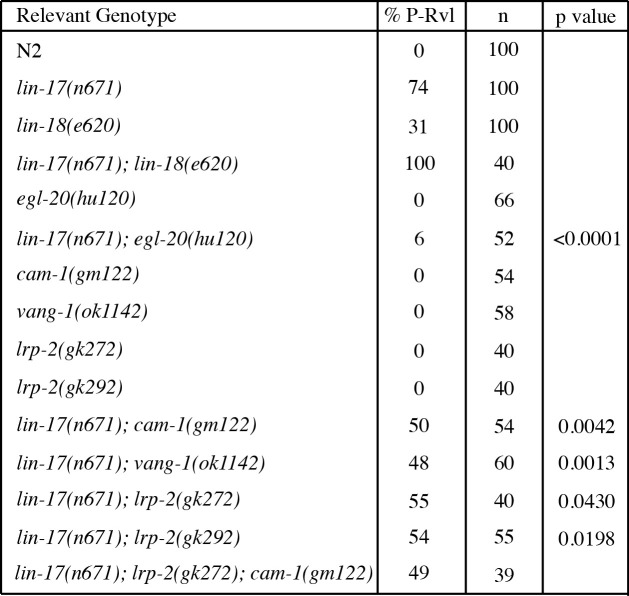
Table 1. LRP-2 functions in ground polarity.
P values in comparison to lin-17(n671) single mutants using Fisher’s Exact Test.
Description
The C. elegans vulva is formed from divisions of three vulval precursor cells (VPCs) – P5.p, P6.p, and P7.p – arranged along the anteroposterior axis in the ventral epithelium (Sulston and Horvitz, 1977). Previous analyses show the orientation of P5.p and P7.p descendants is determined by the interaction of multiple Wnt signals. Specifically, in the absence of all Wnts, the VPCs display a randomized orientation, which is likely the default (Green et al., 2008; Minor et al. 2013). Two separate Wnts from the anchor cell, LIN-44 and MOM-2 acting through receptors LIN-17/Frizzled and LIN-18/Ryk, respectively, regulate P7.p orientation (Ferguson et al., 1987; Sternberg and Horvitz, 1988; Sawa et al., 1996; Inoue et al., 2004; Gleason et al., 2006). In the absence of these signals the orientation of the progeny of P7.p mimic those of P5.p and face toward the posterior of the worm, a phenotype referred to as posterior-reversed vulval lineage (P-Rvl). This posterior orientation is dependent on the instructive signal EGL-20, a Wnt expressed in the tail acting through CAM-1/ROR and VANG-1/Van Gogh, and is referred to as “ground polarity” (Green et al., 2008).
Here we examine the role of a low-density lipoprotein receptor, lrp-2, and its role in controlling the orientation of P7.p daughter cells. To investigate this interaction double mutants were constructed with both alleles of lrp-2 and lin-17(n671) (Table 1). Much like cam-1(gm122) and vang-1(ok1142), both alleles of lrp-2 suppress the lin-17(n671) phenotype from 74 to approximately 50% P-Rvl leading us to hypothesize that lrp-2 functions in the same pathway as cam-1 and vang-1. Furthering this hypothesis we have shown that, like cam-1 and vang-1, lrp-2 controls the localization of SYS-1/b-catenin (Minor and Sternberg, 2019). To ensure that this phenotype was a result of loss of lrp-2 function as opposed to background effects we injected a fosmid (WRM0617cA02) containing the full-length sequence of lrp-2 and found that it does rescue the double mutant phenotype of lin-17(n671); lrp-2(gk272) from 55 to 73%. In order to better test this hypothesis a triple mutant was constructed between lin-17(n671), lrp-2(gk272), and cam-1(gm122) (Table 1). This triple mutant displays the same P-Rvl penetrance as both the lin-17(n671); lrp-2(gk272) and lin-17(n671); cam-1(gm122) double mutants confirming that lrp-2 functions in the same pathway as cam-1.
Methods
Strains were constructed by standard procedures (Ferguson and Horvitz, 1985).
Reagents
Strains:
N2
MT1306: lin-17(n671) (Ferguson and Horvitz, 1985)
PS4818: lin-18(e620)
PS3976: lin-17(n671)/ hT2 [qIs48 (myo-2::gfp etc.)]; +/ hT2 [qIs48]; lin-18(e620)
PS5401: egl-20(hu120)
PS5530: lin-17(n671); egl-20(hu120)
NG2615: cam-1(gm122)
RB1125: vang-1(ok1142)
VC543: lrp-2(gk272)
VC621: lrp-2(gk292)
PS4809: lin-17(n671); cam-1(gm122)
PS5651: lin-17(n671); vang-1(ok1142)
lin-17(n671); lin-18(e620) animal awere segregated from PS3976.
The lin-17(n671); lrp-2(gk292) double mutant was constructed by crossing lrp-2(gk292) males with strain MT1488: lin-17(n671); unc-13(e1091) hermaphrodites.
The lin-17(n671); lrp-2(gk272) double mutant constructed by crossing lrp-2(gk272) males with strain MT1488: lin-17(n671); unc-13(e1091) hermaphrodites.
lin-17(n671); lrp-2(gk272); cam-1(gm122) Triple mutant constructed by crossing lin-17(n671); lrp-2(gk272) hermaphrodites with cam-1(gm122) and sequencing for homozygous triples.
PS6581: qIs95[pSYS-1::VENUS::SYS-1 with pttx-3::dsRed as coninjection marker];
PS6576: lin-18(e620) sem-5(n1779)
PS6580: qIs95; sem-5(n1779)
PS6579: Pegl-17::CWN-1::GFP (unnamed construct) in cwn-1(ok546); lin-18(e620)
Acknowledgments
Funding
Howard Hughes Medical Institute, with whom PWS was an Investigator. The National Institute of Neurological Disorders and Stroke of the National Institutes of Health under award number 1F32NS098658-01A1 awarded to PJM.
References
- Ferguson EL, Horvitz HR. Identification and characterization of 22 genes that affect the vulval cell lineages of the nematode Caenorhabditis elegans. Genetics. 1985 May 01;110(1):17–72. doi: 10.1093/genetics/110.1.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson EL, Sternberg PW, Horvitz HR. A genetic pathway for the specification of the vulval cell lineages of Caenorhabditis elegans. Nature. 1987 Mar 20;326(6110):259–267. doi: 10.1038/326259a0. [DOI] [PubMed] [Google Scholar]
- Gleason JE, Szyleyko EA, Eisenmann DM. Multiple redundant Wnt signaling components function in two processes during C. elegans vulval development. Dev Biol. 2006 Jul 01;298(2):442–457. doi: 10.1016/j.ydbio.2006.06.050. [DOI] [PubMed] [Google Scholar]
- Green JL, Inoue T, Sternberg PW. Opposing Wnt pathways orient cell polarity during organogenesis. Cell. 2008 Aug 22;134(4):646–656. doi: 10.1016/j.cell.2008.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue T, Oz HS, Wiland D, Gharib S, Deshpande R, Hill RJ, Katz WS, Sternberg PW. C. elegans LIN-18 is a Ryk ortholog and functions in parallel to LIN-17/Frizzled in Wnt signaling. Cell. 2004 Sep 17;118(6):795–806. doi: 10.1016/j.cell.2004.09.001. [DOI] [PubMed] [Google Scholar]
- Minor PJ, He TF, Sohn CH, Asthagiri AR, Sternberg PW. FGF signaling regulates Wnt ligand expression to control vulval cell lineage polarity in C. elegans. Development. 2013 Aug 14;140(18):3882–3891. doi: 10.1242/dev.095687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minor Paul J, Sternberg Paul W. LRP-2 controls the localization of C. elegans SYS-1/beta-catenin. microPublication Biology. 2019 Aug 27; doi: 10.17912/micropub.biology.000151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizumoto K, Sawa H. Two betas or not two betas: regulation of asymmetric division by beta-catenin. Trends Cell Biol. 2007 Oct 01;17(10):465–473. doi: 10.1016/j.tcb.2007.08.004. [DOI] [PubMed] [Google Scholar]
- Phillips BT, Kidd AR 3rd, King R, Hardin J, Kimble J. Reciprocal asymmetry of SYS-1/beta-catenin and POP-1/TCF controls asymmetric divisions in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2007 Feb 12;104(9):3231–3236. doi: 10.1073/pnas.0611507104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawa H, Lobel L, Horvitz HR. The Caenorhabditis elegans gene lin-17, which is required for certain asymmetric cell divisions, encodes a putative seven-transmembrane protein similar to the Drosophila frizzled protein. Genes Dev. 1996 Sep 01;10(17):2189–2197. doi: 10.1101/gad.10.17.2189. [DOI] [PubMed] [Google Scholar]
- Sternberg PW, Horvitz HR. lin-17 mutations of Caenorhabditis elegans disrupt certain asymmetric cell divisions. Dev Biol. 1988 Nov 01;130(1):67–73. doi: 10.1016/0012-1606(88)90414-9. [DOI] [PubMed] [Google Scholar]
- Hedgecock EM, White JG. Polyploid tissues in the nematode Caenorhabditis elegans. Dev Biol. 1985 Jan 01;107(1):128–133. doi: 10.1016/0012-1606(85)90381-1. [DOI] [PubMed] [Google Scholar]