Abstract
The post-translational modification of proteins by ubiquitinating enzymes plays a central role in a number of cellular functions, such as cell proteolysis, DNA repair, and cell signaling and communication. Deubiquitinating enzymes (DUBs) disassemble ubiquitin chains and remove ubiquitin moieties from proteins. Targeting DUBs in cancer models has revealed an important role for these enzymes in tumorigenesis, and they therefore have emerged as attractive therapeutic targets. In the present study, the effects of three DUB inhibitors, PR-619, RA-9 and LDN-91946, on a non-small cell lung cancer cell line (A549) and a mesothelioma cell line (H2373) were investigated. PR-619 significantly inhibited cell adhesion and the proliferation of both cell lines. RA-9 exerted an inhibitory effect on the adhesion and proliferation of H2373 cells, whereas it had no effect on A549 cells. Notably, however, while PR-619 attenuated the proliferation of both cell lines, it exerted an opposite effect on cell motility; in the case of A549 cells, there was a significant increase in cell motility, while for the H2373 cells, there was a significant decrease. Furthermore, protein phosphorylation kinetic analyses revealed that the effects were cell line-specific. In H2373 cells, the phosphorylation of only one peptide corresponding to the P85A protein was significantly affected, and while LDN-91946 treatment increased phosphorylation, treatment with RA-9 or PR-619 decreased its phosphorylation compared to the DMSO control. By contrast, in the case of A549 cells, the phosphorylation of 21 peptides was significantly affected by the same compounds. In light of the potential for the negative side-effects of DUB inhibition, such as increased cancer cell motility, the data presented herein underscore the dire need for the development of specific DUB inhibitors and to elucidate the individual role of DUB family members in cancer biology before they can be specifically pharmacologically targeted.
Keywords: ubiquitination, deubiquitinase inhibitor, deubiquitinating enzyme, PR-619, RA-9, LDN-91946, cell proliferation, cell migration, mesothelioma, lung adenocarcinoma
Introduction
Ubiquitination, the process through which the ubiquitin protein is covalently attached to another protein, can alter the functional status of proteins or label them for degradation through either mono- or poly-ubiquitination. Functionally, ubiquitination is instrumental in, among other things, regulating the circadian rhythm, silencing genes and opening ion channels (1). The regulation of this process is often disrupted in cancer cells, and in particular, the polyubiquitin tail that marks proteins for proteasomal degradation can be removed by deubiquinating enzymes (DUBs), allowing the buildup of oncogenic proteins (2). Both ubiquitination and deubiquitination are regulated through a specific set of enzymes. Of interest is the deubiquitinating enzymes (DUBs) as they have been identified as emerging therapeutic targets in cancers.
DUBs activate ubiquitin prior to conjugation, as well as remove it from the ubiquitinated proteins (1). The 79 DUBs tentatively identified in the human genome can be broadly classified as either cysteine proteases or Zn2+ metalloproteases. The DUBs are further divided into 5 subclasses based on the structure of the catalytic domain: Ubiquitin C-terminal hydrolases (UCH), ubiquitin-specific processing proteases (USP or UBP), Machado-Joseph disease protein domain proteases (MJDs), ovarian tumor proteases (OTUs) and (JAB1/MPN/Mov34 metalloenzyme) JAMM motif proteases (3).
Mechanistically, the active sites of UCH and UBP DUBs are reminiscent of the papain proteases; however, these enzymes must undergo a conformational change in the presence of ubiquitin to become catalytically competent. This requirement prevents UCH and UBP DUBs from off target effects when ubiquitin is not present. OTU family DUBs have an almost identical geometry to UCH and UBP family DUBS within their active sites. The primary difference is that while the active site of the OTUs is fully functional even before binding ubiquitin, it remains unable to act due to being sterically blocked by an α-helix. The enzyme can perform its function once ubiquitin binds and moves the α-helix away from the catalytic site. The JAMM motif DUBs have a very similar active site structure to that of a cytidine deaminase. In each case, Zn2+ polarizes a water molecule, which then performs a nucleophilic attack on an isopeptide bond (4).
DUBs are upregulated in several types of cancer, including non-small cell lung cancer (NSCLC) and mesothelioma, and thus represent attractive therapeutic targets (5-7). Several DUB inhibitors have been proposed as anticancer agents based on promising results in cell lines and in vivo models (8). However, highly specific compounds have not yet been developed and none of the current inhibitors have entered the clinic as yet (9). The present study evaluated 3 DUB inhibitors, PR-619, RA-9 and LDN-91946, with different DUB target profiles. These inhibitors exerted differential effects on cell migration and proliferation between the lung carcinoma and mesothelioma cell lines. PR-619 is a wide-spectrum ubiquitin/UbL isopeptidase inhibitor (10). RA-9 is a non-specific inhibitor that irreversibly inhibits DUBs by exposing its carbonyl group to a nucleophilic attack from the SH- group of cysteine (11). In contrast to the two previously described compounds, LDN-91946 is a specific inhibitor of ubiquitin C-terminal hydrolase-L1 (UCH-L1), binding to the complex of the enzyme and the substrate (12).
It was hypothesized that DUB inhibition utilizing these compounds at concentrations both comparable to other studies and in line with concentration recommendations from Selleckchem would affect the growth and/or motility of cancer cell lines (11). The data of the present study indicated that while LDN-91946 and RA-9 attenuated the proliferation of both lung cancer and mesothelioma cell lines, they had no effect on their migration. By contrast, PR-619 attenuated both the proliferation and migration of the mesothelioma cell line. PR-619 also inhibited the proliferation of the lung cancer cell line; surprisingly, however, PR-619 augments the migration of lung cancer cells. Moreover, the effects of these 3 drugs on the kinase activity of the NSCLC cell line, A549, and the malignant pleural mesothelioma (MPM) cell line, H2373, were examined using PamGene's peptide-based microarray platform. A549 cells were selected for analysis along with 3 DUB inhibitors based on previous publications (13,14) that used siRNA library screening to identify 12 DUBs in A549 cells. These DUBs are involved in the activation of MET receptor tyrosine kinase by HGF agonist and the scattering response of A549 cells. As a second target, MPM H2373 cells we selected, which expressed only a single nuclear deubiquitinase, BAP1. Protein kinetic profiles of TRKs showed fewer instances of phosphorylated peptides in H2373 as compared to A549, indicating that the DUBs inhibitors could have a more specific impact on cell function in H2373 cells. The variety of potential interactions between these drugs and a complex set of ubiquitination pathways demonstrated in the present study emphasizes the need to explore the effects of these drugs in further detail in future studies.
Materials and methods
Cell culture and reagents
The mesothelioma cell line, H2373, and the NSCLC cell line, A549, (ATCC) were maintained in RPMI-1640 (Corning Inc.) with 10% fetal bovine serum (Sigma-Aldrich; Merck KGaA) as previously described (15). PR-619, RA-9 and LDN-91946 were purchased commercially (Selleck Chemicals) and used at concentrations of 10, 5 and 10 µM for treatment for 24 h, or unless otherwise noted.
Cell adhesion and proliferation assay
The resistance of cells in culture was measured using ECIS (16). Briefly, cells (7.5×104) were plated in the chambers of the 96W1E+ well plate of single-electrode ECIS arrays (Applied Biophysics) that were pre-coated with fibronectin. Cells were treated either with PR-619, RA-9 and LDN-91946, or dimethyl sulfoxide (DMSO), control. Data were collected at 30 min post-inoculation. Data collection was initiated by passing a small fixed-amplitude alternating current between the electrodes and measuring the potential across them and cell resistance was measured at a frequency of 40 kHz. As cells grew and replicated, an increase in the electrical potential between the electrodes was observed. Therefore, measuring cellular impedance served as a readout for cell attachment and growth (17). Cell attachment, adhesion and proliferation were measured for 24-30 h.
Cell migration assay
Cells were seeded on a 96-well ImageLock (Essen BioScience) plate to reach 90% confluency by the following day. Following cell adherence, 96 uniform wounds were created simultaneously using the WoundMaker (Essen BioScience) tool. Cells were washed once with serum-free medium and replenished with 2% FBS medium with vehicle or drug at a concentration of 10 µM. Low serum medium inhibited cell proliferation as the assay was intended to measure only cell migration. To monitor wound healing, the plate was placed in the IncuCyte S3 Live-Cell Analysis System (Essen BioScience) and images were acquired every hour for 48 h. Data analysis was generated by IncuCyte software using a set confluence mask to measure relative wound density over time.
Protein phosphorylation kinetics
For protein phosphorylation kinetics the PamGene tyrosine kinase array (PamGene) was used along with a 4-array semi-automated system (PamStation® 12, PamGene) designed for processing PamChip®-4 arrays. The assay contains 144 phospho-peptides representing tyrosine kinase substrates. The peptides are immobilized on a porous microarray surface through the N-terminus. Briefly, the array was blocked with 0.2% bovine serum albumin (BSA) for 30 cycles of 30 sec each following which each array was washed thrice for 30 sec with 1X ABL protein tyrosine kinase reaction (PK) buffer solution (New England Biolabs). Thereafter, the arrays were incubated at 30°C with the reaction mix, containing 30 µg cell lysate, 1X PK Buffer, 0.4 µl 1 M dithiothreitol (Sigma-Aldrich; Merck KGaA), 0.4 µl 100X BSA (New England Biolabs), 1 µl of 4 mM ATP (Sigma-Aldrich; Merck KGaA), and 0.3 µl of 1 mg/ml monoclonal anti-phosphotyrosine FITC conjugate (clone PY20, Exalpha Biologicals), adjusted to 40 µl with distilled H2O. The sample was pulsed through the array for 60 cycles, and every fifth pump cycle, a 16-bit TIFF image was acquired with a built-in CCD camera. Spot intensities were normalized to local background signal by subtracting the median background signal and the data were log-transformed to normalize the distribution of intensities. Fold changes were calculated by subtracting mean log control values from mean log treatment values.
Western blot analysis
Cell lines were treated with LD91946 (10 µM), RA9 (5 µM) and PR619 (10 µM) for 24 h, followed by lysing with RIPA buffer. Protein concentration for each sample was measured with Bradford protein assay (Bio-Rad Laboratories, Inc.). A total of 50 µg of protein was loaded into each well of an SDS-polyacrylamide gel (4-20% Mini-Protein TGX precast protein gel, Bio-Rad Laboratories, Inc.) and transferred onto a PVDF membrane using the Bio-Rad Mini Trans-Blot apparatus. The membrane was blocked with 5% milk solution for 1 h while rocking gently. Primary anti-ubiquitin antibody 1:1.000 (ab7780; Abcam) was diluted in 5% milk and applied to a PVDF membrane overnight at 4°C and then incubated with secondary antibodies (HRP-labeled antibody 1:10,000 (ab205718; Abcam) for 1 h at room temperature. β-actin (1:3,000) was used as a loading control (A5441; Sigma-Aldrich; Merck KGaA). To visualize protein bands, PVDF membranes were treated with ECL Western Blotting Detection Reagents and then exposed to X-ray Medical Film before being developed.
Statistical analysis
Mathematica software package (Wolfram Research, Inc.) was used to perform statistical computation. ANOVA followed by Tukey's post hoc test were used to evaluate significant differences in stabilized resistance values of the adhesion and proliferation assay. The same methodology was used for the slopes of the migration assay curves, as well as for the phosphorylation assay. In all three assays the drugs were the factors. P<0.05 was considered to indicate a statistically significant difference.
Results and Discussion
Effect of DUB inhibitors on cell adhesion and proliferation
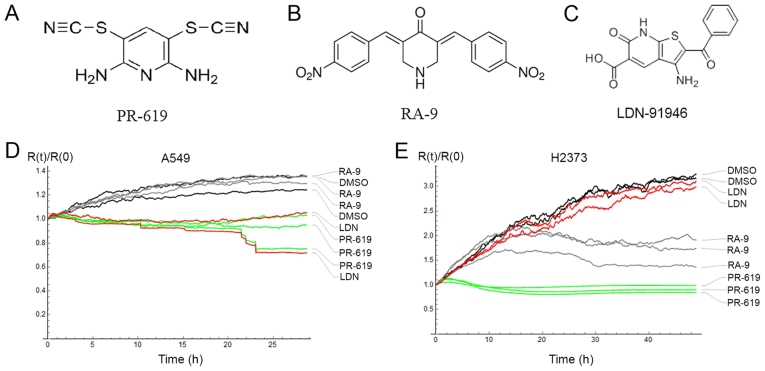
The effect of PR-619, RA-9 and LDN-91946 (Fig. 1A-C) on cell adhesion and proliferation was evaluated by electrical cell-based impedance sensing (ECIS) (16). The resistance time course for control (DMSO) and drug-treated cells was measured by ECIS as a monotonic function of the fraction of the covered electrode surface (18). Therefore, stabilized resistance values for A549 and H2373 cells were compared as a measure of the drug effect. While LDN-91946 and PR-619 attenuated the proliferation of the lung cancer cell line (A549), RA-9 failed to inhibit the proliferation of this cell line. On the other hand, while PR-619 and RA-9 impeded the proliferation of the mesothelioma cells (H2373), LDN-91946 did not. For both cell lines, a statistically significant difference of the drug effect was ascertained by ANOVA with values of P<0.05 (Fig. 1D and E). Furthermore, Tukey's post hoc test indicated significant differences in adhesion and proliferation, as compared with the DMSO control, when treating the A549 cells with LDN-91946 or PR-619, and when treating the H2373 cells with PR-169 or RA-9 (Table I).
Figure 1.
(A-C) DUB inhibitor structures. (D and E) Time course of cell proliferation and adhesion treated with DMSO (control), RA-9 (5 µM), PR-619 (10 µM) and LDN-91946 (10 µM) measured by resistance at 4 kHz; (D) A549 cells; (E) H2373 cells. DUB, deubiquitinating enzyme.
Table I.
Averaged relative resistances at the end point evaluation of the cell adhesion and proliferation assay.
| A549 cells mean R(t)/R(0) at 20-21 h |
H2373 cells mean R(t)/R(0) at 47-48 h |
|
|---|---|---|
| DMSO | 1.284 | 3.166 |
| LDN-91946 | 0.939a | 2.993 |
| PR-619 | 0.944a | 0.909a |
| RA-9 | 1.314 | 1.705a |
P<0.05, statistically significant difference in the post hoc comparison with DMSO.
Consistent with the observations of the differential effects of these DUBs inhibitors on lung cancer and mesothelioma, RA-9 has been observed to inhibit the proliferation of breast, ovarian and cervical cancer cell lines at concentrations comparable to those used in the present study (11). The cytotoxic effect of RA-9 has been attributed to 'proteotoxic stress', where non-degraded ubiquitinated protein accumulates in the cell (19). These experiments establish potent single agent activity of both LDN91946 and PR-619 in the models in the present study. The differential effects of these compounds on the two cancer cell lines is of interest, as it suggests specificity and the involvement of distinct DUBs in this process.
Effect of DUB inhibitors on cell motility
Cell motility was assessed with a wound healing assay. It is important to note the cell proliferation was suppressed (Materials and methods), as is typical in such assays (20), so that the wound closure by cell growth is not a confounding factor. The IncuCyte® Live Cell Analysis System used in the present study measures relative wound density Y(t)
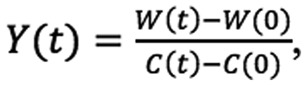 |
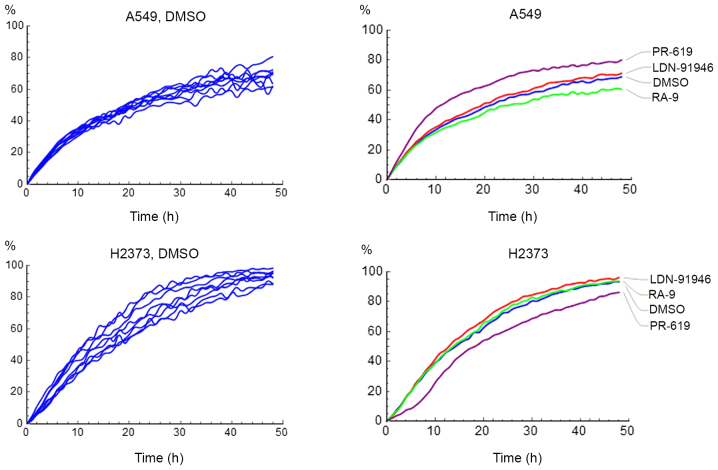
where W(t) and C(t) are the image pixel densities of the wound and non-wounded areas respectively. The assay was replicated 8 times resulting in 8 curves corresponding to wound density vs. time. Representative data of the replicated measurements of the A549 and H2373 cells treated with DMSO, PR-619, or LDN-91946 are shown on Fig. 2.
Figure 2.
Wound healing time course of A549 and H2373 cells treated with DMSO, LDN-91946, RA-9 and PR-619 at 10 µM. Left column, raw data; right column, averaged curves.
In order to compare the curves and to ascertain statistically-significant differences, it was desirable to describe each curve with one parameter, i.e., an average migration rate. To this end, the wound healing process was modelled as follows: The cell migration was a first order process, i.e., the number of migrated cells between the wound and the non-wounded area per unit of time is estimated to be proportional to the number of cells in the non-wounded area. Mathematically, the pixel densities of the wound and the non-wounded areas may be modelled by the following differential equations, which depend on k, the average migration rate:
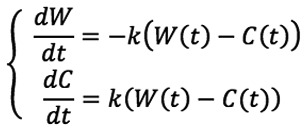 |
Solving the above equations and substituting for Y(t) yields:
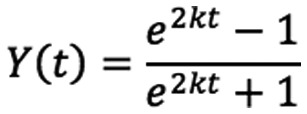 |
First order series expansion yields Y(t)=kt+O(t2). As expected, the data follow a straight line for the first 10 h (Fig. 2). It is also important to note that measuring the initial slope of the curve provides an additional benefit to the analysis. Specifically, given the short time necessary to measure the initial slope, the approach eliminates biases from any residual cell proliferation (if it was not fully suppressed in the experiment).
The slope of the initial parts of the curves was estimated by fitting a straight line with an intercept set to zero. Each experiment was repeated 8 times, and the resulting slopes were further analyzed using a one-way ANOVA. The data are presented in Table II and representative movies of cell migration for the A549 and H2373 cells with and without drug treatments are available as supplementary material (Videos S1-S8). Notably, while LDN-91946 and RA-9 had no detectable effect on THE migration in either cell line, PR-619 significantly attenuated the migration of the mesothelioma cell line (H2373), but increased the migration rate of the lung cancer cell line (A549). To the best of our knowledge, the effects of LDN-91946, PR-619 and RA-9 on the motility of cell lines have not yet been reported. It is believed that the modeling of the wound healing assay evaluated and quantified the cell motility in a statistically meaningful manner.
Table II.
Average migration rate measured by the wound healing assay.
P<0.05, statistically significant difference in the post hoc comparison with DMSO.
Protein phosphorylation
To shed light onto the mechanisms through which these inhibitors may act, protein phosphorylation kinetics were measured using an endpoint PamGene assay. The assay provides surface-immobilized peptides, which are exposed to a cell lysate. Peptide-specific phosphorylation activity of the lysate is measured by immunostaining. For this assay, the cells were grown to 75% confluence, treated with either DMSO (control) or drugs at 10 µM for 5 h. Cell lysates were analyzed by a PamGene chip in triplicate. The extent of phosphorylation of each peptide was measured and compared between the treatments. One-way ANOVA was used to test for significant differences among the treatments. Post hoc tests revealed which treatments significantly affected phosphorylation in pairwise comparisons. The present study focused on those peptides that, amongst the responses to the different drugs, had at least one statistically significant difference in comparison with the control.
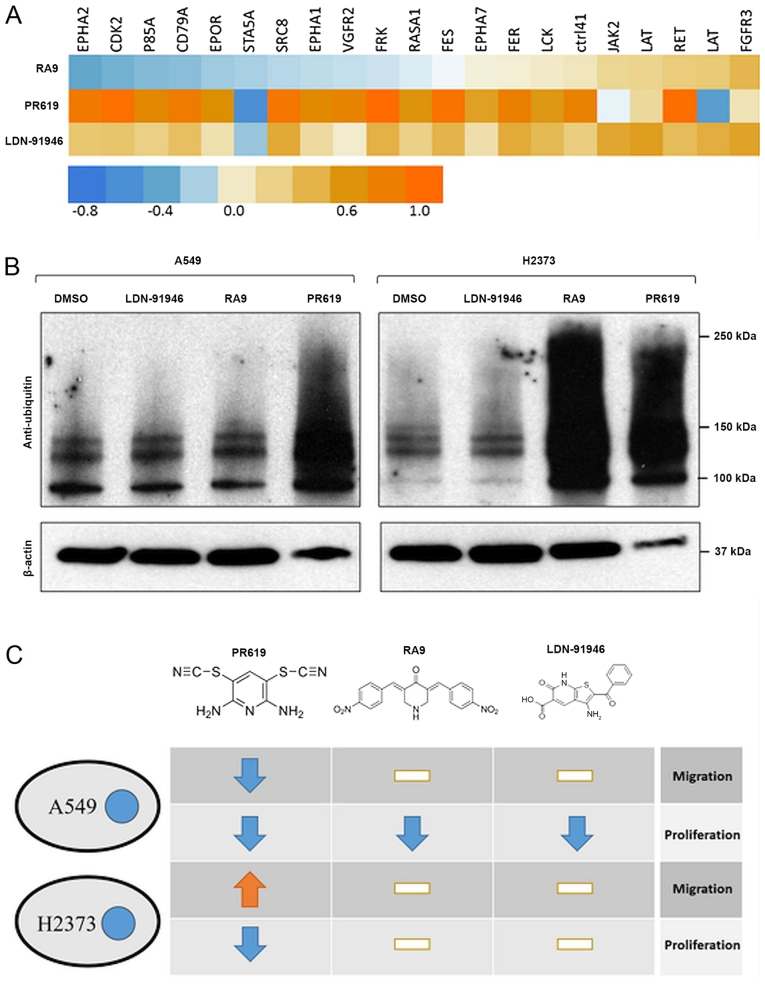
In the case of the mesothelioma cell line (H2373), a single peptide was significantly affected by the treatments. The peptide corresponded to the protein P85A with the phosphorylation site between amino acid positions 600 and 612. Compared to the DMSO control, a statistically significant difference was found in the presence of LDN-91946 (increase), RA-9 (increase) and PR-619 (decrease). P85A is involved in numerous pathways, including the mTOR and PI3K-Akt signaling pathways as found in the KEGG pathway database (KEGG, 21). Similarly, for the A549 cells, >20 peptides were affected, which were involved in 'Pathways in cancer' (hsa05200), including the PI3K-Akt signaling pathway (hsa04151) and the Ras signaling pathway (hsa04014), among others. A visual representation of the data is presented in Fig. 3A.
Figure 3.
(A) Heatmap of log-transformed measurements of the extent of phosphorylation in the A549 cell line. The RA-9, PR-619 and LDN-91946 treatment values are subtracted with DMSO values. Note, LAT appears twice (different peptides). (B) Representative western blot of a ubiqutination assay in A549 and H2373 cell lines treated with LD91946 (10 μM), RA9 (5 μM) and PR619 (10 μM). (C) Tabular representation of the drug effects on the studied cell lines as compared with the DMSO control. White bar indicates no significant change.
Verification of DUB inhibition
Given the diverse effects of the DUB inhibitors on the proliferation and motility of the two cell lines, western blot analysis was performed to verify that the studied compounds had an inhibitory effect on deubiquitination (increase in ubiquitin levels) and whether it was cell line-specific. As shown in Fig. 3B, PR-619, one of the broad spectrum inhibitors, wsa the only inhibitor that resulted in a robust, cell line-independent activity, and its effect on both cell lines was statistically significant (Tables I and II). LDN-91946, being a specific DUB inhibitor, unsurprisingly did not result an easily measurable change in the context of the whole cell ubiquitin levels, making it difficult to determine whether the impact on proliferation in A549 was an on-target effect using this method. RA-9, the other non-specific DUB inhibitor, exhibited a potent inhibitory effect of DUBs on the H2373 cell line, while having very little impact on ubiquitin levels in A549 cells. Of note, this result is in contrast to the results of the proliferation experiment that revealed the inhibition of A549 proliferation, while no inhibition of DUB activity.
Variety of responses
It is interesting to note a seemingly unpredictable response of the cell lines to the drug treatment. One could have studied additional cell lines, which may or may not behave respectively the same as the ones we already studied. Moreover, the current knowledge of the cell lines is not fine grained enough to be able to tease apart the mechanistic effects resulting from the interaction of the cell line peculiarities and the drug. In fact, preliminary studies were conducted with BEAS-2B and Met 5A cell lines and the same drugs to discover equally unexpected outcomes (data not shown). The purpose of the present study, however, is to open up a conversation about the unpredictability of the drug effects on different types of cells.
In conclusion, the findings of the present study demonstrate that while PR-619 acts as a proliferation-inhibitor for both cell lines and exerts a significant effect on overall ubiquitin levels, it affects cell motility quite differently, which points to a need for detailed investigations into the role of ubiquitination in these two important processes (Fig. 3C). Both RA-9 and LDN-91946 exhibited differential effects on proliferation between cell lines, though in the case of RA-9, the proliferation change was in the A549 cells, the cell line that did not exhibit any change in ubiquitin levels. This result is interesting as it not only indicates that a strong decrease in ubiquitin levels can potentially have no effect on cell motility and proliferation, but also indicates that the A549 cell line may have a method for processing or otherwise negating the effects of the RA-9 inhibitor that resulted in reduced proliferation. If RA-9 is being rapidly processed, it could be that the downstream products are toxic. The mechanism responsible for the escape of the A549 cells from the effects RA-9 may prove an interesting avenue of research going forward.
The only noted impact of the change in the phosphorylation state of the H2373 cellss was the increase in migration and a decrease in proliferation found when PR-619 was applied. Increases in the phosphorylation of P85A during exposure to RA-9 and LDN-91946 had little impact on proliferation or migration. Moreover, the larger number of phosphorylation changes observed in the A549 cell line uniformly lead to a reduction in proliferation, though only PR-619, which had the greatest number of increased phosphorylation sites, showed a significant reduction in migration rates. The highly selective effect of altered phosphorylation on P85A in the mesothelioma cell line, versus a much larger number of proteins in the same (PI3K) pathway in the lung cancer cell line, after the application of the same drugs, emphasizes the subtleties with which proliferation and migration are coordinated. The effects of the PI3K-AKT signaling pathway range from feeding into the p53 and NF-κB pathways to cell cycle control and apoptosis while the mTOR pathway impacts regulation of the actin cytoskeleton. Insights regarding how these drugs influence these pathways, and possible off target effects may prove to be both crucial in developing small molecule DUBs inhibitors as targeted cancer therapeutics and critical to avoiding misapplication of these treatments.
Supplementary Data
Acknowledgments
Not applicable.
Funding
The present study was supported by the National Cancer Institute of the National Institutes of Health under Grants nos. P30CA033572, U54CA209978 and R01CA218545.
Availability of data and materials
All data generated or analyzed during this study are included in this published article or are available from the corresponding author on reasonable request.
Authors' contributions
AP and MSN conducted the analysis. TM, YCT, SS, and AN designed and performed the experiments. IM, YHCT, KZ, DR and SS provided reagents and reviewed the manuscript. MWN provided technical guidance. PK, AP, TM and MSN wrote the manuscript. MWN, SKB, MS, IM, YHCT, KZ, DR, SS, PK, MSN and RS conceptualized and designed the experiments and reviewed the manuscript. All authors have read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Kerscher O, Felberbaum R, Hochstrasser M. Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu Rev Cell Dev Biol. 2006;22:159–180. doi: 10.1146/annurev.cellbio.22.010605.093503. [DOI] [PubMed] [Google Scholar]
- 2.Mansour MA. Ubiquitination: Friend and foe in cancer. Int J Biochem Cell Biol. 2018;101:80–93. doi: 10.1016/j.biocel.2018.06.001. [DOI] [PubMed] [Google Scholar]
- 3.Nijman SMB, Luna-Vargas MPA, Velds A, Brummelkamp TR, Dirac AMG, Sixma TK, Bernards R. A genomic and functional inventory of deubiquitinating enzymes. Cell. 2005;123:773–786. doi: 10.1016/j.cell.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 4.Amerik AY, Hochstrasser M. Mechanism and function of deubiquitinating enzymes. Biochim Biophys Acta. 2004;1695:189–207. doi: 10.1016/j.bbamcr.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 5.Poondla N, Chandrasekaran AP, Kim KS, Ramakrishna S. Deubiquitinating enzymes as cancer biomarkers: New therapeutic opportunities? BMB Rep. 2019;52:181–189. doi: 10.5483/BMBRep.2019.52.3.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Patel K, Ahmed ZS, Huang X, Yang Q, Ekinci E, Neslund-Dudas CM, Mitra B, Elnady FA, Ahn YH, Yang H, et al. Discovering proteasomal deubiquitinating enzyme inhibitors for cancer therapy: Lessons from rational design, nature and old drug reposition. Future Med Chem. 2018;10:2087–2108. doi: 10.4155/fmc-2018-0091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kaushal K, Antao AM, Kim KS, Ramakrishna S. Deubiquitinating enzymes in cancer stem cells: Functions and targeted inhibition for cancer therapy. Drug Discov Today. 2018;23:1974–1982. doi: 10.1016/j.drudis.2018.05.035. [DOI] [PubMed] [Google Scholar]
- 8.Yuan T, Yan F, Ying M, Cao J, He Q, Zhu H, Yang B. Inhibition of Ubiquitin-Specific Proteases as a Novel Anticancer Therapeutic Strategy. Front Pharmacol. 2018;9:1080. doi: 10.3389/fphar.2018.01080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harrigan JA, Jacq X, Martin NM, Jackson SP. Deubiquitylating enzymes and drug discovery: Emerging opportunities. Nat Rev Drug Discov. 2018;17:57–78. doi: 10.1038/nrd.2017.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Altun M, Kramer HB, Willems LI, McDermott JL, Leach CA, Goldenberg SJ, Kumar KG, Konietzny R, Fischer R, Kogan E, et al. Activity-based chemical proteomics accelerates inhibitor development for deubiquitylating enzymes. Chem Biol. 2011;18:1401–1412. doi: 10.1016/j.chembiol.2011.08.018. [DOI] [PubMed] [Google Scholar]
- 11.Issaenko OA, Amerik AY. Chalcone-based small-molecule inhibitors attenuate malignant phenotype via targeting deubiquitinating enzymes. Cell Cycle. 2012;11:1804–1817. doi: 10.4161/cc.20174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mermerian AH, Case A, Stein RL, Cuny GD. Structure-activity relationship, kinetic mechanism, and selectivity for a new class of ubiquitin C-terminal hydrolase-L1 (UCH-L1) inhibitors. Bioorg Med Chem Lett. 2007;17:3729–3732. doi: 10.1016/j.bmcl.2007.04.027. [DOI] [PubMed] [Google Scholar]
- 13.D'Arcy P, Wang X, Linder S. Deubiquitinase inhibition as a cancer therapeutic strategy. Pharmacol Ther. 2015;147:32–54. doi: 10.1016/j.pharmthera.2014.11.002. [DOI] [PubMed] [Google Scholar]
- 14.Buus R, Faronato M, Hammond DE, Urbé S, Clague MJ. Deubiquitinase activities required for hepatocyte growth factor-induced scattering of epithelial cells. Curr Biol. 2009;19:1463–1466. doi: 10.1016/j.cub.2009.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lennon FE, Cianci GC, Kanteti R, Riehm JJ, Arif Q, Poroyko VA, Lupovitch E, Vigneswaran W, Husain A, Chen P, et al. Unique fractal evaluation and therapeutic implications of mitochondrial morphology in malignant mesothelioma. Sci Rep. 2016;6:24578. doi: 10.1038/srep24578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Giaever I, Keese CR. Micromotion of mammalian cells measured electrically. Proc Natl Acad Sci U S A. 1991;88:7896–7900. doi: 10.1073/pnas.88.17.7896. [DOI] [PMC free article] [PubMed] [Google Scholar]; Erratum. Proc Natl Acad Sci U S A. 1993;90:1634. [Google Scholar]
- 17.Szulcek R, Bogaard HJ, van Nieuw Amerongen GP. Electric cell-substrate impedance sensing for the quantification of endothelial proliferation, barrier function, and motility. J Vis Exp. 2014;85:e51300. doi: 10.3791/51300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wegener J, Keese CR, Giaever I. Electric cell-substrate impedance sensing (ECIS) as a noninvasive means to monitor the kinetics of cell spreading to artificial surfaces. Exp Cell Res. 2000;259:158–166. doi: 10.1006/excr.2000.4919. [DOI] [PubMed] [Google Scholar]
- 19.Coughlin K, Anchoori R, Iizuka Y, Meints J, MacNeill L, Vogel RI, Orlowski RZ, Lee MK, Roden RB, Bazzaro M. Small-molecule RA-9 inhibits proteasome-associated DUBs and ovarian cancer in vitro and in vivo via exacerbating unfolded protein responses. Clin Cancer Res. 2014;20:3174–3186. doi: 10.1158/1078-0432.CCR-13-2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jonkman JE, Cathcart JA, Xu F, Bartolini ME, Amon JE, Stevens KM, Colarusso P. An introduction to the wound healing assay using live-cell microscopy. Cell Adhes Migr. 2014;8:440–451. doi: 10.4161/cam.36224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kanehisa M, Furumichi M, Tanabe M, Sato Y, Morishima K. KEGG: New perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017;45(D1):D353–D361. doi: 10.1093/nar/gkw1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article or are available from the corresponding author on reasonable request.