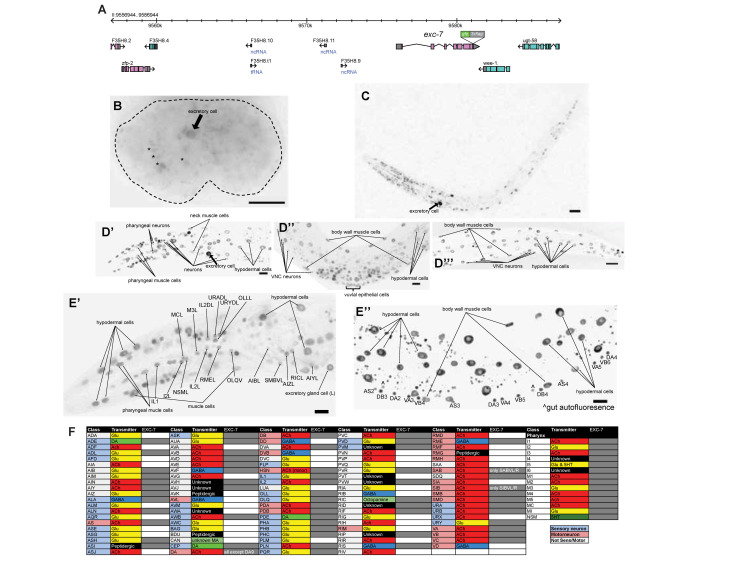
Figure 1.
Expression pattern of exc-7::gfp in the OH16020 strain. exc-7 was endogenously tagged at its genomic locus with a gfp::3xflag tag (A). Embryonic expression (B) was observed in the excretory canal cell (arrow) and other unidentified cells (*). Broad expression was observed in L1 (C) and young adult head (D’), mid body (D’’), and tail (D’’’), including the excretory canal cell at both stages (arrow). In young adult, individual neurons expressing exc-7 were identified in the head (E’) and in the ventral nerve cord (E’’). All exc-7 expressing neurons are listed in panel (F) (grey box). Scale bar, 10 µm.
Description
We are interested in identifying genes that are expressed in a panneuronal manner throughout the nervous system (Stefanakis et al., 2015). The Drosophila elav locus is a panneuronally expressed RNA binding protein (Campos et al., 1987; Robinow and White, 1988). Elav protein staining is routinely used in Drosophila to identify neurons and cis-regulatory control regions from the elav locus are routinely used as panneuronal Gal4 drivers (Berger et al., 2007; Luo et al., 1994; O’Neill et al., 1994; Osterwalder et al., 2001; Robinow and White, 1991). Based on sequence homology, the C. elegans exc-7 locus is the sole C. elegans orthologue of elav (Fujita et al., 2003; Fujita et al., 1999; Loria et al., 2003; Samson, 2008). Previous expression pattern analyses have shown that exc-7 is expressed only in a subset of neurons of the nervous system (the expressing neurons were mostly unidentified) (Fujita et al., 1999; Loria et al., 2003). However, reporter gene constructs previously used to infer exc-7 expression did not contain all intergenic region of the large exc-7 locus. Therefore, the possibility remained that through the use of more distal cis-regulatory elements, C. elegans exc-7 could also be panneuronally expressed, like its fly orthologue. To address this possibility, we tagged endogenous exc-7 with gfp::3xflag at its C-terminus using CRISPR/Cas9 genome engineering (Dokshin et al., 2018) and examined its expression. We cloned gfp from the che-1(ot856[che-1::gfp]) allele (Leyva-Diaz and Hobert, 2019), inserted it into the pMiniT 2.0 vector (NEB), and used that resulting plasmid for subsequent cloning of the gfp tag.
Embryonic expression of exc-7 was first observed at the bean stage. By reverse lineaging with use of SIMI-Biocell software (Schnabel et al., 1997), we confirm the identity of one of the expressing cells at this stage as the excretory canal cell (Fig. 1B, arrow). In L1 animals, broad expression in the head, ventral nerve cord (VNC), and tail was observed (Fig. 1C). In young adults, expression is notably observed in vulva cells (Fig. 1D’’). In the nervous system specifically, expression is observed in many neurons throughout the body (Fig. 1D’-D’’’), but unlike Drosophila Elav, exc-7::gfp it is not panneuronally expressed. We used the NeuroPAL transgene (https://www.biorxiv.org/content/10.1101/676312v1) to individually identify each neuron in which exc-7 is expressed in the young adult worm. Sites of expression are listed in Fig. 1F and some examples of neuronal expression are shown in Fig. 1E’. Expression in all neurons is at least several fold more intense than UPN::NLS::TagRFP-T signal from NeuroPAL. We confirmed previously reported expression in cholinergic VNC MNs, but absence of GABAergic VNC MNs (Fig. 1E’’), consistent with previous reports (Fujita et al., 1999; Loria et al., 2003) and consistent with exc-7 functioning in cholinergic, but not GABAergic neurons to control alternative splicing (Norris et al., 2014). exc-7::gfp is also expressed in some non-neuronal cell types, including muscle and hypodermis, but not in the gut (Fig. 1D’-D’’’). A previous report showed that exc-7 is only transiently and weakly expressed in the excretory cell, which, based on exc-7’s excretory mutant phenotype, has puzzled researchers (Fujita et al., 2003). We find that the gfp tagged exc-7 locus is strongly and continuously expressed in the excretory canal cell (Fig. 1B-D’, arrow). We conclude that unlike its fly orthologue elav, exc-7 is not a panneuronally expressed gene.
Reagents
OH16020: exc-7(ot970[exc-7::gfp::3xflag])
The strain is available through the CGC.
Acknowledgments
Acknowledgments
We thank Chi Chen for expert DNA injection, Eduardo Leyva-Díaz for advice with CRISPR genome editing, and Neda Masoudi for help with embryonic lineaging.
Funding
This work was supported by the HHMI.
References
- Berger C, Renner S, Lüer K, Technau GM. The commonly used marker ELAV is transiently expressed in neuroblasts and glial cells in the Drosophila embryonic CNS. Dev Dyn. 2007 Dec 01;236(12):3562–3568. doi: 10.1002/dvdy.21372. [DOI] [PubMed] [Google Scholar]
- Campos AR, Rosen DR, Robinow SN, White K. Molecular analysis of the locus elav in Drosophila melanogaster: a gene whose embryonic expression is neural specific. EMBO J. 1987 Feb 01;6(2):425–431. doi: 10.1002/j.1460-2075.1987.tb04772.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dokshin GA, Ghanta KS, Piscopo KM, Mello CC. Robust Genome Editing with Short Single-Stranded and Long, Partially Single-Stranded DNA Donors in Caenorhabditis elegans. Genetics. 2018 Sep 13;210(3):781–787. doi: 10.1534/genetics.118.301532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita M, Hawkinson D, King KV, Hall DH, Sakamoto H, Buechner M. The role of the ELAV homologue EXC-7 in the development of the Caenorhabditis elegans excretory canals. Dev Biol. 2003 Apr 15;256(2):290–301. doi: 10.1016/s0012-1606(03)00040-x. [DOI] [PubMed] [Google Scholar]
- Fujita M, Kawano T, Ohta A, Sakamoto H. Neuronal expression of a Caenorhabditis elegans elav-like gene and the effect of its ectopic expression. Biochem Biophys Res Commun. 1999 Jul 14;260(3):646–652. doi: 10.1006/bbrc.1999.0957. [DOI] [PubMed] [Google Scholar]
- Leyva-Díaz E, Hobert O. Transcription factor autoregulation is required for acquisition and maintenance of neuronal identity. Development. 2019 Jun 21;146(13) doi: 10.1242/dev.177378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loria PM, Duke A, Rand JB, Hobert O. Two neuronal, nuclear-localized RNA binding proteins involved in synaptic transmission. Curr Biol. 2003 Aug 01;13(15):1317–1323. doi: 10.1016/s0960-9822(03)00532-3. [DOI] [PubMed] [Google Scholar]
- Luo L, Liao YJ, Jan LY, Jan YN. Distinct morphogenetic functions of similar small GTPases: Drosophila Drac1 is involved in axonal outgrowth and myoblast fusion. Genes Dev. 1994 Aug 01;8(15):1787–1802. doi: 10.1101/gad.8.15.1787. [DOI] [PubMed] [Google Scholar]
- Norris AD, Gao S, Norris ML, Ray D, Ramani AK, Fraser AG, Morris Q, Hughes TR, Zhen M, Calarco JA. A pair of RNA-binding proteins controls networks of splicing events contributing to specialization of neural cell types. Mol Cell. 2014 Jun 01;54(6):946–959. doi: 10.1016/j.molcel.2014.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill EM, Rebay I, Tjian R, Rubin GM. The activities of two Ets-related transcription factors required for Drosophila eye development are modulated by the Ras/MAPK pathway. Cell. 1994 Jul 15;78(1):137–147. doi: 10.1016/0092-8674(94)90580-0. [DOI] [PubMed] [Google Scholar]
- Osterwalder T, Yoon KS, White BH, Keshishian H. A conditional tissue-specific transgene expression system using inducible GAL4. Proc Natl Acad Sci U S A. 2001 Oct 23;98(22):12596–12601. doi: 10.1073/pnas.221303298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinow S, White K. The locus elav of Drosophila melanogaster is expressed in neurons at all developmental stages. Dev Biol. 1988 Apr 01;126(2):294–303. doi: 10.1016/0012-1606(88)90139-x. [DOI] [PubMed] [Google Scholar]
- Robinow S, White K. Characterization and spatial distribution of the ELAV protein during Drosophila melanogaster development. J Neurobiol. 1991 Jul 01;22(5):443–461. doi: 10.1002/neu.480220503. [DOI] [PubMed] [Google Scholar]
- Samson ML. Rapid functional diversification in the structurally conserved ELAV family of neuronal RNA binding proteins. BMC Genomics. 2008 Aug 20;9:392–392. doi: 10.1186/1471-2164-9-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnabel R, Hutter H, Moerman D, Schnabel H. Assessing normal embryogenesis in Caenorhabditis elegans using a 4D microscope: variability of development and regional specification. Dev Biol. 1997 Apr 15;184(2):234–265. doi: 10.1006/dbio.1997.8509. [DOI] [PubMed] [Google Scholar]
- Stefanakis N, Carrera I, Hobert O. Regulatory Logic of Pan-Neuronal Gene Expression in C. elegans. Neuron. 2015 Aug 19;87(4):733–750. doi: 10.1016/j.neuron.2015.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]