Abstract
Objectives
Feline calicivirus (FCV) is a highly variable and globally important feline pathogen for which vaccination has been the mainstay of control. Here, we test whether the continued use of FCV-F9, one of the most frequently used vaccine strains globally, is driving the emergence of vaccine-resistant viruses in the field.
Methods
This study made use of two representative panels of field isolates previously collected from cats visiting randomly selected veterinary practices across the UK as part of separate cross-sectional studies from 2001 and 2013/2014. Phylogenetic analysis and in vitro virus neutralisation tests were used to compare the genetic and antigenic relationships between these populations and FCV-F9.
Results
Phylogenetic analysis showed a typically radial distribution dominated by 52 distinct strains, with strains from both 2001 and 2013/2014 intermingled. The sequence for FCV-F9 appeared to be integral to this phylogeny and there were no significant differences in the genetic distances within each studied population (intra-population distances), or between them (inter-population distances), or between each population and FCV-F9. A 1 in 8 dilution neutralised 97% and 100% of the 2001 and 2013/14 isolates, respectively, and a 1 in 16 dilution neutralised 87% and 75% of isolates, respectively. There was no significant difference either in variance between the FCV-F9 neutralising titres for the two populations, or in the distribution of neutralisation titres across the two populations.
Conclusions and relevance
Although FCV is a highly variable virus, we found no evidence for a progressive divergence of field virus from vaccine strain FCV-F9, either phylogenetically or antigenically, with FCV-F9 antisera remaining broadly and equally cross-reactive to two geographically representative and temporally separated FCV populations. We suggest this may be because the immunodominant region of the FCV capsid responsible for neutralisation may have structural constraints preventing its longer term progressive antigenic evolution.
Keywords: Feline calicivirus, neutralisation, phylogeny, vaccine
Introduction
Feline calicivirus (FCV) is a common pathogen of cats inducing acute oral and upper respiratory tract disease (URTD). 1 As an RNA virus, its genome is often inaccurately replicated, leading to high sequence variability and antigenic variation.2–8 Most commercially available live FCV vaccines are based on FCV-F9,9,10 a strain isolated in 1958 11 and selected for its broad in vitro cross-reactivity. 8 It has been suggested that the continued use of FCV-F9 in this variable genetic background may be driving the emergence of vaccine-resistant strains and that the efficacy of such vaccines may be reducing over time.12–14
We recently reported that FCV-F9 remains broadly cross-reactive to representative FCV isolates collected across Europe between 2013 and 2014. 15 Comparison of the data from this study with results from a previous study performed 12 years earlier 16 suggested that FCV-F9 antiserum neutralised the contemporary isolates as effectively as the earlier isolates. However, although the two studies were performed in a similar manner, the FCV-F9 antisera used for the neutralisation tests were raised in different cats and using different methodologies (infection vs vaccine overdose), making direct comparison of results difficult. Here, we tested the hypothesis that field isolates of FCV are evolving increased resistance to FCV-F9 vaccines by directly comparing the in vitro cross-reactivity of a single FCV-F9 antiserum with representative UK FCV isolates collected in 2001 and 2013/14, together with phylogenetic comparisons of the viruses in our sample populations.
Materials and methods
Viruses
Field isolates were previously collected from veterinary practices recruited randomly across the UK as part of two cross-sectional studies in 2001 and 2013/2014 (see Table 1 in the supplementary material).15,16 Virus stocks were prepared in feline embryo cells. 17 Field isolates were used at passage 4 or less. Vaccine virus (FCV-F9; provided by MSD Animal Health) was used at passage 2.
Phylogenetic analysis
Methods for nucleic acid isolation, reverse transcription, PCR amplification, purification and sequencing were as previously described, 18 leading to a final PCR product of 529 nucleotides of the immunodominant region of the FCV capsid gene, 19 corresponding to residues 6406–6934 of the FCV-F9 genome (Genbank M86379). The resulting sequences were aligned and pairwise p-distances and a neighbour-joining tree calculated (MEGA6).20–22 All ambiguous positions associated with the quasispecies nature of FCV were discounted for each sequence pair.
Virus neutralisation
Sixty field viruses were used in virus neutralisation (VN). Forty isolates from 2001 were used; seven (17.5%) were from cats showing acute disease (URTD and/or mouth ulcers), 30 (75%) were from healthy cats and three (7.5%) had no clinical data (see Table 1 in the supplementary material). Of 48 isolates collected in 2013/14, a subset of 20 was used in this study. To maintain the same approximate clinical ratio as for the 2001 isolates, they were similarly stratified and four (20%) isolates with acute disease and 16 (80%) from healthy cats were randomly selected for inclusion. Viruses from clinically normal cats were included as these were still likely to be virulent. 23
Plasma (hereafter ‘antiserum’) was collected and pooled from four specific-pathogen-free cats vaccinated subcutaneously with 10 commercial doses of Nobivac Tricat Trio (FCV-F9) at 8–9 weeks of age, and again 4 weeks later, as part of a vaccine safety study conducted by the funder; the use and limitations of such ‘high-titre’ antisera is justified elsewhere. 15 Blood samples were taken 3 weeks after the second vaccination. VN tests were performed using a constant virus, varying the antiserum method, as previously described.15,24 An estimated concentration of 100 50% tissue culture infective dose (TCID) of virus was used in each assay and viral back titration was used to ensure the viral titre fell into the accepted range of 32–320 TCID50. 25 Antibody titres were expressed as 50% end points. 26 An internal FCV-F9 homologous control was included in each experiment; individual tests were only considered valid if the neutralising titre for this control was within two-fold of the mean homologous control titre across all experiments. 27 Antibody units (AUs) were also calculated, with 1 AU being the highest dilution of FCV-F9 antiserum that neutralised 32–320 TCID50 of homologous virus in 50% of cultures, based on the average FCV-F9 titre for all experiments.27,28
Statistical analysis
Levene’s homogeneity of variance and Mann–Whitney tests were used to compare the variance and distribution of neutralisation titres across the two populations of isolates (2001 vs 2013/14) and between clinical and non-clinical isolates. Fisher’s exact test was used to compare the proportion of samples neutralised by different levels of AUs. Intra- and/or inter-population genetic distances were compared using the Mann–Whitney test.
Results
Sequencing and phylogenetic analysis
Partial capsids could not be amplified from five of the isolates used in viral neutralisation, probably owing to a mismatch of the primers used.18,29 Sequences for the remaining 55 isolates are available in GenBank (Accession Numbers KX257491–617 [previously published] 15 and MH674290-347) (see Table 2 in the supplementary material). Phylogenetic analysis of the 55 sequences (together with FCV-F9) showed a typically radial distribution dominated by 52 distinct strains (pairwise genetic distance >20%, bootstrap values <80%) 30 (Figure 1), with strains from both studies intermingled. Similarly, the sequence for FCV-F9 appeared integral to this phylogeny. This overall lack of apparent temporal clustering was supported by there being no significant differences in the range of genetic distances produced either within each studied population (intra-population distances for 2001 and 2013/14 isolates), or between them (inter-population distances for comparing 2001 and 2013/14 isolates), or between each population and FCV-F9 (P >0.05 for all comparisons) (see Figure 1 in the supplementary material). Only two clades (‘a’ and ‘b’) were evident, containing possible variants of individual strains (pairwise genetic distance <20%, bootstrap values >80%); clade b contained two isolates from Oxfordshire (isolated 13 years apart), suggesting possible local circulation of this strain over some years.
Figure 1.
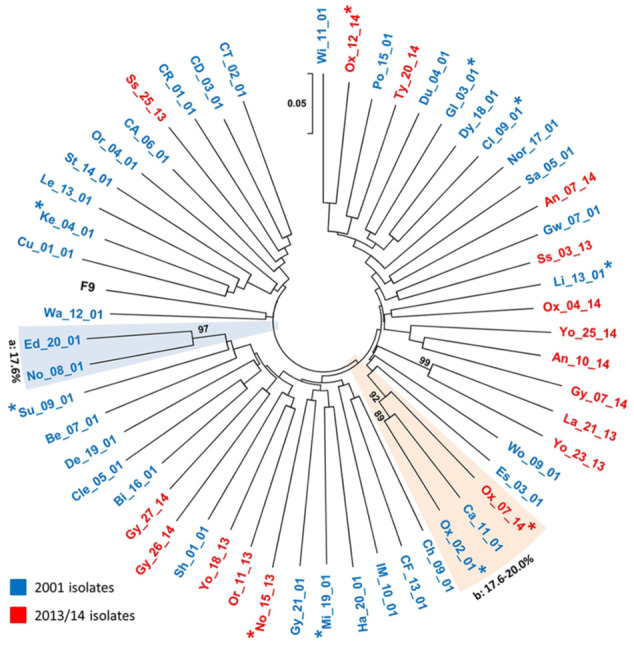
Unrooted neighbour-joining tree of the 56 partial feline calicivirus (FCV) capsid sequences used in this study (including FCV-F9; GenBank Accession Number M86379). The evolutionary distances were computed using the p-distance method, 20 and are in the units of the number of base differences per site (see 0.05 scale bar, which equates to five changes per 100 bases). All codon positions were included. There were 432 nucleotide positions in the final data set. The isolates are numbered as in Table 1 in the supplementary material, including a two-letter code for the geographical area, two digits for the isolate number and two digits for the year of collection; isolates from each of the two studies are also differentiated by colour (see key).
Asterisks represent isolates from cats with acute disease (upper respiratory tract disease ± ulcers). Clades represented by more than a single sequence (<20% capsid divergence, ⩾80% bootstrap values) are boxed, additionally labelled a and b, and the intra-clade diversity indicated. Bootstrap values <80% are not shown
VN
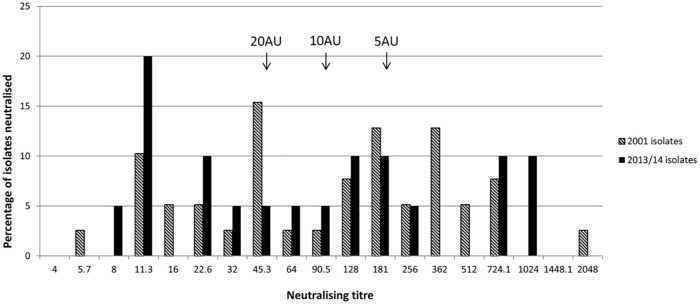
Neutralisation profiles were obtained for 59 of the 60 isolates (see Table 1 and Figure 2 in the supplementary material); one isolate repeatedly failed internal experimental controls. All 59 isolates were neutralised by antiserum at a 1 in 4 dilution. A 1 in 8 dilution neutralised 97% and 100% of the 2001 and 2013/14 isolates, respectively, and a 1 in 16 dilution neutralised 87% and 75% of isolates, respectively. There was no significant difference in variance between the titres for the two populations (2001 vs 2013/14, P = 0.97) and no significant difference in the distribution of titres across these two temporally separated populations (P = 0.46).
Unfortunately, prevaccination negative control plasma was not available to test for non-specific neutralisation, which may sometimes be seen at lower plasma dilutions. However, we observed a wide range of neutralisation titres, most often at high dilutions where non-specific neutralisation would not be expected to occur, suggesting the neutralisation we report is mediated by FCV-specific antibodies. In addition, when isolates that were neutralised at low dilutions (<1 in 8 and <1 in 16), where non-specific neutralisation might occur, were removed from the statistical analysis, there remained no significant difference in the distribution of titres across the two temporally separated populations.
There was no significant difference in the distribution of titres between isolates from clinically affected and clinically normal cats (P = 0.36). The percentage of isolates neutralised by 5, 10 and 20 AUs were 36%, 59% and 64%, respectively, for the 2001 isolates, and 25%, 45% and 55%, respectively, for the 2013/14 isolates (Figure 2). These differences were not statistically significant (P = 0.16).
Figure 2.
Results of virus neutralisation. The percentage of isolates neutralised by each dilution of FCV-F9 antisera in the two-fold dilution series (or by the midpoint between two-fold dilutions as calculated by the Reed–Muench equation) 26 are shown. The results for five isolates fell outside of these groupings (see Table 1 in the supplementary material); in which case the result was rounded down to the nearest grouping. Calculated levels of 5, 10 and 20 antibody units (AU; titres of 1 in 200, 1 in 100 and 1 in 50, respectively) are indicated
Discussion
Some authors have suggested that vaccines containing older FCV strains may be less relevant for the control of current field viruses.12–14 Here, we tested this hypothesis by directly comparing the ability of FCV-F9 antiserum to neutralise two representative UK populations of FCV isolated 12–13 years apart. Our results showed that FCV-F9 antiserum induced broad cross-neutralisation in vitro, showing no significant difference in the neutralisation of either population. These observations would suggest that FCV-F9 antiserum is not becoming less cross-reactive, and are consistent with a challenge experiment in which cats vaccinated with FCV-F9 were broadly protected when challenged with a recent field isolate.31,32 The differences between our results and those of others are likely attributable to variations in methodology, including the use of non-representative isolates. 12 In addition, the majority of earlier studies have not directly compared temporally separated, spatially representative isolates using the same antisera.
How can this apparent antigenic stability be reconciled with both rapid evolution of FCV in acute infection, 6 and the high levels of FCV genetic variability observed, particularly in colonies of cats?29,33 Consistent with previous studies, our phylogenetic analysis highlighted a radial phylogeny containing many strains.3,4,29,34 However, we observed no phylogenetic clustering, with sequences from both time points intermingled in the phylogeny, to which FCV-F9 remained integral.4,29,34,35 Similarly, distance comparisons between both sets of isolates and FCV-F9 showed neither population of field isolates was significantly more variable to FCV-F9 than to itself. These observations suggest that the capsid of FCV is not evolving in a linear (molecular clock-like) fashion, as is typical for some other rapidly evolving viruses such as influenza.15,36 These genetic observations correlate with our neutralisation data, being linked through our use of sequences from the immunodominant region of the FCV capsid, where most known neutralisation epitopes reside.19,37 Our working hypothesis is that while this region can evolve quickly in response to immune selection in individual animals,6,38 structural constraints on the capsid may prevent its continued evolution and broader antigenic escape at the population level. 23 To further understand these observations it will be necessary to expand these analyses to include full FCV genome sequences and older isolates collected closer in time to when FCV-F9 was isolated around 1958.
Using 20 AUs as a cut-off, we observed neutralisation of 64% and 55% of 2001 and 2013/14 isolates, respectively. Older UK studies reported 54% 39 and 74% 27 of field isolates neutralised by FCV-F9 antiserum at the 20 AUs cut-off. Our previous study, using the same antiserum as used here, but with a larger number of isolates, showed that 50% were neutralised by 20 AUs. 15 However, our 2001 study using different antiserum showed only 25% of isolates to be neutralised by 20 AUs, 16 highlighting how results can vary even when AUs are used to correct for differences in antiserum titres. Although AUs are undoubtedly useful for comparing results between studies, the method used here where isolates are directly compared within a single study should, perhaps, be preferable.
Although we failed to identify variation in neutralisation between the two studied time points, a larger study would undoubtedly have greater power to identify smaller changes in neutralisation. It is clear that, as previously suggested, 15 the development of an internationally agreed study protocol, as exists for some other viral vaccines, is necessary to facilitate more comparable studies in the future. Such a protocol would need to include optimal sample size, the selection of cats for sampling and source of control and test antisera.
Conclusions
Our studies demonstrate that although FCV field strains are very variable, they do not appear to be evolving from the FCV-F9 vaccine strain, either antigenically or genetically. Although our in vitro VN study cannot equate with clinical protection, together with the phylogenetic data, it may suggest a mechanism for why FCV-F9 antisera still remain broadly cross-reactive in vitro against FCV field isolates.
Supplemental Material
Evolutionary divergence between sequences
Isolate details and results of virus neutralisation using FCV-F9 antisera
Further isolate details and GenBank accession numbers
Footnotes
Accepted: 1 July 2019
Supplementary material: The following files are available online:
Figure 1: Evolutionary divergence between sequences.
Table 1: Isolate details and results of virus neutralisation using FCV-F9 antisera.
Table 2: Further isolate details and GenBank accession numbers.
This project was funded by MSD-Animal Health, who markets a live attenuated FCV vaccine containing the FCV-F9 virus. The funder was not involved in the study design, collection, analysis or interpretation of data, or in the decision to submit the manuscript for publication.
Funding: This work was supported by MSD-Animal Health (University of Liverpool grant number JXR 12796).
Ethical approval: This work involved the use of non-experimental animals only (owned or unowned) and followed established internationally recognised high standards (best practice) of individual veterinary clinical patient care. Ethical approval from a committee was not necessarily required.
Informed consent: Informed consent (either verbal or written) was obtained from the owner or legal guardian of all animals described in this work for the procedures undertaken. No animals or humans are identifiable within this publication, and therefore additional informed consent for publication was not required.
ORCID iDs: Shirley L Smith  https://orcid.org/0000-0003-1287-4277
https://orcid.org/0000-0003-1287-4277
Alan D Radford  https://orcid.org/0000-0002-4590-1334
https://orcid.org/0000-0002-4590-1334
References
- 1. Radford AD, Addie D, Belak Set al. Feline calicivirus infection. ABCD guidelines on prevention and management. J Feline Med Surg 2009; 11: 556–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Geissler K, Schneider K, Platzer Get al. Genetic and antigenic heterogeneity among feline calicivirus isolates from distinct disease manifestations. Virus Res 1997; 48: 193–206. [DOI] [PubMed] [Google Scholar]
- 3. Glenn M, Radford AD, Turner PCet al. Nucleotide sequence of UK and Australian isolates of feline calicivirus (FCV) and phylogenetic analysis of FCVs. Vet Microbiol 1999; 67: 175–193. [DOI] [PubMed] [Google Scholar]
- 4. Henzel A, Sa e, Silva M, Luo Set al. Genetic and phylogenetic analyses of capsid protein gene in feline calicivirus isolates from Rio Grande do Sul in southern Brazil. Virus Res 2012; 163: 667–671. [DOI] [PubMed] [Google Scholar]
- 5. Kreutz LC, Johnson RP, Seal BS. Phenotypic and genotypic variation of feline calicivirus during persistent infection of cats. Vet Microbiol 1998; 59: 229–236. [DOI] [PubMed] [Google Scholar]
- 6. Radford AD, Turner PC, Bennett Met al. Quasispecies evolution of a hypervariable region of the feline calicivirus capsid gene in cell culture and in persistently infected cats. J Gen Virol 1998; 79: 1–10. [DOI] [PubMed] [Google Scholar]
- 7. Seal BS. Analysis of capsid protein gene variation among divergent isolates of feline calicivirus. Virus Res 1994; 33: 39–53. [DOI] [PubMed] [Google Scholar]
- 8. Kalunda M, Lee KM, Holmes DFet al. Serologic classification of feline caliciviruses by plaque-reduction neutralization and immunodiffusion. Am J Vet Res 1975; 36: 353–356. [PubMed] [Google Scholar]
- 9. Kahn DE, Hoover EA. Feline caliciviral disease: experimental immunoprophylaxis. Am J Vet Res 1976; 37: 279–283. [PubMed] [Google Scholar]
- 10. Pedersen NC, Hawkins KF. Mechanisms for persistence of acute and chronic feline calicivirus infections in the face of vaccination. Vet Microbiol 1995; 47: 141–156. [DOI] [PubMed] [Google Scholar]
- 11. Bittle JL, York CJ, Newberne JWet al. Serologic relationship of new feline cytopathogenic viruses. Am J Vet Res 1960; 21: 547–550. [Google Scholar]
- 12. Addie D, Poulet H, Golder MCet al. Ability of antibodies to two new caliciviral vaccine strains to neutralise feline calicivirus isolates from the UK. Vet Rec 2008; 163: 355–357. [DOI] [PubMed] [Google Scholar]
- 13. Lauritzen A, Jarrett O, Sabara M. Serological analysis of feline calicivirus isolates from the United States and United Kingdom. Vet Microbiol 1997; 56: 55–63. [DOI] [PubMed] [Google Scholar]
- 14. Wensman JJ, Samman A, Lindhe Aet al. Ability of vaccine strain induced antibodies to neutralize field isolates of caliciviruses from Swedish cats. Acta Vet Scand 2015; 57: 86. DOI: 10.1186/513028-015-0178-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Afonso MM, Pinchbeck GL, Smith SLet al. A multi-national European cross-sectional study of feline calicivirus epidemiology, diversity and vaccine cross-reactivity. Vaccine 2017; 35: 2753–2760. [DOI] [PubMed] [Google Scholar]
- 16. Porter CJ, Radford AD, Gaskell RMet al. Comparison of the ability of feline calicivirus (FCV) vaccines to neutralise a panel of current UK FCV isolates. J Feline Med Surg 2008; 10: 32–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jarrett O, Laird HM, Crighton GWet al. Advances in feline leukemia. Bibl Haematol 1968; 30: 244–254. [DOI] [PubMed] [Google Scholar]
- 18. Hou J, Sanchez-Vizcaino F, McGahie Det al. European molecular epidemiology and strain diversity of feline calicivirus. Vet Rec 2016; 178: 114–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Radford AD, Willoughby K, Dawson Set al. The capsid gene of feline calicivirus contains linear B-cell epitopes in both variable and conserved regions. J Virol 1999; 73: 8496–8502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Nei M, Kumar S. Molecular evolution and phylogenetics. Oxford, New York: Oxford University Press, 2000. [Google Scholar]
- 21. Tamura K, Nei M, Kumar S. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc Natl Acad Sci U S A 2004; 101: 11030–11035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tamura K, Stecher G, Peterson Det al. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 2013; 30: 2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Povey RC, Hale CJ. Experimental infections with feline caliciviruses (picornaviruses) in specific-pathogen-free kittens. J Comp Pathol 1974; 84: 245–256. [DOI] [PubMed] [Google Scholar]
- 24. Dawson S, McArdle F, Bennett Det al. Investigation of vaccine reactions and breakdowns after feline calicivirus vaccination. Vet Rec 1993; 132: 346–350. [DOI] [PubMed] [Google Scholar]
- 25. Povey RC, Johnson RH. A standardized serum neutralization test for feline viral rhinotracheitis. II. The virus-serum system. J Comp Pathol 1969; 79: 387–392. [DOI] [PubMed] [Google Scholar]
- 26. Reed LJ, Muench H. A simple method of estimating fifty per cent endpoints. Am J Epidemiol 1938; 27: 493–497. [Google Scholar]
- 27. Dawson S, McArdle F, Bennett Met al. Typing of feline calicivirus isolates from different clinical groups by virus neutralisation tests. Vet Rec 1993; 133: 13–17. [DOI] [PubMed] [Google Scholar]
- 28. Kapikian AZ, Conant RM, Hamparian VVet al. Rhinoviruses: a numbering system. Nature 1967; 213: 761–762. [DOI] [PubMed] [Google Scholar]
- 29. Coyne KP, Christley RM, Pybus OGet al. Large-scale spatial and temporal genetic diversity of feline calicivirus. J Virol 2012; 86: 11356–11367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Radford AD, Bennett M, McArdle Fet al. The use of sequence analysis of a feline calicivirus (FCV) hypervariable region in the epidemiological investigation of FCV related disease and vaccine failures. Vaccine 1997; 15: 1451–1458. [DOI] [PubMed] [Google Scholar]
- 31. Almeras T, Schreiber P, Fournel Set al. Comparative efficacy of the Leucofeligen FeLV/RCP and Purevax RCP FeLV vaccines against infection with circulating feline calicivirus. BMC Vet Res 2017; 13: 300. DOI: 10.1186/s12917-017-1217-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lesbros C, Martin V, Najbar Wet al. Protective efficacy of the calicivirus valency of the Leucofeligen vaccine against a virulent heterologous challenge in kittens. Vet Med Int 2013; 2013: 232397. DOI: 10.1155/2013/232397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Radford AD, Dawson S, Ryvar Ret al. High genetic diversity of the immunodominant region of the feline calicivirus capsid gene in endemically infected cat colonies. Virus Genes 2003; 27: 145–155. [DOI] [PubMed] [Google Scholar]
- 34. Sato Y, Ohe K, Murakami Met al. Phylogenetic analysis of field isolates of feline calcivirus (FCV) in Japan by sequencing part of its capsid gene. Vet Res Commun 2002; 26: 205–219. [DOI] [PubMed] [Google Scholar]
- 35. Prikhodko VG, Sandoval-Jaime C, Abente EJet al. Genetic characterization of feline calicivirus strains associated with varying disease manifestations during an outbreak season in Missouri (1995–1996). Virus Genes 2014; 48: 96–110. [DOI] [PubMed] [Google Scholar]
- 36. Gojobori T, Moriyama EN, Kimura M. Molecular clock of viral evolution, and the neutral theory. Proc Natl Acad Sci U S A 1990; 87: 10015–10018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Tohya Y, Yokoyama N, Maeda Ket al. Mapping of antigenic sites involved in neutralization on the capsid protein of feline calicivirus. J Gen Virol 1997; 78: 303–305. [DOI] [PubMed] [Google Scholar]
- 38. Coyne KP, Gaskell RM, Dawson Set al. Evolutionary mechanisms of persistence and diversification of a calicivirus within endemically infected natural host populations. J Virol 2007; 81: 1961–1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Knowles JO, Dawson S, Gaskell RMet al. Neutralisation patterns among recent British and North American feline calicivirus isolates from different clinical origins. Vet Rec 1990; 127: 125–127. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Evolutionary divergence between sequences
Isolate details and results of virus neutralisation using FCV-F9 antisera
Further isolate details and GenBank accession numbers