Abstract
The genetic material of every organism exists within the context of regulatory networks that govern gene expression, collectively called the epigenome. Epigenetics has taken center stage in the study of diseases such as cancer and diabetes, but its integration into the field of environmental health is still emerging. As the Environmental Mutagenesis and Genomics Society (EMGS) celebrates its 50th Anniversary this year, we have come together to review and summarize the seminal advances in the field of environmental epigenomics. Specifically, we focus on the role epigenetics may play in multigenerational and transgenerational transmission of environmentally induced health effects. We also summarize state of the art techniques for evaluating the epigenome, environmental epigenetic analysis, and the emerging field of epigenome editing. Finally, we evaluate transposon epigenetics as they relate to environmental exposures and explore the role of noncoding RNA as biomarkers of environmental exposures. Although the field has advanced over the past several decades, including being recognized by EMGS with its own Special Interest Group, recently renamed Epigenomics, we are excited about the opportunities for environmental epigenetic science in the next 50 years.
Keywords: environmental epigenetics, transposons, DNA methylation, piRNA, TaRGET II
INTRODUCTION
Epigenetics is the collective term for the study of mitotically heritable and potentially reversible changes in gene expression unrelated to changes to the DNA sequence itself. Epigenetic marks include chromatin modifications (e.g., histone protein acetylation, methylation, and ubiquitination), noncoding long and small RNA (e.g., lncRNA, microRNA [miRNA], and PIWI-interacting RNA [piRNA]), and alterations to DNA itself (e.g., DNA methylation and hydroxymethylation). Epigenetics has taken center stage in the study of chronic diseases such as cancer, obesity, diabetes, and neurodegeneration; however, its integration into the field of environmental health sciences and toxicology is only a few decades old. For example, the developmental origins of health and disease (DOHaD) work of David Barker and colleagues showed that a child’s environment during gestation and soon after birth influences his or her risk of developing disease later in life (Bateson et al., 2004). The molecular basis, however, for this implausible mode of inheritance of disease risk was unknown until 2003, when Drs. Randy Jirtle and Robert Waterland published a seminal paper in Molecular and Cellular Biology (Waterland and Jirtle, 2003). The stunning physical differences in the genetically identical mice seen in Figure 1 are the result of the epigenetic status of a metastable epiallele (shown is the murine Avy viable yellow Agouti locus). In this model, Jirtle and colleagues demonstrated that maternal dietary supplementation with methyl donors, including betaine, methionine, and folic acid, alters the offspring coat color distribution of the offspring. More importantly, they identified DNA methylation of a transposable element (TE) upstream of the Agouti gene as the molecular basis for this shift (Waterland and Jirtle, 2003). This observation marked the advent of a new field of scientific investigation: environmental epigenomics.
Fig. 1.
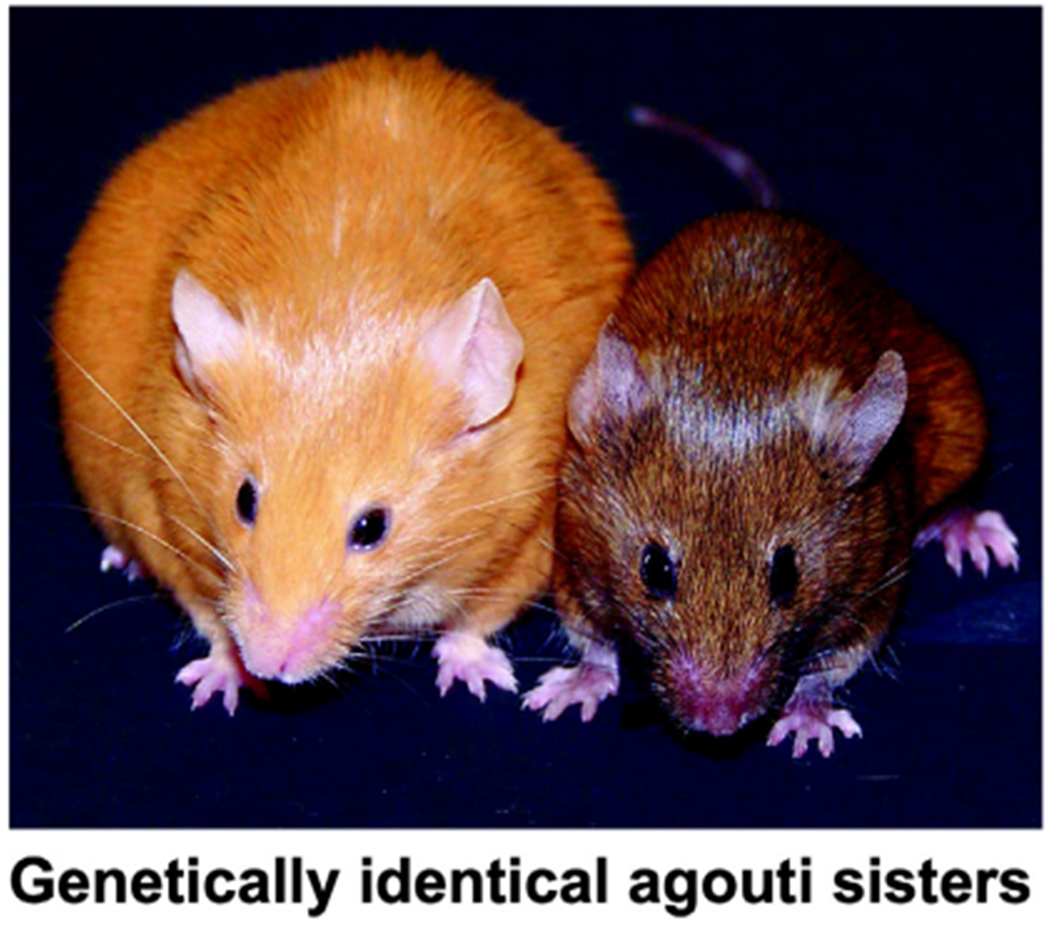
One-year-old female genetically identical viable yellow agouti mice (Avy). Maternal dietary supplementation with methyl donors such as folic acid, choline, and betaine (Waterland and Jirtle, 2003) or the phytoestrogen, genistein (Dolinoy et al., 2006), shifts the coat color of the offspring from yellow to brown and reduces the incidence of obesity, diabetes, and cancer. Furthermore, these maternal dietary supplements can guard the epigenome from the hypomethylating effects of BPA, a common endocrine disrupting chemical used in the production of polycarbonate plastic and epoxy resins (Dolinoy et al., 2007).
Despite a growing interest in the integration of epigenetics into toxicology and translational sciences, recent advances of the field have not yet fully incorporated epigenetics into research, risk assessment, or epigenome editing applications. To honor the 50th Anniversary of the Environmental Mutagenesis and Genomics Society (EMGS), we provide a primer on the principles and practices critical to understanding the role of the epigenome in regulating gene expression and the resulting response of cells, tissues, and individuals to environmental exposures. We first discuss how the work of Jirtle and Waterland has been extended to evaluate multigenerational and transgenerational effects of perinatal exposures. We next focus on the complexity of tissue and even cell type epigenomic specificity by highlighting the National Institute of Environmental Health Sciences’ (NIEHS) multiphased Toxicant Exposures and Responses by Genomic and Epigenomic Regulators of Transcription (TaRGET) Program (Wang et al., 2018). As epigenetics encompasses genes, noncoding elements, regulatory regions, and transposons, we next describe the use of transposons as epigenetic biomarkers. We finish with a discussion of the emerging need to expand beyond DNA methylation in environmental epigenetics research to also include noncoding RNAs (ncRNAs) as biomarkers of exposures and mechanisms of disease risk. Finally, new advances in epigenome editing, including our recent work on the potential of piRNA DNA methylation editing, will be explored for environmental health research (Perera et al., 2019).
MULTIGENERATIONAL AND TRANSGENERATIONAL TRANSMISSION OF ENVIRONMENTALLY INDUCED HEALTH EFFECTS
Gestation as a Sensitive Period for Environmentally Induced Epigenetic Changes
Epigenetic regulation can be modified by the environment, and these changes contribute to growth, development, and disease risk (Faulk and Dolinoy, 2011). The epigenome, and DNA methylation in particular, is especially sensitive to environmental perturbation in the early stages of gestation when epigenetic patterns that can be inherited across subsequent cell divisions are being set up. Hundreds of studies in human cohorts and animal models have shown associations between the gestational environment and epigenetic change in the offspring. These studies provide evidence for epigenetic change as one mechanism underlying the DOHaD hypothesis (Barouki et al., 2018). For example, differential DNA methylation is associated with gestational exposures to toxicants (e.g., lead, arsenic, bisphenol A [BPA], and cigarette smoke; Cardenas et al., 2015; Goodrich et al., 2015; Anderson et al., 2016; Joubert et al., 2016; Wu et al., 2017; Alavian-Ghavanini et al., 2018; Junge et al., 2018), maternal stress (Mulligan et al., 2012; Non et al., 2014), and maternal diet (Waterland and Jirtle, 2003; Dolinoy et al., 2006; Burdge et al., 2007; Amarasekera et al., 2014; Gonzalez-Nahm et al., 2017). Understanding the influence of gestational environmental exposures on the offspring epigenome is thus an important part of risk assessment for toxicants. In addition to epigenetic effects by gestational environment on the F1 offspring, there is evidence for F2 grandoffspring epigenetic changes (Rehan et al., 2012; Mitchell et al., 2016) as the fetal germ cells are directly exposed during this time (Fig. 2) and even F3-F4 transgenerational effects on subsequent generations that were never exposed (Anway et al., 2005; Manikkam et al., 2014). This section will highlight several examples of changes to offspring, grandoffspring, or great-grandoffspring DNA methylation following gestational exposures and will briefly describe common approaches for DNA methylation analysis that can be applied to studies in this area.
Fig. 2.
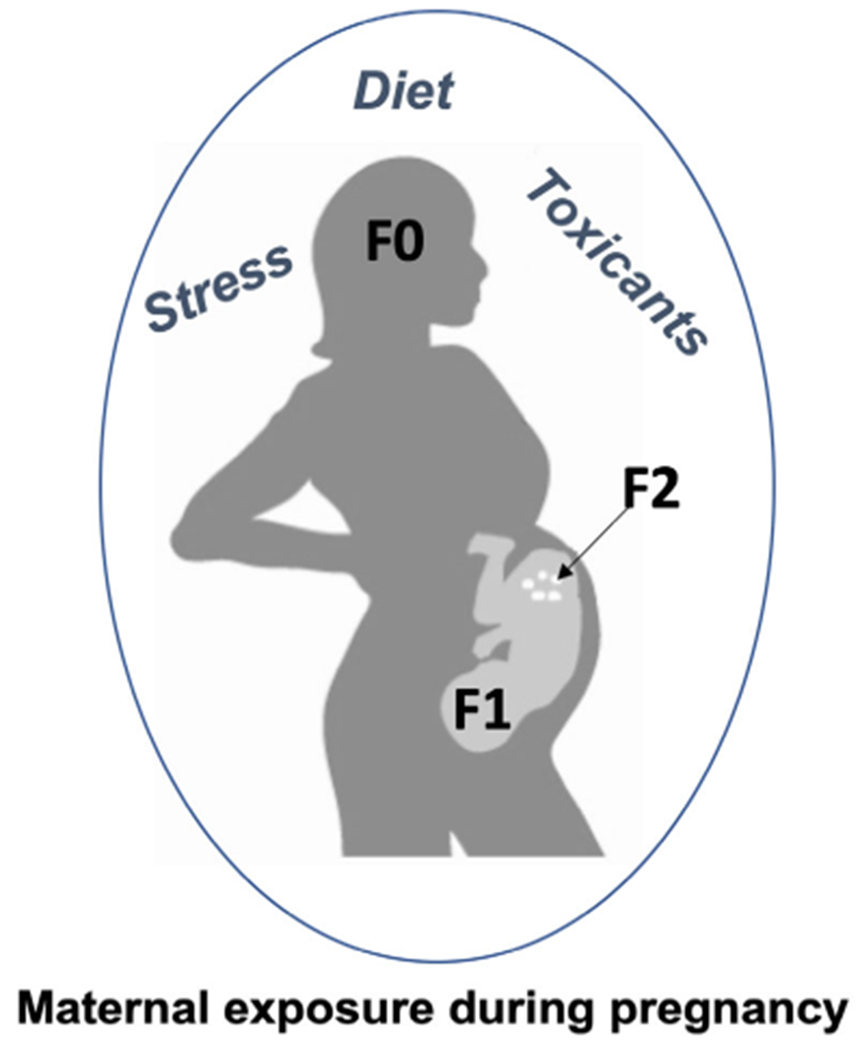
Direct and indirect effects of exposures across multiple generations. Among mammals, maternal exposures during pregnancy may directly influence the mother (F0) and child (F1). Future grandchildren (F2) may be impacted as the primordial germ cells are also directly exposed. Effects are considered transgenerational when they are observed in generations that were not directly exposed (i.e., F3, F4, and beyond in this human example).
Examples of Multigenerational Epigenetic Effects
As mentioned above, perhaps the most striking rodent examples of the impact of the early gestational environment on offspring phenotype through epigenetic change stem from the Avy mouse model (Fig. 1). Avy is a metastable epiallele, an allele that is variably expressed among individuals due to epigenetic modifications established early in development (Rakyan et al., 2002). As such, metastable epialleles are particularly vulnerable to early environmental influences, including diet, toxicants, and radiation. In the Avy mouse model, coat color throughout life and obesity status and tumor susceptibility in adulthood correlate with whether the Avy allele is turned on or off through DNA methylation and histone modifications in early development. The epigenetic status of this locus has been shown to be associated with early gestational exposures to genistein, a methyl-donor-enriched diet, BPA, lead, and other dietary agents or environmental toxicants, all of which continue to impact the offspring’s health later in the life course (Waterland and Jirtle, 2003; Dolinoy et al., 2006; Dolinoy et al., 2007; Kaminen-Ahola et al., 2010; Bernal et al., 2013; Faulk et al., 2013b).
Epidemiological studies have employed genome-wide screening approaches across multiple tissues to identify potential metastable epialleles in humans and to determine their response to the environment (Waterland et al., 2010; Dominguez-Salasetal., 2014; Silver etal., 2015). For example, a natural experiment in rural Gambia compared individuals with periconceptual exposure to the dry vs. rainy season, which largely impacts diet, and found differential methylation of the VTRNA2-1 epiallele, which persisted into later childhood (Silver et al., 2015). Genomic imprinting is an epigenetic process that allows a subset of autosomal genes to be expressed exclusively from one allele, in a parent-of-origin-specific manner. Thus, imprinted genes are another set of candidates to consider in environmental epigenetics due to their unique characteristics, including parent-of-origin-dependent regulation and key functions in early life growth (Lawson et al., 2013) . Importantly, DNA methylation of many imprinted genes is responsive to the gestational environment. For example, DNA methylation at well-characterized differentially methylated regions (DMRs), including the imprinting control regions of IGF2 and H19, are associated with prenatal exposures to smoking, phthalates, folate, malnutrition, and more in epidemiological studies (Heijmans et al., 2008; Murphy et al., 2012; Tobi et al., 2012; Hoyo et al., 2014; LaRocca et al., 2014) . There is also evidence that this epigenetic alteration is persistent—even into late adulthood (Heijmans et al., 2008)— and contributes to adverse health outcomes including low birth weight, adolescent adiposity, and adiposity in postmenopausal women (Murphy et al., 2012; Huang et al., 2012b; Song et al., 2018).
Rodent and human studies have provided evidence for the theory that primordial germ cells are uniquely susceptible to epigenetic change by grandmaternal exposures, and these exposures can thus impact health of not only the F1 but also the F2 generation (Sales et al., 2017). In epidemiological studies, grandmother smoking on the maternal side was associated with autistic traits (Golding et al., 2017), asthma with allergies (Accordini et al., 2018), and obesity (Ding et al., 2017) among F2 grandchildren. In a rat model, following F0 exposure to nicotine during gestation, the F1 and F2 generations both displayed alterations in lung function that were consistent with effects on myofibroblast differentiation. Importantly, changes to both global DNA methylation and histone acetylation were observed in the lung and gonad tissues of the F1, suggesting an epigenetic underpinning for F2 effects (Rehan et al., 2012).
Examples of Transgenerational Epigenetic Effects
Evidence for environmentally induced transgenerational effects via germline epimutations that transmit beyond the exposed generations is recently coming to light (Skinner, 2015). Transgenerational reproductive toxicity including decreased epididymal sperm count and reduced sperm motility were first reported in four generations of male rats following F0 pregnancy exposure to the fungicide vinclozolin and the pesticide methoxyclor (Anway et al., 2005). These transgenerational effects were associated with inherited epigenetic marks, including DNA methylation in the male germline (Anway et al., 2005; Manikkam et al., 2014). Exposure-specific sperm DNA methylation profiles transmitted through the F3 generation have been reported in rats for a pesticide mixture, a plastics mixture, jet fuel, and dioxin (Manikkam et al., 2012). Overall, these studies draw attention to the need to assess whether other common exposures lead to transgenerational epimutations and health effects. The well-characterized model organisms, zebrafish and Caenorhabditis elegans, may serve as valuable screening species for transgenerational effects and their epigenetic underpinnings of a wider array of exposures given the fast generational turnover in these species (Greer et al., 2011; Carvan et al., 2017; Cavalieri and Spinelli, 2017; Camacho et al., 2018).
METHODS FOR DNA METHYLATION ANALYSIS
Candidate Gene or Locus-Specific Analysis
Identifying DNA methylation changes associated with early life exposures has the potential to serve as biomarkers of exposure and/or mechanistic links to increased disease risk across generations and will improve toxicological risk assessment. Common approaches for DNA methylation analysis in human or animal models include quantifying DNA methylation at specific genes, TEs, or across all or most genes (epigenome-wide analyses). Many popular methods for gene-specific analysis rely upon sodium bisulfite treatment of the DNA first, which converts unmethylated cytosine residues to uracil but leaves methylated cytosines unchanged (Grunau et al., 2001). These methods include cloning followed by sequencing (Zhang et al., 2009), methylation-specific quantitative polymerase chain reaction (PCR; Eads et al., 2000), pyrosequencing (Busato et al., 2018), and more recently, targeted bisulfite sequencing, which takes advantage of next-generation sequencing technology (Bernstein et al., 2015; Wendt et al., 2018). In addition to their utility for specific genes, pyrosequencing is widely used to measure both human and mouse TE methylation (e.g., LINE1 and Alu). The EpiTYPER assay is another popular method for gene-specific analysis which involves enzyme digestion after amplification of bisulfite-converted DNA, creating fragments of different mass based on methylation status, which are then analyzed via mass spectrometry (Suchiman et al., 2015).
Epigenome-Wide Analysis
The most common tools used in epigenome-wide association studies in human populations have been the Illumina Infinium series of probe-based arrays that quantify DNA methylation at single CpG site resolution, needing as little as 250 ng of bisulfite-converted DNA as input. The first two versions of this platform, the Infinium HumanMethylation27 and HumanMethylation450 BeadChips provided coverage at >27,000 and >480,000 CpG sites. In December 2015, the latest version of the Infinium array, the MethylationEPIC BeadChip, was released, which quantifies DNA methylation at >850,000 CpG sites across all known genes and in intergenic and key regulatory regions (e.g., gene promoters and enhancers; Moran et al., 2016). Apart from the Infinium arrays, next-generation sequencing-based methods are gaining popularity as these methods can be used for any species with a mapped genome and costs are decreasing (Bock et al., 2010; Zhao et al., 2014; Ziller et al., 2015). In additional to bisulfite-conversion-based sequencing methods, antibody-based methods can be used to semiquantitatively measure methylation at transposons or genome-wide. Referred to as methylated DNA-immunoprecipitation (Me-DIP), these methods involve using anti-5mC antibodies to immunoprecipitate methylated DNA (Zhao et al., 2014; Staunstrup et al., 2016). Antibody-based approaches can also be utilized to evaluate DNA hydroxymethylation, which is also emerging as an environmentally responsive modification (Kochmanski et al., 2018a; Kochmanski et al., 2018c).
THE NIEHS TaRGET II CONSORTIUM AND ENVIRONMENTAL EPIGENOMICS
Current Challenges in Environmental Epigenomics Studies
Although we have made substantial strides in our understanding of how environmental exposures impact the epigenome and human health, several factors continue to pose significant challenges and limitations. These include tissue specificity of environmental exposures, tissue and cellular heterogeneity, sexually dimorphic effects, and the effects of age and stress on exposure-related epigenetic changes and health outcomes. First, human environmental epigenomics studies are limited by tissue accessibility, as access to most disease-relevant tissues targeted by environmental exposures is not possible. Thus, human studies typically rely on surrogate tissues, including the blood, hair, and skin, to gain insight into the effects of environmental exposures on inaccessible target tissues. However, the extent to which epigenetic changes in surrogate tissues reflect those in target tissues is currently unclear. Second, it is increasingly clear that there is significant heterogeneity within tissues, and even within individual cell types, with respect to gene expression and patterns of epigenetic marks (Cheow et al., 2016; Cusanovich et al., 2018; Ben-Moshe and Itzkovitz, 2019). Thus, assessment of tissue-specific effects of environmental exposures may be masked by such heterogeneity. Moreover, environmental exposures may induce changes in the cellular composition of tissues (Trevino and Katz, 2018; Hung et al., 2019), complicating the interpretation of epigenetic studies. These challenges may be addressed through emerging methods in single-cell analyses (Cheow et al., 2016). Third, significant evidence demonstrates that the effects of toxicant exposures are highly sex specific (Faulk et al., 2013b; Bansal et al., 2017; Kundakovic, 2017; Neier et al., 2019; Winterbottom et al., 2019). Given that there are also significant sex disparities in the prevalence and outcomes of many diseases associated with environmental exposures (Graham, 2015; DeSantis et al., 2017), an understanding of how environment differentially affects disease risk in a sex-specific fashion is critical. Finally, although cross-sectional studies have clearly demonstrated environment-induced epigenetic changes (Montrose et al., 2018; Curtis et al., 2019; Winterbottom et al., 2019), whether these changes persist across time in both surrogate and target tissues is not well-understood. Notably, environmental exposures can alter the trajectory of age-related epigenetic changes (Fraga et al., 2005; Faulk et al., 2014; Krauskopf et al., 2018). However, it is currently unclear whether these interactions between environment and aging are uniform across tissues. It will therefore be important to consider age-environment interactions in the design of human toxicoepigenetics studies.
The NIEHS TaRGET Consortium
In order to address these important issues and to aid in the design and analysis of human environmental epigenetics studies, the NIEHS developed the TaRGET program. The first phase of this program, TaRGET I, was formed in 2012 to investigate how environmental exposures affect the cellular machinery critical for establishment of normal epigenetic patterning (Prins et al., 2014; Wang et al., 2016). During the second phase of the project, a multi-institutional consortium, TaRGET II, was established to investigate the conservation of environment-induced epigenetic signatures across multiple target and surrogate tissues and cells (Wang et al., 2018). Using a common mouse model and a well-defined treatment paradigm, consortium members are investigating the effects of several perinatal environmental exposures, including lead (Pb), diethylhexyl phthalate, BPA, arsenic (As), dioxin (TCDD), and particulate matter (PM2.5) on the epigenome and transcriptome in multiple target and surrogate tissues (Wang et al., 2018; Fig. 3). These analyses are conducted at three separate time points during the lifespan of the offspring, including the early postnatal period, early adulthood, and late adulthood, in order to investigate whether perinatal toxicant-induced epigenetic programming persists as the animals age. Collectively, work from this project is anticipated to provide valuable insight into the tissue-, cell-, and sex-specific effects of environmental exposures on epigenetic programming across the lifespan. These findings will aid in the design and interpretation of human population-based epigenomics studies, which will be the focus of the TaRGET III and TaRGET IV phases of the project.
Fig. 3.
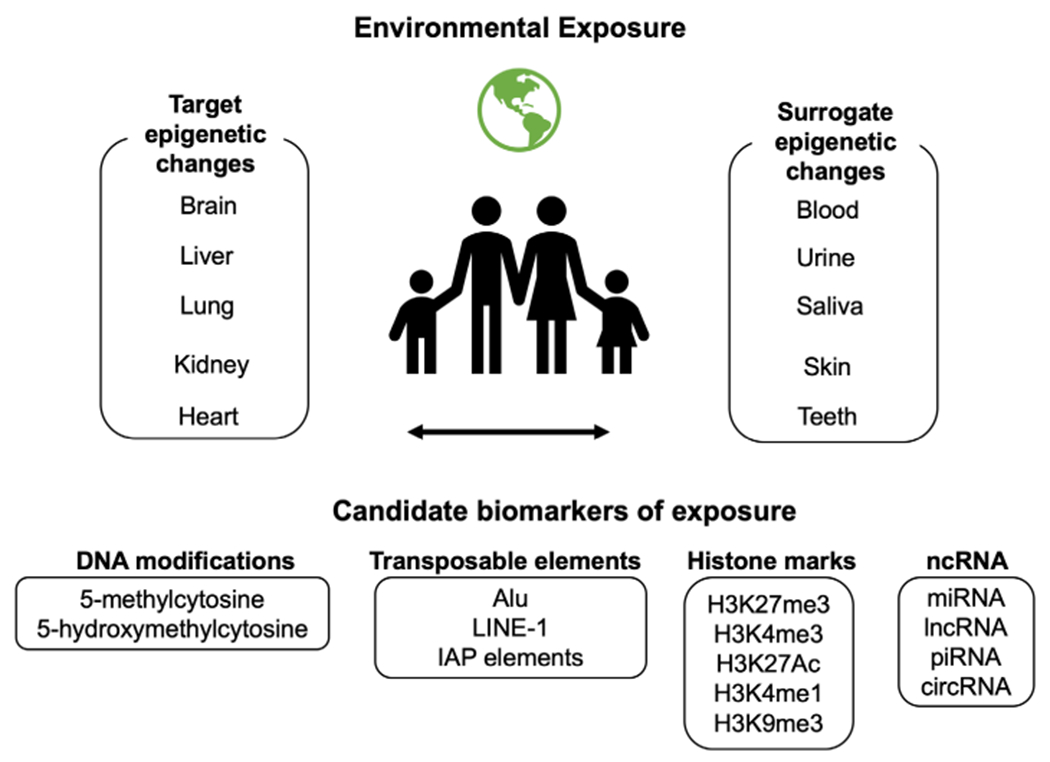
Conceptual diagram for environmental epigenomics studies. Environmental epigenomics studies utilize surrogate tissues as proxies for epigenetic changes in target tissues and determine whether these changes correlate with disease. Several types of epigenetic changes may represent biomarkers of environmental exposures and adverse health outcomes.
Pb Exposure and Tissue-Specific DNA Methylation
As part of the TaRGET consortium, we are investigating the effects of perinatal Pb exposure on epigenetic and transcriptional programming of offspring mice, using the consortium mouse model of environmental exposures. We recently investigated the effects of perinatal Pb exposure on DNA methylation in mouse liver and blood, to determine whether common signatures exist between the two tissues, whether these signatures are sex-specific, and whether they persist into adulthood (Abstract presented at Society of Toxicology Annual Meeting, Baltimore, MD, 2019). For these studies, dams were exposed to Pb acetate (32 ppm) as outlined previously (Faulk et al., 2013b) via drinking water for two weeks prior to mating, and Pb exposure continued throughout pregnancy and lactation. Offspring were weaned at three weeks of age, and Pb exposure ceased at this time point. Between three weeks and five months of age, offspring were given standard chow and Pb-free drinking water. Six male and six female mice were euthanized at five months of age, and the blood and liver were collected according to TaRGET II consortium-approved protocols. Thus, this two-stage study design allows for the assessment of transient as well as persistent epigenetic modifications, as transient changes may readily occur immediately following exposures but may not persist over time when the exposure is removed.
To determine the effects of Pb exposure on DNA methylation, within TaRGET II, we utilized enhanced reduced-representation bisulfite sequencing, which allows the detection of base pair resolution DNA methylation at CpG-rich loci (Garrett-Bakelman et al., 2015). Notably, although Pb exposure ceased at three weeks of age, our studies revealed thousands of DMRs that persisted into adulthood. DMRs were highly sex specific, with few regions overlapping between the blood and liver. In females, three DMRs overlapped between the blood and liver, although the changes in methylation with Pb exposure were in the opposite direction in the two tissues. In males, there were four DMRs that overlapped between the blood and liver, with two loci, mapping to Grifin and Plekhg3, which exhibited concordant changes in methylation with Pb exposure. These findings suggest that perinatal Pb exposure results in stable changes in DNA methylation in the blood and liver of offspring mice, with few sites directly overlapping between the two tissues or across sexes. Ongoing studies are focused on determining whether other epigenetic signatures in the blood may reflect Pb-induced changes in the liver and other target tissues, including the brain.
It is notable that Pb-induced changes in DNA methylation persisted into adulthood, although the functional consequences of these changes are currently unclear. Notably, recent work suggests that early-life environment can induce epigenetic changes that are “silent” but render the target genes hyper-responsive to hormonal cues later in life (Greathouse et al., 2012; Wang et al., 2018). Indeed, Wang et al. demonstrated that early life exposure to BPA induced reprogramming of the activating H3K4me3 histone mark at prostate cancer-associated genes that persisted into adulthood (Wang et al., 2016). Intriguingly, several of the BPA-reprogrammed genes exhibited no change in basal expression; however, they were hyper-responsive to subsequent hormonal stimulation (Wang et al., 2016). These findings suggest that changes in epigenetic marks may provide a more reliable signature of early-life environmental exposures than changes in gene expression. The data also suggest that the functional consequences of toxicant-induced epigenetic programming may only manifest upon subsequent environmental insult. Thus, in future studies, it will be important to investigate how subsequent environmental exposures, nutritional perturbations, and stressors later in life interact with early epigenetic programming to initiate or exacerbate disease. Future research is required to understand the persistence of environmentally induced alterations on epigenome in order to determine transient versus persistent epigenetic changes associated with environmental exposures.
TRANSPOSONS AS EPIGENETIC TARGETS
Transposons
TEs are small regions of DNA that have evolved the ability to copy themselves throughout the genome. Referred to as repetitive elements, mobile DNA, or transposons they are found in all animals. Varying in size from 250 nt to over 7,000 nt, they can encode protein machinery that enables their jumping ability (Huang et al., 2012a). These mobile elements have several origins, including endogenous retroviruses that have incorporated themselves into the genome or gene fusions that have gained the ability to copy themselves. Subsequently, they have spread throughout the genome. They are delineated in classes, such as short or long interspersed elements (SINEs and LINEs) which dominate the genome, and subcategorized into families and subfamilies based on sequence similarity and presumed copying origin (Bao et al., 2015).
Mammalian genomes consist of approximately 50% of sequence derived from transposons, as opposed to just ~1% of sequence coding for genes (Platt et al., 2018). Despite the consistency of this fraction across mammals, the composition of the transposon sequence varies widely between species. Generally, there is a balance between TE proliferation and host genome suppression, often through epigenetic mechanisms (Canapa et al., 2015). Most mammals have at least one actively transposing family of TEs. They are inherited from parent to offspring, rarely transferring horizontally between species. Although “selfish,” due to their ability to promote their own transmission at the expense of other genes, these TEs can often be adapted to serve the host as a source of raw variation and mutation (Dupressoir et al., 2012). Some elements are enriched in CpG sites or other useful features for evolution, so their proliferation throughout the genome can be adaptive.
Humans are illustrative of transposon dynamics in the genome. The most abundant TE by number in the human genome is the Alu element, a short ~300 nt element that does not encode its own transposition machinery and is present in about 1.2 million copies in primate genomes (Deininger, 2011). It relies upon the reverse transcriptase protein for mobilization, encoded by the most abundant transposon by percentage nucleotide in the human genome, the LINE1 element. The LINE1 is >6,000 nt and is present in >100,000 copies in the human genome making up 18% of the genome, but most are 5′ truncated, with around 100 LINE1s suspected to be capable of activity and just 6 “hot” LINEs provide over 80% of the observed transposition in cell culture (McLaughlin, 2018). Both elements can contribute to disease by several mechanisms including transpositional mutation breaking a gene, by causing homology mediated deletions, or, most relevant here, by disruption of chromatin or nearby gene expression (Beck et al., 2011; Rebollo et al., 2011). Alus are primate specific, whereas LINE1s are found across all vertebrates. Both are used as epigenetic biomarkers of exposure and disease status.
Epigenetic Relevance of TE Insertions
In mammals and other organisms, the fifth carbon of cytosines in a CpG context can be methylated. Deamination of methylated cytosine results in a thymine base. Transitions from C to T account for up to 42% of all mutations in the human genome, far more than any other transition or transversion, most occurring at a methylated CpG sites (Gojobori et al., 1982). The rapid turnover of CpG sites to either TpG or CpA has important implications in evolution. Whereas humans have a 1% sequence divergence from chimpanzees on average, at CpG sites the mutation rate rises to 15% (Chimpanzee and Analysis, 2005). Over evolutionary time, mammalian genomes have lost CpG sites from an expected frequency of 6.25% to an observed frequency of less than 1% of dinucleotides. As DNA methylation is crucial for gene regulation, CpG sites at high-density CpG islands (CGIs) near gene promoters must be replenished somehow (McLain and Faulk, 2018). Transposons are typically silenced by DNA methylation at CpG sites to prevent their movement from disrupting the genome. However, nonrandom TE insertions appear to provide new CpG sites via their ability to copy paste sequence throughout the genome. In humans, the Alu element is both enriched in CpG sites and found at higher density near CGIs and therefore replenishes CpG sites, providing flexibility in methylation status near genes (Gu et al., 2016).
Transposons as Epialleles
In mice, the intracisternal A particle (IAP) element is an endogenous retrovirally derived transposon and is responsible for ~10% of de novo mutations found in inbred strains. The high mobilization activity of IAP has resulted in high polymorphism as well, with 60% being private to specific strains (Zhang et al., 2008). The IAP is capped on both ends by long terminal repeats (LTRs) that are relatively CpG dense and have bidirectional promoter activity. Typically, these CpG sites are silenced through DNA methylation, however, several IAPs have been shown to be variably methylated between mice, with consistent methylation across tissues, hallmarks of metastable epialleles. The Avy mouse differential coat colors (Fig. 1) are driven by promoter activity at insufficiently silenced CpG sites within the LTR of an IAP insertion. Generally, most IAPs are completely reset between generations and only rarely escape reprogramming (Kazachenka et al., 2018). Several studies in humans have also identified potential epialleles hosting differential methylation in various classes of TEs (Waterland et al., 2010; Faulk et al., 2016).
Benefits of Transposons as Biomarkers
TEs have several characteristics that make them useful as biomarkers of exposure and disease status. Many classes are found widely throughout the genome, and there are multiple copies per genome. They are not under selective constraint. They are usually heavily methylated, so any change in methylation status is suggestive of strong environmental impact. In practice, their sequence similarity allows thousands of loci to be amplified in a single PCR reaction with a single set of primers. Measuring the DNA methylation of these amplicons gives an average methylation value for all TEs of a specific class, usually Alu in humans or LINE1 in humans and mice. Given their relatively uniform spread across the genome, their methylation serves as a proxy for global methylation across the genome.
Epidemiological Biomarkers in Human Exposure
When studying populations for environmental exposures that are associated with disease, two main transposon assays have been used. The most widely used measure are taken with LINE1 and Alu elements, originally developed by Yang et al. to measure methylation in a PCR pool using primers matching consensus sequences of these two families (Yang et al., 2006). The LINE1 pyrosequencing assay was initially used to measure genomic instability in cancer but has been adapted for epidemiological studies (Estecio et al., 2007). The Alu and LINE1 assays, although both considered global, have different methylation levels by tissue and have distinct responses to environmental exposures (Price et al., 2012). For example, both Alu and LINE 1 correlate to preterm birth (Burris et al., 2012), whereas LINE1 but not Alu was affected by exposure to prenatal air pollution (Breton et al., 2016). While too numerous to list here, these epigenetic biomarkers have shown response to mercury (Narvaez et al., 2017), traffic particles (Baccarelli et al., 2009), industrial environments (Alvarado-Cruz et al., 2017), aging and ischemic heart disease (Baccarelli et al., 2010), carbon nanotubes (Ghosh et al., 2017), and polycyclic aromatic hydrocarbons (Lee et al., 2017a). The Alu assay has been used to study response to persistent organic pollutants in Koreans (Kim et al., 2010; Lee et al., 2017b) and Inuit (Rusiecki et al., 2008). Nearly all studies of toxicant exposures report hypomethylation of LINE1. Fewer studies report Alu methylation responses.
Use as Biomarkers in Mice
There is considerable interest in the use of TEs as biomarkers’ environmental exposures that alter DNA methylation. Studies have examined methylation of a subset (Faulk et al., 2013a) or the entire genomic complement of TE methylation in mice (Kazachenka et al., 2018). Locus-specific measurements of TE DNA methylation allow detection of differential methylation for individual TE insertions and can be considered causal for disruption of nearby gene expression (Rebollo et al., 2011). Potentially, incompletely silenced TEs could transcribe in unintended ways; indeed studies have shown hundreds of protein isoforms that express a fusion of a TE and a native gene (Ekram et al., 2012). Locally, several IAP elements show methylation response to perinatal lead exposure (Montrose et al., 2017) and phthalates (Neier et al., 2019). Globally, IAP elements have been used as genome-wide epigenetic biomarkers, showing hypomethylation in transcription factor binding sites (Shimosuga et al., 2017). Prenatal inflammation can impact murine LINE1 methylation in the brain (Basil et al., 2014). Radiation induces hypomethylation at LINE1 and the SINE B1 in mouse hematopoietic progenitors (Miousse et al., 2014). In contrast, at least one study showed that low dose radiation does not affect methylation of several TEs including IAP, B1, and LINE1 longitudinally (Newman et al., 2014). Similarly, a study by Tommasi et al. found no methylation changes in these same element families upon exposure to secondhand smoke (Tommasi et al., 2012). However, trichloroethylene exposure does alter IAP methylation in immune cells, which is of great interest given its widespread use as an obstetric analgesic (Gilbert et al., 2012). Regarding longitudinal studies, LINE1 and IAP elements have been used to measure epigenetic drift over time (Faulk et al., 2014; Kochmanski et al., 2017), during aging (Barbot et al., 2002) and by dietary exposure (Kochmanski et al., 2018b).
Caveats of TEs as Epigenetic Biomarkers
The nature of repetitive elements produces several caveats that should be considered when analyzing TE methylation data. Most importantly, the lack of conserved CpG sites between various copies of a family of TEs can mask the true methylation level. Assays are built to compare CpG site methylation by using a consensus sequence that is the average of thousands of TEs of a particular family. Often individual TE copies will have mutations at the location of CpG sites when compared to the consensus. In sequencing methods, for example, a read may come from a family member that has a TpG mutation single nucleotide polymorphism at CpG site annotated in the consensus. This position would be erroneously read as an unmethylated CpG site. As methylation is often calculated as percentage C to non-C, over thousands of elements or reads, methylation level can appear anomalously low at specific CpG sites. The true methylation level at CpG sites that exist in TE copies is usually very high in reality. For Alu elements, Yang et al. found only 36% of potential CpG sites maintained in aggregate PCR products (Yang et al., 2004). Of these, 85% were methylated. Another major difficulty arises from the similar, but nonidentical, nature of TE family members. Performing multiple alignments can be difficult, leading to undercounting of true TE copies.
Despite these caveats, methylation assessment between studies is comparable, and deviance from control group methylation level usually indicates an environmental cause. Therefore, the continued use of TEs as epigenetic biomarkers has a bright future, as long as the underlying biology of TEs is understood by those who measure them.
ncRNA AND EPIGENOME EDITINGTOOLS AS POTENTIAL INTERVENTIONS
An Introduction to Noncoding RNA
Although ncRNA was previously considered as products of “junk DNA,” recent scientific advancements have concluded that ncRNA are functional RNA molecules that arise from DNA, which do not translate to specific proteins (Boland, 2017). These ncRNAs are critical for biological functions as they are involved in complex biochemical and epigenetic regulatory mechanisms (Angrish et al., 2018). The genome encodes for lncRNAs, which are nonprotein coding transcripts >200 nucleotides, and small ncRNAs (sncRNAs), which are of much shorter lengths in comparison. lncRNAs are functionally distinct from sncRNA and are involved in biological processes such as genomic imprinting, X-chromosome inactivation, and development (Kung et al., 2013; Dhanoa et al., 2018).
Conversely, sncRNAs include several regulatory RNA species such as miRNA that are involved in mRNA transcription, short interfering RNAs (siRNAs) that are involved in transcriptional repression, PIWI-interacting RNAs (piRNAs) that are involved in transposon silencing, and small nucleolar RNAs that interact with other RNA and proteins for gene regulation. Based on current knowledge, miRNAs and siRNAs are roughly ~20 nucleotides in length and are associated with Ago proteins to silence genes via transcriptional and translational silencing mechanisms within the RNA interference pathway, whereas germline piRNAs are typically 24–32 nucleotides in length, in comparison (Lin, 2007). In contrast, PIWI proteins are important for piRNA biogenesis and forming the piRNA-induced silencing complex to induce DNA methylation for silencing of TEs in the germline (Tan et al., 2015). Therefore, the PIWI-piRNA interactions are functionally distinct from that of other sncRNAs (Han et al., 2017a), including several structural attributions that are distinct from other sncRNAs: preference for a 5′ uridine signature, presence of an adenosine signature at the 10th position, a 2′-O-methylation modification at the 3′ ends and clustering within a 20–90 kb length region (Lin, 2007; Zuo et al., 2016).
Circular RNAs (circRNAs) are another class of ncRNA species, which are a product of back-splicing of linear pre-mRNA that join together a donor site with an upstream acceptor site (Salzman, 2016). In some cases, circRNAs serve as molecular sponges that regulate transcription by removing miRNAs (Carrara et al., 2018). They are associated with cancer, Alzheimer’s disease, diabetes, cellular stress, aging, and genomic imprinting (Qu et al., 2017; Han et al., 2017b; Perera et al., 2018). Although miRNAs are by far the most studied class of sncRNA species in many fields including cancer, environmental toxicology, and risk assessment, future research in other sncRNA species may lead to similar breakthroughs based on their distinct cellular functions.
ncRNA Biomarkers Relevant for Human Health and Disease
The fields of genomics, toxicoepigenetics, and environmental epigenetics have greatly benefitted from the technological advancements and increasing power of sequencing, transcriptomics, and bioinformatics over the past few years. It provides much needed information to better understand the genetics and environmental factors that influence human health and disease. Thus, the discovery of ncRNA biomarkers related to a wide array of human disease stages and environmental exposures contribute toward identifying novel ncRNAs for precision medicine (Karlsson and Baccarelli, 2016; Roy et al., 2018). A few, recent studies in the field of environmental toxicology have provided evidence for lncRNA biomarkers associated with response to environmental stressors such as endocrine disrupting chemicals (EDCs), metals, cigarette smoke extracts, and genotoxic agents (Bhan et al., 2014; Bi et al., 2015; Zhou et al., 2015; Karlsson and Baccarelli, 2016), where genes are regulated by complex cellular and molecular mechanisms which are yet unknown.
SncRNA and circRNA biomarkers are often times preferred over lncRNAs as they are small, versatile, resistant to degradation, and widely available in biospecimens (Bahn et al., 2015). The most extensively studied ncRNA biomarkers are miRNAs, which are associated with EDC exposures (De Felice et al., 2015), metals (Sanders et al., 2015), pollution (Krauskopf et al., 2018), and several human diseases (Siddeek et al., 2014; De Felice et al., 2015; Sanders et al., 2015; Ehrlich et al., 2016; LaRocca et al., 2016; Romano et al., 2017; Krauskopf et al., 2018). The piRNA biomarkers may also be considered for future studies due to their expression in human blood, stability (due to the 2′-O-methylation modification at its 3′ end), and tissue specificity (Yang et al., 2015; Zuo et al., 2016). Although piRNAs are extensively studied and highly expressed from the germline, recent studies have provided evidence for piRNA expression in the soma (Lin, 2007; Zuo et al., 2016; Perera et al., 2019). So far, piRNAs have shown associations with chemical exposures such as dichlorodiphenyltrichloroethane and vinclozolin (Nilsson et al., 2018; Sai et al., 2018; Skinner et al., 2018) and human health and disease (i.e., cancer; Han et al., 2017a; Chalbatani et al., 2019).
CircRNAs are worthy biomarkers as they are stable, resistant to exonucleases, present in liquid biopsies, and highly abundant with specificity toward tissues and stage of development (Carrara et al., 2018; Zhang et al., 2018). Thus, circRNAs are becoming increasingly popular candidates for diagnosis and clinical interventions in recent cancer research and may serve as a potential ncRNA biomarkers of interest for future research (Zhang et al., 2018). Generating reliable ncRNA databases with precise detection methods for tissue-, sex-, and developmental stage-specific ncRNA species (i.e., miRNA, piRNA, and circRNA) is absolutely necessary to improve ncRNA biomarker identification. For instance, the publicly available databases for piRNA mostly consist of germline tissues with high incidences of nonspecific sequences for somatic tissues (Tosar et al., 2018; Perera et al., 2019), thus serving as a limitation for biomarker discovery. The less studied ncRNA species such as lncRNA, piRNA, and circRNA species are potential epigenetic biosensors that may serve as predictors for disease risk and later-life health outcomes that could be developed to improve personalized medicine in the future.
Epigenome Editing Techniques
Epigenome editing/engineering is a technology that manipulates the epigenome without disrupting the actual DNA sequence to regulate desired gene expression, which makes it a safer therapy in comparison to genome editing. Current technologies that act on the epigenome can be categorized as global epigenetic modifiers and target-specific epigenetic modifiers, both of which act on epigenetic readers such as methyl-CpG-binding proteins, histone methylation binding proteins, and histone acetylation binding proteins; writers such as DNA methyltransferases (DNMTs), histone methyltransferases (HMTs), and histone acetyltransferases (HATs); and erasers such as DNA demethylases, histone demethylases, and histone deacetylases (HDACs; Gillette and Hill, 2015).
Global epigenetic modifiers include pharmaceutical agents, such as azacytidine, that are widely used to inhibit DNMTs, resulting in global hypomethylation in dividing cells (Yang et al., 2010; Micevic et al., 2017). HDAC inhibitors, such as vorinostat (suberoylanilide hydroxamic acid), impact chromatin by maintaining an “open chromatin” configuration, facilitating transcriptional activity (Gryder et al., 2012). Advantages of these agents lie in their well-characterized use as human therapeutics and for basic research in cell lines and animals. Disadvantages of the pharmaceutical approach include their pleiotropic effects caused by indiscriminate epigenomic activity and propensity to affect biochemical pathways separate from the epigenome. Despite the desperate need for technologies to precisely modulate the epigenome, it is unlikely that either small-molecule pharmaceuticals or the disruption of chromatin enzymes can be rapidly adapted for locus-specific targeting.
Emergence of Targeted Epigenome Editors
Artificial DNA binding domains (DBDs) fused with catalytic domains of epigenetic modifiers have been developed as useful tools to recognize specific DNA sequences that introduce or remove epigenetic marks (i.e., DNA methylation and histone modifications) at a given locus via zinc-finger proteins (ZFPs), transcriptional activator-like effector nucleases (TALENs), and clustered regulatory interspersed palindromic repeats (CRISPR) systems (Fig. 4; Waryah et al., 2018). A few critical parameters for epigenome editing are specificity and efficiency of the editor, along with stability of the edited state. With its initial discovery in 1985 (Miller et al., 1985), the first practical method used the DBDs of ZFPs to recruit DNA methylation using DNMTs. In this method, iterative phage display selects for a ZFP that recognizes a 9–12 bp motif, and the resulting encoding sequence is ligated to the sequence for the methyltransferase domain (Fig. 4A; Xu and Bestor, 1997). So far, ZFPs have been engineered in combination with other DNMTs (Li et al., 2007; Siddique et al., 2013), DNA demethylases such as ten-eleven translocation (TET) enzymes (Zhao and Chen, 2013; Chen et al., 2014), and histone modifiers (Groner et al., 2010; Falahi et al., 2013). Major drawbacks for this system include high cost for protein domain construction, low specificity due to off-target effects, complexity (every zinc-finger domain must be custom evolved to target a specific sequence), and motif size limitation (Nemudryi et al., 2014; Waryah et al., 2018). Since their discovery in 2009 (Boch et al., 2009; Moscou and Bogdanove, 2009), TALENs are the second method used as potential epigenome editors (Fig. 4B; Konermann et al., 2013), where TET methylcytosine dioxygenase 1 (TET1)/DNMT3a was used to modify DNA methylation (Maeder et al., 2013) and lysine-specific demethylase 1 (LSD1) was used to modify the histones (Mendenhall et al., 2013). The DBDs of the TALEN system contains 7–34 tandem repeats, with each repeat binding to the major groove of DNA. Even though TALENs have higher specificity compared to ZFN, this technology poses significant limitations in assembly and delivery due to its large number of tandem repeats.
Fig. 4.
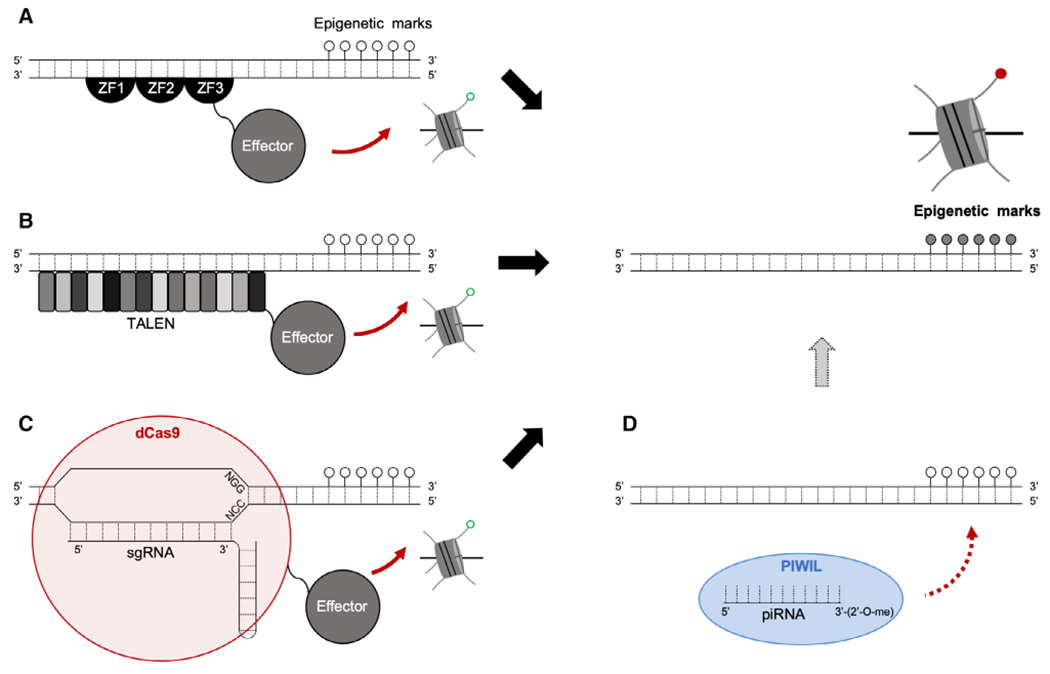
Targeted epigenome editors. Epigenetic marks represent DNA methylation changes and histone modifications. The open popsicles indicate unmethylated DNA, whereas closed popsicles represent methylated DNA, while the unmodified histone (green open dot), is changed to modified histone (red dot) via epigenetic editing. (A) ZFP-based epigenome editing. (B) TALEN-based epigenome editing. (C) CRISPR/dCas9-based epigenome editing (sgRNA, single guide RNA; dCas9, deactivated Cas9; NGG/NCC, PAM sequence). The effector protein signifies a DNA/histone modifying protein that is responsible for the desired epigenetic change. (D) Potential ncRNA-based epigenetic modification. Based on current research that indicates piRNA presence and activity in somatic tissues (Perera et al., 2019), other ncRNA species may be used as potential candidates for epigenome editing. For instance, mature piRNA that contain a 2′-O-methylation modification at its 3′ end and its associated PIWIL proteins may be used to specifically methylate DNA.
The CRISPR and CRISPR-associated (Cas) system is the most recent and impactful genome editing technique (Gasiunas et al., 2012; Jinek et al., 2012), which has recently become desirable for epigenome editing. This system utilizes guide RNAs (gRNAs) that recognize ~20 nucleotide sequences to direct deactivated Cas9 (dCas9) attached to a desired epigenome modifying enzyme to its complementary DNA sequence for desired effects (Fig. 4C; Waryah et al., 2018). The CRISPR/Cas system has been used for targeted demethylation and methylation of genes using dCas9-TET1 and dCas9-DNMT3a, respectively (Liu et al., 2016; Kantor et al., 2018; Xie et al., 2018). The use of dCas9 attached to HATs (p300), HMTs (PRDM3), HDACs, and chromatin-modifying complexes (LSD1 and KRAB) indicate the wide applicability of the CRISPR/Cas9 system (Gilbert et al., 2013; Hilton et al., 2015; Cano-Rodriguez et al., 2016; Waryah et al., 2018). However, high incidence of off-target effects and time taken for screening is one of the main drawbacks of this system (Falahi et al., 2015). Nonetheless, use of ZFP, TALENs, and CRISPR/Cas9 represents a significant advance in the locus-specific manipulation toolkit and are considered standards by which to compare any new methods (Rots and Jeltsch, 2018). Precisely targeting epigenetic marks at specific regions on the genome at a developmental stage and tissue-specific manner still remains a challenge as the molecular mechanisms underlying epigenetic regulation in normal and disease states are still not well-understood.
Epigenetic changes that occur due to environmental exposures are also not well-understood for most exposures. To inform risk assessment of hazardous exposures and intervention efforts to reverse toxic effects and disease risk, systematic evaluation and rigorous analysis of the epigenome by common exposures is needed. This investigation should involve all aspects of epigenetic regulation including DNA methylation at gene promoters, histone modifications, RNA-mediated gene silencing mechanisms, genomic imprinting, and X-inactivation that are induced by sncRNAs (miRNA, piRNA, circRNA, and siRNA). As recent advancements in miRNA- and siRNA-based ncRNA therapeutic approaches have provided much promise (Aagaard and Rossi, 2007; Burnett et al., 2011; Aliabadi et al., 2016), using a similar ncRNA-based technology to modify the epigenome as a potential intervention is indeed an avenue for future studies. For instance, the piRNA class of ncRNA represents a fascinating adaptive mechanism and “ready-made” tool for innovation in locus-specific repression through DNA methylation (Fig. 4D; Mani and Juliano, 2013; Fu et al., 2014). Therefore, it is important to conduct research using in vitro cell cultures, tissue samples, and vivo animal models to evaluate the persistence of epigenetic changes with ncRNA delivery, dosage, and specificity.
CONCLUSIONS AND FUTURE DIRECTIONS
As we have discussed, a comprehensive understanding of the tissue- and even cell-specific epigenetic effects of toxicant exposures, their interactions with other stressors later in life, and their persistence across the lifespan is an important and lofty goal for the next 50 years of the EMGS. We are a Society founded with deep interest and expertise in environmental impacts on mutagenesis, and thus focusing on transposons that make up so much of the genome is crucial to understanding the mechanistic link between the environment and heritable changes in gene expression. Transposons serve as both biomarkers and drivers of epigenetic changes and underlie the most well-known epigenetic phenotypes in animals. Future human studies of environmental exposures should place special emphasis on including transposon epigenetic analysis as both an endpoint and explanatory mechanism. The discovery of disease-associated biomarkers have rapidly moved recent scientific research from basic to translational sciences, which now includes miRNA-based therapies that target-specific interventions (Yu et al., 2018). Meanwhile, the current knowledge of less popular ncRNA biomarkers (lncRNA, piRNA, and circRNA) association in cell-, tissue-, sex-, and developmental stage-specific environmental exposures or disease states still needs to be carefully examined.
As relatively few chemicals have been evaluated for effects on the epigenome, the toxicopigenetic effects of a broader array of environmental exposures, especially exposures during critical periods of development, should be prioritized for comprehensive assessment. The identification of epigenetically labile genes such as additional metastable epialleles or genes that under environmental deflection with age could be prioritized for initial screening. Furthermore, environmentally associated epigenetic changes need to be related to long-term health in order to delineate whether changes are on the causal pathway to toxicity and disease or simply biomarkers of past exposure without phenotypic implications. Similarly, there is the need to acquire knowledge of persistence, duration of persistence, whether altered epigenome status can be revertible to prior pre-exposed status, and temporary versus long-term health or other manifestations of persistent versus revertible effects. Special attention should be paid to the evaluation of multigenerational and transgenerational effects of toxicant exposures. Collectively, this information needs to be incorporated into risk assessment and policy. With respect to epigenetic effects robustly associated with environmental exposures and health outcomes, epigenome-editing interventions could be developed to mitigate health risk.
Acknowledgments
Grant sponsor: University of Michigan (UM) NIEHS/EPA Children’s Environmental Health and Disease Prevention Center; Grant number: P01 ES022844/RD83543601.
Grant sponsor: the TaRGETII Consortium; Grant number: ES026697.
Grant sponsor: the NIH Director’s Transformative Award; Grant number: ES026877.
Grant sponsor: the Michigan Lifestage Environmental Exposures and Disease (M-LEEaD) NIEHS Core Center; Grant number: P30 ES017885.
Grant sponsor: UM Institutional Training Grant; Grant number: T32 ES007062.
Grant sponsor: NIEHS Pathways to Independence Award; Grant number: R00 ES022221.
Footnotes
CONFLICTS OF INTEREST
The authors declare that there are no conflicts of interest.
REFERENCES
- Aagaard L, Rossi JJ. 2007. RNAi therapeutics: Principles, prospects and challenges. Adv Drug Deliv Rev 59(2–3):75–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Accordini S, Calciano L, Johannessen A, Portas L, Benediktsdottir B, Bertelsen RJ, Braback L, Carsin AE, Dharmage SC, Dratva J, et al. 2018. A three-generation study on the association of tobacco smoking with asthma. Int J Epidemiol 47(4):1106–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alavian-Ghavanini A, Lin PI, Lind PM, Risen Rimfors S, Halin Lejonklou M, Dunder L, Tang M, Lindh C, Bornehag CG, Ruegg J. 2018. Prenatal bisphenol A exposure is linked to epigenetic changes in glutamate receptor subunit gene Grin2b in female rats and humans. Sci Rep 8(1):11315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aliabadi HM, Mahdipoor P, Bisoffi M, Hugh JC, Uludag H. 2016. Single and combinational siRNA therapy of cancer cells: Probing changes in targeted and nontargeted mediators after siRNA treatment. Mol Pharm 13(12):4116–4128. [DOI] [PubMed] [Google Scholar]
- Alvarado-Cruz I, Sanchez-Guerra M, Hernandez-Cadena L, De Vizcaya-Ruiz A, Mugica V, Pelallo-Martinez NA, Solis-Heredia MJ, Byun HM, Baccarelli A, Quintanilla-Vega B. 2017. Increased methylation of repetitive elements and DNA repair genes is associated with higher DNA oxidation in children in an urbanized, industrial environment. Mutat Res 813:27–36. [DOI] [PubMed] [Google Scholar]
- Amarasekera M, Martino D, Ashley S, Harb H, Kesper D, Strickland D, Saffery R, Prescott SL. 2014. Genome-wide DNA methylation profiling identifies a folate-sensitive region of differential methylation upstream of ZFP57-imprinting regulator in humans. FASEB J 28 (9):4068–4076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson OS, Kim JH, Peterson KE, Sanchez BN, Sant KE, Sartor MA, Weinhouse C, Dolinoy DC. 2016. Novel epigenetic biomarkers mediating bisphenol A exposure and metabolic phenotypes in female mice. Endocrinology 158(1):31–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angrish MM, Allard P, McCullough SD, Druwe IL, Helbling Chadwick L, Hines E, Chorley BN. 2018. Epigenetic applications in adverse outcome pathways and environmental risk evaluation. Environ Health Perspect 126(4):045001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anway MD, Cupp AS, Uzumcu M, Skinner MK. 2005. Epigenetic transgenerational actions of endocrine disruptors and male fertility. Science 308(5727):1466–1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baccarelli A, Wright RO, Bollati V, Tarantini L, Litonjua AA, Suh HH, Zanobetti A, Sparrow D, Vokonas PS, Schwartz J. 2009. Rapid DNA methylation changes after exposure to traffic particles. Am J Respir Crit Care Med 179(7):572–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baccarelli A, Tarantini L, Wright RO, Bollati V, Litonjua AA, Zanobetti A, Sparrow D, Vokonas PS, Schwartz J. 2010. Repetitive element DNA methylation and circulating endothelial and inflammation markers in the VA normative aging study. Epigenetics 5(3):222–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahn JH, Zhang Q, Li F, Chan TM, Lin X, Kim Y, Wong DT, Xiao X. 2015. The landscape of microRNA, Piwi-interacting RNA, and circular RNA in human saliva. Clin Chem 61(1):221–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bansal A, Rashid C, Xin F, Li C, Polyak E, Duemler A, van der Meer T, Stefaniak M, Wajid S, Doliba N, Bartolomei MS, Simmons RA. 2017. Sex- and dose-specific effects of maternal bisphenol A exposure on pancreatic islets of first- and second-generation adult mice offspring. Environ Health Perspect 125(9):\. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao W, Kojima KK, Kohany O. 2015. Repbase update, a database of repetitive elements in eukaryotic genomes. Mob DNA 6:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbot W, Dupressoir A, Lazar V, Heidmann T. 2002. Epigenetic regulation of an IAP retrotransposon in the aging mouse: Progressive demethylation and de-silencing of the element by its repetitive induction. Nucleic Acids Res 30(11):2365–2373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barouki R, Melen E, Herceg Z, Beckers J, Chen J, Karagas M, Puga A, Xia Y, Chadwick L, Yan W, et al. 2018. Epigenetics as a mechanism linking developmental exposures to long-term toxicity. Environ Int 114:77–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basil P, Li Q, Dempster EL, Mill J, Sham PC, Wong CC, McAlonan GM. 2014. Prenatal maternal immune activation causes epigenetic differences in adolescent mouse brain. Transl Psychiatry 4:e434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateson P, Barker D, Clutton-Brock T, Deb D, D’Udine B, Foley RA, Gluckman P, Godfrey K, Kirkwood T, Lahr MM, et al. 2004. Developmental plasticity and human health. Nature 430(6998):419–421. [DOI] [PubMed] [Google Scholar]
- Beck CR, Garcia-Perez JL, Badge RM, Moran JV. 2011. LINE-1 elements in structural variation and disease. Annu Rev Genomics Hum Genet 12:187–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Moshe S, Itzkovitz S. 2019. Spatial heterogeneity in the mammalian liver. Nat Rev Gastroenterol Hepatol. [DOI] [PubMed] [Google Scholar]
- Bernal AJ, Dolinoy DC, Huang D, Skaar DA, Weinhouse C, Jirtle RL. 2013. Adaptive radiation-induced epigenetic alterations mitigated by antioxidants. FASEB J 27(2):665–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein DL, Kameswaran V, Le Lay JE, Sheaffer KL, Kaestner KH. 2015. The BisPCR2 method for targeted bisulfite sequencing. Epigenetics Chromatin 8(1):27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhan A, Hussain I, Ansari KI, Bobzean SA, Perrotti LI, Mandal SS. 2014. Bisphenol-A and diethylstilbestrol exposure induces the expression of breast cancer associated long noncoding RNA HOTAIR in vitro and in vivo. J Steroid Biochem Mol Biol 141:160–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi H, Zhou J, Wu D, Gao W, Li L, Yu L, Liu F, Huang M, Adcock IM, Barnes PJ, et al. 2015. Microarray analysis of long non-coding RNAs in COPD lung tissue. Inflamm Res 64(2):119–126. [DOI] [PubMed] [Google Scholar]
- Boch J, Scholze H, Schornack S, Landgraf A, Hahn S, Kay S, Lahaye T, Nickstadt A, Bonas U. 2009. Breaking the code of DNA binding specificity of TAL-type III effectors. Science 326(5959):1509–1512. [DOI] [PubMed] [Google Scholar]
- Bock C, Tomazou EM, Brinkman AB, Muller F, Simmer F, Gu H, Jager N, Gnirke A, Stunnenberg HG, Meissner A. 2010. Quantitative comparison of genome-wide DNA methylation mapping technologies. Nat Biotechnol 28(10):1106–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boland CR. 2017. Erratum to: Non-coding RNA: It’s not junk. Dig Dis Sci 62(11):3260. [DOI] [PubMed] [Google Scholar]
- Breton CV, Yao J, Millstein J, Gao L, Siegmund KD, Mack W, Whitfield-Maxwell L, Lurmann F, Hodis H, Avol E, et al. 2016. Prenatal air pollution exposures, DNA methyl transferase genotypes, and associations with newborn LINE1 and Alu methylation and childhood blood pressure and carotid intima-media thickness in the children’s health study. Environ Health Perspect 124(12):1905–1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdge GC, Slater-Jefferies J, Torrens C, Phillips ES, Hanson MA, Lillycrop KA. 2007. Dietary protein restriction of pregnant rats in the F0 generation induces altered methylation of hepatic gene promoters in the adult male offspring in the F1 and F2 generations. Br J Nutr 97(3):435–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnett JC, Rossi JJ, Tiemann K. 2011. Current progress of siRNA/shRNA therapeutics in clinical trials. Biotechnol J 6(9):1130–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burris HH, Rifas-Shiman SL, Baccarelli A, Tarantini L, Boeke CE, Kleinman K, Litonjua AA, Rich-Edwards JW, Gillman MW. 2012. Associations of LINE-1 DNA methylation with preterm birth in a prospective cohort study. J Dev Orig Health Dis 3(3):173–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busato F, Dejeux E, El Abdalaoui H, Gut IG, Tost J. 2018. Quantitative DNA methylation analysis at single-nucleotide resolution by pyrosequencing(R). Methods Mol Biol 1708:427–445. [DOI] [PubMed] [Google Scholar]
- Camacho J, Truong L, Kurt Z, Chen YW, Morselli M, Gutierrez G, Pellegrini M, Yang X, Allard P. 2018. The memory of environmental chemical exposure in C. elegans is dependent on the Jumonji demethylases jmjd-2 and jmjd-3/utx-1. Cell Rep 23(8):2392–2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canapa A, Barucca M, Biscotti MA, Forconi M, Olmo E. 2015. Transposons, genome size, and evolutionary insights in animals. Cytogenet Genome Res 147(4):217–239. [DOI] [PubMed] [Google Scholar]
- Cano-Rodriguez D, Gjaltema RA, Jilderda LJ, Jellema P, Dokter-Fokkens J, Ruiters MH, Rots MG. 2016. Writing of H3K4Me3 overcomes epigenetic silencing in a sustained but context-dependent manner. Nat Commun 7:12284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardenas A, Koestler DC, Houseman EA, Jackson BP, Kile ML, Karagas MR, Marsit CJ. 2015. Differential DNA methylation in umbilical cord blood of infants exposed to mercury and arsenic in utero. Epigenetics 10(6):508–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrara M, Fuschi P, Ivan C, Martelli F. 2018. Circular RNAs: Methodological challenges and perspectives in cardiovascular diseases. J Cell Mol Med 22(11):5176–5187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvan MJ 3rd, Kalluvila TA, Klingler RH, Larson JK, Pickens M, Mora-Zamorano FX, Connaughton VP, Sadler-Riggleman I, Beck D, Skinner MK. 2017. Mercury-induced epigenetic transgenerational inheritance of abnormal neurobehavior is correlated with sperm epimutations in zebrafish. PLoS One 12(5):e0176155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalieri V, Spinelli G. 2017. Environmental epigenetics in zebrafish. Epigenetics Chromatin 10(1):46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalbatani GM, Dana H, Memari F, Gharagozlou E, Ashjaei S, Kheirandish P, Marmari V, Mahmoudzadeh H, Mozayani F, Maleki AR, et al. 2019. Biological function and molecular mechanism of piRNA in cancer. Pract Lab Med 13:e00113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Kazemier HG, de Groote ML, Ruiters MH, Xu GL, Rots MG. 2014. Induced DNA demethylation by targeting ten-eleven translocation 2 to the human ICAM-1 promoter. Nucleic Acids Res 42(3):1563–1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheow LF, Courtois ET, Tan Y, Viswanathan R, Xing Q, Tan RZ, Tan DS, Robson P, Loh YH, Quake SR, et al. 2016. Single-cell multimodal profiling reveals cellular epigenetic heterogeneity. Nat Methods 13(10):833–836. [DOI] [PubMed] [Google Scholar]
- Chimpanzee S, Analysis C. 2005. Initial sequence of the chimpanzee genome and comparison with the human genome. Nature 437 (7055):69–87. [DOI] [PubMed] [Google Scholar]
- Curtis SW, Cobb DO, Kilaru V, Terrell ML, Kennedy EM, Marder ME, Barr DB, Marsit CJ, Marcus M, Conneely KN, et al. 2019. Exposure to polybrominated biphenyl (PBB) associates with genomewide DNA methylation differences in peripheral blood. Epigenetics 14(1):52–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cusanovich DA, Hill AJ, Aghamirzaie D, Daza RM, Pliner HA, Berletch JB, Filippova GN, Huang X, Christiansen L, DeWitt WS, et al. 2018. A single-cell atlas of in vivo mammalian chromatin accessibility. Cell 174(5):1309–1324 e1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Felice B, Manfellotto F, Palumbo A, Troisi J, Zullo F, Di Carlo C, Di Spiezio Sardo A, De Stefano N, Ferbo U, Guida M, et al. 2015. Genome-wide microRNA expression profiling in placentas from pregnant women exposed to BPA. BMC Med Genomics 8:56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deininger P 2011. Alu elements: Know the SINEs. Genome Biol 12 (12):236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeSantis CE, Ma J, Goding Sauer A, Newman LA, Jemal A. 2017. Breast cancer statistics, 2017, racial disparity in mortality by state. CA Cancer J Clin 67(6):439–448. [DOI] [PubMed] [Google Scholar]
- Dhanoa JK, Sethi RS, Verma R, Arora JS, Mukhopadhyay CS. 2018. Long non-coding RNA: Its evolutionary relics and biological implications in mammals: A review. J Anim Sci Technol 60:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding M, Yuan C, Gaskins AJ, Field AE, Missmer SA, Michels KB, Hu F, Zhang C, Gillman MW, Chavarro J. 2017. Smoking during pregnancy in relation to grandchild birth weight and BMI trajectories. PLoS One 12(7):e0179368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolinoy DC, Weidman JR, Waterland RA, Jirtle RL. 2006. Maternal genistein alters coat color and protects Avy mouse offspring from obesity by modifying the fetal epigenome. Environ Health Perspect 114(4):567–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolinoy DC, Huang D, Jirtle RL. 2007. Maternal nutrient supplementation counteracts bisphenol A-induced DNA hypomethylation in early development. Proc Natl Acad Sci U S A 104(32):13056–13061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez-Salas P, Moore SE, Baker MS, Bergen AW, Cox SE, Dyer RA, Fulford AJ, Guan Y, Laritsky E, Silver MJ, et al. 2014. Maternal nutrition at conception modulates DNA methylation of human metastable epialleles. Nat Commun 5:3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupressoir A, Lavialle C, Heidmann T. 2012. From ancestral infectious retroviruses to bona fide cellular genes: Role of the captured syncytins in placentation. Placenta 33(9):663–671. [DOI] [PubMed] [Google Scholar]
- Eads CA, Danenberg KD, Kawakami K, Saltz LB, Blake C, Shibata D, Danenberg PV, Laird PW. 2000. MethyLight: A high-throughput assay to measure DNA methylation. Nucleic Acids Res 28(8):e32–e32, 320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich S, Lambers D, Baccarelli A, Khoury J, Macaluso M, Ho SM. 2016. Endocrine disruptors: A potential risk factor for gestational diabetes mellitus. Am J Perinatol 33(13):1313–1318. [DOI] [PubMed] [Google Scholar]
- Ekram MB, Kang K, Kim H, Kim J. 2012. Retrotransposons as a major source of epigenetic variations in the mammalian genome. Epigenetics 7(4):370–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estecio MR, Gharibyan V, Shen L, Ibrahim AE, Doshi K, He R, Jelinek J, Yang AS, Yan PS, Huang TH, et al. 2007. LINE-1 hypomethylation in cancer is highly variable and inversely correlated with microsatellite instability. PLoS One 2(5):e399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falahi F, Huisman C, Kazemier HG, van der Vlies P, Kok K, Hospers GA, Rots MG. 2013. Towards sustained silencing of HER2/neu in cancer by epigenetic editing. Mol Cancer Res 11(9):1029–1039. [DOI] [PubMed] [Google Scholar]
- Falahi F, Sgro A, Blancafort P. 2015. Epigenome engineering in cancer: Fairytale or a realistic path to the clinic? Front Oncol 5:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faulk C, Dolinoy DC. 2011. Timing is everything: The when and how of environmentally induced changes in the epigenome of animals. Epigenetics 6(7):791–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faulk C, Barks A, Dolinoy DC. 2013a. Phylogenetic and DNA methylation analysis reveal novel regions of variable methylation in the mouse IAP class of transposons. BMC Genomics 14:48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faulk C, Barks A, Liu K, Goodrich JM, Dolinoy DC. 2013b. Early-life lead exposure results in dose- and sex-specific effects on weight and epigenetic gene regulation in weanling mice. Epigenomics 5(5):487–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faulk C, Liu K, Barks A, Goodrich JM, Dolinoy DC. 2014. Longitudinal epigenetic drift in mice perinatally exposed to lead. Epigenetics 9(7):934–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faulk C, Kim JH, Anderson OS, Nahar MS, Jones TR, Sartor MA, Dolinoy DC. 2016. Detection of differential DNA methylation in repetitive DNA of mice and humans perinatally exposed to bisphenol A. Epigenetics 11(7):489–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraga MF, Ballestar E, Paz MF, Ropero S, Setien F, Ballestar ML, Heine-Suner D, Cigudosa JC, Urioste M, Benitez J, et al. 2005. Epigenetic differences arise during the lifetime of monozygotic twins. Proc Natl Acad Sci U S A 102(30):10604–10609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu A, Jacobs DI, Zhu Y. 2014. Epigenome-wide analysis of piRNAs in gene-specific DNA methylation. RNA Biol 11(10):1301–1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrett-Bakelman FE, Sheridan CK, Kacmarczyk TJ, Ishii J, Betel D, Alonso A, Mason CE, Figueroa ME, Melnick AM. 2015. Enhanced reduced representation bisulfite sequencing for assessment of DNA methylation at base pair resolution. J Vis Exp (96):e52246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasiunas G, Barrangou R, Horvath P, Siksnys V. 2012. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc Natl Acad Sci U S A 109(39):E2579–E2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh M, Oner D, Poels K, Tabish AM, Vlaanderen J, Pronk A, Kuijpers E, Lan Q, Vermeulen R, Bekaert B, et al. 2017. Changes in DNA methylation induced by multi-walled carbon nanotube exposure in the workplace. Nanotoxicology 11(9–10):1195–1210. [DOI] [PubMed] [Google Scholar]
- Gilbert KM, Nelson AR, Cooney CA, Reisfeld B, Blossom SJ. 2012. Epigenetic alterations may regulate temporary reversal of CD4(+) T cell activation caused by trichloroethylene exposure. Toxicol Sci 127(1):169–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert LA, Larson MH, Morsut L, Liu Z, Brar GA, Torres SE, Stern-Ginossar N, Brandman O, Whitehead EH, Doudna JA, et al. 2013. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell 154(2):442–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillette TG, Hill JA. 2015. Readers, writers, and erasers: Chromatin as the whiteboard of heart disease. Circ Res 116(7):1245–1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gojobori T, Li WH, Graur D. 1982. Patterns of nucleotide substitution in pseudogenes and functional genes. J Mol Evol 18(5):360–369. [DOI] [PubMed] [Google Scholar]
- Golding J, Ellis G, Gregory S, Birmingham K, Iles-Caven Y, Rai D, Pembrey M. 2017. Grand-maternal smoking in pregnancy and grandchild’s autistic traits and diagnosed autism. Sci Rep 7:46179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Nahm S, Mendez M, Robinson W, Murphy SK, Hoyo C, Hogan V, Rowley D. 2017. Low maternal adherence to a Mediterranean diet is associated with increase in methylation at the MEG3-IG differentially methylated region in female infants. Environ Epigenet 3(2):dvx007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodrich JM, Sanchez BN, Dolinoy DC, Zhang Z, Hernandez-Avila M, Hu H, Peterson KE, Tellez-Rojo MM. 2015. Quality control and statistical modeling for environmental epigenetics: A study on in utero lead exposure and DNA methylation at birth. Epigenetics 10 (1):19–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham G 2015. Disparities in cardiovascular disease risk in the United States. Curr Cardiol Rev 11(3):238–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greathouse KL, Bredfeldt T, Everitt JI, Lin K, Berry T, Kannan K, Mittelstadt ML, Ho SM, Walker CL. 2012. Environmental estrogens differentially engage the histone methyltransferase EZH2 to increase risk of uterine tumorigenesis. Mol Cancer Res 10(4):546–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer EL, Maures TJ, Ucar D, Hauswirth AG, Mancini E, Lim JP, Benayoun BA, Shi Y, Brunet A. 2011. Transgenerational epigenetic inheritance of longevity in Caenorhabditis elegans. Nature 479:365–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groner AC, Meylan S, Ciuffi A, Zangger N, Ambrosini G, Denervaud N, Bucher P, Trono D. 2010. KRAB-zinc finger proteins and KAP1 can mediate long-range transcriptional repression through heterochromatin spreading. PLoS Genet 6(3):e1000869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunau C, Clark SJ, Rosenthal A. 2001. Bisulfite genomic sequencing: Systematic investigation of critical experimental parameters. Nucleic Acids Res 29(13):E65–E65, 665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gryder BE, Sodji QH, Oyelere AK. 2012. Targeted cancer therapy: Giving histone deacetylase inhibitors all they need to succeed. Future Med Chem 4(4):505–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Z, Jin K, Crabbe MJC, Zhang Y, Liu X, Huang Y, Hua M, Nan P, Zhang Z, Zhong Y. 2016. Enrichment analysis of Alu elements with different spatial chromatin proximity in the human genome. Protein Cell 7(4):250–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han YN, Li Y, Xia SQ, Zhang YY, Zheng JH, Li W. 2017a. PIWI proteins and PIWI-interacting RNA: Emerging roles in cancer. Cell Physiol Biochem 44(1):1–20. [DOI] [PubMed] [Google Scholar]
- Han YN, Xia SQ, Zhang YY, Zheng JH, Li W. 2017b. Circular RNAs: A novel type of biomarker and genetic tools in cancer. Oncotarget 8(38):64551–64563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heijmans BT, Tobi EW, Stein AD, Putter H, Blauw GJ, Susser ES, Slagboom PE, Lumey LH. 2008. Persistent epigenetic differences associated with prenatal exposure to famine in humans. Proc Natl Acad SciUSA 105(44):17046–17049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilton IBD’ Ippolito AM, Vockley CM, Thakore PI, Crawford GE, Reddy TE, Gersbach CA. 2015. Epigenome editing by a CRISPR-Cas9-based acetyltransferase activates genes from promoters and enhancers. Nat Biotechnol 33(5):510–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyo C, Daltveit AK, Iversen E, Benjamin-Neelon SE, Fuemmeler B, Schildkraut J, Murtha AP, Overcash F, Vidal AC, Wang F, et al. 2014. Erythrocyte folate concentrations, CpG methylation at genomically imprinted domains, and birth weight in a multiethnic newborn cohort. Epigenetics 9(8):1120–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang CR, Burns KH, Boeke JD. 2012a. Active transposition in genomes. Annu Rev Genet 46:651–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang RC, Galati JC, Burrows S, Beilin LJ, Li X, Pennell CE, van Eekelen J, Mori TA, Adams LA, Craig JM. 2012b. DNA methylation of the IGF2/H19 imprinting control region and adiposity distribution in young adults. Clin Epigenetics 4(1):21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung PH, Van Winkle LS, Williams CJ, Hunt PA, VandeVoort CA. 2019. Prenatal bisphenol A exposure alters epithelial cell composition in the rhesus macaque fetal oviduct. Toxicol Sci 167(2):450–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. 2012. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337(6096):816–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joubert BR, Felix JF, Yousefi P, Bakulski KM, Just AC, Breton C, Reese SE, Markunas CA, Richmond RC, Xu CJ, et al. 2016. DNA methylation in newborns and maternal smoking in pregnancy: Genome-wide consortium meta-analysis. Am J Hum Genet 98(4):680–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junge KM, Leppert B, Jahreis S, Wissenbach DK, Feltens R, Grutzmann K, Thurmann L, Bauer T, Ishaque N, Schick M, et al. 2018. MEST mediates the impact of prenatal bisphenol A exposure on long-term body weight development. Clin Epigenetics 10:58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaminen-Ahola N, Ahola A, Maga M, Mallitt KA, Fahey P, Cox TC, Whitelaw E, Chong S. 2010. Maternal ethanol consumption alters the epigenotype and the phenotype of offspring in a mouse model. PLoS Genet 6(1):e1000811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kantor B, Tagliafierro L, Gu J, Zamora ME, Ilich E, Grenier C, Huang ZY, Murphy S, Chiba-Falek O. 2018. Downregulation of SNCA expression by targeted editing of DNA methylation: A potential strategy for precision therapy in PD. Mol Ther 26:2638–2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsson O, Baccarelli AA. 2016. Environmental health and long noncoding RNAs. Curr Environ Health Rep 3(3):178–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazachenka A, Bertozzi TM, Sjoberg-Herrera MK, Walker N, Gardner J, Gunning R, Pahita E, Adams S, Adams D, Ferguson-Smith AC. 2018. Identification, characterization, and heritability of murine metastable epialleles: Implications for non-genetic inheritance. Cell 175(5):1259–1271.e1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KY, Kim DS, Lee SK, Lee IK, Kang JH, Chang YS, Jacobs DR, Steffes M, Lee DH. 2010. Association of low-dose exposure to persistent organic pollutants with global DNA hypomethylation in healthy Koreans. Environ Health Perspect 118(3):370–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochmanski J, Montrose L, Goodrich JM, Dolinoy DC. 2017. Environmental deflection: The impact of toxicant exposures on the aging epigenome. Toxicol Sci 156(2):325–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochmanski J, Marchlewicz EH, Cavalcante RG, Sartor MA, Dolinoy DC. 2018a. Age-related epigenome-wide DNA methylation and hydroxymethylation in longitudinal mouse blood. Epigenetics 13(7):779–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochmanski J, Marchlewicz EH, Dolinoy DC. 2018b. Longitudinal effects of developmental bisphenol A, variable diet, and physical activity on age-related methylation in blood. Environ Epigenet 4(3):dvy017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochmanski JJ, Marchlewicz EH, Cavalcante RG, Perera BPU, Sartor MA, Dolinoy DC. 2018c. Longitudinal effects of developmental bisphenol A exposure on epigenome-wide DNA hydroxymethylation at imprinted loci in mouse blood. Environ Health Perspect 126(7):077006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konermann S, Brigham MD, Trevino A, Hsu PD, Heidenreich M, Cong L, Platt RJ, Scott DA, Church GM, Zhang F. 2013. Optical control of mammalian endogenous transcription and epigenetic states. Nature 500(7463):472–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krauskopf J, Caiment F, van Veldhoven K, Chadeau-Hyam M, Sinharay R, Chung KF, Cullinan P, Collins P, Barratt B, Kelly FJ, et al. 2018. The human circulating miRNome reflects multiple organ disease risks in association with short-term exposure to traffic-related air pollution. Environ Int 113:26–34. [DOI] [PubMed] [Google Scholar]
- Kundakovic M 2017. Sex-specific epigenetics: Implications for environmental studies of brain and behavior. Curr Environ Health Rep 4 (4):385–391. [DOI] [PubMed] [Google Scholar]
- Kung JT, Colognori D, Lee JT. 2013. Long noncoding RNAs: Past, present, and future. Genetics 193(3):651–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaRocca J, Binder AM, McElrath TF, Michels KB. 2014. The impact of first trimester phthalate and phenol exposure on IGF2/H19 genomic imprinting and birth outcomes. Environ Res 133:396–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaRocca J, Binder AM, McElrath TF, Michels KB. 2016. First-trimester urine concentrations of phthalate metabolites and phenols and placenta miRNA expression in a cohort of U.S. women. Environ Health Perspect 124(3):380–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson HA, Cheverud JM, Wolf JB. 2013. Genomic imprinting and parent-of-origin effects on complex traits. Nat Rev Genet 14(9):609–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Kalia V, Perera F, Herbstman J, Li T, Nie J, Qu LR, Yu J, Tang D. 2017a. Prenatal airborne polycyclic aromatic hydrocarbon exposure, LINE1 methylation and child development in a Chinese cohort. Environ Int 99:315–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MH, Cho ER, Lim JE, Jee SH. 2017b. Association between serum persistent organic pollutants and DNA methylation in Korean adults. Environ Res 158:333–341. [DOI] [PubMed] [Google Scholar]
- Li F, Papworth M, Minczuk M, Rohde C, Zhang Y, Ragozin S, Jeltsch A. 2007. Chimeric DNA methyltransferases target DNA methylation to specific DNA sequences and repress expression of target genes. Nucleic Acids Res 35(1):100–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H 2007. piRNAs in the germ line. Science 316(5823):397. [DOI] [PubMed] [Google Scholar]
- Liu XS, Wu H, Ji X, Stelzer Y, Wu X, Czauderna S, Shu J, Dadon D, Young RA, Jaenisch R. 2016. Editing DNA methylation in the mammalian genome. Cell 167(1):233–247.e217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeder ML, Angstman JF, Richardson ME, Linder SJ, Cascio VM, Tsai SQ, Ho QH, Sander JD, Reyon D, Bernstein BE, et al. 2013. Targeted DNA demethylation and activation of endogenous genes using programmable TALE-TET1 fusion proteins. Nat Biotechnol 31(12):1137–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mani SR, Juliano CE. 2013. Untangling the web: The diverse functions of the PIWI/piRNA pathway. Mol Reprod Dev 80(8):632–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manikkam M, Guerrero-Bosagna C, Tracey R, Haque MM, Skinner MK. 2012. Transgenerational actions of environmental compounds on reproductive disease and identification of epigenetic biomarkers of ancestral exposures. PLoS One 7(2):e31901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manikkam M, Haque MM, Guerrero-Bosagna C, Nilsson EE, Skinner MK. 2014. Pesticide methoxychlor promotes the epigenetic transgenerational inheritance of adult-onset disease through the female germline. PLoS One 9(7):e102091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLain AT, Faulk C. 2018. The evolution of CpG density and lifespan in conserved primate and mammalian promoters. Aging 10(4):561–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaughlin RN Jr. 2018. Reading the tea leaves: Dead transposon copies reveal novel host and transposon biology. PLoS Biol 16(3):e2005470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendenhall EM, Williamson KE, Reyon D, Zou JY, Ram O, Joung JK, Bernstein BE. 2013. Locus-specific editing of histone modifications at endogenous enhancers. Nat Biotechnol 31(12):1133–1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micevic G, Theodosakis N, Bosenberg M. 2017. Aberrant DNA methylation in melanoma: Biomarker and therapeutic opportunities. Clin Epigenetics 9:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J, McLachlan AD, Klug A. 1985. Repetitive zinc-binding domains in the protein transcription factor IIIA from Xenopus oocytes. EMBO J 4(6):1609–1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miousse IR, Shao L, Chang J, Feng W, Wang Y, Allen AR, Turner J, Stewart B, Raber J, Zhou D, et al. 2014. Exposure to low-dose (56)Fe-ion radiation induces long-term epigenetic alterations in mouse bone marrow hematopoietic progenitor and stem cells. Radiat Res 182(1):92–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell E, Klein SL, Argyropoulos KV, Sharma A, Chan RB, Toth JG, Barboza L, Bavley C, Bortolozzi A, Chen Q, et al. 2016. Behavioural traits propagate across generations via segregated iterative-somatic and gametic epigenetic mechanisms. Nat Commun 7:11492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montrose L, Faulk C, Francis J, Dolinoy DC. 2017. Perinatal lead (Pb) exposure results in sex and tissue-dependent adult DNA methylation alterations in murine IAP transposons. Environ Mol Mutagen 58(8):540–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montrose L, Padmanabhan V, Goodrich JM, Domino SE, Treadwell MC, Meeker JD, Watkins DJ, Dolinoy DC. 2018. Maternal levels of endocrine disrupting chemicals in the first trimester of pregnancy are associated with infant cord blood DNA methylation. Epigenetics 13(3):301–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran S, Arribas C, Esteller M. 2016. Validation of a DNA methylation microarray for 850,000 CpG sites of the human genome enriched in enhancer sequences. Epigenomics 8(3):389–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moscou MJ, Bogdanove AJ. 2009. A simple cipher governs DNA recognition by TAL effectors. Science 326(5959):1501. [DOI] [PubMed] [Google Scholar]
- Mulligan CJ, D’Errico NC, Stees J, Hughes DA. 2012. Methylation changes at NR3C1 in newborns associate with maternal prenatal stress exposure and newborn birth weight. Epigenetics 7(8):853–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy SK, Adigun A, Huang Z, Overcash F, Wang F, Jirtle RL, Schildkraut JM, Murtha AP, Iversen ES, Hoyo C. 2012. Gender-specific methylation differences in relation to prenatal exposure to cigarette smoke. Gene 494(1):36–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narvaez DM, Groot H, Diaz SM, Palma RM, Munoz N, Cros MP, Hernandez-Vargas H. 2017. Oxidative stress and repetitive element methylation changes in artisanal gold miners occupationally exposed to mercury. Heliyon 3(9):e00400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neier K, Cheatham D, Bedrosian LD, Dolinoy DC. 2019. Perinatal exposures to phthalates and phthalate mixtures result in sex-specific effects on body weight, organ weights and intracisternal A-particle (IAP) DNA methylation in weanling mice. J Dev Orig Health Dis 10(2):176–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemudryi AA, Valetdinova KR, Medvedev SP, Zakian SM. 2014. TALEN and CRISPR/Cas genome editing systems: Tools of discovery. Acta Nat 6(3):19–40. [PMC free article] [PubMed] [Google Scholar]
- Newman MR, Sykes PJ, Blyth BJ, Bezak E, Lawrence MD, Morel KL, Ormsby RJ. 2014. A single whole-body low dose X-irradiation does not affect L1, B1 and IAP repeat element DNA methylation longitudinally. PLoS One 9(3):e93016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson E, Klukovich R, Sadler-Riggleman I, Beck D, Xie Y, Yan W, Skinner MK. 2018. Environmental toxicant induced epigenetic transgenerational inheritance of ovarian pathology and granulosa cell epigenome and transcriptome alterations: Ancestral origins of polycystic ovarian syndrome and primary ovarian insufiency. Epigenetics 13(8):875–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Non AL, Binder AM, Kubzansky LD, Michels KB. 2014. Genome-wide DNA methylation in neonates exposed to maternal depression, anxiety, or SSRI medication during pregnancy. Epigenetics 9(7):964–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perera BPU, Ghimire S, Kim J. 2018. Circular RNA identified from Peg3 and Igf2r. PLoS One 13(9):e0203850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perera BPU, Tsai ZT, Colwell ML, Jones TR, Goodrich JM, Wang K, Sartor MA, Faulk C, Dolinoy DC. 2019. Somatic expression of piRNA and associated machinery in the mouse identifies short, tissue-specific piRNA. Epigenetics 14(5):504–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platt RN 2nd, Vandewege MW, Ray DA. 2018. Mammalian transposable elements and their impacts on genome evolution. Chromosome Res 26(1–2):25–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price EM, Cotton AM, Penaherrera MS, McFadden DE, Kobor MS, Robinson W. 2012. Different measures of “genome-wide” DNA methylation exhibit unique properties in placental and somatic tissues. Epigenetics 7(6):652–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prins GS, Hu WY, Shi GB, Hu DP, Majumdar S, Li G, Huang K, Nelles JL, Ho SM, Walker CL, et al. 2014. Bisphenol A promotes human prostate stem-progenitor cell self-renewal and increases in vivo carcinogenesis in human prostate epithelium. Endocrinology 155(3):805–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu S, Zhong Y, Shang R, Zhang X, Song W, Kjems J, Li H. 2017. The emerging landscape of circular RNA in life processes. RNA Biol 14(8):992–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakyan VK, Blewitt ME, Druker R, Preis JI, Whitelaw E. 2002. Metastable epialleles in mammals. Trends Genet 18(7):348–351. [DOI] [PubMed] [Google Scholar]
- Rebollo R, Karimi MM, Bilenky M, Gagnier L, Miceli-Royer K, Zhang Y, Goyal P, Keane TM, Jones S, Hirst M, et al. 2011. Retrotransposon-induced heterochromatin spreading in the mouse revealed by insertional polymorphisms. PLoS Genet 7(9):e1002301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehan VK, Liu J, Naeem E, Tian J, Sakurai R, Kwong K, Akbari O, Torday JS. 2012. Perinatal nicotine exposure induces asthma in second generation offspring. BMC Med 10(1):129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano G, Veneziano D, Acunzo M, Croce CM. 2017. Small non-coding RNA and cancer. Carcinogenesis 38(5):485–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rots MG, Jeltsch A. 2018. Editing the Epigenome: Overview, open questions, and directions of future development. Methods Mol Biol 1767:3–18. [DOI] [PubMed] [Google Scholar]
- Roy S, Trautwein C, Luedde T, Roderburg C. 2018. A general overview on non-coding RNA-based diagnostic and therapeutic approaches for liver diseases. Front Pharmacol 9:805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusiecki JA, Baccarelli A, Bollati V, Tarantini L, Moore LE, Bonefeld-Jorgensen EC. 2008. Global DNA hypomethylation is associated with high serum-persistent organic pollutants in Greenlandic Inuit. Environ Health Perspect 116(11):1547–1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sai L, Li L, Hu C, Qu B, Guo Q, Jia Q, Zhang Y, Bo C, Li X, Shao H, et al. 2018. Identification of circular RNAs and their alterations involved in developing male Xenopus laevis chronically exposed to atrazine. Chemosphere 200:295–301. [DOI] [PubMed] [Google Scholar]
- Sales VM, Ferguson-Smith AC, Patti M- E. 2017. Epigenetic mechanisms of transmission of metabolic disease across generations. Cell Metab 25(3):559–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzman J 2016. Circular RNA expression: Its potential regulation and function. Trends Genet 32(5):309–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders AP, Burris HH, Just AC, Motta V, Amarasiriwardena C, Svensson K, Oken E, Solano-Gonzalez M, Mercado-Garcia A, Pantic I, et al. 2015. Altered miRNA expression in the cervix during pregnancy associated with lead and mercury exposure. Epigenomics 7(6):885–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimosuga KI, Fukuda K, Sasaki H, Ichiyanagi K. 2017. Locus-specific hypomethylation of the mouse IAP retrotransposon is associated with transcription factor-binding sites. Mob DNA 8:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddeek B, Inoubli L, Lakhdari N, Rachel PB, Fussell KC, Schneider S, Mauduit C, Benahmed M. 2014. MicroRNAs as potential biomarkers in diseases and toxicology. Mutat Res Genet Toxicol Environ Mutagen 764-765:46–57. [DOI] [PubMed] [Google Scholar]
- Siddique AN, Nunna S, Rajavelu A, Zhang Y, Jurkowska RZ, Reinhardt R, Rots MG, Ragozin S, Jurkowski TP, Jeltsch A. 2013. Targeted methylation and gene silencing of VEGF-A in human cells by using a designed Dnmt3a-Dnmt3L single-chain fusion protein with increased DNA methylation activity. J Mol Biol 425(3):479–491. [DOI] [PubMed] [Google Scholar]
- Silver MJ, Kessler NJ, Hennig BJ, Dominguez-Salas P, Laritsky E, Baker MS, Coarfa C, Hernandez-Vargas H, Castelino JM, Routledge MN, et al. 2015. Independent genomewide screens identify the tumor suppressor VTRNA2-1 as a human epiallele responsive to periconceptional environment. Genome Biol 16:118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skinner MK. 2015. Endocrine disruptors in 2015: Epigenetic transgenerational inheritance. Nat Rev Endocrinol 12(2):68–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skinner MK, Ben Maamar M, Sadler-Riggleman I, Beck D, Nilsson E, McBirney M, Klukovich R, Xie Y, Tang C, Yan W. 2018. Alterations in sperm DNA methylation, non-coding RNA and histone retention associate with DDT-induced epigenetic transgenerational inheritance of disease. Epigenetics Chromatin 11(1):8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song MA, Ernst T, Tiirikainen M, Tost J, Wilkens LR, Chang L, Kolonel LN, Le Marchand L, Lim U. 2018. Methylation of imprinted IGF2 regions is associated with total, visceral, and hepatic adiposity in postmenopausal women. Epigenetics 13(8):858–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staunstrup NH, Starnawska A, Nyegaard M, Christiansen L, Nielsen AL, Borglum A, Mors O. 2016. Genome-wide DNA methylation profiling with MeDIP-seq using archived dried blood spots. Clin Epigenetics 8:81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suchiman HED, Slieker RC, Kremer D, Slagboom PE, Heijmans BT, Tobi EW. 2015. Design, measurement and processing of region-specific DNA methylation assays: The mass spectrometry-based method EpiTYPER. Front Genet 6:287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Y, Liu L, Liao M, Zhang C, Hu S, Zou M, Gu M, Li X. 2015. Emerging roles for PIWI proteins in cancer. Acta Biochim Biophys Sin 47(5):315–324. [DOI] [PubMed] [Google Scholar]
- Tobi EW, Slagboom PE, van Dongen J, Kremer D, Stein AD, Putter H, Heijmans BT, Lumey LH. 2012. Prenatal famine and genetic variation are independently and additively associated with DNA methylation at regulatory loci within IGF2/H19. PLoS One 7(5):e37933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tommasi S, Zheng A, Yoon JI, Li AX, Wu X, Besaratinia A. 2012. Whole DNA methylome profiling in mice exposed to secondhand smoke. Epigenetics 7(11):1302–1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tosar JP, Rovira C, Cayota A. 2018. Non-coding RNA fragments account for the majority of annotated piRNAs expressed in somatic nongonadal tissues. Commun Biol 1(2):2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trevino LS, Katz TA. 2018. Endocrine disruptors and developmental origins of nonalcoholic fatty liver disease. Endocrinology 159(1):20–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Trevino LS, Wong RL, Medvedovic M, Chen J, Ho SM, Shen J, Foulds CE, Coarfa C, O’Malley BW, et al. 2016. Reprogramming of the epigenome by MLL1 links early-life environmental exposures to prostate cancer risk. Mol Endocrinol 30(8):856–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Pehrsson EC, Purushotham D, Li D, Zhuo X, Zhang B, Lawson HA, Province MA, Krapp C, Lan Y, et al. 2018. The NIEHS TaRGET II consortium and environmental epigenomics. Nat Biotechnol 36(3):225–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waryah CB, Moses C, Arooj M, Blancafort P. 2018. Zinc fingers, TALEs, and CRISPR systems: A comparison of tools for epigenome editing. Methods Mol Biol 1767:19–63. [DOI] [PubMed] [Google Scholar]
- Waterland RA, Jirtle RL. 2003. Transposable elements: Targets for early nutritional effects on epigenetic gene regulation. Mol Cell Biol 23(15):5293–5300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterland RA, Kellermayer R, Laritsky E, Rayco-Solon P, Harris RA, Travisano M, Zhang W, Torskaya MS, Zhang J, Shen L, et al. 2010. Season of conception in rural Gambia affects DNA methylation at putative human metastable epialleles. PLoS Genet 6(12):e1001252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendt J, Rosenbaum H, Richmond TA, Jeddeloh JA, Burgess DL. 2018. Targeted bisulfite sequencing using the SeqCap Epi enrichment system. Methods Mol Biol 1708:383–405. [DOI] [PubMed] [Google Scholar]
- Winterbottom EF, Moroishi Y, Halchenko Y, Armstrong DA, Beach PJ, Nguyen QP, Capobianco AJ, Ayad NG, Marsit CJ, Li Z, et al. 2019. Prenatal arsenic exposure alters the placental expression of multiple epigenetic regulators in a sex-dependent manner. Environ Health 18(1):18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Hivert MF, Cardenas A, Zhong J, Rifas-Shiman SL, Agha G, Colicino E, Just AC, Amarasiriwardena C, Lin X, et al. 2017. Exposure to low levels of lead in utero and umbilical cord blood DNA methylation in project viva: An epigenome-wide association study. Environ Health Perspect 125(8):087019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie N, Zhou Y, Sun Q, Tang B. 2018. Novel epigenetic techniques provided by the CRISPR/Cas9 system. Stem Cells Int 2018:7834175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu GL, Bestor TH. 1997. Cytosine methylation targetted to predetermined sequences. Nat Genet 17(4):376–378. [DOI] [PubMed] [Google Scholar]
- Yang AS, Estecio MR, Doshi K, Kondo Y, Tajara EH, Issa JP. 2004. A simple method for estimating global DNA methylation using bisulfite PCR of repetitive DNA elements. Nucleic Acids Res 32(3):e38–e338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang AS, Doshi KD, Choi SW, Mason JB, Mannari RK, Gharybian V, Luna R, Rashid A, Shen L, Estecio MR, et al. 2006. DNA methylation changes after 5-aza-2′-deoxycytidine therapy in patients with leukemia. Cancer Res 66(10):5495–5503. [DOI] [PubMed] [Google Scholar]
- Yang X, Lay F, Han H, Jones PA. 2010. Targeting DNA methylation for epigenetic therapy. Trends Pharmacol Sci 31(11):536–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Cheng Y, Lu Q, Wei J, Yang H, Gu M. 2015. Detection of stably expressed piRNAs in human blood. Int J Clin Exp Med 8(8):13353–13358. [PMC free article] [PubMed] [Google Scholar]
- Yu AM, Jian C, Yu AH, Tu MJ. 2018. RNA therapy: Are we using the right molecules. Pharmacol Ther 196:91–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Maksakova IA, Gagnier L, van de Lagemaat LN, Mager DL. 2008. Genome-wide assessments reveal extremely high levels of polymorphism of two active families of mouse endogenous retroviral elements. PLoS Genet 4(2):e1000007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Rohde C, Tierling S, Stamerjohanns H, Reinhardt R, Walter J, Jeltsch A. 2009. DNA methylation analysis by bisulfite conversion, cloning, and sequencing of individual clones In: Tost J, editor. DNA Methylation: Methods and Protocols. Totowa, NJ: Humana Press; pp. 177–187. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Yang T, Xiao J. 2018. Circular RNAs: Promising biomarkers for human diseases. EBioMedicine 34:267–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Chen T. 2013. Tet family of 5-methylcytosine dioxygenases in mammalian development. J Hum Genet 58(7):421–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao MT, Whyte JJ, Hopkins GM, Kirk MD, Prather RS. 2014. Methylated DNA immunoprecipitation and high-throughput sequencing (MeDIP-seq) using low amounts of genomic DNA. Cell Reprogram 16(3):175–184. [DOI] [PubMed] [Google Scholar]
- Zhou Z, Liu H, Wang C, Lu Q, Huang Q, Zheng C, Lei Y. 2015. Long non-coding RNAs as novel expression signatures modulate DNA damage and repair in cadmium toxicology. Sci Rep 5:15293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziller MJ, Hansen KD, Meissner A, Aryee MJ. 2015. Coverage recommendations for methylation analysis by whole genome bisulfite sequencing. Nat Methods 12(3):230–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo LJ, Wang ZR, Tan YL, Chen XN, Luo XG. 2016. piRNAs and their functions in the brain. Int J Human Genet 16(1–2):53–60. [DOI] [PMC free article] [PubMed] [Google Scholar]


