Abstract
During αβ T cell development, T cell antigen receptor (TCR) engagement transduces biochemical signals through a protein-protein interaction (PPI) network that dictates dichotomous cell fate decisions. It remains unclear how signal specificity is communicated, instructing either positive selection to advance cell differentiation or death by negative selection. Early signal discrimination might occur by PPI signatures differing qualitatively (customized, unique PPI combinations for each signal), quantitatively (graded amounts of a single PPI series), or kinetically (speed of PPI pathway progression). Using a novel PPI network analysis, we found that early TCR-proximal signals distinguishing positive from negative selection appeared to be primarily quantitative in nature. Furthermore, the signal intensity of this PPI network was used to find an antigen dose that caused a classic negative selection ligand to induce positive selection of conventional αβ T cells, suggesting that the quantity of TCR triggering was sufficient to program selection outcome. Because previous work had suggested that positive selection might involve a qualitatively unique signal through CD3δ, we reexamined the block in positive selection observed in CD3δ0 mice. We found that CD3δ0 thymocytes were inhibited but capable of signaling positive selection, generating low numbers of MHC-dependent αβ T cells that expressed diverse TCR repertoires and participated in immune responses against infection. We conclude that the major role for CD3δ in positive selection is to quantitatively boost the signal for maximal generation of αβ T cells. Together, these data indicate that a quantitative network signaling mechanism through the early proximal TCR signalosome determines thymic selection outcome.
INTRODUCTION
For conventional αβ T cells, central tolerance involves positive selection of clones bearing T cell antigen receptors (TCRs) with weak reactivity to self-peptide/major histocompatibility complexes (pMHCs) and negative selection by deletion of clones bearing TCRs with strong reactivity to self-pMHCs within the thymus (1, 2). During selection, TCR engagement initiates biochemical signals through six associated CD3 subunits (γ, δ, 2ε, and 2ζ), each containing one to three immuno-receptor tyrosine-based activation motifs (ITAMs) and other signaling sequences (3, 4). CD3 activation initiates a protein-protein interaction (PPI) signaling cascade involving LCK, ZAP70, LAT, SLP76, and other enzymes and adaptor proteins that together form a highly interactive network, termed the proximal TCR signalosome (5-8). This signalosome can transmit more than one type of signal (3, 9-13), which may arise from the conditional interconnectivity of its multiprotein, modular network. Yet, how these TCR/CD3-intrinsic and -extrinsic proximal proteins form discrete network signatures that program the cellular response for positive versus negative selection is incompletely understood.
It has been proposed that signaling proteins form complexes like letters form words, with specific combinations and quantities instructing cells to perform specific functions (14-16). As a starting theoretical framework, basic categories of network PPI patterns that could instruct dichotomous responses in T cell selection are those in which signatures differ qualitatively, quantitatively, or kinetically (fig. S1). In general, qualitative and kinetic signaling models are currently favored in the field (2, 17-19), whereas a former quantitative model (20, 21) is now thought to apply to innate-like and regulatory agonist-selected T cells more than conventional αβ T cells (22-26), although there is some controversy over this issue (27-30).
Supporting the qualitative signal model, CD3δ was shown to play a specific role in positive selection (31, 32). Mutation of TCR α-connecting peptide motif (αCPM) hindered association with CD3δ and blocked thymocyte development at the preselection CD4+CD8+ (double positive, DP) stage, preventing positive selection but leaving negative selection intact (33, 34). CD3δ0 mice presented a phenotype with similarities to αCPM mutant mice, including blockade of T cell development at the prepositive selection CD4+CD8+ DP stage (31, 32). All other CD3 subunits (ε, γ, and ζ) are required for γδ TCR and pre-TCR signaling, making δ the only CD3 subunit that is not required until αβ TCRs are first expressed to audition for selection at the DP stage (31). In addition, in fetal thymic organ culture (FTOC) experiments using wild-type OT1 thymocytes, the nominal antigenic ligand [ovalbumin (OVA) peptide in H2-Kb] was tested over ten 10-fold dilutions, in which all functional signals induced negative selection, none induced positive selection of conventional αβ T cells, and antigen dilutions that were too low for negative selection induced no selection response (27, 35). These and other studies showed that positive versus negative selection signals led to critical kinetic differences in Ras-Raf-ERK activation and subcellular localization downstream of the TCR signalosome (1, 2, 32, 34-38), supporting the proposal that TCR decides between positive and negative selection by initiating qualitatively distinct signaling pathways. Thus, the current conventional model is (i) kinetic proofreading governs TCR engagement (39), which determines (ii) a currently undefined quality of activity in the signalosome that specifies positive versus negative selection, leading to (iii) differential Ras-Raf-ERK activation kinetics, subcellular localization, and downstream events that program selection outcome.
The present work was dedicated to elucidating the TCR-proximal signaling step (ii) above, to determine the network mechanism at play very early as protein complexes of the signalosome begin to specify selection outcome. To assess this protein network model of signal specificity in the TCR signalosome, we used a recently described multiplex microsphere-based approach called PiSCES (for proteins in shared complexes detected by exposed surface epitopes) (40), for analysis of interactions between 20+ cellular proteins. On the basis of previous knowledge and current interactome data (6), we assembled a panel of immunoprecipitation (IP) antibodies (Abs) covalently coupled to specific microsphere classes, each defined by a unique proportion of two dyes within the polystyrene latex material base. After various stimulatory or control conditions, cells were lysed, multiplex IP was performed, and co-associated proteins were probed with fluorochrome-labeled Abs and analyzed by flow cytometry (fig. S1C and table S1). In previous studies, PiSCES analysis identified both qualitatively and quantitatively distinct network PPI patterns when comparing human TCR versus CD28 signaling pathways and antigenic signaling in skin from autoimmune alopecia areata versus control patient T cells (40). Therefore, PiSCES analysis appeared well suited to address how the TCR signalosome responds to different functional signals in the context of thymocyte selection.
RESULTS
Qualitative and quantitative signaling differences can be observed in human T cell lines
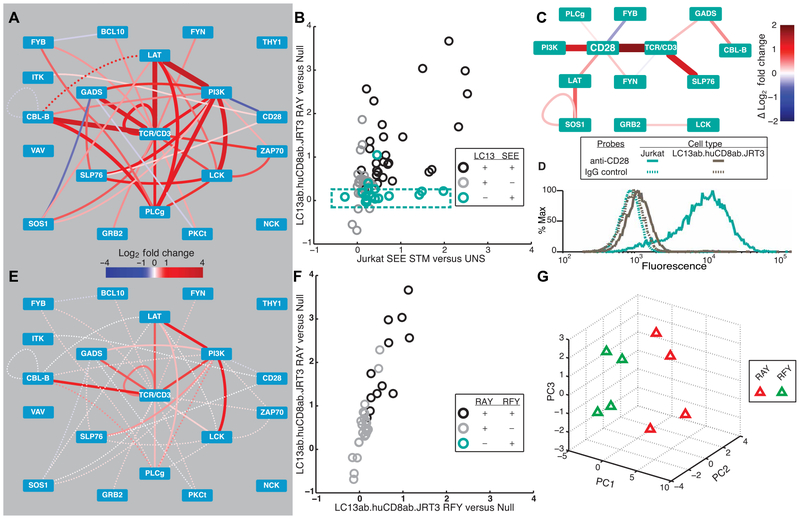
To examine differences in PiSCES signatures between functionally distinct TCR signals, we began with the LC13 system, a human TCR whose responses to agonist and antagonist pMHC ligands are well characterized (41-43). Whereas LC13 TCR responds to the Epstein-Barr virus–derived FLRGRAYGL (RAY) peptide loaded in HLA-*B08:01 as an agonist, the single-peptide–substituted FLRGRFYGL (RFY) in HLA-*B08:01 has no agonist activity, but strong antagonist activity, despite both ligands binding LC13 TCR with similar half-lives (41). A Jurkat-based cell line system was used, because (i) these cells represent transformed human thymocytes, originating from T cell acute lymphoblastic leukemia (44), and (ii) in mice, previously defined antagonist ligands were shown to induce positive selection in thymocytes, whereas agonists induced negative selection (27, 35, 45, 46). Therefore, the LC13 TCR and human CD8 coreceptor were retrovirally expressed in JRT3 [a TCRβ-deficient derivative of Jurkat (47)] to create LC13ab.huCD8ab.JRT3 cells, which were stimulated with agonist peptide RAY-loaded C1R.B0801 antigen-presenting cells (APCs). After 5 min, T cells were lysed and subjected to PiSCES analysis, revealing induction of a rich network signature of multiprotein signaling complexes (Fig. 1A). To examine the extent to which two different strong agonist signals in related cell lines would produce similar PiSCES network signatures, we compared the LC13 RAY–induced PiSCES signature with our previously published dataset, in which Jurkat T cells had been stimulated for 5 min with staphylococcal enterotoxin E (SEE) superantigen (40). We found that many of the same protein pairs joined complexes in both sets of experiments, and we plotted the average fold induction for protein pairs that were hits in either dataset, with SEE-Jurkat stimulation on the x axis and agonist LC13-JRT3 stimulation on the y axis (Fig. 1B). In a perfect correlation, all points would have fallen linearly along the 45° angle, which would have indicated that all signaling complexes were induced to the same degree in both datasets, but this was not the case. Among the differences noted, some protein pairs joined complexes that were almost exclusively induced in the SEE dataset only (Fig. 1B; teal box), in a subnetwork that appeared to be centered around CD28 (Fig. 1C). Pursuing possible reasons for this difference, we found that unlike Jurkat, LC13ab.huCD8ab.JRT3 cells expressed low surface CD28, and thus fewer copies were available to respond to stimulation (Fig. 1D). These data provide an example where PiSCES analysis revealed qualitative and quantitative differences in cell signaling responses between two stimulatory conditions.
Fig. 1. PiSCES signatures reveal differential qualitative and quantitative TCR-proximal signaling activities.
(A) PiSCES signature of LC13ab.huCD8ab.JRT3 cells stimulated with agonist peptide (FLRGRAYGL) for 5 min (mean log2 fold change, stimulated/basal; dotted lines indicate trend of nonsignificant protein pairs that appear as hits for data comparisons shown ahead). (B) Comparison of mean log2 fold changes in abundance of protein pair hits induced by agonist pMHC in LC13ab.huCD8ab. JRT3 cells versus SEE superantigen in Jurkat cells. Statistically significant hits that occurred in both stimuli are black, hits in LC13 system only are gray, and hits in SEE-Jurkat system are teal. (C) Subnetwork [from (B), teal points within the teal dashed box] visualized as the difference in mean log2 fold change in SEE versus LC13. (D) Surface staining with anti-CD28 or immunoglobulin G (IgG) negative control for Jurkat versus LC13ab.huCD8ab.JRT3 cells. (E) PiSCES signature of LC13ab.huCD8ab.JRT3 cells stimulated with antagonist peptide (FLRGRFYGL) for 5 min (mean log2 fold change, stimulated/basal; dotted lines indicate trend of nonsignificant protein pairs). (F) Comparison of mean log2 fold changes in abundance of protein pair hits induced by LC13 agonist versus antagonist stimulation. Compared with (B), note lack of teal “antagonist-only” hits. (G) PCA of four independent experiments shows separation of 5-min agonist versus antagonist PiSCES signatures.
Human LC13 TCR signalosome activity differs quantitatively in response to agonist versus antagonist pMHC ligands
We next assessed how TCR-proximal signaling protein complexes would respond to antagonist stimulation. Initially, PiSCES network visualization of LC13 antagonist peptide (RFY) stimulation (Fig. 1E) appeared to present a lower-intensity version of the agonist (RAY)–induced signature (Fig. 1A). Comparing fold change intensities between agonist (RAY) and antagonist (RFY) stimuli, protein complexes were induced by the agonist to a quantitatively greater degree in an approximately linear fashion weighted toward agonist stimulation, and the antagonist did not appear to induce customized complexes that would indicate a qualitatively unique antagonist signature (Fig. 1F). The differences in response were sufficient to allow principal component analysis (PCA) to separate agonist (RAY)– from antagonist (RFY)–stimulated LC13 data (Fig. 1G). Thus, in contrast to that observed relative to SEE-stimulated Jurkat cells, quantitative but not qualitative differences in PiSCES signatures were evident when comparing agonist versus antagonist pMHC stimuli of LC13ab. huCD8ab.JRT3 cells.
Primary OT1 TCR signalosome activity differs quantitatively in response to stimulation by positive versus negative selection pMHC ligands
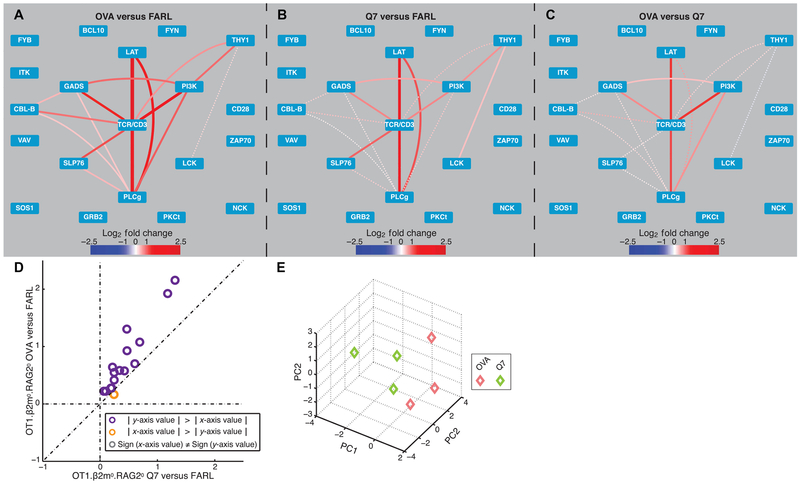
To assess PiSCES signatures of the TCR-proximal signalosome in primary preselection DP thymocytes, we used a mouse genotype (OT1.RAG20.β2m0) that had previously generated data in support of the qualitative signaling model for thymic selection, and for which functionally distinct pMHC ligands have been characterized (27, 35). Thymocytes were co-incubated for 1 min with C1R.Kb APCs presenting either OVA (negative selection), Q7 (positive selection), or FARL (null, no selection) peptides (see Materials and Methods for sequences), in vitro. After cell lysis, multiplex IP and PiSCES analysis were performed, by normalizing both OVA (Fig. 2A) and Q7 stimulation (Fig. 2B) to the null control condition or, alternatively, by dividing the average median fluorescence intensities (MFIs) in OVA by those of Q7 directly (Fig. 2C). With OVA stimulation, a clear network signature included shared protein complexes already being induced at 1 min between TCR/CD3, LAT, SLP76, GADS, PI3K, PLCγ, and others (Fig. 2A), whereas Q7 stimulation presented a similar network pattern but was generally weaker in both fold change and statistical significance (Fig. 2B). Dividing average MFI intensities of hits in OVA stimulation by those of Q7 showed that fold changes were greater in OVA stimulation without appearance of a clear subnetwork of protein complexes that might be balanced toward the Q7 positive selection stimulus (Fig. 2C). Comparing fold change intensities between negative and positive selection stimuli, protein complexes were induced by OVA to a quantitatively greater degree in an apparent linear fashion, and Q7 did not appear to induce customized complexes that would indicate a qualitatively unique positive selection signature (Fig. 2D). These differences were sufficient for PCA to separate the response to the two stimuli (Fig. 2E). Thus, similar to the patterns observed when comparing agonist and antagonist stimulations of the human LC13 TCR, primary mouse preselection thymocytes responded to positive or negative selection ligands primarily via quantitatively distinct PPI activities through the proximal TCR signalosome. In contrast, if nonphysiologic stimuli, such as pervanadate or H2O2, were administered to OT1.RAG20.β2m0 preselection thymocytes, qualitatively distinct PiSCES signatures were produced (figs. S2 and S3 and Supplementary Results and Discussion), supporting the interpretation that the primarily quantitative- only network PPI differential occurred specifically in response to positive versus negative selection pMHC ligands.
Fig. 2. Predominant quantitative difference in PiSCES signatures comparing OT1 thymocyte response to positive and negative selection pMHC ligands.
PiSCES signature of preselection OT1.β2m0.RAG20 thymocytes stimulated for 1 min with (A) a negative selection peptide, OVA (mean log2 fold change, OVA/FARL conditions), or (B) a positive selection peptide, Q7 (mean log2 fold change, Q7/FARL conditions). (C) PiSCES signature when response to OVA is directly normalized to the response to Q7 (mean log2 fold change, OVA/Q7 conditions). Dotted lines in (A) to (C) indicate trend of nonsignificant protein pairs that appear as hits in any of the three experimental comparisons performed. (D) Comparison of mean log2 fold changes in abundance of protein pair hits induced by OVA versus Q7 in preselection OT1.β2m0.RAG20 thymocytes. Data points are displayed for hits that were statistically significant in any of the OVA versus FARL, Q7 versus FARL, or OVA versus Q7 comparisons. A separate trajectory of orange points ∣x-axis value∣ > ∣y-axis value∣ that would clearly indicate positive selection-specific protein complexes is not observed. (E) PCA of three independent experiments shows separation of 1-min OVA/FARL versus Q7/FARL PiSCES signatures.
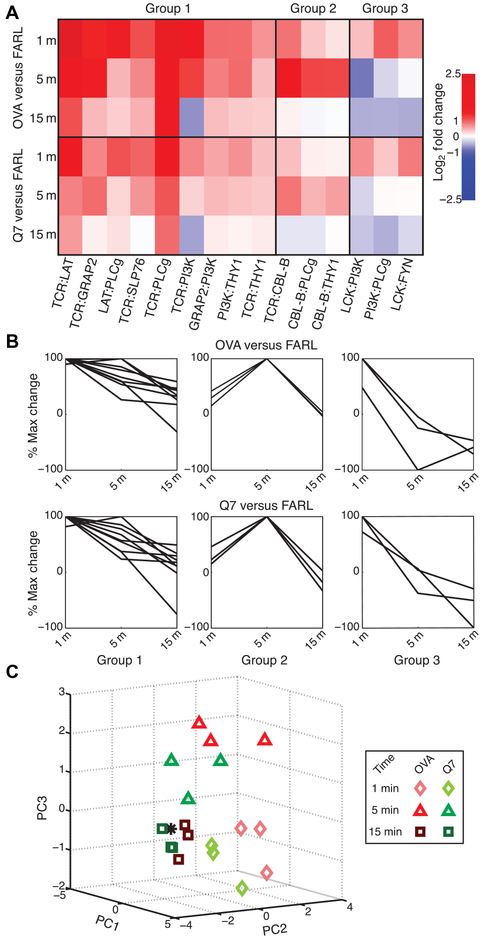
We next determined the extent to which positive and negative selection pMHC stimuli induced differences in the early kinetics of PiSCES data. For OT1.RAG20.β2m0 thymocytes, PiSCES signatures began by 1 min (Fig. 2), showing continued high activity and progression of the signature at 5 min (fig. S4, A to D), with waning toward early TCR-proximal signal dissolution by 15 min (fig. S4, E to H). At each time point, a quantitative trend was observed where the magnitude of average fold change for induced protein pairs was greater in response to OVA than to Q7 (Fig. 2D and fig. S4, D and H). Throughout the time course, PiSCES signatures appeared to evolve similarly in response to OVA or Q7 stimulation. For example, for OVA stimulation, maximal TCR:LAT appearing in shared complexes occurred at 1 min (Fig. 2A) and was reducing by 5 min (fig. S4A), by which time Cbl-b was maximally induced in shared complexes with TCR, PLCγ, and Thy1, and TCR clustering began to be observed [fig. S4A; the loop around TCR/CD3 indicates appearance of multiple copies of TCR/CD3 in shared complexes (40)]. By comparison, in response to Q7 stimulation, the same kinetic trends were observed, but changes were of decreased relative magnitude (Fig. 2B and fig. S4B). K-means cluster analysis of the top 15 protein pair hits revealed three kinetic behavior profiles in response to OVA stimulation; when applied to matching protein pairs from the Q7 stimulation condition, the same kinetic trends were preserved (Fig. 3, A and B). Finally, PCA showed that at each time point, the two stimulation conditions could be separated, with OVA response appearing the farthest from a zero-stimulation point (Fig. 3C). These data showed that the kinetics of early PPI progression in response to positive or negative selection pMHC ligands were similar, with the main difference in PiSCES signature at each time point being one of magnitude.
Fig. 3. Kinetics of PiSCES data for positive and negative selection stimuli.
PiSCES data from preselection OT1.β2m0.RAG20 thymocytes stimulated for 1, 5, or 15 min with null peptide FARL, positive selection peptide Q7, or negative selection peptide OVA. (A and B) K-means clustering was performed using percent maximum log2 fold changes to define three kinetic patterns observed among the top 15 hits in response to OVA stimulation, categorized in groups 1 to 3. (A) A kinetic heat map of the log2 fold changes is shown for these hits defined by the OVA stimulation condition. The matching data points in response to Q7 stimulation were observed to display similar kinetic behavior, but lower intensity fold changes than those induced by OVA stimulation. (B) K-means clustering data displayed as percent maximum log2 fold change show that the three kinetic behavior groups defined by response to OVA stimulation (top) also described the overall kinetic behavior of the same protein pairs in response to Q7 stimulation (bottom). (C) With experimental n = 3 per time point, data across 1-, 5-, and 15-min time points were used to generate a kinetic PCA matrix. Subjectively, it appears that data for response to OVA versus Q7 are distinguishable but relatively close to each other at each time point, with the time point of stimulation playing a major role in data placement in three-dimensional analysis space, and OVA data appearing farther than Q7 data from a zero-stimulation point (*).
Because primary thymocytes responded more weakly to stimuli than did Jurkat cells, we performed the same kinetic experiments and analyses in LC13ab.huCD8ab.JRT3 cells (figs. S5 and S6) and cells transduced to express OT1 and mouse CD8 transgenes, OT1ab. muCD8ab.JRT3 cells (figs. S7 and S8). These cells were hyperresponsive when compared with primary cells, but within each cell system the same general trends were observed: Quantitative differences predominated between agonist (negative selection) and antagonist (positive selection) stimuli, whereas substantial qualitative and early kinetic differences were not evident. Collectively, the greatest difference observed between positive and negative selection stimuli was the magnitude of change in a common set of early TCR-proximal protein complexes (signal strength), rather than formation of unique sets of protein complexes (signal composition) or different speeds of pathway progression (signal kinetic).
PiSCES network activity predicts and confirms that the quantity of antigenic engagement is sufficient to specify thymic selection outcome
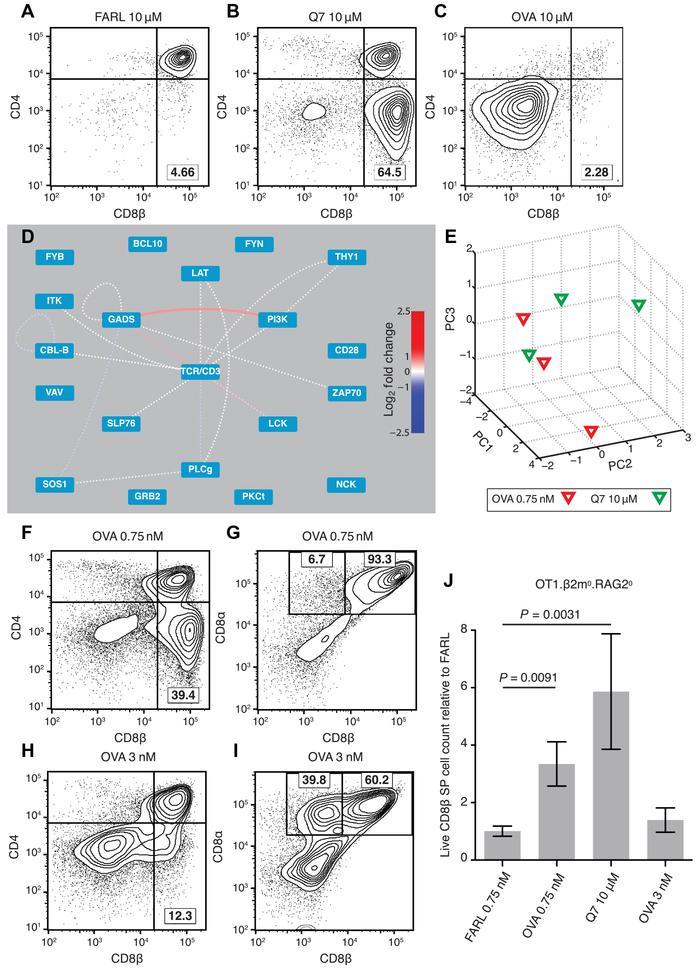
These data led to the prediction that a classic negative selection peptide should induce positive selection if the dose of its signal in responding thymocytes could be placed in the range of positive selection. To test this prediction, we used a published FTOC system that had been previously used for this purpose and over specific titration ranges had not produced evidence to support the hypothesis (27, 35). OT1. RAG20.β2m0 fetal thymi were supplied exogenous β2m to restore MHC class I antigen presentation in conjunction with OT1-nonspecific FARL peptide for no selection (Fig. 4A), Q7 peptide for positive selection (Fig. 4B), or OVA peptide for negative selection (Fig. 4C). To directly test the quantitative hypothesis predicted by PiSCES analysis, we identified a dilute concentration of OVA (0.75 nM) that, after 5 min of stimulation, induced an approximately indistinguishable PiSCES signature from that of full-dose Q7 (10 μM). This rough equivalence was determined by dividing the response to dilute OVA by the response to full-dose Q7 and observing that the two signals virtually canceled out (Fig. 4D) and could not be distinguished by PCA (Fig. 4E). We found that using this concentration of OVA in FTOC induced positive selection of conventional CD8αβ single-positive (SP) T cells (Fig. 4, F and G). Increasing that dose by fourfold was sufficient to induce substantial generation of unconventional CD8α+ CD8β− SP T cells (Fig. 4, H and I), correlating with preferential CD8αα homodimer expression (fig. S9 and Supplementary Results and Discussion), an observation consistent with the idea that CD8αα SP T cells are generated by signals stronger than those that generate conventional CD8αβ SP T cells (24, 25). In FTOCs that favored conventional positive selection, enhanced CD8αβ SP T cell viability was observed (Fig. 4J). Furthermore, when FTOC cells from 20 μM Q7 or 0.75 nM OVA conditions were subsequently stimulated with OVA-bearing APCs, T cell proliferation and development of cytotoxic T lymphocyte activity were observed (fig. S10). Overall, these data support the hypothesis that quantitative network activity through the TCR signalosome provides an early instruction to specify positive versus negative selection in developing thymocytes.
Fig. 4. PiSCES signature makes quantitative signaling prediction, which passes functional test in FTOC.
(A to C) Fetal thymi of genotype OT1.RAG20.β2m0 were cultured for 7 days in the presence of exogenous β2m and specific peptides at the stated concentrations. (A) FARL peptide (10 μM) loads with high affinity into H-2Kb but has no functional affinity for the OT1 TCR and represents the background no-selection condition. (B) Q7 (10 μM) induced positive selection of a substantial portion of CD8 SP cells. (C) OVA (10 μM) induced deletion and loss of CD8+ cells. (D and E) Lowering the dose of OVA peptide to 0.75 nM made its PiSCES signature almost indistinguishable from that of 10 μM Q7. (D) When PiSCES data resulting from 5-min stimulation of preselection OT1.β2m0.RAG20 thymocytes with 0.75 nM OVA were normalized to data from stimulation with 10 μM Q7, the signatures virtually cancelled out, eliminating almost all hits (mean log2 fold change, OVA/Q7 comparison; dotted lines indicate trend of nonsignificant protein pairs). (E) PCA of PiSCES data in four experiments described in (D), where separation of the two stimulatory conditions was no longer observable. (F) OVA (0.75 nM) in FTOC induced positive selection of CD8 SP cells (lower right quadrant). (G) Gating the data in (F) on CD4(−) cells, the positively selected CD8 SP cells were largely conventional σβ T cells marked by CD8αβ expression. (H) Raising OVA concentration to 3 nM in FTOC still induced positive selection of CD8 SP cells. (I) Gating the data in (H) on CD4(−) cells, many of the positively selected cells were seen to represent unconventional CD8α+ CD8β− cells. (J) Live CD8β+ SP T cell counts from two independent experiments [including the experiment depicted in (A) to (C) and (F) to (I)], with each FTOC normalized to the mean of its corresponding FARL 0.75 nM condition and reported as fold change. Number of thymus lobes per condition: 9 (FARL 0.75 nM), 9 (OVA 0.75 nM), 4 (Q7 10 μM), and 6 (OVA 3 nM). Unpaired two-tailed Student’s t test, P < 0.01.
Some positive selection occurs in CD3δ0 mice
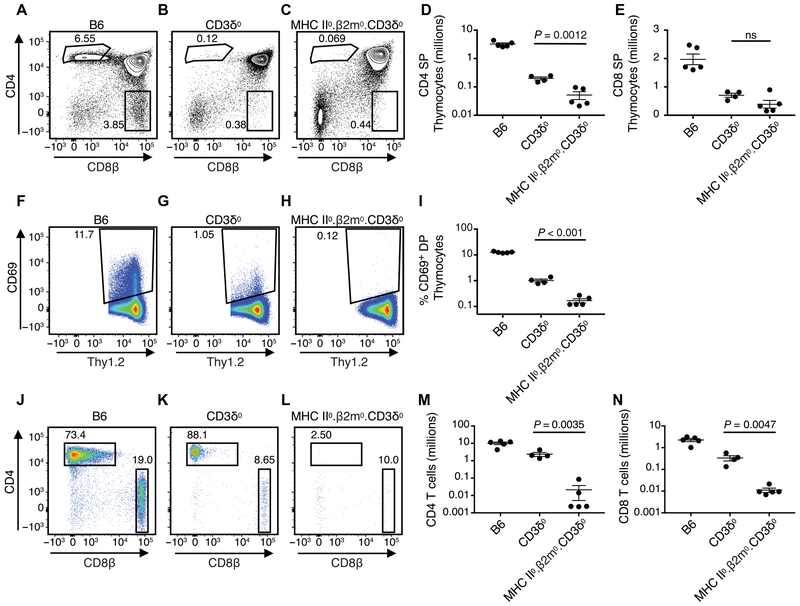
Because previous work had suggested that positive selection might involve a qualitatively unique signal through CD3δ (31-34), we reexamined the block in positive selection observed in CD3δ0 mice to determine whether CD3δ’s contribution might instead be quantitative, amplifying TCR/CD3 signals toward a selection threshold. As previously reported, relative to wild-type C57BL/6 (B6) mice, a severe block in positive selection and generation of CD4 or CD8 SP αβ T cells was observed upon analysis by flow cytometry of thymocytes from CD3δ0 mice (Fig. 5, A and B). To determine whether some residual positive selection was occurring in CD3δ0 mice, we crossed them with MHC II0.β2m0 to generate MHC II0.β2m0.CD3δ0 mice. A barely perceptible difference was observed in the percentage of CD4 SP thymocytes between the two mutant genotypes (Fig. 5, B and C and fig. S11, A and B). CD3δ0 mice that were MHC+ had a higher average number of live SP thymocytes than MHC II0.β2m0.CD3δ0 mice, a difference that only reached significance for CD4 SP but not for CD8 SP subsets (Fig. 5, D and E). Consistent with the possibility that a few SP cells were being generated by positive selection, surface CD69 expression was observed on a greater percentage of thymocytes that were CD3δ0 than MHC II0.β2m0.CD3δ0 at stages DP (Fig. 5, F to I) and CD4 or CD8 SP (fig. S11, C to J). To isolate the ability of the thymus to generate SP cells, we performed FTOC, where a greater percentage of CD4 SP cells was generated in CD3δ0 than MHC II0.β2m0. CD3δ0 thymi (fig. S11, K to O). Analysis of CD4 and CD8 splenic T cells confirmed that CD3δ0 mice had low cell numbers relative to B6, but such cells were absent in MHC II0.β2m0.CD3δ0 mice (Fig. 5, J to N). Compared with B6, CD3δ0 splenic T cells expressed less CD5, indicative of having undergone weaker selection signals (48), and most CD3δ0 T cells were CD24-low and PNA-low staining, consistent with the progressive differentiation expected to occur after selection (fig. S12). Because signaling occurred in CD3δ0 thymocytes (resulting in CD69 induction) in an MHC-dependent manner to generate low-count SP cells in the thymus and periphery, we conclude that a low level of positive selection still occurred in these mice.
Fig. 5. MHC-dependent signaling generates residual αβ T cells in CD3δ0 mice.
Live Thy1.2+ thymocytes were analyzed for CD4 and CD8 surface expression by flow cytometry from (A) wild-type B6, (B) CD3δ0, and (C) MHC II0.β2m0.CD3δ0 mice, and counts were obtained for (D) CD4 SP cells and (E) CD8 SP cells. The percentage of CD4+CD8+ DP thymocytes that were Thy1.2+ and CD69+ was obtained from the same three genotypes (F to I). Live Thy1.2+ splenocytes were analyzed for CD4 and CD8 surface expression by flow cytometry from the same three genotypes (J to L), and live counts were calculated for (M) CD4 T cells and (N) CD8 T cells. Mouse n was ≥ 4 per genotype. Statistical significance was determined by unpaired two-tailed t test, P < 0.05.
To test the hypothesis that a specific TCR lacking CD3δ could mediate some amount of positive selection in vivo, we generated OT1.RAG20.CD3δ0 and OT1. RAG20.β2m0.CD3δ0 mice. Compared with OT1.RAG20 mice, OT1.RAG20.CD3δ0 mice showed a reduction in peripheral blood T cells; however, their low T cell number was dependent on MHC class I–mediated survival, because peripheral blood T cells were absent in OT1. RAG20.β2m0.CD3δ0 mice (fig. S13). We conclude that a low amount of MHC- dependent positive selection still appeared to be active when single-TCR–expressing cells lacked CD3δ.
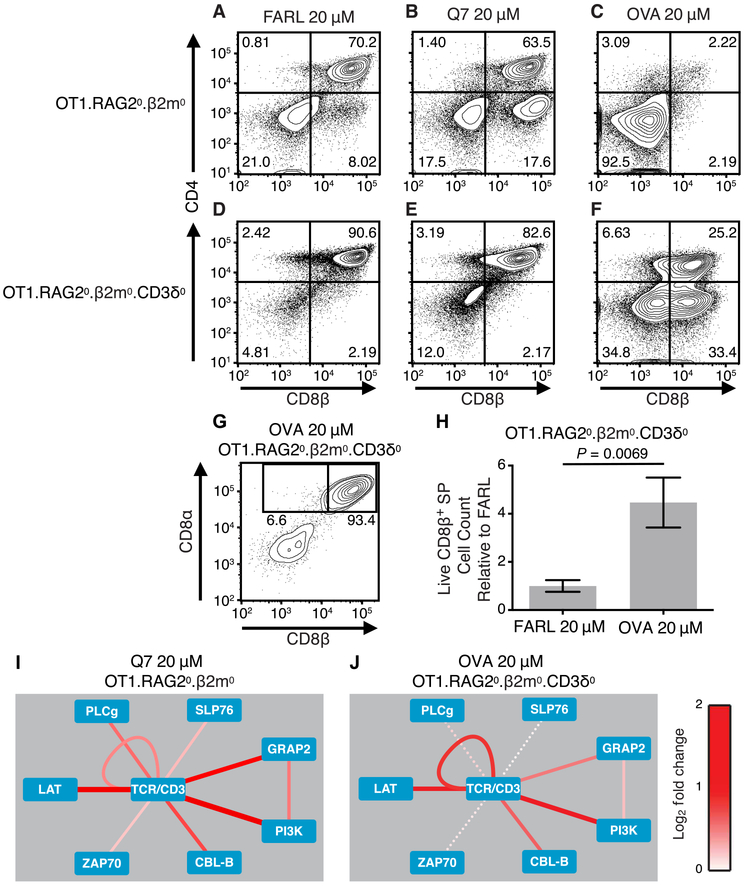
To directly test the hypothesis that a specific TCR lacking CD3δ could transduce a positive selection signal under conditions of controlled, specific peptide presentation, we compared OT1. RAG20.β2m0 and OT1.RAG20.β2m0. CD3δ0 mice in FTOC. As expected for OT1.RAG20.β2m0 thymocytes, FARL peptide induced no selection (Fig. 6A), Q7 peptide induced positive selection (Fig. 6B), and OVA peptide induced negative selection (Fig. 6C). In contrast, OT1.RAG20.β2m0.CD3δ0 thymocytes failed selection with Q7 peptide (Fig. 6, D and E) but underwent positive selection in response to OVA peptide (Fig. 6F), producing conventional CD8αβ SP T cells (Fig. 6, G and H). Having observed these selection outcomes, we examined the ability of preselection OT1 thymocytes to induce TCR signal transduction with or without CD3δ in response to positive selection pMHC ligands. To maximize sensitivity to small signaling changes (by minimizing multiple-hypothesis statistical correction) in the severely low TCR–expressing OT1.RAG20.β2m0.CD3δ0 thymocytes, we limited these PiSCES experiments to a small collection of strongly inducible protein pairs. We found that 5-min stimulation of preselection OT1.RAG20.β2m0 thymocytes with Q7 peptide presented by C1R.Kb APCs induced multiple copies of TCR/CD3 to join shared complexes, an expected consequence of receptor cross-linking (visualized as a loop around TCR/CD3; Fig. 6I). TCR also appeared in shared complexes with LAT, GADS, PI3K, Cbl-b, and others. By comparison, stimulation of preselection OT1.RAG20.β2m0.CD3δ0 thymocytes with OVA peptide also induced those protein complexes (Fig. 6J); however, the network signature of CD3δ0 cells responding to OVA was mostly weaker, with fewer statistically significant hits (statistically nonsignificant trends shown with dashed lines; Fig. 6J), with most heteroprotein associations being of weaker intensity than those seen for OT1.RAG20.β2m0 thymocytes responding to Q7 peptide. We conclude that the OT1 TCR was capable of transducing a positive selection signal with or without CD3δ, although when there was severely low expression of both TCR (CD3δ0) and MHC (β2m0) in FTOC, positive selection required a strong pMHC ligand.
Fig. 6. Positive selection signaling in the context of CD3δ0.
(A to G) Fetal thymi were cultured for 7 days in the presence of exogenous β2m (5 μg/ml) and specific peptides. For OT1.RAG20.β2m0 thymocytes (A to C), (A) 20 μM FARL peptide loads with high affinity into H-2Kb but has no functional affinity for the OT1 TCR and represents the background no-selection condition. (B) Q7 peptide (20 μM) induced positive selection of a substantial portion of CD8 SP cells. (C) OVA peptide (20 μM) induced deletion and loss of CD8+ cells. For OT1.β2m0.RAG20.CD3δ0 thymocytes (D to G), (D) 20 μM FARL peptide was inert and (E) Q7 peptide was inert, whereas (F) 20 μM OVA induced positive selection of a substantial portion of CD8 SP cells (lower right quadrant). (G) The positively selected CD8 SP cells from the OT1.β2m0.RAG20.CD3δ0 genotype were largely conventional αβ T cells as marked by CD8αβ expression. (H) Live CD8β+ SP T cell counts from two independent experiments [including the experiment depicted in (A) to (G)], with each FTOC normalized to the mean of its corresponding FARL 20 μM condition and reported as fold change. Number of thymus lobes per condition: 7 (FARL 20 μM) and 7 (OVA 20 μM). Unpaired two-tailed Student’s t test, P = 0.0069. (I and J) PiSCES analysis of signaling proteins that joined shared complexes in response to positive selection pMHC antigens. Dotted lines indicate trend of nonsignificant protein pairs. (I) Induction of protein pairs from OT1. RAG20.β2m0 thymocytes when assessing 5-min stimulation with 20 μM Q7 peptide over negative-control FARL peptide. (J) Induction of protein pairs from OT1.β2m0.RAG20.CD3δ0 thymocytes when assessing 5-min stimulation with 20 μM OVA peptide over negative-control FARL peptide.
The αβ TCR repertoire in peripheral CD3δ0 T cells is diverse
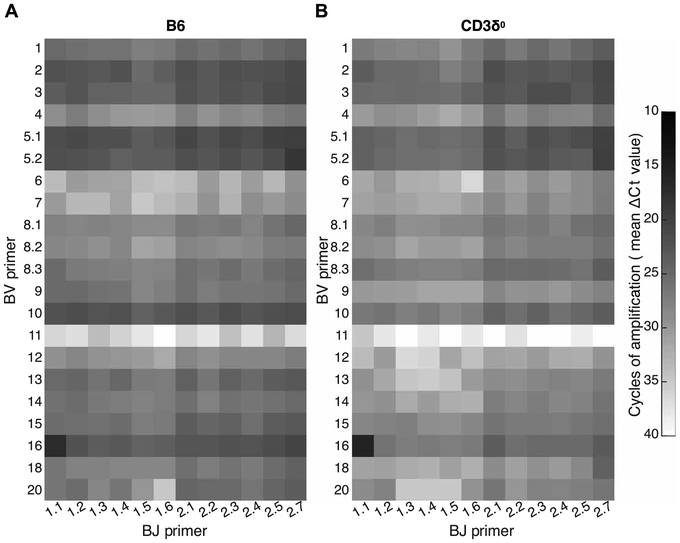
Perhaps CD3δ supplied a qualitative positive selection–specific signal that, as a general rule, was required by the vast majority of T cell clones; in that case, in the absence of CD3δ, residual positive selection could be explained if it were due to a few clones with rare, unusual CD3δ-independent TCRs. If correct, then peripheral, polyclonal CD3δ0 T cells should display a relatively limited TCR repertoire. Alternatively, if the main role of CD3δ was to quantitatively enhance positive selection signals, then peripheral CD3δ0 T cells might display a diverse TCR repertoire even though there were few T cells, because selection signals from all clones would be inhibited equally. To distinguish between these two disparate predictions, we used a mouse TCRβ repertoire diversity matrix system in which real-time polymerase chain reactions (PCRs) were performed for 252 fully nested BV-BJ combinations of 21 BV primers and 12 BJ primers (49). This system allowed repertoire diversity of T cell populations to be assessed at a substantially deeper level than standard V gene spectratyping, by comprehensively assessing all possible V-J combinations. We found that specific TCRβ transcripts tended to be decreased in quantity from CD3δ0 relative to B6, as expected due to their decreased peripheral T cell number in the mutant, but both CD3δ0 and B6 were capable of expressing highly diverse repertoires (Fig. 7, averages; fig. S14, individual mice). The diversities of the 252 BV:BJ combinations were estimated by scaled Shannon entropy under conditions where B6 input RNA was decreased relative to CD3δ0 RNA so that samples would provide comparable T cell representation despite the lower T cell numbers in mutant spleens. Scaled Shannon entropy is a value whose range is 0 to 1, where 0 represents minimum diversity exhibited by a monoclonal T cell population and 1 represents maximal repertoire diversity when all BV:BJ combinations are expressed equally (49). Entropy values for two B6 mice were 0.74 and 0.64, whereas those of two CD3δ0 mice were in a similar range, 0.67 and 0.68, indicating that mice with either genotype were capable of generating a diverse TCRβ repertoire in the periphery. These data did not support the hypothesis that in CD3δ0 mice, only a few T cell clones underwent positive selection.
Fig. 7. Diverse TCR repertoire in B6 and CD3δ0 mice.
A qPCR matrix-based method was used to assess relative representation of all possible TCRβ V-J combinations expressed as transcripts in splenocytes from two (A) B6 or (B) CD3δ0 mice. To approximate the maximum diversity generation potential per mouse for each genotype, the mean cycle threshold (Ct) value from the two mice with highest Shannon entropy for each genotype is displayed (top 2 of four B6 mice and top 2 of six CD3δ0 mice tested, when 15-ng total splenic RNA was used as input). As expected, because of higher T cell representation among splenocytes, B6 transcripts for TCR were more abundant than was found in CD3δ0. Greater quantities of specific transcripts appear darker due to fewer PCR amplification cycles required to reach Ct.
Because the TCRα subunit and CD3δ are part of the same positive selection signaling axis (33, 34), we also assessed peripheral TCRα diversity with a spectratype survey of 16 Vα genes using previously published primers and methods (50, 51) and observed similar expression patterns between CD3δ0 and B6 genotypes (fig. S15). We conclude that positive selection in the absence of CD3δ occurs for a population of T cells expressing substantial repertoire diversity with usage of numerous TCRα and TCRβ genes. These data do not provide evidence that CD3δ supplies a qualitatively distinctive signal that would be required as a general rule for positive selection. Instead, it appears that diverse TCRs can signal positive selection without CD3δ, but clones expressing these TCRs have a poor success rate, implying that the role of CD3δ is to quantitatively enhance positive selection to maximize the generation of peripheral T cells.
CD3δ0 αβ T cells have immune function
To test the function of the diverse MHC-dependent αβ T cell pool that was positively selected in CD3δ0 mice, they were challenged with two infections known to require αβ T cells for survival. These were (i) Pneumocystis fungal pulmonary infection causing pneumonia (PCP), an infection that is fatal in CD4-depleted hosts such as patients with AIDS (52), or (ii) Theiler’s murine encephalomyelitis virus (TMEV), an infection whose clearance in mice normally requires CD4 and CD8 T cells to prevent acute encephalitis and paralysis (53).
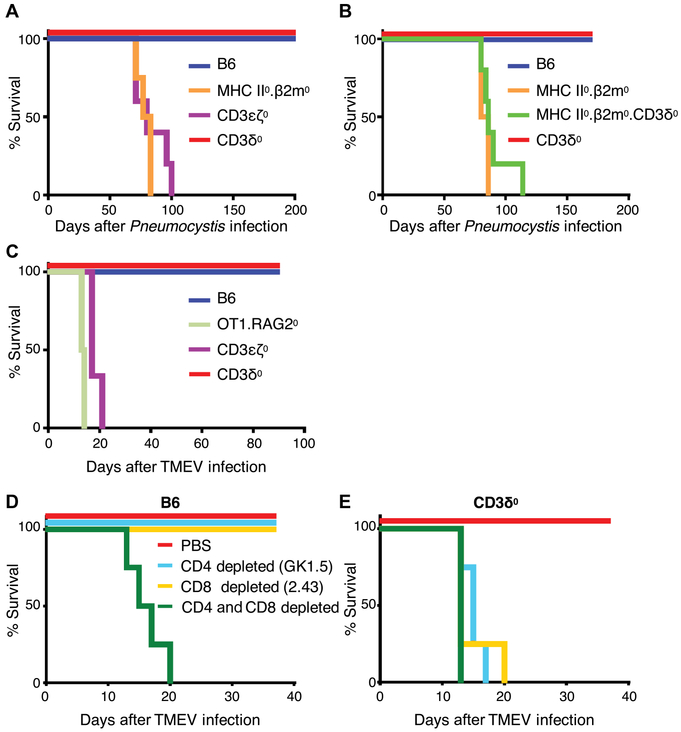
Mice from the following genotypes were infected with Pneumocystis murina: B6, MHC II0.β2m0 (MHC deficient), CD3ε0ζ0 [T cell and TCR/CD3 deficient, lacking expression of all four CD3 genes (54)], and CD3δ0. Both B6 and CD3δ0 genotypes were resistant to PCP with 100% survival, whereas MHC-deficient and other T cell-deficient mice were susceptible and succumbed (Fig. 8A), with accompanying symptoms of pneumonia including weight loss (fig. S16A); increased lung weight, indicative of fluid accumulation (fig. S16B); and decreased blood oxygen saturation, indicating loss of respiratory function (fig. S16C). To test the hypothesis that CD4 T cells were required for PCP resistance in CD3δ0 mice, we compared infection of CD3δ0 mice that were depleted of CD4 T cells through weekly injections of monoclonal antibody (mAb) GK1.5 anti-CD4, with CD3δ0 mice that had received control phosphate-buffered saline (PBS) injections. We found that CD4-depleted CD3δ0 mice were susceptible and succumbed to PCP, similar to other susceptible genotypes, including T cell-deficient CD3ε0ζ0 mice, and OT1.RAG20 mice expressing a single PCP-irrelevant transgenic TCR, whereas PBS-injected CD3δ0 mice and B6 mice were resistant with 100% survival (fig. S16, D and E). Antigen presentation was required for PCP resistance, because MHC II0.β2m0 and MHC II0.β2m0.CD3δ0 mice were susceptible and succumbed (Fig. 8B) with symptoms of pneumonia (fig. S16, F to H). We conclude that the low numbers of positively selected CD4 T cells in CD3δ0 mice were able to express MHC-dependent immune activity.
Fig. 8. T cells in CD3δ0 mice provide immune activity against PCP and TMEV.
(A) To assess the extent of T cell immune activity in a CD3δ0 setting, we infected mice from the listed genotypes with P. murina (mouse n ≥ 4 per genotype). Kaplan-Meier curves display survival defined by mice being euthanized upon loss of 20% weight, where CD3δ0 was statistically different from susceptible genotypes (P ≤ 0.004) by log-rank Mantel-Cox tests. (B) To test the role of MHC in mediating protection from PCP in CD3δ0 setting, mice from the listed genotypes were infected with PCP (mouse n ≥ 4 per genotype). Kaplan-Meier curves display survival defined by mice being euthanized upon loss of 20% weight, where CD3δ0 and MHC II0.β2m0.CD3δ0 were statistically different (P = 0.0018) by log-rank Mantel-Cox test. (C) Mice from the listed genotypes were infected with TMEV [mouse n ≥ 4 for all genotypes, except for CD3ε0ζ0 (n = 3)]. Kaplan-Meier curves display survival defined by mice being euthanized upon immobilization due to functional deficit or loss of 20% weight, where CD3δ0 was statistically different from susceptible genotypes (P ≤ 0.01) by log-rank Mantel-Cox tests. (D and E) To assess CD4 and CD8 T cell immune activity, B6 and CD3δ0 mice were either depleted of CD4 cells with GK1.5 anti-CD4 mAb injections, depleted of CD8 cells with 2.43 anti-CD8 mAb injections, depleted of both CD4 and CD8 cells, or PBS-control injected as indicated, and were infected with TMEV [mouse n = 4 for all groups, except for CD3δ0 + PBS (n = 3)]. Kaplan-Meier curves display survival defined by mice being euthanized upon immobilization due to functional deficit or loss of 20% weight, where CD3δ0 + PBS was statistically different from any of the three subset-depleted conditions (P < 0.02) by log-rank Mantel-Cox tests.
To assess CD8 T cell function, we infected mice from the following genotypes with TMEV: B6, CD3δ0, CD3ε0ζ0 (T cell deficient), and OT1.RAG20 (containing a single TMEV-irrelevant TCR transgenic CD8 T cell population). Both B6 and CD3δ0 genotypes were resistant to TMEV infection with 100% survival, whereas T cell–deficient and OT1.RAG20 mice were susceptible (Fig. 8C), suffering functional deficit that was measured by loss of RotaRod performance (fig. S17A). To determine which T cell subsets contributed to protection from TMEV of B6 and CD3δ0 mice, we compared infection of mice that were (i) depleted of CD4 T cells through weekly injections of mAb GK1.5 anti-CD4, (ii) depleted of CD8 T cells through weekly injections of mAb 2.43 anti-CD8, (iii) both CD4 and CD8 T cell depleted, or (iv) nondepleted, control PBS-injected. Whereas B6 mice only succumbed upon depletion of both CD4 and CD8 T cells, as expected (53), CD3δ0 mice succumbed upon depletion of either subset (Fig. 8, D and E, and fig. S17, B to E). These data suggest that CD3δ0 mice are immune-compromised relative to wild-type B6 mice, as expected, but the low-count peripheral CD3δ0 CD4 and CD8 T cells participate in protective immune activity against infection.
DISCUSSION
Life versus death represents a qualitative difference, which creates the expectation that a qualitative difference in signal transduction must specify these outcomes when they are programmed cellular responses. We have presented the possibility of qualitative, quantitative, or kinetic differences in early TCR-proximal network PPI signaling activity in a framework that considers them to be opposing models to be distinguished from each other. Historically, that original framework was useful and warranted, because the signals that decide thymic selection have been proposed to originate from TCR-proximal signaling elements with qualitative properties, including special subunits such as CD3δ (31, 32), motifs such as TCR αCPM (33, 34), patterns of ITAM phosphorylation (10, 55-58), and other possible candidates that might specify selection outcome (59, 60). The present data suggest that a major role in selection for players such as these may be to affect the initial quantity of signal transmitted through the proximal TCR signalosome, rather than to create unique combinations of the most TCR-proximal protein complex intermediates during early selection signals. The data are compatible with a quantitative signaling model that has been proposed (61, 62), where a single TCR signaling signature may be active at subthreshold level in the basal state, and different signals are transduced as (i) CD3 subunit multiplicity and activity mostly provide quantitative information of ligand binding strength, (ii) the balance of signaling activity is perturbed away from the basal state, and (iii) a series of quantitative thresholds demarcates separate functional instructions.
The present data imply that early signal translation between two kinetic parameters (initially TCR engagement, later Ras-Raf-ERK activity) might be mediated by this quantitative network parameter that specifies positive versus negative selection based on the accumulation or frequency of a single-series (and single-kinetic) network PPI signature that propagates through the early proximal TCR signalosome upon productive engagement by a pMHC ligand. This hypothesis was generated by PiSCES analysis, which identifies signatures of targeted subnetworks, but does not comprehensively assess all proteins, lipids, and other molecules that contribute to the signal (63). Therefore, it is possible that key qualitatively distinct molecular species that instruct selection early are expressing activity that PiSCES did not observe, if such key species do not affect the association of proteins in the subnetwork being measured. Furthermore, downstream and later signaling activity may develop qualitative and kinetic characteristics that are critical for instructing ultimate selection outcome. Therefore, we reasoned that the only way to determine whether selection outcome was dependent on the early TCR-proximal, quantitative signaling principle predicted by PiSCES was to test that principle functionally. Upon doing so, we identified a specific, low concentration of OVA, a classic negative selection peptide in the OT1 system, which induced positive selection of conventional T cells. This principle was corroborated a second way; reducing TCR signaling strength in the context of CD3δ0 and β2m0 allowed high-dose OVA to induce positive selection of conventional OT1 T cells. These data support the conclusion that a quantitative network signaling mechanism provides an early instruction that culminates in determination of thymic selection outcome.
Previously, there were already data in which OVA peptide did not induce positive selection of conventional OT1 T cells across 10-fold dilutions in FTOC (27, 35). This implied that signaling quantity might be insufficient to explain the difference between positive and negative selection. In the present work, the specific dilution of OVA peptide that induced positive selection of conventional αβ T cells in FTOC falls almost midway between the two closest log dilutions that were previously published. Thus, the present data are compatible with the previous conclusion that pMHC:TCR affinity is the master driver of selection outcome across a wide range of concentrations. However, the new lesson learned is that a key early proximal network PPI signaling parameter that is controlled by pMHC:TCR affinity is quantitative in nature.
CD3δ plays a critical role in positive selection, and in the absence of this subunit, a poorly populated peripheral αβ T cell pool is generated that results in immune compromise in mice. That CD3δ might provide a qualitative subunit-specific distinctive signal required for positive selection was an attractive possibility for many reasons, including the fact that if one wished to control positive selection and central tolerance pharmacologically, pathways could be more easily targeted with specificity if qualitatively distinctive intermediates were involved. Our data began to diverge from this possibility when it was found that the few αβ T cells known to be present in CD3δ0 mice (31) did not represent a secondary “leakiness” effect in the face of a strong developmental block, but rather these cells were products of residual signaling- and MHC-dependent positive selection (Figs. 5 and 6 and fig. S11). CD3δ0 and wild-type B6 mice both exhibited high diversity in rearranged TCRβ and TCRα genes in the peripheral T cell pool (Fig. 7 and figs. S14 and S15), and those T cells participated actively in CD4-dependent and CD8-dependent T cell responses to infection (Fig. 8 and figs. S16 and S17). These data are most easily explained if, as a general rule, CD3δ does not provide a subunit-specific qualitatively distinctive signal required for positive selection.
The model most consistent with the data is that CD3δ quantitatively boosts the positive selection signal to increase the number of T cells generated from a diverse repertoire of candidate αβ TCR–bearing clones (64). This function could be partially due to the fact that the presence of CD3δ improves TCR/CD3 folding and increases surface expression, which may contribute to quantitatively increasing signals. This possibility is consistent with the previously reported observation that positive selection was rescued in CD3δ0 mice by a CD3δ-mutant transgene that restored surface TCR/CD3 expression levels but carried a nonfunctional ITAM (32). Under a qualitative signaling model, the non-ITAM CD3δ motif that might provide the specificity for positive selection remained unidentified, but in a quantitative model, provision by CD3δ of optimal native TCR/CD3 assembly and expression could play a major role. Furthermore, models in which CD3δ acts in concert with αCPM and CD4 or CD8 coreceptors to mediate positive selection (65) are compatible with the present proposal that a main outcome of these interactions is to boost signal intensity. We conclude that the main functional signal for positive selection contributed by CD3δ is not qualitatively distinctive, but rather involves quantitative amplification to maximize the number of αβ T cells that populate the peripheral immune system.
MATERIALS AND METHODS
Study design
The aim of this study was to determine how a network of interacting proteins proximal to αβ TCR transmits dichotomous signals for either positive or negative selection during thymocyte development. Either human thymic leukemia cells or primary mouse thymocytes were stimulated with strong or weak pMHC ligands, after which network protein interaction signatures were assessed by a multiplex PiSCES analysis. Results generated a prediction that was tested functionally using the same primary mouse thymocytes in FTOC, with validation using an additional primary cell system based on ex vivo and in vivo experimentation and analysis of CD3δ0 mice. The size of the experimental groups is specified in figure captions, this section, and the Supplementary Materials.
Cell lines
JRT3-T3.5 (ATCC TIB-153) cells were purchased from the American Type Culture Collection (ATCC). The LC13 system, including LC13ab. huCD8ab.JRT3 cells and C1R.HLA-B*08:01 (C1R.B0801) APCs, were described previously (41-43, 66, 67). We generated OT1ab. muCD8ab.JRT3 cells using retroviral transduction of previously published OT1 sequences (68) that were provided by D. Vignali (then at St. Jude Children’s Research Hospital, Memphis, TN) and mouse CD8 genes isolated from cDNA. All cells were grown in RPMI media (Life Technologies) with 10% Cosmic Calf Serum (HyClone), 2 mM L-glutamine (Life Technologies), and penicillin (100 U/ml)/streptomycin (100 μg/ml) (Life Technologies), and maintained in a 37°C incubator with 5% CO2.
Mice
All mice were B6 background. OT1 mice were originally provided by L.R.P. (Mayo Clinic). Mice were screened by flow cytometry of peripheral blood and also by standard genomic PCR for the OT1 transgene using primers and protocols described on the Jackson Laboratory webpage for the C57BL/6-Tg(TcraTcrb)1100Mjb/J mouse strain. CD3δ0 mice originated from the Fox Chase Cancer Center. CD3ε0ζ0 mice, which lack expression of all four CD3 genes (54), were originally provided by D. Vignali (then at St. Jude Children’s Research Hospital, Memphis, TN) with permission from C. Terhorst (Beth Israel Deaconess Medical Center, Harvard Medical School, Boston, MA). β2m0 mice were obtained from the Jackson Laboratory, as were MHC II0 mice in which all conventional MHC class II genes are deleted (69). All genetically engineered mouse strains were bred and maintained at the Mayo Clinic and University of Missouri-Columbia, and all animals were housed in specific pathogen-free facilities. In general, mice were used at 6 to 12 weeks of age and, within experiments, were age- and sex-matched between experimental groups. Individual mice within 15 days of age were accepted as age-matched. Mice used in infection experiments were age- and sex-matched between experimental groups, and began experiments at 8 to 16 weeks of age, except for OT1.RAG20 mice, which served as infection-susceptible controls without age and sex matching. All mouse care and experimentation adhered to institutional guidelines and the NIH Guide for the Care and Use of Laboratory Animals.
Abs and staining reagents
For cellular flow cytometry, mAbs included the following: Thy1.2/CD90 (30-H12, BioLegend), TCRβ (H57, eBioscience), Vα2 (B20.1, BD), CD3ε (145-2C11, BioLegend), CD8α (53.6.7, BioLegend), CD8β (53-5.8, BioLegend), CD4 (GK1.5, BD Biosciences), CD69 (H1.2F3, BioLegend), CD24 (M1/69, BioLegend), and CD5 (53-7.3, BioLegend). Other binding reagents included the following: peanut agglutinin (PNA)-FITC (Sigma-Aldrich), streptavidin-PE and streptavidin-APC-Cy7 (BioLegend), H-2Kb-SIINFEKL (OVA) tetramer, and thymic leukemia antigen (TLA) tetramers (NIH tetramer core facility). For multiplex IP and PiSCES analysis, antimouse Abs are listed in table S1, whereas a similar table for antihuman Abs used in conjunction with Jurkat and JRT3 cell lines was previously published (40).
Organ processing and flow cytometry
Whole thymocyte or splenocyte single-cell suspensions were prepared for analysis, as previously described (70). Cells were analyzed by flow cytometry using either BD Biosciences LSR II or Accuri C6 instruments. The gating strategy used for this analysis is shown in fig. S18.
Cell stimulation and lysis
To prepare APCs for T cell stimulation, we incubated transgenic C1R cells with specific peptides for 2 hours in serum-free RPMI at 37°C. For the LC13 system, C1R.B0801 cells were used as APCs, with agonist peptide RAY (FLRGRAYGL), antagonist peptide RFY (FLRGRFYGL), or no peptide. For OT1 systems, C1R.Kb cells were used as APCs, with peptides FARL (SSIEFARL), Q7 (SIINFEQL), or OVA (SIINFEKL). Peptide-loaded APCs were washed once in PBS and then fixed by resuspension in 0.05% glutaraldehyde in PBS, as previously described (34). After 30 s, fixation was halted by the addition of 200 mM glycine/PBS, and fixed cells were washed three times in PBS. Responding T cells included LC13ab.huCD8ab.JRT3, OT1ab.muCD8ab.JRT3, or primary preselection DP thymocytes of OT1.β2m0.RAG20 or OT1.β2m0. RAG20.CD3δ0 genotypes. JRT3-based cell lines were used after having been placed in fresh media at 0.35 × 106/ml in tissue culture the previous evening. The day of stimulation, 15 × 106 T cells were resuspended in 200 μl of ice-cold PBS, mixed with 20 × 106 APCs in an equal volume of ice-cold PBS, and centrifuged for 5 min at 300g at 4°C to facilitate cell conjugation. Supernatant was discarded, and stimulation commenced as the T cell/APC pellet was placed in a 37°C water bath for the indicated amount of time before being flash-frozen in liquid nitrogen. Control unstimulated T cells were mixed with non-loaded (LC13 system) or null peptide-loaded (OT1 system) fixed APCs in parallel. Frozen cell pellets were either lysed immediately or stored briefly at −80°C before being lysed in lysis buffer [150 mM NaCl, 50 mM tris (pH 7.4), 1% digitonin (High Purity, Millipore), 1× Halt protease/phosphatase inhibitors (Pierce), 10 mM NaF, 2 mM sodium orthovanadate, and 10 mM iodoacetamide].
Multiplex capture and probe of protein complexes
In the TCR signalosome, complete analysis involved 20 signaling proteins that were targeted in a minimum 20 × 20 matrix of capture/ probe Abs (table S1). This generated a minimum of 400 protein co-association measurements, including 210 total unique protein co-associations: {[(20 capture × 20 probe) – 20 homotypic combinations]/2} + 20 homotypic combinations = 210. In some experiments where noted, only eight proteins were targeted, with commensurate reduction in the matrix. Some experiments included capture/probe combinations targeting additional proteins beyond the 20 described in table S1, but their data are not visualized because they were not assessed in all experiments. Procedures followed published protocols (40). Briefly, a master mix containing equal numbers of each Ab-coupled Luminex bead class was prepared and distributed into postnuclear lysate samples in duplicate. Protein complexes were immunoprecipitated from samples overnight, washed twice, and distributed into as many wells of a 96-well plate as there were probes. Probe Abs were added and incubated for 1 hour, with gentle agitation at 500 rpm in a cold room, followed by three more washes. Biotinylated probe Ab wells were then incubated 30 min with streptavidin-PE. After three final washes, microbeads were resuspended in 125 μl and fluorescence data were acquired on a Bio-Plex 200 instrument that was calibrated according to manufacturer recommendations and run on the “high RP target” setting. Data files were exported in both Microsoft Excel and XML formats for further processing.
Fetal thymic organ culture
Timed matings were performed, as previously described (71). Briefly, mouse cages were divided in half using an acrylic divider, with adequate food and water available on each side. One male and one female resided on each side of a divided cage for ~72 hours, after which the divider was removed in late afternoon to make mating possible overnight. FTOC was performed as previously described (35). On embryonic day (e)15, carbon dioxide was used to euthanize mothers, and fetal thymi were harvested. Each fetal thymic lobe was placed on sterile mixed cellulose ester gridded filter paper (Millipore) on top of a Gelfoam sterile sponge (Pfizer), in HyClone CCM1 serum-free media (GE Life Sciences) in one well of a 48- or 96-well plate. Some cultures were supplemented with exogenous human β2m (5 μg/ml, Sigma) and peptides FARL, Q7, or OVA at the indicated concentrations. Tissue culture occurred at 37°C for 7 days before harvest and assessment of thymic selection by flow cytometry.
TCR BVJ repertoire analysis
The diversity of TCRβ repertoires was evaluated by a previously described matrix that assesses all possible BV-BJ combinations (49). Briefly, TCRβ transcripts were reverse-transcribed from total RNA with a biotinylated BC region reverse primer and amplified with pools of BV-specific forward primers. The resulting amplicons were mixed with streptavidin-coated magnetic beads to enrich products that included the biotinylated BC region primer. The bead-enriched products were delivered to microtiter wells for amplification in real-time PCRs using 252 fully nested BV-BJ combinations of 21 BV primers and 12 BJ primers (see Supplementary Materials and Methods).
P. murina infection causing pneumonia
Mice were fed mouse chow containing trimethoprim-sulfamethoxazole for 3 weeks to clear possible previous infections (72) and then allowed 2 weeks to clear the medication (73). Throughout experimentation, mice were maintained on an antibiotic regimen (500 mg/liter) in drinking water, alternating monthly between cephalexin and amoxicillin. Mice were intranasally infected with 0.5 × 106 P. murina cysts (ATCC) intranasally on day 0 and day 7 and monitored for 20% weight loss and subjective moribund characteristics resulting from PCP. Before sacrifice, blood oxygen saturation was measured to indicate compromise of pulmonary function and active pneumonia via the MouseOx System (Starr Life Sciences, Oakmont, PA). Upon sacrifice via carbon dioxide asphyxiation, lungs were weighed to assess general fluid influx into lung parenchyma. Mice that did not reach a terminal endpoint were allowed to live a minimum of 150 days. Depletion of CD4 T cells was accomplished by intraperitoneal injection of 0.3 mg GK1.5 mAb (BioXcell, New Lebanon, NH; or PBS control) on days −4 and −1 before infection, and weekly thereafter. Depletion status was monitored by flow cytometry analysis of blood periodically throughout the experiment.
TMEV infection
Age- and sex-matched mice were weighed and trained on a RotaRod apparatus daily for 3 days before intracranial infection with 2 million plaque-forming units of TMEV (Daniel’s strain), as previously described (74). Mice were regularly monitored for 20% weight loss, moribund characteristics, and declining neurological function via decreased RotaRod performance. Depletion of CD4 and/or CD8 T cells was accomplished by intraperitoneal injection of 0.3 mg of each mAb (GK1.5 anti-CD4, 2.43 anti-CD8, BioXcell, New Lebanon, NH; or PBS control) on days −3 and −1 before infection, and weekly thereafter. Depletion status was monitored by flow cytometry analysis of blood periodically throughout the experiment.
PiSCES analysis
PiSCES analysis was previously described in detail (40). XML output files were parsed to acquire the raw data for use in MATLAB, R statistical package, Cytoscape, and other analysis/visualization platforms. For each well from an experiment’s data acquisition plate, data were processed to (i) eliminate doublets based on the doublet discriminator intensity (>5000 and <25,000 arbitrary units, Bio-Plex 200), (ii) identify specific bead classes within the bead regions used, and (iii) pair individual bead PE fluorescence measurements with their corresponding bead region. This processing generated a distribution of PE intensity values for each pairwise protein PiSCES measurement. Adaptive nonparametric with empirical cutoff analysis was used as the primary method to determine statistical significance of PiSCES data, as previously described (40). Briefly, differences in specific protein-pair PiSCES measurements were considered hits if they were consistently identified as significant in 100% of experiments, with the minimum number of experiments per experimental condition set as three. An overall type I error of 0.05 was set as the cutoff for statistical significance, which incorporated a Bonferroni adjustment for multiple hypotheses. PCA was performed using the built-in function in MATLAB, using as input variables the log2 MFI of each PiSCES measurement performed, where each stimulation condition in one experiment constituted an observation. PiSCES analysis by PCA used complete datasets from all experiments. In contrast, node-edge diagrams only visualized subsets of data and were generated using the publicly free open network resource, Cytoscape. For protein pairs that had multiple measurements targeting different epitope combinations, the measurement with the greatest mean log2 fold change was selected for node-edge diagram visualizations.
Other statistical analyses
For kinetic analysis, the mean absolute value log2 fold change across each time point measured and across both stimulation conditions (OVA/FARL and Q7/FARL, or RAY/Null and RFY/Null) was used to select the top 15 to 20 protein pair hits. K-means clustering was performed in MATLAB using Squared Euclidean as the distance measurement on log2 fold changes expressed as % max change across the kinetic. The cluster number was selected to maximize the number of distinct kinetic patterns for a given experimental system. Clustering was performed on the negative selection ligand or agonist stimulation, and then data from the positive selection ligand or antagonist stimulation were mapped onto these clusters to compare the overall signaling kinetics. Statistical analysis of FTOC data was performed using two-tailed Student’s t tests with GraphPad Prism software. Paired data originated from single fetal thymi whose two lobes were separated for culture in OVA peptide or matched-dose FARL peptide. Other statistical analyses between groups were completed using unpaired one- or two-tailed Student’s t tests. Survival analysis was completed using the log-rank (Mantel-Cox) statistical test. Statistical significance was defined as P < 0.05 α cutoff.
Supplementary Material
Fig. S1. Theoretical framework to categorize network patterns of TCR-proximal signaling protein complexes that could specify positive versus negative selection.
Fig. S2. The response of preselection OT1.β2m0.RAG20 DP thymocytes to H2O2, and comparison with PV stimulation.
Fig. S3. The response of preselection OT1.β2m0.RAG20 DP thymocytes to PV stimulation.
Fig. S4. Kinetics of PiSCES signatures for positive and negative selection stimuli.
Fig. S5. Kinetics of PiSCES signatures for agonist and antagonist stimuli in LC13ab.huCD8ab. JRT3 cells.
Fig. S6. Clustering analysis for top hits observed across the kinetic for agonist versus antagonist stimuli in LC13ab.huCD8ab.JRT3 cells.
Fig. S7. Kinetics of PiSCES signatures for agonist and antagonist stimuli in OT1ab.muCD8ab. JRT3 cells.
Fig. S8. Clustering analysis for top hits observed across the kinetic for OVA versus Q7 stimuli in OT1ab.muCD8ab.JRT3 cells.
Fig. S9. Assessment of CD8β and CD8αα (TLA+) expression in OT1.RAG20.β2m0 FTOC cells.
Fig. S10. FTOCs from positive selection conditions can be induced by antigenic stimulation to proliferate and kill target cells.
Fig. S11. MHC-dependent signaling generates residual αβ T cells in CD3δ0 mice.
Fig. S12. Developmental and maturation markers on peripheral T cells of wild-type and mutant mice.
Fig. S13. The few peripheral T cells in OT1.RAG20.CD3δ0 mice require MHC class I for their generation/survival.
Fig. S14. Diverse TCR repertoire in individual B6 and CD3δ0 mice.
Fig. S15. Multiple peripheral TCRα transcripts in B6 and CD3δ0 mice.
Fig. S16. T cells in CD3δ0 mice provide immune activity against PCP.
Fig. S17. T cells in CD3δ0 mice provide immune activity against TMEV.
Fig. S18. Example gating used for flow cytometry data.
Table S1. Validated Ab pairs used to identify each mouse protein target.
Table S2. Raw data for experiments with n ≤ 25.
Acknowledgments:
We thank M. Bell, L. Zoecklein, S. Bharati, and M. Zhang for technical assistance; M. Blanco, VMD, for veterinary consultation; and the NIH tetramer core facility at Emory University for providing tetramers.
Funding: This work was supported by the University of Missouri-Columbia and the Mayo Foundation (to D.G. and A.G.S.); NIH grants R01GM103841 (to A.G.S.), R01AI097187 (to D.G.), and R21NS094765 (to A.J.J.); BD Biosciences Research Award (to A.G.S.); NIH training grants T32AI7425 and T32AI007386 (to S.C.N.) and T32GM65841 (to K.M.W.); Consortium of Multiple Sclerosis Centers Foundation Student Research Scholarship Program (to K.M.W.); and the generous philanthropic partnership of B. Fineman with the Mayo Clinic (to A.G.S.).
Footnotes
SUPPLEMENTARY MATERIALS
immunology.sciencemag.org/cgi/content/full/4/32/eaal2201/DC1
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper or in the Supplementary Materials.
REFERENCES AND NOTES
- 1.Hogquist KA, Baldwin TA, Jameson SC, Central tolerance: Learning self-control in the thymus. Nat. Rev. Immunol. 5, 772–782 (2005). [DOI] [PubMed] [Google Scholar]
- 2.Werlen G, Hausmann B, Naeher D, Palmer E, Signaling life and death in the thymus: Timing is everything. Science 299, 1859–1863 (2003). [DOI] [PubMed] [Google Scholar]
- 3.Guy CS, Vignali DAA, Organization of proximal signal initiation at the TCR:CD3 complex. Immunol. Rev. 232, 7–21 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Borroto A, Abia D, Alarcon B, Crammed signaling motifs in the T-cell receptor. Immunol. Lett. 161, 113–117 (2014). [DOI] [PubMed] [Google Scholar]
- 5.Werlen G, Palmer E, The T-cell receptor signalosome: A dynamic structure with expanding complexity. Curr. Opin. Immunol. 14, 299–305 (2002). [DOI] [PubMed] [Google Scholar]
- 6.Roncagalli R, Hauri S, Fiore F, Liang Y, Chen Z, Sansoni A, Kanduri K, Joly R, Malzac A, Lähdesmäki H, Lahesmaa R, Yamasaki S, Saito T, Malissen M, Aebersold R, Gstaiger M, Malissen B, Quantitative proteomics analysis of signalosome dynamics in primary T cells identifies the surface receptor CD6 as a Lat adaptor–independent TCR signaling hub. Nat. Immunol. 15, 384–392 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Balagopalan L, Kortum RL, Coussens NP, Barr VA, Samelson LE, The linker for activation of T cells (LAT) signaling hub: From signaling complexes to microclusters. J.Biol. Chem. 290, 26422–26429 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pageon SV, Tabarin T, Yamamoto Y, Ma Y, Nicovich PR, Bridgeman JS, Cohnen A, Benzing C, Gao Y, Crowther MD, Tungatt K, Dolton G, Sewell AK, Price DA, Acuto O, Parton RG, Gooding JJ, Rossy J, Rossjohn J, Gaus K, Functional role of T-cell receptor nanoclusters in signal initiation and antigen discrimination. Proc. Natl. Acad. Sci. U.S.A. 113, E5454–E5463 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sloan-Lancaster J, Evavold BD, Allen PM, Induction of T-cell anergy by altered T-cell-receptor ligand on live antigen-presenting cells. Nature 363, 156–159 (1993). [DOI] [PubMed] [Google Scholar]
- 10.Madrenas J, Wange RL, Wang JL, Isakov N, Samelson LE, Germain RN, Zeta phosphorylation without ZAP-70 activation induced by TCR antagonists or partial agonists. Science 267, 515–518 (1995). [DOI] [PubMed] [Google Scholar]
- 11.Teixeiro E, Daniels MA, Hausmann B, Schrum AG, Naeher D, Luescher I, Thome M, Bragado R, Palmer E, T cell division and death are segregated by mutation of TCRbeta chain constant domains. Immunity 21, 515–526 (2004). [DOI] [PubMed] [Google Scholar]
- 12.Altan-Bonnet G, Germain RN, Modeling T cell antigen discrimination based on feedback control of digital ERK responses. PLOS Biol. 3, e356 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Teixeiro E, Daniels MA, Hamilton SE, Schrum AG, Bragado R, Jameson SC, Palmer E, Different T cell receptor signals determine CD8+ memory versus effector development. Science 323, 502–505 (2009). [DOI] [PubMed] [Google Scholar]
- 14.Komarova NL, Zou X, Nie Q, Bardwell L, A theoretical framework for specificity in cell signaling. Mol. Syst. Biol. 1, 2005.0023 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jin J, Pawson T, Modular evolution of phosphorylation-based signalling systems. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 367, 2540–2555 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schrum AG, Gil D, Robustness and specificity in signal transduction via physiologic protein interaction networks. Clin. Exp. Pharmacol. 2, S3.001 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Elliott JI, T cell repertoire formation displays characteristics of qualitative models of thymic selection. Eur. J. Immunol. 27, 1831–1837 (1997). [DOI] [PubMed] [Google Scholar]
- 18.Starr TK, Jameson SC, Hogquist KA, Positive and negative selection of T cells. Annu. Rev. Immunol. 21, 139–176 (2003). [DOI] [PubMed] [Google Scholar]
- 19.Melichar HJ, Ross JO, Herzmark P, Hogquist KA, Robey EA, Distinct temporal patterns of T cell receptor signaling during positive versus negative selection in situ. Sci. Signal. 6, ra92 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ashton-Rickardt PG, Tonegawa S, A differential-avidity model for T-cell selection. Immunol. Today 15, 362–366 (1994). [DOI] [PubMed] [Google Scholar]
- 21.Sebzda E, Wallace VA, Mayer J, Yeung RS, Mak TW, Ohashi PS, Positive and negative thymocyte selection induced by different concentrations of a single peptide. Science 263, 1615–1618 (1994). [DOI] [PubMed] [Google Scholar]
- 22.Hu Q, Bazemore Walker CR, Girao C, Opferman JT, Sun J, Shabanowitz J, Hunt DF, Ashton-Rickardt PG, Specific recognition of thymic self-peptides induces the positive selection of cytotoxic T lymphocytes. Immunity 7, 221–231 (1997). [DOI] [PubMed] [Google Scholar]
- 23.Ober BT, Hu Q, Opferman JT, Hagevik S, Chiu N, Wang C-R, Ashton-Rickardt PG, Affinity of thymic self-peptides for the TCR determines the selection of CD8+ T lymphocytes in the thymus. Int. Immunol. 12, 1353–1363 (2000). [DOI] [PubMed] [Google Scholar]
- 24.Leishman AJ, Gapin L, Capone M, Palmer E, MacDonald HR, Kronenberg M, Cheroutre H, Precursors of functional MHC class I- or class II-restricted CD8αα+ T cells are positively selected in the thymus by agonist self-peptides. Immunity 16, 355–364 (2002). [DOI] [PubMed] [Google Scholar]
- 25.Yamagata T, Mathis D, Benoist C, Self-reactivity in thymic double-positive cells commits cells to a CD8αα lineage with characteristics of innate immune cells. Nat. Immunol. 5, 597–605 (2004). [DOI] [PubMed] [Google Scholar]
- 26.Wyss L, Stadinski BD, King CG, Schallenberg S, McCarthy NI, Lee JY, Kretschmer K, Terracciano LM, Anderson G, Surh CD, Huseby ES, Palmer E, Affinity for self antigen selects Treg cells with distinct functional properties. Nat. Immunol. 17, 1093–1101 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hogquist KA, Jameson SC, Bevan MJ, Strong agonist ligands for the T cell receptor do not mediate positive selection of functional CD8+ T cells. Immunity 3, 79–86 (1995). [DOI] [PubMed] [Google Scholar]
- 28.Sebzda E, Mariathasan S, Ohteki T, Jones R, Bachmann MF, Ohashi PS, Selection of the T cell repertoire. Annu. Rev. Immunol. 17, 829–874 (1999). [DOI] [PubMed] [Google Scholar]
- 29.Mariathasan S, Jones RG, Ohashi PS, Signals involved in thymocyte positive and negative selection. Semin. Immunol. 11, 263–272 (1999). [DOI] [PubMed] [Google Scholar]
- 30.Mintern JD, Maurice MM, Ploegh HL, Schott E, Thymic selection and peripheral activation of CD8 T cells by the same class I MHC/peptide complex. J. Immunol. 172, 699–708 (2004). [DOI] [PubMed] [Google Scholar]
- 31.Dave VP, Cao Z, Browne C, Alarcon B, Fernandez-Miguel G, Lafaille J, de la Hera A, Tonegawa S, Kappes DJ, CD3 delta deficiency arrests development of the alpha beta but not the gamma delta T cell lineage. EMBO J. 16, 1360–1370 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Delgado P, Fernández E, Dave V, Kappes D, Alarcón B, CD3delta couples T-cell receptor signalling to ERK activation and thymocyte positive selection. Nature 406, 426–430 (2000). [DOI] [PubMed] [Google Scholar]
- 33.Bäckström BT, Müller U, Hausmann B, Palmer E, Positive selection through a motif in the αβ T cell receptor. Science 281, 835–838 (1998). [DOI] [PubMed] [Google Scholar]
- 34.Werlen G, Hausmann B, Palmer E, A motif in the αβ T-cell receptor controls positive selection by modulating ERK activity. Nature 406, 422–426 (2000). [DOI] [PubMed] [Google Scholar]
- 35.Daniels MA, Teixeiro E, Gill J, Hausmann B, Roubaty D, Holmberg K, Werlen G, Hollander GA, Gascoigne NRJ, Palmer E, Thymic selection threshold defined by compartmentalization of Ras/MAPK signalling. Nature 444, 724–729 (2006). [DOI] [PubMed] [Google Scholar]
- 36.Swan KA, Alberola-Ila J, Gross JA, Appleby MW, Forbush KA, Thomas JF, Perlmutter RM, Involvement of p21ras distinguishes positive and negative selection in thymocytes. EMBO J. 14, 276–285 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Alberola-Ila J, Hogquist KA, Swan KA, Bevan MJ, Perlmutter RM, Positive and negative selection invoke distinct signaling pathways. J. Exp. Med. 184, 9–18 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.O’Shea CC, Crompton T, Rosewell IR, Hayday AC, Owen MJ, Raf regulates positive selection. Eur. J. Immunol. 26, 2350–2355 (1996). [DOI] [PubMed] [Google Scholar]
- 39.McKeithan TW, Kinetic proofreading in T-cell receptor signal transduction. Proc. Natl. Acad. Sci. U.S.A. 92, 5042–5046 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smith SEP, Neier SC, Reed BK, Davis TR, Sinnwell JP, Eckel-Passow JE, Sciallis GF, Wieland CN, Torgerson RR, Gil D, Neuhauser C, Schrum AG, Multiplex matrix network analysis of protein complexes in the human TCR signalosome. Sci. Signal. 9, rs7 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ely LK, Green KJ, Beddoe T, Clements CS, Miles JJ, Bottomley SP, Zernich D, Kjer-Nielsen L, Purcell AW, McCluskey J, Rossjohn J, Burrows SR, Antagonism of antiviral and allogeneic activity of a human public CTL clonotype by a single altered peptide ligand: Implications for allograft rejection. J. Immunol. 174, 5593–5601 (2005). [DOI] [PubMed] [Google Scholar]
- 42.Beddoe T, Chen Z, Clements CS, Ely LK, Bushell SR, Vivian JP, Kjer-Nielsen L, Pang SS, Dunstone MA, Liu YC, Macdonald WA, Perugini MA, Wilce MCJ, Burrows SR, Purcell AW, Tiganis T, Bottomley SP, McCluskey J, Rossjohn J, Antigen ligation triggers a conformational change within the constant domain of the αβ T cell receptor. Immunity 30, 777–788 (2009). [DOI] [PubMed] [Google Scholar]
- 43.Macdonald WA, Chen Z, Gras S, Archbold JK, Tynan FE, Clements CS, Bharadwaj M, Kjer-Nielsen L, Saunders PM, Wilce MC, Crawford F, Stadinsky B, Jackson D, Brooks AG, Purcell AW, Kappler JW, Burrows SR, Rossjohn J, McCluskey J, T cell allorecognition via molecular mimicry. Immunity 31, 897–908 (2009). [DOI] [PubMed] [Google Scholar]
- 44.Abraham RT, Weiss A, Jurkat T cells and development of the T-cell receptor signalling paradigm. Nat. Rev. Immunol. 4, 301–308 (2004). [DOI] [PubMed] [Google Scholar]
- 45.Hogquist KA, Jameson SC, Heath WR, Howard JL, Bevan MJ, Carbone FR, T cell receptor antagonist peptides induce positive selection. Cell 76, 17–27 (1994). [DOI] [PubMed] [Google Scholar]
- 46.Stefanski HE, Mayerova D, Jameson SC, Hogquist KA, A low affinity TCR ligand restores positive selection of CD8+ T cells in vivo. J. Immunol. 166, 6602–6607 (2001). [DOI] [PubMed] [Google Scholar]
- 47.Weiss A, Stobo JD, Requirement for the coexpression of T3 and the T cell antigen receptor on a malignant human T cell line. J. Exp. Med. 160, 1284–1299 (1984). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Azzam HS, Grinberg A, Lui K, Shen H, Shores EW, Love PE, CD5 expression is developmentally regulated by T cell receptor (TCR) signals and TCR avidity. J. Exp. Med. 188, 2301–2311 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wettstein P, Strausbauch M, Therneau T, Borson N, The application of real-time PCR to the analysis of T cell repertoires. Nucleic Acids Res. 36, e140 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Johnston SL, Wettstein PJ, T cell receptor diversity in CTLs specific for the CTT-1 and CTT-2 minor histocompatibility antigens. J. Immunol. 159, 2606–2615 (1997). [PubMed] [Google Scholar]
- 51.Wettstein PJ, Strausbauch M, Borson N, Repertoires of T cell receptors expressed by graft-infiltrating T cells evolve during long-term recall responses to single minor histocompatibility antigens. Int. Immunol. 19, 523–534 (2007). [DOI] [PubMed] [Google Scholar]
- 52.Carmona EM, Limper AH, Update on the diagnosis and treatment of Pneumocystis pneumonia. Ther. Adv. Respir. Dis. 5, 41–59 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Murray PD, Pavelko KD, Leibowitz J, Lin X, Rodriguez M, CD4+ and CD8+ T cells make discrete contributions to demyelination and neurologic disease in a viral model of multiple sclerosis. J. Virol. 72, 7320–7329 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ferrer A, Schrum AG, Gil D, A PCR-based method to genotype mice knocked out for all four CD3 subunits, the standard recipient strain for retrogenic TCR/CD3 bone marrow reconstitution technology. Biores. Open Access 2, 222–226 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sloan-Lancaster J, Allen PM, Altered peptide ligand-induced partial T cell activation: Molecular mechanisms and role in T cell biology. Annu. Rev. Immunol. 14, 1–27 (1996). [DOI] [PubMed] [Google Scholar]
- 56.La Face DM, Couture C, Anderson K, Shih G, Alexander J, Sette A, Mustelin T, Altman A, Grey HM, Differential T cell signaling induced by antagonist peptide-MHC complexes and the associated phenotypic responses. J. Immunol. 158, 2057–2064 (1997). [PubMed] [Google Scholar]
- 57.Kersh EN, Shaw AS, Allen PM, Fidelity of T cell activation through multistep T cell receptor ζ phosphorylation. Science 281, 572–575 (1998). [DOI] [PubMed] [Google Scholar]
- 58.Kersh GJ, Kersh EN, Fremont DH, Allen PM, High- and low-potency ligands with similar affinities for the TCR: The importance of kinetics in TCR signaling. Immunity 9, 817–826 (1998). [DOI] [PubMed] [Google Scholar]
- 59.Rabinowitz JD, Beeson C, Wülfing C, Tate K, Allen PM, Davis MM, McConnell HM, Altered T cell receptor ligands trigger a subset of early T cell signals. Immunity 5, 125–135 (1996). [DOI] [PubMed] [Google Scholar]
- 60.Morris GP, Allen PM, How the TCR balances sensitivity and specificity for the recognition of self and pathogens. Nat. Immunol. 13, 121–128 (2012). [DOI] [PubMed] [Google Scholar]
- 61.Zikherman J, Jenne C, Watson S, Doan K, Raschke W, Goodnow CC, Weiss A, CD45-Csk phosphatase-kinase titration uncouples basal and inducible T cell receptor signaling during thymic development. Immunity 32, 342–354 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schoenborn JR, Tan YX, Zhang C. o., Shokat KM, Weiss A, Feedback circuits monitor and adjust basal Lck-dependent events in T cell receptor signaling. Sci. Signal. 4, ra59 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang F, Beck-García K, Zorzin C, Schamel WWA, Davis MM, Inhibition of T cell receptor signaling by cholesterol sulfate, a naturally occurring derivative of membrane cholesterol. Nat. Immunol. 17, 844–850 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Stepanek O, Prabhakar AS, Osswald C, King CG, Bulek A, Naeher D, Beaufils-Hugot M, Abanto ML, Galati V, Hausmann B, Lang R, Cole DK, Huseby ES, Sewell AK, Chakraborty AK, Palmer E, Coreceptor scanning by the T cell receptor provides a mechanism for T cell tolerance. Cell 159, 333–345 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Palmer E, Naeher D, Affinity threshold for thymic selection through a T-cell receptor-coreceptor zipper. Nat. Rev. Immunol. 9, 207–213 (2009). [DOI] [PubMed] [Google Scholar]
- 66.Kjer-Nielsen L, Clements CS, Purcell AW, Brooks AG, Whisstock JC, Burrows SR, McCluskey J, Rossjohn J, A structural basis for the selection of dominant alphabeta T cell receptors in antiviral immunity. Immunity 18, 53–64 (2003). [DOI] [PubMed] [Google Scholar]
- 67.Rossjohn J, Gras S, Miles JJ, Turner SJ, Godfrey DI, McCluskey J, T cell antigen receptor recognition of antigen-presenting molecules. Annu. Rev. Immunol. 33, 169–200 (2015). [DOI] [PubMed] [Google Scholar]
- 68.Holst J, Vignali KM, Burton AR, Vignali DAA, Rapid analysis of T-cell selection in vivo using T cell-receptor retrogenic mice. Nat. Methods 3, 191–197 (2006). [DOI] [PubMed] [Google Scholar]
- 69.Madsen L, Labrecque N, Engberg J, Dierich A, Svejgaard A, Benoist C, Mathis D, Fugger L, Mice lacking all conventional MHC class II genes. Proc. Natl. Acad. Sci. U.S.A. 96, 10338–10343 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schrum AG, Turka LA, The proliferative capacity of individual naive CD4+ T cells is amplified by prolonged T cell antigen receptor triggering. J. Exp. Med. 196, 793–803 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Stiles RJ, Schrum AG, Gil D, A co-housing strategy to improve fecundity of mice in timed matings. Lab. Anim. 42, 62–65 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lobo ML, Esteves F, de Sousa B, Cardoso F, Cushion MT, Antunes F, Matos O, Therapeutic potential of caspofungin combined with trimethoprim-sulfamethoxazole for Pneumocystis pneumonia: A pilot study in mice. PLOS ONE 8, e70619 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lund FE, Hollifield M, Schuer K, Lines JL, Randall TD, Garvy BA, B cells are required for generation of protective effector and memory CD4 cells in response to Pneumocystis lung infection. J. Immunol. 176, 6147–6154 (2006). [DOI] [PubMed] [Google Scholar]
- 74.Deb C, Lafrance-Corey RG, Zoecklein L, Papke L, Rodriguez M, Howe CL, Demyelinated axons and motor function are protected by genetic deletion of perforin in a mouse model of multiple sclerosis. J. Neuropathol. Exp. Neurol. 68, 1037–1048 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Schieven GL, Mittler RS, Nadler SG, Kirihara JM, Bolen JB, Kanner SB, Ledbetter JA, ZAP-70 tyrosine kinase, CD45, and T cell receptor involvement in UV- and H2O2-induced T cell signal transduction. J. Biol. Chem. 269, 20718–20726 (1994). [PubMed] [Google Scholar]
- 76.Lambolez F, Kronenberg M, Cheroutre H, Thymic differentiation of TCRαβ+ CD8αα+ lELs. Immunol. Rev. 215, 178–188 (2007). [DOI] [PubMed] [Google Scholar]
- 77.Gangadharan D, Lambolez F, Attinger A, Wang-Zhu Y, Sullivan BA, Cheroutre H, Identification of pre- and postselection TCRαβ+ intraepithelial lymphocyte precursors in the thymus. Immunity 25, 631–641 (2006). [DOI] [PubMed] [Google Scholar]
- 78.Fink PJ, Hendricks DW, Post-thymic maturation: Young T cells assert their individuality. Nat. Rev. Immunol. 11, 544–549 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pavelko KD, Bell MP, Karyampudi L, Hansen MJ, Allen KS, Knutson KL, Pease LR, The epitope integration site for vaccine antigens determines virus control while maintaining efficacy in an engineered cancer vaccine. Mol. Ther. 21, 1087–1095 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Johnson AJ, Njenga MK, Hansen MJ, Kuhns ST, Chen L, Rodriguez M, Pease LR, Prevalent class l-restricted T-cell response to the Theiler’s virus epitope Db:VP2121-130 in the absence of endogenous CD4 help, tumor necrosis factor alpha, gamma interferon, perforin, or costimulation through CD28. J. Virol. 73, 3702–3708 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Schrum AG, Gil D, Turka LA, Palmer E, Physical and functional bivalency observed among TCR/CD3 complexes isolated from primary T cells. J. Immunol. 187, 870–878 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Stewart JJ, Lee CY, lbrahim S, Watts P, Shlomchik M, Weigert M, Litwin S, A Shannon entropy analysis of immunoglobulin and T cell receptor. Mol. Immunol. 34, 1067–1082 (1997). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Theoretical framework to categorize network patterns of TCR-proximal signaling protein complexes that could specify positive versus negative selection.
Fig. S2. The response of preselection OT1.β2m0.RAG20 DP thymocytes to H2O2, and comparison with PV stimulation.
Fig. S3. The response of preselection OT1.β2m0.RAG20 DP thymocytes to PV stimulation.
Fig. S4. Kinetics of PiSCES signatures for positive and negative selection stimuli.
Fig. S5. Kinetics of PiSCES signatures for agonist and antagonist stimuli in LC13ab.huCD8ab. JRT3 cells.
Fig. S6. Clustering analysis for top hits observed across the kinetic for agonist versus antagonist stimuli in LC13ab.huCD8ab.JRT3 cells.
Fig. S7. Kinetics of PiSCES signatures for agonist and antagonist stimuli in OT1ab.muCD8ab. JRT3 cells.
Fig. S8. Clustering analysis for top hits observed across the kinetic for OVA versus Q7 stimuli in OT1ab.muCD8ab.JRT3 cells.
Fig. S9. Assessment of CD8β and CD8αα (TLA+) expression in OT1.RAG20.β2m0 FTOC cells.
Fig. S10. FTOCs from positive selection conditions can be induced by antigenic stimulation to proliferate and kill target cells.
Fig. S11. MHC-dependent signaling generates residual αβ T cells in CD3δ0 mice.
Fig. S12. Developmental and maturation markers on peripheral T cells of wild-type and mutant mice.
Fig. S13. The few peripheral T cells in OT1.RAG20.CD3δ0 mice require MHC class I for their generation/survival.
Fig. S14. Diverse TCR repertoire in individual B6 and CD3δ0 mice.
Fig. S15. Multiple peripheral TCRα transcripts in B6 and CD3δ0 mice.
Fig. S16. T cells in CD3δ0 mice provide immune activity against PCP.
Fig. S17. T cells in CD3δ0 mice provide immune activity against TMEV.
Fig. S18. Example gating used for flow cytometry data.
Table S1. Validated Ab pairs used to identify each mouse protein target.
Table S2. Raw data for experiments with n ≤ 25.