Abstract
Background:
The early identification of treatment effect is wanted in several settings, including the management of metastatic colorectal cancer (mCRC). A potential universal marker is circulating tumor DNA (ctDNA). Our prospective study explored the association between progression-free survival (PFS) and overall survival (OS), and early change of ctDNA after one cycle of chemotherapy in patients with mCRC.
Methods:
The study included mCRC patients receiving standard first line combination chemotherapy with 5-Fluorouracil (FU), oxaliplatin, and bevacizumab. Hypermethylated neuropeptide Y (NPY) ctDNA (meth-ctDNA) served as a marker analyzed by droplet digital polymerase chain reaction (PCR). The meth-ctDNA level was analyzed in plasma before treatment start and again before cycle two. The patients were divided into two groups according to the dynamics of meth-ctDNA. Low ctDNA (LctDNA) included patients with zero or values of meth-ctDNA decreasing to a level including zero in the 95% confidence interval. High ctDNA (HctDNA) included all other patients (stable, increasing, or slightly decreasing values). The two groups were compared as to PFS and OS.
Results:
The study included 123 patients. The PFS in the two groups differed significantly with a median of 9.2 and 6.7 months in LctDNA and HctDNA, respectively (p = 0.0005). This translated into a 12-month difference in OS with a median of 25.4 and 13.5 months, respectively (p = 0.0001).
Conclusions:
Early therapeutic reconsideration is of utmost importance. A low level of meth-ctDNA after one cycle of chemotherapy in the first line setting is a potential marker for excellent clinical outcomes. The clinical utility should be confirmed in randomized clinical trials.
Keywords: blood-based biomarker, colorectal cancer, drug resistance, liquid biopsy, prognostic biomarker
Introduction
Early identification of treatment benefit is desirable in several settings, including the management of colorectal cancer (CRC). Clinicians rely on scans and biomarkers of limited sensitivity and specificity to evaluate and predict time to treatment resistance in order to save patients from ineffective and sometimes even harmful treatments. A rational basis for early treatment reorientation would be a major step forward.
Liquid biopsies allow for minimally invasive and repetitive biomarker analyses. They provide easily accessible material and facilitate monitoring of biomarker development with a minimum of inconvenience to the patient. Circulating tumor DNA (ctDNA) has great potential as a biomarker and is intensively investigated in the plasma.1,2 Circulating tumor DNA is small, tumor-specific DNA fragments detectable in body fluids, which can be determined by the presence of genetic or epigenetic alterations. RAS and RAF mutated ctDNA (mut-ctDNA) are widely investigated in CRC, and mut-ctDNA seems to be an early marker of treatment effect.3 The great diversity of activating mutations, however, is challenging and is not detectable in all CRC patients even under optimal conditions.
Aberrant methylation is an epigenetic change of the DNA, which often occurs in malignant neoplasms. Hypermethylation is an early event in carcinogenesis and it has the potential to identify ctDNA. Recent publications show that ctDNA with tumor-specific hypermethylation signatures (meth-ctDNA) can be measured in the vast majority of metastatic CRC (mCRC) patients.4,5 In addition, the stability of methylation changes makes them relevant biomarkers for early diagnosis.6 In CRC, hypermethylation of the neuropeptide Y (NPY) promoter region has been associated with inactivation of gene expression and thereby carcinogenesis, and hypermethylation of the NPY gene promoter is currently advocated as a blood-based biomarker.2,4,7 Meth-ctDNA can be detected by droplet digital polymerase chain reaction (ddPCR) offering a relatively short turnover time from blood sample collection to data output, high sensitivity, and low costs.8 Considering these advantages, the clinical application of meth-ctDNA in the early detection of treatment effect in mCRC patients appears feasible.
We have previously published results on the utility of mutated ctDNA dynamics.9,10 The present approach included RAS/RAF mutated as well as wild-type patients to investigate the quality of meth-ctDNA as an early general marker of treatment benefit in first line chemotherapy of mCRC patients.
Materials and methods
Study population
One hundred and twenty-three mCRC patients receiving first line treatment were included in a prospective biomarker study at the Danish Colorectal Cancer Center South, Vejle University Hospital, Denmark, between March 2010 and November 2015. The inclusion criteria were adenocarcinoma in the colon or rectum, recurrent or primary metastatic disease, measureable disease according to The Response Evaluation Criteria in Solid Tumors (RECIST), planned treatment with first line chemotherapy (capecitabine and oxaliplatin) and bevacizumab, age above 18 years, and Eastern Cooperative Oncology Group (ECOG) performance status (PS) 0–2. Treatment was discontinued in the case of progressive disease (PD), unacceptable toxicity, death, patient request, or as decided by the treating physician.
Plasma samples from all patients were collected at baseline (T0) and after the first treatment cycle (T1). Treatment effect was evaluated by a computed tomography (CT) scan of the chest and abdomen according to RECIST version 1.1 at baseline and every three treatment cycles during the whole treatment course.
Written and oral informed consent was obtained from all patients regarding translational research according to the Helsinki II Declaration. The study was approved by the Regional Committees on Health Research Ethics for Southern Denmark (S-20100005) and the investigation was conducted in accordance with the Reporting Recommendations for Tumor Marker Prognostic Studies (REMARK) criteria.11
Blood sample analysis
Nine millilitre blood samples were collected in ethylenediamine tetraacetic acid (EDTA) tubes. Plasma was isolated by centrifugation at 2000 g for 10 minutes within four hours and stored at –80°C until use. The plasma was centrifuged again at 10,000 g for 10 minutes before purification and cysteine-rich polycomb-like protein 1 (CPP1) DNA fragments were added as an exogenous internal control.12 The DNA from patients was purified from up to 4 ml plasma on the QIAsymphony SP instrument using QIAsymphony DSP circulating DNA kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions.
Tumor-specific methylated DNA was defined as DNA with methylation of the NPY gene promoter.7 Prior to the ddPCR methylation analysis bisulfite conversion of circulating DNA was performed and the converted DNA was analyzed with a methylation-specific assay and a control assay (Albumin) using the BioRad Droplet Digital PCR system QX100 (BioRad, Hercules, CA, USA). The purified DNA was concentrated to 20 µl on Amicon Ultra Centrifugal filter units (Millipore, Burlington, MA, USA) and bisulfite converted in a 150 µl reaction with the EZ DNA Methylation lightning spin-column kit (Zymo research, Irvine, CA, USA) and eluted in 15 µl. The Albumin/NPY duplex analysis was performed in two wells with 5 µl converted DNA per well in 20 µl reactions. ddPCR supermix for probes (no Deoxyuridine Triphosphate) and NPY/ALB assays were applied (Supplemental Table 1). Droplets were generated on the QX200 automated droplet generator from BioRad. PCR was completed on the Veriti PCR-device. The droplets were counted in the QX100 droplet reader from BioRad. Data analysis was performed with QuantaSoft version 1.7.4 (BioRad).
Water and a pool of lymphocyte DNA from non-cancer individuals were used as negative controls and universal human methylated control DNA (Zymo Research) and EpiTech control DNA (Qiagen) were used as positive controls. Negative controls and universal human methylated control DNA was bisulfite converted together with the samples and all controls were included in each round of digital PCR.
Analyses were performed according to the digital The Minimum Information for Publication of Quantitative Real-Time PCR Experiments (MIQE) guidelines.13
Data management
The meth-ctDNA level was defined as the fraction of meth-ctDNA and expressed as the proportion of NPY methylated alleles in the total circulating cell-free DNA (NPY methylated alleles/albumin alleles) and calculated by Quantasoft software. The variation of the meth-ctDNA level is reported with a 95% confidence interval (CI) based on Poisson statistics. After the first cycle of chemotherapy the patients were divided into two groups; low-level methylated ctDNA (LctDNA) and high-level methylated ctDNA (HctDNA). The LctDNA group constituted patients with a level of zero at baseline as well as those decreasing to a level with zero included in the 95% CI at T1. The HctDNA group included all other patients. At the first radiological evaluation, response was defined as partial or complete, and patients with non-evaluable or unknown response status were not included in the analysis. The ctDNA analyses were performed blinded to radiological assessments.
Statistical analysis
Survival analyses were calculated from the second cycle of treatment using the Kaplan–Meier method. Progression-free survival (PFS) was calculated to the date of progression, death, or censored at last hospital contact. Overall survival (OS) was calculated to the date of death from any cause. Patients still alive were censored at the last known date alive. Survival curves were compared using the log-rank test providing a hazard ratio (HR). Baseline characteristics were compared using Fisher’s exact test for binary values and the Wilcoxon Mann–Whitney test for numerical values. All reported p values were two sided, and p < 0.05 was considered statistically significant. All statistical tests were performed using the NCSS 10 Statistical Software (2015) (NCSS, LLC, Kaysville, UT, USA, ncss.com/software/ncss).
Results
The methylation analysis was performed on both the T0 and T1 blood samples of all 123 patients. Baseline characteristics and patient characteristics according to dynamics in ctDNA are shown in Table 1. No significant difference was seen in the parameters according to the two subgroups.
Table 1.
Baseline characteristics of all patients with complete analyses of meth-ctDNA (n = 123).
| Parameter | All patients | Meth-ctDNA zero at T0 (n = 16) |
Elevated meth-ctDNA at T0 (n = 107) |
p values comparing the two groups |
|---|---|---|---|---|
| n (%) | n (% of all patients) | n (% of all patients) | ||
| Age, years | ||||
| Median (range) | 67 (32–80) | 69 (50–77) | 67 (32–80) | ns |
| Gender | ||||
| Female | 45 (37) | 6 (38) | 39 (36) | ns |
| Male | 78 (63) | 10 (63) | 68 (64) | |
| PS | ||||
| 0–1 | 117 (95) | 15 (94) | 102 (95) | ns |
| 2 | 6 (5) | 1 (6) | 5 (5) | |
| Location of primary tumor | ||||
| Right/transverse colon | 32 (26) | 2 (13) | 30 (28) | |
| Left colon | 40 (33) | 6 (38) | 34 (32) | ns |
| Rectum | 51 (41) | 8 (50) | 43 (40) | |
| Primary disseminated disease | 99 (80) | 11 (69) | 88 (82) | ns |
| Recurrent disease | 24 (20) | 5 (4) | 19 (15) | |
| RAS/RAF mutated tumor | 63 (50)* | 13 (11) | 50 (42) | – |
| Number of metastatic sites | ||||
| ⩽2 | 97 (79) | 15 (94) | 82 (77) | ns |
| >2 | 26 (21) | 1 (6) | 25 (23) | |
meth-ctDNA, hypermethylated neuropeptide Y (NPY) circulating tumor DNA; n, number; ns, non-significant; PS, performance status.
11 Patients did not have their tumor tissue analyzed due to insufficient material.
Sixteen patients (13%) were meth-ctDNA negative at both time points as shown in Table 2. The median level of meth-ctDNA at baseline was 9.5% (range 0–95%) and 0.59% (range 0–42.5%) at T1. LctDNA and HctDNA was detected in 55 patients (45%) and 68 patients (55%), respectively. The median level and range of meth-ctDNA in the LctDNA group was 1.9 (0–40.5) and 0 (0–0.97) at T0 and T1, respectively. The corresponding data in the HctDNA group were 23.3 (0.61–95) and 3 (0.25–40), respectively.
Table 2.
Number of patients with different ctDNA dynamics.
| Number of patients (%) | |
|---|---|
| Zero at baseline (T0) to zero after the first treatment (T1) | 16 (13) |
| Zero at T0 to elevated at T1 | 0 (0) |
| Elevated at T0 to zero at T1 | 39 (32) |
| Elevated at T0 to elevated at T1 | 68 (55) |
ctDNA, circulating tumor DNA.
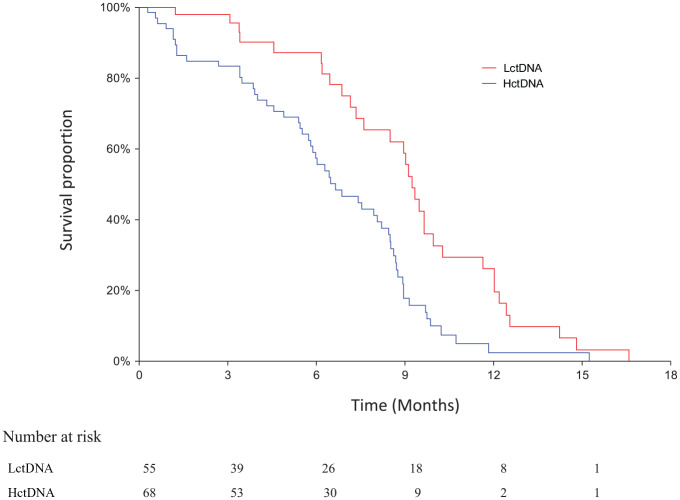
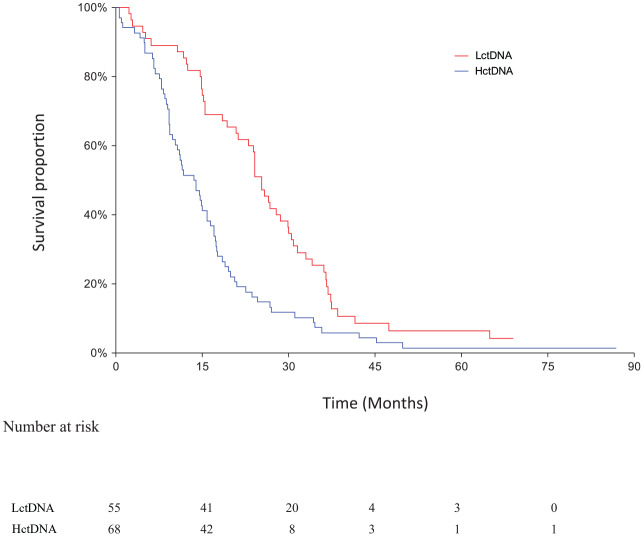
The clinical database was last updated on 7 September 2018. Median follow-up time for OS was 17.8 months and for PFS 6.7 months as estimated by inverse Kaplan–Meier analysis. The OS analysis was based on 118 events in the whole cohort. Of the 123 patients, 101 had a first response evaluation (82%). The response rate in patients with LctDNA and HctDNA was 40% (16/40) and 33% (20/61), respectively (p = 0.38). There was no significant difference in PFS and OS according to the median meth-ctDNA level at baseline (data not shown). The PFS in the LctDNA and HctDNA patients is shown in Figure 1. The median values were 9.2 months (95% CI 7.6–9.6 months) and 6.7 months (95% CI 5.8–8.2 months), respectively (HR = 0.48, 95% CI 0.32–0.74, p = 0.0005). A significant difference also applied to OS. The median values in LctDNA and HctDNA were 25.4 months (95% CI 24.1–28.6 months) and 13.5 months (95% CI 10.4–15.8 months), respectively (HR = 0.50, 95% CI 0.35–0.72, p = 0.0001) (Figure 2).
Figure 1.
Progression-free survival according to methylation response.
The median values in the two groups.
Red: LctDNA: 9.2 months (95% CI 7.6–9.6 months).
Blue: HctDNA: 6.7 months (95% CI 5.8–8.2 months).
Log rank test: HR = 0.48, 95% CI 0.32–0.74, p = 0.0005.
CI, confidence interval; HctDNA, high-level methylated ctDNA; HR, hazard ratio; LctDNA, low-level methylated ctDNA.
Figure 2.
Overall survival according to methylation response.
The median values in the two groups.
Red: LctDNA: 25.4 months (95% CI 21.2–28.6 months).
Blue: HctDNA: 13.5 months (95% CI 10.4–15.8 months).
Log rank test: HR = 0.50, 95% CI 0.35–0.72, p = 0.0001.
CI, confidence interval; HctDNA, high-level methylated ctDNA; HR, hazard ratio; LctDNA, low-level methylated ctDNA.
Discussion
This prospective study of mCRC patients receiving first line chemotherapy illustrated a potential correlation between the initial dynamics in meth-ctDNA and the early prediction of treatment benefit.
Mutated ctDNA is intensively investigated and seems to hold prognostic value and feasibility as to the early monitoring of treatment effect.14 However, this requires knowledge of tumor-specific mutations. In order to overcome this limitation meth-ctDNA is being studied.2,5 Methylated ctDNA is accessible in patients with unknown mutational status and is relevant when the qualitative mutation analysis is not feasible. Thus, meth-ctDNA is a possible, minimally invasive tool in predicting the effectiveness of treatment in the general population of mCRC patients planned to be given first line chemotherapy with capecitabine, oxaliplaitin, and bevacizumab.
A recent study by Barault et al. included 45 mCRC patients with longitudinal follow-up based on meth-ctDNA and found 13% to be baseline negative,5 which is in line with our results. In agreement with our study, the authors demonstrated the possibility to analyze meth-ctDNA instead of mut-ctDNA for monitoring treatment effect, and they suggest meth-ctDNA as a supplement to radiological assessment to improve the monitoring of mCRC. However, their study differs from ours in several important aspects. Importantly, they only investigated eight patients receiving conventional chemotherapy. Furthermore, the time span between ctDNA analyses was longer than ours. The importance of harboring ctDNA at baseline is debated. As ctDNA is shed in the circulation by several active and passive mechanisms,15 the transition from tissue to blood may result from tumor size and site or grade of malignancy, but the issue needs further clarification. In agreement with our findings, Boeckx et al. found a high correlation between NPY methylated ctDNA and radiological assessment in 24 mCRC patients treated with first line chemotherapy.2 Our study validates their findings in a larger cohort based on the same methylation marker and blood samples drawn at similar time points. However, they detected a steep decrease in the ctDNA level immediately after the start of treatment in all patients but one. In our study, seven patients had an initial increase, which seems reasonable considering the larger cohort analyzed. The correlation between ctDNA response and therapeutic effect, and hence survival, was also found in the PLACOL study by Garlan et al. based on 73 mCRC patients.16 The authors found an early increase of ctDNA in seven patients. Also, in accordance with our study, they found a significant correlation between PFS and a decrease below a negligible threshold of ctDNA rather than just a decrease. Unlike our study, their cohort was heterogeneous including first and second line chemotherapy with or without targeted treatment.
Table 1 shows the baseline characteristics and the characteristics of the patients with and without meth-ctDNA at T0. The two subgroups did not differ significantly in any parameter. The subgroup without ctDNA at baseline was small and hence calculations should be performed with caution. The hypothesis of a correlation between tumor burden and dynamics of ctDNA (the larger tumor burden, the more chemotherapy might be needed to see a decrease to zero) was not supported in this study.
Our study has limitations. First of all there is no validation cohort and the sample size is rather small. The protocol was initiated in 2010 and the first line treatment has changed since then so the results presented here might not reflect the mCRC patients receiving first line treatment as of now.
The data presented here do not allow us to distinguish formally between the prognostic and predictive value of meth-ctDNA.
In conclusion, meth-ctDNA may reflect treatment benefit and provide an opportunity for early therapeutic reorientation. It also has the additional advantage of facilitating follow-up in mCRC patients with unknown mutational status.
Supplemental Material
Supplemental material, Supplementary_material_3 for Early identification of treatment benefit by methylated circulating tumor DNA in metastatic colorectal cancer by Caroline B. Thomsen, Torben F. Hansen, Rikke F. Andersen, Jan Lindebjerg, Lars H. Jensen and Anders Jakobsen in Therapeutic Advances in Medical Oncology
Acknowledgments
The authors would like to thank the study participants and the staff at Vejle University Hospital who collected samples for this study. Also, they gratefully acknowledge the technicians Lone Hartmann Hansen, Pia Nielsen, and Tina Brandt Christensen. Finally, they express gratitude to Karin Larsen for linguistic editing of the manuscript.
Footnotes
Conflict of interest: The authors declare that there is no conflict of interest.
Funding: The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: Financial support was received from the Regional Strategic Council for Research in the Region of Southern Denmark.
ORCID iD: Caroline B. Thomsen  https://orcid.org/0000-0003-4481-6274
https://orcid.org/0000-0003-4481-6274
Supplemental material: Supplemental material for this article is available online.
Contributor Information
Caroline B. Thomsen, Danish Colorectal Cancer Center South, Vejle University Hospital, Beriderbakken 4, DK-7100 Vejle, Denmark.
Torben F. Hansen, Danish Colorectal Cancer Center South, Vejle University Hospital, Vejle, Denmark Institute of Regional Health Research, University of Southern Denmark, Odense, Denmark
Rikke F. Andersen, Danish Colorectal Cancer Center South, Vejle University Hospital, Vejle, Denmark
Jan Lindebjerg, Danish Colorectal Cancer Center South, Vejle University Hospital, Vejle, Denmark Institute of Regional Health Research, University of Southern Denmark, Odense, Denmark.
Lars H. Jensen, Danish Colorectal Cancer Center South, Vejle University Hospital, Vejle, Denmark Institute of Regional Health Research, University of Southern Denmark, Odense, Denmark
Anders Jakobsen, Danish Colorectal Cancer Center South, Vejle University Hospital, Vejle, Denmark Institute of Regional Health Research, University of Southern Denmark, Odense, Denmark.
References
- 1. Bettegowda C, Sausen M, Leary RJ, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med 2014; 6: 224ra24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Boeckx N, Op de Beeck K, Beyens M, et al. Mutation and methylation analysis of circulating tumor DNA can be used for the follow-up of metastatic colorectal cancer patients. Clin Colorectal Cancer. Epub ahead of print 22 Febuary 2018. DOI: 10.1016/j.clcc.2018.02.006. [DOI] [PubMed] [Google Scholar]
- 3. Thierry AR, Pastor B, Jiang ZQ, et al. Circulating DNA demonstrates convergent evolution and common resistance mechanisms during treatment of colorectal cancer. Clin Cancer Res 2017; 23: 4578–4591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Garrigou S, Perkins G, Garlan F, et al. A study of hypermethylated circulating tumor DNA as a universal colorectal cancer biomarker. Clin Chem 2016; 62: 1129–1139. [DOI] [PubMed] [Google Scholar]
- 5. Barault L, Amatu A, Siravegna G, et al. Discovery of methylated circulating DNA biomarkers for comprehensive non-invasive monitoring of treatment response in metastatic colorectal cancer. Gut. Epub ahead of print 5 October 2017. DOI: 10.1136/gutjnl-2016-313372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Warton K, Samimi G. Methylation of cell-free circulating DNA in the diagnosis of cancer. Front Mol Biosci 2015; 2: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Roperch JP, Incitti R, Forbin S, et al. Aberrant methylation of NPY, PENK, and WIF1 as a promising marker for blood-based diagnosis of colorectal cancer. BMC Cancer 2013; 13: 566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Thierry AR, El Messaoudi S, Mollevi C, et al. Clinical utility of circulating DNA analysis for rapid detection of actionable mutations to select metastatic colorectal patients for anti-EGFR treatment. Ann Oncol 2017; 28: 2149–2159. [DOI] [PubMed] [Google Scholar]
- 9. Thomsen CB, Hansen TF, Andersen RF, et al. Monitoring the effect of first-line treatment in RAS/RAF mutated metastatic colorectal cancer by serial analysis of tumor specific DNA in plasma. J Exp Clin Cancer Res 2018; 37: 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Thomsen CB, Andersen RF, Lindebjerg J, et al. Plasma dynamics of RAS/RAF mutations in patients with metastatic colorectal cancer receiving chemotherapy and anti-EGFR treatment. Clin Colorectal Cancer. Epub ahead of print 24 October 2018. DOI: 10.1016/j.clcc.2018.10.004. [DOI] [PubMed] [Google Scholar]
- 11. McShane LM, Altman DG, Sauerbrei W, et al. Reporting recommendations for tumor marker prognostic studies. J Clin Oncol 2005; 23: 9067–9072. [DOI] [PubMed] [Google Scholar]
- 12. Pallisgaard N, Spindler KL, Andersen RF, et al. Controls to validate plasma samples for cell free DNA quantification. Clin Chim Acta 2015; 446: 141–146. [DOI] [PubMed] [Google Scholar]
- 13. Huggett JF, Foy CA, Benes V, et al. The digital MIQE guidelines: minimum information for publication of quantitative digital PCR experiments. Clin Chem 2013; 59: 892–902. [DOI] [PubMed] [Google Scholar]
- 14. Tie J, Kinde I, Wang Y, et al. Circulating tumor DNA as an early marker of therapeutic response in patients with metastatic colorectal cancer. Ann Oncol 2015; 26: 1715–1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mouliere F, Thierry AR. The importance of examining the proportion of circulating DNA originating from tumor, microenvironment and normal cells in colorectal cancer patients. Expert Opin Biol Ther 2012; 12 (Suppl. 1): S209–S215. [DOI] [PubMed] [Google Scholar]
- 16. Garlan F, Laurent-Puig P, Sefrioui D, et al. Early evaluation of circulating tumor DNA as marker of therapeutic efficacy in metastatic colorectal cancer patients (PLACOL study). Clin Cancer Res 2017; 23: 5416–5426. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, Supplementary_material_3 for Early identification of treatment benefit by methylated circulating tumor DNA in metastatic colorectal cancer by Caroline B. Thomsen, Torben F. Hansen, Rikke F. Andersen, Jan Lindebjerg, Lars H. Jensen and Anders Jakobsen in Therapeutic Advances in Medical Oncology