induced gene. Furthermore, using both Transcription Factor database (TRANSFAC) data hosted by the interferome database, as well as chromatin immunoprecipitation sequencing (ChIP-seq) data (provided by the ENCODE Factorbook repository), we found evidence for STAT1, STAT3, IRF8, and IRF1 binding sites within −1500–500 bp of the transcription start site of ACE2 (all in human studies, Figure S4B) (Gerstein et al., 2012, Matys et al., 2003, Wang et al., 2012, Wang et al., 2013). This finding is supportive of our current hypothesis that ACE2 represents a previously unappreciated ISG in epithelial cells within barrier tissues.
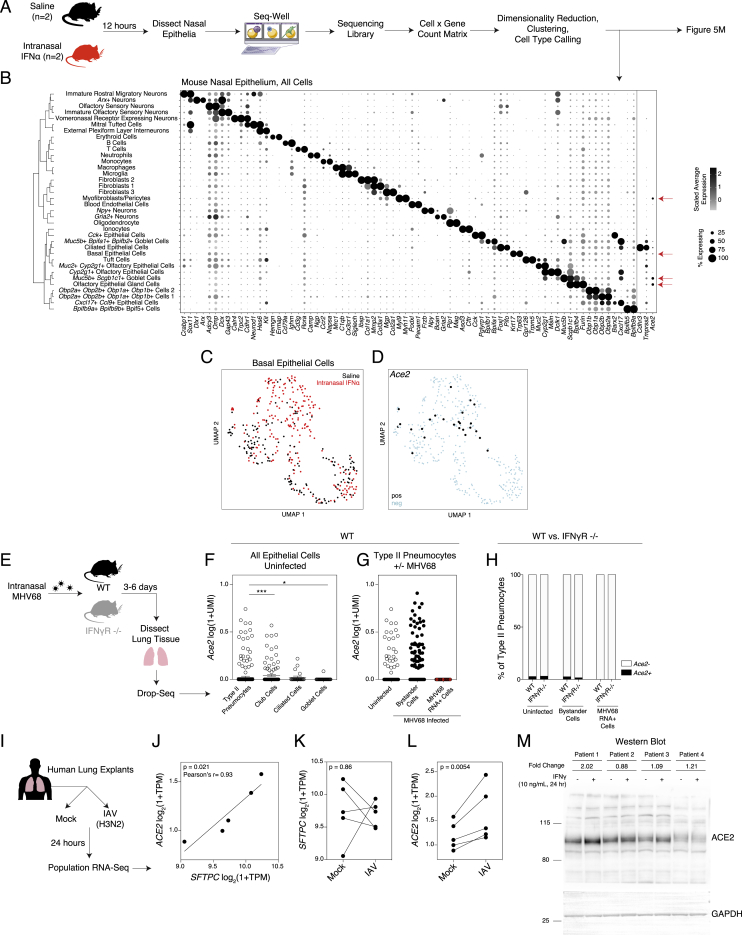
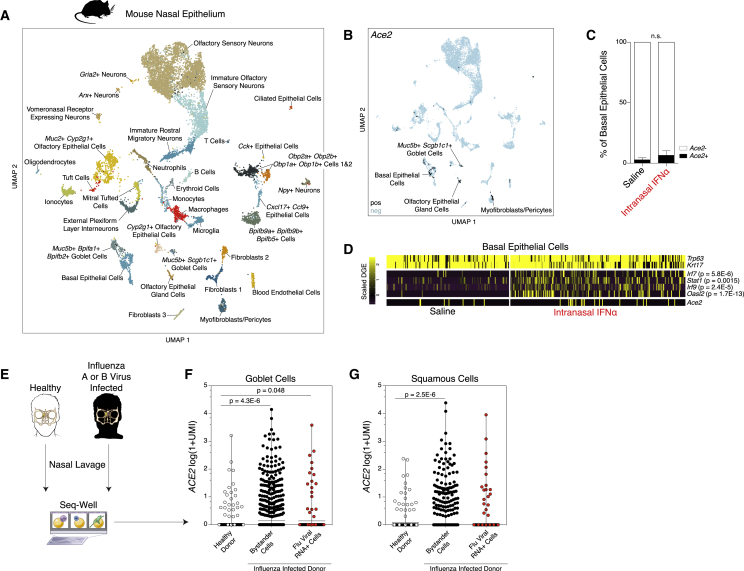
Given minimal upregulation of Ace2 among primary mouse basal cells in vitro, we were curious as to whether Ace2 represented a murine ISG in vivo. We treated two mice intranasally with saline and two mice intranasally with 10,000 units of IFN-α (Guerrero-Plata et al., 2005). After 12 h, we isolated the nasal mucosa, consisting of both respiratory and olfactory epithelium, with underlying lamina propria, and performed scRNA-seq using Seq-Well S3 (Figure S5A). We collected from both tissue sites because of early reports of anosmia in COVID-19 (Lechien et al., 2020). We recovered 11,358 single cells, including epithelial, stromal, neuronal, and immune cell types, generating the largest single-cell atlas of mouse respiratory and olfactory mucosa to date (Figures 6A and S5B). We annotated all 36 clusters, focusing our attention on epithelial cell clusters, given that we noted enrichment for Ace2 and Tmprss2 within epithelial cell subsets, consistent with our human and NHP results (Table S8). Specifically, we found Ace2 enriched within olfactory epithelial gland cells, Muc5b+Scgb1c1+ goblet cells, basal epithelial cells, and myofibroblasts/pericytes (Bonferroni-corrected p < 0.01) (Figures 6B and S5B) (Brann et al., 2020, Dear et al., 1991, Montoro et al., 2018, Tepe et al., 2018). Notably, Furin was enriched within olfactory epithelial gland cells (Table S8). Next, we asked whether a 12 h stimulation with IFN-α would upregulate Ace2 in vivo. Focusing on basal epithelial cells, which contain the highest abundance of Ace2+ cells, we found that despite robust upregulation of canonical murine ISGs, Ace2 expression was only slightly elevated after IFN-α treatment (Figures 6C, 6D, S5C, and S5D).
Figure S5.
Mouse Nasal Epithelium Following Interferon-α Exposure Related to Figure 6
(A). Schematic: mice were exposed to 10,000 units of IFN-α or saline by intranasal application (n = 2 per group). After 12 h, animals were sacrificed and nasal epithelium was dissected and dissociated for scRNA-seq using Seq-Well S3.
(B). Dot plot of 2 defining genes for each cell type, with Ace2, Tmprss2, and Cdhr3. Dot size represents fraction of cells within cell type expressing, and color intensity binned count-based expression level (log(scaled UMI+1)) among expressing cells. All cluster defining genes are provided in Table S8. Red arrows: cell types with largest proportion of Ace2+ cells. Dendrogram (left) by person correlation over differentially expressed genes with Ward clustering.
(C). UMAP of Basal Epithelial Cells (380 cells) across 4 mice. Black: Saline-treated mouse; red: IFN-α treated.
(D). UMAP of Basal Epithelial Cells as in C, points colored by detection of Ace2. Black: RNA positive, blue: RNA negative (6.6% Ace2+, Bonferroni-adjusted p = 1.1E-10 for Basal Epithelial Cell expression versus all other cells).
(E). Schematic: wildtype (WT) and IFNγ-receptor knockout (IFNγR−/−) mice were infected intranasally with murine gamma-herpesvirus-68 (MHV68). Cells from whole lung were digested for scRNA-seq using Drop-seq (yielding 5,558 Epcam+ cells).
(F). Expression of Ace2 by epithelial cell type, wild type (WT) mice. Statistical significance by Wilcoxon rank sum test with Bonferroni correction.
(G). Expression of Ace2 among type II pneumocytes binned by infection status in WT mice. All pairwise comparisons non-significant (p > 0.05) by Wilcoxon rank sum test.
(H). Percent of Ace2+ cells by infection condition (uninfected, bystander cells in MHV68-infected mouse, MHV68 RNA+ cells) and mouse genotype (WT, IFNγR −/−). Black bars: Ace2+ positive cells; white bars: Ace2- cells.
(I). Schematic of RNA-Seq data from (Matos et al., 2019) of human lung explants (n = 5 donors) exposed to influenza A virus (IAV, H3N2) at 24 h post infection.
(J). Expression of SFTPC (surfactant protein C, a marker of type II pneumocytes) versus ACE2 among mock-infected lung explants. Statistical significance assessed by Pearson’s correlation, r = 0.93, p = 0.021. TPM: transcripts per million.
(K). SFTPC expression among matched donors following mock or IAV infection for 24 h. Statistical significance assessed by ratio paired t test, p = 0.86.
(L). ACE2 expression among matched donors following mock or IAV infection for 24 h. Statistical significance assessed by ratio paired t test, p = 0.0054.
(M). Western blot of fully-differentiated air-liquid interface cultures from bronchial cells derived from 4 human donors with asthma. Cells from each donor were treated with 10 ng/mL IFNγ for 24 h, and compared to a matched untreated condition. ACE2 protein: AF933 (R&D). Fold changes quantified for IFNγ treated versus untreated for each patient donor following normalization to GAPDH.
Figure 6.
In Vivo Administration of Interferons in Mice Does Not Induce Ace2, and ACE2 Is Induced in Goblet Secretory Cells during Human Influenza Infection
(A) UMAP of 11,358 single cells from mouse nasal epithelium (n = 4).
(B) UMAP projection as in (A), points colored by detection of Ace2 (SARS-CoV-2 receptor homolog). Color coding is as follows: black, RNA positive; blue, RNA negative.
(C) Percent of Ace2+ cells by treatment condition (n = 4 arrays per condition; n = 2 arrays per mouse). Black bars indicate Ace2+ cells; white bars indicate Ace2− cells. p = 0.4 by Student’s t test.
(D) Heatmap of cell-type-defining genes (Trp63 and Krt17), interferon-induced genes (Irf7, Stat1, Irf9, and Oasl2), and Ace2 among basal epithelial cells, separated by cells derived from saline-treated mice (left) and IFN-α-treated mice (right). Statistical significance by likelihood-ratio test with Bonferroni correction is shown. A full list of differentially expressed genes can be found in Table S8.
(E) Schematic for sampling cells derived from nasal washes of n = 18 human donors with and without current influenza A or B infection for Seq-Well v1 (35,840 single cells). See Cao et al., (2020).
(F and G) ACE2 expression among goblet cells (F) and squamous cells (G) by infection status. Shown are Healthy Donor cells from influenza-negative donors (white); Bystander Cells from influenza A (IAV)- or influenza B (IBV)-infected donors, no intracellular viral RNA detected (black); Flu Viral RNA+ Cells with detectable intracellular influenza A or B viral RNA (red). Statistical significance by Wilcoxon test with Bonferroni correction, n.s. for Bystander versus Flu Viral RNA+.
This observation was supported by analysis of scRNA-seq data from 5,558 epithelial cells from the lungs of mice 3–6 days after intranasal infection with murine gamma herpesvirus-68 (MHV68) (Figure S5E). Here, we found significant enrichment of Ace2+ cells within type II pneumocytes, in line with our data from NHP and human lungs (Figures S5F). We did not observe changes in Ace2 expression among viral-transcript-positive cells or “bystander” type II pneumocytes (those without detectable cell-associated viral RNA in MHV68-infected animals), nor did we see significant alterations in Ace2+ cell abundance among MHV68-infected mice lacking IFN-γR (Figure S5G and S5H). These observations were in agreement with our in vitro murine basal cell assay (Figure 5A and 5E).
Finally, we sought to validate our hypothesis that ACE2 is upregulated in human epithelial cells during upper airway viral infections, which are known to induce a robust IFN response (Bailey et al., 2014, Everitt et al., 2012, Iwasaki and Pillai, 2014, Jewell et al., 2010, Russell et al., 2018, Steuerman et al., 2018). We re-analyzed a publicly available dataset of RNA-seq from human lung explants isolated after surgical resections that were infected with influenza A virus ex vivo for 24 h. Here, we found that ACE2 expression was significantly correlated with that of SFTPC, supporting our hypothesis that ACE2 is expressed within type II pneumocytes (Figures 1C, 2C, S5I, and S5J) (Matos et al., 2019). Furthermore, although the abundance of SFTPC was not significantly altered by influenza A virus infection, ACE2 expression was significantly upregulated after viral exposure (p = 0.0054, ratio paired t test) (Figures S5K and S5L). This suggests that influenza A virus infection increases ACE2 expression. Nevertheless, these population-level analyses are not able to definitively resolve specific cell subsets of relevance, nor whether they are directly infected cells or bystanders of infection.
In order to address these questions, we leveraged an ongoing scRNA-seq study of nasal washes from 18 individuals with confirmed influenza A virus or influenza B virus infection or healthy controls collected with Seq-Well v1, which yielded 35,840 cells resolved into 17 distinct cell types (Figure 6E; STAR Methods) (Cao et al., 2020). We investigated the cell types with greatest enrichment for ACE2 and TMPRSS2 in non-infected controls and individuals with influenza A and B. Strikingly, ACE2 was most upregulated in samples from influenza-virus-infected individuals within bystander goblet or squamous cells not directly infected by virus (Figures 6F and 6G). ACE2+TMPRSS2+ goblet cells during influenza infection exhibited enrichment for canonical ISGs such as the CXCL9/CXCL10/CXCL11 gene cluster; correspondence with ACE2+TMPRSS2+ goblet cells in healthy and allergic nasal scrapings; and a shared overlap in ISGs including GBP2, ZNFX1, ADAR, and ACE2 (significantly differentially expressed gene lists) (Table S6). Together, our data suggest that ACE2 is an ISG in vitro and in vivo in human primary upper airway epithelial basal cells, but that the murine homolog Ace2 is not in airway epithelial basal cells or pulmonary epithelial cells in vitro or in vivo. Collectively, our findings suggest that careful considerations of animal and cellular models will be needed for assessing therapeutic interventions targeting the IFN system when studying ACE2/Ace2-associated biology.
Finally, because our in vivo and in vitro work indicate that IFN might promote human cellular targets for SARS-CoV-2 infection in the human upper airway by inducing ACE2, we attempted to extend our transcriptomic data on IFN-driven expression of ACE2 to protein-level induction of ACE2. As testing of various commercially available polyclonal antibody preparations found broad evidence for non-specific or inconclusive staining in histological immunofluorescent based readouts (data not shown), we assessed whether IFN-γ-stimulated human bronchial air-liquid interface cultures induced ACE2 within 24 h. Our results show that cells from one patient robustly induced ACE2 (+2.02x), cells from another mildly induced ACE2 (+1.21x) and two patient’s cells showed minor changes (+/−1.12x) (Figure S5M). We provide a note of caution as these cells were derived from asthmatic patients, and the overall changes did not reach significance. Furthermore, we could not determine cell surface localization of ACE2 but do note that these results align with our transcriptomic data.
Discussion
Here, we utilize scRNA-seq across various barrier tissues and model organisms to identify the potential initial cellular targets of SARS-CoV-2 infection. To review the data presented: (1) we found that expression of the cellular entry receptor for SARS-CoV-2, ACE2, is primarily restricted to type II pneumocytes in the lung, absorptive enterocytes within the gut, and goblet secretory cells of the nasal mucosa; (2) ACE2 and TMPRSS2 co-expression in respiratory tissues is consistently found only among a rare subset of epithelial cells; (3) we observed similarities in the cellular identities and frequencies of putative SARS-CoV-2 target cells across human and NHP cohorts; (4) we observe increased expression of ACE2 during SHIV and TB infection of NHPs, and HIV/TB co-infection and influenza infection of humans compared with that in matched controls but caution that none of the datasets presented here were designed to answer this specific query. Specific targeting of these cell subsets has only been described for a handful of viruses, including the following: goblet cells by human adenovirus-5p and enterovirus 71, type II pneumocytes by H5N1 avian influenza, and absorptive enterocytes by rotavirus (Fleming et al., 2014, Good et al., 2019, Holly and Smith, 2018, Weinheimer et al., 2012).
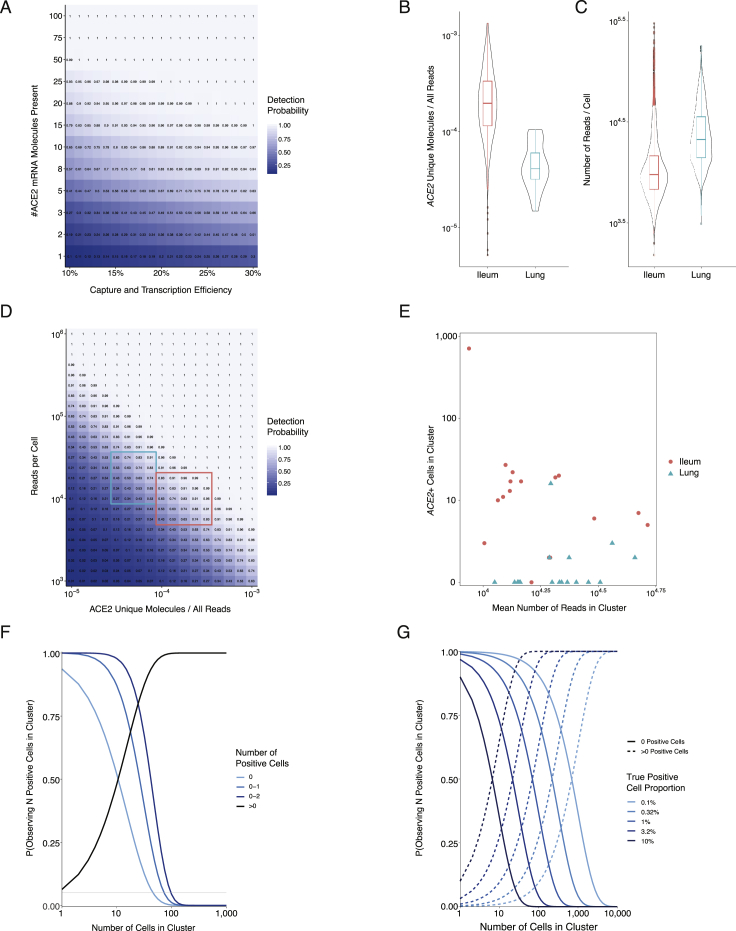
Additionally, we provide an overall note of caution when interpreting scRNA-seq data for low abundance transcripts like ACE2 and TMPRSS2 because detection inefficiencies might result in an underestimation of the actual frequencies of ACE2+ or ACE2+TMPRSS2+ cells in a tissue. Moreover, the protein amounts of each might differ from their mRNA abundances (Genshaft et al., 2016, Jovanovic et al., 2015, Rabani et al., 2011, Shalek et al., 2013). We also present datasets separately, given that each study differed in its methods of tissue processing and collection, which can influence the frequency of recovered cell subsets (STAR Methods). We provide Table S9 as a summary of ACE2+ and ACE2+TMPRSS2+ cells across various datasets. Moreover, we present Figure S6, which describes statistical modeling and power calculations underlying detection and dropout of ACE2, to help guide interpretation of these data. This includes an examination of the probability to detect a lowly expressed transcript like ACE2 within a cell, as well as upper bound estimates on the percentage of positive cells within a cluster, considering the effects of transcript counts, sequencing depth, and cell numbers in these calculations (STAR Methods).
Figure S6.
Power Calculations and Statistical Modeling of ACE2 Capture and Dropout Related to STAR Methods
(A). Probability of capturing and transcribing at least 1 ACE2 cDNA molecule, as a function of the capture/reverse transcription efficiency for a single molecule and the number of ACE2 molecules expressed in an individual cell. Note that Drop-Seq provides a capture/transcription efficiency of approximately 11-13%, setting a floor on this parameter, and the experimental platforms used in this study are either equivalent or superior (Macosko et al., 2015).
(B). Distribution of ACE2 fractional abundance within individual cells’ cDNA libraries (i.e., ACE2 UMIs / total number of reads), across non-human primate lung and ileum cell populations (see Figures 1 and 3). Mean fractional abundance among ACE2+ lung cells = 5.0E-5; mean fractional abundance among ACE2+ ileum cells = 2.7E-4.
(C). Distribution of the number of reads within non-human primate lung and ileum cell populations (see Figures 1 and 3). Mean ± SEM reads among all lung cells = 28,512 ± 344; ACE2+ lung cells = 28,553 ± 2,988; all ileum cells = 14,864 ± 288; ACE2+ ileum cells = 10,591 ± 441.
(D). Probability of observing at least one transcript for a gene of interest (e.g., ACE2) within an individual cell, as a function of sequencing depth and the gene’s fractional abundance (i.e., ACE2 reads / all reads) within the cell’s cDNA library. Fractional abundance provides the probability that a single read corresponds to the gene of interest, and presented heatmap indicates the probability that at least one read in the total number of reads allocated to the cell (i.e., from 103 to 106) originates from the gene of interest. Mean read depths and ACE2 fractional abundances for each tissue produce a 93.7% probability of detecting at least 1 ACE2 read in ileum cells, and a 76.0% chance for lung cells. Outlined rectangles highlight the regimes where cells from lung (turquoise) and ileum (pink) samples typically lie.
(E). Number of ACE2+ cells within each cluster, as a function of average read depth for all cells in that cluster. Number of cells detected as ACE2+ is not correlated with read depth, even across relatively wide ranges of average read depths (Pearson’s r = −0.31, n.s.).
(F). Probability of observing a particular number of cells positive for a gene of interest within a cluster, as a function of number of cells in the cluster. Probabilities were calculated under a negative binomial distribution with parameter p = 0.063 (the proportion of ACE2+ cells among type II pneumocytes presented in Figure 1; STAR Methods). The horizontal gray line indicates the arbitrary cut-off value of p = 0.05.
(G). Given a population of cells with a known proportion that are positive for a gene of interest, probability of observing no positive cells (i.e., false negative identification of the cluster; solid lines) and probability of observing at least one positive cell as a function of cluster size.
Whether ACE2 and TMPRSS2 are needed on the same cell or soluble proteases can activate SARS-CoV-2 S protein to invade ACE2 single-positive cells is an area of active inquiry (Coutard et al., 2020, Letko et al., 2020). Importantly, rapidly evolving literature has identified that SARS-CoV-2-S might have a furin cleavage site, leading to a broader set of host proteases that could mediate S protein activation (Bugge et al., 2009, Coutard et al., 2020, Walls et al., 2020). However, because an active S protein has a finite lifetime to find a target cell membrane, the timing and cellular location of S protein activation is key to consider. Activation events proximal to the plasma membrane have been shown to be most effective for SARS-CoV entry (Shulla et al., 2011).
Our study finds that type I IFNs, and to a lesser extent type II IFNs, upregulate ACE2. This is based on several lines of evidence: (1) we identified a human goblet secretory cell subset in upper airway nasal epithelium enriched for ACE2 expression to have the highest IFN-α-induced gene signature; (2) we found that IFN-α, and to a lesser extent IFN-β or IFN-γ, induced ACE2 expression in a published dataset of air-liquid interface cultures derived from human nasal epithelial cells (Giovannini-Chami et al., 2012, Ordovas-Montanes et al., 2018); (3) we extended our search through the Interferome database (Rusinova et al., 2013) and found that, in epithelial barrier tissues, type I IFNs upregulate ACE2 in multiple studies, especially in primary bronchial cells and keratinocytes (Rusinova et al., 2013); (4) we found two STAT1 binding sites in the promoter of ACE2; (5) in our unpublished atlas of SHIV-infected macaques, known to have elevated amounts of chronic IFN signaling, we found ACE2 upregulation in absorptive enterocytes; (6) we directly provided evidence for IFN-α, and to some extent IFN-γ, inducing ACE2 expression in primary human upper airway basal cells; and (7) influenza infection in humans, a known inducer of the IFN pathway, leads to increased ACE2 expression in goblet secretory cells of the nasal epithelium (Cao et al., 2020).
Altogether, our own and publicly available data highlight that ACE2 might have been missed as a canonical ISG because of its notable absence in peripheral blood mononuclear cell datasets and in lung-derived transformed cell lines such as the A549 cell line (Butler et al., 2018, Letko et al., 2020, Rusinova et al., 2013). Importantly, other groups have independently analyzed publicly available datasets, some referenced in our work, and observed ACE2’s behavior as an ISG (Wang and Cheng, 2020). Furthermore, we found weak IFN- or virally driven induction of Ace2 in murine cells and tissues. This highlights the importance of studying primary human epithelial cells and the careful consideration of appropriately selected gene lists and in vitro models of in vivo cellular systems for understanding human biology (Jonsdottir and Dijkman, 2016, Mead and Karp, 2019, Regev et al., 2017).
As SARS-CoV-S leads to ACE2-receptor-mediated internalization, the host IFN response could thus promote the ability for SARS-CoV and SARS-CoV-2 to maintain cellular targets in neighboring human upper airway epithelial cells. Altogether along with a study of HCoV-OC43, which co-opts IFN-inducible transmembrane 2 (IFITM2) and IFITM3 to promote viral entry, this adds to the growing evidence that coronaviruses, as well as other viruses, have evolved to leverage features of the human IFN pathway (Fung and Liu, 2019, Mar et al., 2018, Zhao et al., 2014). Whether type I IFNs are net protective or detrimental to the host might depend on the stage of infection; cell subsets in question; the SARS viral clade (Channappanavar et al., 2016, Channappanavar et al., 2019, Channappanavar and Perlman, 2017, Davidson et al., 2015); and other factors such as co-infection, age, gender, and co-morbidities, among others. Understanding the specific host restriction factors targeting SARS-CoV-2 and identifying specific drivers of these genes in the absence of ACE2 upregulation might provide strategies to dissociate the dual roles of IFN in certain coronavirus infections. Whether IFNs upregulate ACE2 in putative target cell subsets in vivo will be of significant interest to define in future work once current COVID-19-related restrictions on basic scientific inquiry are lifted (Qian et al., 2013).
ACE2 is a central component of the renin-angiotensin system, which has emerged as a key regulator of sterile- or microbially induced lung pathology (Imai et al., 2005). In brief, ACE cleaves angiotensin I to generate angiotensin II (Skeggs et al., 1980). Angiotensin II then acts to drive acute lung injury through various mechanisms, including increased vascular permeability (Imai et al., 2005). Amounts of angiotensin II in humans and mice are elevated during influenza infection, and ACE2 exerts tissue-protective functions by reducing amounts of angiotensin II (Zou et al., 2014). Binding of SARS-CoV-S to mouse ACE2 in vivo reduced ACE2 expression leading to acute acid-aspiration-induced lung failure (Kuba et al., 2005). Depending on the questions asked in future work, there are mouse models available on the basis of transgenic expression of human ACE2 (required for overt infectious pathology of SARS-CoV in mice), there are established NHP models available of SARS-CoV infection in M. fascicularis and C. aethiops, and early reports suggest symptomatic infection in M. mulatta and M. fascicularis models for SARS-CoV-2 (Bao et al., 2020, McCray et al., 2007, Munster et al., 2020, Rockx et al., 2020, Smits et al., 2011). For example, examining the efficacy of recombinant human ACE2 to act as a decoy receptor or the effect of “ACE inhibitors” in patients with, or at risk for, COVID-19 will require careful experimentation in appropriate models together with well-controlled clinical trials (Hofmann et al., 2004, Monteil et al., 2020, Vaduganathan et al., 2020).
IFN responses that induce ISGs are essential for host antiviral defense in mice, NHPs, and humans (Bailey et al., 2014, Dupuis et al., 2003, Everitt et al., 2012). Canonical ISGs function by directly restricting viruses and reducing burden (Schneider et al., 2014). More recently, disease tolerance to equivalent pathogen burden by factors that increase the ability of the host to tolerate tissue damage has been identified as part of a combined host defense strategy (Iwasaki et al., 2017, Iwasaki and Pillai, 2014, Medzhitov et al., 2012, Schneider and Ayres, 2008). Disease tolerance factors in the lung include IL-22 and amphiregulin (Iwasaki et al., 2017). During acute infection in the respiratory system, ACE2 is critical for early tissue tolerance responses to respiratory infection, including H5N1 influenza (Huang et al., 2014, Zou et al., 2014). However, our discovery that ACE2 is an ISG in human epithelial cells, along with SARS-CoV-2 utilizing host ACE2 to gain entry to cells, suggests that SARS-CoV and SARS-CoV-2 might exploit the ACE2-mediated tissue-protective response to provide further cellular targets for entry. This potential strategy employed by SARS-CoV-2 could present a unique challenge for the human host and is distinct from HCoV-OC43, which targets the two restriction factors IFITM2 and IFITM3 (Zhao et al., 2014). Our study provides motivation to understand the specific role and balance of type I and type II IFNs, as well as type III IFNs, in tissue protection during, and host restriction of, SARS-CoV-2 infection. Key experiments to understand ACE2 as an ISG in tissue protection or genuine tolerance will require the appropriate mouse, NHP, or other model in BSL3 or BSL4 facilities to execute SARS-CoV-2 viral infections and measure host tissue health along with viral loads. Further work will also be needed to understand how co-infections, as well as other host factors, might affect both the susceptibility to, and dynamics of, host SARS-CoV-2 infection. Moreover, carefully controlled clinical trials will be essential to determine the overall effects of different IFNs (Prokunina-Olsson et al., 2020).
Altogether, we anticipate that comprehensive characterization of the putative cellular targets of SARS-CoV-2 will be critical to understand basic mechanisms of viral tropism and disease pathophysiology, inform differential susceptibility among vulnerable populations, and potentially suggest unanticipated targets for drug inhibitors of viral infection. The cellular targets we nominate will need to be confirmed by specific reagents for SARS-CoV-2, as done for SARS-CoV (Ding et al., 2004). Furthermore, the transcriptional response to the virus will need to be rigorously characterized in appropriate in vitro and in vivo model systems (Blanco-Melo et al., 2020). We provide gene lists associated with target cells in specific tissues and diseases to aid the community in understanding this emergent disease. A concurrent HCA Lung Biological Network study assessing ACE2 and TMPRSS2 across more tissues also identified enrichment in nasal goblet and ciliated cells (Sungnak et al., 2020). Other studies are considering additional tissues; co-variates such as age, sex, and co-infection state; and represent a large coordinated international effort to the ongoing crisis (Pinto et al., 2020). One study in particular identified upregulation of ACE2 by respiratory viruses and TMPRSS2 by IL-13 in a pediatric cohort, suggesting further links to how underlying allergic conditions or co-infections might modulate these two SARS-CoV-2-related host factors (Sajuthi et al., 2020).
During the preparation of this manuscript, several papers have been posted to bioRxiv assessing patterns of ACE2+ and TMPRSS2+ cells in barrier tissues (Brann et al., 2020, Lukassen et al., 2020, Qi et al., 2020, Wu et al., 2020, Zhang et al., 2020). At a high level, these studies are largely in agreement with our report. Furthermore, another study appeared on medRxiv profiling bronchoalveolar lavage fluid from 3 severe and 3 mild COVID-19 patients, though they were unable to profile sufficient numbers of epithelial cells (Liao et al., 2020).
Our study highlights the power of scRNA-seq datasets, both existing and novel, to derive hypotheses relevant to human disease that might differ from paradigms established by using cell lines. Further work will be critical to determine how SARS-CoV-2 influences temporal dynamics of host responses at single-cell resolution and which host factors might affect this (Kazer et al., 2020). Given the unappreciated complexities of host-pathogen interactions between humans and SARS-CoV-2, the best measures to combat this pandemic continue to be surveillance and avoidance—especially given that a deep understanding of the full spectrum of resistance and tolerance mechanisms will require the concerted efforts of scientists around the globe (Amanat et al., 2020, Chu et al., 2020, Hadfield et al., 2018). Here, we seek to share our initial findings and data so that other groups might build on this discovery of ACE2 as an ISG and further consider the careful balance between tissue tolerance and viral infection needed at the human airway epithelium.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Biological Samples | ||
| M. mulatta lung, bone marrow, brain, colon, ileum, jejunum, liver, lung, peripheral blood, spleen, thymus, tonsil, and lymph nodes from various sites | Washington National Primate Research Center | N/A |
| Human lung tissue from surgical excess | University of KwaZulu-Natal | IRB Code: BE024/09 |
| Human non-inflamed ileal pinch biopsies | Multi-center clinical study, approved by the Institutional Review Board at Boston Children’s Hospital | IRB Code: IRB-P00030890 |
| Human nasal lavage | University of Massachusetts Medical School | N/A |
| Human nasal scraping, polyp scrapings, ethmoid sinus surgical tissue samples | Partners HealthCare Institute | N/A |
| M. fascicularis lung and granulomatous tissue | University of Pittsburgh School of Medicine | N/A |
| Antibodies | ||
| anti-ACE2 human antibody, goat polyclonal | R&D | Cat#AF933 |
| Bacterial and Virus Strains | ||
| MHV-68 | Adler et al., 2000 | N/A |
| Mycobacterium Tuberculosis, Modified Erdman Strain | Martin et al., 2017 | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| 2-Mercaptoethanol | Sigma | Cat#M3148-25ML |
| RLT Buffer | QIAGEN | Cat#79216 |
| dNTP | New England BioLabs | Cat#N0447L |
| RNase Inhibitor | Fisher Scientific | Cat#AM2696 |
| Maxima RNaseH-minus RT Enzyme | Fisher Scientific | Cat#EP0753 |
| MgCl2 | Sigma | Cat#63069-100ML |
| Betaine | Sigma | Cat#B0300-5VL |
| AMPure RNAClean XP RNA-SPRI beads | Beckman Coulter | Cat#A63987 |
| AMPure XP SPRI beads | Beckman Coulter | Cat#A63881 |
| Guanidinium thiocyanate | Sigma | Cat#AM9422 |
| Sarkosyl | Sigma | Cat#L7414 |
| Exonuclease I | New England BioLabs | Cat#M0293S |
| Klenow Fragment | New England BioLabs | Cat#M0212L |
| DNase I | Roche | Cat#10104159001 |
| Collagenase IV | Life Technologies | Cat#17104019 |
| Collagenase D | Roche | Cat#11088858001 |
| Liberase TM | Roche | Cat#5401119001 |
| TrypLE | Thermo Fisher | Cat#12604013 |
| ACK Buffer | Thermo Fisher | Cat#A1049201 |
| IFN-α | Biolegend | Cat#752802 |
| Dispase II | Thermo Fisher | Cat#17105041 |
| Elastase | Worthington Biochem | Cat#LS002292 |
| Pneumacult-Ex serum-free media | StemCell Technologies, Inc. | Cat#05040 |
| IL-4, human | Biolegend | Cat#574002 |
| IL17A, human | Biolegend | Cat#570502 |
| IFNγ, human | Biolegend | Cat#570202 |
| IFNγ, mouse | Peprotech | Cat#315-05 |
| IFNα, human | Biolegend | Cat#592702 |
| IFNα, mouse | Biolegend | Cat#752802 |
| IFNβ, mouse | R&D Systems | Cat#8234-MB-010 |
| Critical Commercial Assays | ||
| Nextera XT DNA Library Preparation Kit | Illumina | Cat#FC-131-1096 |
| High Sensitivity D5000 ScreenTape | Agilent | Cat#5067-5592 |
| Qubit dsDNA High-Sensitivity kit | ThermoFisher | Cat#Q32854 |
| NextSeq 500/550 High Output v2 (75 cycles) | Illumina | Cat#FC-404-2005 |
| NovaSeq 6000 S2 (100 cycles) | Illumina | Cat#20012862 |
| Kapa HiFi HotStart ReadyMix | Kapa Biosystems | Cat#KK2602 |
| MACOSKO-2011-10 mRNA Capture Beads | ChemGenes | Cat#NC0927472 |
| Tumor Dissociation Kit, Human | Miltenyi Biotec | Cat#130-095-929 |
| Chromium Single Cell 3′ v2 | 10X Genomics | Cat#120237 |
| Deposited Data | ||
| scRNA-seq Processed Data | This paper | https://singlecell.broadinstitute.org/single_cell?scpbr=the-alexandria-project |
| scRNA-seq Processed Data | This paper | https://drive.google.com/drive/folders/1bxCIqNeZ7wLuVOT16gphwj98_cc9KhrV?usp=sharing |
| scRNA-seq Processed Data | This paper | https://chanzuckerberg.github.io/cellxgene/posts/cellxgene_cziscience_com |
| scRNA-seq Processed Data | This paper | https://singlecell.broadinstitute.org/single_cell/covid19 |
| scRNA-seq Processed Data (all species) and FASTQ files (for NHP and murine datasets) | This paper | GEO: GSE148829 |
| scRNA-seq data from human nasal mucosa | Ordovas-Montanes et al., 2018 | https://singlecell.broadinstitute.org/single_cell/study/SCP253/allergic-inflammatory-memory-in-human-respiratory-epithelial-progenitor-cells |
| Human reference genome NCBI build 38 (GRCh38) | Genome Reference Consortium | http://www.ncbi.nlm.nih.gov/projects/genome/assembly/grc/human/ |
| Human reference genome NCBI build 19 | Genome Reference Consortium | http://www.ncbi.nlm.nih.gov/projects/genome/assembly/grc/human/ |
| Mouse reference genome NCBI build 10 | Genome Reference Consortium | http://www.ncbi.nlm.nih.gov/projects/genome/assembly/grc/mouse/ |
| Macaca mulatta reference genome assembly 8.0.1, annotation 102 | NCBI Eukaryotic Genome Annotation Pipeline | https://www.ncbi.nlm.nih.gov/genome/annotation_euk/Macaca_mulatta/102/ |
| Macaca fascicularis reference genome assembly 5, annotation 101 | NCBI Eukaryotic Genome Annotation Pipeline | https://www.ncbi.nlm.nih.gov/genome/annotation_euk/Macaca_fascicularis/101/ |
| Interferome Database | Rusinova et al., 2013 | http://www.interferome.org/ |
| RNA-seq from human lung explants ± ex vivo IAV infection | Matos et al., 2019 | GEO: GSE135069 |
| RNA-seq from human nasal epithelial cells | Giovannini-Chami et al., 2012 | GEO: GSE19190, GSE22147 |
| Experimental Models: Cell Lines | ||
| Human: Passage 4 BEAS-2B | ATCC | CRL-9609 |
| Experimental Models: Organisms/Strains | ||
| Mouse: C57BL/6J | The Jackson Laboratory | Cat#000664 |
| Mouse: C57BL/6, IFNγR−/− B6.129S7-Ifngr1tm1Agt/J | The Jackson Laboratory | Cat#003288 |
| Oligonucleotides | ||
| SMART-seq2 2 3′ Oligo-dT Primer: /5Biosg/AAG CAG TGG TAT CAA CGC AGA GTA CTT TTT TTT TTT TTT TTT TTT TTT TTT TTT TVN | Integrated DNA Technologies | N/A |
| SMART-seq2 5′ TSO: AAG CAG TGG TAT CAA CGC AGA GTA CAT rGrGrG | Integrated DNA Technologies | N/A |
| SMART-seq2 and Seq-Well ISPCR: AAG CAG TGG TAT CAA CGC AGA GT |
Integrated DNA Technologies | N/A |
| Custom Read 1 Primer: GCC TGT CCG CGG AAG CAG TGG TAT CAA CGC AGA GTA C | Integrated DNA Technologies | N/A |
| Seq-Well 5′ TSO: AAG CAG TGG TAT CAA CGC AGA GTG AAT rGrGrG | Integrated DNA Technologies | N/A |
| Seq-Well Custom P5-SMART PCR hybrid oligo: AAT GAT ACG GCG ACC ACC GAG ATC TAC ACG CCT GTC CGC GGA AGC AGT GGT ATC AAC GCA GAG TAC | Integrated DNA Technologies | N/A |
| Seq-Well dN-SMRT oligo: AAG CAG TGG TAT CAA CGC AGA GTG ANN NGG NNN B | Integrated DNA Technologies | N/A |
| Software and Algorithms | ||
| R | R Core Team | https://www.r-project.org |
| R package – Seurat v2.3.4 and v3.1.0 | Github | https://github.com/satijalab/seurat |
| Scanpy | Wolf et al., 2018 | https://github.com/theislab/scanpy |
| R package – SCDE | Bioconductor | http://bioconductor.org/packages/scde/ |
| Prism 6 | GraphPad Software | https://www.graphpad.com/scientific-software/prism/ |
| STAR | Github | https://github.com/alexdobin/STAR |
| Uniform Manifold Approximation and Projection | Github | https://github.com/lmcinnes/umap |
| Rtsne | CRAN | https://cran.r-project.org/web/packages/Rtsne/ |
Resource Availability
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by Dr. Jose Ordovas-Montanes (jose.ordovas-montanes@childrens.harvard.edu).
Materials Availability
This study did not generate new unique reagents.
Data and Code Availability
In Table S9, we provide a guide to all datasets analyzed in this paper as well as links to each individual dataset for download with the main landing page here: https://singlecell.broadinstitute.org/single_cell?scpbr=the-alexandria-project. To download the data from the portal, follow the link to the visualization page, sign in a free account in the portal using a Google apps enabled email address, and select the ‘Download’ tab in the study. Downloadable datasets include both raw and normalized cell x gene matrices, as well as relevant metadata. These datasets are additionally available here to facilitate downloading: https://drive.google.com/drive/folders/1bxCIqNeZ7wLuVOT16gphwj98_cc9KhrV?usp=sharing. We have also posted these cell x gene matrices to Chan Zuckerberg Initiative cellxgene (https://chanzuckerberg.github.io/cellxgene/posts/cellxgene_cziscience_com) and the Broad Institute Single Cell COVID-19 portal (https://singlecell.broadinstitute.org/single_cell/covid19) as leading community efforts. FASTQ files and cell x gene matrices for NHP and murine datasets, and cell x gene matrices for human datasets, are available at GEO: GSE148829.
In this same table, we further highlight four access types. 1. published datasets where everything is available (1 study); 2. unpublished datasets where everything is available (2 studies, 19,670 new cells for download), 3. unpublished datasets where ACE2+ cell subsets, and the necessary subsets to contextualize those cells (i.e., epithelial cells for type II pneumocytes) are fully available (5 studies, 17,986 new cells for download); and, 4. those unpublished datasets where expression is shared for ACE2/TMPRSS2 (2 studies, 9,112 new cells). For those unpublished datasets where only specific subsets of cells or genes are available, full expression matrices are available upon request for COVID-19 related questions.
All data included in the present study can be visualized using the following web viewer:
https://singlecell.broadinstitute.org/single_cell?scpbr=the-alexandria-project.
As we gain further insight and feedback from our own groups, collaborators, and investigators, we will continue to provide updates on our resource websites, including the utility of in vitro systems, such as organoids (Mead et al., 2018), for the study of SARS-CoV-2: http://shaleklab.com/resource/covid-19-resources/ and www.ordovasmontaneslab.com/covid-19-resources/. We also note that there are several ongoing efforts unified together through the HCA Lung Biological Network group that we will reference and to which we will link as they become available.
No custom code was used to analyze these data and all methods and packages used are cited in the Method Details section.
Experimental Model and Subject Details
Human Intestinal Biopsies
For human intestinal biopsies from the terminal ileum, the subjects were enrolled on a multi-center clinical study, which was approved by the Institutional Review Board at Boston Children’s Hospital (protocol number: IRB-P00030890). Full information related to subject age/developmental stage and sex found in metadata associated with provided raw datasets.
Human Lungs, Surgical Excess
Samples were obtained through indicated lung lobe resection or diagnostic procedures in collaboration with clinicians at the Department of Cardiothoracic Surgery at Inkosi Albert Luthuli Central Hospital in Durban, South Africa. Informed consent was obtained from each participant. The study protocol was approved by the University of KwaZulu-Natal Institutional Review Board (approval BE024/09). Full information related to subject age/developmental stage and sex found in metadata associated with provided raw datasets.
Human Nasal Polyps and Scrapings
For inferior turbinate nasal scrapings, polyp scrapings, and ethmoid sinus surgical tissue samples, the Partners HealthCare Institutional Review Board (Boston, Massachusetts), approved the study and all subjects provided written informed consent (Ordovas-Montanes et al., 2018). Full information related to subject age/developmental stage and sex found in metadata associated with provided raw datasets.
Human Nasal Washes, Healthy and Influenza Infected
The Institutional Review Board of the University of Massachusetts Medical School (Worcester, Massachusetts) approved the study and all subjects provided written informed consent.
Cell Culture of Primary Basal Cells and Cell Lines
Human basal cells from non-polyp surgical resections from ethmoid sinus, BEAS-2B cells (ATCC), or mouse tracheal basal cells were placed into culture at a number of 10,000 cells seeded at passage 3 and cultured at confluence in 96 well flat-bottom collagen-coated tissue culture plates (Corning 3596) for 48 h in Pneumacult-Ex serum-free media (StemCell Technologies, Inc.). All cells were incubated at 37°C and 5% CO2.
Non-Human Primates (M. mulatta)
Healthy and SHIV-infected non-human primate (M. mulatta) work was conducted at the Washington National Primate Research Center (WaNPRC), an AAALAC accredited program, in accordance with the regulations detailed in the U.S. Department of Agriculture Animal Welfare Act and in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. It was approved by University of Washington Institutional Animal Care and Use Committee. Expanded cohort characteristics described previously (Colonna et al., 2018). Full information related to subject age/developmental stage and sex found in metadata associated with provided raw datasets.
Non-Human Primates (M. fascicularis)
Tissues from Mycobacterium tuberculosis-infected non-human primates (M. fascicularis) were conducted at the University of Pittsburgh School of Medicine, an AAALAC accredited program, in accordance with the regulations detailed in the U.S. Department of Agriculture Animal Welfare Act and in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. Full information related to subject age/developmental stage and sex found in metadata associated with provided raw datasets.
Mouse Nasal and Olfactory Epithelium and Tracheal Cells
C57BL/6J mice purchased from Jackson laboratory (Bar Harbor, ME, USA) were maintained within Ragon Institute’s HPPF barrier facility and all experiments were conducted with institutional IACUC approval. In this study, mice were 8-10 weeks of age, representing male and female animals.
Mouse Lungs, MHV68 Infection
C57BL/6 mice were purchased from Charles River Laboratories (Sulzfeld, Germany). IFNγR−/− mice on C57BL/6 background (C57BL/6, IFNγR−/− B6.129S7-Ifngr1tm1Agt/J) were originally obtained from the Jackson Laboratory (Bar Harbor, ME, USA) and subsequently bred and propagated under SPF conditions at the Helmholtz Zentrum München. Animals with different genotypes were kept in the same animal room for the time of the experiment including an adaptation period prior to the start of the experiment. All animal experiments were in compliance with the German Animal Welfare Act (German Federal Law §8 Abs. 1 TierSchG), and the protocols were approved by the local Animal Care and Use Committee.
Method Details
Methods of Sample Collection and Tissue Preparation for Single-Cell RNA-Seq
NHP Ileum, Jejunum, Colon, Liver, Tonsil, Thymus, and Lung Tissue
Animals were perfused with 0.5 L of PBS/kg immediately following euthanasia, tissues were isolated and placed in RPMI + 10% FBS and kept on ice until dissociation. Tissue sections were digested by mincing and incubating with collagenase IV (Life Technologies) and DNase I (Roche) at 37°C for 1 h with agitation. Digested tissue was passed through a 100 μm metal strainer, cells were pelleted by centrifugation at 300 g, rinsed with RPMI + 10% FBS, counted, and prepared as a single cell suspension for scRNA-seq using Seq-Well v1 (see below).
NHP Lymphoid Organs, Bone Marrow, PBMCs
All lymph nodes, spleen, and bone marrow were ground through a metal strainer, transferred to a conical in RPMI + 10% FBS, and pelleted by centrifugation at 400 g x 10 min. LN-derived cells were resuspended in RPMI + 10% FBS, counted and prepared as a single cell suspension. Spleen, bone marrow, and PBMCs were subjected to ACK lysis for 10 min at room temperature, quenched with RPMI + 10% FBS. PBMCs and bone marrow derived cells were purified over a ficoll gradient (GE Healthcare) by centrifuging at 400 g for 20 min at room temperature with no brake. Cells were then resuspended in RPMI + 10% FBS, counted, and diluted for scRNA-seq using Seq-Well v1 (see below).
NHP Tuberculosis Infected Lung and Granuloma
Ten Mycobacterium tuberculosis infected (Martin et al., 2017) adult non-human primates (M. fascicularis) were included in this study. A piece of lung tissue (without any grossly visible pathology) and 4 individual TB lung granulomas per animal were excised at necropsy and enzymatically dissociated using the GentleMacs system (Tumor dissociation kit, human; Miltenyi Biotec). Single cell suspensions were resuspended in RPMI + 10% FBS, counted and diluted for scRNA-seq using Seq-Well S3 (see below).
Human Lung Tissue
Surgical samples from diseased lung tissue (n = 3 TB+HIV+; n = 3 TB+; n = 2 non-infected patients) were processed as described in (Ardain et al., 2019). Briefly, each sample was collected into cold RP-10 (RPMI (Sigma-Aldrich) + 10% FBS), minced, and incubated for 25-30 min at 37°C with digestion buffer containing collagenase D (Sigma-Aldrich), DNase I (Sigma-Aldrich) in RPMI 1640 (Sigma-Aldrich) with 10% FBS (Hyclone). Following incubation, samples were homogenized using a GentleMACS, filtered using a 70 μm metal strainer, and pelleted by centrifugation at 400 g for 5 min. After obtaining the pellet, cells were resuspended in RP-10, passed through another 70μm strainer (Corning), stained with trypan blue, and then counted and diluted for scRNA-seq using Seq-Well S3 (see below).
Human Ileum
Single-cell suspensions were collected from biopsies as described (Smillie et al., 2019). Briefly, biopsies were rinsed in cold PBS, the epithelial layer was separated from the underlying lamina propria by end over end rotation for 15 min. The lamina propria and epithelial fractions were digested separately, using Liberase TM (Roche) and DNase I (Roche) for the lamina propria, and TrypLE (ThermoFisher) for the epithelial fraction. Following digestion, cells were pelleted by centrifugation, subjected to ACK lysis for 3 min, and filtered through a 40 μm strainer. Following centrifugation, the cells were counted and prepared as a single cell suspension for scRNA-seq using 10X 3′ v2 (10X Genomics).
Nasal Mucosa and Nasal Scrapings
Surgical samples from ethmoid sinus and nasal scraping of the inferior turbinate were processed as described (Ordovas-Montanes et al., 2018). Briefly, each sample was collected into cold RPMI (Corning), minced and incubated for 30 min (15 min for nasal scrapings) at 37°C with digestion buffer containing collagenase IV (Worthington), DNase I (Roche) in RPMI with 10% FBS. Samples were triturated and digestion quenched with EDTA. Cells were filtered using a 70 μm metal strainer and pelleted by centrifugation at 500 g, rinsed with PBS, and subjected to red blood cell (RBC) lysis using ACK buffer (ThermoFisher) for 3 min on ice, and finally pelleted prepared as a single cell suspension for scRNA-seq using Seq-Well v1 or S3 (see below).
Interferon Treatment of Mouse Nasal Mucosa
Mice received either 200ng of IFNα (Biolegend 752802) or saline intranasally (each group n = 2 mice), and were sacrificed 12 h later. Respiratory and olfactory mucosa were isolated as in (Davidson et al., 2004, Dunston et al., 2013). Briefly, using surgical tools under a dissecting microscope, the skull bones surrounding the nasal tissue of skinned mouse heads were removed. The respiratory and olfactory mucosa were collected in RPMI media with 10% FBS. Cells were digested in media containing Liberase TM (Roche) and DNase I (Roche) for 30 min at 37°C with agitation. Cells were filtered using a 70 μm strainer, washed with EDTA-containing media to quench enzymatic digestion, and pelleted by centrifugation. RBCs were lysed using ACK buffer (ThermoFisher) for 2 min, cells were again pelleted, counted, and prepared as a diluted single cell suspension for scRNA-seq using Seq-Well S3.
MHV68 Infected Mouse Lung
Mice were housed in individually ventilated cages during the MHV68 infection period. MHV68 stocks were grown and quantified by plaque assay as previously described (Adler et al., 2000). Mice were infected intranasally (i.n.) with 5 × 10∗4 plaque forming units of MHV68 diluted in PBS in a total volume of 30 μl. Prior to i.n. infection, mice were anesthetized with medetomidine–midazolam–fentanyl. At the predetermined time points, mice were sacrificed by cervical dislocation and lung tissue was processed for subsequent experiments. All lobes were removed, minced and transferred for mild enzymatic digestion for 20-30 min at 37°C in an enzymatic mix containing Dispase (50 caseinolytic U/mL), Collagenase (2 mg/mL), Elastase (1 mg/mL), and DNase I (30 μg/mL). Single cells were harvested by straining the digested tissue suspension through a 70μm strainer. After centrifugation at 300 x g for 5 min, single cells were counted, and prepared as a single cell suspension. For Drop-seq, cells were aliquoted in PBS supplemented with 0.04% of bovine serum albumin at a final concentration of 100 cells/μl.
Nasal Washes during Influenza Infection
Nasal washes were obtained from adult healthy controls and from adults with diagnosis of acute influenza A or B by rapid antigen test (Flu A or B antigen, direct fluorescence antigen test) and/or by respiratory virus panel (PCR testing for influenza A, influenza A H1, influenza A H3, influenza B, adenovirus, metapneumovirus, respiratory syncytial virus A, respiratory syncytial virus B, rhino/enterovirus, parainfluenza 1, parainfluenza 2, parainfluenza 3), who show symptoms up to seven days (Cao et al., 2020). Samples were obtained by irrigation of each naris with up to 10 mL of saline, and collected in a single container. The sample was then transported to the research laboratory for processing. Upon receipt, the sample was immediately stored on ice and 10 mL cell growth media (DMEM or RPMI1640 with 10% fetal bovine serum) was added. The material was strained using a 40 μm nylon cell strainer (Corning) into a 50 mL centrifuge tube. Cells were pelleted at 1300 rpm for 10 min at 4°C. All but 1 mL of supernatant was discarded, the pellet resuspended in the remaining 1 mL of supernatant, and material was transferred to an Eppendorf tube and pelleted at 2000 rpm for 5 min. If the pellet contained visible blood, 200 μL of RBC lysis solution (ACK buffer, Thermo Fisher) was added to resuspend the pellet and incubated at room temperature for 2 min, after which 1 mL of cell media was added, and the cells were pelleted at 2000 rpm for 5 min. The final pellet was resuspended in up to 1 mL of media and quantified before performing scRNA-seq with Seq-Well v1.
Methods to Generate Single-Cell and Bulk RNA-seq Libraries
Seq-Well v1
Seq-Well was performed as described (Gierahn et al., 2017). Single cells were diluted to 15,000 cells in 200 μL RPMI + 10% FBS and deposited onto a pre-functionalized PDMS array. 15,000 cells were deposited onto the top of each PDMS array and let settle by gravity into distinct wells. The array was gently washed with PBS, and sealed using a functionalized polycarbonate membrane. Seq-Well arrays were sealed in a dry 37°C oven for 40 min, and submerged in a lysis buffer containing guanidium thiocyanate (Sigma), EDTA, 1% beta-mercaptoethanol and sarkosyl (Sigma) for 20 min at room temperature. Arrays were transferred to hybridization buffer containing NaCl (Fisher Scientific) and agitated for 40 min at room temperature, mRNA capture beads with mRNA hybridized were collected from each Seq-Well array, and beads were resuspended in a master mix for reverse transcription containing Maxima H Minus Reverse Transcriptase and buffer, dNTPs, RNase inhibitor, a 5′ template switch oligonucleotide, and PEG for 30 min at room temperature, and overnight at 52°C with end-over-end rotation. Exonuclease digestion and PCR were carried out as described. Post-whole transcriptome amplification workup involved AMPure XP SPRI bead cleanup occurred at a 0.6 x volume ratio, followed by 0.8x. Library size was analyzed using an Agilent Tapestation hsD5000 kit, confirming the expected peak at ∼1000 bp, and absence of smaller peaks corresponding to primer. Libraries were quantified using Qubit High-Sensitivity DNA kit and prepared for Illumina sequencing using Nextera XT DNA Sample Preparation kit using 900 pg of cDNA library as input to tagmentation reactions. Amplified final libraries were purified twice with AMPure XP SPRI beads as before, with a volume ratio of 0.6x followed by 0.8x. Libraries from 2-3 Seq-Well arrays were pooled and sequenced together using a NextSeq 500/550 High Output v2 kit (75 cycles) using a paired end read structure with custom read 1 primer: read 1: 20 bases, read 2: 50 bases, read 1 index: 8 bases.
Seq-Well S3
Seq-Well S3 modified the following protocol steps from v1, above (Hughes et al., 2019). First, hybridization buffer was supplanted with 8% (v/v) polyethylene glycol (PEG, Sigma). Second, after exonuclease digestion, bead-associated cDNA was denatured for 5 min in 0.2 mM NaOH with end over end rotation. Next, beads were washed with TE + 0.01% tween-20, and second strand synthesis was carried out by resuspending beads in a master mix containing Klenow Fragment (NEB), dNTPs, PEG, and the dN-SMRT oligonucleotide to enable random priming off of the beads.
10X v2 3′
Single cells were loaded onto 3′ library chips as per the manufacturers protocol for Chromium Single Cell 3′ Library (v2) (10X Genomics). Each biopsy was sequenced on two channels of the 10X Chromium Single Cell Platform, one for the epithelial fraction and the other for the lamina propria fraction in order to recover sufficient numbers of epithelial and lamina propria cells for downstream analyses. An input of 6,000 single cells was added to each channel with a recovery rate of approximately 2,000 cells.
Drop-seq
Drop-seq experiments were performed according to the original protocol (Macosko et al., 2015). Briefly, single cells (100/μl) were co-encapsulated in droplets with barcoded beads (120/μl, ChemGenes) at rates of 4000 μl/h. Droplet emulsions were collected for 10-20 min/each prior to droplet breakage by perfluorooctanol (Sigma-Aldrich). After breakage, beads were harvested and the hybridized mRNA transcripts reverse transcribed (Maxima RT, Thermo Fisher). Exonuclease digestion and PCR were carried out as described (12 PCR cycles). For each sample, 1 ng of pre-amplified cDNA from an estimated 1000 cells was tagmented by Nextera XT (Illumina) with a custom P5-primer (Integrated DNA Technologies). Single-cell libraries were sequenced in a 100 bp paired-end run on the Illumina HiSeq4000.
Smart-Seq2 for Bulk RNA-Seq
Population RNA-seq was performed as described (Ordovas-Montanes et al., 2018, Trombetta et al., 2014). Briefly, RNA from population lysates was purified using AMPure RNA Clean Spri beads (Beckman Coulter) at a 2.2x volume ratio, and mixed with oligo-dT primer, dNTPs (NEB), and RNase inhibitor (Fisher Scientific) at 72°C for 3 min on a thermal cycler to anneal the 3′ primer to polyadenylated mRNA. Reverse transcription was carried out in a master mix of Maxima RNaseH-minus RT enzyme and buffer (Fisher Scientific), MgCl2 (Sigma), Betaine (Sigma), RNase inhibitor, and a 5′ template switch oligonucleotide, and PCR was carried out using KAPA HiFi HotStart ReadyMix (Kapa Biosystems) and IS PCR primer and amplified for 18 cycles. Libraries were purified using AMPure XP SPRI beads at a volume ratio of 0.8x followed by 0.9x. Library size was assessed using a High-Sensitivity DNA chip (Agilent Bioanalyzer), confirming the expected size distribution of ∼1,000-2,000 bp. Tagmentation reactions were carried out with the Nextera XT DNA Sample Preparation Kit (Illumina) using 250 pg of cDNA per single cell as input, with modified manufacturer’s instructions as described. Libraries were purified twice with AMPure XP SPRI beads at a volume ratio of 0.9x, size distribution assessed using a High Sensitivity DNA chip (Agilent Bioanalyzer) and Qubit High-Sensitivity DNA kit (Invitrogen). Libraries were pooled and sequenced using NextSeq500/550 High Output v2 kits (75 cycles, Illumina) using 30-30 paired end sequencing with 8-mer dual indexing.
Human and Mouse Basal Cell Cytokine Stimulation
Data represented in Figures 5A–5L: Cytokines were added for 12 h overnight at increasing doses (0, 0.1, 0.5, 1, 2, 5, 10 ng/mL) of IL-4 (human: Biolegend 574002), IL-17A (human: Biolegend 570502), IFNγ (human: Biolegend 570202; mouse: Peprotech 315-05), IFNα (human: Biolegend 592702; mouse: Biolegend 752802), or IFNβ (mouse: R&D Systems 8234-MB-010). Each condition was run as a biological triplicate. Data represented in Figure S3C-K: cytokines were added for 12 h overnight at increasing doses (0, 0.1, 0.5, 1, 5, 10 ng/mL) of human IL-4 (Biolegend 574004), IL-13 (Biolegend 571104), IFNα (Biolegend 592704), IFNγ (Biolegend 570204), IL-17A (Biolegend 570504), or IL-1β (Biolegend 579404) (each condition run as a biological quadruplicate). All populations were lysed in 50 μL lysis buffer (RLT + 1% BME, QIAGEN and Sigma, respectively) and snap frozen on dry ice. Bulk RNA-seq was performed as described previously and summarized above (Ordovas-Montanes et al., 2018). Populations were sequenced to an average ± SEM read depth of 3.95 ± 0.11 million reads per sample, with an average ± SEM alignment percentage to either hg19 or mm10 reference transcriptomes of 71 ± 0.3%. All samples met quality thresholds regarding genomic and transcriptomic alignment.
Western blot for human ACE2
Established air-liquid interface cultures from bronchial brushings of four asthmatic patients were treated with 10ng/μL of human IFNγ for 24 h. Protein lysates were prepared, and anti-ACE2 human antibody (AF933 R&D goat polyclonal) was used to probe for ACE2 by western blot. Bands were normalized to GAPDH as loading control, and fold change was computed based on normalized ACE2 values.
Quantification and Statistical Analysis
Non-Human Primate Lung and Ileum
Libraries corresponding to 7 animals (variable number of tissues per animal) were sequenced using Illumina NextSeq. Reads were aligned to the M. mulatta genome assembly 8.0.1 annotation version 102 and processed according to the Drop-Seq Computational Protocol v2.0 (https://github.com/broadinstitute/Drop-seq). Data was normalized and scaled using the Seurat R package v2.3.4 (https://satijalab.org/seurat/): transforming the data to loge(UMI+1) and applying a scale factor of 10,000. To identify major axes of variation within our data, we first examined only highly variable genes across all cells, yielding approximately 1,000-3,000 variable genes with average expression > 0.1 log-normalized UMI across all cells. An approximate principal component analysis was applied to the cells to generate 100 principal components (PCs). Using the JackStraw function within Seurat, we identified significant PCs to be used for subsequent clustering and further dimensionality reduction. For 2D visualization and cell type clustering, we used a Uniform Manifold Approximation and Projection (UMAP) dimensionality reduction technique (https://github.com/lmcinnes/umap) with “min_dist” set to 0.5 and “n_neighbors” set to 30. To identify clusters of transcriptionally similar cells, we employed unsupervised clustering as described above using the FindClusters tool within the Seurat R package with default parameters and k.param set to 10 and resolution set to 0.5. Each cluster was sub-clustered to identify more granular cell types, requiring each cell type to express > 25 significantly upregulated genes by differential expression test (FindMarkers implemented in Seurat, setting “test.use” to “bimod,” Bonferroni-adjusted p value cutoff < 0.001). Differential expression tests between cells from ACE2+ versus ACE2- Type II Pneumocytes were conducted using the SCDE R package with default parameters (Kharchenko et al., 2014). Expression data for epithelial cells and enterocytes included in this dataset can be visualized and downloaded here: https://singlecell.broadinstitute.org/single_cell/study/SCP807?scpbr=the-alexandria-project#study-summary.
Human Lung Tissue
Libraries corresponding to 8 donors were sequenced using Illumina NextSeq. Reads were aligned to the hg19 genome assembly and processed according to the Drop-Seq Computational Protocol v2.0 (https://github.com/broadinstitute/Drop-seq). Data was normalized and scaled using the Seurat R package v3.1.0 (https://satijalab.org/seurat/), transforming the data to loge(UMI+1) and applying a scale factor of 10,000. For each array, we assessed the quality of constructed libraries by examining the distribution of reads, genes and transcripts per cell. Variable gene selection, principal components analysis, and selection of significant principal components was performed as above. We visualized our results in a two-dimensional space using UMAP (https://github.com/lmcinnes/umap), and annotated each cluster based on the identification of highly expressed genes. To further characterize substructure within cell types (for example, T cells), we performed dimensionality reduction (PCA) and clustering over those cells alone. Sub-clusters (i.e., clusters within broad cell type classifications) were annotated by cross-referencing cluster-defining genes with curated gene lists and online databases SaVanT (http://newpathways.mcdb.ucla.edu/savant-dev/) and GSEA/MsigDB (https://www.gsea-msigdb.org/gsea/msigdb/index.jsp). Proliferating cells from the human lung (Figure 2C) express high levels of mitotic markers, such as MKI67, and represent primarily T cells (CD3D, CD3E), B cells/antibody-secreting cells (IGJ, MZB1, IGHG1), and myeloid cells (CD14, APOE) and represent a composite cell cluster. Differential expression analysis between ACE2+ TMPRSS2+ and negative type II pneumocytes was performed in Seurat using a likelihood-ratio test (FindMarkers implemented in Seurat, setting “test.use” to bimod). Expression data for epithelial cells included in this dataset can be visualized and downloaded here: https://singlecell.broadinstitute.org/single_cell/study/SCP814?scpbr=the-alexandria-project#study-summary.
Human Ileum
Libraries corresponding to 13 donors were sequenced using Illumina NovaSeq S2 with a Read 1 26bp, Read 2 91bp, Index 1 8bp configuration before reads were aligned to GRCh38. Each sample was filtered individually for low quality cells and genes by analyzing distributions of reads, transcripts, percent reads mapped to mitochondrial genes, and complexity per cell, then merged as an outer join to create a single dataset. Clustering and differential expression tests were processed using Seurat v3.1.0 (https://satijalab.org/seurat/). Normalization and variable gene selection was processed with SCTransform (https://github.com/ChristophH/sctransform). Clustering for major cell types was performed using Louvain clustering on dimensionally reduced PCA space with resolution set via grid search optimizing for maximum average silhouette score. Due to the scale of the dataset, a randomized subsampling from across the dataset was used to calculate the silhouette score. We annotated clusters based on highly expressed genes, then sub-clusters were characterized by performing PCA dimensionality reduction and clustering over those cells alone, and annotated based on highly expressed genes found via one-versus-rest differential expression test (Wilcoxon) within the major cell type. Differential expression analysis between ACE2+TMPRSS2+ and negative epithelial cells was performed in Seurat using a Wilcoxon test and Bonferroni p value correction. Expression data for epithelial cells included in this dataset can be visualized and downloaded here: https://singlecell.broadinstitute.org/single_cell/study/SCP812?scpbr=the-alexandria-project#study-summary.
Human Adult Nasal Mucosa
Sample processing, sequencing, and analysis was performed as in (Ordovas-Montanes et al., 2018). Briefly, scRNA-seq cell suspensions were freshly processed using Seq-Well v1 and Seurat v2.3.4 was utilized for computational analyses presented here (Butler et al., 2018, Satija et al., 2015). Cell by gene matrix and R code for initialization of object available to download as Supplemental Data and Supplementary Tables here https://www.nature.com/articles/s41586-018-0449-8 and here:
http://shaleklab.com/resource/mapping-allergic-inflammation/? and visualized here: https://singlecell.broadinstitute.org/single_cell/study/SCP253?scpbr=the-alexandria-project#study-summary. Scores for various cytokines acting on human airway epithelial cells were calculated based on gene lists derived for (Ordovas-Montanes et al., 2018), calculated using AddModuleScore function Seurat, and effect size calculated by Cohen’s d, as previously reported.
Granulomatous Tissue from Mycobacterium Tuberculosis Infected NHPs
Libraries corresponding to 10 animals (variable number of tissues/animal) were sequenced using Illumina NovaSeq S2. Data was aligned using the Dropseq-tools pipeline on Terra (app.terra.bio) to M. fascicularis reference genome assembly 5, annotation version 101. Clustering was performed using Leiden clustering in the Scanpy (scanpy.readthedocs.io) package (Wolf et al., 2018). Cell type labels were assigned using known marker genes. In this analysis, we include all epithelial cell subsets (secretory, multiciliated, type II pneumocytes, and type I pneumocytes) from all samples. Differential expression between ACE2+TMPRSS2+ cells and other cells of the matched cell subtype (e.g., Secretory Cells) were performed using the “bimod” likelihood-ratio test within each cell subtype and filtered on Benjamini-Hochberg-corrected p value < 0.05. Expression data for epithelial cells included in this dataset can be visualized and downloaded here:
Basal Cell Cytokine Stimulation
Libraries corresponding to 279 populations were sequenced using Illumina NextSeq. Reads were aligned to the hg19 or mm10 genome assembly using the cumulus platform https://cumulus-doc.readthedocs.io/en/0.12.0/smart_seq_2.html and output as TPM using RSEM v1.3.2. Populations were transformed to transcripts per 10K reads and log2(1+TP10K) transformed. ACE2 expression by stimulation condition and dose were assessed using one-way ANOVA with post hoc testing using a Bonferroni correction. Plots were generated using ggplot2, and transcriptome-wide differential expression was calculated using the Seurat R package v3.1.0 (https://satijalab.org/seurat), function FindMarkers with test.use = ”bimod.” Expression data can be visualized and downloaded here:
https://singlecell.broadinstitute.org/single_cell/study/SCP822?scpbr=the-alexandria-project.
Interferon Treatment of Mouse Nasal Mucosa
Libraries corresponding to 4 mice, with 2 Seq-Well arrays per mouse were sequenced using Illumina NextSeq as described (Gierahn et al., 2017, Hughes et al., 2019). Reads were aligned to the mm10 genome and processed according to the Drop-Seq Computational Protocol v2.0 (https://github.com/broadinstitute/Drop-seq). Data was normalized and scaled using the Seurat R package v2.3.4 (https://satijalab.org/seurat/): transforming the data to loge(UMI+1) and applying a scale factor of 10,000. Cells with fewer than 1000 UMIs and 500 unique genes were removed. To identify major axes of variation within our data, we first examined only highly variable genes across all cells, yielding approximately 5,000 variable genes. An approximate principal component analysis was applied to the cells to generate 200 principal components (PCs). Using a combination of the Jackstraw function in Seurat and observing the “elbow” of the standard deviations of PCs, we chose the top 70 PCs for subsequent clustering and visualization. For 2D visualization, we used a Uniform Manifold Approximation and Projection (UMAP) dimensionality reduction technique (https://github.com/lmcinnes/umap) with “min_dist” set to 0.3 and “n_neighbors” set to 50. To identify clusters of transcriptionally similar cells, we employed unsupervised clustering as described above using the FindClusters tool within the Seurat R package with default parameters and k.param set to 10. Resolution was chosen based on maximization of the average silhouette width across all cells. Clusters were merged if a cell type expressed fewer than 25 significantly upregulated genes by differential expression test (FindAllMarkers implemented in Seurat, setting “test.use” to “bimod,” Bonferroni-adjusted p value cutoff < 0.001). Differential expression tests between cells from saline-treated or IFNa-treated mice were assessed using the FindMarkers function with “test.use” set to “bimod. This dataset can be visualized and downloaded here:
Lung from MHV68-Infected WT and IFNγR KO Mice
Libraries corresponding to 14 mice were aligned to a custom reference genome encompassing both murine (mm10) and herpes virus genes: 84 known genes from MHV68 were retrieved from NCBI (NCBI: txid33708) and added to the mm10 mouse genome. Reads were aligned to the custom joint genome and processed according to the Drop-Seq Computational Protocol v2.0 (https://github.com/broadinstitute/Drop-seq). Barcodes with < 200 unique genes, > 20,000 UMI counts, and > 30% of transcript counts derived from mitochondrially encoded genes were discarded. Data analysis was performed using the Scanpy Package following the common procedure, the expression matrices were normalized using scran’s size factor based approach and log transformed via scanpy’s pp.log1p() function (Lun et al., 2016, Wolf et al., 2018). SoupX was utilized to reduce ambient RNA bias, using default parameters with pCut set to 0.3, and was applied to each sample before merging the count matrices (Young and Behjati, 2020). UMI per cell and cell cycle were regressed out. Highly variable genes were selected by running pp.highly_variable_genes() for each sample separately, returning the top 4,000 variable genes per sample, and genes identified in variable in > 5 samples were retained, yielding 14,305 genes. Next, only Epcam+ cells were considered, principal components (PCs) were calculated using only the selected variable genes, and 6 PCs were used to perform unsupervised Louvain clustering. Type I Pneumocytes were excluded from this analysis based on uniformly negative expression of Ace2, resulting in a final dataset subset of 5,558 cells. Cells were identified as infected if at least one viral read was detected.
Nasal Washes during Influenza Infection
Sample processing, sequencing, and analysis was performed as in (Cao et al., 2020). Reads were aligned to the GRCh37 reference genome combined with influenza genomes. Mapped reads from each sample were then corrected for Drop-seq barcode synthesis error using the Drop-seq core computational tools developed by the McCarroll Lab (Macosko et al., 2015). Genes were quantified using End Sequence Analysis Toolkit (ESAT, github/garber-lab/ESAT) with parameters -wlen 100 -wOlap 50 -wExt 0 -scPrep (Derr et al., 2016). Finally, UMIs that likely result from sequencing errors were corrected by merging any UMIs that were observed only once and have 1 hamming distance from a UMI detected by two or more aligned reads. Only cell barcodes with more than 1,000 UMIs were analyzed. Cell barcodes with mostly erythrocyte genes (HBA, HBB) were removed. From here on, the remaining cell barcodes in the matrix would be referred to as cells. The final gene by cell matrix was normalized using the scran package v3.10 (Lun et al., 2016). The normalized matrix was used for dimensionality reduction by first selecting variable genes that had a high coefficient of variance (CV) and were expressed (> = 1 UMI) by more than three cells. Influenza viral genes, interferon stimulated genes, and cell cycle related genes were removed from the variable gene list in order to minimize the impact of viral responses and mitosis on clustering and cell type identification. This resulted in the selection of 2484 variable genes. t-distributed stochastic neighbor embedding (tSNE) was applied to the first ten principal components (PCs), which explained 95% of the total data variance. Density clustering (Rodriguez and Laio, 2014) was performed on the resulting tSNE coordinates and identified four major clusters: epithelial cells, neutrophils, macrophages and leukocytes. The epithelial cell cluster and the leukocyte cluster were then re-clustered independently, as described above, to identify populations within each metacluster. Specifically, the epithelial cell cluster was re-embedded using 2629 variable genes selected by the same criteria mentioned in the previous section and 13 PCs that explained 95% of the variance. Density clustering over the epithelial cell subset revealed ten clusters. Differential gene expression analysis using edgeR (Robinson et al., 2010) was performed to identify marker genes for each cluster. Influenza-infected and bystander cells were identified after correcting for sample-specific distribution of ambient influenza mRNA contamination and predicted cells most likely to be infected identified using a hurdle zero inflated negative binomial (ZINB) model and a support vector machine (SVM) classifier.
Power Calculations for Detection of Rare Transcripts
We conducted the following statistical analysis to estimate the effects of various factors on our ability to make confident claims regarding the presence/absence of transcripts of interest (e.g., ACE2), both within individual cells and clusters (Figure S6). Specifically, we investigated the roles of capture/reverse transcription efficiency, ACE2 expression level, sequencing depth, and cell numbers. Taken together, the results of this power analysis are in agreement with other efforts to model biological and technical sources of zero-inflation within scRNA-seq data (e.g., https://satijalab.org/howmanycells and Kharchenko et al., 2014, Svensson, 2020).
We began by quantifying how likely we are to capture and transcribe at least one ACE2 mRNA molecule, as a function of the number ACE2 mRNA molecules per cell and a protocol’s efficiency (Figure S6A). Drop-Seq has a capture/transcription efficiency of ∼10% (as estimated using ERCC spike ins; see (Macosko et al., 2015), and the experimental platforms used in this study are either equivalent (e.g., Seq-Well v1, (Gierahn et al., 2017) or superior (e.g., 10-fold better unique molecule detection, 5-fold better gene detection using Seq-Well S3;(Hughes et al., 2019)). Most relevant to this context, inferior turbinate scrapings were processed using both Seq-Well v1 and Seq-Well S3 (Figure S3B). Importantly, Seq-Well S3 provided > two-fold increase in the detection frequency of rare ACE2 transcripts (i.e., ACE2+: 4.7% for v1 versus 9.8% for S3), making it reasonable to expect that such improvements in single-cell experimental technologies have yielded corresponding improvements in capture and transcription efficiency. Based on Drop-Seq’s 10% efficiency, even if ACE2 is expressed at the low level of 5 mRNA molecules per cell (a reasonable order-of-magnitude estimate, given that non-human primate ileum cells had a maximum of 10 ACE2 unique molecules per cell observed via sequencing and an average of 1.93 molecules per cell in expressing cells, see Figures 3B and 3C), our experimental platforms have a minimum likelihood of 41% to capture and reverse transcribe at least one ACE2 mRNA molecule in any given individual cell. This likelihood rapidly increases if we estimate higher efficiencies for improved scRNA-seq technologies (e.g., 67% likelihood within any individual cell at 20% capture/transcription efficiency, 76% likelihood at 25% efficiency, Figure S6A). Thus, while transcript drop-out may reduce the fraction of positive cells, with the capture and transcription efficiencies of improved single-cell technologies, the impact is likely to be minor (reads are likely underestimated by up to a factor of ∼2.5x), given a sufficient depth of sequencing (see below). We note that this impacts both clusters deemed to contain and not contain ACE2+ cells, and suggests our percentages are likely lower bounds for true expression (within a factor of ∼2.5x).
Next, we examined the probability of sequencing an ACE2 transcript as a function of read depth and ACE2’s fractional abundance in each single cell within our sequencing libraries. First, across two different tissues (non-human primate ileum and lung, representing a high expresser of ACE2 and low expresser, respectively), we calculated the proportion of unique ACE2 molecules in our ACE2+ cells (defined as any cell with at least 1 UMI aligning to ACE2) as a fraction of total reads within individual cells to provide an order-of-magnitude estimate for average ACE2 abundance in our single-cell sequencing libraries (i.e., the probability that a read within a cell corresponds to a unique molecule of ACE2, Figure S6B). We highlight that by calculating probabilities based on ACE2 unique molecules divided by an individual cell’s total reads, we are providing a conservative estimate for the probability of observing ACE2 as a function of sequencing depth (e.g., as compared to basing these probabilities on ACE2 non-UMI-collapsed reads divided by total reads). Next, we obtained information on the number of reads in these cell populations to provide estimates of average sequencing depths (Figure S6C). Using the mean fractional abundances of ACE2 from each tissue (Figure S6B) and the mean read depths for all genes (Figure S6C), we calculated the probability of detecting at least 1 ACE2 molecule (i.e., P(detecting > 0 ACE2 molecules) = 1 - (1 - ACE2 fractional abundance)Read depth). This results in a 93.7% probability in ileum-derived cell libraries that contain ACE2, and a 76.0% probability for lung-derived cell libraries, indicating that our sequencing depths are sufficient to detect ACE2+ cells (Figure S6D).
To further evaluate whether our ability to detect ACE2+ cells was an artifact of sequencing depth, we compared the number of ACE2+ cells in a cluster to the mean number of reads across all cells in that same cluster (Figure S6E). We did not observe any significant correlation: the ileum cell cluster with the highest number of ACE2+ cells had the lowest sequencing depth of all ileum clusters, and the lung cell cluster with the highest number of ACE2+ cells was approximately average in its read depth (on a log-log scale, Pearson’s r = −0.31, non-significant). Further, when comparing ACE2+ cells to ACE2- cells within a given tissue, we did not observe a positive correlation between read depth and ACE2 status (i.e., mean ± standard error of the mean, SEM, reads among all lung cells = 28,512 ± 344; mean ± SEM reads among ACE2+ lung cells = 28,553 ± 2,988; mean ± SEM reads among all ileum cells = 14,864 ± 288; mean ± SEM reads among ACE2+ ileum cells = 10,591 ± 441, full statistics on cell depth among ACE2+ cells compared to ACE2- cells of the same cell type can be found in Table S9). Thus, we can be confident that the observed differences in ACE2+ proportions across clusters are not driven by differences in sequencing depth.
Finally, we investigated how observed differences in ACE2+ proportions across clusters might be affected by cell sampling. Using the proportion of ACE2+ cells in a “typical” cluster annotated as being ACE2 positive (i.e., 6.8% in non-human primate type II pneumocytes, Figure 1), we calculated the cluster sizes needed to be confident that the probability of observing zero to a few positive cells is unlikely to have arisen by random chance (probabilities calculated under a negative binomial distribution with parameter p = 0.068, Figure S6E). We found that as cluster sizes approach and exceed 100 cells, the probability of observing zero to a few positive cells rapidly approaches zero, if we assume 6.8% of cells are positive. Further, to examine our confidence in estimating an approximate upper bound (ignoring the impact of protocol inefficiencies discussed above) for the fraction of cells positive in a cluster as a function of the number of cells in that cluster, we also calculated the probability of observing zero (and its complement, probability of observing at least 1) ACE2+ cells as a function of cluster size across true positive proportions ranging from 0.1% to 10% (probabilities calculated under a negative binomial distribution with parameter p = 0.001 to 0.1, representing hypothetical proportions of ACE2+ cells Figure S6F). Given our typical cluster sizes (on the order of hundreds of cells, exact values provided in Table S9), we find that for us to observe 0 ACE2+ cells in a cluster due to sampling artifacts, the fraction of true positives must be ∼1% or less. Thus, these complementary approaches demonstrate that our observed variations in ACE2+ cell proportions across clusters likely reflect underlying biological differences, rather than random chance.
Statistical Testing
Parameters such as sample size, number of replicates, number of independent experiments, measures of center, dispersion, and precision (mean ± SEM) and statistical significances are reported in Figures and Figure Legends. A p value less than 0.05 was considered significant. Where appropriate, a Bonferroni or FDR correction was used to account for multiple tests, alternative correction methods are noted in the figure legends or Methods. All statistical tests corresponding to differential gene expression are described above and completed using R language for Statistical Computing.
Consortia
The members of HCA Lung Biological Network are Nicholas E. Banovich, Pascal Barbry, Alvis Brazma, Tushar Desai, Thu Elizabeth Duong, Oliver Eickelberg, Christine Falk, Michael Farzan, Ian Glass, Muzlifah Haniffa, Peter Horvath, Deborah Hung, Naftali Kaminski, Mark Krasnow, Jonathan A. Kropski, Malte Kuhnemund, Robert Lafyatis, Haeock Lee, Sylvie Leroy, Sten Linnarson, Joakim Lundeberg, Kerstin B. Meyer, Alexander Misharin, Martijn Nawijn, Marko Z. Nikolic, Jose Ordovas-Montanes, Dana Pe’er, Joseph Powell, Stephen Quake, Jay Rajagopal, Purushothama Rao Tata, Emma L. Rawlins, Aviv Regev, Paul A. Reyfman, Mauricio Rojas, Orit Rosen, Kourosh Saeb-Parsy, Christos Samakovlis, Herbert Schiller, Joachim L. Schultze, Max A. Seibold, Alex K. Shalek, Douglas Shepherd, Jason Spence, Avrum Spira, Xin Sun, Sarah Teichmann, Fabian Theis, Alexander Tsankov, Maarten van den Berge, Michael von Papen, Jeffrey Whitsett, Ramnik Xavier, Yan Xu, Laure-Emmanuelle Zaragosi, and Kun Zhang. Pascal Barbry, Alexander Misharin, Martijn Nawijn, and Jay Rajagopal serve as the coordinators.
Acknowledgments
We are grateful to the study participants who made this work possible. We would like to thank Bruce Horwitz, Ivan Zanoni, Matt Sampson, Michael Retchin, Peter Winter, Andrew Navia, Jamie Cohen, and Audrey Sporrij for discussions. Mengyang (Vicky) Li Horst, Timothy Tickle, Jonathan Bistline, Jean Chang, Eric Weitz, Eno-Abasi Augustine-Akpan, and Devon Bush for development and support of the Broad Institute Single Cell Portal. This work was supported in part by the Searle Scholars Program, the Beckman Young Investigator Program, the Pew-Stewart Scholars Program for Cancer Research, a Sloan Fellowship in Chemistry, the MIT Stem Cell Initiative through Fondation MIT, the NIH (5U24AI118672 and BAA-NIAID-NIHAI201700104), and the Bill and Melinda Gates Foundation to A.K.S., as well as NIH R56 AI139053 to J.L.F and P.L.L., and the Aeras Foundation to J.L.F. B.B. and S.K.N. are partially supported by NIH 5R01GM081871. We acknowledge support from the Damon Runyon Cancer Research Foundation (DRG-2274-16) and Richard and Susan Smith Family Foundation to J.O.-M; from a National Science Foundation Graduate Research Fellowship (1122374) to S.K.N., S.J.A., and C.N.T.; from a Fannie and John Hertz Foundation Fellowship to C.N.T.; by T32GM007753 from the National Institute of General Medical Sciences to C.G.K.Z. This work was further supported by the UMass Center for Clinical and Translational Science Project Pilot Program; and the Office of the Assistant Secretary of Defense for Health Affairs, through the Peer Reviewed Medical Research Program (W81XWH-15-1-0317) to R.W.F. We also acknowledge support from NIH grants AI078908, HL111113, HL117945, R37AI052353, R01AI136041, R01HL136209, and U19AI095219 to J.A.B.; by grants from the NIH and National Heart, Lung, and Blood Institute (U19 HL129902) to H.P.K and L.S.K; from National Institute of Allergy and Infectious Diseases (UM1 AI126623) to H.P.K.; and to P.B. from the Fondation pour la Recherche Médicale (DEQ20180339158), and the Agence Nationale pour la Recherche (ANR-19-CE14-0027); and by the following grants to L.S.K: NIH/NIAID U19 AI051731, NIH/NHLBI R01 HL095791 NIH/NIAID R33-AI116184, NIH/NIAID U19 AI117945, and DHHS/NIH 1UM1AI126617. B.E.M. was supported by the Massachusetts Institute of Technology - GlaxoSmithKline (MIT-GSK) Gertrude B. Elion Postdoctoral Fellowship; T.M.L. by the NIH/NHLBI 1R01HL128241-01, K.M.B. by NIH/NIAID K23AI139352; and D.L. by NIH R01AI137057, DP2DA042422, and R01AI124378. This publication is part of the Human Cell Atlas (www.humancellatlas.org/publications).
Author Contributions
Document S1 details contributions of all authors.
Declaration of Interests
A.R. is an SAB member of ThermoFisher Scientific, Neogene Therapeutics, Asimov, and Syros Pharmaceuticals; a co-founder of and equity holder in Celsius Therapeutics; and an equity holder in Immunitas Therapeutics. A.K.S. reports compensation for consulting and/or SAB membership from Merck, Honeycomb Biotechnologies, Cellarity, Cogen Therapeutics, Orche Bio, and Dahlia Biosciences. L.S.K. is on the SAB for HiFiBio; she reports research funding from Kymab Limited, Bristol Meyers Squibb, Magenta Therapeutics, BlueBird Bio, and Regeneron Pharmaceuticals and consulting fees from Equillium, FortySeven, Inc, Novartis, Inc, EMD Serono, Gilead Sciences, and Takeda Pharmaceuticals. A.S. is an employee of Johnson and Johnson. N.K. is an inventor on a patent using thyroid hormone mimetics in acute lung injury that is now being considered for intervention in COVID-19 patients. J.L. is a scientific consultant for 10X Genomics, Inc. O.R.R, is a co-inventor on patent applications filed by the Broad Institute to inventions relating to single-cell genomics applications, such as in PCT/US2018/060860 and US Provisional Application No. 62/745,259. S.T. in the last three years was a consultant at Genentech, Biogen, and Roche and is a member of the SAB of Foresite Labs. M.H.W. is now an employee of Pfizer. F.J.T. reports receiving consulting fees from Roche Diagnostics GmbH and ownership interest in Cellarity, Inc. P.H. is a co-inventor on a patent using artificial intelligence and high-resolution microscopy for COVID-19 infection testing based on serology.
Published: April 27, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.cell.2020.04.035.
Contributor Information
Alex K. Shalek, Email: shalek@mit.edu.
Jose Ordovas-Montanes, Email: jose.ordovas-montanes@childrens.harvard.edu.
HCA Lung Biological Networklung-network@humancellatlas.org:
Nicholas Banovich, Pascal Barbry, Alvis Brazma, Tushar Desai, Thu Elizabeth Duong, Oliver Eickelberg, Christine Falk, Michael Farzan, Ian Glass, Muzlifah Haniffa, Peter Horvath, Deborah Hung, Naftali Kaminski, Mark Krasnow, Jonathan A. Kropski, Malte Kuhnemund, Robert Lafyatis, Haeock Lee, Sylvie Leroy, Sten Linnarson, Joakim Lundeberg, Kerstin Meyer, Alexander Misharin, Martijn Nawijn, Marko Z. Nikolic, Jose Ordovas-Montanes, Dana Pe’er, Joseph Powell, Stephen Quake, Jay Rajagopal, Purushothama Rao Tata, Emma L. Rawlins, Aviv Regev, Paul A. Reyfman, Mauricio Rojas, Orit Rosen, Kourosh Saeb-Parsy, Christos Samakovlis, Herbert Schiller, Joachim L. Schultze, Max A. Seibold, Alex K. Shalek, Douglas Shepherd, Jason Spence, Avrum Spira, Xin Sun, Sarah Teichmann, Fabian Theis, Alexander Tsankov, Maarten van den Berge, Michael von Papen, Jeffrey Whitsett, Ramnik Xavier, Yan Xu, Laure-Emmanuelle Zaragosi, and Kun Zhang
Supplemental Information
Statistics and Gene Lists Related to Figure 1 (Non-Human Primate Lung) A. Cluster defining genes for cell types presented in Figure 1. B. ACE2 and TMPRSS2 statistics by cell type. C. Differentially expressed genes between ACE2+ and ACE2- Type II Pneumocytes.
Statistics and Gene Lists Related to Figure 2 (Human Lung) A. Cluster defining genes for cell types presented in Figure 2. B. ACE2 and TMPRSS2 statistics by cell type. C. Differentially expressed genes between ACE2+TMPRSS2+ versus rest Type II Pneumocytes.
Statistics and Gene Lists Related to STAR Methods and Figure S1 (NHP Tuberculosis Infected Lung and Granuloma) A. Cluster defining genes for cell types presented in Figure S1 B. ACE2 and TMPRSS2 statistics by cell type C. Abundances of ACE2+TMPRSS2+ cells by cell type and tissue of origin D. Differentially expressed genes between ACE2+TMPRSS2+ versus rest Type II Pneumocytes
Statistics and Gene Lists Related to Figure 3 (Non-Human Primate Ileum) A. Cluster defining genes for cell types presented in Figure 3B B. ACE2 and TMPRSS2 statistics by cell type. C. Differentially expressed genes between ACE2+TMPRSS2+ versus rest Absorptive Enterocytes
Statistics and Gene Lists Related to Figure 3 (Human Ileum) A. Cluster defining genes for cell types presented in Figure 3D B. ACE2 and TMPRSS2 statistics by cell type. C. Differentially expressed genes between ACE2+TMPRSS2+ Absorptive Enterocytes versus rest Enterocytes D. Differentially expressed genes between ACE2+TMPRSS2+ Absorptive Enterocytes versus rest Absorptive Enterocytes
Statistics and Gene Lists Related to Figure 4 and 5 (Human Nasal Mucosa and Human Influenza) A. ACE2 and TMPRSS2 statistics by cell type presented in Figure 4B. B. Cluster defining genes for cell types presented in Figure 4C and 4D. C. Cluster defining genes for cell types presented in Figure 4C and 4F. D. ACE2 and TMPRSS2 statistics by cell type presented in Figure 4C. E. ACE2 and TMPRSS2 statistics by cell type presented in Figure 4D. F. Differentially expressed genes between ACE2+TMPRSS2+ epithelial cells versus rest epithelial cells in Figure 4E G. Differentially expressed genes between ACE2+TMPRSS2+ epithelial cells versus rest secretory epithelial cells in Figure 4E H. Raw data for cell type abundances by tissue and disease, Figure 4H I. Differentially expressed genes between ACE2+TMPRSS2+ versus Goblet Cells from Human Influenza dataset (Figure 6E-G).
Differentially Expressed Genes in Human and Mouse Basal Epithelial Cells Treated with Various Cytokines and Interferons, Related to Figure 5 A. Mouse Untreated versus IFNβ B. Mouse Untreated versus IFNα2 C. Mouse Untreated versus IFNγ D. BEAS-2B Untreated versus IL4 E. BEAS-2B Untreated versus IL17A F. BEAS-2B Untreated versus IFNγ G. BEAS-2B Untreated versus IFNα2 H. Human Donor 1 Untreated versus IL4 I. Human Donor 1 Untreated versus IL17A J. Human Donor 1 Untreated versus IFNγ K. Human Donor 1 Untreated versus IFNα2 L. Human Donor 2 Untreated versus IL4 M. Human Donor 2 Untreated versus IL17A N. Human Donor 2 Untreated versus IFNγ O. Human Donor 2 Untreated versus IFNα2
Statistics and Gene Lists Related to Figure 6 and Figure S5 (Mouse Nasal Epithelium) A. Cluster defining genes for cell types presented in Figure 6A B. Ace2 and Tmprss2 statistics by cell type C. Differentially expressed genes among Basal Epithelial Cells treated with Saline versus Intranasal IFNα
Summary of Datasets Analyzed in this Study with Links to Data, Related to STAR Methods
References
- Adler H., Messerle M., Wagner M., Koszinowski U.H. Cloning and mutagenesis of the murine gammaherpesvirus 68 genome as an infectious bacterial artificial chromosome. J. Virol. 2000;74:6964–6974. doi: 10.1128/jvi.74.15.6964-6974.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amanat F., Nguyen T., Chromikova V., Strohmeier S., Stadlbauer D., Javier A., Jiang K., Asthagiri-Arunkumar G., Polanco J., Bermudez-Gonzalez M. A serological assay to detect SARS-CoV-2 seroconversion in humans. medRxiv. 2020 doi: 10.1101/2020.03.17.20037713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ardain A., Domingo-Gonzalez R., Das S., Kazer S.W., Howard N.C., Singh A., Ahmed M., Nhamoyebonde S., Rangel-Moreno J., Ogongo P. Group 3 innate lymphoid cells mediate early protective immunity against tuberculosis. Nature. 2019;570:528–532. doi: 10.1038/s41586-019-1276-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey C.C., Zhong G., Huang I.C., Farzan M. IFITM-Family Proteins: The Cell’s First Line of Antiviral Defense. Annu. Rev. Virol. 2014;1:261–283. doi: 10.1146/annurev-virology-031413-085537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao L., Deng W., Gao H., Xiao C., Liu J., Xue J., Lv Q., Liu J., Yu P., Xu Y. Reinfection could not occur in SARS-CoV-2 infected rhesus macaques. bioRxiv. 2020 doi: 10.1101/2020.03.13.990226. [DOI] [Google Scholar]
- Blanco-Melo D., Nilsson-Payant B.E., Liu W.-C., Møller R., Panis M., Sachs D., Albrecht R.A., tenOever B.R. SARS-CoV-2 launches a unique transcriptional signature from in vitro, ex vivo, and in vivo systems. bioRxiv. 2020 doi: 10.1101/2020.03.24.004655. [DOI] [Google Scholar]
- Böttcher-Friebertshäuser E., Klenk H.D., Garten W. Activation of influenza viruses by proteases from host cells and bacteria in the human airway epithelium. Pathog. Dis. 2013;69:87–100. doi: 10.1111/2049-632X.12053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brann D., Tsukahara T., Weinreb C., Logan D.W., Datta S.R. Non-neural expression of SARS-CoV-2 entry genes in the olfactory epithelium suggests mechanisms underlying anosmia in COVID-19 patients. bioRxiv. 2020 doi: 10.1101/2020.03.25.009084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun E., Sauter D. Furin-mediated protein processing in infectious diseases and cancer. Clin. Transl. Immunology. 2019;8:e1073. doi: 10.1002/cti2.1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broggi A., Granucci F., Zanoni I. Type III interferons: Balancing tissue tolerance and resistance to pathogen invasion. J. Exp. Med. 2020;217:e20190295. doi: 10.1084/jem.20190295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugge T.H., Antalis T.M., Wu Q. Type II transmembrane serine proteases. J. Biol. Chem. 2009;284:23177–23181. doi: 10.1074/jbc.R109.021006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler A., Hoffman P., Smibert P., Papalexi E., Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 2018;36:411–420. doi: 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y., Guo Z., Vangala P., Donnard E., Liu P., McDonel P., Ordovas-Montanes J., Shalek A.K., Finberg R.W., Wang J.P. Single-cell analysis of upper airway cells reveals host-viral dynamics in influenza infected adults. bioRxiv. 2020 doi: 10.1101/2020.04.15.042978. [DOI] [Google Scholar]
- Channappanavar R., Perlman S. Pathogenic human coronavirus infections: causes and consequences of cytokine storm and immunopathology. Semin. Immunopathol. 2017;39:529–539. doi: 10.1007/s00281-017-0629-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Channappanavar R., Fehr A.R., Vijay R., Mack M., Zhao J., Meyerholz D.K., Perlman S. Dysregulated Type I Interferon and Inflammatory Monocyte-Macrophage Responses Cause Lethal Pneumonia in SARS-CoV-Infected Mice. Cell Host Microbe. 2016;19:181–193. doi: 10.1016/j.chom.2016.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Channappanavar R., Fehr A.R., Zheng J., Wohlford-Lenane C., Abrahante J.E., Mack M., Sompallae R., McCray P.B., Jr., Meyerholz D.K., Perlman S. IFN-I response timing relative to virus replication determines MERS coronavirus infection outcomes. J. Clin. Invest. 2019;130:3625–3639. doi: 10.1172/JCI126363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu H.Y., Boeckh M., Englund J.A., Famulare M., Lutz B.R., Nickerson D.A., Rieder M.J., Starita L.M., Adler A., Brandstetter E. The Seattle Flu Study: a multi-arm community-based prospective study protocol for assessing influenza prevalence, transmission, and genomic epidemiology. medRxiv. 2020 doi: 10.1101/2020.03.02.20029595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colonna L., Peterson C.W., Schell J.B., Carlson J.M., Tkachev V., Brown M., Yu A., Reddy S., Obenza W.M., Nelson V. Evidence for persistence of the SHIV reservoir early after MHC haploidentical hematopoietic stem cell transplantation. Nat. Commun. 2018;9:4438. doi: 10.1038/s41467-018-06736-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coronaviridae Study Group of the International Committee on Taxonomy of V. The species Severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020 doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coutard B., Valle C., de Lamballerie X., Canard B., Seidah N.G., Decroly E. The spike glycoprotein of the new coronavirus 2019-nCoV contains a furin-like cleavage site absent in CoV of the same clade. Antiviral Res. 2020;176:104742. doi: 10.1016/j.antiviral.2020.104742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson D.J., Gray M.A., Kilanowski F.M., Tarran R., Randell S.H., Sheppard D.N., Argent B.E., Dorin J.R. Murine epithelial cells: isolation and culture. J. Cyst. Fibros. 2004;3(Suppl 2):59–62. doi: 10.1016/j.jcf.2004.05.013. [DOI] [PubMed] [Google Scholar]
- Davidson S., Maini M.K., Wack A. Disease-promoting effects of type I interferons in viral, bacterial, and coinfections. J. Interferon Cytokine Res. 2015;35:252–264. doi: 10.1089/jir.2014.0227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lang A., Osterhaus A.D., Haagmans B.L. Interferon-gamma and interleukin-4 downregulate expression of the SARS coronavirus receptor ACE2 in Vero E6 cells. Virology. 2006;353:474–481. doi: 10.1016/j.virol.2006.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dear T.N., Boehm T., Keverne E.B., Rabbitts T.H. Novel genes for potential ligand-binding proteins in subregions of the olfactory mucosa. EMBO J. 1991;10:2813–2819. doi: 10.1002/j.1460-2075.1991.tb07830.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deeks S.G., Odorizzi P.M., Sekaly R.P. The interferon paradox: can inhibiting an antiviral mechanism advance an HIV cure? J. Clin. Invest. 2017;127:103–105. doi: 10.1172/JCI91916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derr A., Yang C., Zilionis R., Sergushichev A., Blodgett D.M., Redick S., Bortell R., Luban J., Harlan D.M., Kadener S. End Sequence Analysis Toolkit (ESAT) expands the extractable information from single-cell RNA-seq data. Genome Res. 2016;26:1397–1410. doi: 10.1101/gr.207902.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y., He L., Zhang Q., Huang Z., Che X., Hou J., Wang H., Shen H., Qiu L., Li Z. Organ distribution of severe acute respiratory syndrome (SARS) associated coronavirus (SARS-CoV) in SARS patients: implications for pathogenesis and virus transmission pathways. J. Pathol. 2004;203:622–630. doi: 10.1002/path.1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong E., Du H., Gardner L. An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect. Dis. 2020 doi: 10.1016/S1473-3099(20)30120-1. S1473-3099(20)30120-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong L., Hu S., Gao J. Discovering drugs to treat coronavirus disease 2019 (COVID-19) Drug Discov. Ther. 2020;14:58–60. doi: 10.5582/ddt.2020.01012. [DOI] [PubMed] [Google Scholar]
- Dunston D., Ashby S., Krosnowski K., Ogura T., Lin W. An effective manual deboning method to prepare intact mouse nasal tissue with preserved anatomical organization. J. Vis. Exp. 2013;(78) doi: 10.3791/50538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupuis S., Jouanguy E., Al-Hajjar S., Fieschi C., Al-Mohsen I.Z., Al-Jumaah S., Yang K., Chapgier A., Eidenschenk C., Eid P. Impaired response to interferon-alpha/beta and lethal viral disease in human STAT1 deficiency. Nat. Genet. 2003;33:388–391. doi: 10.1038/ng1097. [DOI] [PubMed] [Google Scholar]
- Everitt A.R., Clare S., Pertel T., John S.P., Wash R.S., Smith S.E., Chin C.R., Feeley E.M., Sims J.S., Adams D.J., GenISIS Investigators. MOSAIC Investigators IFITM3 restricts the morbidity and mortality associated with influenza. Nature. 2012;484:519–523. doi: 10.1038/nature10921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming F.E., Böhm R., Dang V.T., Holloway G., Haselhorst T., Madge P.D., Deveryshetty J., Yu X., Blanchard H., von Itzstein M., Coulson B.S. Relative roles of GM1 ganglioside, N-acylneuraminic acids, and α2β1 integrin in mediating rotavirus infection. J. Virol. 2014;88:4558–4571. doi: 10.1128/JVI.03431-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung T.S., Liu D.X. Human Coronavirus: Host-Pathogen Interaction. Annu. Rev. Microbiol. 2019;73:529–557. doi: 10.1146/annurev-micro-020518-115759. [DOI] [PubMed] [Google Scholar]
- Genshaft A.S., Li S., Gallant C.J., Darmanis S., Prakadan S.M., Ziegler C.G., Lundberg M., Fredriksson S., Hong J., Regev A. Multiplexed, targeted profiling of single-cell proteomes and transcriptomes in a single reaction. Genome Biol. 2016;17:188. doi: 10.1186/s13059-016-1045-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstein M.B., Kundaje A., Hariharan M., Landt S.G., Yan K.K., Cheng C., Mu X.J., Khurana E., Rozowsky J., Alexander R. Architecture of the human regulatory network derived from ENCODE data. Nature. 2012;489:91–100. doi: 10.1038/nature11245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gierahn T.M., Wadsworth M.H., 2nd, Hughes T.K., Bryson B.D., Butler A., Satija R., Fortune S., Love J.C., Shalek A.K. Seq-Well: portable, low-cost RNA sequencing of single cells at high throughput. Nat. Methods. 2017;14:395–398. doi: 10.1038/nmeth.4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannini-Chami L., Marcet B., Moreilhon C., Chevalier B., Illie M.I., Lebrigand K., Robbe-Sermesant K., Bourrier T., Michiels J.F., Mari B. Distinct epithelial gene expression phenotypes in childhood respiratory allergy. Eur. Respir. J. 2012;39:1197–1205. doi: 10.1183/09031936.00070511. [DOI] [PubMed] [Google Scholar]
- Glowacka I., Bertram S., Müller M.A., Allen P., Soilleux E., Pfefferle S., Steffen I., Tsegaye T.S., He Y., Gnirss K. Evidence that TMPRSS2 activates the severe acute respiratory syndrome coronavirus spike protein for membrane fusion and reduces viral control by the humoral immune response. J. Virol. 2011;85:4122–4134. doi: 10.1128/JVI.02232-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good C., Wells A.I., Coyne C.B. Type III interferon signaling restricts enterovirus 71 infection of goblet cells. Sci Adv. 2019 doi: 10.1126/sciadv.aau4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griggs T.F., Bochkov Y.A., Basnet S., Pasic T.R., Brockman-Schneider R.A., Palmenberg A.C., Gern J.E. Rhinovirus C targets ciliated airway epithelial cells. Respir. Res. 2017;18:84. doi: 10.1186/s12931-017-0567-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grove J., Marsh M. The cell biology of receptor-mediated virus entry. J. Cell Biol. 2011;195:1071–1082. doi: 10.1083/jcb.201108131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan W.J., Ni Z.Y., Hu Y., Liang W.H., Ou C.Q., He J.X., Liu L., Shan H., Lei C.L., Hui D.S.C., China Medical Treatment Expert Group for Covid-19 Clinical Characteristics of Coronavirus Disease 2019 in China. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrero-Plata A., Baron S., Poast J.S., Adegboyega P.A., Casola A., Garofalo R.P. Activity and regulation of alpha interferon in respiratory syncytial virus and human metapneumovirus experimental infections. J. Virol. 2005;79:10190–10199. doi: 10.1128/JVI.79.16.10190-10199.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadfield J., Megill C., Bell S.M., Huddleston J., Potter B., Callender C., Sagulenko P., Bedford T., Neher R.A. Nextstrain: real-time tracking of pathogen evolution. Bioinformatics. 2018;34:4121–4123. doi: 10.1093/bioinformatics/bty407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamming I., Timens W., Bulthuis M.L., Lely A.T., Navis G., van Goor H. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J. Pathol. 2004;203:631–637. doi: 10.1002/path.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmer D., Gilbert M., Borman R., Clark K.L. Quantitative mRNA expression profiling of ACE 2, a novel homologue of angiotensin converting enzyme. FEBS Lett. 2002;532:107–110. doi: 10.1016/s0014-5793(02)03640-2. [DOI] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.-H., Nitsche A. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann H., Geier M., Marzi A., Krumbiegel M., Peipp M., Fey G.H., Gramberg T., Pöhlmann S. Susceptibility to SARS coronavirus S protein-driven infection correlates with expression of angiotensin converting enzyme 2 and infection can be blocked by soluble receptor. Biochem. Biophys. Res. Commun. 2004;319:1216–1221. doi: 10.1016/j.bbrc.2004.05.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holly M.K., Smith J.G. Adenovirus Infection of Human Enteroids Reveals Interferon Sensitivity and Preferential Infection of Goblet Cells. J. Virol. 2018;92:e00250-18. doi: 10.1128/JVI.00250-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holshue M.L., DeBolt C., Lindquist S., Lofy K.H., Wiesman J., Bruce H., Spitters C., Ericson K., Wilkerson S., Tural A., Washington State 2019-nCoV Case Investigation Team First Case of 2019 Novel Coronavirus in the United States. N. Engl. J. Med. 2020;382:929–936. doi: 10.1056/NEJMoa2001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang F., Guo J., Zou Z., Liu J., Cao B., Zhang S., Li H., Wang W., Sheng M., Liu S. Angiotensin II plasma levels are linked to disease severity and predict fatal outcomes in H7N9-infected patients. Nat. Commun. 2014;5:3595. doi: 10.1038/ncomms4595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes T.K., Wadsworth M.H., Gierahn T.M., Do T., Weiss D., Andrade P.R., Ma F., de Andrade Silva B.J., Shao S., Tsoi L.C. Highly Efficient, Massively-Parallel Single-Cell RNA-Seq Reveals Cellular States and Molecular Features of Human Skin Pathology. bioRxiv. 2019 doi: 10.1101/689273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai Y., Kuba K., Rao S., Huan Y., Guo F., Guan B., Yang P., Sarao R., Wada T., Leong-Poi H. Angiotensin-converting enzyme 2 protects from severe acute lung failure. Nature. 2005;436:112–116. doi: 10.1038/nature03712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki A., Pillai P.S. Innate immunity to influenza virus infection. Nat. Rev. Immunol. 2014;14:315–328. doi: 10.1038/nri3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki A., Foxman E.F., Molony R.D. Early local immune defences in the respiratory tract. Nat. Rev. Immunol. 2017;17:7–20. doi: 10.1038/nri.2016.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata-Yoshikawa N., Okamura T., Shimizu Y., Hasegawa H., Takeda M., Nagata N. TMPRSS2 Contributes to Virus Spread and Immunopathology in the Airways of Murine Models after Coronavirus Infection. J. Virol. 2019;93:e01815-18. doi: 10.1128/JVI.01815-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jewell N.A., Cline T., Mertz S.E., Smirnov S.V., Flaño E., Schindler C., Grieves J.L., Durbin R.K., Kotenko S.V., Durbin J.E. Lambda interferon is the predominant interferon induced by influenza A virus infection in vivo. J. Virol. 2010;84:11515–11522. doi: 10.1128/JVI.01703-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonsdottir H.R., Dijkman R. Coronaviruses and the human airway: a universal system for virus-host interaction studies. Virol. J. 2016;13:24. doi: 10.1186/s12985-016-0479-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jovanovic M., Rooney M.S., Mertins P., Przybylski D., Chevrier N., Satija R., Rodriguez E.H., Fields A.P., Schwartz S., Raychowdhury R. Immunogenetics. Dynamic profiling of the protein life cycle in response to pathogens. Science. 2015;347:1259038. doi: 10.1126/science.1259038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazer S.W., Aicher T.P., Muema D.M., Carroll S.L., Ordovas-Montanes J., Miao V.N., Tu A.A., Ziegler C.G.K., Nyquist S.K., Wong E.B. Integrated single-cell analysis of multicellular immune dynamics during hyperacute HIV-1 infection. Nat. Med. 2020;26:511–518. doi: 10.1038/s41591-020-0799-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharchenko P.V., Silberstein L., Scadden D.T. Bayesian approach to single-cell differential expression analysis. Nat. Methods. 2014;11:740–742. doi: 10.1038/nmeth.2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krischuns T., Günl F., Henschel L., Binder M., Willemsen J., Schloer S., Rescher U., Gerlt V., Zimmer G., Nordhoff C. Phosphorylation of TRIM28 Enhances the Expression of IFN-β and Proinflammatory Cytokines During HPAIV Infection of Human Lung Epithelial Cells. Front. Immunol. 2018;9:2229. doi: 10.3389/fimmu.2018.02229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuba K., Imai Y., Rao S., Gao H., Guo F., Guan B., Huan Y., Yang P., Zhang Y., Deng W. A crucial role of angiotensin converting enzyme 2 (ACE2) in SARS coronavirus-induced lung injury. Nat. Med. 2005;11:875–879. doi: 10.1038/nm1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucharski A.J., Russell T.W., Diamond C., Liu Y., Edmunds J., Funk S., Eggo R.M., Centre for Mathematical Modelling of Infectious Diseases COVID-19 working group Early dynamics of transmission and control of COVID-19: a mathematical modelling study. Lancet Infect. Dis. 2020 doi: 10.1016/S1473-3099(20)30144-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechien J.R., Chiesa-Estomba C.M., De Siati D.R., Horoi M., Le Bon S.D., Rodriguez A., Dequanter D., Blecic S., El Afia F., Distinguin L. Olfactory and gustatory dysfunctions as a clinical presentation of mild-to-moderate forms of the coronavirus disease (COVID-19): a multicenter European study. Eur. Arch. Otorhinolaryngol. 2020 doi: 10.1007/s00405-020-05965-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei J., Li J., Li X., Qi X. CT Imaging of the 2019 Novel Coronavirus (2019-nCoV) Pneumonia. Radiology. 2020;295:18. doi: 10.1148/radiol.2020200236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letko M., Marzi A., Munster V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat. Microbiol. 2020;5:562–569. doi: 10.1038/s41564-020-0688-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G., De Clercq E. Therapeutic options for the 2019 novel coronavirus (2019-nCoV) Nat. Rev. Drug Discov. 2020;19:149–150. doi: 10.1038/d41573-020-00016-0. [DOI] [PubMed] [Google Scholar]
- Li W., Moore M.J., Vasilieva N., Sui J., Wong S.K., Berne M.A., Somasundaran M., Sullivan J.L., Luzuriaga K., Greenough T.C. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao M., Liu Y., Yuan J., Wen Y., Xu G., Zhao J., Chen L., Li J., Wang X., Wang F. The landscape of lung bronchoalveolar immune cells in COVID-19 revealed by single-cell RNA sequencing. medRxiv. 2020 doi: 10.1101/2020.02.23.20026690. [DOI] [Google Scholar]
- Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., Wang W., Song H., Huang B., Zhu N. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukassen S., Chua R.L., Trefzer T., Kahn N.C., Schneider M.A., Muley T., Winter H., Meister M., Veith C., Boots A.W. SARS-CoV-2 receptor ACE2 and TMPRSS2 are predominantly expressed in a transient secretory cell type in subsegmental bronchial branches. bioRxiv. 2020 doi: 10.1101/2020.03.13.991455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lun A.T., McCarthy D.J., Marioni J.C. A step-by-step workflow for low-level analysis of single-cell RNA-seq data with Bioconductor. F1000Res. 2016;5:2122. doi: 10.12688/f1000research.9501.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macosko E.Z., Basu A., Satija R., Nemesh J., Shekhar K., Goldman M., Tirosh I., Bialas A.R., Kamitaki N., Martersteck E.M. Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell. 2015;161:1202–1214. doi: 10.1016/j.cell.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mar K.B., Rinkenberger N.R., Boys I.N., Eitson J.L., McDougal M.B., Richardson R.B., Schoggins J.W. LY6E mediates an evolutionarily conserved enhancement of virus infection by targeting a late entry step. Nat. Commun. 2018;9:3603. doi: 10.1038/s41467-018-06000-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin C.J., Cadena A.M., Leung V.W., Lin P.L., Maiello P., Hicks N., Chase M.R., Flynn J.L., Fortune S.M. Digitally Barcoding Mycobacterium tuberculosis Reveals In Vivo Infection Dynamics in the Macaque Model of Tuberculosis. MBio. 2017;8:e00312-17. doi: 10.1128/mBio.00312-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matos A.D.R., Wunderlich K., Schloer S., Schughart K., Geffers R., Seders M., Witt M., Christersson A., Wiewrodt R., Wiebe K. Antiviral potential of human IFN-α subtypes against influenza A H3N2 infection in human lung explants reveals subtype-specific activities. Emerg. Microbes Infect. 2019;8:1763–1776. doi: 10.1080/22221751.2019.1698271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuyama S., Nagata N., Shirato K., Kawase M., Takeda M., Taguchi F. Efficient activation of the severe acute respiratory syndrome coronavirus spike protein by the transmembrane protease TMPRSS2. J. Virol. 2010;84:12658–12664. doi: 10.1128/JVI.01542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matys V., Fricke E., Geffers R., Gössling E., Haubrock M., Hehl R., Hornischer K., Karas D., Kel A.E., Kel-Margoulis O.V. TRANSFAC: transcriptional regulation, from patterns to profiles. Nucleic Acids Res. 2003;31:374–378. doi: 10.1093/nar/gkg108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCray P.B., Jr., Pewe L., Wohlford-Lenane C., Hickey M., Manzel L., Shi L., Netland J., Jia H.P., Halabi C., Sigmund C.D. Lethal infection of K18-hACE2 mice infected with severe acute respiratory syndrome coronavirus. J. Virol. 2007;81:813–821. doi: 10.1128/JVI.02012-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mead B.E., Karp J.M. All models are wrong, but some organoids may be useful. Genome Biol. 2019;20:66. doi: 10.1186/s13059-019-1677-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mead B.E., Ordovas-Montanes J., Braun A.P., Levy L.E., Bhargava P., Szucs M.J., Ammendolia D.A., MacMullan M.A., Yin X., Hughes T.K. Harnessing single-cell genomics to improve the physiological fidelity of organoid-derived cell types. BMC Biol. 2018;16:62. doi: 10.1186/s12915-018-0527-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medzhitov R., Schneider D.S., Soares M.P. Disease tolerance as a defense strategy. Science. 2012;335:936–941. doi: 10.1126/science.1214935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteil V., Kwon H., Prado P., Hagelkryus A., Wimmer R.A., al e. Inhibition of SARS-CoV-2 infections in engineered human tissues using clinical-grade soluble human ACE2. Cell. 2020 doi: 10.1016/j.cell.2020.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montoro D.T., Haber A.L., Biton M., Vinarsky V., Lin B., Birket S.E., Yuan F., Chen S., Leung H.M., Villoria J. A revised airway epithelial hierarchy includes CFTR-expressing ionocytes. Nature. 2018;560:319–324. doi: 10.1038/s41586-018-0393-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller U., Steinhoff U., Reis L.F., Hemmi S., Pavlovic J., Zinkernagel R.M., Aguet M. Functional role of type I and type II interferons in antiviral defense. Science. 1994;264:1918–1921. doi: 10.1126/science.8009221. [DOI] [PubMed] [Google Scholar]
- Munster V.J., Feldmann F., Williamson B.N., van Doremalen N., Pérez-Pérez L., Schulz J., Meade-White K., Okumura A., Callison J., Brumbaugh B. Respiratory disease and virus shedding in rhesus macaques inoculated with SARS-CoV-2. bioRxiv. 2020 doi: 10.1101/2020.03.21.001628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy T.L., Tussiwand R., Murphy K.M. Specificity through cooperation: BATF-IRF interactions control immune-regulatory networks. Nat. Rev. Immunol. 2013;13:499–509. doi: 10.1038/nri3470. [DOI] [PubMed] [Google Scholar]
- Ordovas-Montanes J., Dwyer D.F., Nyquist S.K., Buchheit K.M., Vukovic M., Deb C., Wadsworth M.H., 2nd, Hughes T.K., Kazer S.W., Yoshimoto E. Allergic inflammatory memory in human respiratory epithelial progenitor cells. Nature. 2018;560:649–654. doi: 10.1038/s41586-018-0449-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ordovas-Montanes J., Beyaz S., Rakoff-Nahoum S., Shalek A.K. Distribution and storage of inflammatory memory in barrier tissues. Nat. Rev. Immunol. 2020 doi: 10.1038/s41577-019-0263-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paules C.I., Marston H.D., Fauci A.S. Coronavirus Infections-More Than Just the Common Cold. JAMA. 2020 doi: 10.1001/jama.2020.0757. [DOI] [PubMed] [Google Scholar]
- Pinto B.G., Oliveira A.E., Singh Y., Jimenez L., Goncalves A.N., Ogava R.L., Creighton R., Peron J.P., Nakaya H.I. ACE2 Expression is Increased in the Lungs of Patients with Comorbidities Associated with Severe COVID-19. medRxiv. 2020 doi: 10.1101/2020.03.21.20040261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prokunina-Olsson L., Alphonse N., Dickenson R.E., Durbin J.E., Glenn J.S., Hartmann R., Kotenko S.V., Lazear H.M., O’Brien T.R., Odendall C. COVID-19 and emerging viral infections: The case for interferon lambda. J. Exp. Med. 2020;217:e20200653. doi: 10.1084/jem.20200653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi F., Qian S., Zhang S., Zhang Z. Single cell RNA sequencing of 13 human tissues identify cell types and receptors of human coronaviruses. bioRxiv. 2020 doi: 10.1101/2020.02.16.951913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian Z., Travanty E.A., Oko L., Edeen K., Berglund A., Wang J., Ito Y., Holmes K.V., Mason R.J. Innate immune response of human alveolar type II cells infected with severe acute respiratory syndrome-coronavirus. Am. J. Respir. Cell Mol. Biol. 2013;48:742–748. doi: 10.1165/rcmb.2012-0339OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabani M., Levin J.Z., Fan L., Adiconis X., Raychowdhury R., Garber M., Gnirke A., Nusbaum C., Hacohen N., Friedman N. Metabolic labeling of RNA uncovers principles of RNA production and degradation dynamics in mammalian cells. Nat. Biotechnol. 2011;29:436–442. doi: 10.1038/nbt.1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regev A., Teichmann S.A., Lander E.S., Amit I., Benoist C., Birney E., Bodenmiller B., Campbell P., Carninci P., Clatworthy M., Human Cell Atlas Meeting Participants The Human Cell Atlas. eLife. 2017;6:e27041. doi: 10.7554/eLife.27041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockx B., Kuiken T., Herfst S., Bestebroer T., Lamers M.M., Oude Munnink B.B., de Meulder D., van Amerongen G., van den Brand J., Okba N.M.A. Comparative pathogenesis of COVID-19, MERS, and SARS in a nonhuman primate model. Science. 2020 doi: 10.1126/science.abb7314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez A., Laio A. Machine learning. Clustering by fast search and find of density peaks. Science. 2014;344:1492–1496. doi: 10.1126/science.1242072. [DOI] [PubMed] [Google Scholar]
- Ruiz García S., Deprez M., Lebrigand K., Cavard A., Paquet A., Arguel M.J., Magnone V., Truchi M., Caballero I., Leroy S. Novel dynamics of human mucociliary differentiation revealed by single-cell RNA sequencing of nasal epithelial cultures. Development. 2019;146:dev177428. doi: 10.1242/dev.177428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusinova I., Forster S., Yu S., Kannan A., Masse M., Cumming H., Chapman R., Hertzog P.J. Interferome v2.0: an updated database of annotated interferon-regulated genes. Nucleic Acids Res. 2013;41:D1040–D1046. doi: 10.1093/nar/gks1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell A.B., Trapnell C., Bloom J.D. Extreme heterogeneity of influenza virus infection in single cells. eLife. 2018;7:e32303. doi: 10.7554/eLife.32303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sainz B., Jr., Mossel E.C., Peters C.J., Garry R.F. Interferon-beta and interferon-gamma synergistically inhibit the replication of severe acute respiratory syndrome-associated coronavirus (SARS-CoV) Virology. 2004;329:11–17. doi: 10.1016/j.virol.2004.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sajuthi S.P., DeFord P., Jackson N.D., Montgomery M.T., Everman J.L., Rios C.L., Pruesse E., Nolin J.D., Plender E.G., Wechsler M.E. Type 2 and interferon inflammation strongly regulate SARS-CoV-2 related gene expression in the airway epithelium. bioRxiv. 2020 doi: 10.1101/2020.04.09.034454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanda C., Weitzel P., Tsukahara T., Schaley J., Edenberg H.J., Stephens M.A., McClintick J.N., Blatt L.M., Li L., Brodsky L., Taylor M.W. Differential gene induction by type I and type II interferons and their combination. J. Interferon Cytokine Res. 2006;26:462–472. doi: 10.1089/jir.2006.26.462. [DOI] [PubMed] [Google Scholar]
- Satija R., Farrell J.A., Gennert D., Schier A.F., Regev A. Spatial reconstruction of single-cell gene expression data. Nat. Biotechnol. 2015;33:495–502. doi: 10.1038/nbt.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiller H.B., Montoro D.T., Simon L.M., Rawlins E.L., Meyer K.B., Strunz M., Vieira Braga F.A., Timens W., Koppelman G.H., Budinger G.R.S. The Human Lung Cell Atlas: A High-Resolution Reference Map of the Human Lung in Health and Disease. Am. J. Respir. Cell Mol. Biol. 2019;61:31–41. doi: 10.1165/rcmb.2018-0416TR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider D.S., Ayres J.S. Two ways to survive infection: what resistance and tolerance can teach us about treating infectious diseases. Nat. Rev. Immunol. 2008;8:889–895. doi: 10.1038/nri2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider W.M., Chevillotte M.D., Rice C.M. Interferon-stimulated genes: a complex web of host defenses. Annu. Rev. Immunol. 2014;32:513–545. doi: 10.1146/annurev-immunol-032713-120231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalek A.K., Satija R., Adiconis X., Gertner R.S., Gaublomme J.T., Raychowdhury R., Schwartz S., Yosef N., Malboeuf C., Lu D. Single-cell transcriptomics reveals bimodality in expression and splicing in immune cells. Nature. 2013;498:236–240. doi: 10.1038/nature12172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shulla A., Heald-Sargent T., Subramanya G., Zhao J., Perlman S., Gallagher T. A transmembrane serine protease is linked to the severe acute respiratory syndrome coronavirus receptor and activates virus entry. J. Virol. 2011;85:873–882. doi: 10.1128/JVI.02062-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skeggs L.T., Dorer F.E., Levine M., Lentz K.E., Kahn J.R. The biochemistry of the renin-angiotensin system. Adv. Exp. Med. Biol. 1980;130:1–27. doi: 10.1007/978-1-4615-9173-3_1. [DOI] [PubMed] [Google Scholar]
- Smillie C.S., Biton M., Ordovas-Montanes J., Sullivan K.M., Burgin G., Graham D.B., Herbst R.H., Rogel N., Slyper M., Waldman J. Intra- and Inter-cellular Rewiring of the Human Colon during Ulcerative Colitis. Cell. 2019 doi: 10.1016/j.cell.2019.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smits S.L., van den Brand J.M., de Lang A., Leijten L.M., van Ijcken W.F., van Amerongen G., Osterhaus A.D., Andeweg A.C., Haagmans B.L. Distinct severe acute respiratory syndrome coronavirus-induced acute lung injury pathways in two different nonhuman primate species. J. Virol. 2011;85:4234–4245. doi: 10.1128/JVI.02395-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stetson D.B., Medzhitov R. Type I interferons in host defense. Immunity. 2006;25:373–381. doi: 10.1016/j.immuni.2006.08.007. [DOI] [PubMed] [Google Scholar]
- Steuerman Y., Cohen M., Peshes-Yaloz N., Valadarsky L., Cohn O., David E., Frishberg A., Mayo L., Bacharach E., Amit I. Dissection of Influenza Infection In Vivo by Single-Cell RNA Sequencing. Cell Syst. 2018 doi: 10.1016/j.cels.2018.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sungnak W., Huang N., Becavin C., Berg M., Network H.L.B. SARS-CoV-2 Entry Genes Are Most Highly Expressed in Nasal Goblet and Ciliated Cells within Human Airways. Nat. Med. 2020 doi: 10.1038/s41591-020-0868-6. [DOI] [Google Scholar]
- Svensson V. Droplet scRNA-seq is not zero-inflated. Nat. Biotechnol. 2020;38:147–150. doi: 10.1038/s41587-019-0379-5. [DOI] [PubMed] [Google Scholar]
- Tepe B., Hill M.C., Pekarek B.T., Hunt P.J., Martin T.J., Martin J.F., Arenkiel B.R. Single-Cell RNA-Seq of Mouse Olfactory Bulb Reveals Cellular Heterogeneity and Activity-Dependent Molecular Census of Adult-Born Neurons. Cell Rep. 2018 doi: 10.1016/j.celrep.2018.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trombetta J.J., Gennert D., Lu D., Satija R., Shalek A.K., Regev A. Preparation of Single-Cell RNA-Seq Libraries for Next Generation Sequencing. Curr Protoc Mol Biol. 2014 doi: 10.1002/0471142727.mb0422s107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utay N.S., Douek D.C. Interferons and HIV Infection: The Good, the Bad, and the Ugly. Pathog. Immun. 2016;1:107–116. doi: 10.20411/pai.v1i1.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaduganathan M., Vardeny O., Michel T., McMurray J.J.V., Pfeffer M.A., Solomon S.D. Renin-Angiotensin-Aldosterone System Inhibitors in Patients with Covid-19. N. Engl. J. Med. 2020 doi: 10.1056/NEJMsr2005760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira Braga F.A., Kar G., Berg M., Carpaij O.A., Polanski K., Simon L.M., Brouwer S., Gomes T., Hesse L., Jiang J. A cellular census of human lungs identifies novel cell states in health and in asthma. Nat. Med. 2019;25:1153–1163. doi: 10.1038/s41591-019-0468-5. [DOI] [PubMed] [Google Scholar]
- Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell. 2020;181:281–292.e6. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P.-H., Cheng Y. Increasing Host Cellular Receptor—Angiotensin-Converting Enzyme 2 (ACE2) Expression by Coronavirus may Facilitate 2019-nCoV Infection. bioRxiv. 2020 doi: 10.1101/2020.02.24.963348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Zhuang J., Iyer S., Lin X., Whitfield T.W., Greven M.C., Pierce B.G., Dong X., Kundaje A., Cheng Y. Sequence features and chromatin structure around the genomic regions bound by 119 human transcription factors. Genome Res. 2012;22:1798–1812. doi: 10.1101/gr.139105.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Zhuang J., Iyer S., Lin X.Y., Greven M.C., Kim B.H., Moore J., Pierce B.G., Dong X., Virgil D. Factorbook.org: a Wiki-based database for transcription factor-binding data generated by the ENCODE consortium. Nucleic Acids Res. 2013;41:D171–D176. doi: 10.1093/nar/gks1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Xu Y., Gao R., Lu R., Han K., Wu G., Tan W. Detection of SARS-CoV-2 in Different Types of Clinical Specimens. JAMA. 2020 doi: 10.1001/jama.2020.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinheimer V.K., Becher A., Tönnies M., Holland G., Knepper J., Bauer T.T., Schneider P., Neudecker J., Rückert J.C., Szymanski K. Influenza A viruses target type II pneumocytes in the human lung. J. Infect. Dis. 2012;206:1685–1694. doi: 10.1093/infdis/jis455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf F.A., Angerer P., Theis F.J. SCANPY: large-scale single-cell gene expression data analysis. Genome Biol. 2018;19:15. doi: 10.1186/s13059-017-1382-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wölfel R., Corman V.M., Guggemos W., Seilmaier M., Zange S., Müller M.A., Niemeyer D., Jones T.C., Vollmar P., Rothe C. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020 doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C., Zheng S., Chen Y., Zheng M. Single-cell RNA expression profiling of ACE2, the putative receptor of Wuhan 2019-nCoV, in the nasal tissue. medRxiv. 2020 doi: 10.1101/2020.02.11.20022228. [DOI] [Google Scholar]
- Xiao F., Tang M., Zheng X., Liu Y., Li X., Shan H. Evidence for gastrointestinal infection of SARS-CoV-2. Gastroenterology. 2020 doi: 10.1053/j.gastro.2020.02.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y., Li X., Zhu B., Liang H., Fang C., Gong Y., Guo Q., Sun X., Zhao D., Shen J. Characteristics of pediatric SARS-CoV-2 infection and potential evidence for persistent fecal viral shedding. Nat. Med. 2020;26:502–505. doi: 10.1038/s41591-020-0817-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young M.D., Behjati S. SoupX removes ambient RNA contamination from droplet based single-cell RNA sequencing data. bioRxiv. 2020 doi: 10.1101/303727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Kang Z., Gong H., Xu D., Wang J., Li Z., Cui X., Xiao J., Meng T., Zhou W. The digestive system is a potential route of 2019-nCov infection: a bioinformatics analysis based on single-cell transcriptomes. bioRxiv. 2020 doi: 10.1101/2020.01.30.927806. [DOI] [Google Scholar]
- Zhao X., Guo F., Liu F., Cuconati A., Chang J., Block T.M., Guo J.T. Interferon induction of IFITM proteins promotes infection by human coronavirus OC43. Proc. Natl. Acad. Sci. USA. 2014;111:6756–6761. doi: 10.1073/pnas.1320856111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng B., He M.L., Wong K.L., Lum C.T., Poon L.L., Peng Y., Guan Y., Lin M.C., Kung H.F. Potent inhibition of SARS-associated coronavirus (SCOV) infection and replication by type I interferons (IFN-alpha/beta) but not by type II interferon (IFN-gamma) J. Interferon Cytokine Res. 2004;24:388–390. doi: 10.1089/1079990041535610. [DOI] [PubMed] [Google Scholar]
- Zou Z., Yan Y., Shu Y., Gao R., Sun Y., Li X., Ju X., Liang Z., Liu Q., Zhao Y. Angiotensin-converting enzyme 2 protects from lethal avian influenza A H5N1 infections. Nat. Commun. 2014;5:3594. doi: 10.1038/ncomms4594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou L., Ruan F., Huang M., Liang L., Huang H., Hong Z., Yu J., Kang M., Song Y., Xia J. SARS-CoV-2 Viral Load in Upper Respiratory Specimens of Infected Patients. N. Engl. J. Med. 2020;382:1177–1179. doi: 10.1056/NEJMc2001737. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Statistics and Gene Lists Related to Figure 1 (Non-Human Primate Lung) A. Cluster defining genes for cell types presented in Figure 1. B. ACE2 and TMPRSS2 statistics by cell type. C. Differentially expressed genes between ACE2+ and ACE2- Type II Pneumocytes.
Statistics and Gene Lists Related to Figure 2 (Human Lung) A. Cluster defining genes for cell types presented in Figure 2. B. ACE2 and TMPRSS2 statistics by cell type. C. Differentially expressed genes between ACE2+TMPRSS2+ versus rest Type II Pneumocytes.
Statistics and Gene Lists Related to STAR Methods and Figure S1 (NHP Tuberculosis Infected Lung and Granuloma) A. Cluster defining genes for cell types presented in Figure S1 B. ACE2 and TMPRSS2 statistics by cell type C. Abundances of ACE2+TMPRSS2+ cells by cell type and tissue of origin D. Differentially expressed genes between ACE2+TMPRSS2+ versus rest Type II Pneumocytes
Statistics and Gene Lists Related to Figure 3 (Non-Human Primate Ileum) A. Cluster defining genes for cell types presented in Figure 3B B. ACE2 and TMPRSS2 statistics by cell type. C. Differentially expressed genes between ACE2+TMPRSS2+ versus rest Absorptive Enterocytes
Statistics and Gene Lists Related to Figure 3 (Human Ileum) A. Cluster defining genes for cell types presented in Figure 3D B. ACE2 and TMPRSS2 statistics by cell type. C. Differentially expressed genes between ACE2+TMPRSS2+ Absorptive Enterocytes versus rest Enterocytes D. Differentially expressed genes between ACE2+TMPRSS2+ Absorptive Enterocytes versus rest Absorptive Enterocytes
Statistics and Gene Lists Related to Figure 4 and 5 (Human Nasal Mucosa and Human Influenza) A. ACE2 and TMPRSS2 statistics by cell type presented in Figure 4B. B. Cluster defining genes for cell types presented in Figure 4C and 4D. C. Cluster defining genes for cell types presented in Figure 4C and 4F. D. ACE2 and TMPRSS2 statistics by cell type presented in Figure 4C. E. ACE2 and TMPRSS2 statistics by cell type presented in Figure 4D. F. Differentially expressed genes between ACE2+TMPRSS2+ epithelial cells versus rest epithelial cells in Figure 4E G. Differentially expressed genes between ACE2+TMPRSS2+ epithelial cells versus rest secretory epithelial cells in Figure 4E H. Raw data for cell type abundances by tissue and disease, Figure 4H I. Differentially expressed genes between ACE2+TMPRSS2+ versus Goblet Cells from Human Influenza dataset (Figure 6E-G).
Differentially Expressed Genes in Human and Mouse Basal Epithelial Cells Treated with Various Cytokines and Interferons, Related to Figure 5 A. Mouse Untreated versus IFNβ B. Mouse Untreated versus IFNα2 C. Mouse Untreated versus IFNγ D. BEAS-2B Untreated versus IL4 E. BEAS-2B Untreated versus IL17A F. BEAS-2B Untreated versus IFNγ G. BEAS-2B Untreated versus IFNα2 H. Human Donor 1 Untreated versus IL4 I. Human Donor 1 Untreated versus IL17A J. Human Donor 1 Untreated versus IFNγ K. Human Donor 1 Untreated versus IFNα2 L. Human Donor 2 Untreated versus IL4 M. Human Donor 2 Untreated versus IL17A N. Human Donor 2 Untreated versus IFNγ O. Human Donor 2 Untreated versus IFNα2
Statistics and Gene Lists Related to Figure 6 and Figure S5 (Mouse Nasal Epithelium) A. Cluster defining genes for cell types presented in Figure 6A B. Ace2 and Tmprss2 statistics by cell type C. Differentially expressed genes among Basal Epithelial Cells treated with Saline versus Intranasal IFNα
Summary of Datasets Analyzed in this Study with Links to Data, Related to STAR Methods
Data Availability Statement
In Table S9, we provide a guide to all datasets analyzed in this paper as well as links to each individual dataset for download with the main landing page here: https://singlecell.broadinstitute.org/single_cell?scpbr=the-alexandria-project. To download the data from the portal, follow the link to the visualization page, sign in a free account in the portal using a Google apps enabled email address, and select the ‘Download’ tab in the study. Downloadable datasets include both raw and normalized cell x gene matrices, as well as relevant metadata. These datasets are additionally available here to facilitate downloading: https://drive.google.com/drive/folders/1bxCIqNeZ7wLuVOT16gphwj98_cc9KhrV?usp=sharing. We have also posted these cell x gene matrices to Chan Zuckerberg Initiative cellxgene (https://chanzuckerberg.github.io/cellxgene/posts/cellxgene_cziscience_com) and the Broad Institute Single Cell COVID-19 portal (https://singlecell.broadinstitute.org/single_cell/covid19) as leading community efforts. FASTQ files and cell x gene matrices for NHP and murine datasets, and cell x gene matrices for human datasets, are available at GEO: GSE148829.
In this same table, we further highlight four access types. 1. published datasets where everything is available (1 study); 2. unpublished datasets where everything is available (2 studies, 19,670 new cells for download), 3. unpublished datasets where ACE2+ cell subsets, and the necessary subsets to contextualize those cells (i.e., epithelial cells for type II pneumocytes) are fully available (5 studies, 17,986 new cells for download); and, 4. those unpublished datasets where expression is shared for ACE2/TMPRSS2 (2 studies, 9,112 new cells). For those unpublished datasets where only specific subsets of cells or genes are available, full expression matrices are available upon request for COVID-19 related questions.
All data included in the present study can be visualized using the following web viewer:
https://singlecell.broadinstitute.org/single_cell?scpbr=the-alexandria-project.
As we gain further insight and feedback from our own groups, collaborators, and investigators, we will continue to provide updates on our resource websites, including the utility of in vitro systems, such as organoids (Mead et al., 2018), for the study of SARS-CoV-2: http://shaleklab.com/resource/covid-19-resources/ and www.ordovasmontaneslab.com/covid-19-resources/. We also note that there are several ongoing efforts unified together through the HCA Lung Biological Network group that we will reference and to which we will link as they become available.
No custom code was used to analyze these data and all methods and packages used are cited in the Method Details section.