Figure 1.
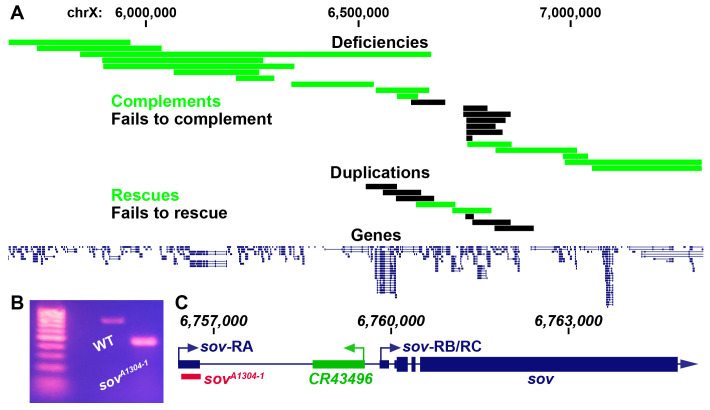
A) Complementation mapping of fs(1)A13041. Boxes represent either the portion of the chromosome deleted or duplicated. For deficiencies, green indicates complementing deletions and black indicates non-complementing deletions. For duplications, green indicates rescuing fragments while black indicates non-rescuing fragments. Numbers indicate genomic coordinates in bases along the X chromosome. B) Genomic PCR of wildtype (WT) and sovA1304-1 flies. Primers were designed to amplify genomic DNA encoding the 5′ UTR region of the sov-RA transcript. C) Cartoon of the sov locus. Dark blue represents the sov gene region with the left arrow representing the sov-RA transcriptional start site and right arrow representing the sov-RB/RC transcriptional start site. Green represents the CR43496 gene region with the arrow representing the transcriptional start site. Red box represents the deleted segment in sovA1304-1 flies. Small rectangles represent untranslated regions while large boxes represent translated regions. Numbers indicate genomic coordinates in bases along the X chromosome.
Description
X-linked female sterile screens in Drosophila have led to a tremendous increase in our understanding of the genetic control of oogenesis (Gans et al. 1975; Mohler 1977; Komitopoulou et al. 1983). However, many of the loci in these screens have not been mapped to a single gene and therefore remain a rich resource for further elucidating the genetic control of female fertility. fs(1)A13041 is one such allele that is germline dependent and results in a degenerative ovary phenotype (Gans et al. 1975; Khipple and King 1976; Mulligan 1981; Wieschaus et al. 1981; Mulligan and Rasch 1985; Lamnissou and Gelti-Douka 1985). We were interested in determining the mutation that leads to sterility in fs(1)A13041 females. Previous recombination mapping had placed fs(1)A13041 at 19±2 cM on the X chromosome (Gans, Audit, and Masson 1975; Khipple and King 1976). We confirmed the previous mapping interval by meiotically mapping fs(1)A13041 to the right of crossveinless (12 cM) and to the left of singed (22 cM). We began complementation tests for female sterility with known deficiencies tiling the crossveinless and singed region and placed the lesion within a roughly 235 kb region (Figure 1A, non-complementing Df(1)BSC276, BSC285, BSC286, BSC297, BSC351, BSC535, and sov) (Parks et al. 2004; Cook et al. 2012). Two duplications within this narrow region rescued fs(1)A13041 sterility and thus further narrowed down the possible location of the causal mutation (Figure 1A, Dp(1;3)DC486 and Dp(1;3)DC026) (Venken et al. 2010). The mapping results were somewhat ambiguous within this narrow region (discussed below). However, the smallest non-complementing deficiency, Df(1)sov, contains only the protein coding gene small ovary (sov) and non-coding RNA gene CR43496. We therefore decided to complementation test fs(1)A13041 with known alleles of sov. Flies homozygous for hypomorphic alleles of sov show a similar female sterility phenotype to flies bearing fs(1)A13041 while amorphic sov alleles are embryonic lethal (Wayne et al. 1995; Jankovics et al. 2018; Benner et al. 2019). We found that amorphic alleles sovEA42 and sovML150 failed to complement fs(1)A13041 female sterility while the hypomorphic sov2 complemented fs(1)A13041 sterility. Collectively this indicates that fs(1)A13041 is a sov allele (sovA1304-1).
To determine the molecular lesion, we performed paired-end DNA sequencing on sovA1304-1 females. The sov locus contains three annotated transcripts; sov-RA has an annotated upstream transcriptional start site while sov-RB/RC are annotated to use a downstream transcriptional start site (Thurmond et al. 2019). Our sequencing data suggested that sovA1304-1 flies contained a deletion within the sov gene region that would delete a majority of the sov-RA 5′ UTR. Genomic PCR of this potential deletion confirmed the presence of a deletion in sovA1304-1 flies (Figure 1B). Sanger sequencing of the sovA1304-1 genomic PCR product showed that there was a 324 nucleotide deletion (chrX:6,756,385-6,756,709) and a 10 nucleotide insertion (TCAACCTTCG) in the sov-RA 5′ UTR and would therefore remove most of the annotated 5′ UTR and donor splice site (Figure 1C).
We are unsure why a duplication (Dp(1;3)DC026) and a deficiency (Df(1)BSC535) to the left of the sov region rescued and failed to complement sovA1304-1, respectively. We also found that the small duplication of just sov and CR43496 (Dp(1;3)sovtCH322-191E24) failed to rescue. We were not able to find any deleterious mutations or structural variants in our sequencing data to the left of sov that might indicate the presence of a second-site suppressor or long-range genomic interactions with the sov locus that are necessary for its proper expression. It is interesting that sovA1304-1 had not been previously mapped to sov since the Mohler and Gans X-linked female sterile collections had been previously complementation tested inter se (Perrimon et al. 1986). We found that one of the original Mohler alleles, sov2, complemented sovA1304-1sterility and is thus possible that the other two Mohler alleles, sov1 and sov3, behaved similarly, providing an explanation as to why sovA1304-1 was not previously recognized as belonging to the sov locus. It would be interesting to determine if the 5′ UTR deletion of the sov-RA transcript found in sovA1304-1 flies affects sov activity in other tissues of the body other than the ovary. There is no indication that sov-RA, or sov-RB/RC, is differentially expressed in the ovary or other adult tissues (Benner et al. 2019). Pole cell transplantation studies of sovA1304-1 indicated that defects are germline dependent (Wieschaus et al. 1981; Lamnissou and Gelti-Douka 1985), however, sov is an essential gene that has been shown to dominantly suppress position-effect variegation in tissues such as the eye (Jankovics et al. 2018; Benner et al. 2019). It is possible that the deletion solely affects sov-RA and that the Drosophila ovary is more sensitive to loss of sov-RA, or sov transcripts in general, in comparison to other tissues since sovA1304-1 females are viable but sterile. However, we have not directly measured the deletions effects on sov-RB/RC transcript levels, which might also be perturbed. The nature of the sovA1304-1 deletion therefore provides a unique mechanism to further elucidate the function of Sov at potentially both the transcript and regulatory level in Drosophila.
Methods
Flies were cultured on ‘Fly Food A’ (LabExpress, Ann Arbor, MI) under standard laboratory conditions at 25°C. Genomic DNA was extracted from 30 homozygous fs(1)A13041 flies with a Qiagen DNeasy Blood and Tissue Kit (Hilden, Germany) according to the manufacturers insect protocol. DNA-sequencing libraries were made with Illumina Nextera DNA Library Prep Kit (San Diego, CA). 50 nucleotide paired-end sequencing was performed (Illumina HiSeq 2500, CASAVA base calling). Sequencing reads were mapped with Hisat2 to the FlyBase r6.25 genome and are available at the SRA (SRP238927) (Kim et al. 2015; Thurmond et al. 2019). Variant calling was completed with mpileup and bcftools from SAMtools within the X chromosome region 6625450-6860753 (Li et al. 2009; Li 2011) followed with variant annotation software snpEFF (Cingolani et al. 2012). For structural variant calling, we used BreakDancer software (Chen et al. 2009). Sanger sequencing was completed by Genewiz (Plainfield, NJ).
Reagents
Deficiencies and duplications in order as they appear in Figure 1 (top to bottom).
Deficiencies:
Df(1)ED6802 = BDSC 8949 (or FBst0008949)
Df(1)BSC654 = BDSC 26506 (or FBst0026506)
Df(1)dx81 = BDSC 5281 (or FBst0005281)
Df(1)ED418 = BDSC 8032 (or FBst0008032)
Df(1)ED6829 = BDSC 8947 (or FBst0008947)
Df(1)Exel6238 = BDSC 7712 (or FBst0007712)
Df(1)BSC640 = BDSC 25730 (or FBst0025730)
Df(1)Exel6239 = BDSC 7713 (or FBst0007713)
Df(1)Exel6240 = BDSC 7714 (or FBst0007714)
Df(1)e02477-d06059 = BDSC 39617 (or FBst0039617)
Df(1)BSC535 = BDSC 25063 (or FBst0025063)
Df(1)BSC285 = BDSC 23670 (or FBst0023670)
Df(1)BSC351 = BDSC 24375 (or FBst0024375)
Df(1)BSC297 = BDSC 23681 (or FBst0023681)
Df(1)BSC286 = BDSC 23671 (or FBst0023671)
Df(1)BSC276 = BDSC 23661 (or FBst0023661)
Df(1)sov = Benner et al., 2019
Df(1)ED6878 = BDSC 9625 (or FBst0009625)
Df(1)BSC882 = BDSC 30587 (or FBst0030587)
Df(1)BSC867 = BDSC 29990 (or FBst0029990)
Df(1)Sxl-bt = BDSC 3196 (or FBst0003196)
Df(1)SxlfP7B0 = BDSC 58489 (or FBst0058489)
Duplications:
Dp(1;3)DC158 = BDSC 30296 (or FBst0030296)
Dp(1;3)DC159 = BDSC 32268 (or FBst0032268)
Dp(1;3)DC160 = BDSC 30297 (or FBst0030297
Dp(1;3)DC026 = BDSC 30226 (or FBst0030226)
Dp(1;3)DC486 = BDSC 32306 (or FBst0032306)
Dp(1;3)sovtCH322-191E24 = Venken et al., 2010 (or FBal0243261)
Dp(1;3)DC163 = BDSC 32269 (or FBst0032269)
Dp(1;3)DC164 = BDSC 32270 (or FBst0032270)
Alleles:
fs(1)A13041 (sovA1304-1) = BDSC 4314 (or FBst0004314)
sov2 = BDSC 4611 (or FBst0004611)
sovEA42 (synonymous with l(1)6Dc3) = FBal0007068
sovML150 = BDSC 4591 (or FBst0004591)
Primer fs(1)A13041 Forward = TGACCATGTTGCATCTAAGCCA
Primer fs(1)A13041 Reverse = AGTAGAGCTCGCAATACGCC
Acknowledgments
Acknowledgments
Stocks obtained from the Bloomington Drosophila Stock Center (NIH P40OD018537) were used in this study. Sequencing was performed by the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) Genomics Core, under the direction of Harold Smith. Genetic and genomic information was obtained from FlyBase (U41 HG-000739). This work utilized the computational resources of the NIH High-Performance Computing Biowulf cluster (http://hpc.nih.gov).
Funding
This research was supported in part by the Intramural Research Program of the NIH, National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) to BO and by an Undergraduate Research and Inquiry grant from The University of Tampa to MH.
References
- Benner L, Castro EA, Whitworth C, Venken KJT, Yang H, Fang J, Oliver B, Cook KR, Lerit DA. Drosophila Heterochromatin Stabilization Requires the Zinc-Finger Protein Small Ovary. Genetics. 2019 Sep 26;213(3):877–895. doi: 10.1534/genetics.119.302590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K, Wallis JW, McLellan MD, Larson DE, Kalicki JM, Pohl CS, McGrath SD, Wendl MC, Zhang Q, Locke DP, Shi X, Fulton RS, Ley TJ, Wilson RK, Ding L, Mardis ER. BreakDancer: an algorithm for high-resolution mapping of genomic structural variation. Nat Methods. 2009 Aug 01;6(9):677–681. doi: 10.1038/nmeth.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cingolani P, Platts A, Wang le L, Coon M, Nguyen T, Wang L, Land SJ, Lu X, Ruden DM. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 1970 Jan 01;6(2):80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook RK, Christensen SJ, Deal JA, Coburn RA, Deal ME, Gresens JM, Kaufman TC, Cook KR. The generation of chromosomal deletions to provide extensive coverage and subdivision of the Drosophila melanogaster genome. Genome Biol. 2012;13(3):R21–R21. doi: 10.1186/gb-2012-13-3-r21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gans M, Audit C, Masson M. Isolation and characterization of sex-linked female-sterile mutants in Drosophila melanogaster. Genetics. 1975 Dec 01;81(4):683–704. doi: 10.1093/genetics/81.4.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jankovics F, Bence M, Sinka R, Faragó A, Bodai L, Pettkó-Szandtner A, Ibrahim K, Takács Z, Szarka-Kovács AB, Erdélyi M. Drosophila small ovary gene is required for transposon silencing and heterochromatin organization, and ensures germline stem cell maintenance and differentiation. Development. 2018 Dec 01;145(23) doi: 10.1242/dev.170639. [DOI] [PubMed] [Google Scholar]
- Khipple, Pamela, and Robert C. King. 1976. “Oogenesis in the Female Sterile (1) 1304 Mutant of Drosophila Melanogaster Meigen (Diptera: Drosophilidae).” <i>International Journal of Insect Morphology and Embryology</i> 5 (2): 127–35.
- Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. 2015 Mar 01;12(4):357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komitopoulou K, Gans M, Margaritis LH, Kafatos FC, Masson M. Isolation and Characterization of Sex-Linked Female-Sterile Mutants in DROSOPHILA MELANOGASTER with Special Attention to Eggshell Mutants. Genetics. 1983 Dec 01;105(4):897–920. doi: 10.1093/genetics/105.4.897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamnissou, Klea M., and Helen Gelti-Douka. 1985. “Analysis of the Drosophila Female Sterile mutation fs(1) 1304 by Pole Cell Transplantation Experiments.” <i>Developmental Genetics</i>, Carnegie Inst, 6 (4): 239–46.
- Li H. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics. 2011 Sep 01;27(21):2987–2993. doi: 10.1093/bioinformatics/btr509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009 Jun 01;25(16):2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohler JD. Developmental genetics of the Drosophila egg. I. Identification of 59 sex-linked cistrons with maternal effects on embryonic development. Genetics. 1977 Feb 01;85(2):259–272. doi: 10.1093/genetics/85.2.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulligan PK. Characterization of the female sterile (1) 1304 mutant of Drosophila melanogaster: pattern of RNA metabolism in the ovary. J Exp Zool. 1981 Jul 01;217(1):109–118. doi: 10.1002/jez.1402170112. [DOI] [PubMed] [Google Scholar]
- Mulligan PK, Rasch EM. Determination of DNA content in the nurse and follicle cells from wild type and mutant Drosophila melanogaster by DNA-Feulgen cytophotometry. Histochemistry. 1985;82(3):233–247. doi: 10.1007/BF00501400. [DOI] [PubMed] [Google Scholar]
- Parks AL, Cook KR, Belvin M, Dompe NA, Fawcett R, Huppert K, Tan LR, Winter CG, Bogart KP, Deal JE, Deal-Herr ME, Grant D, Marcinko M, Miyazaki WY, Robertson S, Shaw KJ, Tabios M, Vysotskaia V, Zhao L, Andrade RS, Edgar KA, Howie E, Killpack K, Milash B, Norton A, Thao D, Whittaker K, Winner MA, Friedman L, Margolis J, Singer MA, Kopczynski C, Curtis D, Kaufman TC, Plowman GD, Duyk G, Francis-Lang HL. Systematic generation of high-resolution deletion coverage of the Drosophila melanogaster genome. Nat Genet. 2004 Feb 22;36(3):288–292. doi: 10.1038/ng1312. [DOI] [PubMed] [Google Scholar]
- Perrimon N, Mohler D, Engstrom L, Mahowald AP. X-linked female-sterile loci in Drosophila melanogaster. Genetics. 1986 Jul 01;113(3):695–712. doi: 10.1093/genetics/113.3.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurmond J, Goodman JL, Strelets VB, Attrill H, Gramates LS, Marygold SJ, Matthews BB, Millburn G, Antonazzo G, Trovisco V, Kaufman TC, Calvi BR, FlyBase Consortium. FlyBase 2.0: the next generation. Nucleic Acids Res. 2019 Jan 01;47(D1):D759–D765. doi: 10.1093/nar/gky1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venken KJ, Popodi E, Holtzman SL, Schulze KL, Park S, Carlson JW, Hoskins RA, Bellen HJ, Kaufman TC. A molecularly defined duplication set for the X chromosome of Drosophila melanogaster. Genetics. 2010 Sep 27;186(4):1111–1125. doi: 10.1534/genetics.110.121285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wayne S, Liggett K, Pettus J, Nagoshi RN. Genetic characterization of small ovaries, a gene required in the soma for the development of the Drosophila ovary and the female germline. Genetics. 1995 Mar 01;139(3):1309–1320. doi: 10.1093/genetics/139.3.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieschaus E, Audit C, Masson M. A clonal analysis of the roles of somatic cells and germ line during oogenesis in Drosophila. Dev Biol. 1981 Nov 01;88(1):92–9103. doi: 10.1016/0012-1606(81)90221-9. [DOI] [PubMed] [Google Scholar]