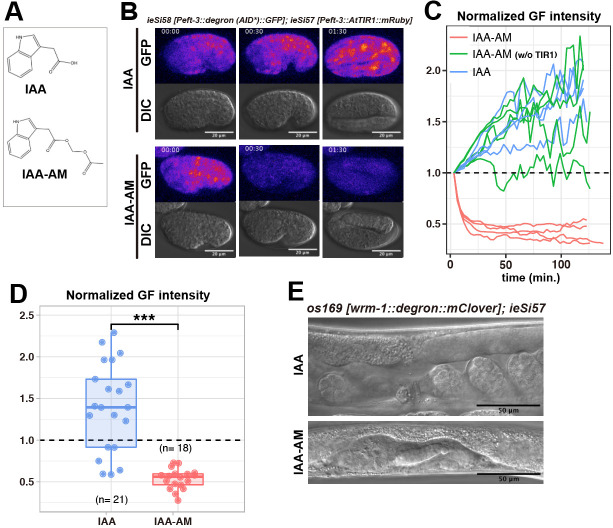
Figure 1.
(A) Chemical structural formula of indole-3-acetic acid (IAA) and acetoxymethyl indole-3-acetic acid (IAA-AM). (B) Frames from time lapse videos of ieSi58 [Peft-3::degron (AID*)::GFP]; ieSi57 [Peft–3::AtTIR1::mRuby] laid embryos in IAA (1 mM) or IAA-AM (1 mM); elapsed time (hours:minutes) from the beginning of recording is indicated. GFP panels show maximum intensity projection images, and green fluorescent signal is colored with “Fire” look-up-table. Scale bar, 20 μm. (C) Plots of green fluorescent (GF) intensity in timelapse observation. Red lines: IAA-AM treatment of ieSi58 ieSi57 embryos, green lines: IAA-AM treatment of ieSi58 embryos (no TIR1 present), blue lines: IAA treatment of ieSi58 ieSi57 embryos. Mean values of GF intensity in eggshells were obtained every three minutes, and normalized by the intensity at 0 min. (D) Comparison of GF depletion between IAA and IAA-AM treatment of ieSi58; ieSi57 embryos. Mean values of GF intensity within eggshells were obtained after each treatment (two hours), and normalized with that of the beginning, *** indicates p < 0.01, p= 1.6 x 10-8 (Wilcoxon rank sum test). (E) Representative images of os169 ieSi57 adults, which are IAA or IAA-AM treated during embryos development (two hours).
Description
The targeted protein degradation systems in which a protein accompanying with specific tags can be degraded are developed as an approach of conditional loss of function analyses (Natsume and Kanemaki 2017). The insertion of tags into the gene loci by the CRISPR/Cas9 system allows us to deplete endogenous proteins in stage and cell specific manners. So far, four systems can be used to deplete the tagged proteins in C . elegans (Armenti et al. 2014; Zhang et al. 2015; Wang et al. 2017; Wu et al. 2017). The auxin-inducible degradation (AID) system can degrade the tagged protein by administration of the phytohormone auxin. In the AID system, a plant-derived TIR1 F-box protein can form an E3 ubiquitin ligase complex with the endogenous Skp and Cullin proteins, and in the presence of auxin, the complex interacts with a degron tag derived from the IAA17 transcriptional repressor (Nishimura et al. 2009; Yesbolatova et al. 2019). Consequently, the degron-tagged proteins are polyubiquitinated for degradation by the proteasome. With the controlled expression of TIR1 and administration of auxin, the AID system can allow us to perform spaciotemporal protein depletion. Although this system can be a powerful tool for conditional loss of function analyses, it appears to be difficult to degrade proteins in embryos, especially at late embryonic stages, since efficiencies of degradation in laid embryos surrounded by the eggshell is low compared to those in hatched larvae or adults (Zhang et al. 2015).
Here, we report that acetoxymethyl indole-3-acetic acid (IAA-AM), a cell permeable analog of natural auxin IAA (Figure1A), is more effective in triggering the degradation of tagged proteins in embryos. To test the efficiency of protein degradation, we used the previously reported strain ubiquitously expressing both a 44-amino acid (-aa) degron derived from the Arabidopsis IAA17 protein (known as AID*) fused with GFP and Arabidopsis TIR1 (AtTIR1) fused with mRuby (ieSi58 [Peft-3::degron (AID*)::GFP] and ieSi57 [Peft–3::AtTIR1::mRuby])(Morawska and Ulrich 2013; Zhang et al. 2015). When we put laid embryos into the drop of M9 buffer containing 1 mM IAA-AM (AM form of IAA), we observed rapid decrease of green fluorescence (GF) intensity (Figure1B, C and D). In contrast to IAA-AM, the embryos treated with IAA showed increased GF intensity due to elevated expression of degron-tagged GFP (Figure1B, C and D). We also confirmed that IAA-AM did not cause the decreased GF intensity without the AtTIR1 transgene (Figure1C).
Next, we tried to deplete the endogenous protein in laid embryos. WRM-1 is the C. elegans homolog of β-catenin and is essential for asymmetry of most cell divisions (Mizumoto and Sawa 2007). Since WRM-1 is required for asymmetric division of EMS in the 4-cell stage embryo (Rocheleau et al. 1997), conditional knockdown approaches are required for the investigation of its function in later stages. To test whether the AID system with IAA-AM allows us to perform conditional knockdown of wrm-1 in embryos, we inserted a fusion tag composed of the mini-auxin inducible degron (mAID), a 68-aa degron derived from the Arabidopsis IAA17 protein that showed similar degradation efficiency with that of AID* (Li et al. 2019), and mClover at the 3’ end of wrm-1 endogenous locus by the CRISPR/Cas9 system (Natsume et al 2016) (Dickinson et al. 2015). We put laid embryos of the strain carrying wrm-1::mAID::mClover (os169) and Peft-3::AtTIR1::mRuby (ieSi57) into drops of 1 mM IAA-AM or IAA (both with 0.2% DMSO in M9 buffer) for two hours. After washing with M9 buffer two times, we placed embryos on NGM plates without the inducing ligand. At the young adult stage, we scored elongation of gonadal arms guided by the distal tip cells (DTCs) whose production requires wrm-1-regulated asymmetric divisions (Siegfried et al. 2004). When we treated os169 ieSi57 embryos with 1 mM IAA-AM, all animals (13/13) shows no gonadal extension. In contrast, treating with 1 mM IAA caused this phenotype only in 2/17 animals (15/17 showed normal gonad) (Figure 1E). In addition, all os169 ieSi57 embryos treated with 0.2% DMSO (16/16) or ieSi58 ieSi57 embryos treated with 1mM IAA-AM (19/19) produced animals with normal gonads. These results show that the AM modification of IAA induces efficient protein degradation in laid embryos with the AID system. Since the AM modification is widely used to improve cell membrane permeability (Schultz 2003), IAA-AM is likely to penetrate into the eggshell more easily than IAA. After penetration into cells, endogenous cytoplasmic esterases are known to cleave the AM bond, making the compound again impermeant to cell membrane (Schultz 2003). We note that the chemical cleavage reaction of the AM bond produces formaldehyde (Schultz 2003), which may be toxic to cells at high concentrations. In this study, however, we found the application of IAA-AM at a concentration of 1 mM for two hours allowed normal development including gonad elongation. Our data presented here show IAA-AM will broaden the possible applications of the AID system for conditional loss of function analyses in C. elegans.
Reagents
Plasmid construction: For insertion of degron tag and mClover into 3’ end of wrm-1 locus by CRISPR/Cas9 system, the fragment of mAID::mClover was amplified with the primers (F: 5’-GCCTCAGGAGCATCGGGATCCGGTGCAGGCGCCAAGG-3’, R: 5’- AAAGTACAGATTCTCCTTGTACAGCTCGTCCATGCCA-3’) from pMK290 (Addgene: #72828). The fragment of self-excising cassette (SEC) described in Dickinson et al., 2015 was amplified with the primers (F: 5’-CGATGCTCCTGAGGCTCCCGAT-3’, R: 5’- GAGAATCTGTACTTTCAATCCG-3’) from pDD287 (Addgene #70685). These fragments were assembled by InFusion HD (Takara Clontech). Then, 5’ and 3’ wrm-1 homology arms were amplified from C. elegans genome with the following primers:
wrm-1RightL: 5’-AGCGAGGAAGACTTGTGAATGAATCTTTGTGCGGGTA-3’,
wrm-1RightR: 5’-CTATGACCATGTTATAACTGGTGGTGATCGTGCTTGG-3’
wrm-1LeftL: 5’-AACGACGGCCAGTCGTTTTGTTGAATGCAAATATGTG-3’
wrm-1LeftR: 5’-GGATCCCGATGCTCcCATTAGTTGTCGATGATGCTGC-3’.
Both fragments of homology arms were assembled into the backbone and SEC fragments amplified from the above-described construct with the primers (backbone-F: 5’-CGACTGGCCGTCGTTTTACAAC-3’, backbone-R: 5’- ATAACATGGTCATAGCTGTTTC-3’ and SEC-F: 5’-GGAGCATCGGGATCCGGTGCAG-3’, SEC-R: 5’-CAAGTCTTCCTCGCTGATCAAC-3’) by InFusion HD (Takara Clontech). The vector expressing the guide RNA for wrm-1 was constructed with the primers including the target sequence (F: 5′-[ATGTGAATGAATCTTTGTGC]GTTTTAGAGCTAGAAATAGCAA-3′, R: 5′-AAGATTCATTCACATAAACATTTAGATTTGCAATTCA-3′, brackets indicate the target sequence) by inverse PCR with PU6::unc-119_sgRNA (Addgene #46469).
Strain construction: All C. elegans strains were cultured by standard methods. Plasmids (50 ng/µl mAID::mClover with wrm-1 homology arms construct, 50 ng/µl wrm-1 guide RNA construct, 50 ng/µl sur-5::GFP co-injection marker and Cas9 expression construct (pDD162, Addgene #47549)) were microinjected into N2. After picking an animal displaying a roller phenotype, self-excising cassette was excised as described (Dickinson et al. 2015). Then, we crossed this strain to the Peft-3::AtTIR1::mRuby strain.
Observation: In time lapse observation, embryos were mounted on slides with 20 μm diameter polystyrene beads (Polyscience #18329) diluted 1:30 in M9 buffer containing 1 mM IAA or IAA-AM (stocks 500 mM in DMSO)(Bao and Murray 2011). In the endogenous protein degradation experiments, laid embryos were put in drops of M9 buffer containing 0.2% DMSO, 1 mM IAA or IAA-AM for two hours, and transferred into M9 buffer two times before hatching. Washed embryos were allowed to develop on new NGM plates. All imaging was performed with Zeiss LSM700 microscope using a 63x N.A. 1.40 oil-immersion objective. Acquired images were processed by Fiji (Schindelin et al. 2012).
Strains:
CA1202: ieSi57 (Peft-3::AtTIR1::mRuby) II ; ieSi58 (Peft-3::degron (AID*)::GFP) IV, CGC
HS3280: ieSi58 (Peft-3::degron (AID*)::GFP) IV , this study
HS3176: ieSi57 (Peft-3::AtTIR1::mRuby) II; wrm-1(os169 [wrm-1::mAID::mClover]) III, this study
IAA and IAA-AM:
IAA (indole-3-acetic acid) was purchased from Nacalai tesque (#19119-61). IAA-AM was custom synthesized by Tokyo Chemical Industry Co., Ltd. Both ligands were dissolved in DMSO to make a 500 mM stock solution before storing at -20˚C.
Acknowledgments
Funding
This work was supported by Ministry of Education, Culture, Sports, Science, and Technology of Japan Grants-in Aid for Scientific Research to MTK (18H02170) and HS (16KT0078).
References
- Armenti ST, Lohmer LL, Sherwood DR, Nance J. Repurposing an endogenous degradation system for rapid and targeted depletion of C. elegans proteins. Development. 2014 Nov 01;141(23):4640–4647. doi: 10.1242/dev.115048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Echteld CJ, Kirkels JH, Eijgelshoven MH, van der Meer P, Ruigrok TJ. Intracellular sodium during ischemia and calcium-free perfusion: a 23Na NMR study. J Mol Cell Cardiol. 1991 Mar 01;23(3):297–307. doi: 10.1016/0022-2828(91)90066-u. [DOI] [PubMed] [Google Scholar]
- Dickinson DJ, Pani AM, Heppert JK, Higgins CD, Goldstein B. Streamlined Genome Engineering with a Self-Excising Drug Selection Cassette. Genetics. 2015 Jun 01;200(4):1035–1049. doi: 10.1534/genetics.115.178335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Prasanna X, Salo VT, Vattulainen I, Ikonen E. An efficient auxin-inducible degron system with low basal degradation in human cells. Nat Methods. 2019 Aug 26;16(9):866–869. doi: 10.1038/s41592-019-0512-x. [DOI] [PubMed] [Google Scholar]
- Mizumoto K, Sawa H. Two betas or not two betas: regulation of asymmetric division by beta-catenin. Trends Cell Biol. 2007 Oct 01;17(10):465–473. doi: 10.1016/j.tcb.2007.08.004. [DOI] [PubMed] [Google Scholar]
- Morawska M, Ulrich HD. An expanded tool kit for the auxin-inducible degron system in budding yeast. Yeast. 2013 Jul 23;30(9):341–351. doi: 10.1002/yea.2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natsume T, Kanemaki MT. Conditional Degrons for Controlling Protein Expression at the Protein Level. Annu Rev Genet. 2017 Nov 27;51:83–8102. doi: 10.1146/annurev-genet-120116-024656. [DOI] [PubMed] [Google Scholar]
- Natsume T, Kiyomitsu T, Saga Y, Kanemaki MT. Rapid Protein Depletion in Human Cells by Auxin-Inducible Degron Tagging with Short Homology Donors. Cell Rep. 2016 Mar 24;15(1):210–218. doi: 10.1016/j.celrep.2016.03.001. [DOI] [PubMed] [Google Scholar]
- Nishimura K, Fukagawa T, Takisawa H, Kakimoto T, Kanemaki M. An auxin-based degron system for the rapid depletion of proteins in nonplant cells. Nat Methods. 2009 Nov 15;6(12):917–922. doi: 10.1038/nmeth.1401. [DOI] [PubMed] [Google Scholar]
- Rocheleau CE, Downs WD, Lin R, Wittmann C, Bei Y, Cha YH, Ali M, Priess JR, Mello CC. Wnt signaling and an APC-related gene specify endoderm in early C. elegans embryos. Cell. 1997 Aug 22;90(4):707–716. doi: 10.1016/s0092-8674(00)80531-0. [DOI] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez JY, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012 Jun 28;9(7):676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz C. Prodrugs of biologically active phosphate esters. Bioorg Med Chem. 2003 Mar 20;11(6):885–898. doi: 10.1016/s0968-0896(02)00552-7. [DOI] [PubMed] [Google Scholar]
- Siegfried KR, Kidd AR 3rd, Chesney MA, Kimble J. The sys-1 and sys-3 genes cooperate with Wnt signaling to establish the proximal-distal axis of the Caenorhabditis elegans gonad. Genetics. 2004 Jan 01;166(1):171–186. doi: 10.1534/genetics.166.1.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Tang NH, Lara-Gonzalez P, Zhao Z, Cheerambathur DK, Prevo B, Chisholm AD, Desai A, Oegema K. A toolkit for GFP-mediated tissue-specific protein degradation in C. elegans. Development. 2017 Jun 15;144(14):2694–2701. doi: 10.1242/dev.150094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Q, Ploegh HL, Truttmann MC. Hepta-Mutant Staphylococcus aureus Sortase A (SrtA7m) as a Tool for in Vivo Protein Labeling in Caenorhabditis elegans. ACS Chem Biol. 2017 Jan 18;12(3):664–673. doi: 10.1021/acschembio.6b00998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yesbolatova A, Natsume T, Hayashi KI, Kanemaki MT. Generation of conditional auxin-inducible degron (AID) cells and tight control of degron-fused proteins using the degradation inhibitor auxinole. Methods. 2019 Apr 24;164-165:73–80. doi: 10.1016/j.ymeth.2019.04.010. [DOI] [PubMed] [Google Scholar]
- Zhang L, Ward JD, Cheng Z, Dernburg AF. The auxin-inducible degradation (AID) system enables versatile conditional protein depletion in C. elegans. Development. 2015 Nov 01;142(24):4374–4384. doi: 10.1242/dev.129635. [DOI] [PMC free article] [PubMed] [Google Scholar]