Abstract
The new era of artificial intelligence (AI) has introduced revolutionary data-driven analysis paradigms that have led to significant advancements in information processing techniques in the context of clinical decision-support systems. These advances have created unprecedented momentum in computational medical imaging applications and have given rise to new precision medicine research areas. Radiogenomics is a novel research field focusing on establishing associations between radiological features and genomic or molecular expression in order to shed light on the underlying disease mechanisms and enhance diagnostic procedures towards personalized medicine. The aim of the current review was to elucidate recent advances in radiogenomics research, focusing on deep learning with emphasis on radiology and oncology applications. The main deep learning radiogenomics architectures, together with the clinical questions addressed, and the achieved genetic or molecular correlations are presented, while a performance comparison of the proposed methodologies is conducted. Finally, current limitations, potentially understudied topics and future research directions are discussed.
Keywords: medical imaging/radiogenomics, artificial intelligence, deep learning, machine learning, precision medicine, oncology
1. Introduction
Radiogenomics is an emerging research field focusing on establishing multi-scale associations between medical imaging and gene expression data. Deciphering the interplay of radiological and genetic/molecular features is of utmost importance in oncology and can be achieved through the fusion of selected genomic and radiomics features in a unified feature-space leading to more precise decision support systems. Furthermore, radiogenomics can enhance diagnosis (1), the non-invasive prediction of molecular background (2) and survival prediction (3) in oncology by associating genomic data with radiomics features acquired in a non-invasive fashion shedding light on underling oncogenic mechanisms.
Despite advances in both multimodal-imaging technologies involving novel agents, powerful protocols and computer-aided diagnostic tools, there is a critical knowledge gap between imaging information at the tissue level and the underlying molecular and genetic disease biomarkers. This information gap has important socio-economic consequences since in a number of cases, only imaging information is available in the diagnostic setting, rendering decisions such as the administration of expensive treatments challenging, while risking under- or overtreatment. For this reason, the vision of precision medicine is closely linked with the understanding of the interplay and the joint effect of multi-scale pathophysiological disease biomarkers. To this end, radiogenomics approaches aim to link the imaging phenotype with underlying molecular/genetic characteristics of disease entities, particularly in oncologic applications in order to enhance precision in the early diagnosis and management of patients. In this context, several studies have tried to establish statistically significant associations of image-derived features, such as the shape and texture of a lesion with molecular or genetic information based on usually small, but information-rich patient cohorts, where both imaging [e.g., ultrasound (US), computed tomography (CT) magnetic resonance imaging (MRI) and positron emission tomography (PET)/CT] and molecular or genetic information [e.g., deoxyribonucleic (DNA) microarrays, micro-ribonucleic acid (miRNA), ribonucleic acid sequencing (RNA-seq)] are available.
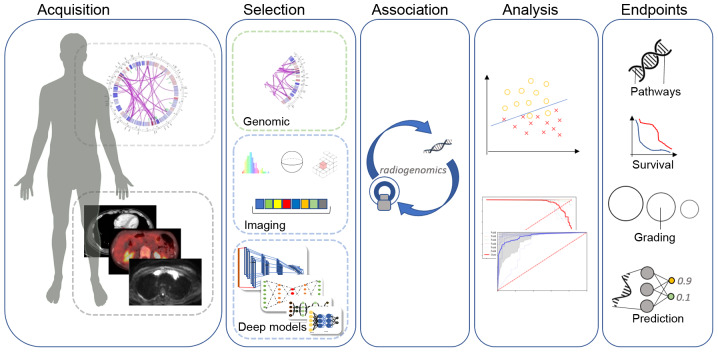
The fundamental hypothesis in radiogenomics research is that certain molecular/genetic alterations induce alterations at the tissue level that can be computationally detected in terms of radiological appearance by properties, such as tissue shape and texture (Fig. 1). Such changes can however be very subtle or even invisible to the human eye. For this reason, artificial intelligence (AI) techniques, such as deep learning have provided the means to detect and decode these changes/patterns in medical images and associate them with molecular/genetic characteristics, paving the way for the development of robust decision support systems and hybrid prognostic, predictive and diagnostic signatures. In particular, in recent years, the introduction and widespread availability in oncologic care of hybrid clinical diagnostic imaging systems, such as PET/CT and PET/MRI provides rich opportunities for AI-based strategies aiming to enhance effectiveness and precision in the management of oncologic patients (4-6).
Figure 1.
The radiogenomics pipeline includes a predefined data acquisition protocol for genomic and imaging data, feature extraction (radiomics, deep learning and traditional image processing) and feature selection based on statistical analysis. The association and modeling of these highly discriminative biomarkers holds the potential to enhance robustness in decision support systems.
Deep learning models are data-driven architectures with state-of-the-art performance in almost all modern data-processing tasks. These innovative approaches offer end-to-end, fully automated data analysis pipelines by computing exhaustively discriminative features to achieve the highest performance on the given task. The model's parameters are learned only from the data by updating its inner representation during the optimization process aiming to approximate the optimal convergence state. Additionally, multi-path architectures enable diverse data types to coexist in the same model providing new opportunities to combine imaging, clinical, laboratory, molecular and genetic data towards unified AI decision-support systems extending the available frameworks. Although the majority of radiogenomics studies are based on traditional radiomics approaches (7) i.e., high throughput hand-crafted image feature extraction followed by association with genomic/molecular markers, the application of AI techniques, such as deep learning in the field is still limited, due to the lack of large multi-modal cross-sectional and/or longitudinal datasets. The main aims of the present review were to summarize the most important results concerning the application of deep learning in radiogenomics, to highlight the clinical significance of associating molecular or genetic information with imaging phenotype, to emphasize the potential for the discovery of novel, highly discriminative combined biomarkers and at the same time, to discuss the main limitations concerning the state-of-the-art research in the field. Furthermore, representative DL-based studies focusing on cancer tissue differentiation or therapy prediction solely from medical images have been included. This is due to the fact that such clinical issues are attractive for radiogenomics research, since they involve molecular/genetic mechanisms and have been also studied with in vitro tumor phenotyping involving the in vitro assay of tumor biopsy material. As an example, a previous study (8) provided a deep learning study based on a convolutional neural network (CNN), which predicts the neoadjuvant therapy response in esophageal cancer with fluorodeoxyglucose positron emission tomography (18F-FDG PET) imaging data and achieves 80% sensitivity and specificity. In a gene expression study (9) on the same clinical problem, a 17-molecule was found to be predictive of complete response to neoadjuvant chemotherapy for esophageal cancer with docetaxel, cisplatin and 5-fluorouracil, while a recent study reported that on-treatment genetic biomarkers can predict complete response in neoadjuvant breast cancer (10). Similarly, it has been reported (11) that tumor mutational burden information can differentiate between primary and metastatic breast cancer and for this reason, the present review includes deep learning efforts to differentiate primary from metastatic cancer directly from medical imaging data [such as previously done (12) for liver cancer]. In addition, apart from the limited deep learning-based radiogenomics studies available, representative deep learning studies on cancer type classification or therapy prediction in oncology are included, since these studies can be extended towards associating imaging phenotype and genetic/molecular markers.
2. Research methodology
The search was performed in widely used databases, such as PubMed and Google Scholar. Initially, articles including a subset or a combination of the terms noted in the title or abstract (Table I) were identified. The Abstracts and conclusions of 123 studies were examined during the first screening process. Although the bulk of these manuscripts included a subset or a combination of the examined keywords, the second screening revealed that the methodology of a number of these papers was not relevant to deep learning radiogenomics analysis and these were therefore excluded. The exclusion criteria used in the present review are summarized as follows: i) Studies focusing on radiomics, deep learning segmentation with radiomics, handcrafted features associated with genomic data or traditional image analysis with machine learning techniques; ii) studies where deep learning was used only for data augmentation and synthetic data generation paired with radiomics or other analysis; and iii) studies on tissue classification (normal, benign, malignant tissue), scoring [Gleason, breast imaging reporting and data system (BI-RADS)] or genomic data analyses without involving image-derived features.
Table I.
Keywords used in the present review article.
| Deep learning | Cancer | Magnetic resonance imaging | Radiogenomics |
|---|---|---|---|
| Artificial intelligence | Carcinoma | Positron emission tomography | Therapy response |
| Convolutional neural networks | Computer tomography | Treatment response | |
| Recurrent neural networks | Medical imaging | Tissue classification | |
| Molecular signatures |
As a result, 25 studies were selected for the presented deep learning radiogenomics in-depth review. Information related to the examined malignancy, patient cohort size, imaging modalities and deep network architecture type is presented in Table II. The objective and performance of the examined studies are described in Table III, including evaluation results in terms of accuracy (ACC) and area under the curve (AUC) scores.
Table II.
Overview of the studied malignancies, imaging modalities and proposed deep architectures.
| Author (Refs.) | Anatomical area | Patient cohort | Imaging modality | Deep architecture type |
|---|---|---|---|---|
| Chang et al (18) | Brain | 496 | MRI: FLAIR, T2, T1 pre-contrast, T1 post-contrast | Multiple residual CNN with decision fusion |
| Grinband et al (19) | Brain | 155 | MRI: T1, FLAIR | Residual CNN |
| Li et al (20) | Brain | 119 | MRI: T1, T2 | ROI-only CNN |
| Liang et al (21) | Brain | 167 | MRI: T1, T2, T1Gd, FLAIR | Multi-channel ROI-only 3D DenseNet |
| Korfiatis et al (22) | Brain | 155 | MRI: T2 | ResNet50 |
| Akkus et al (23) | Brain | 159 | MRI: T1C, T2 | Multi-kernel CNN |
| Bonte et al (25) | Brain | 285 | MRI: T1ce | VGG-11 |
| Smedley et al (26) | Brain | 528 | MRI: T1WI+c, T2WI, FLAIR | AE, DNN |
| Zhou et al (27) | Brain | 233 | MRI: T1, T2, FLAIR | DenseNet AE, LSTM |
| Momeni et al (28) | Brain | 47 | H&E, MRI: T1C, FLAIR | DenseNet, 3D-CNN |
| Afshar et al (29) | Brain | 233 | MRI | ROI-only Capsule Network |
| Yu et al (31) | NSCLC | 684 | CT | Patch-2D CNN |
| Zhu et al (34) | Breast | 270 | DCE-MRI | GooLeNet, VGG, pre-trained, fine-tuned |
| Ha et al (35) | Breast | 216 | Pre-contrast MRI | Residual CNN |
| Yoon et al (36) | Breast | 213 | MRI: T1WI | CNN with feature fusion (image and genetic) |
| Zhu et al (37) | Breast | 131 | MRI: T1WI | Pre-trained GooLeNet |
| Ypsilantis et al (8) | Esophagus | 107 | 18F-FDG PET | Multi-channel 3-slice CNN |
| Bibault et al (38) | Rectal | 95 | CT | DNN |
| Chen et al (39) | Pancreas | 48 | MRI: T1, T2 | ROI detection faster-RCNN, ROI-only inception layers |
| Trivizakis et al (12) | Liver | 130 | MRI: b1000 | Patch-2D, 2D and 3D CNN |
| Cha et al (40) | Bladder | 82 | CT | CNN transfer-learning |
| Cha et al (41) | Bladder | 170 | CT | CNN and radiomics |
| Banerjee et al (42) | RMS | 21 | DWI, T1W | CNN transfer-learning |
| Zhou et al (43) | Head and neck | 31 | PET and CT | Evidential reasoning: 3D CNN and radiomics |
NSCLC, non-small cell lung cancer; RMS, rhabdomyosarcoma; MRI, magnetic resonance imaging; CT, computerized tomography; PET, positron emission tomography; 18F-FDG PET, fluorodeoxyglucose positron emission tomography; FLAIR, fluid-attenuated inversion recovery; CNN, convolutional neural network; AE, autoencoder; DNN, deep neural network; VGG, video geometry group; ROI, region of interest.
Table III.
Aggregated results for comparison of the reviewed articles.
| Author (Refs.) | Study objective | Performance (ACC/AUC %) |
|---|---|---|
| Chang et al (18) | IDH1 mutation | 85.7-89.1/94-95 |
| Grinband et al (19) | IDH1 mutation | 94/91 |
| Li et al (20) | IDH1 mutation | 82.4-92.4/92-95 |
| Liang et al, (21) | IDH1 mutation | 84.6,91.4/85.7,94.8 |
| Korfiatis et al (22) | MGMT state | 94.9/- |
| Grinband et al (19) | MGMT state | 83/84 |
| Akkus et al (23) | 1p19q codeletion status | 87.7/- |
| Grinband et al (19) | 1p19q codeletion status | 92/88 |
| Bonte et al (25) | Glioma grading | 91.1,93.5/82,86.1 |
| Zhou et al (27) | Metastatic/glioma/meningioma | 92.1/- |
| Momeni et al (28) | Oligendroglioma/astrocytoma | 85/92 |
| Afshar et al (29) | Glioma/pituitary/meningioma | 86.6/- |
| Yu et al (31) | EGFR mutation status | 76.1/82.8 |
| Wang et al (32) | EGFR mutation status | 73.9/81 |
| Zhu et al (34) | Luminal A vs others | -/58-65 |
| Ha et al (35) | Luminal A vs. B vs. HER2+ vs. Basal | 70/87.1 |
| Yoon et al (36) | Pathological state, ER, PR, HER2 | -/69.7, 97.6, 89.9, 84.2 |
| Zhu et al (37) | Occult invasive disease status | -/70 |
| Ypsilantis et al (8) | Neoadjuvant chemotherapy response | 73.4/66.3 |
| Bibault et al (38) | Neoadjuvant chemoradiation response | 80/72 |
| Chen et al (39) | Subtype prediction | 80, voting: 92.3/- |
| Trivizakis et al (12) | Primary/metastasis | 83/80 |
| Cha et al (40) | Chemotherapy response | -/62-77 |
| Cha et al (41) | Chemotherapy response | -/62-79 |
| Banerjee et al (42) | Subtype prediction | 85/- |
| Zhou et al (43) | Lymph node metastasis | 72.7-93/65-92 |
IDH1, isocitrate dehydrogenase isozyme 1; MGMT, methylguanine methyltransferase; EGFR, epidermal growth factor receptor; ER, estrogen receptor; PR, progesterone receptor; HER2, human epidermal growth factor receptor 2; ACC, accuracy; AUC, area under the curve.
3. Deep architectures used in current radiogenomics studies
Deep learning methodologies are very attractive for advancing radiogenomics studies, particularly when taking into consideration the large search-space due to the abundance of features in both images and genetic/molecular data. Critically, in contrast to handcrafted feature-based machine learning techniques, the whole process is fully automated and through exhaustive analysis, non-intuitive correspondences between medical image features and genetic/molecular information can be established. Thus, a higher-level modeling is achieved through computational analysis of the high complexity raw radiogenomics data, allowing focusing on particular disease properties for outcome prediction by leveraging pathways, molecular or meta-gene convergence objectives. The main deep learning architectures in the reviewed studies are listed below:
Convolutional neural network (CNNs)
CNNs were originally designed by LeCun et al, as a fully automated image analysis network for classifying handcrafted digits (13). The fundamental principle of this deep architecture is to massively compute and combine feature maps inferring non-linear associations between the input signal and the targeted output. This type of network is popular for the automatic extraction, selection and reduction of discriminative features from high-dimensional input data providing state-of-the-art classification performance in demanding fields.
Recurrent neural networks (RNNs)
RNNs exhibit similar functionality with the regular feedforward neural networks, where every hidden state will be fed as an input for the next state but in addition, they integrate internal memory. This short-term memory allows recurrent networks to remember information from the previously analyzed states, a perfect fit for sequential signal analysis and predictive models.
CapsuleNet
A capsule is a set of neurons that models a part of an object of the input by activating a small subset of its properties. The CapsuleNet (14) consists of independent sets of capsules instead of kernels propagating information to the successive higher-level capsules through the routing-by-agreement process. This architecture is the newest evolvement in the deep learning field and to date, has not been extensively tested by the medical research community.
Autoencoders (AEs)
AEs learn a compact representation of their input by reconstructing it. The encoder part of the AE has a decreasing number of neurons after every successive hidden layer, reducing the dimensionality of the incoming state. The decoder part reconstructs the resulted compact representation to an approximation of the initial input by backpropagating the reconstruction error.
Artificial neural networks (ANNs)
ANNs or deep neural networks (DNNs) were inspired by the alleged learning functionality of the biological brain. The ANN constitutes of a set of fully-connected nodes modelling the stimuli propagation of brain synapses -fire or not- across the neural network to perform a specific task. It can be used for feature selection, classification or dimensionality reduction as a submodule of a deeper architecture (CNN and AE) or as a stand-alone module with hand-crafted features.
Multi-model decision fusion
A meta-analysis of a set of models can accumulate the predictive power of a number of diverse models built on different data types, but aiming at a single objective. Decision fusion combines the outcome of multiple classifiers into a singular final prediction forming a meta-estimator by utilizing statistical methods to amplify the individual classifiers. This leads to an improved accumulated predictive power and can resolve uncertainties or disagreements among singular analyses.
Pre-trained models
A source model trained with widely available 'natural' images can be transferred to a target model that will perform similar tasks but in the medical imaging domain. The learnt feature detectors of these deep architectures as a result of their low-level status can be an alternative and viable option for tasks with small dataset. In this context two major methodologies can be followed: Off-the-self models (for classification or feature extraction) and fine-tuned models. There are several available pre-trained models [video geometry group (VGG)-16 (http://www.robots.ox.ac.uk/~vgg/research/very_deep/), Inception (15), DenseNet (16), Mask R-CNN (17)] and more employed by several authors, claiming mixed results for the off-the-self method with fine-tuning being the most promising due to its supplementary adaptation to the targeted model.
4. Clinical applications of deep learning-based radiogenomics
Brain neoplasms
Isocitrate dehydrogenase (IDH) isozyme mutation status
Patients with glioma carrying IDH1 or IDH2 mutations have a relatively favorable survival, when compared to glioma patients with wild-type IDH1/2 genes. For this reason, the non-invasive prediction of the IDH1 mutation status at the pre-treatment stage is critical for treatment planning. Discriminating between wild-type and mutation status of IDH1 can be challenging. The architecture proposed by Chang et al (18), consists of multiple residual convolutional neural networks, one for each MRI modality, with decision fusion evaluated on multi-institutional data of grade II-IV glioma. A traditional CNN approach trained on segmented regions of interest (ROI) of wild-type vs. mutation 2D slices was applied by Grinband et al (19) and a hybrid architecture was proposed by Li et al (20), including a convolutional network trained with patches of ROI/no ROI, a fisher vector module for encoding the extracted salience maps and a support vector machine classifier for predicting the IDH1 mutation status. During inference the convolutional-only network with multi-scale input produces feature maps encoded by the fisher vector module in a compact representation similar to bag of visual words. The patches and ROIs were extracted from multi-modal low-grade glioma MRIs. Liang et al (21), utilized a multi-channel 3D DenseNet architecture for predicting the IDH1 status trained on segmented 3D ROIs with multi-modal [T1, T2, T1Gd and fluid-attenuated inversion recovery (FLAIR)].
Methylguanine methyltransferase (MGMT) methylation state
MGMT is a gene responsible for repairing the mismatched DNA molecules and its methylation status. Clinically, it is used as a predictive biomarker for response to chemotherapy with temozolamide, which is the standard for glioblastoma multiforme (GBM) following therapy. A 50-layer ResNet was previously used (22) and a CNN trained with ROIs-only (19) as mentioned earlier.
1p19q chromosome
The co-deletion status (the combined loss of the short arm chromosome 1 and the long arm of chromosome 19) of 1p19q in low-grade glioma has been associated with an improved response to therapy and a higher survival rate. A proposed multi-kernel CNN (23) predicts the 1p19q chromosomal expression from multi-modal (T1C and T2) low-grade glioma MRI. In a previous study (19), a similar approach was followed to the previously mentioned IDH1 and MGMT by utilizing the same deep architecture to predict the 1p19q codeletion.
Glioma grading
Glioma is a type of tumor that begins in glial cells and is the most common type of brain tumor associated with a poor prognosis (24). The classification of the glioma type (e.g., glioblastoma, astrocytoma and oligodendroglioma), and grade is critical for the treatment plan, which consists of external radiation therapy, surgery, chemotherapy, or a combination of these. In a previous study (25), a pre-trained VGG-11 CNN with a random forest classifier was used for glioma grading.
Genetic association with MR imaging features
A novel application of radiogenomics analysis by the deep learning reconstruction of genetic data and MR imaging features in patients with GBM has been previously proposed (26). An AutoEncoder architecture was trained to match the tumor gene expression with the extracted morphological MRI features including volume, surface area, surface area to volume (SA:V) ratio, sphericity, spherical disproportion, max diameter, major and minor axes and compactness for both cancer region and edema. According to the authors of that study, the network was able to capture predictive radiogenomics associations, linking the genetic profile to MRI morphology of the examined lesion better than traditional statistical methods, such as linear regression or pairwise comparison.
Tissue characterization
Furthermore, selected studies dealing with cancer tissue characterization have been included in the current review, since in such cases, both differences in imaging phenotype and gene-expression have been linked with cancer tissue characterization. In particular, for brain tumor deferential diagnosis, several deep learning architectures have been proposed, including a DenseNet AutoEnconder combined with Long-Short Term Memory (27) for metastatic vs. glioma vs. meningioma, a two-path CNN with a dropout meta-estimator for combined histopathology and radiology images discrimination between oligendroglioma vs. astrocytoma (28) and CapsuleNet (29) for meningioma vs. pituitary vs. glioma differentiation.
Non-small cell lung cancer (NSCLC)
NSCLC is the most common type of lung cancer (up to 85% globally) with a high mortality rate among the cases diagnosed (30). Effective staging is challenging for therapy decision therefore imaging (CT and PET), laboratory and molecular biomarkers are of utmost importance. epithelial growth factor receptor (EGFR) is a protein involved in cell growth located on the cell surface and is associated with malignancy. The prediction of the EGFR mutation profile can lead to better, more effective and targeted treatment. Yu et al (31), proposed a 2D CNN trained on CT nodule patches labeled by the mutation state of EGFR. The study identified deep learning imaging features related to the EGFR profile of NSCLC, verifying the mutational status determined by biopsy with the additive benefit of a non-invasive examination
Wang et al (32), used the DenseNet architecture and in particular the first 20 layers with weights acquired from the ImageNet dataset in a transfer learning scheme. Additionally, fine-tuning was applied with a dataset comprised of approximately 15,000 CT images towards identifying the EGFR mutation status (mutation vs. wild-type). The authors also presented a framework for visualizing the areas with high probability of lesion presence extracted from feature maps of the deep architecture as an additional level for evaluating the AI predictions. The aim of extending the main classification framework is to provide the clinicians with interpretable visual cues and novel attention maps of the inference path followed by the deep model.
Breast cancer
Discriminating molecular subtypes of breast cancer has a significant effect on the clinical management of patients. Some of these subtypes include luminal A and B, the expression of human epidermal growth factor receptor 2 (HER2) and triple-negative types [estrogen receptor (ER)-, progesterone receptor (PR)- and HER2-]. Breast cancer subtypes can be determined by genetic testing or by immunohistochemistry markers. The catalyst for promoting breast cancer research is the availability of open-access data, such as The Cancer Genomic Atlas-Breast Invasive Carcinoma (TCGA-BRCA) (33) database which offers interdisciplinary research opportunities for the medical and the computational/AI research community. Deep learning molecular subtype classification has been introduced in several studies, such as in a previous study (34) where GoogLeNet architectures (pre-trained and fine-tuned, from scratch, off-the-shelf) and inception-based residual CNN (35) were used for differentiating luminal and other subtypes based on dynamic contrast enhanced magnetic resonance imaging (DCE-MRI) and pre-contrast MRI data, respectively. Yoon et al (36), incorporated 3D CNN feature maps from MRI data and the corresponding gene expression at the fully-connected neural part of the deep network through feature space fusion to predict several clinical associations such as the pathological stage, ER status, PR status and HER2. Zhu et al (37), proposed a pre-trained GooLeNet architecture fine-tuned using data augmentation techniques based on MR image patches extracted from the examined region of interest. Additionally, an 'off-the-shelf' deep learning module for feature extraction was combined with an SVM classifier to predict the status of occult invasive disease following the diagnosis of ductal carcinoma in situ (DCIS).
Oral cancer
Ypsilantis et al (8), investigated the therapeutic response and survivability of examined esophageal cancer patients by stratifying them into responders vs. non-responders. The proposed 3-slice CNN (3sCNN) was trained with a subset of the 18F-FDG PET examinations targeting the metabolic activity of the malignancy. This was achieved by selecting from each examination three slices representative of the intra-tumor region and applying concatenation in the color space to introduce them as a unified image to the network.
Colorectal cancer
The rectal cancer therapeutic strategy may include a combination of chemotherapy, radiation and surgery. The non-invasive prediction of complete response of neo-adjuvant therapy can affect the therapeutic algorithm, avoiding unnecessary medical procedures. This application can significantly improve the quality of life and/or survival rate of the patients, spare organ function, reduce the cost of treatment and minimize the toxicity risk or local and distant recurrences. A deep neural network (38) trained on radiomics, clinical and radiological semantic features was used to predict complete therapy response following chemoradiation in colorectal cancer patients.
Pancreatic cancer
Tissue classification for pancreatic neoplasms from routine imaging data can lead to therapy response prediction. In particular, based on the type of cystic lesions different strategies for treatment and/or follow-up are followed. An inception-style architecture with feature space fusion (39) was trained on multi-modal (T1 and T2) ROI images of pancreatic cystic neoplasms (PCNs) to discriminate tissue sub-types such as mucinous cystic neoplasms, intraductal papillary mucinous neoplasm and serous cystic neoplasms. Given the difficulties and morbidity of biopsy procedure in the pancreas, the importance of this application is critically high.
In addition, over the past decade, effective systemic treatment protocols have been developed, and more accurate, effective and less toxic radiotherapeutic strategies have been applied. Due to the poor prognosis of pancreatic cancer, there is a growing effort being made to improve the efficacy of neoadjuvant chemotherapy or chemoradiotherapy, particularly in 'marginally' resectable or even in clearly resectable pancreatic adenocarcinomas. Radiogenomics may play a crucial role in the selection of patients with a higher probability of response to these approaches.
Liver cancer
The diagnosis of liver cancer with traditional machine learning techniques is extremely challenging considering its multifocal distribution. Tissue discrimination between primary and metastatic liver cancer was performed by utilizing 3D-CNN (12) with no pre-processing or segmentation from raw high b-value MRI volumes (b=1,000 sec/mm2) achieving state-of-the-art performance through a fully automated analysis.
Bladder cancer
The analysis of the pre and post-treatment routine imaging examinations elucidates the effectiveness of the followed treatment. Bladder chemotherapy response prediction was previously performed using contrast enhanced CT of pre-treated cancer patients following two different transfer learning approaches (40) and applying feature fusion from deep models combined with radiomic signatures (41).
Rhabdomyosarcoma (RMS)
2D CNN with transfer-learning and multimodal MRI was previously proposed by Banerjee et al (42), to determine embryonal (ERMS) or alveolar (ARMS) genetic expression of RMS contributing to individualized therapy decision making and survival prediction for patients with the aforementioned subtypes.
Head and neck cancer
Zhou et al (43), combined 3D CNN and image processing features through evidential reasoning to predict lymph node metastasis (LNM) of head and neck cancer patients from routine PET and CT. This multi-modal image analysis framework comprised of novel AI models and commonly accepted radiomics for establishing a robust model. The authors of that study claimed an improved diagnosis with the advantage of a non-invasive method sparing patients from unnecessary or ineffective medical procedures.
5. Limitations of radiogenomic research
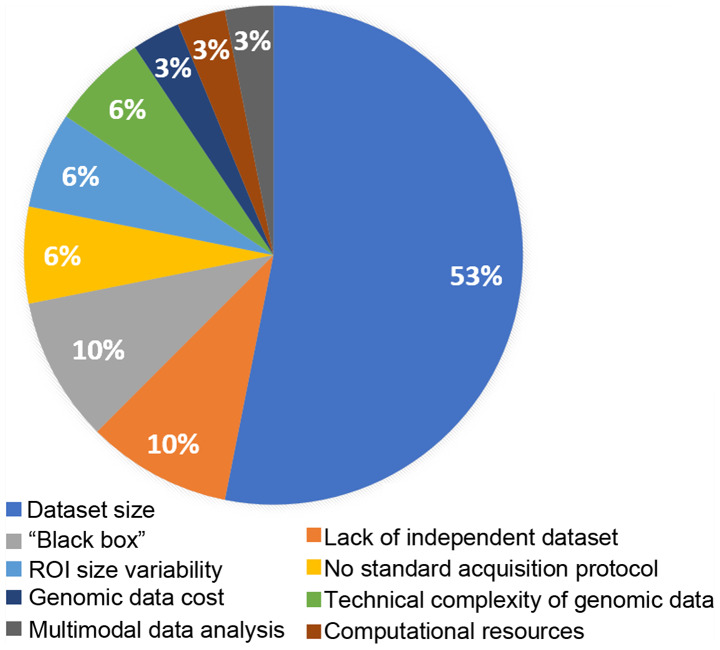
The most pronounced limitation for deep learning radiogenomics, as highlighted in Fig. 2, is related to the size of the available datasets. The lack of the required volume of data can lead to inadequate stratification (19,35,37) among training, validation and testing datasets compromising the model adaptation, optimization and evaluation process respectively. For this reason, the numbers of studies on diseases with limited datasets such as rectal cancer, rhabdomyosarcoma, head and neck malignancies, pancreatic and esophageal cancers included in the present review are limited. For comparison, the study of one of the most common types of cancer, NSCLC, examines 684 cases, brain and breast studies on average include >200 patients in contrast to other cancer types, where the samples vary from 21 to 170 cases at most, as evidenced in Table II. Additionally, limited-size datasets can be a source of concern regarding the fitting status of deep neural networks, potentially leading to a high risk of model overfitting and poor generalization ability due to the wide biological variability of cancer (tumor heterogeneity). This can lead to incompetent decision support systems with high false-positive rate for examinations from diagnostic centers with different imaging protocols or devices. Open-access, curated and high-quality public benchmark databases with complete genomic and imaging data across disease types need to be made available in order to promote radiogenomic research and introduce comparable metrics among studies. For instance, a large number of radiogenomics publications examine brain (44) and breast (45) cancer as widely available open-access databases are available.
Figure 2.
Limitations addressed by the authors of the reviewed publications. ROI, region of interest.
The infamous 'black box' (19,29,37) systems, as commonly deep learning and ANN are known, compute and propagate high-throughput features calculated during a continuously evolving learning process producing an inner representation uninterpretable by traditional statistical analysis or other mathematical methods. A deep learning model throughout the training process incorporates changes on tens of millions of parameters adapted solely in analyzing the retrospective input data with no a-priory knowledge. The fully automated nature of the analysis poses a great drawback in the medical domain where each decision is led by a diverse set of various laboratory, clinical, imaging data in multiple time points, knowledge of malignancy biology and accumulated empirical patient outcomes. Moreover, translating deep learning technologies to clinical practice poses significant challenges considering the 'explainability' of clinical requirements for AI decision support tools (46). The recommendation of a personalized therapeutic care plan can be challenging even for expert clinicians. For this reason, novel methods, algorithms and tools for supporting explainable AI are needed in order to accelerate the clinical evaluation and translation of DL-based radiogenomics technologies.
The demand for computational power and high-throughput infrastructure (28) in these types of models can also be a limiting factor regarding the available resources, particularly in an ensemble or meta-model environment where inference with different data analysis paths may coincide. Investing in specialized infrastructure for enabling access to large data repositories or patient registries by highly qualified scientific personnel is required for conducting AI research and deploying scalable models towards advancing traditional oncology to data-driven precision medicine.
Contemporary artificial intelligence decision support systems have limitations related to the 'ground truth' for the studied region of interest (26,27). The differences in pixel-wise labeling (inter-observer variability and bias) for lesion delineation, uncertainty in the examined anatomical areas of malignances (surrounding area, necrosis), disregarding location-based information of the tumor and dependence on morphological features from ROIs may introduce additional variability and misdirection during the convergence process of a fully automated data-driven AI model. Fusing imaging modalities (21) with no a-priori knowledge or evidence about their optimal combination for the targeted clinical question can lead to unnecessary, redundant analysis with a negative effect on the final decision. Thus, clinicians should provide insight and be in active cooperation with the data science engineers regarding specific lesions attributes with respect to the followed diagnosis protocols. Other types of data, such as laboratory (blood exam results), anthropomorphic (height and weight), demographic (age and sex) and supplementary imaging modalities can introduce diversity and complementarity towards achieving better problem formulation, improved predictive power and a robust decision support process (21).
Finally, a limited number of laboratories (22) can conduct genomic research since it is a challenging and costly task (35) requiring adequate expertise and laboratory certifications. At the same time, in a number of cases, genomic and molecular analyses take place outside the hospital environment in different laboratories, rendering data integration challenging due to the implementation of different proprietary genome sequencing technology by commercialized platforms. Such limiting factors may partially explain the lack of combined genomic and imaging databases which in turn limits current deep learning radiogenomics efforts.
6. Discussion and future directions
The initial successful paradigm of DL in natural images paved the way for the clinical research community to adopt this novel methodology, leading to unprecedented advancements related to computer aided diagnosis or detection (CAD or CADe) systems. Furthermore, advanced data analysis methodologies on medical images of cancer patients can provide insight or associations with molecular and genetic characteristics of the disease. Such radiogenomics analyses can lead to the identification of novel hybrid biomarkers based on the association of the radiomic signature with genomic information of a lesion, sparing patients from unnecessary invasive procedures. Several methods, such as statistical analysis, machine learning and deep learning, aim to find statistically significant correlations or patterns among these high-throughput and high-dimensional features.
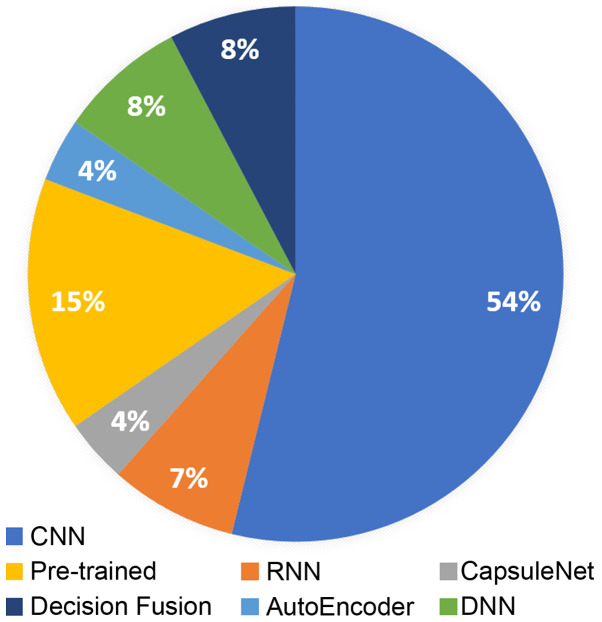
The present review summarizes recent literature in radiogenomics focusing on deep learning methodologies. Deep learning-based radiogenomics is a relatively understudied field, as it has become evident by the limited literature. However, the rise of AI and the association of diverse data, such as imaging phenotypes/features with genomic information can lead to a positive outlook regarding the clinical value of the radiogenomic models. CNNs are by far the most popular architecture in the literature (Fig. 3), due to their efficiency in learning meaningful representations that can demonstrate state-of-the-art results in many image analysis tasks. Additionally, pre-trained models are widely used due to the lack of large medical imaging and genomic datasets providing a valuable alternative in the small dataset setting. Brain and breast are the most commonly studied anatomical areas mainly because of the availability of publicly open and standardized databases.
Figure 3.
Deep architectures used in radiogenomics studies. CNN, convolutional neural network; RNN, recurrent neural network; DNN, deep neural network.
The most important limiting factors as referred by the examined studies concern the number of available patients with both imaging and genomic data available and lack of independent benchmark databases that can provide a performance baseline for comparing different approaches or modeling methodologies. Furthermore, an important concern especially in the clinical practice is the lack of explainability and transparency regarding the fully automated inference in modern AI techniques. This can pose trust and legal issues by either the clinicians or the health authorities respectively. In addition, other concerns raised by the authors of the examined studies included the availability of computational resources, the lack of standardized methodologies for multimodal data analysis, interobserver variability in regions of interest for the studied problem, variations in image acquisition protocol, and issues related to the cost and technical challenges in genomic data acquisition.
Despite all the aforementioned drawbacks, exceptional performance has been achieved in studies regarding the prediction of IDH1 expression with an AUC of 91-94% (18-21), as well as a codeletion of 1p19q (19) with an AUC of 94% in brain gliomas with residual CNN. Korfiatis et al (22), reported an ACC of 94.9% for MGMT methylation status prediction by applying residual learning. The subtyping of invasive breast cancer was investigated by Yoon et al (36), demonstrating an AUC performance between 84 and 97.6%. Finally, the challenging tissue classification in lymph node metastasis was addressed by Zhou et al (43), achieving an AUC performance up to 92% with the combinations of 3D CNN and radiomics on PET/CT examinations. These studies highlight the discriminative ability of deep learning architectures and its robustness as computational framework supporting radiogenomics studies.
In data-driven models it is crucial to transparently describe the methodology for the data handling by providing details about subject stratification and data augmentation increasing trust in the proposed experimental protocol and promoting reproducibility of the results. In this regard, certain studies present limited evidence regarding the fitting status of the examined models. In particular, in a subset of the examined binary classification problems (22,23,42) the performance analysis is assessed only in terms of accuracy, raising concerns for the employed model evaluation method. Deep learning models are prone to overfitting and memorization especially in small datasets or due to the adopted experimental protocol. Moreover, details about the proposed data stratification process in a subject or sample basis for splitting the original dataset into training, validation and testing set are crucial information with impact on the validity of the study.
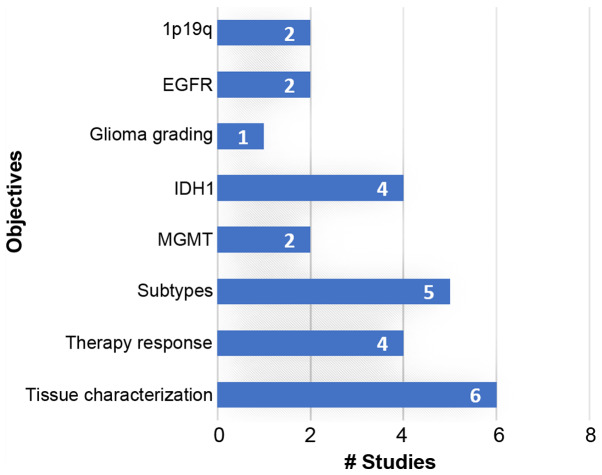
Specialized and open-access databases with multimodal data (imaging, genomics, clinical and laboratory) suitable for radiogenomics analysis could increase the interest of the scientific community in this field. Novel research paradigms are expected to exploit such high-dimensional data by discovering novel associations and interplays among pathways and imaging phenotypes contributing towards robust and accurate computer-aided diagnosis systems. The majority of the current studies investigate brain, breast and lung cancer for deep learning-based radiogenomic analyses, as evidenced in Table II and Fig. 4, mainly due to the high prevalence of these cancers and the availability of open-access databases. More efforts should be made to collect and investigate such combined datasets, also from rare malignancies, in order to enhance the role of DL-based radiogenomics in oncology decision support systems.
Figure 4.
Research objectives of the examined publications. EGFR, epidermal growth factor receptor; IDH1, isocitrate dehydrogenase isozyme 1; MGMT, methylguanine methyltransferase.
Acknowledgments
Not applicable.
Abbreviations
- RMS
rhabdomyosarcoma
- ERMS
embryonal rhabdomyosarcoma
- ARMS
alveolar rhabdomyosarcoma
- NSCLC
non-small cell lung cancer
- GBM
glioblastoma multiforme
- DCIS
ductal carcinoma in situ
- PCN
pancreatic cystic neoplasms
- LNM
lymph node metastasis
- IDH
isocitrate dehydrogenase MGMT methylguanine methyltransferase
- EGFR
epidermal growth factor receptor
- HER2
human epidermal growth factor receptor 2
- ER
estrogen receptor
- PR
progesterone receptor
- DNA
deoxyribonucleic
- miRNA
micro-ribonucleic acid
- RNA-seq
ribonucleic acid sequencing
- H&E
hematoxylin and eosin
- DCE-MRI
dynamic contrast enhanced magnetic resonance imaging
- DWI
diffusion weighted imaging
- MRI
magnetic resonance imaging
- FLAIR
fluid-attenuated inversion recovery
- 18F-FDG PET
fluorodeoxyglucose positron emission tomography
- PET
positron emission tomography
- CT
computerized tomography
- AI
artificial intelligence
- CNN
convolutional neural networks
- AE
autoencoders
- RNN
recurrent neural networks
- LSTM
long-short term memory
- ANN
artificial neural network
- DNN
deep neural network
- VGG
video geometry group
- ResNet
residual network
- SVM
support vector machine
- ROI
region of interest
- ACC
accuracy
- AUC
area under the curve
- SA:V
surface area to volume
- BI-RADS
breast imaging reporting and data system
- TCGA-BRCA
The Cancer Genome Atlas Breast Invasive Carcinoma
Funding
Part of the present review was financially supported by the Stavros Niarchos Foundation within the framework of the project ARCHERS ('Advancing Young Researchers' Human Capital in Cutting Edge Technologies in the Preservation of Cultural Heritage and the Tackling of Societal Challenges').
Availability of data and materials
Not applicable.
Authors' contributions
ET and KM conceived and designed the study. ET and KM researched the literature, performed analysis and interpretation of data and drafted the manuscript. GZP, IS, AHK, LK, NP, DAS and AT critically revised the article for important intellectual content, and assisted in the literature search for this review article. All authors agree to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated, and finally approved the version of the manuscript to be published.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
DAS is the Editor-in-Chief for the journal, but had no personal involvement in the reviewing process, or any influence in terms of adjudicating on the final decision, for this article. All the other authors declare that they have no competing interests.
References
- 1.Woodard GA, Ray KM, Joe BN, Price ER. Qualitative Radiogenomics: Association between Oncotype DX Test Recurrence Score and BI-RADS Mammographic and Breast MR Imaging Features. Radiology. 2018;286:60–70. doi: 10.1148/radiol.2017162333. [DOI] [PubMed] [Google Scholar]
- 2.Gevaert O, Echegaray S, Khuong A, Hoang CD, Shrager JB, Jensen KC, Berry GJ, Guo HH, Lau C, Plevritis SK, et al. Predictive radiogenomics modeling of EGFR mutation status in lung cancer. Sci Rep. 2017;7:41674. doi: 10.1038/srep41674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhou M, Leung A, Echegaray S, Gentles A, Shrager JB, Jensen KC, Berry GJ, Plevritis SK, Rubin DL, Napel S, et al. Non-small cell lung cancer radiogenomics map identifies relationships between molecular and imaging phenotypes with prognostic implications. Radiology. 2018;286:307–315. doi: 10.1148/radiol.2017161845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hussein S, Green A, Watane A, Reiter D, Chen X, Papadakis GZ, Wood B, Cypess A, Osman M, Bagci U. Automatic segmentation and quantification of white and brown adipose tissues from PET/CT scans. IEEE Trans Med Imaging. 2017;36:734–744. doi: 10.1109/TMI.2016.2636188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nitipir C, Niculae D, Orlov C, Barbu MA, Popescu B, Popa AM, Pantea AMS, Stanciu AE, Galateanu B, Ginghina O, et al. Update on radionuclide therapy in oncology. Oncol Lett. 2017;14:7011–7015. doi: 10.3892/ol.2017.7141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.El-Maouche D, Sadowski SM, Papadakis GZ, Guthrie L, Cottle-Delisle C, Merkel R, Millo C, Chen CC, Kebebew E, Collins MT. 68Ga-DOTATATE for tumor localization in tumor-induced osteomalacia. J Clin Endocrinol Metab. 2016;101:3575–3581. doi: 10.1210/jc.2016-2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jansen RW, van Amstel P, Martens RM, Kooi IE, Wesseling P, de Langen AJ, Menke-Van der Houven van Oordt CW, Jansen BHE, Moll AC, Dorsman JC, et al. Non-invasive tumor genotyping using radiogenomic biomarkers, a systematic review and oncology-wide pathway analysis. Oncotarget. 2018;9:20134–20155. doi: 10.18632/oncotarget.24893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ypsilantis PP, Siddique M, Sohn HM, Davies A, Cook G, Goh V, Montana G. Predicting response to neoadjuvant chemotherapy with PET imaging using convolutional neural networks. PLoS One. 2015;10:e0137036. doi: 10.1371/journal.pone.0137036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fujishima H, Fumoto S, Shibata T, Nishiki K, Tsukamoto Y, Etoh T, Moriyama M, Shiraishi N, Inomata M. A 17-molecule set as a predictor of complete response to neoadjuvant chemotherapy with docetaxel, cisplatin, and 5-fluorouracil in esophageal cancer. PLoS One. 2017;12:e0188098. doi: 10.1371/journal.pone.0188098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bownes RJ, Turnbull AK, Martinez-Perez C, Cameron DA, Sims AH, Oikonomidou O. On-treatment biomarkers can improve prediction of response to neoadjuvant chemotherapy in breast cancer. Breast Cancer Res. 2019;21:73. doi: 10.1186/s13058-019-1159-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Angus L, Smid M, Wilting SM, van Riet J, Van Hoeck A, Nguyen L, Nik-Zainal S, Steenbruggen TG, Tjan-Heijnen VCG, Labots M, et al. The genomic landscape of metastatic breast cancer highlights changes in mutation and signature frequencies. Nat Genet. 2019;51:1450–1458. doi: 10.1038/s41588-019-0507-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Trivizakis E, Manikis GC, Nikiforaki K, Drevelegas K, Constantinides M, Drevelegas A, Marias K. Extending 2D convolutional neural networks to 3D for advancing deep learning cancer classification with application to MRI liver tumor differentiation. IEEE J Biomed Health Inform. 2018;23:923–930. doi: 10.1109/JBHI.2018.2886276. [DOI] [PubMed] [Google Scholar]
- 13.Lecun Y, Bottou L, Bengio Y, Haffner P. Gradient-based learning applied to document recognition. Proceedings of the IEEE. 1998;86:2278–2324. doi: 10.1109/5.726791. [DOI] [Google Scholar]
- 14.Sabour S, Frosst N, Hinton GE. Dynamic routing between capsules. Proceedings of the 31st International Conference on Neural Information Processing Systems; Red Hook, NY. Curran Associates Inc; 2017. pp. 3856–3866. [Google Scholar]
- 15.Szegedy C, Vanhoucke V, Ioffe S, Shlens J, Wojna Z. Rethinking the inception architecture for computer vision. Proceedings of the Conference on Computer Vision and Pattern Recognition (CVPR); Las Vegas, NV. 2016. pp. 2818–2826. [Google Scholar]
- 16.Huang G, Liu Z, van der Maaten L, Weinberger KQ. Densely connected convolutional networks. Proceedings of the Conference on Computer Vision and Pattern Recognition (CVPR); Honolulu, HI. 2017. pp. 2261–2269. [Google Scholar]
- 17.He K, Gkioxari G, Dollar P, Girshick R. Mask R-CNN. Proceedings of the IEEE International Conference on Computer Vision. IEEE International Conference on Computer Vision (ICCV); 2017. pp. 2980–2988. [Google Scholar]
- 18.Chang K, Bai HX, Zhou H, Su C, Bi WL, Agbodza E, Kavouridis VK, Senders JT, Boaro A, Beers A, et al. Residual convolutional neural network for the determination of IDH status in low- and high-grade gliomas from mr imaging. Clin Cancer Res. 2018;24:1073–1081. doi: 10.1158/1078-0432.CCR-17-2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chang P, Grinband J, Weinberg BD, Bardis M, Khy M, Cadena G, Su MY, Cha S, Filippi CG, Bota D, et al. Deep-learning convolutional neural networks accurately classify genetic mutations in gliomas. AJNR Am J Neuroradiol. 2018;39:1201–1207. doi: 10.3174/ajnr.A5667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li Z, Wang Y, Yu J, Guo Y, Cao W. Deep Learning based Radiomics (DLR) and its usage in noninvasive IDH1 prediction for low grade glioma. Sci Rep. 2017;7:5467. doi: 10.1038/s41598-017-05848-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liang S, Zhang R, Liang D, Song T, Ai T, Xia C, Xia L, Wang Y. Multimodal 3D densenet for IDH genotype prediction in gliomas. Genes (Basel) 2018;9:1–17. doi: 10.3390/genes9080382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Korfiatis P, Kline TL, Lachance DH, Parney IF, Buckner JC, Erickson BJ. Residual deep convolutional neural network predicts MGMT methylation status. J Digit Imaging. 2017;30:622–628. doi: 10.1007/s10278-017-0009-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Akkus Z, Ali I, Sedlar J, Kline TL, Agrawal JP, Parney IF, Giannini C, Erickson BJ. Predicting 1p19q chromosomal deletion of low-grade gliomas from MR Images using deep learning. doi: 10.1007/s10278-017-9984-3. arXiv:1611.06939. Accessed November 21, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gupta T, Sarin R. Poor-prognosis high-grade gliomas: Evolving an evidence-based standard of care. Lancet Oncol. 2002;3:557–564. doi: 10.1016/S1470-2045(02)00853-7. [DOI] [PubMed] [Google Scholar]
- 25.Decuyper M, Bonte S, van Holen R. Binary glioma grading: Radiomics versus pre-trained CNN features. Proceedings of the 21st International Conference. Part III; Granada, Spain. 2018. pp. 498–505. [Google Scholar]
- 26.Smedley NF, Hsu W. Using deep neural networks for radiogenomic analysis. Proceedings of the 15th International Symposium on Biomedical Imaging (ISBI 2018); Washington, DC. 2018. pp. 1529–1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou Y, Li Z, Zhu H, Chen C, Gao M, Xu K, Xu J. Holistic brain tumor screening and classification based on densenet and recurrent neural network. Proceedings of the International MICCAI Brainlesion Workshop; Cham. Springer; 2018. pp. 208–217. [Google Scholar]
- 28.Momeni A, Thibault M, Gevaert O. Dropout-enabled ensemble learning for multi-scale biomedical data. Proceedings of the International MICCAI Brainlesion Workshop; Cham: Springer; 2018. pp. 407–415. [Google Scholar]
- 29.Afshar P, Mohammadi A, Plataniotis KN. Brain tumor type classification via capsule networks. Proceedings of the International Conference on Image Processing, ICIP; IEEE; 2018. pp. 3129–3133. [Google Scholar]
- 30.Molina JR, Yang P, Cassivi SD, Schild SE, Adjei AA. Non-small cell lung cancer: Epidemiology, risk factors, treatment, and survivorship. Mayo Clin Proc. 2008;83:584–594. doi: 10.1016/S0025-6196(11)60735-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yu D, Zhou M, Yang F, Dong D, Gevaert O, Liu Z, Shi J, Tian J. Convolutional neural networks for predicting molecular profiles of non-small cell lung cancer. Proceedings of the 14th International Symposium on Biomedical Imaging (ISBI 2017); Melbourne, VIC. 2017. pp. 569–572. [Google Scholar]
- 32.Wang S, Shi J, Ye Z, Dong D, Yu D, Zhou M, Liu Y, Gevaert O, Wang K, Zhu Y, et al. Predicting EGFR mutation status in lung adenocarcinoma on CT image using deep learning. Eur Respir J. 2019;53:1800986. doi: 10.1183/13993003.00986-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lingle W, Erickson BJ, Zuley ML, Jarosz R, Bonaccio E, Filippini J, Gruszauskas N. Radiology data from the cancer genome atlas breast invasive carcinoma [TCGA-BRCA] collection. Cancer Imaging Arch. doi: 10.7937/K9/TCIA.2016.AB2NAZRP. [DOI] [Google Scholar]
- 34.Zhu Z, Albadawy E, Saha A, Zhang J, Harowicz MR, Mazurowski MA. Deep learning for identifying radiogenomic associations in breast cancer. Comput Biol Med. 2019;109:85–90. doi: 10.1016/j.compbiomed.2019.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ha R, Mutasa S, Karcich J, Gupta N, Pascual Van Sant E, Nemer J, Sun M, Chang P, Liu MZ, Jambawalikar S. Predicting breast cancer molecular subtype with MRI dataset utilizing convolutional neural network algorithm. J Digit Imaging. 2019;32:276–282. doi: 10.1007/s10278-019-00179-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yoon H, Ramanathan A, Alamudun F, Tourassi G. Deep radiogenomics for predicting clinical phenotypes in invasive breast cancer. Proceedings of the 14th International Workshop on Breast Imaging (IWBI 2018); 2018. p. 75. [Google Scholar]
- 37.Zhu Z, Harowicz M, Zhang J, Saha A, Grimm LJ, Hwang ES, Mazurowski MA. Deep learning analysis of breast MRIs for prediction of occult invasive disease in ductal carcinoma in situ. Comput Biol Med. 2019;115:103498. doi: 10.1016/j.compbiomed.2019.103498. [DOI] [PubMed] [Google Scholar]
- 38.Bibault JE, Giraud P, Durdux C, Taieb J, Berger A, Coriat R, Chaussade S, Dousset B, Nordlinger B, Burgun A. Deep Learning and Radiomics predict complete response after neo-adjuvant chemoradiation for locally advanced rectal cancer. Sci Rep. 2018;8:1–8. doi: 10.1038/s41598-018-30657-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen W, Ji H, Feng J, Liu R, Yu Y, Zhou R, Zhou J. Classification of pancreatic cystic neoplasms based on multimodality images. Proceedings of the International Workshop on Machine Learning in Medical Imaging (MLMI 2018); 2018. pp. 161–169. [Google Scholar]
- 40.Cha KH, Hadjiiski LM, Chan H-P, Samala RK, Cohan RH, Caoili EM, Paramagul C, Alva A, Weizer AZ. Bladder cancer treatment response assessment using deep learning in CT with transfer learning. Proceedings Volume 10134, Medical Imaging 2017: Computer-Aided Diagnosis; 2017. p. 1013404. [Google Scholar]
- 41.Cha KH, Hadjiiski L, Chan HP, Weizer AZ, Alva A, Cohan RH, Caoili EM, Paramagul C, Samala RK. Bladder cancer treatment response assessment in CT using radiomics with deep-learning. Sci Rep. 2017;7:8738. doi: 10.1038/s41598-017-09315-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Banerjee I, Crawley A, Bhethanabotla M, Daldrup-Link HE, Rubin DL. Transfer learning on fused multiparametric MR images for classifying histopathological subtypes of rhabdomyosarcoma. Comput Med Imaging Graph. 2018;65:167–175. doi: 10.1016/j.compmedimag.2017.05.002. [DOI] [PubMed] [Google Scholar]
- 43.Zhou Z, Chen L, Sher D, Zhang Q, Shah J, Pham NL, Jiang S, Wang J. Predicting lymph node metastasis in head and neck cancer by combining many-objective radiomics and 3-dimensioal convolutional neural network through evidential reasoning. doi: 10.1109/EMBC.2018.8513070. arXiv180507021. Accessed May 18, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pedano N, Flanders AE, Scarpace L, Mikkelsen T, Eschbacher JM, Hermes B, Ostrom Q. Radiology data from The Cancer Genome Atlas Low Grade Glioma [TCGA-LGG] Collection. Cancer Imaging Arch. doi: 10.7937/K9/TCIA.2016.L4LTD3TK. [DOI] [Google Scholar]
- 45.Newitt D, Hylton N. Multi-center breast DCE-MRI data and segmentations from patients in the I-SPY 1/ACRIN 6657 trials. Cancer Imaging Arch. doi: 10.1186/s40644-018-0145-9. [DOI] [Google Scholar]
- 46.London AJ. Artificial intelligence and black-box medical decisions: Accuracy versus explainability. Hastings Cent Rep. 2019;49:15–21. doi: 10.1002/hast.973. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.