Abstract
Dynamic DNA structures, a type of DNA construct built using programmable DNA self-assembly, have the capability to reconfigure their conformations in response to environmental stimulation. A general strategy to design dynamic DNA structures is to integrate reconfigurable elements into conventional static DNA structures that may be assembled from a variety of methods including DNA origami and DNA tiles. Commonly used reconfigurable elements range from strand displacement reactions, special structural motifs, target-binding DNA aptamers, and base stacking components, to DNA conformational change domains, etc. Morphological changes of dynamic DNA structures may be visualized by imaging techniques or may be translated to other detectable readout signals (e.g., fluorescence). Owing to their programmable capability of recognizing environmental cues with high specificity, dynamic DNA structures embody the epitome of robust and versatile systems that hold great promise in sensing and imaging biological analytes, in delivering molecular cargos, and in building programmable systems that are able to conduct sophisticated tasks.
Keywords: DNA walkers and circuits, dynamic DNA structures, nanorobotic transportation of molecular cargos, self-assembly, sensing and imaging
1. Introduction
Deoxyribonucleic acid (DNA) is the genetic blueprint for the vast majority of life forms on Earth. It is a biomacromolecule composed of nucleotides carrying one of four different types of nucleobases, adenine (A), thymine (T), cytosine (C), or guanine (G); a deoxyribose; and a phosphate group. Watson-Crick base pairing determines the interactions between DNA bases, where A pairs with T and C pairs with G via hydrogen bonding.[1] The specificity and predictability of Watson-Crick base pairing make DNA a powerful and programmable material for nanoscale engineering. This sets the foundation of DNA nanotechnology, a field where DNA is used as building blocks to construct diverse static and dynamic nanostructures via prescribed assembly of DNA strands. The concept of structural DNA nanotechnology can be traced back to 1982 when Nadrian Seeman proposed to use 3D DNA crystals to aid the crystallization of proteins for crystallography studies.[2] After four decades of development, though no protein structures have been solved with the help of DNA crystals, structural DNA nanotechnology has largely outgrown Seeman’s initial scope and has become a multidisciplinary technology for fabricating nanoscale structures with unprecedented controllability and versatility.[3] Many strategies have been developed for designing DNA structures. The simplest DNA structure may be a single-stranded DNA (ssDNA) with known sequence, secondary structure, and function (Figure 1a). A more rigid double-stranded DNA (dsDNA) structure would form if two partially or fully complementary ssDNAs are joined (Figure 1b). Multistranded structures of higher structural complexity may be assembled from several DNA strands of designed sequence complementarity, such as one of the most popularly used: the tetrahedral structure (Figure 1c).[4] The realization of DNA tile-based assembly for complex DNA structures represents one of the milestones in the history of structural DNA nanotechnology. The design concept is that a DNA tile is first constructed from one or multiple DNA strands of designed length and sequence. Sticky-end mediated hierarchical assembly of DNA tiles leads to the formation of finite- or infinite-sized DNA structures composed of repeating tiles (Figure 1d).[5] A special type of DNA tile design is called DNA brick (Figure 1e),[6,7] where each tile has a unique sequence. Such uniqueness grants modularity to DNA bricks to allow assembly of arbitrary DNA objects by simply sculpting unwanted bricks from the pool. Another powerful method to assemble structures of arbitrary geometry is DNA origami (Figure 1f),[8] which was invented prior to the DNA brick method. In DNA origami, a long ssDNA (scaffold DNA) is folded along prescribed paths by hundreds of short ssDNAs (staple DNA) to form predesigned DNA structures. Figure 1g,h shows some representative DNA crystal structures[5,9–13] and polyhedral[14–17] structures assembled from multiarm DNA tiles. Two-dimensional (2D)[7] and three-dimensional (3D)[6] objects assembled from DNA bricks are illustrated in Figure 1i. DNA structures constructed from a single DNA origami, either in closely packed-lattice style or wire-frame style are shown in Figure 1j,[8,18–22] while hierarchically assembled large DNA structures from DNA origami tiles are shown in Figure 1k.[23–25] It is worth noting that DNA structures presented in Figure 1 are static structures without any dynamic capability. To build dynamic DNA structures that are able to reconfigure upon external stimulation, a general strategy is to incorporate reconfigurable elements into static DNA structures. The next section presents a brief introduction to commonly used reconfigurable elements that may be used for dynamic DNA structures.
Figure 1.

Assembly strategies and representative DNA structures. a–f) Assembly strategies for constructing DNA structures including single-stranded DNA, double-stranded DNA duplex, multistranded DNA structures, multiarm tiles, DNA bricks, and DNA origami. g) Infinite-sized 2D lattices and 3D crystal assembled multiarm DNA tiles. Reproduced with permission.[5,9–13] Copyright 1998, Springer Nature; 2005, ACS; 2003, AAAS; 2008, NAS; 2006, ACS; 2009, Springer Nature. h) 3D polyhedral structures assembled from multiarm DNA tiles. Reproduced with permission.[14–17] Copyright 2008, Springer Nature; 2014, Wiley-VCH.; 2016, ACS; 2009, ACS. i) 2D and 3D objects assembled from DNA bricks. Reproduced with permission.[6,7] Copyright 2012, Springer Nature; 2012, AAAS. j) 2D and 3D objects assembled from DNA origami. Reproduced with permission.[8,18–22] Copyright 2006, Springer Nature; 2009, Springer Nature; 2011, AAAS; 2015, Springer Nature; 2009, AAAS; 2011, Springer Nature and k) enlarged DNA origami structures via hierarchical assembly of DNA origami units. Reproduced with permission.[23–25] Copyright 2017, Springer Nature; 2017, Springer Nature; 2011, Wiley-VCH.
2. Reconfigurable Elements for Constructing Dynamic DNA Structures
Reconfigurable elements responsive to environmental stimuli are essential for designing dynamic DNA structures. Such elements may be embedded into static DNA structures to achieve dynamic motion. Commonly used reconfigurable elements are summarized in Figure 2. For instance, hydrogen bond–mediated strand association and dissociation between ssDNA and dsDNA represents one of the most conventional methods to induce dynamic motion (Figure 2a). The switch between these two states may be induced via changing environmental parameters such as temperature, light exposure (e.g., UV versus visible light for azobenzene modified DNA),[26] or ionic concentration[27] (e.g., metal ions, pH value). Another method involves enzymatic reactions to DNA strands (Figure 2b), such as strand degradation, cleavage, or ligation.[28,29] Strand displacement is probably the most commonly used strategy to make dynamic DNA structures, in which one strand is displaced from a double stranded DNA, typically mediated by a toehold design (Figure 2c).[30] Strand displacement may also happen if one of the strands has higher affinity to another target (e.g., proteins). DNA strands with such capability are designated as DNA aptamers (Figure 2d).[31] Certain special structural motifs are reconfigurable as well, such as guanine- and cytosine-rich sequences that can fold into stable quadruplexes in the presence of specific metal ions and low pH (Figure 2e).[32,33] Base stacking between blunt ends of DNA helices are ion- and temperature-sensitive (Figure 2f), and are, therefore, able to reconfigure upon stimulation.[34] DNA duplexes of certain sequences may transit between B-form and Z-form conformation in response to environmental ionic strength (Figure 2g).[35] For example, the dynamic antijunction has recently been demonstrated to be able to switch between two conformations controlled by molecular triggers or temperature (Figure 2h).[36] A special type of dynamic DNA structure called mechanical joints contains no reconfigurable elements but instead utilizes mobile components (Figure 2i).[37] These structures have mechanically flexible parts that can rotate or slide if external forces are applied via sources such as electric field[37] magnetic field[38] or thermoresponsive polymers.[39]
Figure 2.
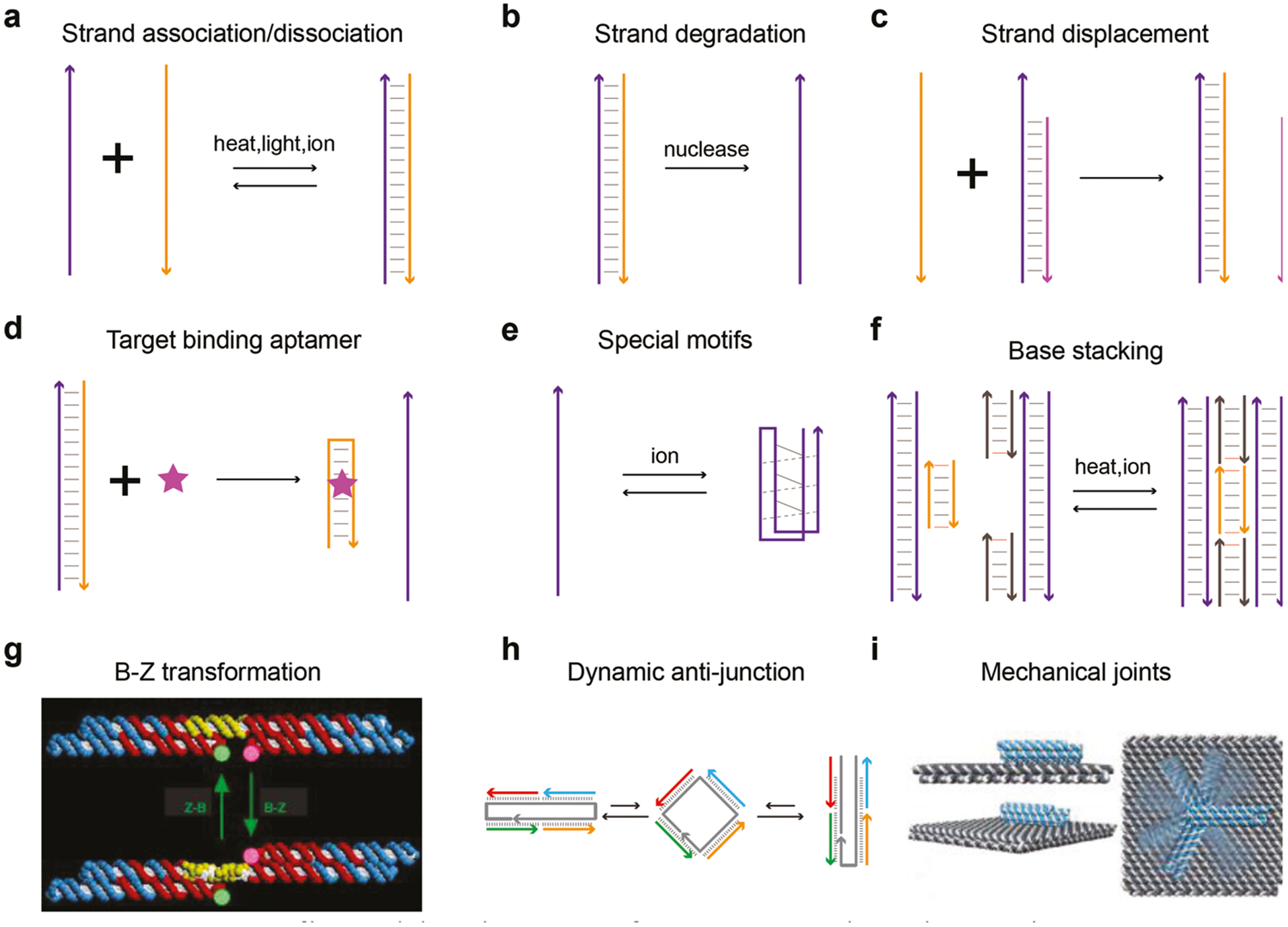
Reconfigurable elements for constructing dynamic DNA structures. a) DNA strands associate or dissociate upon environmental stimulation. b) Degradation of DNA strands. c) Toehold mediated strand displacement. d) Target binding DNA aptamers induced strand displacement. e) Special DNA motifs that change secondary structures in response to environmental cues. f) DNA blunt-end mediated base stacking. g) DNA duplex transits between B-form and Z-form. Reproduced with permission.[35] Copyright 1999, Springer Nature. h) Dynamic antijunction unit. Reproduced with permission.[36] Copyright 2017, AAAS and i) flexible mechanical joint that rotates by electric force. Reproduced with permission.[37] Copyright 2018, AAAS.
3. Readout Signals of Dynamic DNA Structures
Reconfiguration of dynamic DNA structures leads to nano- or macroscale structural changes. These changes may be unambiguously visualized by imaging techniques. For instance, Ke and colleagues demonstrated that DNA structures composing of dynamic DNA antijunctions can transform from one state to another with highly different aspect ratio when molecular triggers are added (Figure 3a).[36] The structural change of units can propagate along designated pathways through information relay to complete structural transformation of the whole structure. Such nanoscale structural transformation was readily captured by high-resolution imaging modalities such as atomic force microscopy (AFM). Macroscopic structural change may also be realized by dynamic DNA structures. Schulman and colleagues incorporated hybridization chain reaction (HCR) active components into macroscopic polyacrylamide-based hydrogels with DNA serving as crosslinkers (Figure 3b).[40] HCR of DNA strands within the macroscopic hydrogel leads to ≈100-fold volume expansion. They later demonstrated that such change may also be induced by molecular triggers such as adenosine triphosphate (ATP).[41]
Figure 3.
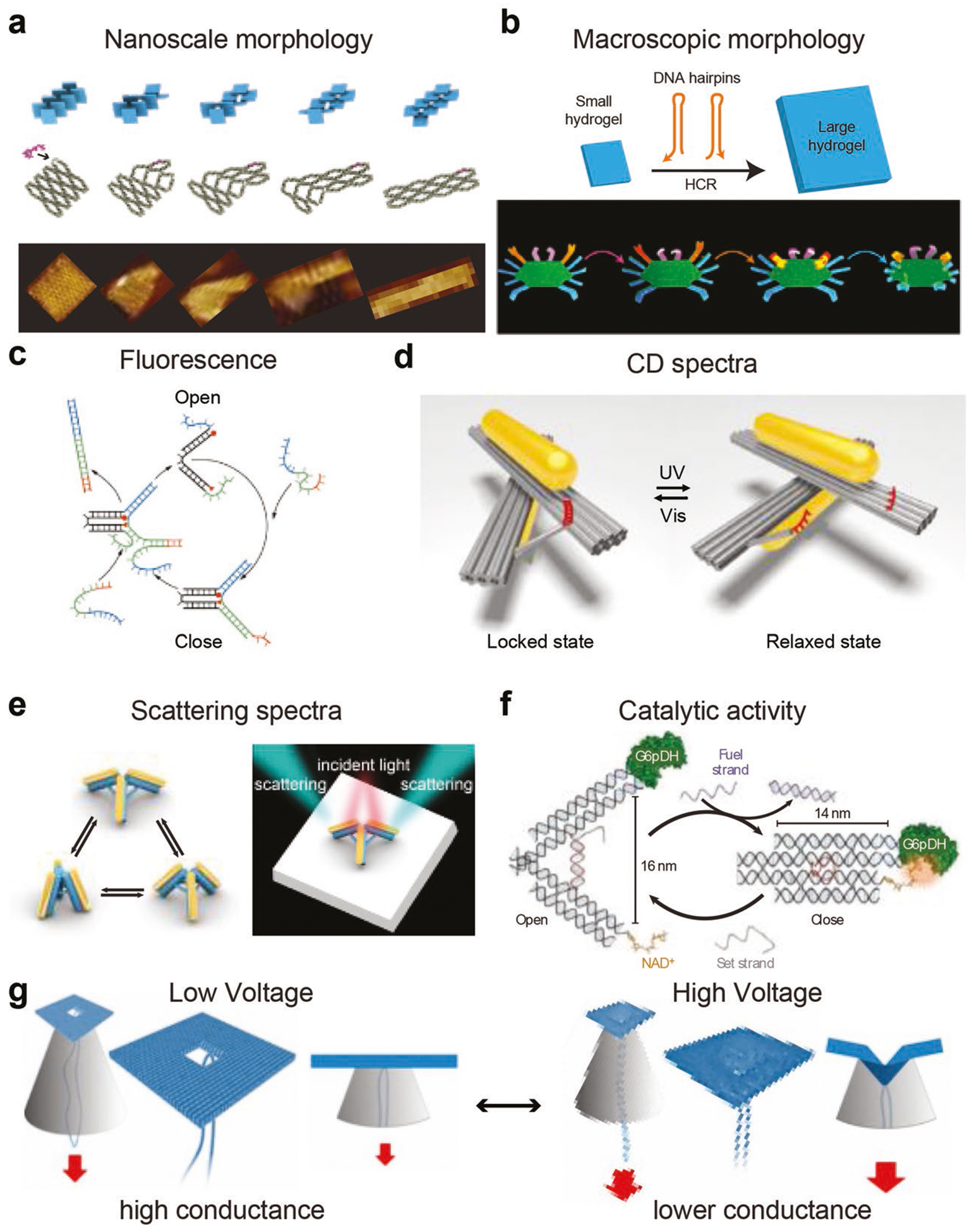
Readout signals of dynamic DNA structures. a) Nanoscale morphology change of dynamic DNA structures composing of antijunctions. Reproduced with permission.[36] Copyright 2017, AAAS. b) DNA hybridization chain reaction induced macroscopic morphology change in polyacrylamide hydrogels. Reproduced with permission.[40] Copyright 2017, AAAS. c) Open and close of a DNA tweezer regulates FRET efficiency. Reproduced with permission.[30] Copyright 2000, Springer Nature. d) A chiral gold nanorod nanodevice with photo-tunable CD signal. Reproduced with permission.[26] Copyright 2016, Springer Nature. e) Dynamic gold nanorod tripod with tunable scattering property. Reproduced with permission.[42] Copyright 2017, ACS. f) The catalytic activity of an enzyme mediated by underlying dynamic DNA structure. Reproduced with permission.[43] Copyright 2013, Springer Nature and g) the conductance of DNA origami nanopore controlled by the voltage-dependent dynamic deformation of origami structure. Reproduced with permission.[46] Copyright 2014, ACS.
Dynamic DNA structures may carry optical reporters, whose optical readouts could reflect the structural change of the structure. Organic fluorophores are among typical optical reporters used in dynamic DNA structures due to ease of preparation and detection. For instance, the opening and closing of a DNA tweezer regulates the energy transfer between two fluorophores and, therefore, the fluorescent readout of the system (Figure 3c).[30] Metallic nanoparticles are another type of optical reporters. A pair of gold nanorods (AuNR) may be assembled onto DNA origami templates, which can switch between left-handed and right-handled chirality when the underlying DNA origami templates undergo structural change as induced by optical stimuli (Figure 3d).[26] In addition to chiral properties, the scattering spectra of AuNRs organized on DNA origami templates is also able to reflect the structural change of the template since the relative angle between AuNRs largely affects the scattering light when the incident light comes from a certain direction (Figure 3e).[42] Proximity-sensitive catalytic components may be assembled on reconfigurable DNA structures (Figure 3f);[43] thus, the catalytic activity of the assembled system is dependent on the structural state of the DNA structures, which could in turn serve as the readout signal of the dynamic DNA structure. DNA nanostructures have also been used as important elements for making nanopore sensing devices.[44,45] DNA-based nanopores may exhibit dynamic characteristics in response to external stimuli such as electrophoretic force. For instance, higher voltage induced mechanical deformation of a DNA origami nanopore led to a “buckled conformation” with an increased ionic current blockage (Figure 3g).[46]
4. Biological Sensing and Imaging
Dynamic DNA structures have great promise in serving as biological sensors since they are able to specifically recognize biological analytes (e.g., molecules, ions) of interest. Biological sensors are composed of target recognition elements and signal transducers integrated into DNA structures. Dynamic DNA structures can reconfigure in response to target recognition, where shape change–inducing events are used as signal readouts, typically in the form of fluorescence. For instance, Krishnan and colleagues built an i-motif–based DNA switch whose open or closed state is dependent on environmental pH (Figure 4a).[47] This switch was used as a fluorescent reporter to measure and map intracellular pH changes in living cells with spatial and temporal resolution. They later developed a series of DNA nanomachines that can sense and measure important physiological ions (e.g., Ca2+, Cl−1) in intracellular compartments.[48,49] In addition to ions, biological molecules in living cells such as ATP may be probed as well. Li et al. recently reported an upconversion luminescence activated DNA nanodevice that can sense ATP in living cells (Figure 4b).[50] A DNA aptamer with ATP binding capability was incorporated into a simple duplex structure with the aptamer initially protecting from ATP attack. Once illuminated, the protecting strand was cleaved, rendering the aptamer available for ATP binding while enabling detection of fluorescence. Biological nucleic acids such as mRNAs, miRNA, and viral RNAs are important indicators to interpret physiological and pathological conditions. Liedl et al. constructed a chiral optical nanodevice by placing a pair of AuNRs onto a DNA origami template (Figure 4c).[51] Once viral RNA of as low as pico-molar concentration was introduced, circular dichroism (CD) spectrophotometer recordings of nanodevice chirality was reversed accordingly. Bellot et al. developed a nanoactuator using DNA origami (Figure 4d), which can sense and actuate in response to a number of different biological cues such as metal ions (K+), enzymes (BamHI), or nucleic acids (miR-210).[52] Keyser et al. constructed voltage-dependent dynamic DNA nanopores with single-molecule fluorescence resonance energy transfer (FRET) as the optical readout for voltage detection, which has the application potential for live-cell imaging of transmembrane potentials (Figure 4e).[53] Super-resolution imaging was achieved by utilizing the dynamic and transient binding of fluorophore-tagged short DNA strands onto targets, a technique called DNA points accumulation for imaging in nanoscale topography (DNA-PAINT) (Figure 4f). With DNA-PAINT, a sub-10 nm resolution of multiplex capability could be achieved by using fluorescent microscopy, which was able to clearly image cellular compartments in 3D.[54]
Figure 4.
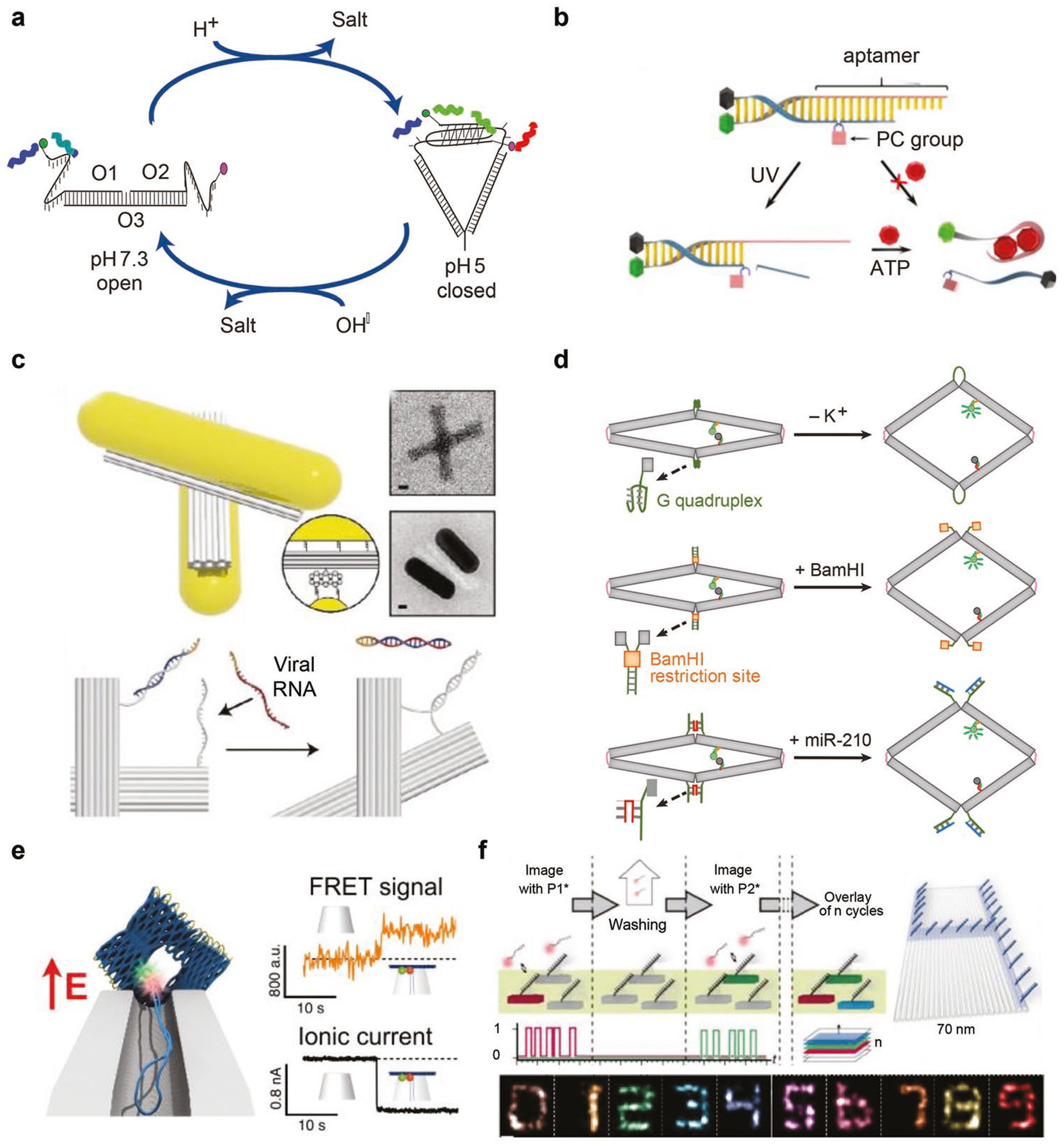
Dynamic DNA structures for biological sensing and imaging. a) A i-motif–based pH sensor that is capable of mapping the intracellular pH. Reproduced with permission.[47] Copyright 2009, Springer Nature. b) A light activatable, aptamer-based sensor for intracellular ATP sensing. Reproduced with permission.[50] Copyright 2018, ACS. c) A chiroptical metamolecule for viral RNA sensing in vitro. Reproduced with permission.[51] Copyright 2018, Wiley-VCH. d) Biosensing of ions, proteins, or nucleic acids via allosteric activation of nanoactuator. Reproduced with permission.[52] Copyright 2016, Springer Nature. e) Optical voltage sensing device based on DNA origami nanopore with responsive deformation upon eletrical stimulation. Reproduced with permission.[53] Copyright 2018, ACS and f) transient binding of fluorophore DNA (DNA-PAINT) for optical imaging at sub-10 nm resolution. Reproduced with permission.[54] Copyright 2014, Springer Nature.
5. Nanorobotic Systems for Molecular Cargo Transportation
Transportation of molecular cargos represents one of the most promising applications of dynamic DNA structures. For instance, Keyser et al. demonstrated that a DNA nanostructure with cholesterol tags was able to dock into biological membranes, and effectively catalyze the transport of lipid molecules between the inner and outer leaflets, outperforming the corresponding biological archetypes by three orders of magnitude (Figure 5a).[55] Numerous studies have proposed strategies to load, deliver, and release molecular drugs for gene silencing, immunostimulation, and photodynamic therapy using static DNA structures.[56–58] In order to achieve active and responsive delivery of drugs into targeting sites with minimal off-target-associated systemic toxicity, nano robotic delivery systems are urgently needed and may play important roles in next-generation medicine. Though there have been only a few reports so far, dynamic DNA structures have shown great promise in serving as drug delivery nanorobots. Douglas and colleagues designed a DNA origami barrel that was locked by a DNA aptamer (Figure 5b).[59] Upon sensing targets on the cell membrane, DNA aptamer preferred binding to the target instead of locking, which led to opening of the barrel and releasing of the docked molecular payloads. Logic-gated designs may be integrated into the locking mechanism to enable cargo releasing in the presence of multiple cellular targets at the same time (and gate), minimizing false-positive events. Gu et al. constructed a telomerase-responsive DNA icosahedral structure loaded with platinum nanodrugs (Figure 5c).[60] Telomerase is reportedly overexpressed in the majority of cancer cells while nearly absent in normal cells. In vitro and in vivo delivery of Pt-encapsulated DNA icosahedral structures exhibited higher efficacy on inhibiting cancer cell growth—probably due to telomerase-mediated higher releasing efficiency of Pt nanodrugs into cancer cells. Ding et al. recently developed a DNA origami tubular robot capable of opening into planar rectangular structures to subsequently release encapsulated thrombin upon recognition of a molecular cancer biomarker called nucleolin (Figure 5d).[61] Thrombin induced blood coagulation cuts off nutrient supplies to cancer cells and thus inhibits tumor growth in both xenograft and ortho-topic tumor models in mice and Bama miniature pigs.
Figure 5.
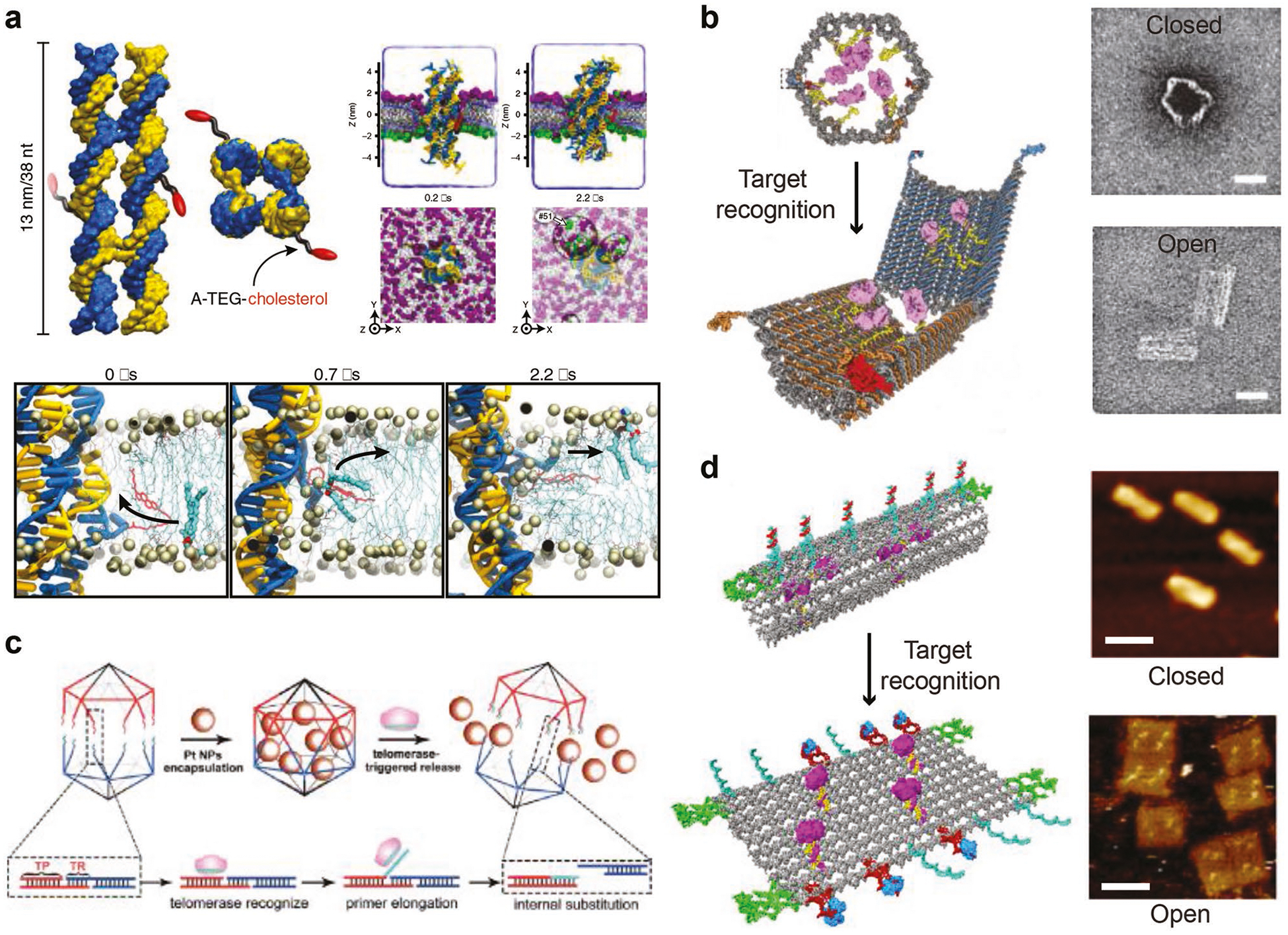
Dynamic DNA structures for molecular cargo transportation. a) A DNA nanostructure with cholesterol tags that scrambles lipid molecules between interior and outerior membranes at high speed. Reproduced with permission.[55] Copyright 2018, Springer Nature. b) A DNA origami barrel that opens and releases molecular payloads upon recognizing receptors on cell membrane. Reproduced with permission.[59] Copyright 2012, AAAS. c) A DNA icosahedron that releases Pt nanoparticles once triggered by telomerase in vitro and in vivo. Reproduced with permission.[60] Copyright 2018, Wiley-VCH. d) A tubular DNA origami robot that opens and releases thrombin in vivo upon stimulation by a molecular cancer biomarker nucleolin. Reproduced with permission.[61] Copyright 2018, Springer Nature.
6. Programmable DNA Walkers and Circuits
The programmability of DNA molecules makes them suitable for fabricating dynamic structures capable of conducting sophisticated jobs. Yan and co-workers designed a molecular spider composed of a streptavidin body and DNA legs (Figure 6a).[62] They demonstrated that such walkers can achieve directional movement by sensing and modifying tracks of substrate molecules laid out on a 2D DNA origami landscape. With an appropriately designed landscape, the molecular spiders autonomously carry out sequences of actions including “start,” “follow,” “turn,” and “stop.” Dynamic DNA probes may also serve as circuits to report biological events in living cells. Tan and co-workers anchored DNA probes onto the exterior membrane of cells (Figure 6b).[63] Once the probes encounter other probes, strand displacement reactions take place, and a fluorescent readout may be recorded. Using this system, they successfully investigated the dynamic and transient encounter events of biological molecules on membranes of living cells. Programmable DNA walkers could serve as templates to precisely direct stepwise organic synthesis. Liu and co-workers used a DNA walker to walk along a prescribed template docked with three reactive organic molecules (Figure 6c).[64] This walker picks up these organic molecules step by step via chemical reactions while walking along the path, yielding a final product composed of three precursor molecules. Intelligent dynamic DNA walkers may serve as robots to conduct sophisticated tasks such as cargo sorting (Figure 6d).[65] Qian et al. designed a DNA robot and docked it onto a DNA origami field. This robot is capable of randomly walking along tracks within the field to pick up molecular cargos (fluorophore-tagged DNA) and transport them to designated sites. More strikingly, the robot is able to sort and transport cargos to wherever they belong, all programmed into the robots. In addition to cargo sorting, molecular neural circuits by dynamic DNA structures may bring task complexity to unprecedented levels. The same group developed a winner-take-all neural network out of DNA motifs (Figure 6e).[66] This neural network, after training, is able to recognize molecular patterns of arbitrary handwritten digits “1” to “9.” The network successfully classified test patterns with up to 30 of the 100 bits flipped relative to the digit patterns “remembered” during training, suggesting that molecular circuits can robustly accomplish the sophisticated task of classifying highly complex and noisy information on the basis of similarity to a memory. DNA walkers are also capable of maze solving. Fan and co-workers[67] demonstrate a DNA navigator system that can perform single-molecule parallel depth-first search on a 2D DNA origami platform. Pathfinding by the DNA navigators exploits a localized strand exchange cascade, which is initiated at a unique trigger site on the origami with subsequent automatic progression along paths defined by DNA hairpins containing a universal traversal sequence. A specific solution path connecting a given pair of start and end vertices can then be easily extracted from the set of all paths taken by the navigators collectively.
Figure 6.
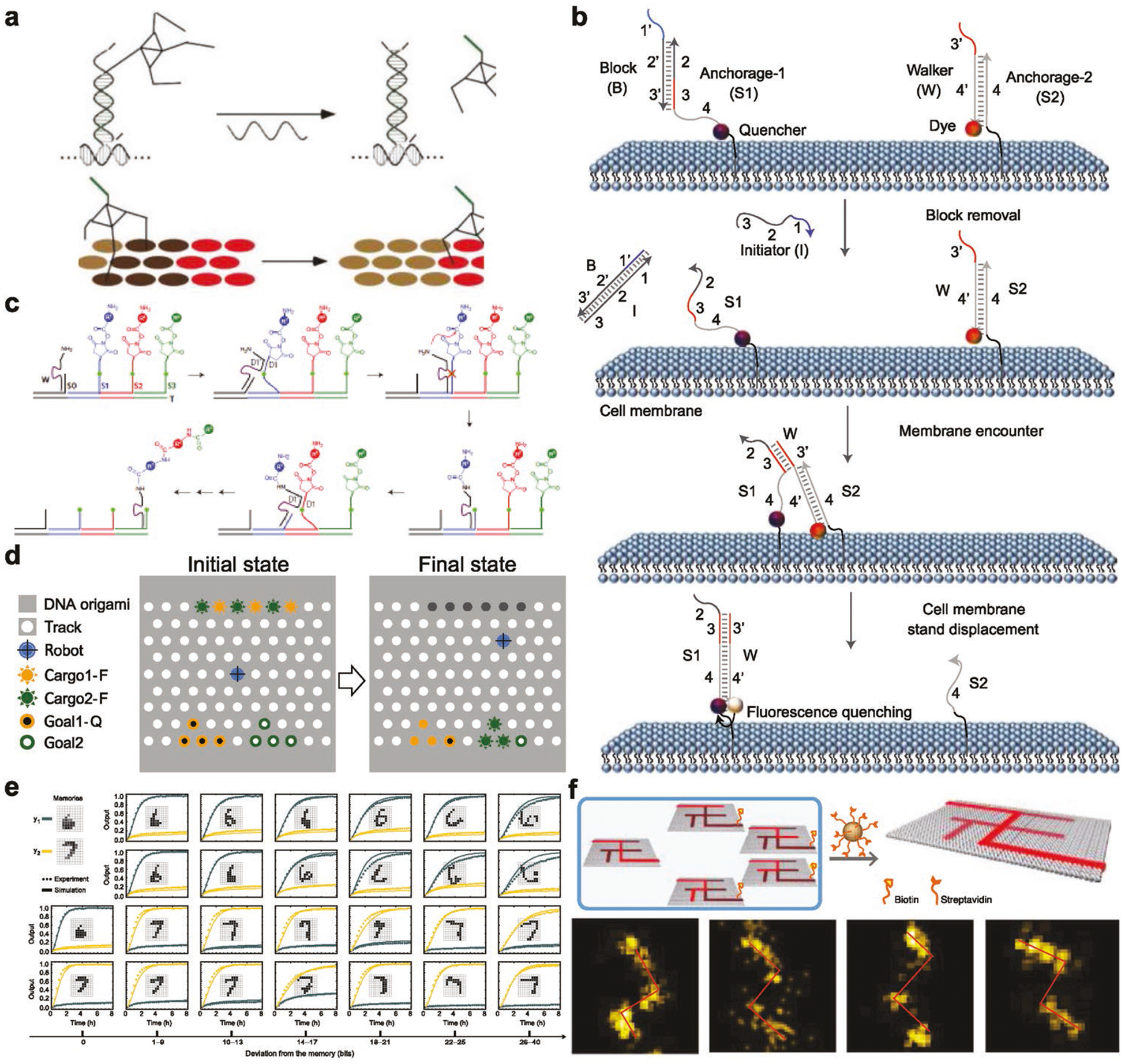
Programmable DNA walkers and circuits. a) A DNA walker that walks along a prescriptive landscape. Reproduced with permission.[62] Copyright 2010, Springer Nature. b) DNA probes for monitoring dynamic and transient molecular encounters on live cell membranes. Reproduced with permission.[63] Copyright 2017, Springer Nature. c) DNA walker enables precise and step-wise chemical synthesis. Reproduced with permission.[64] Copyright 2010, Springer Nature. d) A cargo-sorting robot that pick up and transport a prescribed cargo to a designated site on a DNA origami field. Reproduced with permission.[65] Copyright 2017, AAAS. e) DNA circuits that are trained to recognize molecular patterns of high accuracy. Reproduced with permission.[66] Copyright 2018, Springer Nature and f) a DNA navigator that is capable of maze solving. Reproduced with permission.[67] Copyright 2019, Springer Nature.
7. Conclusion and Outlook
Over the last several decades, dynamic DNA structures have been continuously evolving at a fast pace toward smart systems with unprecedented capability of conducting sophisticated tasks at the molecular level, both in vitro and in vivo. Though various stimulus response mechanisms have been developed, major challenges remain in controlling the performance of dynamic DNA structures. For instance, the response times of current systems are drastically slower than inherent fluctuation times or dynamic biological events, suggesting there is large room to improve the response rate of dynamic DNA structures. Second, the ability to achieve stepwise and continuous control over a wide range of states remains a grand challenge. New design strategies or external control mechanisms need to be developed in order to rival the complexity of biological systems. Integrating synthetic DNA structures with dynamic biological components might serve the purpose toward this direction. Lastly, the operation of dynamic DNA structures in vivo remains difficult. Though a few reports have shown that dynamic DNA structures have undergone reconfiguration in vivo, the complex physiological environment posts a hurdle on thoroughly investigating the performance of such systems since DNA structures face a series of challenges in vivo to drag their use, including nuclease degradation, protein opsonization, biological barriers, etc. Methods to minimize these interfering factors would largely benefit the in vivo applications of dynamic DNA structures.
Acknowledgements
Y.K. acknowledges NSF support under grant DMR-1654485 and grant ECCS-1807568, and NIH support under grant 1R21AI135753-01. Y.Z. thanks the support from Natural Science Fundation of China (grants 51672022 and 51302010).
Biographies

Yingwei Zhang is an associated professor at Beijing University of Chemical Technology, China. She obtained her BS and Ph.D. degrees in materials chemistry and inorganic chemistry from Jilin University, China. Her current research interests focus on DNA nanotechnology and its applications in biosensing and the programmable and controllable assembly of nano/mesoscale functional materials.

Pengfei Wang is a professor at the Institute of Molecular Medicine (IMM), Shanghai Jiao Tong University School of Medicine, China. He obtained his BS and MS degrees in materials science and engineering from Tianjin University, China, and his Ph.D. degree in biomedical engineering from Purdue University, USA. His research interests include nucleic acid nanotechnology and its applications in fabricating functional materials, disease diagnosis and therapy, and synthetic biology.

Yonggang Ke is an assistant professor at the Wallace H. Coulter Department of Biomedical Engineering at Emory University and Georgia Institute of Technology. He received his Ph.D. degree in chemistry from Arizona State University in 2009. He then worked as a postdoctoral fellow in the Department of Cancer Biology at Harvard University from 2009 to 2014. His current research focuses on programmable self-assembly of complex nanostructures and dynamic nanomachines using DNA and other biomolecules, functional nanomaterial self-assembly, drug delivery, and single-molecule biophysics.
Footnotes
Conflict of Interest
The authors declare no conflict of interest.
Contributor Information
Yingwei Zhang, Institute of Molecular Medicine, Renji Hospital, Shanghai Jiao Tong University School of Medicine, Shanghai 200127, China; State Key Laboratory of Chemical Resource Engineering, College of Materials Science and Engineering, Beijing University of Chemical Technology, Beijing 100029, China.
Victor Pan, Wallace H. Coulter Department of Biomedical Engineering, Georgia Institute of Technology and Emory University, Department of Chemistry, Emory University, Atlanta, GA 30322, USA.
Yonggang Ke, Wallace H. Coulter Department of Biomedical Engineering, Georgia Institute of Technology and Emory University, Department of Chemistry, Emory University, Atlanta, GA 30322, USA.
References
- [1].Watson JD, Crick FH, Nature 1953, 171, 737. [DOI] [PubMed] [Google Scholar]
- [2].Seeman NC, J. Theor. Biol 1982, 99, 237. [DOI] [PubMed] [Google Scholar]
- [3].Hong F, Zhang F, Liu Y, Yan H, Chem. Rev 2017, 117, 12584. [DOI] [PubMed] [Google Scholar]
- [4].Goodman RP, Berry RM, Turberfield AJ, Chem. Commun 2004, 0, 1372. [DOI] [PubMed] [Google Scholar]
- [5].Yan H, Park SH, Finkelstein G, Reif JH, LaBean TH, Science 2003, 301, 1882. [DOI] [PubMed] [Google Scholar]
- [6].Ke YG, Ong LL, Shih WM, Yin P, Science 2012, 338, 1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Wei B, Dai MJ, Yin P, Nature 2012, 485, 623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Rothemund PWK, Nature 2006, 440, 297. [DOI] [PubMed] [Google Scholar]
- [9].Winfree E, Liu F, Wenzler LA, Seeman NC, Nature 1998, 394, 539. [DOI] [PubMed] [Google Scholar]
- [10].He Y, Chen Y, Liu H, Ribbe AE, Mao C, J. Am. Chem. Soc 2005, 127, 12202. [DOI] [PubMed] [Google Scholar]
- [11].He Y, Tian Y, Ribbe AE, Mao C, J. Am. Chem. Soc 2006, 128, 15978. [DOI] [PubMed] [Google Scholar]
- [12].Zhang C, Su M, He Y, Zhao X, Fang PA, Ribbe AE, Jiang W, Mao C, Proc. Natl. Acad. Sci. USA 2008, 105, 10665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Zheng J, Birktoft JJ, Chen Y, Wang T, Sha R, Constantinou PE, Ginell SL, Mao C, Seeman NC, Nature 2009, 461, 74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].He Y, Ye T, Su M, Zhang C, Ribbe AE, Jiang W, Mao C, Nature 2008, 452, 198. [DOI] [PubMed] [Google Scholar]
- [15].Tian C, Li X, Liu Z, Jiang W, Wang G, Mao C, Angew. Chem., Int. Ed 2014, 53, 8041. [DOI] [PubMed] [Google Scholar]
- [16].Wang P, Wu S, Tian C, Yu G, Jiang W, Wang G, Mao C, J. Am. Chem. Soc 2016, 138, 13579. [DOI] [PubMed] [Google Scholar]
- [17].Zhang C, Ko SH, Su M, Leng Y, Ribbe AE, Jiang W, Mao C, J. Am. Chem. Soc 2009, 131, 1413. [DOI] [PubMed] [Google Scholar]
- [18].Andersen ES, Dong M, Nielsen MM, Jahn K, Subramani R, Mamdouh W, Golas MM, Sander B, Stark H, Cristiano LPO, Pedersen JS, Birkedal V, Besenbacher F, Gothelf KV, Kjems J, Nature 2009, 459, 73. [DOI] [PubMed] [Google Scholar]
- [19].Dietz H, Douglas SM, Shih WM, Science 2009, 325, 725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Castro CE, Kilchherr F, Kim DN, Shiao EL, Wauer T, Wortmann P, Bathe M, Dietz H, Nat. Methods 2011, 8, 221. [DOI] [PubMed] [Google Scholar]
- [21].Han D, Pal S, Nangreave J, Deng Z, Liu Y, Yan H, Science 2011, 332, 342. [DOI] [PubMed] [Google Scholar]
- [22].Zhang F, Jiang S, Wu S, Li Y, Liu Y, Mao C, Nat. Nanotechnol 2015, 10, 779. [DOI] [PubMed] [Google Scholar]
- [23].Liu W, Zhong H, Wang R, Seeman NC, Angew. Chem., Int. Ed 2011, 50, 264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Tikhomirov G, Petersen P, Qian L, Nature 2017, 552, 67. [DOI] [PubMed] [Google Scholar]
- [25].Wagenbauer KF, Sigl C, Dietz H, Nature 2017, 552, 78. [DOI] [PubMed] [Google Scholar]
- [26].Kuzyk A, Yang Y, Duan X, Stoll S, Govorov AO, Sugiyama H, Endo M, Liu N, Nat. Commun 2016, 7, 10591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Day HA, Pavlou P, Waller ZA, Bioorg. Med. Chem 2014, 22, 4407. [DOI] [PubMed] [Google Scholar]
- [28].Chen Y, Wang M, Mao C, Angew. Chem., Int. Ed 2004, 43, 3554. [DOI] [PubMed] [Google Scholar]
- [29].Yin P, Yan H, Daniell XG, Turberfield AJ, Reif JH, Angew. Chem., Int. Ed 2004, 43, 4906. [DOI] [PubMed] [Google Scholar]
- [30].Yurke B, Turberfield AJ, Mills AP Jr., Simmel FC, Neumann JL, Nature 2000, 406, 605. [DOI] [PubMed] [Google Scholar]
- [31].Shangguan D, Li Y, Tang Z, Cao ZC, Chen HW, Mallikaratchy P, Sefah K, Yang CJ, Tan W, Proc. Natl. Acad. Sci. USA 2006, 103, 11838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Henderson E, Hardin CC, Walk SK, Tinoco I Jr., Blackburn EH, Cell 1987, 51, 899. [DOI] [PubMed] [Google Scholar]
- [33].Zeraati M, Langley DB, Schofield P, Moye AL, Rouet R, Hughes WE, Bryan TM, Dinger ME, Christ D, Nat. Chem 2018, 10, 631. [DOI] [PubMed] [Google Scholar]
- [34].Gerling T, Wagenbauer KF, Neuner AM, Dietz H, Science 2015, 347, 1446. [DOI] [PubMed] [Google Scholar]
- [35].Mao C, Sun W, Shen Z, Seeman NC, Nature 1999, 397, 144. [DOI] [PubMed] [Google Scholar]
- [36].Song J, Li Z, Wang P, Meyer T, Mao C, Ke Y, Science 2017, 357, eaan3377. [DOI] [PubMed] [Google Scholar]
- [37].Kopperger E, List J, Madhira S, Rothfischer F, Lamb DC, Simmel FC, Science 2018, 359, 296. [DOI] [PubMed] [Google Scholar]
- [38].Lauback S, Mattioli KR, Marras AE, Armstrong M, Rudibaugh TP, Sooryakumar R, Castro CE, Nat. Commun 2018, 9, 1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Turek VA, Chikkaraddy R, Cormier S, Stockham B, Ding T, Keyser UF, Baumberg JJ, Adv. Funct. Mater 2018, 28, 1706410. [Google Scholar]
- [40].Cangialosi A, Yoon C, Liu J, Huang Q, Guo J, Nguyen TD, Gracias DH, Schulman R, Science 2017, 357, 1126. [DOI] [PubMed] [Google Scholar]
- [41].Fern J, Schulman R, Nat. Commun 2018, 9, 3766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Zhan P, Dutta PK, Wang P, Song G, Dai M, Zhao SX, Wang ZG, Yin P, Zhang W, Ding B, Ke Y, ACS Nano 2017, 11, 1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Liu M, Fu J, Hejesen C, Yang Y, Woodbury NW, Gothelf K, Liu Y, Yan H, Nat. Commun 2013, 4, 2127. [DOI] [PubMed] [Google Scholar]
- [44].Hernández-Ainsa S, Bell NAW, Thacker VV, Göpfrich K, Misiunas K, Fuentes-Perez ME, Moreno-Herrero F, Keyser UF, ACS Nano 2013, 7, 6024. [DOI] [PubMed] [Google Scholar]
- [45].Langecker M, Arnaut V, Martin TG, List J, Renner S, Mayer M, Dietz H, Simmel FC, Science 2012, 338, 932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Hernández-Ainsa S, Misiunas K, Thacker VV, Hemmig EA, Keyser UF, Nano Lett. 2014, 14, 1270. [DOI] [PubMed] [Google Scholar]
- [47].Modi S, Swetha MG, Goswami D, Gupta GD, Mayor S, Krishnan Y, Nat. Nanotechnol 2009, 4, 325. [DOI] [PubMed] [Google Scholar]
- [48].Leung K, Chakraborty K, Saminathan A, Krishnan Y, Nat. Nanotechnol 2019, 14, 176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Narayanaswamy N, Chakraborty K, Saminathan A, Zeichner E, Leung K, Devany J, Krishnan Y, Nat. Methods 2019, 16, 95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Zhao J, Gao J, Xue W, Di Z, Xing H, Lu Y, Li L, J. Am. Chem. Soc 2018, 140, 578. [DOI] [PubMed] [Google Scholar]
- [51].Funck T, Nicoli F, Kuzyk A, Liedl T, Angew. Chem., Int. Ed 2018, 57, 13495. [DOI] [PubMed] [Google Scholar]
- [52].Ke Y, Meyer T, Shih WM, Bellot G, Nat. Commun 2016, 7, 10935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Hemmig EA, Fitzgerald C, Maffeo C, Hecker L, Ochmann SE, Aksimentiev A, Tinnefeld P, Keyser UF, Nano Lett. 2018, 18, 1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Jungmann R, Avendano MS, Woehrstein JB, Dai M, Shih WM, Yin P, Nat. Methods 2014, 11, 313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Ohmann A, Li CY, Maffeo C, Nahas AK, Baumann KN, Gopfrich K, Yoo J, Keyser UF, Aksimentiev A, Nat. Commun 2018, 9, 2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Jiang Q, Liu S, Liu J, Wang ZG, Ding B, Adv. Mater 2018, 1804785. [DOI] [PubMed] [Google Scholar]
- [57].Rahman MA, Wang P, Zhao Z, Wang D, Nannapaneni S, Zhang C, Chen Z, Griffith CC, Hurwitz SJ, Chen ZG, Ke Y, Shin DM, Angew. Chem., Int. Ed 2017, 56, 16023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Wang P, Rahman MA, Zhao Z, Weiss K, Zhang C, Chen Z, Hurwitz SJ, Chen ZG, Shin DM, Ke Y, J. Am. Chem. Soc 2018, 140, 2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Douglas SM, Bachelet I, Church GM, Science 2012, 335, 831. [DOI] [PubMed] [Google Scholar]
- [60].Ma Y, Wang Z, Ma Y, Han Z, Zhang M, Chen H, Gu Y, Angew. Chem., Int. Ed 2018, 57, 5389. [DOI] [PubMed] [Google Scholar]
- [61].Li S, Jiang Q, Liu S, Zhang Y, Tian Y, Song C, Wang J, Zou Y, Anderson GJ, Han JY, Chang Y, Liu Y, Zhang C, Chen L, Zhou G, Nie G, Yan H, Ding B, Zhao Y, Nat. Biotechnol 2018, 36, 258. [DOI] [PubMed] [Google Scholar]
- [62].Lund K, Manzo AJ, Dabby N, Michelotti N, Johnson-Buck A, Nangreave J, Taylor S, Pei R, Stojanovic MN, Walter NG, Winfree E, Yan H, Nature 2010, 465, 206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].You M, Lyu Y, Han D, Qiu L, Liu Q, Chen T, Wu SC, Peng L, Zhang L, Bao G, Tan W, Nat. Nanotechnol 2017, 12, 453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].He Y, Liu DR, Nat. Nanotechnol 2010, 5, 778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Thubagere AJ, Li W, Johnson RF, Chen Z, Doroudi S, Lee YL, Izatt G, Wittman S, Srinivas N, Woods D, Winfree E, Qian L, Science 2017, 357, eaan6558. [DOI] [PubMed] [Google Scholar]
- [66].Cherry KM, Qian L, Nature 2018, 559, 370. [DOI] [PubMed] [Google Scholar]
- [67].Chao J, Wang J, Wang F, Ouyang X, Kopperger E, Liu H, Li Q, Shi J, Wang L, Hu J, Wang L, Huang W, Simmel FC, Fan C, Nat. Mater 2019, 18, 273. [DOI] [PubMed] [Google Scholar]


