Figure 1.
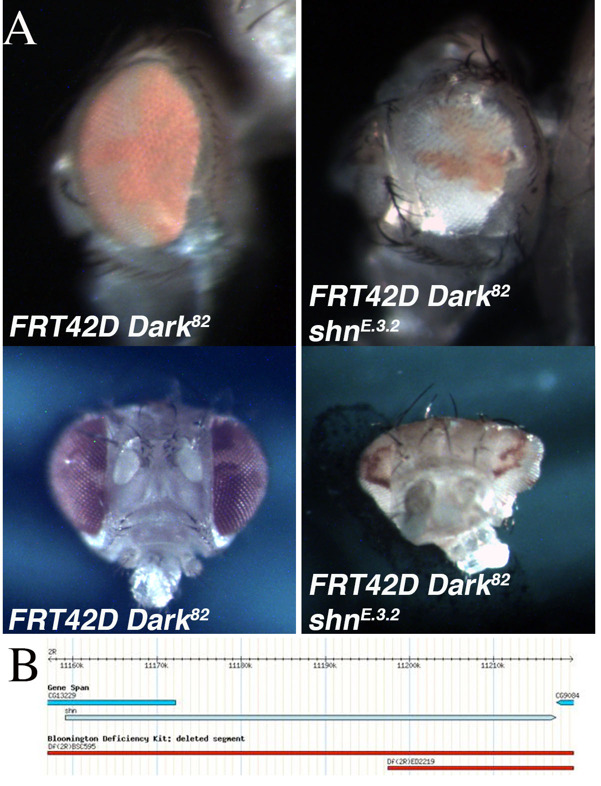
A. Mosaic control (FRT42D Dark82) and mosaic mutant E.3.2 (FRT42D Dark82shnE.3.2) eyes. In both genotypes, homozygous mutant eye tissue is pigmented (w+mC). The top panels are representative images of the lateral view and bottom panels the medial view. B. Genomic region of chromosome 2R in which mutant E.3.2 failed to complement with deficiency mapping (2R:11,186,047..11,665,391). The shn gene falls within this region. B. Image adapted from flybase.org (Gramates et al., 2017).
Description
An EMS screen was conducted utilizing the Flp/FRT system to identify mutations that lead to phenotypic alterations in the size of the eye, the ratio of mutant to wild type tissue (red over white), or the developmental patterning of the mosaic eye. This screen was completed in the genetic background of blocked apoptosis in the homozygous mutant cells to identify conditional regulators of cell growth and eye development (Kagey et al., 2012). The block in apoptosis in the mosaic mutant tissue was achieved by using the FRT42D Dark82chromosome, which retains the w+mC(pigmentation), as a starting point for the EMS mutagenesis (Akdemir et al., 2006). One of the mutants identified was mutant E.3.2. The mutant mosaic phenotype, generated from the cross FRT42D Dark82 E.3.2 XEy>Flp; FRT42D, resulted in a range of phenotypes. This included, gross eye pattern disruption, abnormal shape, and antennal overgrowth (see bottom image) when compared to the FRT42D Dark82 mosaic controls (Figure 1A). The control mosaic phenotype had a characteristic 60:40 red:white ratio as compared to the mutant mosaic phenotype of approximately 30:70 red:white ratio. This indicates a reduction in mutant tissue, but included abnormal cranial, eye, and antennal development (Figure 1A).
The genetic mapping of the location of E.3.2 on 2R was completed by three independent groups of undergraduate researchers at Nevada State College, Albion College, and Ohio Northern University as part of the Fly-CURE consortium (Bieser et al., 2018, Stamm et al., 2019). Virgin females from the FRT42D E.3.2 Dark82/CyO stock were mated in series to male flies from the 87 deficiency stocks that comprise the Bloomington Stock Center 2R Deficiency Kit (only stocks distal to the FRT42D site were used for mapping) (Cook et al., 2012). Mutant E.3.2 failed to complement deficiencies Df(2R)BSC595 (2R:10,385,967..11,288,578), Df(2R)ED2155 (2R:10,894,096..11,397,442), and Df(2R)ED2219 (2R:11,197,412..11,665,391), while complementing the flanking deficiency Df(2R)Exel6059 (2R:10,874,385..11,186,047). This resulted in the genomic region of 2R:11,186,047..11,665,391 failing to complement (Figure 1B). A lethal allele for the candidate gene Schnurri, shn[1],was crossed independently to E.3.2 to test for complementation. E.3.2 failed to complement a previously defined strong hypomorphic allele, shn[1] (Horsfield et al., 1998), indicating that E.3.2 is a novel shn allele, shnE.3.2. Shn is a zinc-finger transcription factor known to act in complex with Mothers Against Dpp (mad) in response to the BMP-related Decapentaplegic (Dpp) signaling pathway (Dai et al., 2000; Udagawa et al., 2000). Dpp, a member of the TGFβ superfamily, is a Drosophila morphogen critical for directing early embryonic patterning and regulating cell growth. Dpp mutants have shown that Dpp signaling is necessary for proper patterning in the eye-antenna disc in the L2 and L3 wandering larval stages of Drosophila development (Won et al., 2015). The current work is in agreement with previous findings and indicate that perturbations of shn, a downstream target of Dpp, is involved in regulation of cell growth and developmental patterning in vivo.
Reagents
FRT42D Dark82/CyO (Akdemir et al., 2006)
FRT42D Dark82 shnE.3.2/CyO (this manuscript)
Ey>Flp; FRT42D (BDSC 5616)
Bloomington Drosophila Stock Center 2R Deficiency Kit (Cook et al., 2012)
w1118;Df(2R)BSC595/CyO (BDSC 25428)
w1118;Df(2R)ED2219,P{w[+mW.Scer\FRT.hs3]=3’.RS5+3.3’}ED2219/Sm6a (BDSC 8910)
w1118;Df(2R)ED2155,P{w[+mW.Scer\FRT.hs3]=3’.RS5+3.3’}ED2155/SM6a (BDSC 9344)
w1118;Df(2R)Exel6059,P{w[+mC]=XP-U}Exel6059/CyO (BDSC 7541)
cn[1] shn[1] bw[1] sp[1]/CyO (BDSC 3008)
Acknowledgments
Acknowledgments
Stocks obtained from the Bloomington Drosophila Stock Center (NIH P40OD018537) were used in this study.
Funding
None
References
- Akdemir F, Farkas R, Chen P, Juhasz G, Medved'ová L, Sass M, Wang L, Wang X, Chittaranjan S, Gorski SM, Rodriguez A, Abrams JM. Autophagy occurs upstream or parallel to the apoptosome during histolytic cell death. Development. 2006 Mar 15;133(8):1457–1465. doi: 10.1242/dev.02332. [DOI] [PubMed] [Google Scholar]
- Bieser K, Stamm J, Aldo A, Bhaskara S, Clairborne M, Coronel Gómez J, Dean R, Dowell A, Dowell E, Eissa M, Fawaz A, Fouad-Meshriky M, Godoy D, Gonzalez K, Hachem M, Hammoud M, Huffman A, Ingram H, Jackman A, Karki B, Khalil N, Khalil H, Ha TK, Kharel A, Kobylarz I, Lomprey H, Lonnberg A, Mahbuba S, Massarani H, Minster M, Molina K, Molitor L, Murray T, Patel P, Pechulis S, Raja A, Rastegari G, Reeves S, Sabu N, Salazar R, Schulert D, Senopole M, Sportiello K, Torres C, Villalobos J, Wu J, Zeigler S, Kagey J. The mapping of Drosophila melanogaster mutant A.4.4. MicroPubl Biol. 2018 Dec 17;2018 doi: 10.17912/micropub.biology.000069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook RK, Christensen SJ, Deal JA, Coburn RA, Deal ME, Gresens JM, Kaufman TC, Cook KR. The generation of chromosomal deletions to provide extensive coverage and subdivision of the Drosophila melanogaster genome. Genome Biol. 2012;13(3):R21–R21. doi: 10.1186/gb-2012-13-3-r21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai H, Hogan C, Gopalakrishnan B, Torres-Vazquez J, Nguyen M, Park S, Raftery LA, Warrior R, Arora K. The zinc finger protein schnurri acts as a Smad partner in mediating the transcriptional response to decapentaplegic. Dev Biol. 2000 Nov 15;227(2):373–387. doi: 10.1006/dbio.2000.9901. [DOI] [PubMed] [Google Scholar]
- Gramates LS, Marygold SJ, Santos GD, Urbano JM, Antonazzo G, Matthews BB, Rey AJ, Tabone CJ, Crosby MA, Emmert DB, Falls K, Goodman JL, Hu Y, Ponting L, Schroeder AJ, Strelets VB, Thurmond J, Zhou P, the FlyBase Consortium. FlyBase at 25: looking to the future. Nucleic Acids Res. 2016 Oct 30;45(D1):D663–D671. doi: 10.1093/nar/gkw1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horsfield J, Penton A, Secombe J, Hoffman FM, Richardson H. decapentaplegic is required for arrest in G1 phase during Drosophila eye development. Development. 1998 Dec 01;125(24):5069–5078. doi: 10.1242/dev.125.24.5069. [DOI] [PubMed] [Google Scholar]
- Kagey JD, Brown JA, Moberg KH. Regulation of Yorkie activity in Drosophila imaginal discs by the Hedgehog receptor gene patched. Mech Dev. 2012 Jun 15;129(9-12):339–349. doi: 10.1016/j.mod.2012.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamm Joyce, Joshi Gnanda S, Anderson MA, Bussing Katie, Houchin Colton, Elinsky Amber C, Flyte Jacob T, Husseini Nadine, Jarosz Dominika, Johnson Chelsea L, Johnson Abby F, Jones Christina E, Kooner Taj P, Myhre Daniel, Rafaill Thomas N, Sayed Sarah, Swan Kirby W, Toma Jonathan, Kagey Jacob D. Genetic mapping of EgfrL.3.1 in Drosophila melanogaster. microPublication Biology. 2019 Apr 26; doi: 10.17912/micropub.biology.000098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udagawa Y, Hanai J, Tada K, Grieder NC, Momoeda M, Taketani Y, Affolter M, Kawabata M, Miyazono K. Schnurri interacts with Mad in a Dpp-dependent manner. Genes Cells. 2000 May 01;5(5):359–369. doi: 10.1046/j.1365-2443.2000.00328.x. [DOI] [PubMed] [Google Scholar]
- Won JH, Tsogtbaatar O, Son W, Singh A, Choi KW, Cho KO. Cell type-specific responses to wingless, hedgehog and decapentaplegic are essential for patterning early eye-antenna disc in Drosophila. PLoS One. 2015 Apr 01;10(4):e0121999–e0121999. doi: 10.1371/journal.pone.0121999. [DOI] [PMC free article] [PubMed] [Google Scholar]


