Abstract
Aims
Human embryonic stem cells (hESCs) can be used to generate scalable numbers of cardiomyocytes (CMs) for studying cardiac biology, disease modelling, drug screens, and potentially for regenerative therapies. A fluorescence-based reporter line will significantly enhance our capacities to visualize the derivation, survival, and function of hESC-derived CMs. Our goal was to develop a reporter cell line for real-time monitoring of live hESC-derived CMs.
Methods and results
We used CRISPR/Cas9 to knock a mCherry reporter gene into the MYH6 locus of hESC lines, H1 and H9, enabling real-time monitoring of the generation of CMs. MYH6:mCherry+ cells express atrial or ventricular markers and display a range of cardiomyocyte action potential morphologies. At 20 days of differentiation, MYH6:mCherry+ cells show features characteristic of human CMs and can be used successfully to monitor drug-induced cardiotoxicity and oleic acid-induced cardiac arrhythmia.
Conclusion
We created two MYH6:mCherry hESC reporter lines and documented the application of these lines for disease modelling relevant to cardiomyocyte biology.
Keywords: Cardiomyocyte, Cardiotoxicity testing, MYH6, hESC reporter, Disease model
1. Introduction
Loss or impairment of cardiomyocyte function can result in various forms of cardiovascular disease, the leading cause of morbidity and mortality in the world representing 31% of all global deaths.1 Human pluripotent stem cells (hPSCs), including human embryonic stem cells (hESCs) and induced pluripotent stem cells (hiPSCs), have the unique capacity to differentiate into cell types representing all three germ layers, including mesoderm-derived cardiomyocytes (CMs). Hence, hPSCs can provide in principle essentially unlimited materials for studying cardiac biology, including lineage differentiation, disease modelling, drug screening, and development of regenerative therapeutics. Indeed, directed differentiation techniques starting with hPSCs have been successfully used to generate many different specific cell types by recapitulating normal developmental programmes that function during embryogenesis, including for relatively efficient generation of CMs.2,3
However, differentiation is never entirely successful, and for many applications, it may be important to purify the CMs. Several surface markers have been utilized in order to enrich CMs. For instance, TNNT2, a CM-specific maker, has been used for this purpose, but since it is intracellular, it cannot be used to purify live CMs. Alternative approaches to isolate pure hPSC-derived CMs have been applied to overcome this hurdle. Two surface markers, SIRPA (signal regulatory protein-α) and VCAM-1 (vascular cell adhesion molecule 1), have been used to successfully enrich the CM population.4–6 However, these markers are expressed on many other cell types, including in the brain, lung, and blood vessels.7 Some non-genetic manipulation approaches including the fluorescent mitochondrial dye TMRM (tetramethylrhodamine methyl ester perchlorate) and magnetic bead isolation by fluorescence-activated cell sorting (FACS) analysis or Percoll gradient separation have been used to isolate CMs.8–10 However, these strategies have limitations for purity and scalability. Furthermore, several research groups used glucose-free medium with lactate or fatty acid supplement to enrich hPSC-CMs by metabolic purification methods.11,12 However, it is not clear whether metabolic purification methods impact the survival or function of CMs in the context of disease modelling or cardiotoxicity studies. Finally, several cardiac-specific promoters have been used to direct expression of reporter genes, including regulatory sequences for genes that encode NKX2.5 (NK2 homeobox 5),5 ISL1 (Islet-1),13 MYL2 (myosin light chain 2, also known as MLC2v),14 and MYH6 (known as α-MHC),15,16 but these approaches also have limitations. NKX2.5 and ISL1 identify both cardiac progenitors and CMs. The reported transgenic approaches using MYL2 or MYH6 created lines by cloning a portion of the promoter region or with lentiviral transduction.15,16 Prior to development of CRISPR/Cas9 technology, these transgenic CM reporters have been used for purification of CMs and to study cardiac biology.17,18 However, the transgenes may not fully recapitulate the endogenous cardiac programme and could impart genetic alterations.
A fluorescence-based reporter line should significantly enhance our capacity to visualize the derivation, survival, and function of hESC-derived CMs. About 90% of promising pharmaceutical compounds eventually fail and are withdrawn from the market primarily due to drug-induced cardiovascular toxicity.19,20 While several groups have used hPSC-derived CMs as a model for testing cardiotoxicity,21–27 it remains challenging to monitor in real-time cardiomyocyte arrhythmia or death, which is important to further probe mechanism of the cell-type-specific toxicities. Therefore, we sought to establish a fluorescence-based reporter line to purify CMs and monitor cardiomyocyte function and survival.
We applied CRISPR/Cas9 technology to generate a cardiomyocyte reporter hESC line with a knockin of mCherry at the MYH6 locus. MYH6 is expressed in both atria and ventricles during human embryonic development. After birth, MYH6 is predominantly expressed in atrial CMs and at lower levels in ventricular CMs.28,29 Mutations in MYH6 have been reported to cause hypertrophic cardiomyopathy, dilated cardiomyopathy, atrial septal defects, and sick sinus syndrome.30–32 Notably, based on antibody staining and transmission electron microscopy (TEM), the hESC-derived MYH6:mCherry+ cells co-express MYH6 and additional cardiac atrial and ventricular markers, and display well-organized sarcomeric structures. Moreover, global gene expression profiles of MYH6:mCherry+ cells cluster together with relatively mature hESC-derived CMs. Finally, we successfully used this reporter system to quantitatively evaluate induced cardiotoxicity in living cells.
2. Methods
2.1 hESC culture and differentiation
Human H1 and H9 ESCs were purchased from WiCell. hESCs were grown on a Matrigel-coated plate in mTeSR medium (Stem Cell Technologies, USA) at 37°C in 5% CO2. The culture medium was exchanged daily. About 0.5 mM EDTA was used for routine passage of hESCs. Accutase was used for single cell dissociation. For cardiac differentiation, MYH6:mCherry hESCs were cultured in hESC medium to about 80% confluence. At differentiation Day 0, hESCs were treated with 12 μM CHIR99021 (CHIR, Stemgent) in RPMI (Cellgro, USA) supplemented with B27 minus insulin, 2 mM GlutaMAX and 100 U/mL Pen/Strep for 24 h. The next day, CHIR was removed. At Day 2, differentiated cells were treated with 5 µM WNT antagonist I (IWR-1, Stemgent, USA) for 3 days. At Day 5, IWR1 was removed. At Day 7, B27 minus insulin in cardiac differentiation medium was changed to complete B27. Beating cells were typically observed around Day 8.
2.2 Immunofluorescence
Cells were fixed with 4% paraformaldehyde for 10 min at room temperature. Cells were blocked in 5% horse serum (Invitrogen, USA), 0.3% Triton X in PBS for one hour at room temperature, followed by primary antibody incubation overnight. The following antibodies were used: mouse anti-MYH6 (1:500, R&D, USA, MAB8979), rabbit anti-MYL2 (1:500, Santa Cruz, USA, SC-34490), goat anti-MYL7 (1:500, Santa Cruz, USA, SC-365255), rabbit anti-GATA4 (1:500, Abcam, USA, ab61767), mouse anti-TNNT2 (1:1000, Invitrogen, USA, MA5-12960), mouse anti-Sarcomeric Alpha Actinin (1:100, Sigma, USA, A7811), and rabbit anti-cleaved-caspase3 (1:400, Cell Signaling, USA, 9661). Antibodies were detected with Alexa-488-, Alexa-555-, and Alexa-647-conjugated donkey secondary antibodies against mouse, goat, or rabbit (1:500, Invitrogen, USA). Nuclei were counterstained with DAPI.
2.3 Generation of the MYH6:mCherry hESC reporter line
sgRNA sequences (Table 1) were designed using the website http://crispr.mit.edu/ and targeted the sequence 5′-GCAGCAAAAAATGCACGATG-3′. Surveyor assays were used to evaluate guide efficiency as described.33 The sgRNA sequence was cloned into the PX330 vector (Addgene, plasmid #42230).34,35 For constructing the MYH6:mCherry donor plasmid, 901-bp left (Primers: HL-F and HL-R) and 971-bp right (Primers: HR-F and HR-R) homology arms were PCR amplified from H9 genomic DNA and cloned into the pGolden-Hyg plasmid (Addgene, plasmid #51423). P2A-mCherry was PCR amplified from the IRES-puro-p2A-mCherry reporter cassette that was developed in-house. Loxp-Puro inserts were PCR amplified from the OCT4-2A-eGFP-PGK-Puro plasmid (Addgene: 31938) using LoxP-Puro-F and LoxP-Puro-R primers, digested and cloned into the pGolden-Hyg plasmid to generate the MYH6:mCherry donor plasmid. The donor plasmid was further confirmed by DNA sequencing.
Table 1.
Primer lists
| Name of primer | Sequence (5′–3′) |
|---|---|
| HR_F | GGATCCTGACACTGCCTCGGGAACCTCACT |
| HR_R | GGTACCTAGAAGAGGCTCCACTTGGGGAGT |
| HL_F | CTCGAGGGTGGAATTGGGCCTAGAAGGGGA |
| HL_R | TCTAGACTCCTCATCGTGCATTTTTTGCTG |
| P2A-mCherry_F | TCTAGAGGAAGCGGAGCTACTAACTTCAGC |
| P2A-mCherry_R | CATATGTCATCTGAGTCCGGACTTGTACAGCTC |
| SgRNA#1_F | CACCGCAGCAAAAAATGCACGATG |
| SgRNA#1_R | AAACCATCGTGCATTTTTTGCTGC |
| SgRNA#2_F | CACCGTGGCAAGAGTGAGGTTCCCG |
| SgRNA#2_R | AAACCCGGGAACCTCACTCTTGCCA |
| loxp puro_F | CATATG CGATCATATTCAATAACCCTTAATATAACTTCG |
| loxp puro_R | GGATCCTCTTCGAGAGTGAACCTGGACCTAATAAC |
| Genotype_F | GCTTGCCCTATATGTAGGCAGTTCTG |
| Genotype_R | GATGGCCATGTTATCCTCCTCGC |
| Middle of LoxP-Puro | CTGCAAGAACTCTTCCTCACGCGC |
| Outside HR_R1 | GCACAGTCCATTCTCATGGTCTC |
| Outside HL_F | GGCACCACCTACAAGCTGCTTAC |
| Outside HR_R2 | CAGCTCCCAAGTCCTGCCCGAAC |
| Sequencing primer_F(HL) | CACTTCTCCCTCTTGCCAGCTGC |
| Sequencing primer_F(mCherry) | GCTGAGGTCAAGACCACCTACAAG |
| Px330 sequence | GGACTATCATATGCTTACCG |
| GAPDH_F | GAAGGTGAAGGTCGGAGTC |
| GAPDH_R | GAAGATGGTGATGGGATTT |
| RYR2_F | CGAAGACGAGATCCAGTTCC |
| RYR2_R | CAAATCCTTCTGCTGCCAAG |
| KCNA4_F | CAGCCGGTGGATTTTCTTTA |
| KCNA4_R | CAGAGCATTCTTCAGCCAAA |
| KCNA2_F | TTACATGCCTCTGTACCCCC |
| KCNA2_R | TGACTCAGCTGACATCCAGA |
| BAX_F | ACCAAGAAGCTGAGCGAGTGTCACC |
| BAX-R | ACAAAGATGGTCACGGTCTGCC |
| ACTC1_F | GCTGTCTTCCCGTCCATC |
| ACTC1_R | CATGCTCGATGGGATACTTCAG |
| TNNI3_F | TTTGACCTTCGAGGCAAGTTT |
| TNNI3_R | TGCAGAGATCCTCACTCTCCG |
| MYH7_F | TCGTGCCTGATGACAAACAGGAGT |
| MYH7_R | ATACTCGGTCTCGGCAGTGACTTT |
| MYH6_F | CGGTGCTTTTCAACCTCAAGG |
| MYH6_R | GGACTGGTTCTCCCGATCTGT |
H1 and H9 hESCs were dissociated into single cells with Accutase for 5–7 min. Cells were electroporated using a human stem cell nucleofector kit2 (Lonza, VPH-5022) according to the manufacturer’s guidelines. Briefly, 1 million cells were resuspended in 100 µL nucleofector mix to which was added 4 µg CRISPR targeting plasmid and 2-µg donor plasmid. Cells were re-plated on Matrigel-coated plates with 10 µM ROCK inhibitor. Two days after electroporation cells were treated with puromycin (0.5 µg/mL) for 2 days. Two weeks after electroporation, 80 clones were picked and each clone was separated into two wells of a 96 well-plate. When cells were 80–90% confluent, we added lysis buffer (Sigma, Cat# L3289) and neutralization buffer (Sigma, Cat# N9784) to isolate genomic DNA for genotyping (internal primers listed in Table 1). After this initial genotyping, we chose six random clones and performed PCR using a pair of primers that were located (forward) in the middle of the LoxP-Puro cassette and (reverse) outside of the HA-R cassette in the untargeted genomic DNA. Then, targeted clones were electroporated with a vector expressing Cre recombinase by the same experimental procedure described above, in order to remove the Loxp-Puro selection cassette. Positive clones bearing mCherry knock-in at the correct locus were then verified using primers both located outside of HL and HR arms (primers listed in Table 1). PCR products were confirmed by DNA sequencing. The sequences for all primers used for generating the donor plasmids are listed in Table 1.
2.4 FACS and flow cytometry
MYH6:mCherry hESC reporter cells were differentiated into CMs as described above. Cells were dissociated with Accutase for 5 min at 37°C. Cells were centrifuged at 1400 rpm for 4 min and resuspended in FACS buffer containing DMEM without phenol red (Life Technologies, 21063-029), 1 mM EDTA, 25 mM HEPES, and 5% foetal bovine serum. Cells were collected using a BD FACSARIA cell sorter. For the flow analysis, ∼5000 single live-cell events were recorded and analysed per sample on a C6 flow cytometer (Accuri) and analysed using FCS express (De novo Software).
2.5 RNA sequencing and data analysis
Total RNA was isolated using the Qiagen RNeasy mini kit according to manufacturer’s instructions. The quality of RNA samples was examined by Agilent Bioanalyzer (Agilent). cDNA libraries were generated using TruSeq RNA Sample Preparation kits (Illumina). Each library was sequenced using single-end reads in HiSeq4000 (Illumina). Gene expression levels were analysed with TopHat and Cufflinks by the Weill Cornell Genomic Core facility. RNA-seq data are deposited in the GEO database and can be accessed with the GEO accession number GSE111365. Raw data were normalized with negative cells as fold-change. Heat maps were analysed with MeV (Multiple Experiment Viewer) and gene lists were filtered based on expression level differences ≥4 or ≤ −4. To perform principle component analysis (PCA), raw read counts on protein-coding genes were collected from our RNA-seq data and from public data set GSE93841 and were normalized by performing regularized log transformation with DESeq2 package. Batch effects introduced by experiments were corrected using remove Batch Effect from limma package. PCA was then performed using plot PCA from DESeq2 package. GSEA was performed using GSEA software. Genesets were selected based on genes that are >10-fold up-regulated from MYH6:mCherry+ samples. Global human adult atrial, ventricular myocyte, and hESC-derived cardiomyocyte gene expression data were obtained from a published database (GSE64189).36 Enriched genesets are selected based on statistical significance (FDR q value <0.25 and/or NOM P value <0.05). Enriched genes were grouped into functional categories based on Gene Ontology classifications using the PANTHER site (http://www.pantherdb.org) (PMID:12520017).
2.6 Cellular electrophysiology
Action potentials were recorded using an AM-Systems (WA, USA) model 2400 patch-clamp amplifier in current-clamp mode using the Real-Time eXperiment Interface (RTXI; www.rtxi.org).37,38 Cells were superfused at 35°C with a Tyrode’s solution containing: 137 mM NaCl, 5.4 mM KCl, 2 mM CaCl2, 1 mM MgSO4, 10 mM HEPES, and 10 mM glucose, with pH 7.35 (NaOH). The perforated patch-clamp technique was used to gain whole cell access by 480 µg/µL Amphotericin-B (Sigma-Aldrich Corp, MO, USA) in the pipette solution, which also contained: 5 mM NaCl, 20 mM KCl, 120 mM K-aspartate, and 10 mM HEPES, with pH 7.2 (KOH). Pipettes were pulled from 1.5 mm capillary tubes (AM-Systems, WA, USA) to a resistance between 2.5 and 3.5 MΩ, and corrected for a liquid junction potential of 14 mV. Recordings consisted of 10 s of no stimulation, followed by 10 s of 2 Hz stimulation with 3–5 ms suprathreshold current injection, and another 10 s of no stimulation.
2.7 Subtype characterization
Characterization was based on action potentials from a 10 s recording. At minimum, the 10 s recording contained at least three action potentials with spontaneous recordings prioritized (n = 13/15) over stimulated recordings (n = 2/15). Action potentials were analysed using MATLAB (MathWorks, Natick, MA, USA), and cells were subdivided into three subtypes based on upstroke velocity (dV/dtmax) and action potential shape (APD90/30 ratio). Criteria for ventricular-like action potentials were a fast upstroke (dV/dtmax > 30) and long plateau phase (APD90/30 ratio < 1.6). For atrial-like action potentials, criteria were a fast upstroke (dV/dtmax > 30) and absence of a plateau phase (APD90/30 ratio > 1.6). For nodal-like action potentials, spontaneity was required along with a slow upstroke (dV/dtmax < 30).
2.8 Calcium imaging
hESC-derived CMs were plated on ibidi plates coated with 0.1% gelatine. The cells were loaded with 2 μM Fluo-4 AM dissolved in 1:1 (v/v) amount of 20% Pluronic®-F127 and DMSO with stock concentration of 1 mM for 45 min at room temperature in Tyrode solution consisting of: 140 mM NaCl, 5.4 mM KCl, 1 mM MgCl2, 1.8 mM CaCl2, 10 mM glucose, and 10 mM HEPES at pH 7.4. The calcium transients of hESC-derived beating cardiac clusters were recorded on a heated stage using a confocal microscope (Zeiss LSM 710) at intervals of 200 ms (five frames per second). The images were then quantified as the background subtracted fluorescence intensity changes normalized to the background subtracted baseline fluorescence using MetaXpress software.
2.9 qRT-PCR
RNA was isolated using an RNase Mini Kit (Qiagen, 74106), and 2 μg of total RNA was used to synthesize cDNA (Thermofisher, 18080400). Real-time qPCR was performed in a CFX384 (Bio-Rad) machine with LightCycler DNA Master SYBR Green I reagents (Roche). Differences between samples and controls were calculated based on the 2−ΔΔCT method and normalized to GAPDH. Statistical significance was determined using a two-tailed Student’s t-test (P < 0.05). The primers are listed in Table 1.
2.10 Drug-induced cardiotoxicity
MYH6:mCherry-derived CMs (differentiation Day 20) were treated with 20 µM doxorubicin (Dox, Cat# 2252, Tocris) for 24 h. For live-cell imaging experiments, CMs were treated with Dox for 48 h. At Day 20, CMs were treated with 8 mM of Oleic acid (Cat# O1257, sigma) for 48 h. Cells were then harvested for further analyses. For the maturation experiment, Day 15 CMs were treated with 100 µM palmitic acid (Cat# P9767, Sigma) for 10 days, following by 48 h OA treatment.
2.11 Transmission electron microscopy
MYH6:mCherry-derived CMs (differentiation Day 15) were replated on Matrigel-coated Aclar embedding film for 5 days. Cells were fixed with 2.5% glutaraldehyde, 2% paraformaldehyde, 0.1% tannic acid in 0.1 M cacodylate buffer (pH 7.2) for 30 min. Then, cells were washed three times with 0.2 M sucrose, 0.1% calcium chloride in 0.1 M cacodylate buffer (pH 7.2), and post-fixed in 1% OsO4 in 0.1 M cacodylate buffer (pH 7.2) for 30 min. Cells were washed with ddH2O three times and stained cells with 1% uranyl acetate for 30 min. Cells were then washed with ddH2O three times and dehydrated with gradient ethanol (30%, 50%, 70%, 90%, and 100%) for 5 min each. Samples were infiltrated with 1:1, 2:1 (EPON:100% EtOH) and pure EPON resin. All procedures were done at room temperature. Samples were embedded in EPON at 60°C for 48 h. After slicing, the images of ultrastruture were captured by a Tecnai G2 Spirit TWIN electron microscope (FEI) equipped with a Gatan CCD Camera (794.10.BP2 MultiScan) and acquisition software DigitalMicrograph (Gatan).
2.12 Statistics
A two-tailed Student’s t-test was used to calculate statistical significance.
3. Results
3.1 Generation of knock-in MYH6:mCherry hESC reporter lines using CRISPR/Cas9-based gene targeting
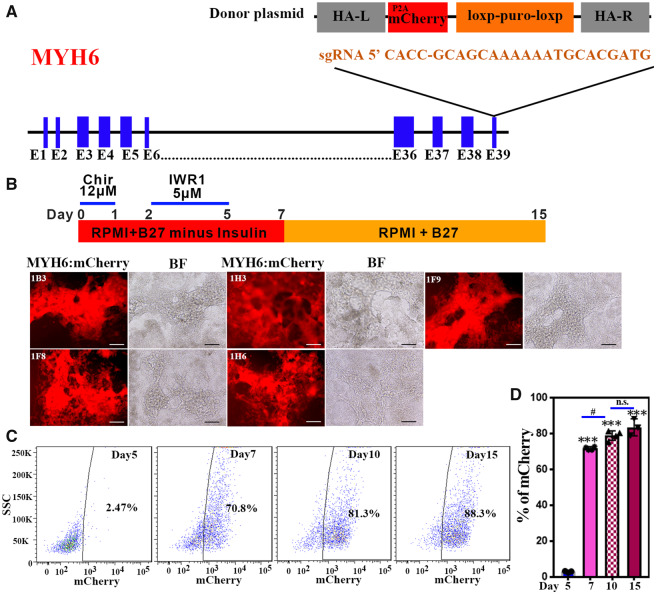
To generate the knock-in reporter lines, we first designed two single guide RNAs (sgRNAs) and cloned the sequences (Table 1) into the pX330 vector that also supports expression of Cas9. Each sgRNA targets a location close to the stop codon of the MYH6 locus. A surveyor assay was used to evaluate the cutting efficiency in 293 T cells. Both sgRNAs were able to target the MYH6 locus, and we chose sg#1 because the targeting efficiency appeared to be slightly higher than for sg#2 (Supplementary material online, Figure S1A). To construct a donor plasmid, left and right homology arms were generated by PCR of genomic DNA and cloned into the pGolden-Hyg plasmid. A fluorescent reporter P2A-mCherry cassette was fused in frame into the left homology arm along with a selection cassette, comprising a loxP-flanked puromycin gene controlled by the PGK promotor (Figure 1A). After validating the targeting and donor plasmids by DNA sequencing, H9 hESCs were transfected with both the sgRNA and donor plasmids by electroporation. Two days later, cells were selected with puromycin for 48 h. Approximately 10 days later, 80 colonies were picked, expanded, and genotyped using primers internal to the donor plasmid (Supplementary material online, Figure S1B, black arrows and Table 1), which generates a 497 bp PCR product. Among 80 clones tested, 75 (94%) positive clones were obtained (Supplementary material online, Figure S1C). After this initial genotyping, we chose six random clones and performed PCR using a pair of primers that were located (forward) in the middle of the LoxP-Puro cassette and (reverse) outside of the HA-R cassette in the untargeted genomic DNA (Supplementary material online, Figure S1B, red arrows and Table 1), which generates a 2086 bp PCR product. All six clones were positive (Supplementary material online, Figure S1D), suggesting that the mCherry construct was inserted to the correct location in most clones. To further demonstrate that the mCherry reporter is knocked into the MYH6 locus after removing LoxP-Puro cassette, we performed PCR using a pair of primers that are located outside of the left and right homology arms (Supplementary material online, Figure S1B, blue arrows and Table 1). We identified heterozygous clones that displayed PCR products for both wildtype and targeted loci (e.g. shown in Supplementary material online, Figure S1E). The targeted allele was further verified by DNA sequencing the larger PCR product (data not shown). To examine whether mCherry insertion affects chromosomal stability, the reporter line was karyotyped and found to have normal chromosomes (Supplementary material online, Figure S1F). To validate the MYH6:mCherry reporter activity, we examined five selected reporter clones using a human cardiac differentiation protocol described previously.3 Notably, mCherry fluorescence was observed starting around Days 6 and 7 of differentiation using each of these selected clones (Figure 1B). The mCherry+ cells showed beating activity at Day 7 of differentiation. Additionally, we typically obtained about 70–90% CM differentiation efficiency (Figure 1C and D). To confirm fidelity of the reporter, we generated a second MYH6:mCherry reporter, exactly as described above, using the distinct (male) H1 hESC line (Supplementary material online, Figure S1G). Again, all the beating cells were mCherry+ and co-expressed TNNT2 (Supplementary material online, Figure S1H).
Figure 1.
MYH6 gene targeting strategy and evaluation of an MYH6:mCherry hESC knock-in reporter line. (A) A sgRNA sequence targets a region close to the stop codon of the MYH6 gene. A P2A-mCherry fluorescence reporter cassette together with an excisable selection marker (PGK-Puro) was inserted using homologous recombination. After genotyping hESC MYH6:mCherry clones, the selection cassette was excised by expression of Cre recombinase. (B) Evaluation of the hESC MYH6:mCherry knock-in reporter line following human cardiomyocyte differentiation. Five independent MYH6:mCherry clones show mCherry fluorescent signals at differentiation Day 15. Scale bar: 100 µm. (C) Percentage of mCherry populations analysed by flow cytometry at different cardiac differentiation stages at Days 5, 7, 10, and 15, respectively. (D) Quantification of the mCherry+ population. Results represent three independent experiments. A two-tailed Student’s t-test was used to calculate statistical significance. Error bars show SD. Statistical significance is indicated: #P < 0.05, ∗∗∗P < 0.001. n.s., not significant.
3.2 Characterization of the MYH6:mCherry expressing cells
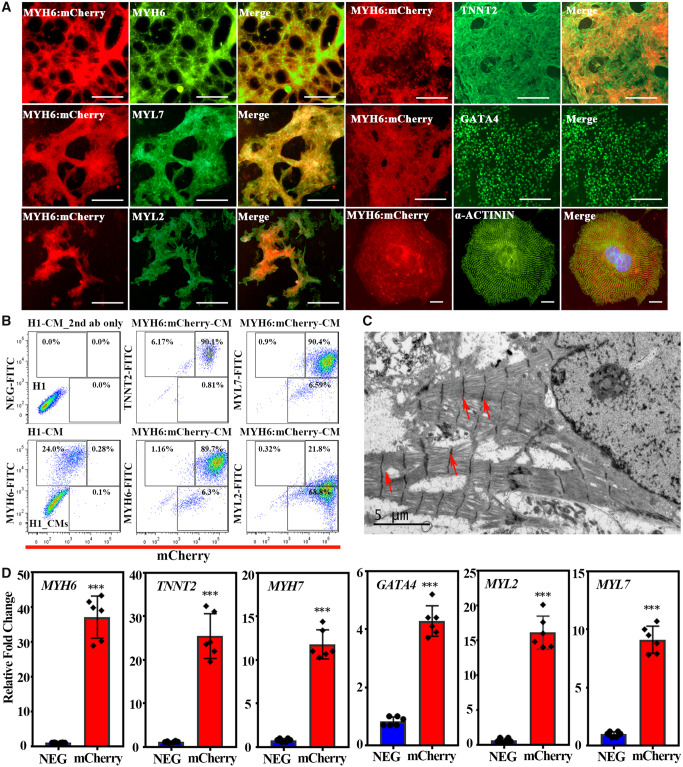
To further characterize the MYH6:mCherry hESC reporter line, immunofluorescence staining was performed using additional cardiac markers, including MYH6, MYL7, MYL2, TNNT2, and GATA4 (Figure 2A). All MYH6:mCherry+ cells labelled positively with an antibody against MYH6, which validates the reporter (Figure 2A). MYH6:mCherry+ cells also express atrial and ventricular markers, MYL7 and MYL2 (Figure 2A), suggesting that the reporter line can identify both types of CMs. Moreover, all MYH6:mCherry+ cells co-express cardiac regulatory transcription factor GATA4 and the differentiation muscle marker TNNT2 (Figure 2A). Flow cytometry was used to show that more than 90% of the MYH6:mCherry+ cells co-expressed TNNT2, MYH6, and MYL7 (Figure 2B). The MYL2 staining was less convincing by flow cytometry either due to limitations of the antibodies or relatively lower expression levels in the ESC-derived cells. We stained mCherry+ cells for the sarcomeric marker alpha ACTININ (ACTN2) and examined cells by TEM to evaluate sarcomeric structures. Notably, the mCherry+ cells displayed organized sarcomeric structures typical of hESC-derived CMs (Figure 2A and C). Additionally, mCherry+ cells were isolated by FACS, and qRT-PCR assays demonstrated robust enrichment compared to mCherry-negative cells for CM markers, including MYH6, TNNT2, MYH7, GATA4, MYL2, and MYL7 (Figure 2D). To further demonstrate that MYH6:mCherry+ cells are CMs, immunostaining was used to confirm that mCherry+ cells do not express the endothelial marker (CD31) or a haematopoietic marker (CD34) (Supplementary material online, Figure S2A). Flow cytometry further demonstrated that mCherry+ cells do not express CD31 or the fibroblast marker CD90, but mCherry+ cells do express cardiomyocyte marker VCAM-1 (Supplementary material online, Figure S2B). Taken together, hESC-derived MYH6:mCherry+ cells express cardiac markers MYH6, MYL7, GATA4, and TNNT2 and are relatively well differentiated as they display organized sarcomeric structures.
Figure 2.
MYH6:mCherry+ cells express cardiac markers and display well-organized sarcomeric structure. (A) MYH6:mCherry+ cells co-express cardiac markers MYH6, MYL7, MYL2, TNNT2, GATA4, and ACTN2 at differentiation Day 15. In the merged images, MYH6:mCherry are shown in red and individual cardiac makers are coloured by green. Scale bar: 100 µm, except for ACTN2: 10 µm. (B) Flow cytometry analysis of MYH6:mCherry+ CMs with co-expressed cardiac markers. (C) Transmission electron micrograph demonstrating the sarcomeric structures in MYH6:mCherry+ CMs. Red arrows indicate Z-lines. (D) Expression of cardiac genes in MYH6:mCherry sorted cells. Results represent data from three independent experiments. A two-tailed Student’s t-test was used to calculate statistical significance. Error bars show SD. Statistical significance is indicated: ∗∗∗P < 0.001.
3.3 Electrophysiological analysis of MYH6:mCherry ESC-derived CMs
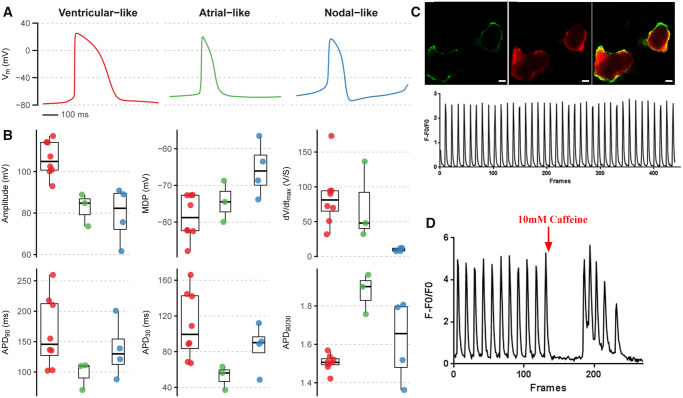
The patch-clamp technique was used to determine electrophysiological properties of CMs derived from the MYH6:mCherry hESC reporter line. The mCherry+ cells are electrically active and display typical profiles found in hESC-derived CMs. Action potentials from a 10 s recording were analysed using MATLAB, and cells clustered naturally into three subtypes based on upstroke velocity (dV/dtmax) and action potential shape (APD90/30 ratio). Criteria for ventricular-like action potentials were a fast upstroke (dV/dtmax > 30) and long plateau phase (APD90/30 ratio < 1.6). For atrial-like action potentials, criteria were a fast upstroke (dV/dtmax > 30) and absence of a plateau phase (APD90/30 ratio > 1.6). For nodal-like action potentials, spontaneity was required along with a slow upstroke (dV/dtmax < 30) (Figure 3A and B). Following 25 days of differentiation, mCherry+ cells were sorted and replated for analysis. Out of 15 patch-clamped cells, eight (53%) showed ventricular-like action potential, three (20%) displayed an atrial-like action potential, and the remaining four (27%) were classified as nodal-like cells (Supplementary material online, Figure S3, Table 2). Moreover, intracellular calcium imaging in these mCherry+ cells revealed spontaneous calcium transients (Figure 3C), and the cells responded robustly to caffeine stimulation (Figure 3D).
Figure 3.
MYH6:mCherry+ cells display atrial-like, ventricular-like, or nodal-like action potential morphology. (A) Examples of the mean spontaneous action potential recorded using whole-cell patch clamp from each of the three subtypes characterized by upstroke velocity (dV/dtmax) and action potential shape (APD90/30 ratio). (B) Action potential parameters from all recorded cells (n = 15) separated by subtype. Boxplot whiskers represent maximum and minimum values within 1.5*inter-quartile range. (C) Images of calcium transients of MYH6:mCherry expressing cells. Scale bar: 50 µm. (D) MYH6:mCherry+ cells respond to caffeine stimulation. Data represent cells analysed from three independent experiments.
Table 2.
Electrophysiological analysis of MYH6:mCherry expressing cells
| Amp (mV) | Amp SE (mV) | APD90 (mV) | APD90 SE (mV) | APD30 (mV) | APD30 SE (mV) | APD90/30 (mV) | dV/dt (V/S) | MDP (mV) | MDP SE (mV) | Category |
|---|---|---|---|---|---|---|---|---|---|---|
| 100.93 | 1.02 | 135.17 | 51.28 | 104.47 | 6.03 | 1.51 | 31.71 | −72.70 | 0.57 | Ventricular |
| 114.18 | 3.74 | 210.65 | 12.80 | 170.05 | 6.58 | 1.48 | 69.79 | −87.93 | 3.30 | Ventricular |
| 102.48 | 0.64 | 102.42 | 1.50 | 82.64 | 1.49 | 1.50 | 72.40 | −82.47 | 0.23 | Ventricular |
| 114.03 | 0.38 | 136.55 | 2.31 | 109.33 | 2.42 | 1.52 | 89.92 | −75.38 | 0.52 | Ventricular |
| 92.99 | 0.77 | 154.86 | 1.51 | 128.68 | 1.03 | 1.42 | 173.14 | −72.58 | 0.76 | Ventricular |
| 100.09 | 1.88 | 103.24 | 4.03 | 82.07 | 2.69 | 1.54 | 94.16 | −82.32 | 1.04 | Ventricular |
| 117.20 | 4.28 | 217.66 | 5.03 | 176.17 | 3.55 | 1.51 | 94.58 | −72.68 | 0.58 | Ventricular |
| 107.32 | 0.77 | 260.00 | 18.64 | 203.03 | 17.33 | 1.57 | 50.86 | −82.19 | 1.13 | Ventricular |
| 61.64 | 1.83 | 87.73 | 2.87 | 61.73 | 1.38 | 1.81 | 7.76 | −56.52 | 0.53 | Atrial |
| 90.83 | 0.98 | 120.97 | 3.20 | 103.04 | 2.67 | 1.36 | 11.85 | −73.83 | 0.42 | Nodal |
| 84.81 | 2.04 | 70.46 | 4.01 | 47.41 | 2.10 | 1.90 | 47.97 | −79.96 | 0.71 | Nodal |
| 88.89 | 0.33 | 110.98 | 0.81 | 81.28 | 0.71 | 1.76 | 136.44 | −68.73 | 0.13 | Nodal |
| 75.63 | 0.60 | 200.79 | 3.80 | 147.52 | 2.64 | 1.79 | 7.57 | −63.50 | 0.27 | Nodal |
| 89.00 | 1.36 | 138.85 | 4.90 | 113.33 | 4.16 | 1.52 | 12.04 | −68.66 | 0.54 | Nodal |
| 73.59 | 1.21 | 110.03 | 3.27 | 74.44 | 1.78 | 1.96 | 32.03 | −74.49 | 0.36 | Nodal |
| Averaged data | ||||||||||
| 106.15 | 2.20 | 165.07 | 19.98 | 132.06 | 7.61 | 1.25 | 84.57 | −78.53 | 1.36 | Ventricular |
| 61.64 | 1.83 | 87.73 | 2.87 | 61.73 | 1.91 | 1.42 | 7.76 | −56.52 | 0.54 | Atrial |
| 83.79 | 1.22 | 125.35 | 3.56 | 94.50 | 3.03 | 1.35 | 41.32 | −71.53 | 0.45 | Nodal |
3.4 Global gene expression profile of CMs derived from MYH6:mCherry hESCs
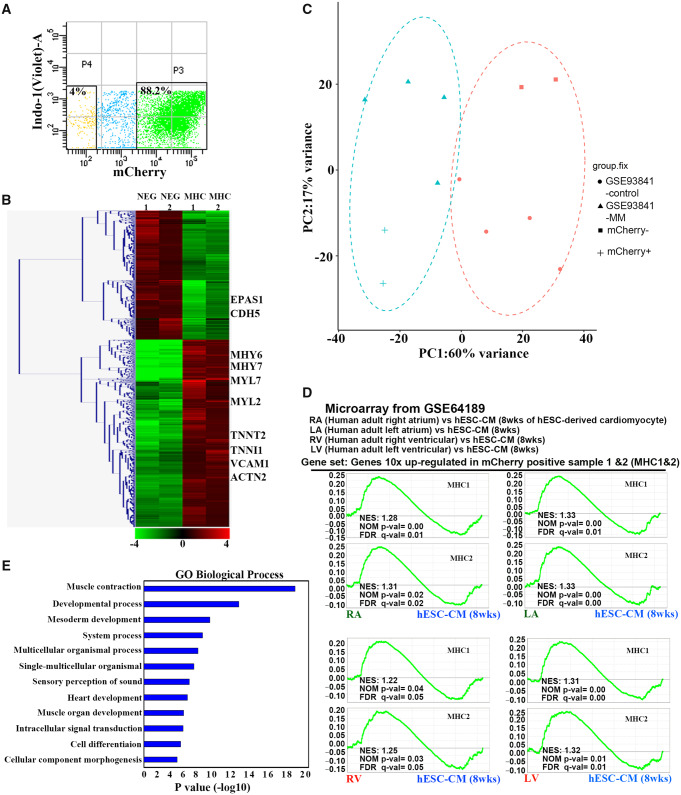
To further characterize the phenotype of MYH6:mCherry+ cells, they were isolated by FACS at differentiation Day 20, following ∼80–90% differentiation efficiency (Figure 4A). RNA sequencing was performed to compare the global gene expression profiles in mCherry− and mCherry+ cell populations. The gene expression levels in the mCherry+ cell population were normalized by the expression levels in the negative cell population. As expected, hierarchical clustering analysis revealed a large geneset containing cardiac-restricted genes that are enriched in the mCherry+ cell population (Figure 4B). MYH7 is expressed predominately in the human foetal and adult ventricle. MYH6 is more restricted to adult atria with minor expression in adult ventricle.39,40 Notably, both MYH6 and MYH7 are highly expressed in mCherry+ cells (Figure 4B). Moreover, atrial and ventricular markers MYL7 and MYL2, and classic CM markers TNNT2, TNNI1, ACTN2, and VCAM1 are all highly expressed in mCherry+ cells (Figure 4B). In addition, we observed that endothelial marker CDH5 (also known as VE-cadherin) and EPAS1 (Endothelial PAS domain-containing protein1) are highly expressed in the negative cell population, but not in the MYH6:mCherry+ population (Figure 4B). PCA was performed to compare the whole genome transcriptome of our samples and a published database of hESC-derived CMs (GSE93841). The MYH6:mCherry+ cells clustered together with hESC-derived CMs matured in a high palmitate medium,41 while the MYH6:mCherry− cell profiles clustered with control cells, which include both undifferentiated progenitors and immature cells (Figure 4C).
Figure 4.
ESC-derived MYH6:mCherry+ cells display cardiac gene expression profiles. (A) A representative FACS plot of MYH6:mCherry+ and MYH6:mCherry− cell populations. Cells were harvested at differentiation Day 20. (B) Hierarchical clustering analysis of altered gene expression based on RNA sequencing from MYH6:mCherry+ and MYH6:mCherry− populations. (C) PCA analysis of the transcriptome of MYH6:mCherry+ and MYH6:mCherry− cells and previously published hESC-derived cardiomyocytes (GSE93841). GSE93841-MM: hESC-derived CMs cultured in maturation medium; GSE93841-control: hESC-derived cells cultured in control medium; (D) GSEA analysis indicates that genesets of two independent MYH6:mCherry+ samples (MHC1 and MHC2) are both significantly enriched in adult human right/left atrial and ventricular cells compared to hESC-CMs (at 8 weeks). A published microarray dataset (GSE64189) and the genesets that are >10-fold up-regulated from MYH6:mCherry+ populations were used for comparison. Enriched genesets are selected based on statistical significance (FDR q value <0.25 and/or NOM P value <0.05). MHC 1 and MHC2: two independent MYH6:mCherry+ samples. RA/LA: human adult right/left atrium. RV/LV: human adult right/left ventricular cells. hESC-CM (8 weeks): hESC-derived cardiomyocytes harvested at differentiation Week 8. (E) GO biological process analyses of the genes that are significantly enriched in human adult right/left atrial samples. Sorted cells are from two independent experiments.
Geneset enrichment analysis (GSEA) was performed to assess if the global gene expression profiles of MYH6:mCherry+ cells are comparable to human heart cells. The gene list was filtered based on expression levels that are at least 10-fold changed in MYH6:mCherry+ cells compared to the negative cells. The gene list was compared with published microarray datasets (GSE64189) for gene expression levels from hESC-derived CMs at different stages: 2, 4, or 8 weeks, in addition to isolated human adult right atrium, left atrium, left ventricle, or right ventricle. The MYH6:mCherry+ cell geneset is significantly enriched in each of these adult human cardiac data sets (Figure 4D), which is consistent with the PCA analysis described above. We further compared the gene list with two genesets derived from profiling adult atrial myocytes and ventricular myocytes. Interestingly, the results are not statistically enriched in either atrial or ventricular myocytes (Supplementary material online, Figure S4), again consistent with MYH6:mCherry+ cells as pan-cardiomyocyte fated. Gene ontology (GO) biological analyses were performed using the gene lists that are significantly enriched in human adult atrial or ventricular myocytes. As expected, genes that are enriched in both atrial and ventricular myocyte sets are involved in various processes of muscle development and cardiac cell development (Figure 4E). Taken together, transcript profiling indicates that MYH6:mCherry+ cells are a highly enriched pan-cardiomyocyte population. Furthermore, RNA-seq analysis suggested that MYH6:mCherry+ cell profiles cluster with relatively mature hESC-derived CM profiles.
3.5 Monitoring drug-induced cardiotoxicity
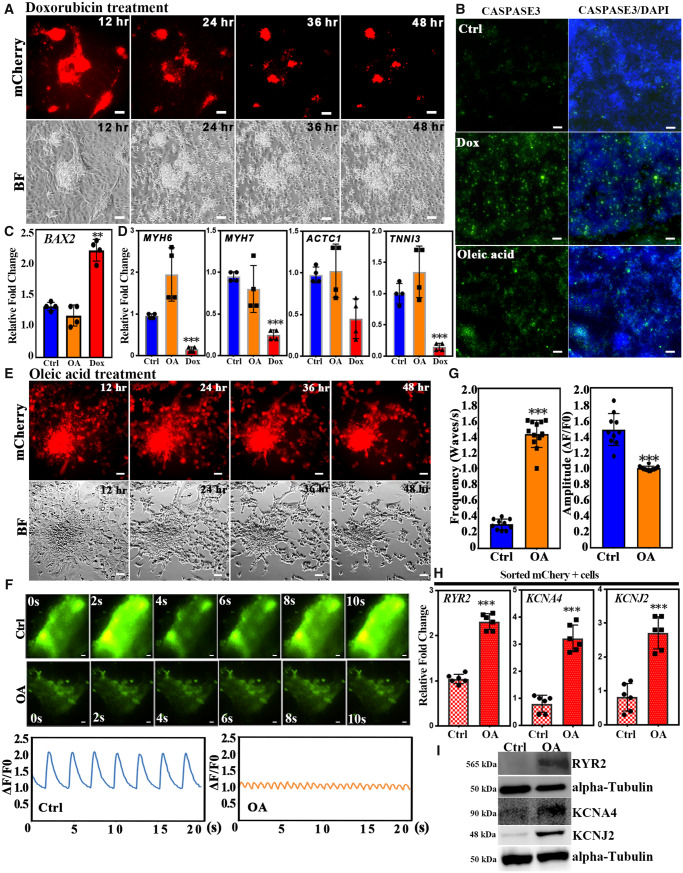
Drug-induced cardiotoxicity has caused failure or withdrawal of more than 90% of potential drug candidates.19,20 Therefore, a simple and robust platform to test drug-induced cardiotoxicity is an important goal for drug development. To evaluate whether the MYH6:mCherry reporter line provides a good cell-based model for cardiotoxicity, we treated Day 20 hESC-derived CMs with doxorubicin (Dox), a known anti-cancer drug that can cause cardiotoxicity.42,43 To quantitatively visualize the progress of cardiotoxicity, we carried out live-cell imaging to monitor Dox-treated cells for 48 h (Supplementary material online, Movie S1). Notably, cells started to die around 12 h after Dox treatment (Figure 5A). Dox-induced cardiotoxicity is known to correlate with apoptosis, which was monitored in treated cells with apoptosis marker cleaved-caspase3. Dox-treated cells were harvested after 24 h treatment. As expected, Dox significantly increased the number of cells positive for cleaved-CASPASE3 compared with the control group (Figure 5B). This result was further confirmed by western blotting experiments (Supplementary material online, Figure S5). Furthermore, expression levels of the pro-apoptotic gene BAX2 were significantly enhanced following Dox treatment (Figure 5C). We also examined if Dox treatment affects the expression of cardiac genes. Based on qPCR experiments, expression of cardiomyocyte genes was significantly decreased (e.g. MYH6, MYH7, ACTC1, and TNNI3,) following 24 h Dox treatment (Figure 5D). Taken together, we validated the capacity for the MYH6:mCherry reporter line to provide a good platform for testing cardiotoxicity.
Figure 5.
Establishment of a platform for cardiotoxicity testing. (A) Day 20 differentiated CMs were treated with 20 µM doxorubixin (Dox) and monitored for 48 h. Shown are representative live images. CMs started to die after 12 h Dox treatment. (B) CMs were harvested and stained with the apoptotic marker cleaved-CASPASE 3 after Dox treatment for 24 h and OA treatment for 48 h, respectively. Green fluorescence indicates cleaved-CASPASE 3+ cells. Scale bar: 50 µm. (C) The expression levels of BAX2 were measured by qRT-PCR in control, OA, and Dox treatments. (D) The expression levels of cardiac genes were determined by qRT-PCR. (E) Day 20 differentiated CMs were treated with 8 mM OA and monitored for 48 h. Shown are representative live images. (F) Shown are representative images of calcium transits of control (Ctrl) or cells showing OA-induced tachycardia. Scale bar: 50 µm. Low panel: quantification of calcium transits. (G) Frequency and amplitude quantification of calcium transits. (H) OA induces the expression levels of transcripts encoding calcium-handling genes RYR2, KCNA4, and KCNJ2. MYH6:mCherry+ cells were sorted from control and OA-treated cells. (I) Representative western blot of RYR2, KCNA4, and KCNJ2 in lysates from control or OA-treated cells. Error bars show SD. Results are from three independent experiments. A two-tailed Student’s t-test was used to calculate statistical significance. Statistical significance is indicated: ∗∗P < 0.01, ∗∗∗P < 0.001.
3.6 Monitoring oleic acid-induced cardiomyocyte arrhythmia
Obesity and metabolic syndrome are risk factors for atrial fibrillation.44 High plasma concentrations of mono-unsaturated fatty acids, including oleic acid (OA), are frequently noted in patients with obesity or other metabolic syndromes. To determine the effect of OA on hESC-derived CMs, MYH6:mCherry-derived CMs were treated at differentiation Day 20 with different doses of OA. In contrast to the Dox-treated condition, OA treatment at these doses neither caused cell death (Figure 5E and Supplementary material online, Movie S2) nor induced a significant increase in cleaved-CASPASE3 (Figure 5B and Supplementary material online, Figure S5) and BAX2 expression levels (Figure 5C). In addition, 48 h OA treatment did not affect the expression of cardiac genes MYH6, MYH7, ACTC1, and TNNI3 (Figure 5D). Instead, the beating rate of OA-treated MYH6:mCherry+ cells was significantly increased after 48 h OA treatment (Supplementary material online, Movie S3A and 3B), and consistent with this observation, the frequency of spontaneous calcium activity was significantly increased following OA treatment compared with the control group (Figure 5F and G). We further used FACS to isolate mCherry+ cells from control and OA-treated groups, respectively. This OA-induced tachycardia was associated with increased expression levels for genes that are involved in intracellular calcium handling for cardiomyocyte contraction including RYR2, KCNA4, and KCNJ2 (Figure 5H), suggesting that OA-induced tachycardia may act through activation of these calcium-handling genes. This result was further confirmed by western blotting (Figure 5I). Mills et al.41 showed that palmitate can promote maturation of hESC-CMs. Thus, to rule out that OA-induced arrhythmia was caused by hESC-CMs immaturity, we first treated hESC-CMs at differentiation Day 15 with 100 µM palmitate for 10 days. As expected, after treating hESC-CMs with palmitate, the proliferation markers Ki67 and PHH3 were significantly decreased (Supplementary material online, Figure S6A and B), consistent with maturation. We treated these hESC-CMs with OA to test if OA can still induce arrhythmia. Notably, OA not only enhanced beating rate (Supplementary material online, Figure S6C) but also increased expression of intracellular calcium-handling genes RYR2, KCNA4 and KCNJ2 (Supplementary material online, Figure S6D), suggesting that OA-induced arrhythmia was independent of maturation status.
4. Discussion
Although relatively efficient directed differentiation protocols have been developed to generate cardiac cells from hPSCs, optimized tools to isolate human CMs with high purity have been limited. Thus, while hPSC-derived CMs provide an outstanding platform for studying cardiac biology, drug toxicity, and development of regenerative therapeutics, the lack of robust tools to image and purify live CM populations presents a gap in their application. We successfully generated MYH6:mCherry hESC reporter lines, using CRISPR/Cas9. Using an established directed differentiation protocol, all of the beating cells are mCherry+. These MYH6:mCherry+ cells can be observed during early differentiation stages already by around Day 6–7 and co-express cardiac markers, MYH6, MYH7, TNNT2, GATA4, and atrial and ventricular myocyte markers, MYL2 and MYL7. Moreover, the mCherry+ cells express the sarcomeric marker ACTN2 and display organized sarcomere structures. The MYH6:mCherry+ cell population, using a standard pan-CM directed differentiation protocol, is comprised of cells with atrial-like, ventricular-like, and nodal-like action potential morphology. Additionally, the global gene expression profiling suggested that these MYH6:mCherry closely resemble relatively mature hESC-derived CMs. However, as is known from many previous studies, hESC-derived CMs are immature compared to adult human CMs. A subset of the mCherry+ CMs express both atrial and ventricular myocyte markers, MYL2 and MYL7, also indicative of an immature status for the derived CMs. Using the MYH6:mCherry reporter line, we could monitor Dox-induced cardiotoxicity and oleic acid-induced cardiomyocyte arrhythmia. We could demonstrate that Dox or OA caused cardiomyocyte defects through distinct mechanisms. Therefore, the reporter is able to distinguish the mechanism of action for different toxins on hESC-derived CMs. It provides a convenient platform to systematically examine cardiac toxins. Therefore, the MYH6:mCherry hESC reporter line should serve as a useful tool for disease modelling and drug development relevant to cardiomyocyte biology.
Interestingly, OA-induced tachycardia is associated with alteration of the expression of calcium-handling genes RYR2, KCNA4, and KCNJ2. These three genes are involved in cardiac excitation-contraction coupling.45 Moreover, mutation of RYR2 has been found in catecholaminergic polymorphic ventricular tachycardia (CPVT) patients. RYR2 is the regulator for sarcoplasmic release of calcium and plays an essential role in regulating heart rate. Patients with RYR2 mutations suffer cardiac arrhythmias.46,47 A gain-of-function mouse model also leads to cardiac arrhythmias and sudden cardiac death.48 Defects in KCNJ2 have been shown to cause short-QT-syndrome and Anderson–Tawil syndrome, including symptoms that induce cardiac arrhythmias.49,50 In addition, fatty acids have been shown to affect Ca2+ homoeostasis, stimulation of mitochondrial damage and induction of cardiac arrhythmia.44,51,52 Therefore, OA-induced tachycardia may function through regulation of these genes and thereby altered Ca2+ concentration in hESC-CMs.
Even after passing clinical trials, many drugs are found to cause cardiovascular toxicity and need to be withdrawn from the market. Since hPSC-CMs can be generated efficiently in scalable amounts using current differentiation protocols, they provide an ideal cell-based model for cardiotoxicity screening. Although several groups have used hPSC-CMs as a platform for cardiotoxicity testing, the interpretation can be limited due to impure cell populations and/or the use of transgenic manipulation. Cardiac differentiation protocols generate mixed cell populations (e.g. endothelial cells, smooth muscle cells, cardiac conduction cells, and CMs), which can complicate interpretations for what cell types are targeted by the drugs. Here, we established a platform for cardiotoxicity testing using an MYH6:mCherry hESC reporter line to distinguish chemical-induced cardiomyocyte death and arrhythmia. This reporter line can be used to easily isolate pure populations of live CMs and as a platform for drug evaluation. In the future, it can also be used for modelling cardiomyopathy disease, coupling gene editing approaches to introduce defined genetic variation and isolation of isogenic mutant CMs for studying pathological mechanisms and drug screening.
Authors’ contribution
S.C., T.E., and S.T. designed the project and provides the funding, S.T., Z.G., Z.B., H.W., Z.H., M.B., and R.K., performed most tissue culture work. S.A. and H.W. performed Ca2+ imaging. F.A.O. and D.J.C. performed electrophysiological experiments. S.C., T.E., and S.T. wrote the manuscript.
Supplementary Material
Acknowledgements
The authors are grateful for technical support and advice provided by Jenny Xiang in the Genomics Resources Core Facility at Weill Cornell Medical College. They appreciate expert assistance from the Flow Cytometry Core Facility at Weill Cornell. They thank Chien-Yuen Pan for help with calcium transient experiments at National Taiwan University and Hsueh-Kai Chang and IBMS electrophysiology lab at Academia Sinica for technical support. They also thank the Technology Commons, College of Life Science, National Taiwan University for technical support.
Conflict of interest: none declared.
Funding
This work was supported by a contract from the Empire State Stem Cell Research Program [NYSTEM, #C028115 to T.E. and S.C.], the National Institutes of Health [R35-HL135778 to T.E.], American Heart Association [18CSA34080171 to S.C. and T.E.], and the Ministry of Science and Technology, Taiwan [MOST 105-2320-B-002-065-MY2 and 107-2320-B-002-026 to S.-Y.T.].
Time for primary review: 52 days
Translational perspective
Cardiovascular disease is a leading cause of death in the world. Moreover, 90% of pharmaceutical compounds eventually fail and are withdrawn from the market primarily due to drug-induced cardiovascular toxicity. Thus, it is compelling needed to establish a platform to overcome this hurdle. Here, we report to generate a fluorescence-based cardiomyocyte reporter, the benefit of studying underlying mechanism of heart disease as well as providing a platform for cardiotoxicity screening.
References
- 1. Mozaffarian D, Benjamin EJ, Go AS, Arnett DK, Blaha MJ, Cushman M, de Ferranti S, Despres JP, Fullerton HJ, Howard VJ, Huffman MD, Judd SE, Kissela BM, Lackland DT, Lichtman JH, Lisabeth LD, Liu S, Mackey RH, Matchar DB, McGuire DK, Mohler ER 3rd, Moy CS, Muntner P, Mussolino ME, Nasir K, Neumar RW, Nichol G, Palaniappan L, Pandey DK, Reeves MJ, Rodriguez CJ, Sorlie PD, Stein J, Towfighi A, Turan TN, Virani SS, Willey JZ, Woo D, Yeh RW, Turner MB.. Heart disease and stroke statistics–2015 update: a report from the American Heart Association. Circulation 2015;131:17. [DOI] [PubMed] [Google Scholar]
- 2. Burridge PW, Matsa E, Shukla P, Lin ZC, Churko JM, Ebert AD, Lan F, Diecke S, Huber B, Mordwinkin NM, Plews JR, Abilez OJ, Cui B, Gold JD, Wu JC.. Chemically defined generation of human cardiomyocytes. Nat Methods 2014;11:855–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lian X, Hsiao C, Wilson G, Zhu K, Hazeltine LB, Azarin SM, Raval KK, Zhang J, Kamp TJ, Palecek SP.. Robust cardiomyocyte differentiation from human pluripotent stem cells via temporal modulation of canonical Wnt signaling. Proc Natl Acad Sci U S A 2012;109:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dubois NC, Craft AM, Sharma P, Elliott DA, Stanley EG, Elefanty AG, Gramolini A, Keller G.. SIRPA is a specific cell-surface marker for isolating cardiomyocytes derived from human pluripotent stem cells. Nat Biotechnol 2011;29:1011–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Elliott DA, Braam SR, Koutsis K, Ng ES, Jenny R, Lagerqvist EL, Biben C, Hatzistavrou T, Hirst CE, Yu QC, Skelton RJ, Ward-van Oostwaard D, Lim SM, Khammy O, Li X, Hawes SM, Davis RP, Goulburn AL, Passier R, Prall OW, Haynes JM, Pouton CW, Kaye DM, Mummery CL, Elefanty AG, Stanley EG.. NKX2-5(eGFP/w) hESCs for isolation of human cardiac progenitors and cardiomyocytes. Nat Methods 2011;8:1037–1040. [DOI] [PubMed] [Google Scholar]
- 6. Skelton RJ, Costa M, Anderson DJ, Bruveris F, Finnin BW, Koutsis K, Arasaratnam D, White AJ, Rafii A, Ng ES, Elefanty AG, Stanley EG, Pouton CW, Haynes JM, Ardehali R, Davis RP, Mummery CL, Elliott DA.. SIRPA, VCAM1 and CD34 identify discrete lineages during early human cardiovascular development. Stem Cell Res 2014;13:172–179. [DOI] [PubMed] [Google Scholar]
- 7. Osborn L, Hession C, Tizard R, Vassallo C, Luhowskyj S, Chi-Rosso G, Lobb R.. Direct expression cloning of vascular cell adhesion molecule 1, a cytokine-induced endothelial protein that binds to lymphocytes. Cell 1989;59:1203–1211. [DOI] [PubMed] [Google Scholar]
- 8. Schwach V, Passier R.. Generation and purification of human stem cell-derived cardiomyocytes. Differentiation 2016;91:126–138. [DOI] [PubMed] [Google Scholar]
- 9. Hattori F, Chen H, Yamashita H, Tohyama S, Satoh YS, Yuasa S, Li W, Yamakawa H, Tanaka T, Onitsuka T, Shimoji K, Ohno Y, Egashira T, Kaneda R, Murata M, Hidaka K, Morisaki T, Sasaki E, Suzuki T, Sano M, Makino S, Oikawa S, Fukuda K.. Nongenetic method for purifying stem cell-derived cardiomyocytes. Nat Methods 2010;7:61–66. [DOI] [PubMed] [Google Scholar]
- 10. Louch WE, Sheehan KA, Wolska BM.. Methods in cardiomyocyte isolation, culture, and gene transfer. J Mol Cell Cardiol 2011;51:288–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tohyama S, Hattori F, Sano M, Hishiki T, Nagahata Y, Matsuura T, Hashimoto H, Suzuki T, Yamashita H, Satoh Y, Egashira T, Seki T, Muraoka N, Yamakawa H, Ohgino Y, Tanaka T, Yoichi M, Yuasa S, Murata M, Suematsu M, Fukuda K.. Distinct metabolic flow enables large-scale purification of mouse and human pluripotent stem cell-derived cardiomyocytes. Cell Stem Cell 2013;12: 127–137. [DOI] [PubMed] [Google Scholar]
- 12. Lin B, Lin X, Stachel M, Wang E, Luo Y, Lader J, Sun X, Delmar M, Bu L.. Culture in glucose-depleted medium supplemented with fatty acid and 3,3',5-Triiodo-l-Thyronine facilitates purification and maturation of human pluripotent stem cell-derived cardiomyocytes. Front Endocrinol 2017;8:253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bu L, Jiang X, Martin-Puig S, Caron L, Zhu S, Shao Y, Roberts DJ, Huang PL, Domian IJ, Chien KR.. Human ISL1 heart progenitors generate diverse multipotent cardiovascular cell lineages. Nature 2009;460:113–117. [DOI] [PubMed] [Google Scholar]
- 14. Huber I, Itzhaki I, Caspi O, Arbel G, Tzukerman M, Gepstein A, Habib M, Yankelson L, Kehat I, Gepstein L.. Identification and selection of cardiomyocytes during human embryonic stem cell differentiation. FASEB J 2007;21:2551–2563. [DOI] [PubMed] [Google Scholar]
- 15. Ritner C, Wong SS, King FW, Mihardja SS, Liszewski W, Erle DJ, Lee RJ, Bernstein HS.. An engineered cardiac reporter cell line identifies human embryonic stem cell-derived myocardial precursors. PLoS One 2011;6:e16004.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kita-Matsuo H, Barcova M, Prigozhina N, Salomonis N, Wei K, Jacot JG, Nelson B, Spiering S, Haverslag R, Kim C, Talantova M, Bajpai R, Calzolari D, Terskikh A, McCulloch AD, Price JH, Conklin BR, Chen HS, Mercola M.. Lentiviral vectors and protocols for creation of stable hESC lines for fluorescent tracking and drug resistance selection of cardiomyocytes. PLoS One 2009;4:e5046.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wong SS, Ritner C, Ramachandran S, Aurigui J, Pitt C, Chandra P, Ling VB, Yabut O, Bernstein HS.. miR-125b promotes early germ layer specification through Lin28/let-7d and preferential differentiation of mesoderm in human embryonic stem cells. PLoS One 2012;7:24.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Maass K, Shekhar A, Lu J, Kang G, See F, Kim EE, Delgado C, Shen S, Cohen L, Fishman GI.. Isolation and characterization of embryonic stem cell-derived cardiac Purkinje cells. Stem Cells 2015;33:1102–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Schuster D, Laggner C, Langer T.. Why drugs fail—a study on side effects in new chemical entities. Curr Pharm Des 2005;11:3545–3559. [DOI] [PubMed] [Google Scholar]
- 20. Singh SS. Preclinical pharmacokinetics: an approach towards safer and efficacious drugs. Curr Drug Med 2006;7:165–182. [DOI] [PubMed] [Google Scholar]
- 21. Zhao Q, Wang X, Wang S, Song Z, Wang J, Ma J.. Cardiotoxicity evaluation using human embryonic stem cells and induced pluripotent stem cell-derived cardiomyocytes. Stem Cell Res Ther 2017;8:54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Koci B, Luerman G, Duenbostell A, Kettenhofen R, Bohlen H, Coyle L, Knight B, Ku W, Volberg W, Woska JR Jr, Brown MP.. An impedance-based approach using human iPSC-derived cardiomyocytes significantly improves in vitro prediction of in vivo cardiotox liabilities. Toxicol Appl Pharmacol 2017;329:121–127. [DOI] [PubMed] [Google Scholar]
- 23. Yokoo N, Baba S, Kaichi S, Niwa A, Mima T, Doi H, Yamanaka S, Nakahata T, Heike T.. The effects of cardioactive drugs on cardiomyocytes derived from human induced pluripotent stem cells. Biochem Biophys Res Commun 2009;387:482–488. [DOI] [PubMed] [Google Scholar]
- 24. Andersson H, Steel D, Asp J, Dahlenborg K, Jonsson M, Jeppsson A, Lindahl A, Kagedal B, Sartipy P, Mandenius CF.. Assaying cardiac biomarkers for toxicity testing using biosensing and cardiomyocytes derived from human embryonic stem cells. J Biotechnol 2010;150:175–181. [DOI] [PubMed] [Google Scholar]
- 25. Braam SR, Tertoolen L, van de Stolpe A, Meyer T, Passier R, Mummery CL.. Prediction of drug-induced cardiotoxicity using human embryonic stem cell-derived cardiomyocytes. Stem Cell Res 2010;4:107–116. [DOI] [PubMed] [Google Scholar]
- 26. Zeevi-Levin N, Itskovitz-Eldor J, Binah O.. Cardiomyocytes derived from human pluripotent stem cells for drug screening. Pharmacol Ther 2012;134:180–188. [DOI] [PubMed] [Google Scholar]
- 27. Burridge PW, Li YF, Matsa E, Wu H, Ong SG, Sharma A, Holmstrom A, Chang AC, Coronado MJ, Ebert AD, Knowles JW, Telli ML, Witteles RM, Blau HM, Bernstein D, Altman RB, Wu JC.. Human induced pluripotent stem cell-derived cardiomyocytes recapitulate the predilection of breast cancer patients to doxorubicin-induced cardiotoxicity. Nat Med 2016;22:547–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. England J, Loughna S.. Heavy and light roles: myosin in the morphogenesis of the heart. Cell Mol Life Sci 2013;70:1221–1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Warkman AS, Whitman SA, Miller MK, Garriock RJ, Schwach CM, Gregorio CC, Krieg PA.. Developmental expression and cardiac transcriptional regulation of Myh7b, a third myosin heavy chain in the vertebrate heart. Cytoskeleton (Hoboken) 2012;69:324–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Carniel E, Taylor MR, Sinagra G, Di Lenarda A, Ku L, Fain PR, Boucek MM, Cavanaugh J, Miocic S, Slavov D, Graw SL, Feiger J, Zhu XZ, Dao D, Ferguson DA, Bristow MR, Mestroni L.. Alpha-myosin heavy chain: a sarcomeric gene associated with dilated and hypertrophic phenotypes of cardiomyopathy. Circulation 2005;112:54–59. [DOI] [PubMed] [Google Scholar]
- 31. Ching YH, Ghosh TK, Cross SJ, Packham EA, Honeyman L, Loughna S, Robinson TE, Dearlove AM, Ribas G, Bonser AJ, Thomas NR, Scotter AJ, Caves LS, Tyrrell GP, Newbury-Ecob RA, Munnich A, Bonnet D, Brook JD.. Mutation in myosin heavy chain 6 causes atrial septal defect. Nat Genet 2005;37:423–428. [DOI] [PubMed] [Google Scholar]
- 32. Ishikawa T, Jou CJ, Nogami A, Kowase S, Arrington CB, Barnett SM, Harrell DT, Arimura T, Tsuji Y, Kimura A, Makita N.. Novel mutation in the alpha-myosin heavy chain gene is associated with sick sinus syndrome. Circ Arrhythm Electrophysiol 2015;8: 400–408. [DOI] [PubMed] [Google Scholar]
- 33. Qiu P, Shandilya H, D'Alessio JM, O'Connor K, Durocher J, Gerard GF.. Mutation detection using Surveyor nuclease. Biotechniques 2004;36:702–707. [DOI] [PubMed] [Google Scholar]
- 34. Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F.. Multiplex genome engineering using CRISPR/Cas systems. Science 2013;339:819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F.. Genome engineering using the CRISPR-Cas9 system. Nat Protoc 2013;8: 2281–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zhang M, Schulte JS, Heinick A, Piccini I, Rao J, Quaranta R, Zeuschner D, Malan D, Kim K-P, Röpke A, Sasse P, Araúzo-Bravo M, Seebohm G, Schöler H, Fabritz L, Kirchhof P, Müller FU, Greber B.. Universal cardiac induction of human pluripotent stem cells in two and three-dimensional formats: implications for in vitro maturation. Stem Cells 2015;33:1456–1469. [DOI] [PubMed] [Google Scholar]
- 37. Ortega FA, Butera RJ, Christini DJ, White JA, Dorval AD 2nd. Dynamic clamp in cardiac and neuronal systems using RTXI. Methods Mol Biol 2014;1183:327–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Patel YA, George A, Dorval AD, White JA, Christini DJ, Butera RJ.. Hard real-time closed-loop electrophysiology with the Real-Time eXperiment Interface (RTXI). PLoS Comput Biol 2017;13:e1005430.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Everett AW. Isomyosin expression in human heart in early pre- and post-natal life. J Mol Cell Cardiol 1986;18:607–615. [DOI] [PubMed] [Google Scholar]
- 40. Kurabayashi M, Tsuchimochi H, Komuro I, Takaku F, Yazaki Y.. Molecular cloning and characterization of human cardiac alpha- and beta-form myosin heavy chain complementary DNA clones. Regulation of expression during development and pressure overload in human atrium. J Clin Invest 1988;82:524–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Mills RJ, Titmarsh DM, Koenig X, Parker BL, Ryall JG, Quaife-Ryan GA, Voges HK, Hodson MP, Ferguson C, Drowley L, Plowright AT, Needham EJ, Wang QD, Gregorevic P, Xin M, Thomas WG, Parton RG, Nielsen LK, Launikonis BS, James DE, Elliott DA, Porrello ER, Hudson JE.. Functional screening in human cardiac organoids reveals a metabolic mechanism for cardiomyocyte cell cycle arrest. Proc Natl Acad Sci USA 2017;114:E8372–E8381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lefrak EA, Pitha J, Rosenheim S, Gottlieb JA.. A clinicopathologic analysis of adriamycin cardiotoxicity. Cancer 1973;32:302–314. [DOI] [PubMed] [Google Scholar]
- 43. Von HDD, Layard MW, Basa P, Davis HL Jr, Von HA, Rozencweig M, Muggia FM.. Risk factors for doxorubicin-induced congestive heart failure. Ann Intern Med 1979;91:710–717. [DOI] [PubMed] [Google Scholar]
- 44. Nystrom PK, Carlsson AC, Leander K, de Faire U, Hellenius ML, Gigante B.. Obesity, metabolic syndrome and risk of atrial fibrillation: a Swedish, prospective cohort study. PLoS One 2015;10:e0127111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Preininger MK, Jha R, Maxwell JT, Wu Q, Singh M, Wang B, Dalal A, McEachin ZT, Rossoll W, Hales CM, Fischbach PS, Wagner MB, Xu C.. A human pluripotent stem cell model of catecholaminergic polymorphic ventricular tachycardia recapitulates patient-specific drug responses. Dis Model Mech 2016;9: 927–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Priori SG, Napolitano C, Tiso N, Memmi M, Vignati G, Bloise R, Sorrentino V, Danieli GA.. Mutations in the cardiac ryanodine receptor gene (hRyR2) underlie catecholaminergic polymorphic ventricular tachycardia. Circulation 2001;103:196–200. [DOI] [PubMed] [Google Scholar]
- 47. Priori SG, Chen SR.. Inherited dysfunction of sarcoplasmic reticulum Ca2+ handling and arrhythmogenesis. Circ Res 2011;108:871–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Lehnart SE, Mongillo M, Bellinger A, Lindegger N, Chen BX, Hsueh W, Reiken S, Wronska A, Drew LJ, Ward CW, Lederer WJ, Kass RS, Morley G, Marks AR.. Leaky Ca2+ release channel/ryanodine receptor 2 causes seizures and sudden cardiac death in mice. J Clin Invest 2008;118:2230–2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ai T, Fujiwara Y, Tsuji K, Otani H, Nakano S, Kubo Y, Horie M.. Novel KCNJ2 mutation in familial periodic paralysis with ventricular dysrhythmia. Circulation 2002;105:2592–2594. [DOI] [PubMed] [Google Scholar]
- 50. Priori SG, Pandit SV, Rivolta I, Berenfeld O, Ronchetti E, Dhamoon A, Napolitano C, Anumonwo J, di Barletta MR, Gudapakkam S, Bosi G, Stramba-Badiale M, Jalife J.. A novel form of short QT syndrome (SQT3) is caused by a mutation in the KCNJ2 gene. Circ Res 2005;96:800–807. [DOI] [PubMed] [Google Scholar]
- 51. Wanahita N, Messerli FH, Bangalore S, Gami AS, Somers VK, Steinberg JS.. Atrial fibrillation and obesity—results of a meta-analysis. Am Heart J 2008;155:310–315. [DOI] [PubMed] [Google Scholar]
- 52. Zhang Y, Ren J.. Role of cardiac steatosis and lipotoxicity in obesity cardiomyopathy. Hypertension 2011;57:148–150. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.