Abstract
Glioma is the most common primary tumour of the central nervous system, and is associated with a high postoperative recurrence rate and resistance to chemotherapy. High-grade glioblastoma in particular has a very poor prognosis and poses a serious threat to human health. Related studies have confirmed that the occurrence and development of gliomas are closely associated with the abnormal expression and regulation of genes. Moreover, the number of studies on the association of the expression of non-coding RNAs [linear RNAs, microRNAs and circular RNAs (circRNAs)] in human cells with glioma has been gradually increasing in recent years. Among those, circRNAs, previously considered to be 'splicing errors', have been shown to be highly expressed in eukaryotic cells and regulate the biological behaviour of gliomas. circRNAs are highly abundant and stable, and have become a research hotspot in the field of glioma molecular biology. The aim of the present review was to focus on the research progress regarding the association between circRNA expression and gliomas, and to provide a theoretical basis according to the currently available literature for further exploring this association. The present study may be of value for the early diagnosis, pathological grading, targeted therapy and prognostic evaluation of gliomas.
Keywords: circular RNAs, glioma, glioblastoma, function, mechanism, central nervous system, biomarker, gene therapy, correlation, research progress
1. Introduction
Glioma is the most common primary tumour of the central nervous system, and its incidence accounts for ~80% of primary brain malignancies. Glioma is a high-grade malignancy and has a poor prognosis. Despite various high-intensity treatments, including surgery combined with chemoradiation, the median survival time of patients with glioblastoma (GBM) is only 12-15 months, and only 3-5% of the patients survive for >3 years (1-3). In recent years, genetic diagnosis and targeted therapy have been attracting widespread attention as emerging research hotspots, and studies have confirmed that the targeting of non-coding RNAs in glioma may be more efficacious. Non-coding RNAs are regulatory factors that participate in embryonic development, inflammatory response, metabolism and chemotherapy resistance. Additionally, tumour development, cell invasion, proliferation and apoptosis are also closely associated with the expression and regulation of non-coding RNAs (4-6).
Circular RNAs (circRNAs) are important members of the non-coding RNA family, and they are single-stranded closed circular RNA molecules without a 5′-end cap or a 3′-end poly(A) tail, which are formed by covalent bonding. Due to their special circular stable structure, circRNAs cannot be degraded by RNase R, and they are highly evolutionarily conserved. When circRNAs were first discovered in the 1970s, they were considered as 'noise' resulting from incorrect splicing. However, with the rapid development of high-throughput sequencing-based molecular biotechnology in recent years, several circRNAs have been detected in eukaryotic cells. As regards the gene expression of human cells, circRNA molecules have more general expression characteristics compared with their linear counterparts (7,8). Moreover, circRNAs are abundant and stable. A large number of studies have confirmed that circRNAs are differentially expressed in various tumour cells (9,10). Their regulatory effects on tumourigenesis, tumour development and corresponding biological behaviours require further in-depth research. The aim of the present review was to focus on the research progress in this field, in order to provide a theoretical and literature basis for further exploring the association between the expression of circRNAs and gliomas. The findings of the study may prove to be of value for the early diagnosis, pathological grading, targeted therapy and prognostic evaluation of gliomas.
2. History of circRNAs
In the 1970s, Sanger et al discovered the presence of circRNAs in RNA viruses (11). In 1979, Hsu and Coca-Prados first observed, by means of electron microscopy, that RNA in the cytoplasm of eukaryotic cells may exist in a circular form (12). One year later, Arnberg et al also observed the presence of circRNAs while studying the components of yeast mitochondria (13). In 1993, Cocquerelle et al reported that there were several exon-derived circRNAs in human cell transcripts (14). During the early years of circRNA discovery, circRNAs were considered non-functional, lowly expressed RNA molecules resulting from 'mis-splicing' of exon transcripts. Due to this interpretation, the depth and breadth of circRNA research has been inconsistent. Up until the beginning of the 21st century, scientists had identified no more than 10 types of circRNAs. However, in recent years, with the rapid development of molecular biology technology and bioinformatics analysis based on RNA sequencing (RNA-seq), scientists have identified several exon-derived transcripts that form circRNAs by non-linear reverse splicing or gene rearrangements. These transcripts account for a large proportion of the entire splicing transcript. In 2012, Salzman et al discovered hundreds of circRNAs and established that they are closely associated with human gene expression (15). Jeck and Sharpless identified ~25,000 circRNAs through RNA detection in human fibroblasts (16). Memczak et al compared the RNA-seq results with the human leukocyte database and found 1,950 human circRNAs, 1,903 mouse circRNAs (81 circRNAs were the same as human circRNAs), and 724 nematode circRNAs (17). Guo et al conducted deep sequencing on 39 biological samples related to human cell lines and found >7,000 circRNAs (18). In 2013, two major studies on circRNAs were published in Nature (17,19). Since then, numerous related studies have been published, and circRNAs have come to represent a new direction in the field of non-coding RNA worldwide.
3. Basic characteristics of circRNAs
Novel and unique circRNAs
circRNAs are generated from variable splicing. The majority are formed by the circularization of exons, and a few are derived from introns (Fig. 1). The majority of circRNAs are located in the cytoplasm of eukaryotic cells, but a small proportion are located in the nucleus (mainly intron-derived circRNAs). They are specific per tissue type, disease type and chronological order; overall, they are highly evolutionarily conserved, although there are also certain evolutionary changes (20,21).
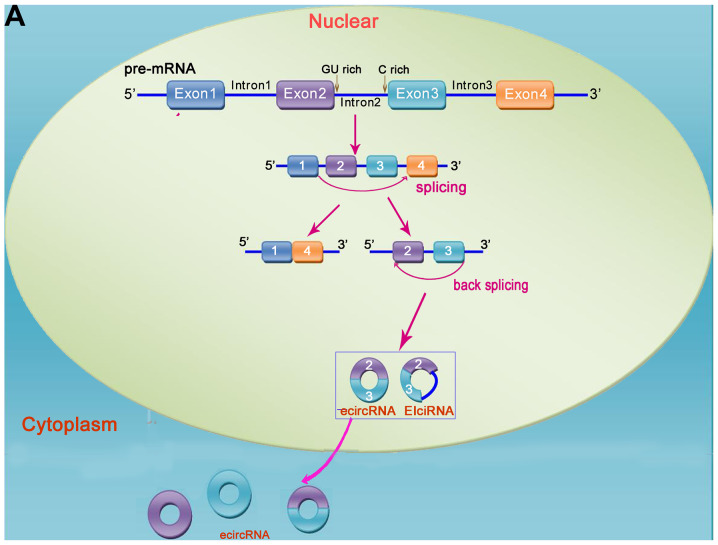
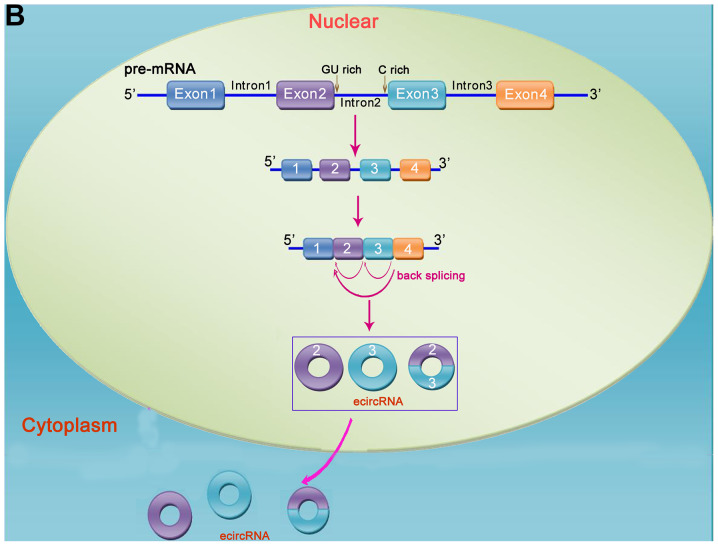
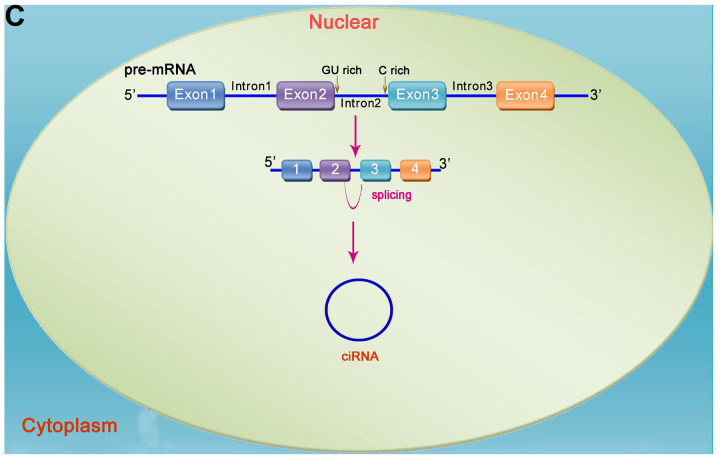
Figure 1.
Mechanism of circRNA formation. circRNAs are formed via reverse splicing and include three main types, namely ecircRNA (exons only), ciRNA (introns only), and EIciRNA (introns inserted between two exons). (A) In pre-mRNA transcripts, non-adjacent exons close to each other can form lariat intermediates, and ecircRNA or EIciRNA may form via exon skipping. (B) Pre-mRNA is processed into mature mRNA by splicing, and ecircRNA forms via reverse splicing and cyclization. ecircRNA is transferred from the nucleus to the cytoplasm, where it exerts its function. (C) ciRNA is formed by a lariat intermediate containing exons. circRNA, circular RNA.
'Tailless' circRNAs
The conventional 5′-end cap and 3′-end poly(A) tail structure in linear RNA molecules are absent in circular RNAs due to their closed circular structure. As one of the key steps in classical RNA detection methods (RNA extraction) the principle of isolating RNA depends on the structure of the poly(A) tail. This may be an important reason that the research on circRNA molecules was overlooked prior to the rapid development of sequencing technology.
Non-translated circRNAs
A number of circRNAs are produced by related genes with the ability to encode proteins; however, to date, the vast majority of circRNAs are still classified as new non-coding RNAs.
Localization and stability of circRNAs
The majority of circRNAs are located in the cytoplasm and, occasionally, their expression level is >10 times that of the corresponding linear isomers. The increased expression may be explained by the fact that RNase R acts by recognizing the end sites of linear RNA structures, and circRNAs have a special 'end-to-end' closed loop structure, which makes them highly resistant to nucleases and degradation.
circRNAs act as competitive endogenous RNAs (ceRNAs)
circRNAs are enriched in the cytoplasm and have the same transcription sequence as their corresponding linear counterparts. This characteristic also indirectly indicates that circRNAs are likely to play biological roles by regulating the corresponding linear RNAs. In fact, given the high abundance and stability of circRNAs in specific tissues, they exhibit extremely strong ceRNA activity when they interact with linear RNAs. It was recently demonstrated that the newly discovered circRNA-CDR1as (also referred to as ciRS-7) has >60 conserved binding sites for miR-7. Experiments in zebrafish were later conducted, and their results revealed that the expression of ciRS-7 may damage the development of the midbrain. This effect was the same as that of knocking out miR-7 (22).
4. The present and future of the biological functions of cir-cRNAs
circRNAs are mainly located in the cytoplasm or exosomes, and exhibit high stability and strong specificity in terms of tissue, disease and timing. In recent years, with the in-depth study of gene function and mechanisms, an increasing number of studies have demonstrated that circRNAs have important regulatory functions in the growth and development of organisms, immune responses, inflammatory responses, and tumour development. circRNAs are also becoming prominent in the field of disease diagnostic markers in biomedical medicine. They have great potential for clinical application, but the types of biological functions and underlying mechanisms of action of circRNAs remain unknown. To date, circRNAs have been found to have several biological functions as follows (Fig. 2): i) miRNA sponging: circRNAs contain abundant miRNA-binding sites, which are the structural basis for their molecular sponge action, and inhibit the interaction between the 3′ untranslated region of miRNAs and mRNAs, thereby indirectly affecting the expression of downstream target genes and proteins of miRNAs. However, other studies reported that most identified human and mouse circRNAs rarely contain miRNA target sites, indicating that these circRNAs may function in other manners that have yet to be determined. Therefore, it was inferred that the main mode of circRNA action may not be miRNA sponging, or that there may be other unknown mechanisms (23). ii) Binding of regulatory proteins: circRNAs can interact with RNA-binding proteins (RBPs), which are regulated by mRNAs, and change the splicing mode or regulate the stability of mRNAs. iii) Regulation of gene transcription: circRNAs can interact with RNA polymerase II, or exon-intron circRNA can first bind with a small ribonucleoprotein and then with RNA polymerase II to play a role in regulating gene transcription. iv) Potential encoding capabilities: Although circRNAs were originally classified as non-coding genes, certain circRNAs can be translated by ribosomes and encode polypeptides based on the latest protein translation research. Moreover, some circRNAs already have the ability to translate proteins and then play roles corresponding to their related proteins. These findings have opened up a new and fascinating perspective in functional studies of circRNAs (24-26).
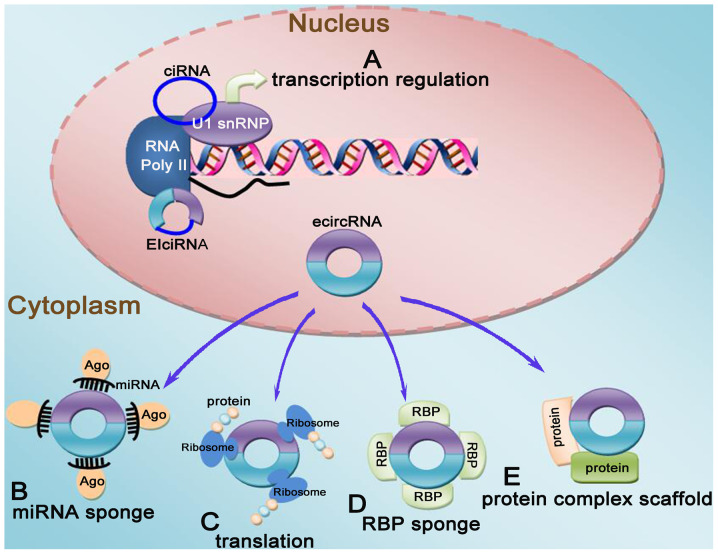
Figure 2.
Functional summary of circRNAs. (A) Transcriptional regulation: In the nucleus, EIciRNA and ciRNA can regulate transcription by directly interacting with RNA polymerase II and U1 snRNP. (B) miRNA molecular sponging: ecircRNA that contains miRNA-binding sites can block the binding between miRNA and its target mRNA and prevent the inhibitory effect of miRNA on the target protein. (C) Regulation of translation: circRNAs contain an internal ribosome entry site, which can bind to the ribosome to regulate translation. (D) RBP sponging: circRNAs with an RBP-binding site can act as a protein sponge, thereby regulating protein activity. (E) circRNAs may function as protein scaffold. circRNA, circular RNA; RBP; RNA-binding protein; ecircRNA, exonic circular RNA; ciRNA, circular intronic RNA; EIciRNA, exon-intron circular RNA.
5. circRNAs in the central nervous system
Compared with other tissues, circRNAs are more diverse and abundant in the central nervous system. Rybak-Wolf et al reported high-level dynamic expression of circRNAs in mammalian brain tissue. The authors analysed the relevant data based on RNA-seq in separate brain regions, and discovered 65,731 brain-specific circRNAs in primary neurons and isolated synaptic samples (27). In addition, some researchers have collected specific circRNAs in human and mouse tissues and identified 89,137 brain-specific circRNAs in human foetuses (28). In 2014, Westholm et al analysed the structure and distribution characteristics of circRNAs in the brain of Drosophila, and found that the formation of circRNAs tends to surround the long intron regions of brain tissue and to aggregate in ageing brain tissue (29). In 2015, in a report published by Professor You in the Journal of Nature Neuroscience, his research team analysed the basic characteristics of central nervous system circRNAs and confirmed that, based on animal experiments, the expression of circRNAs in the central nervous system is indeed higher compared with that in other systems, and that the genes associated with synapses are more easily circularized. Immediately after that, scientists used optimized high-resolution fluorescence in situ hybridization (FISH) to prove that circRNAs are indeed highly localized in the dendrites of neurons (30). In the same year, Rybak-Wolf et al reported the same conclusion in his studies (27). By analysing the expression of circRNAs in mammalian brain tissue, that research group confirmed the abundance of circRNA molecules in brain tissue and that the expression of circRNAs was positively associated with developmental brain processes, particularly in synaptic structures. They subsequently hypothesized that the expression of circRNAs may differ among different anatomical structures of the brain, and proceeded to analyse the expression of circRNAs in different brain-related structures (striatum, prefrontal cortex, olfactory cortex, cerebellum and hippocampal tissue). As predicted, the expression profiles of circRNAs in different neural tissues were indeed different (27). A report published in Science in 2017 demonstrated that the loss of mammalian circRNA loci directly led to miRNA imbalance, which affected brain function (31).
In addition, the miRNA-mRNA-circRNA interaction network may form the epigenetic basis of neurological diseases. For example, miRNA-7 acts as a regulator of α-synuclein and ubiquitin protein ligase A, which is a causative agent of chronic neurodegenerative diseases, such as Alzheimer's disease, Parkinson's disease, amyotrophic lateral sclerosis, multiple sclerosis and multiple system atrophy. In addition, circRNAs also play an important role in secondary brain damage following acute central nervous system insults, such as stroke and neonatal hypoxia-ischemia. Recent studies have also indicated a potential link between depressive disorder or Moyamoya disease and circRNA regulation (32,33). As regards why circRNAs are more likely to be enriched in brain tissue, as mentioned earlier, circRNAs are more likely to form near long introns; in the central nervous system, important genes carry longer introns, which may be the genetic basis for the formation and enrichment of circRNAs (34). In addition, nerve-specific RBPs are likely to be closely associated with the formation of circRNAs. However, the mechanisms of action of some known RBPs, such as splicing factor muscleblind, Quaking and adenosine-to-inosine acting on RNA enzyme 1, which are associated with circRNA formation, appear to contradict this hypothesis. Therefore, scientists inferred that other unknown RBPs, which also play important roles in promoting the formation and enrichment of circRNAs, may be present in the central nervous system.
As regards the role of circRNAs in the nervous system, an in vitro differentiation model was established to systematically analyse the change trends in the expression of circRNAs during the differentiation processes. The results demonstrated that the expression of circRNAs suddenly increases during the formation of synapses (33). A related study revealed that an abrupt increase in circRNA expression levels occurs during the transition from P10 to P30 in mice (30). Another study on circRNAs in the human brain reported the same findings. Compared with the amount of circRNAs during embryonic development, their levels increased significantly in foetal and adult brains (27,28). The synaptogenic function of circRNAs was also supported by their host genes that are associated with synaptic function. On the other hand, a number of brain-enriched circRNAs were found to be associated with neurotransmitter function, neuron maturation and synaptic activity. For example, circFoxo3 was shown to be highly active in binding and deactivating cyclin-dependent kinase 2, leading to disrupted cell cycle progression (35). In addition, circHomer 1 was proven to modulate some of the structural changes at the synapse during neuronal plasticity and development (30). Furthermore, it has been demonstrated that 58% of cerebral circRNAs are developmentally regulated, while only 2% of homologous linear isoforms have exhibited this trend (36). Interestingly, several developmentally regulated circRNAs also exhibited sexual dimorphism in the brain and were observed to target ageing-associated mRNAs (32,33).
6. circRNA expression in gliomas
In recent years, several non-coding RNAs associated with the occurrence and development of gliomas have been identified. The pattern and characteristics of circRNA expression in gliomas have been attracting increasing attention in the field of non-coding RNA research (37,38).
Glioma is a term referring to a large group of central nervous system tumours, and its pathological classification is variable, so the expression and regulation of circRNAs are also different (39). Song et al included 20 fresh-frozen de novo GBM samples, 19 normal brain tissue samples and 7 samples of other oligodendrogliomas (3 WHO grade III and 4 WHO grade II) as the basis of library preparation and sequencing (40). The researchers found 572 highly expressed circRNAs in the 46 samples, 476 of which were differentially expressed between GBM and normal brain tissue, and the expression of 468 circRNAs in normal brain tissue was significantly upregulated compared with that in GBM tissue. Even in normal brain tissue, the researchers observed differential expression profiles of circRNAs among different anatomical locations. The findings mentioned above provided an important molecular and theoretical basis for the tissue- and disease-specific expression patterns of circRNAs. Moreover, studies on the correlation between the differential expression of circRNAs and the pathological grade or prognosis of glioma also confirmed the tissue specificity (41,42).
Wang et al analysed 33 pairs of matched isocitrate dehydrogenase wild-type GBM and paracancerous tissues by microarray (43). The results demonstrated that, compared with paracancerous tissues, there were 254 upregulated circRNAs and 361 downregulated circRNAs (>1.5-fold) in GBM tissues. In addition, 3 pairs of matched GBM and para-cancerous tissues were also selected for microarray analysis (lncRNAs, miRNAs and circRNAs), and 548 upregulated circRNAs and lncRNAs (>1.5-fold) were identified. The expression levels of circRNAs were further verified between 10 pairs of matched gliomas and adjacent normal brain tissue and 2,709 differentially expressed circRNAs were identified. In total, 105 differentially expressed circRNAs were later discovered by cross-matching the two abovementioned data sets (43). Xu et al downloaded 3 pairs of RNA-seq-related data for gliomas and normal brain tissue from the Gene Expression Omnibus database and analysed the expression of circRNAs. A total of 5 different circRNA analysis tools were used and 12 universally expressed circRNAs were identified (44).
In addition to the abovementioned studies based on high-throughput screening technology, other studies have also reported that circRNAs may be potential biomarkers for gliomas. Barbagallo et al studied 56 paraffin-embedded GBM biopsy samples and 7 normal brain tissue samples. The expression level of circSMARCA5 was determined in the control group. The results revealed that the expression of circ-SMARCA5 in GBM was significantly lower compared with that in the control group, but there was no significant difference in the linear mRNA level of the host gene SMARCA5. Furthermore, the research team also analysed the expression of miR-671 in 45 GBM samples and 5 GBM cell lines. The results revealed the presence of the miR-671/CDR1as/miR-7 axis in GBM, and that miR-671-5p overexpression significantly increased the migration capacity of GBM cells, but exerted a less prominent effect on cell proliferation (45). In addition, some researchers analysed 31 glioma tumour samples and matched adjacent normal tissues. Based on the obtained data and biological information analysis, circCFH was found to be upregulated in WHO grade I-II and III-IV glioma samples (46). Xie also investigated the expression of hsa-circ-0012129 in 31 matched glioma tumours and adjacent normal tissues, and the results revealed that the expression of hsa-circ-0012129 was significantly higher in glioma tissues compared with that in adjacent tissues (47).
Bioinformatics predictive analysis and biotin-labelled miR-442a pull-down assays have confirmed that the blinding of miR-442a to circNT5E is significantly upregulated in glioma samples. In a study by Wang et al, the interaction between miR-442a and circNT5E was verified using RNA pull-down and FISH techniques. The researchers had previously demonstrated that circNT5E can bind to miR-442a and inhibit its function (48). In this case, the expression levels of miR-442a and circNT5E were negatively correlated. This represents the adsorption and inhibitory action of typical circRNAs as miRNA sponges. However, in the earliest miRNA sponge models of CDR1as and miR-7, the expression level of CDR1as was not affected by miR-7, but was regulated by miR-671. The same research group also observed an interaction between circNFIX and miR-34a-5p through RNA co-immunoprecipitation, and confirmed that NOTCH1 is a target of miR-34a-5p, i.e., when circNFIX is inhibited by siRNA, the expression level of miR-34a-5p is upregulated, while NOTCH1 is inhibited (44).
Thus far, although most studies have focused on the differential expression of circRNAs in gliomas and some emerging molecular circRNA species have also been discovered and identified, the expression patterns of circRNAs in gliomas exhibit high specificity and diversity among individuals. This indicates that the expression pattern of circRNAs in glioma and normal brain tissues requires a larger-sample in-depth study to verify.
7. circRNAs are involved in regulating the life activities of glioma cells
circRNAs and the glioma cell cycle
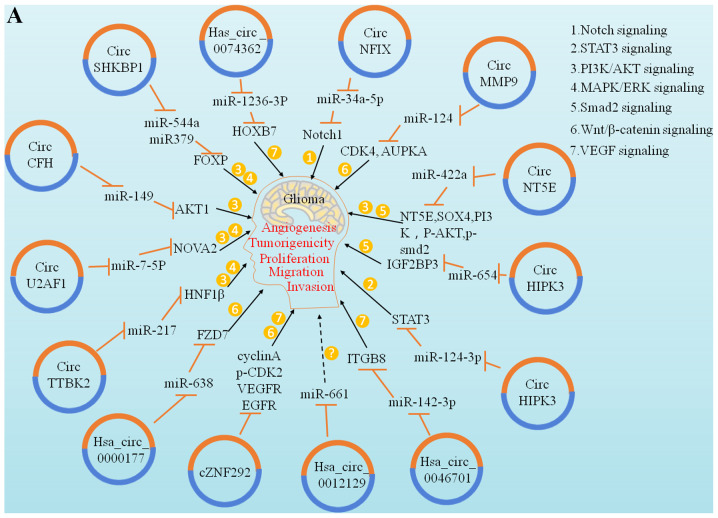
The ability of tumour cells to proliferate indefinitely is the most important characteristic that distinguishes them from normal cells. The overexpression of genes and proliferation of tumour cells play important roles in the regulation of the cell cycle. Numerous studies have demonstrated that circRNAs may play the role of competitive endogenous RNAs (ceRNAs) in tumour proliferation control. A study by Kumar et al reported that has-circ-0046701 is over-expressed in glioma tissues and cell lines. If has-circ-0046701 is knocked out, it can significantly inhibit the proliferation and invasion of tumour cells; further investigation revealed that it can promote the expression of ITGβ8, a target of miR-142-3p, and regulate the proliferation and invasion of glioma cells. Therefore, the has-circ-0046701/miR-142-3p/ITGβ8 loop is considered to be involved in regulating the occurrence and development of gliomas (49). The study of glioma cells by Liu et al and Yang et al revealed that the cZNF292 gene is associated with angiogenesis and that it is an important carcinogenic circRNA that participates in the important process of tumour cell lumen formation, whereas cZNF292 gene knockout can inhibit cell proliferation and causes cell cycle arrest via the Wnt/β-catenin signalling pathway (50,51) (Fig. 3).
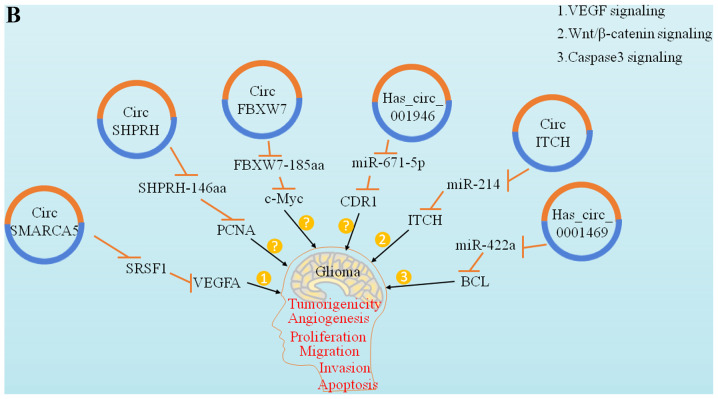
Figure 3.
Functions and clinical significance of circRNAs in glioma. (A) Representative diagram of circRNAs involved in the regulation of glioma cell proliferation, migration, invasion, etc. Multiple identified circRNAs act as miRNA sponges and subsequently upregulate the relevant target gene expression level. These target genes or proteins further regulate downstream factors associated with cancer signaling pathways via acting as transcriptional factors or regulatory proteins and other mechanisms. For example, circ-NFIX activates the Notch signaling pathway through miR-34a-5p. (B) Representative diagram of circRNAs involved in the regulation of glioma cell proliferation, migration, invasion, etc. Multiple identified circRNAs act as miRNA sponges and subsequently downregulate the relevant target gene expression level. These target genes or proteins further regulate downstream factors associated with cancer signaling pathways via acting as transcriptional factors or regulatory proteins and other mechanisms. For example, cir-ITCH activates the Wnt/β-catenin signaling pathway through miR-214. circRNA, circular RNA.
circRNAs and glioma cell apoptosis
Loss of apoptosis is another important characteristic of tumour cells. It was recently reported that some circRNAs are involved in regulating tumour cell apoptosis (52). The Bcl-2 protein family plays a key role in the apoptotic pathway. The anti-apoptotic protein Bcl-2 is located in the outer mitochondrial membrane and can inhibit the secretion and release of pro-apoptotic factors, such as cytochrome c. Bcl-2 is a target of miR-143. circRNAs also play an important role in regulating the apoptotic pathway as miRNA molecular sponges (53). A study by Zhang et al revealed that the circular form of the snf2 histone linker PHD cyclohelicase (circ-SHPRH) can encode SHPRH-146aa and protects SHPRH from degradation by ubiquitin protease. Both are widely present and highly expressed in the normal human brain. However, circ-SHPRH expression is significantly reduced in GBM. Therefore, the low expression of SHPRH-146aa may promote the malignant behaviour and tumourigenicity of tumour cells in vivo (54). circ-SHPRH generates 'UGA' stop codons to promote the translation of SHPRH-146aa. It is hypothesized that circ-SHPRH and SHPRH-146aa are tumour suppressors of human GBM and promote glioma cell apoptosis (Fig. 3).
circRNAs and glioma cell invasion and migration
Invasion and migration are hallmarks of malignant tumours. Epithelial-to-mesenchymal transition (EMT) plays an important role in regulating early invasion and migration in several tumours. miRNAs are involved in the regulation of EMT (55). circRNAs, acting as miRNA sponges, may also participate in the regulation of EMT through endogenous competitive mechanisms to affect tumour cell invasion and migration. A study by Zheng et al demonstrated that hepatocyte nuclear factor 1 (HNF1) is a direct target of miR-217 and is an important carcinogen at the glioma molecular level. In addition, circ-TTBK2 is a circRNA gene that can bind to and inactivate miR-217, thereby promoting the activation of the phosphatidylinositol 3-kinase/protein kinase B-extracellular signal-regulated kinase pathway through HNF-1β. Activation can trigger the invasion of glioma cells, leading to a further increase in tumour malignancy. When circ-TTBK2 is knocked out, miR-217 overexpression can inhibit tumour cell invasion and migration in vivo and promote glioma cell apoptosis (56). Li et al observed that has-circ-0007534 is highly expressed in glioma tissues and that its knockdown can also inhibit the occurrence and development of gliomas (57). ZIC5 has also been found to be a target of miR-761 (57). Has-circ-0007534 can promote the expression of ZIC5 by inhibiting miR-761 in glioma cells, thereby markedly enhancing the proliferation and migration ability of glioma cells. It has been reported that the expression level of has-circ-0007534 is positively correlated with advanced glioma grade (WHO III/IV) (57). In addition, the degradation of has-circ-0007534 significantly inhibited the proliferation and migration of glioma cells, indicating that circRNAs may also play a role in the differentiation of glioma cells (Fig. 3).
8. circRNAs promote the occurrence and development of gliomas
circRNAs act as promoters in glioma
Song et al collected 46 samples of glioma tissues and normal brain tissues for gene chip analysis. Among 572 measurable circRNAs, 476 exhibited a significantly different expression between glioma and normal brain tissue. The expression of circ_CDR1, circ_ATRNL1, circ_AKT3, circ_SPTAN1, circ_ZNF483, circ_RIMS1, circ_FKBP8 and circ_UNC13C was low in glioma tissues, while the expression of circ_COL1A2, circ_PTN, circ_VCAN, circ_SMO, circ_PLOD2, circ_GLIS3 and circ_EPHB4 and circ_GLIP2 was high compared with that in normal brain tissues (40).
Due to the strong specificity of the expression pattern of circRNAs in gliomas, their regulatory effects on the occurrence and development of gliomas have also attracted significant attention. A series of circRNAs implicated in carcinogenesis are involved in initiating the malignant progression of glioma. For example, circNFIX is upregulated in GBM tissues and cells. The inhibition of circNFIX can inhibit cell migration and proliferation by downregulating the NOTCH1 pathway. This also indicates that circNFIX may be involved in the occurrence and development of gliomas (44). In addition, circNT5E affects the proliferation, invasion and migration ability of GBM cells in the interaction system by stimulating miR-422a. This also provides further evidence of circRNAs acting as molecular sponges (48).
It has been reported that the expression of circ-TTBK2 is significantly increased in glioma tumour tissues and corresponding cell lines, while the RNA expression of TTBK2, corresponding to the linear structure, is not significantly different. It may be hypothesized that the overexpression of circ-TTBK2 plays a role in the proliferation, invasion and migration of tumour cells. This also confirms that circ-TTBK2 plays a key role in the progression of gliomas (56). A study by Chen and Duan demonstrated that the inhibitory effect of miRNA on hsa_circ_0000177 can significantly inhibit cell proliferation and invasion, which affects the participation of the FZD7/Wnt7 pathway in the progression of gliomas through sponge adsorption of miR-638 (58). In addition to the abovementioned circRNAs, other studies have also reported that hsa_circ_0046701 and circHIPK3 can promote the proliferation and migration of glioma cells (59,60).
In addition to the role of circRNAs in promoting cell proliferation and migration, circRNAs can also promote angiogenesis. It was demonstrated that, compared with endothelial cells exposed to astrocytes, the corresponding SHKBP1 mRNA of circ-SHKBP1 is upregulated in U87 glioma-exposed endothelial cells (GECs), and knocking out circ-SHKBP1 can significantly inhibit GEC proliferation, migration and angiogenesis. circ-SHKBP1 affects the proliferation of GECs through the miR-544a/FOXP1 and miR-379/FOXP2 pathways (61). In another study, Yang et al reported that cZNF292 may be involved in the angiogenesis of human gliomas and that cZNF292 circRNA inhibits glioma angiogenesis via the Wnt/β-catenin pathway (51).
circRNAs as inhibitors in gliomas
The regulatory effect of several circRNAs on gliomas is mediated via tumour suppression. A study by Zhang et al demonstrated that circ-FBXW7 encodes FBXW7-185aa, a protein consisting of 185 amino acids that can significantly inhibit tumour cell processes in vivo and reduce the half-life of c-Myc by antagonizing the stabilization effect of c-Myc induced by USP28. An in situ GBM mouse model also demonstrated the tumour-suppressive effect of FBXW7-185aa through IRES mutation. In addition, circ-SHPRH can be translated into a 146-aa peptide called SHPRH-146aa. Compared with paracancerous tissues, the expression of SHPRH-146aa in tumour tissues was found to be significantly lower (62). Other findings have demonstrated that SHPRH-146aa overexpression can significantly inhibit the growth of gliomas in xenograft mouse models; in addition, it can protect the full-length SHPRH protein from degradation by the ubiquitin proteasome system (54). In addition to the common tumour suppressor genes mentioned above, recent studies have also reported other tumour suppressor genes associated with gliomas. circ-SMARCA5 is downregulated in glioma cells, and circ-SMARCA5 overexpression can increase SRSF3 expression and effectively control the excessive proliferation of glioma cells (45). In addition, other researchers have confirmed that circ-ITCH and hsa_circ_0001649 act as tumour suppressors in gliomas, and they may prove beneficial for the treatment of gliomas in the future (63,64).
9. Emergence of circRNAs in the clinical treatment of glio-mas
Relevant research shows that circRNAs are widely present in body fluids, such as plasma, cerebrospinal fluid and breast milk, particularly in exosomes. The circular closed structure of circRNAs protects them from degradation and confers stability (65). Therefore, their potential application for gene diagnosis and targeted therapy appears to be a promising approach.
It was demonstrated that hsa_circ_0133159 and circ_ZEB1 are significantly overexpressed in glioma tissues and cell lines, and that they may be used as biomarkers in the differential diagnosis and prognostic evaluation of gliomas (66).
miR-24-3p is overexpressed in glioma cells and promotes tumour cell proliferation, migration and invasion by down-regulating bone morphogenetic protein 3 (67). Recent studies have revealed that endothelial monocyte-activating polypeptide II (EMAP-II) can inhibit angiogenesis and promote endothelial cell apoptosis, and can directly promote tumour cell autophagy and apoptosis. The binding of EMAP-II to temozolomide (TMZ) reduces the expression of miR-24-3p, thereby enhancing the cytotoxic effect of TMZ on the proliferation, migration and invasion of germline stem cells in vitro and in vivo, further enhancing its therapeutic efficacy. circRNAs play important roles in the regulation of this process (68). Combined treatment with miR-24-3p (-), EMAP-II and TMZ can reduce tumour malignancy and increase the survival rate, whereas it has also been reported that EMAP-II can promote tumour necrosis factor receptor 1 apoptosis, thereby exerting an antitumour effect (68). In addition, the permeability of the blood-brain barrier is increased to further promote tumour cell apoptosis.
Wesselhoeft et al invented a new method for expressing proteins through engineered circRNA vectors. As circRNAs are relatively more stable compared with normal linear RNA, protein expression based on circRNAs can greatly increase production efficiency. The results revealed that exogenous circRNAs can produce high-quality and high-yield proteins, and indicated that circRNAs are good and efficient RNA molecules that can be used for protein expression in the future (69). Meganck et al constructed a tissue-specific circRNA expression vector based on a recombinant adeno-associated virus (AAV) vector. The results demonstrated that the vector efficiently expressed and translated circRNAs in mouse brain, eyes and heart (70). Based on the research results mentioned above, in the near future, protein-based expression systems will likely be used to design circRNAs that target specific brain areas to treat glioma.
In addition, the molecular sponge effect of circRNAs may also be used in the treatment of gliomas in the future. It was recently reported that a circRNA with 5 miR-21-binding sites, scRNA21, can significantly reduce the expression level of miR-21 and inhibit the proliferation of gastric cancer cell lines (71). Although that study addressed gastric cancer treatment, the concept of designing and synthesizing circRNAs for specific miRNAs may hold promise in the treatment of gliomas. miR-21 and several other miRNAs play important roles in the development of gliomas. This also provides a basis for gene therapy for gliomas, and scRNA21 may also be used to interfere with these miRNAs in gliomas. AAV-based circRNA expression vectors are a promising method and experimental basis for this novel treatment concept (72).
TMZ is a first-line chemotherapy agent for glioma treatment. According to tumour histopathology, immunohistochemistry and molecular diagnosis, for patients with high-grade (WHO classification) gliomas, particularly GBM, who undergo surgical treatment, the routine use of TMZ after surgery is crucial for preventing glioma recurrence and prolonging patient survival. However, tumour resistance to chemotherapeutic drugs is an important cause of treatment failure (73). Ding et al found that exosomal circNFIX was upregulated in the serum of TMZ-resistant patients and predicted a poor prognosis. In addition, TMZ-resistant cell-derived circNFIX conferred TMZ resistance to recipient sensitive cells by regulating cell migration, invasion and apoptosis following TMZ exposure. Further analyses revealed that following the knockdown of circNFIX, this enhanced TMZ sensitivity in resistant glioma cells by upregulating miR-132. Therefore, in terms of the circRNA molecular sponge effect, circNFIX can induce a sponge effect of miR-132, thereby enhancing TMZ resistance in glioma. (74,75).
Finally, the latest study by Yang et al in 2017 revealed that circ-FBXW7 has the ability to translate and encode proteins. The 185-amino acid circ-FBXW7-185aa encoded by circRNA-FBXW7 has a marked antitumour effect. At the animal and clinical levels, after analysing the correlation between the expression of circRNA-FBXW7 and overall survival, it was observed that its expression level was positively associated with overall survival (62). Therefore, circRNAs have potential application prospects as biomarkers for evaluating the prognosis of patients with glioma.
10. Final remarks
The role of RNAs in cellular activities is multi-layered and multi-faceted. Despite being the most recently discovered non-coding RNA molecules, circRNAs still follow the basic rules of RNA. Current studies indicate that, in the human genome, circRNAs are functional molecules in various tissues and cells (10). They are differentially expressed across different functional tissues and cells, which indicates that circRNAs may regulate different biological functions. circRNAs may play important roles in the proliferation, apoptosis and invasion of glioma cells through functions such as competitive inhibition, alternative splicing, transcriptional regulation, protein coding, antiviral immune responses and miRNA sponging; however, the specific mechanisms remain elusive (76). The continuous development of bioinformatics analysis technology may further promote the exploration of circRNAs in the field of glioma.
Although the popularity of circRNAs in the research field is increasing, glioma is a highly heterogeneous disease that has different pathological manifestations at different stages of the disease and in different anatomic locations, in the same patient or in different patients. Therefore, the association between circRNAs and the clinical characteristics of gliomas is complex and remains largely elusive, and the mechanisms underlying glioma-specific circRNA expression patterns require further investigation. Thus far, circRNA knockout remains challenging. As the majority of circRNAs form from genes with coding ability, knockout or downregulation of the corresponding circRNAs will inevitably affect the host genes. In addition, widespread low-abundance expression levels may also hinder protein translation of circRNAs in gliomas and other human diseases.
Finally, high-throughput sequencing technology, also referred to as 'next-generation' sequencing technology, removes technical obstacles to reveal a large number of unknown circRNAs. However, research on the function and mechanism of circRNAs is in its early stages (77,78). In terms of glioma research at the cellular and molecular biology level, the concept of studying a certain gene, protein or pathway alone is outdated. In the future, the key stages of glioma development must be further investigated to identify specific circRNAs, in order to establish a complete gene-protein interaction network and to determine their function and underlying mechanism of action by comprehensively using both in vitro and in vivo experiments and clinical trials. In the foreseeable future, circRNAs are expected to become a focus in the field of non-coding RNA research. Related studies will be required to confirm that circRNAs play key roles in glioma and other diseases, thereby providing new perspectives for the early diagnosis, pathological grading, targeted therapy and prognosis of this disease.
Finally, relevant websites on circRNA biological information analysis and databases for researchers have been organized (Tables I and II).
Table I.
Software for circRNA identification.
| Name | URL | Trait | Refs. |
|---|---|---|---|
| MapSplice | http://www.netlab.uky.edu/p/bioinfo/MapSplice | Two-step method of tag matching and splicing inference | (79) |
| CIRCfinder | https://github.com/YangLab/CIRCfinder | Identification of circRNA in introns | (20) |
| circRNA_finder | https://github.com/orzechoj/circRNA_finder.git | High accuracy. Runs independently of gene annotation information | (29) |
| segemehl | www.bioinf.uni-leipzig.de/Software/segemehl/ | Detects splicing, trans-splicing and gene fusion events | (80) |
| KNIFE | https://github.com/lindaszabo/KNIFE | Static modelling and detection of circRNA. | (81) |
| DCC | https://github.com/dieterich-lab/DCC | Assesses expression between circRNA and host genes | (82) |
| Acfs | https://code.google.com/p/acfs/ | Allows de novo sequencing to accurately identify circRNA and quantify abundance | (83) |
| UROBORUS | http://uroborus.openbioinformatics.org/ | Accurately predicts circRNA with low expression | (40) |
| Circseq_cup | http://ibi.zju.edu.cn/bioinplant/ | Assembly of full-length circRNA sequences | (84) |
| FUCHS | https://github.com/dieterich-lab/FUCHS | Analyses alternative splicing in circRNA, single and double breakpoint events and read coverage of circRNA | (85) |
| CIRI-full | https://sourceforge.net/projects/ciri-full/ | Quantifies alternative splicing products of circRNA | (86) |
| CircExplorer2 | https://github.com/YangLab/CIRCexplorer2 | De novo assembly of full-length circRNA transcripts | (87) |
| CIRI2 | https://sourceforge.net/projects/ciri/files/CIRI2 | Filters false positives from repeated sequences and mapping errors | (88) |
| CircView | http://gb.whu.edu.cn/CircView/ or https://github.com/GeneFeng/CircView | Allows users to view the regulatory elements of circRNA and predict potential function | (89) |
| PRAPI | http://www.bioinfor.org/bioinfor/tool/PRAPI/ | Vector graphics of circRNA | (90) |
| CircSplice | http://gb.whu.edu.cn/CircSplice/ or https://github.com/GeneFeng/CircSplice | Comparison of differences between groups of alternative spliced circRNA | (91) |
circRNA, circular RNA.
Table II.
Databases for circRNA deposition.
| Tool | URL | Trait | Refs. |
|---|---|---|---|
| circAtlas | http://circatlas.biols.ac.cn | Includes circRNA sequence information for humans and other animals. Inquire about binding of circRNA to miRNA or RBPs. | (92) |
| circBase | http://www.circbase.org/ | Includes all identified circRNA data for humans and other animals | (93) |
| CIRCpedia | http://www.picb.ac.cn/rnomics/circpedia | Alternative reverse splicing and alternative splicing of human circRNA | (87) |
| TSCD | http://gb.whu.edu.cn/TSCD | Storage of tissue-specific circRNA from humans and mice | (94) |
| CircInteractome | http://circinteractome.nia.nih.gov | Query binding sites of interacting proteins and related miRNAs on human circRNA | (95) |
| CircNet | http://circnet.mbc.nctu.edu.tw/ | circRNA-miRNA-mRNA interaction regulatory network | (96) |
| circRNADb | http://reprod.njmu.edu.cn/circrnadb | Describes 46 circRNAs with coding potential | (97) |
| CSCD | http://gb.whu.edu.cn/CSCD | Describes 87 cancer-specific circRNAs | (98) |
| circR2Disease | http://bioinfo.snnu.edu.cn/circR2Disease/ | Correlation between 100 human circRNAs and 661 diseases. | (99) |
| MiOncoCirc | https://nguyenjoshvo.github.io/ | Description of targets that can be used for cancer diagnosis or treatment | (100) |
circRNA, circular RNA.
Acknowledgments
The authors would like to give special thanks to Ruijuan Ren, the curator of Hebei University library, for her guidance and assistance in the literature retrieval process of this manuscript.
Funding
The present study was financially supported by the government-funded Provincial Clinical Medicine Talent Programs in 2017; the High-level Talents Scientific Research Launched Project of Hebei University (grant no. 521000981301); the Science and Technology Capacity Improvement Projects of Hebei University of Chinese Medicine in 2019 (grant no. KTZ2019019); the Outstanding Student Scientific Research Ability Improvement Projects of Hebei University of Chinese Medicine in 2019 (grant no. YXZ2019002); the Graduate Innovative Ability Training Projects of Hebei University of Chinese Medicine in 2020 (grant no. XCXZZBS2020002); and the Graduate Innovative Ability Training Projects of Hebei University in 2020 (grant no. hbu2020bs003).
Availability of data and materials
The datasets used and analysed during the present study are available from the corresponding author on reasonable request.
Authors' contributions
YZ and XL were major contributors to the writing and revision of the manuscript. XG, LS, QL and FL collected the related references and participated in the discussions. CF and HWwere involved in the conception and design of the study. All authors have read and approved the final version of the manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
All the authors declare that they have no competing interests.
References
- 1.Pace A, Dirven L, Koekkoek JAF, Golla H, Fleming J, Rudà R, Marosi C, Rhun EL, Grant R, Oliver K, et al. European Association of Neuro-Oncology palliative care task force: European Association for Neuro-Oncology (EANO) guidelines for palliative care in adults with glioma. Lancet Oncol. 2017;18:e330–e340. doi: 10.1016/S1470-2045(17)30345-5. [DOI] [PubMed] [Google Scholar]
- 2.Weller M, van den Bent M, Tonn JC, Stupp R, Preusser M, Cohen-Jonathan-Moyal E, Henriksson R, Rhun EL, Balana C, Chinot O, et al. European Association for Neuro-Oncology (EANO) Task Force on Gliomas: European Association for Neuro-Oncology (EANO) guideline on the diagnosis and treatment of adult astrocytic and oligodendroglial gliomas. Lancet Oncol. 2017;18:e315–e329. doi: 10.1016/S1470-2045(17)30194-8. [DOI] [PubMed] [Google Scholar]
- 3.Bush NA, Chang SM, Berger MS. Current and future strategies for treatment of glioma. Neurosurg Rev. 2017;40:1–14. doi: 10.1007/s10143-016-0709-8. [DOI] [PubMed] [Google Scholar]
- 4.Calore F, Lovat F, Garofalo M. Non-coding RNAs and cancer. Int J Mol Sci. 2013;14:17085–17110. doi: 10.3390/ijms140817085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shukla GC, Gupta S. Hallmarks of cancer- focus on RNA metabolism and regulatory noncoding RNAs. Cancer Lett. 2018;420:208–209. doi: 10.1016/j.canlet.2018.01.063. [DOI] [PubMed] [Google Scholar]
- 6.Bach DH, Lee SK, Sood AK. Circular RNAs in Cancer. Mol Ther Nucleic Acids. 2019;16:118–129. doi: 10.1016/j.omtn.2019.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang Y, Lu T, Wang Q, Liu J, Jiao W. Circular RNAs: Crucial regulators in the human body (Review) Oncol Rep. 2018;40:3119–3135. doi: 10.3892/or.2018.6733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Patop IL, Wüst S, Kadener S. Past, present, and future of circRNAs. EMBO J. 2019;38:e100836. doi: 10.15252/embj.2018100836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arnaiz E, Sole C, Manterola L, Iparraguirre L, Otaegui D, Lawrie CH. circRNAs and cancer: Biomarkers and master regulators. Semin Cancer Biol. 2019;58:90–99. doi: 10.1016/j.semcancer.2018.12.002. [DOI] [PubMed] [Google Scholar]
- 10.Patop IL, Kadener S. circRNAs in Cancer. Curr Opin Genet Dev. 2018;48:121–127. doi: 10.1016/j.gde.2017.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sanger HL, Klotz G, Riesner D, Gross HJ, Kleinschmidt AK. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc Natl Acad Sci USA. 1976;73:3852–3856. doi: 10.1073/pnas.73.11.3852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hsu MT, Coca-Prados M. Electron microscopic evidence for the circular form of RNA in the cytoplasm of eukaryotic cells. Nature. 1979;280:339–340. doi: 10.1038/280339a0. [DOI] [PubMed] [Google Scholar]
- 13.Arnberg AC, Van Ommen GJ, Grivell LA, Van Bruggen EF, Borst P. Some yeast mitochondrial RNAs are circular. Cell. 1980;19:313–319. doi: 10.1016/0092-8674(80)90505-X. [DOI] [PubMed] [Google Scholar]
- 14.Cocquerelle C, Mascrez B, Hétuin D, Bailleul B. Mis-splicing yields circular RNA molecules. FASEB J. 1993;7:155–160. doi: 10.1096/fasebj.7.1.7678559. [DOI] [PubMed] [Google Scholar]
- 15.Salzman J, Gawad C, Wang PL, Lacayo N, Brown PO. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One. 2012;7:e30733. doi: 10.1371/journal.pone.0030733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat Biotechnol. 2014;32:453–461. doi: 10.1038/nbt.2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 18.Guo JU, Agarwal V, Guo H, Bartel DP. Expanded identification and characterization of mammalian circular RNAs. Genome Biol. 2014;15:409. doi: 10.1186/s13059-014-0409-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 20.Zhang Y, Zhang XO, Chen T, Xiang JF, Yin QF, Xing YH, Zhu S, Yang L, Chen LL. Circular intronic long noncoding RNAs. Mol Cell. 2013;51:792–806. doi: 10.1016/j.molcel.2013.08.017. [DOI] [PubMed] [Google Scholar]
- 21.Hansen T. Biogenesis and function of circRNAs. FEBS Open Bio. 2019;9:39. [Google Scholar]
- 22.Shen Y, Guo X, Wang W. Identification and characterization of circular RNAs in zebrafish. FEBS Lett. 2017;591:213–220. doi: 10.1002/1873-3468.12500. [DOI] [PubMed] [Google Scholar]
- 23.Huang G, Li S, Yang N, Zou Y, Zheng D, Xiao T. Recent progress in circular RNAs in human cancers. Cancer Lett. 2017;404:8–18. doi: 10.1016/j.canlet.2017.07.002. [DOI] [PubMed] [Google Scholar]
- 24.Pamudurti NR, Bartok O, Jens M, Ashwal-Fluss R, Stottmeister C, Ruhe L, Hanan M, Wyler E, Perez-Hernandez D, Ramberger E, et al. Translation of circRNAs. Mol Cell. 2017;66:9–21.e7. doi: 10.1016/j.molcel.2017.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu XQ, Gao YB, Zhao LZ, Cai YC, Wang HY, Miao M, Gu LF, Zhang HX. Biogenesis, research methods, and functions of circular RNAs. Yi Chuan. 2019;41:469–485. doi: 10.16288/j.yczz.19-061. In Chinese. [DOI] [PubMed] [Google Scholar]
- 26.Harper KL, Mcdonnell E, Whitehouse A. circRNAs: From anonymity to novel regulators of gene expression in cancer (Review) Int J Oncol. 2019;55:1183–1193. doi: 10.3892/ijo.2019.4904. [DOI] [PubMed] [Google Scholar]
- 27.Rybak-Wolf A, Stottmeister C, Glažar P, Jens M, Pino N, Giusti S, Hanan M, Behm M, Bartok O, Ashwal-Fluss R, et al. Circular RNAs in the Mammalian Brain Are Highly Abundant, Conserved, and Dynamically Expressed. Mol Cell. 2015;58:870–885. doi: 10.1016/j.molcel.2015.03.027. [DOI] [PubMed] [Google Scholar]
- 28.Chen BJ, Huang S, Janitz M. Changes in circular RNA expression patterns during human foetal brain development. Genomics. 2019;111:753–758. doi: 10.1016/j.ygeno.2018.04.015. [DOI] [PubMed] [Google Scholar]
- 29.Westholm JO, Miura P, Olson S, Shenker S, Joseph B, Sanfilippo P, Celniker SE, Graveley BR, Lai EC. Genome-wide analysis of drosophila circular RNAs reveals their structural and sequence properties and age-dependent neural accumulation. Cell Rep. 2014;9:1966–1980. doi: 10.1016/j.celrep.2014.10.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.You X, Vlatkovic I, Babic A, Will T, Epstein I, Tushev G, Akbalik G, Wang M, Glock C, Quedenau C, et al. Neural circular RNAs are derived from synaptic genes and regulated by development and plasticity. Nat Neurosci. 2015;18:603–610. doi: 10.1038/nn.3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Piwecka M, Glažar P, Hernandez-Miranda LR, Memczak S, Wolf SA, Rybak-Wolf A, Filipchyk A, Klironomos F, Cerda Jara CA, Fenske P, et al. Loss of a mammalian circular RNA locus causes miRNA deregulation and affects brain function. Science. 2017;357:357. doi: 10.1126/science.aam8526. [DOI] [PubMed] [Google Scholar]
- 32.Wang Q, Qu L, Chen X, Zhao YH, Luo Q. Progress in Understanding the Relationship Between Circular RNAs and Neurological Disorders. J Mol Neurosci. 2018;65:546–556. doi: 10.1007/s12031-018-1125-z. [DOI] [PubMed] [Google Scholar]
- 33.Mehta SL, Dempsey RJ, Vemuganti R. Role of circular RNAs in brain development and CNS diseases. Prog Neurobiol. 2020;186:101746. doi: 10.1016/j.pneurobio.2020.101746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen LL, Yang L. Regulation of circRNA biogenesis. RNA Biol. 2015;12:381–388. doi: 10.1080/15476286.2015.1020271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Du WW, Yang W, Chen Y, Wu ZK, Foster FS, Yang Z, Li X, Yang BB. Foxo3 circular RNA promotes cardiac senescence by modulating multiple factors associated with stress and senescence responses. Eur Heart J. 2017;38:1402–1412. doi: 10.1093/eurheartj/ehw001. [DOI] [PubMed] [Google Scholar]
- 36.Mahmoudi E, Fitzsimmons C, Geaghan MP, Shannon Weickert C, Atkins JR, Wang X, Cairns MJ. Circular RNA biogenesis is decreased in postmortem cortical gray matter in schizophrenia and may alter the bioavailability of associated miRNA. Neuropsychopharmacology. 2019;44:1043–1054. doi: 10.1038/s41386-019-0348-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rynkeviciene R, Simiene J, Strainiene E, Stankevicius V, Usinskiene J, Miseikyte Kaubriene E, Meskinyte I, Cicenas J, Suziedelis K. Non-Coding RNAs in Glioma. Cancers (Basel) 2018;11:17. doi: 10.3390/cancers11010017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu J, Zhao K, Huang N, Zhang N. Circular RNAs and human glioma. Cancer Biol Med. 2019;16:11–23. doi: 10.20892/j.issn.2095-3941.2018.0425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Louis DN, Perry A, Reifenberger G, von Deimling A, Figarella-Branger D, Cavenee WK, Ohgaki H, Wiestler OD, Kleihues P, Ellison DW. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: A summary. Acta Neuropathol. 2016;131:803–820. doi: 10.1007/s00401-016-1545-1. [DOI] [PubMed] [Google Scholar]
- 40.Song X, Zhang N, Han P, Moon BS, Lai RK, Wang K, Lu W. Circular RNA profile in gliomas revealed by identification tool UROBORUS. Nucleic Acids Res. 2016;44:e87. doi: 10.1093/nar/gkw075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sun J, Li B, Shu C, Ma Q, Wang J. Functions and clinical significance of circular RNAs in glioma. Mol Cancer. 2020;19:34. doi: 10.1186/s12943-019-1121-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhu J, Ye J, Zhang L, Xia L, Hu H, Jiang H, Wan Z, Sheng F, Ma Y, Li W, et al. Differential Expression of Circular RNAs in Glioblastoma Multiforme and Its Correlation with Prognosis. Transl Oncol. 2017;10:271–279. doi: 10.1016/j.tranon.2016.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang HX, Huang QL, Shen JY, Xu T, Hong F, Gong ZY, Li F, Yan Y, Chen JX. Expression profile of circular RNAs in IDH-wild type glioblastoma tissues. Clin Neurol Neurosurg. 2018;171:168–173. doi: 10.1016/j.clineuro.2018.06.020. [DOI] [PubMed] [Google Scholar]
- 44.Xu H, Zhang Y, Qi L, Ding L, Jiang H, Yu H. NFIX circular RNA promotes glioma progression by regulating miR-34a-5p via notch signaling pathway. Front Mol Neurosci. 2018;11:225. doi: 10.3389/fnmol.2018.00225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Barbagallo D, Caponnetto A, Cirnigliaro M, Brex D, Barbagallo C, D'Angeli F, Morrone A, Caltabiano R, Barbagallo GM, Ragusa M, et al. circSMARCA5 inhibits migration of glioblastoma multiforme cells by regulating a molecular axis involving splicing factors SRSF1/SRSF3/PTB. Int J Mol Sci. 2018;19:480. doi: 10.3390/ijms19020480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bian A, Wang Y, Liu J, Wang X, Liu D, Jiang J, Ding L, Hui X. Circular RNA complement factor H (CFH) promotes glioma progression by sponging mir-149 and regulating AKT1. Med Sci Monit. 2018;24:5704–5712. doi: 10.12659/MSM.910180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xie G. Circular RNA hsa-circ-0012129 promotes cell proliferation and invasion in 30 cases of human glioma and human glioma cell lines U373, A172, and SHG44, by targeting MicroRNA-661 (miR-661) Med Sci Monit. 2018;24:2497–2507. doi: 10.12659/MSM.909229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang R, Zhang S, Chen X, Li N, Li J, Jia R, Pan Y, Liang H. CircNT5E acts as a sponge of miR-422a to promote glioblastoma tumorigenesis. Cancer Res. 2018;78:4812–4825. doi: 10.1158/0008-5472.CAN-18-0532. [DOI] [PubMed] [Google Scholar]
- 49.Kumar V, Soni UK, Maurya VK, Singh K, Jha RK. Integrin beta8 (ITGB8) activates VAV-RAC1 signaling via FAK in the acquisition of endometrial epithelial cell receptivity for blastocyst implantation. Sci Rep. 2017;7:1885. doi: 10.1038/s41598-017-01764-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu Z, Hu G, Zhao Y, Xiao Z, Yan M, Ren M. Silence of cZNF292 suppresses the growth, migration, and invasion of human esophageal cancer Eca-109 cells via upregulating miR-206. J Cell Biochem. 2020;121:2354–2362. doi: 10.1002/jcb.29458. [DOI] [PubMed] [Google Scholar]
- 51.Yang P, Qiu Z, Jiang Y, Dong L, Yang W, Gu C, Li G, Zhu Y. Silencing of cZNF292 circular RNA suppresses human glioma tube formation via the Wnt/beta-catenin signaling pathway. Oncotarget. 2016;7:63449–63455. doi: 10.18632/oncotarget.11523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bi W, Huang J, Nie C, Liu B, He G, Han J, Pang R, Ding Z, Xu J, Zhang J. circRNA circRNA_102171 promotes papillary thyroid cancer progression through modulating CTNNBIP1-dependent activation of β-catenin pathway. J Exp Clin Cancer Res. 2018;37:275. doi: 10.1186/s13046-018-0936-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ouyang YB, Giffard RG. MicroRNAs affect BCL-2 family proteins in the setting of cerebral ischemia. Neurochem Int. 2014;77:2–8. doi: 10.1016/j.neuint.2013.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang M, Huang N, Yang X, Luo J, Yan S, Xiao F, Chen W, Gao X, Zhao K, Zhou H, et al. A novel protein encoded by the circular form of the SHPRH gene suppresses glioma tumori-genesis. Oncogene. 2018;37:1805–1814. doi: 10.1038/s41388-017-0019-9. [DOI] [PubMed] [Google Scholar]
- 55.Chaffer CL, San Juan BP, Lim E, Weinberg RA. EMT, cell plasticity and metastasis. Cancer Metastasis Rev. 2016;35:645–654. doi: 10.1007/s10555-016-9648-7. [DOI] [PubMed] [Google Scholar]
- 56.Zheng J, Liu X, Xue Y, Gong W, Ma J, Xi Z, Que Z, Liu Y. TTBK2 circular RNA promotes glioma malignancy by regulating miR-217/HNF1β/Derlin-1 pathway. J Hematol Oncol. 2017;10:52. doi: 10.1186/s13045-017-0422-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li GF, Li L, Yao ZQ, Zhuang SJ. Hsa_circ_ 0007534/miR-761/ZIC5 regulatory loop modulates the proliferation and migration of glioma cells. Biochem Biophys Res Commun. 2018;499:765–771. doi: 10.1016/j.bbrc.2018.03.219. [DOI] [PubMed] [Google Scholar]
- 58.Chen Z, Duan X. hsa_circ_0000177-miR-638-FZD7-Wnt Signaling Cascade Contributes to the Malignant Behaviors in Glioma. DNA Cell Biol. 2018;37:791–797. doi: 10.1089/dna.2018.4294. [DOI] [PubMed] [Google Scholar]
- 59.Li G, Yang H, Han K, Zhu D, Lun P, Zhao Y. A novel circular RNA, hsa_circ_0046701, promotes carcinogenesis by increasing the expression of miR-142-3p target ITGB8 in glioma. Biochem Biophys Res Commun. 2018;498:254–261. doi: 10.1016/j.bbrc.2018.01.076. [DOI] [PubMed] [Google Scholar]
- 60.Jin P, Huang Y, Zhu P, Zou Y, Shao T, Wang O. circRNA circHIPK3 serves as a prognostic marker to promote glioma progression by regulating miR-654/IGF2BP3 signaling. Biochem Biophys Res Commun. 2018;503:1570–1574. doi: 10.1016/j.bbrc.2018.07.081. [DOI] [PubMed] [Google Scholar]
- 61.He Q, Zhao L, Liu Y, Liu X, Zheng J, Yu H, Cai H, Ma J, Liu L, Wang P, et al. circ-SHKBP1 Regulates the Angiogenesis of U87 Glioma-Exposed Endothelial Cells through miR-544a/FOXP1 and miR-379/FOXP2 Pathways. Mol Ther Nucleic Acids. 2018;10:331–348. doi: 10.1016/j.omtn.2017.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yang Y, Gao X, Zhang M, Yan S, Sun C, Xiao F, Huang N, Yang X, Zhao K, Zhou H, et al. Novel role of FBXW7 circular RNA in repressing glioma tumorigenesis. J Natl Cancer Inst. 2018;110:304–315. doi: 10.1093/jnci/djx166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li F, Ma K, Sun M, Shi S. Identification of the tumor-suppressive function of circular RNA ITCH in glioma cells through sponging miR-214 and promoting linear ITCH expression. Am J Transl Res. 2018;10:1373–1386. [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 64.Wang Y, Sui X, Zhao H, Cong L, Li Y, Xin T, Guo M, Hao W. Decreased circular RNA hsa_circ_0001649 predicts unfavorable prognosis in glioma and exerts oncogenic properties in vitro and in vivo. Gene. 2018;676:117–122. doi: 10.1016/j.gene.2018.07.037. [DOI] [PubMed] [Google Scholar]
- 65.Bai H, Lei K, Huang F, Jiang Z, Zhou X. ExocircRNAs: A new paradigm for anticancer therapy. Mol Cancer. 2019;18:56. doi: 10.1186/s12943-019-0986-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Edwards LA, Li A, Berel D, Madany M, Kim N-H, Liu M, Hymowitz M, Uy B, Jung R, Xu M, et al. ZEB1 regulates glioma stemness through LIF repression. Sci Rep. 2017;7:69. doi: 10.1038/s41598-017-00106-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chen J, Liu L, Liu Y, Liu X, Qu C, Meng F, Ma J, Lin Y, Xue Y. Low-Dose Endothelial-Monocyte-Activating Polypeptide-II Induced Autophagy by Down-Regulating miR-20a in U-87 and U-251 Glioma Cells. Front Cell Neurosci. 2016;10:128. doi: 10.3389/fncel.2016.00128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhang J, Liu L, Xue Y, Ma Y, Liu X, Li Z, Li Z, Liu Y. Endothelial Monocyte-Activating Polypeptide-II Induces BNIP3-Mediated Mitophagy to Enhance Temozolomide Cytotoxicity of Glioma Stem Cells via Down-Regulating miR-24-3p. Front Mol Neurosci. 2018;11:92. doi: 10.3389/fnmol.2018.00092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wesselhoeft RA, Kowalski PS, Anderson DG. Engineering circular RNA for potent and stable translation in eukaryotic cells. Nat Commun. 2018;9:2629. doi: 10.1038/s41467-018-05096-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Meganck RM, Borchardt EK, Castellanos Rivera RM, Scalabrino ML, Wilusz JE, Marzluff WF, Asokan A. Tissue-dependent expression and translation of circular RNAs with recombinant AAV vectors in vivo. Mol Ther Nucleic Acids. 2018;13:89–98. doi: 10.1016/j.omtn.2018.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Liu X, Abraham JM, Cheng Y, Wang Z, Wang Z, Zhang G, Ashktorab H, Smoot DT, Cole RN, Boronina TN, et al. Synthetic circular RNA functions as a miR-21 sponge to suppress gastric carcinoma cell proliferation. Mol Ther Nucleic Acids. 2018;13:312–321. doi: 10.1016/j.omtn.2018.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Møller HG, Rasmussen AP, Andersen HH, Johnsen KB, Henriksen M, Duroux M. A systematic review of microRNA in glioblastoma multiforme: Micro-modulators in the mesenchymal mode of migration and invasion. Mol Neurobiol. 2013;47:131–144. doi: 10.1007/s12035-012-8349-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Annovazzi L, Caldera V, Mellai M, Riganti C, Battaglia L, Chirio D, Melcarne A, Schiffer D. The DNA damage/repair cascade in glioblastoma cell lines after chemotherapeutic agent treatment. Int J Oncol. 2015;46:2299–2308. doi: 10.3892/ijo.2015.2963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ding C, Yi X, Wu X, Bu X, Wang D, Wu Z, Zhang G, Gu J, Kang D. Exosome-mediated transfer of circRNA CircNFIX enhances temozolomide resistance in glioma. Cancer Lett. 2020;479:1–12. doi: 10.1016/j.canlet.2020.03.002. [DOI] [PubMed] [Google Scholar]
- 75.Geng X, Jia Y, Zhang Y, Shi L, Li Q, Zang A, Wang H. Circular RNA: Biogenesis, degradation, functions and potential roles in mediating resistance to anticarcinogens. Epigenomics. 2020;12:267–283. doi: 10.2217/epi-2019-0295. [DOI] [PubMed] [Google Scholar]
- 76.Hao Z, Hu S, Liu Z, Song W, Zhao Y, Li M. Circular RNAs: Functions and Prospects in Glioma. J Mol Neurosci. 2019;67:72–81. doi: 10.1007/s12031-018-1211-2. [DOI] [PubMed] [Google Scholar]
- 77.Chaabane M, Rouchka EC, Park JW. Circular RNA Detection from High-throughput Sequencing. Proceedings of the International Conference on Research in Adaptive and Convergent Systems; September 20-23, 2017; Krakow, Poland. 2017. pp. 19–24. [Google Scholar]
- 78.Fischer JW, Leung AKL. circRNAs: A regulator of cellular stress. Crit Rev Biochem Mol Biol. 2017;52:220–233. doi: 10.1080/10409238.2016.1276882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wang K, Singh D, Zeng Z, Coleman SJ, Huang Y, Savich GL, He X, Mieczkowski P, Grimm SA, Perou CM, et al. MapSplice: Accurate mapping of RNA-seq reads for splice junction discovery. Nucleic Acids Res. 2010;38:e178. doi: 10.1093/nar/gkq622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hoffmann S, Otto C, Doose G, Tanzer A, Langenberger D, Christ S, Kunz M, Holdt LM, Teupser D, Hackermüller J, et al. A multi-split mapping algorithm for circular RNA, splicing, trans-splicing and fusion detection. Genome Biol. 2014;15:R34. doi: 10.1186/gb-2014-15-2-r34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Szabo L, Morey R, Palpant NJ, Wang PL, Afari N, Jiang C, Parast MM, Murry CE, Laurent LC, Salzman J. Statistically based splicing detection reveals neural enrichment and tissue-specific induction of circular RNA during human fetal development. Genome Biol. 2015;16:126. doi: 10.1186/s13059-015-0690-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Cheng J, Metge F, Dieterich C. Specific identification and quantification of circular RNAs from sequencing data. Bioinformatics. 2016;32:1094–1096. doi: 10.1093/bioinformatics/btv656. [DOI] [PubMed] [Google Scholar]
- 83.You X, Conrad TO. Acfs: Accurate circRNA identification and quantification from RNA-Seq data. Sci Rep. 2016;6:38820. doi: 10.1038/srep38820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ye CY, Zhang X, Chu Q, Liu C, Yu Y, Jiang W, Zhu QH, Fan L, Guo L. Full-length sequence assembly reveals circular RNAs with diverse non-GT/AG splicing signals in rice. RNA Biol. 2017;14:1055–1063. doi: 10.1080/15476286.2016.1245268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Metge F, Czaja-Hasse LF, Reinhardt R, Dieterich C. FUCHS-towards full circular RNA characterization using RNAseq. PeerJ. 2017;5:e2934. doi: 10.7717/peerj.2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zheng Y, Ji P, Chen S, Hou L, Zhao F. Reconstruction of full-length circular RNAs enables isoform-level quantification. Genome Med. 2019;11:2. doi: 10.1186/s13073-019-0614-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhang XO, Dong R, Zhang Y, Zhang JL, Luo Z, Zhang J, Chen LL, Yang L. Diverse alternative back-splicing and alternative splicing landscape of circular RNAs. Genome Res. 2016;26:1277–1287. doi: 10.1101/gr.202895.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gao Y, Zhang J, Zhao F. Circular RNA identification based on multiple seed matching. Brief Bioinform. 2018;19:803–810. doi: 10.1093/bib/bbx014. [DOI] [PubMed] [Google Scholar]
- 89.Feng J, Xiang Y, Xia S, Liu H, Wang J, Ozguc FM, Lei L, Kong R, Diao L, He C, et al. CircView: A visualization and exploration tool for circular RNAs. Brief Bioinform. 2018;19:1310–1316. doi: 10.1093/bib/bbx070. [DOI] [PubMed] [Google Scholar]
- 90.Gao Y, Wang H, Zhang H, Wang Y, Chen J, Gu L. PRAPI: Post-transcriptional regulation analysis pipeline for Iso-Seq. Bioinformatics. 2018;34:1580–1582. doi: 10.1093/bioinformatics/btx830. [DOI] [PubMed] [Google Scholar]
- 91.Feng J, Chen K, Dong X, Xu X, Jin Y, Zhang X, Chen W, Han Y, Shao L, Gao Y, et al. Genome-wide identification of cancer-specific alternative splicing in circRNA. Mol Cancer. 2019;18:35. doi: 10.1186/s12943-019-0996-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ji P, Wu W, Chen S, Zheng Y, Zhou L, Zhang J, Cheng H, Yan J, Zhang S, Yang P, et al. Expanded expression landscape and prioritization of circular RNAs in mammals. Cell Rep. 2019;26:3444–3460.e5. doi: 10.1016/j.celrep.2019.02.078. [DOI] [PubMed] [Google Scholar]
- 93.Glažar P, Papavasileiou P, Rajewsky N. circBase: A database for circular RNAs. RNA. 2014;20:1666–1670. doi: 10.1261/rna.043687.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Xia S, Feng J, Lei L, Hu J, Xia L, Wang J, Xiang Y, Liu L, Zhong S, Han L, et al. Comprehensive characterization of tissue-specific circular RNAs in the human and mouse genomes. Brief Bioinform. 2017;18:984–992. doi: 10.1093/bib/bbw081. [DOI] [PubMed] [Google Scholar]
- 95.Dudekula DB, Panda AC, Grammatikakis I, De S, Abdelmohsen K, Gorospe M. CircInteractome: A web tool for exploring circular RNAs and their interacting proteins and microRNAs. RNA Biol. 2016;13:34–42. doi: 10.1080/15476286.2015.1128065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Liu YC, Li JR, Sun CH, Andrews E, Chao RF, Lin FM, Weng SL, Hsu SD, Huang CC, Cheng C, et al. CircNet: A database of circular RNAs derived from transcriptome sequencing data. Nucleic Acids Res. 2016;44D:D209–D215. doi: 10.1093/nar/gkv940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Chen X, Han P, Zhou T, Guo X, Song X, Li Y. circRNADb: A comprehensive database for human circular RNAs with protein-coding annotations. Sci Rep. 2016;6:34985. doi: 10.1038/srep34985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Xia S, Feng J, Chen K, Ma Y, Gong J, Cai F, Jin Y, Gao Y, Xia L, Chang H, et al. CSCD: A database for cancer-specific circular RNAs. Nucleic Acids Res. 2018;46D:D925–D929. doi: 10.1093/nar/gkx863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Fan C, Lei X, Fang Z, Jiang Q, Wu FX. circR2Disease: A manually curated database for experimentally supported circular RNAs associated with various diseases. Database (Oxford) 2018;2018:bay044. doi: 10.1093/database/bay044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Vo JN, Cieslik M, Zhang Y, Shukla S, Xiao L, Zhang Y, Wu YM, Dhanasekaran SM, Engelke CG, Cao X, et al. The landscape of circular RNA in cancer. Cell. 2019;176:869–881.e13. doi: 10.1016/j.cell.2018.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and analysed during the present study are available from the corresponding author on reasonable request.