Abstract
Heparanase (HPSE) is a multifunctional protein endowed with many non-enzymatic functions and a unique enzymatic activity as an endo-β-d-glucuronidase. The latter allows it to serve as a key modulator of extracellular matrix (ECM) via a well-regulated cleavage of heparan sulfate side chains of proteoglycans at cell surfaces. The cleavage and associated changes at the ECM cause release of multiple signaling molecules with important cellular and pathological functions. New and emerging data suggest that both enzymatic as well as non-enzymatic functions of HPSE are important for health and illnesses including viral infections and virally induced cancers. This review summarizes recent findings on the roles of HPSE in activation, inhibition, or bioavailability of key signaling molecules such as AKT, VEGF, MAPK–ERK, and EGFR, which are known regulators of common viral infections in immune and non-immune cell types. Altogether, our review provides a unique overview of HPSE in cell-survival signaling pathways and how they relate to viral infections.
Keywords: Heparanase, Herpesvirus, AKT, VEGF, ERK, EGFR
Introduction to HPSE
Heparanase (HPSE) is an endo-β-d-endoglycosidase that is the only known mammalian enzyme able to cleave heparan sulfate (HS) moieties at certain positions [1]. HPSE plays an important role in the degradation and modification of the extracellular matrix (ECM) [2]. It is a 58 kDa heterodimer composed of 50 kDa and 8 kDa subunits which bind noncovalently [2]. The enzyme is initially synthesized in the endoplasmic reticulum as a 68 kDa precursor protein, modified in the Golgi apparatus to become a 65 kDa proenzyme, and transported to the exterior of the cell [3]. Once there, it can bind to heparan sulfate proteoglycans (HSPGs), low-density lipoprotein-receptor-related protein (LRP), or mannose 6-phosphate [3]. This binding causes the complex to become endocytosed and transported to a lysosome for processing [3]. The acidic pH of the lysosome activates the cathepsin L protease which cleaves a 6 kDa linker region in the HPSE enzyme and converts HPSE into its active heterodimer form [4]. From there, HPSE can participate in a variety of roles: secretion into the exterior of a cell where it cleaves HS side chains; inside the cell, it complexes with autophagosomes and enables autophagy, binds to exosomes and induce their exit from the cell, and enters the nucleus to influence gene transcription [2].
Active HPSE has been implicated in a variety of diseases, most notably cancer [5]. Most tumors display increased levels of HPSE expression [6]. Indeed, increasing HPSE levels have been correlated in enhanced tumor progression, size, metastasis, and angiogenesis [7]. Because of its role in promoting autophagy and exosome formation, HPSE has been shown to increase chemoresistance and longevity in cancer cells [8]. By degrading HS moieties, HPSE releases important growth factors, which were bound to HS, such as vascular endothelial growth factor (VEGF) and epidermal growth factor (EGF). The cleavage of HSPGs also releases numerous cytokines and chemokines that can affect cell-signaling pathways and induce inflammatory responses [9]. Because of the variety of roles, HPSE can play in a cell and its emerging implications in many types of viral diseases, there is a greater need to elucidate the cellular mechanisms and signaling pathways by which HPSE performs its major functions. Our review of existing literature is designed to develop a more concise understanding of the signaling networks in which HPSE participates and thus, directly or indirectly, regulates viral infections. We also highlight new therapeutic targets and approaches that have the potential to translate into new clinical breakthroughs against a variety of viral infections.
Heparanase–Akt signaling
Akt, also known as protein kinase B (PKB), is a serine/threonine kinase that plays a key role in cell growth, metabolism, and survival [10]. Three isoforms of Akt have been reported in the literature thus far: AKT1, AKT2, and AKT3 [11]. Akt has four phosphorylation sites: Ser-124, Thr-308, Thr-450, and Ser-473 [12]. However, phosphorylation of only two of the sites, Thr-308 and Ser-473, contributes to AKT activation [13]. Akt functions downstream of phosphoinositide 3-kinase (PI3K) [14]. Activation of a receptor tyrosine kinase (RTK) or a G-protein-coupled receptor (GPCR) can recruit and activate PI3K with the aid of the Ras family of GTPases [15]. The activation of PI3K converts phosphatidylinositol-4,5-biphosphate (PIP2) to phosphatidylinositol-3,4,5-triphosphate (PIP3) which is required for the translocation of inactive Akt to the plasma membrane [16]. Phosphoinositide-dependent protein kinase 1 (PDK1) will then bind to the Akt-PIP3 complex and phosphorylate Akt at the Thr-308 position, thereby activating it [17]. Mammalian target of rapamycin complex 2 (mTORC2) can then phosphorylate Akt at Ser-473 which is required for its maximal activation [18]. Akt phosphorylates over 100 different proteins, which can (1) activate them, stimulating growth and survival responses or (2) inactivate them, preventing them from stimulating apoptotic responses [19]. For example, phosphorylation of FOXO and GSK3 proteins by Akt inhibits them, which promotes cell survival, proliferation, and metabolism [20]. Phosphorylation of TSC2 by Akt allows the downstream mTORC1 to become activated and initiate metabolism and growth [21]. Other targets of Akt include transcription factors, cell cycle regulators, metabolic enzymes, and regulators of protein and vesicle trafficking [20].
Termination of this pathway can be achieved by multiple kinds of phosphatases. The first uses the protein phosphatase and tensin homolog (PTEN) to dephosphorylate PIP3 back to PIP2, preventing Akt from being recruited to the plasma membrane [22]. The second method uses protein phosphatase 2A (PP2A) and PH domain leucine-rich repeat protein phosphatases (PHLPPs) to dephosphorylate Akt at the Thr-308 and Ser-473 positions, respectively [23].
Cellular apoptosis is one method by which organisms fend off viral infections. Induction of apoptosis reduces the amount of replication which a virus can achieve in a host cell, choking off the supply of viral progeny [24]. It is, therefore, in the interest of a virus to prolong the survival of the host cell. Accordingly, multiple types of viruses have been shown to stimulate Akt phosphorylation, including adenoviruses, poxviruses, HIV, papilloma viruses, herpesviruses, and influenza A [24–27]. There appears to be a trend of mammalian DNA viruses utilizing the Akt pathway for their own benefit, leveraging its role as a master regulator of signaling [21]. For example, in the herpesvirus family, human cytomegalovirus (HCMV), Epstein–Barr virus (EBV), and Kaposi’s sarcoma-associated herpesvirus (KSHV) each encode a unique protein that activates Akt [28, 29]. Activation of Akt signaling allows an infected cell with a latent virus to avoid apoptosis, immortalizing the cell in certain cases [30]. Herpes simplex virus type 1 (HSV-1) activates Akt initially and later encodes for an Akt mimetic protein with its US3 gene [31]. The mimetic protein was able to phosphorylate many of the same substrates as Akt, including tuberous sclerosis complex 2 (TSC2), FOXO1, and GSK3 [32]. In addition, the HSV-1 immediate–early protein ICP0 stimulates the formation of the EIF4F complex, which is downstream of Akt and necessary for the onset of translation [21]. Thus, even if the host cell successfully inhibits the PI3K–Akt axis, HSV-1 can still translate proteins necessary for its survival. However, the inhibition of Akt results in a reduction of viral load in certain viruses, highlighting the importance of the pathway in a productive infection [25]. Viruses have been shown to modify the PI3K pathway at nearly every point from PI3K to eukaryotic initiation factor 4F (EIF4F) [21]. Although many DNA viruses activate Akt during their life cycles, some RNA viruses benefit from Akt downregulation. For example, the membrane protein of the severe acute respiratory syndrome coronavirus (SARS-CoV) inhibits Akt activation, which causes cytopathic effects in infected cells [33, 34].
Many viruses have been shown to up-regulate and exploit host HPSE during infection [9]. One method by which HPSE may aid them is the stimulation of Akt signaling. HPSE has been shown to induce Akt phosphorylation in fibroblasts, endothelial cells, macrophages, and numerous tumor-derived cell lines [14, 35–38]. In addition, HPSE and Akt activation have been correlated in glioma cells, T cells, and lymphoma cells [2, 39]. However, in one study, the addition of HPSE alone to macrophages was unable to stimulate Akt phosphorylation [40]. The mechanism by which HPSE increases Akt phosphorylation levels is still under investigation, but it has been shown to be independent of mannose 6-phosphate receptor (MPR), low-density lipoprotein-receptor-related protein (LRP), heparan sulfate proteoglycans (HSPGs), and even HPSE enzymatic activity [14, 35]. Non-enzymatic activity is likely localized to the C-terminal domain of the HPSE protein [41]. However, it appears to be mediated in part by lipid rafts: changes in lipid raft structure via lovastatin or methyl-beta-cyclodextrin (MβCD) have been shown to abrogate Akt phosphorylation despite the addition of HPSE in fibroblasts [35]. In addition, the effects of HPSE on Akt phosphorylation were enhanced following the addition of heparin or heparan sulfate to the medium [14]. HPSE was shown to mediate Akt phosphorylation at the Ser473 residue in anmTORC2-dependent manner as an siRNA targeting RICTOR, a subunit of mTORC2, diminished phospho-Akt levels [36].
Two studies demonstrated that PI3K is necessary for HPSE-induced Akt activation [14, 36]. HPSE-induced Akt phosphorylation did require the presence of the subunit p110α of PI3K, the integrins αVβ3 or α5β1, and one of the tyrosine kinases FAK or PYK2 [36]. These results regarding the tyrosine kinases are consistent with the literature, as both FAK-dependent and independent mechanisms of Akt phosphorylation have been described [42, 43]. The replaceable nature of the integrins and FAK/PYK2 suggests that the pathway has significant flexibility in terms of the specific proteins used. Additionally, since Akt phosphorylation is enhanced when both components of a pair (the integrins αVβ3/α5β1 or FAK/PYK2) are functioning, these molecular players may exhibit synergistic effects or independently promote p-Akt formation. A 2009 review indicated that HPSE-induced Akt phosphorylation may require firmly attached cells, which may explain why certain integrins were necessary to activate the pathway [44].
In two human cancer cell lines, the SRC family kinase (SFK) inhibitor PP2 was demonstrated to prevent HPSE from activating Akt [36, 39]. The mechanism by which this occurs did not involve syndencan-4 or the following kinases in the SRC family: Src, YES, and FYN [36]. Because these three proteins are members of the SrcA subfamily, it may be the case that the mechanism is mediated by a member of the SrcB subfamily: Lck, Hck, Blk, or Lyn [45, 46]. It may also be the case that PP2 inhibits an off-target member of the HPSE–Akt pathway.
Other players in the HPSE–AKT pathway may include the GTPase Ras and the growth factor TGF-β. Cells with a mutant Ras gene were more receptive to the inhibition of Akt phosphorylation when given both HPSE and an HPSE inhibitor PG545 [47]. Finally, TGF-β was reported to have dose-dependent effects on HPSE-induced Akt phosphorylation as it stimulated Akt activity at lower concentrations, but inhibited it at higher concentrations in tumor cells [48].
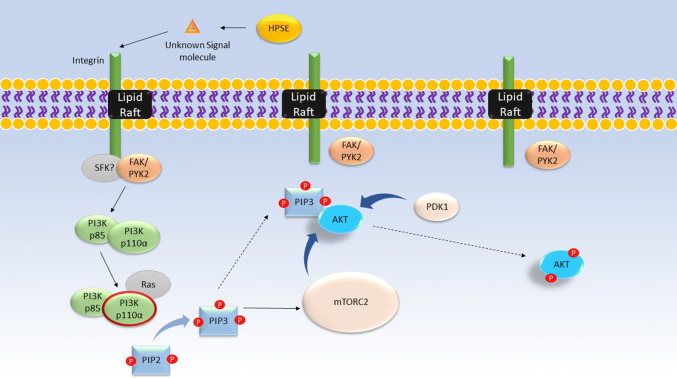
The results of the above studies suggest a potential mechanism by which HPSE activates Akt (Fig. 1). Through an unknown mechanism that is independent of its enzymatic activity, HPSE activates the αVβ3 and α5β1 integrins. Because lipid rafts have been shown to play a role in the regulation and activity of β1 integrins [49], it may be the case that the αVβ3 and α5β1 integrins must either be associated with a lipid raft in order to activate or cluster in a lipid raft upon activation. In either case, these integrins then recruit FAK which autophosphorylates itself at Tyr397 and phosphorylates PI3K at its p85 subunit [50]. Similarly, the integrins recruit and induce the autophosphorylation of PYK2at Tyr402 [51]. The interaction between integrins and PYK2 may be mediated by SFK. The SFK inhibitor PP2 has been shown to inhibit the phosphorylation of FAK and the p85 subunit of PI3K, along with p-Akt [50, 52]. The p85 subunit contains Src homology 2 (SH2) domain, and since the p-FAK may bind to the p85 subunit of PI3K, both FAK and PI3K may be targets of the PP2 inhibitor [50, 53]. Activation of the p85 subunit is required for the activation of the p110α subunit of PI3K, which has been shown to be the essential catalytic site for the downstream phosphorylation of Akt [36, 50]. PYK2 may also activate the p85 subunit of PI3K as it shares a similar structure as FAK, even containing an SH2 domain [54]. Additionally, since inhibition of PI3K reduces Akt phosphorylation, the mechanism of PYK2 is likely to be upstream of or acting on PI3K. However, since PYK2 is expressed in only a few types of cells [55], it may not impact HPSE-induced signaling in many cell lines which is supported by studies showing significant loss of p-Akt with FAK inhibition alone [50]. Once activation of the p110α subunit of PI3K is achieved, PI3K converts PIP2 to PIP3 which has been shown to activate mTORC2 and bind to Akt at the plasma membrane [56]. In certain situations, Ras may interact with PI3K to amplify downstream Akt phosphorylation [57]. Once PIP3 is bound to Akt, PDK1 phosphorylates Akt at Thr308 in its “activation loop”, partially activating it [58]. Finally, mTORC2 then phosphorylates Akt at Ser473, fully activating it [18].
Fig. 1.
Proposed mechanism by which HPSE activates Akt phosphorylation. Proteins shown in gray have roles that are not fully understood. HPSE uses an unknown signaling molecule to bind and activate an integrin bound to a lipid raft. The proteins FAK or PYK2, with a possible role for a Src family kinase, activate the p85 subunit of PI3K. The p85 subunit activates the p110α subunit of PI3K, possibly with the aid of Ras. Activation of PI3K results in the phosphorylation of PIP2 to PIP3. PIP3 plays a dual role in recruiting Akt to the plasma membrane and activating mTORC2. PDK1 and mTORC2 phosphorylate Akt at its Thr-308 and Ser-473 sites, respectively. Phosphorylation of both sites is necessary for maximal activation of Akt
Viral activation of HPSE may stimulate Akt phosphorylation, in part, through the above mechanism. Although some viruses encode proteins which activate Akt independently of HPSE, the Akt–HPSE axis may constitute a positive feedback loop. For example, the tegument protein of HSV-1 VP11/12 induces Akt activation and the US3 kinase phosphorylates multiple Akt substrates [59]. Akt has been shown to regulate NF-ĸB, which is necessary for HSV-1-induced HPSE upregulation [60, 61]. If HPSE leads to the activation of Akt by the proposed mechanism, then it will sustain a prolonged pro-survival state of the cell that is advantageous to the virus.
There are still components of the HPSE–Akt pathway that remain under investigation: (1) How do lipid rafts and the αVβ3/α5β1 interact, if at all? (2) Do the integrins activate FAK directly or is there an unknown SFK mediating the interaction? (3) Can the unknown SFK directly phosphorylate PI3K? and (4) What is the mechanism by which Ras and TGF-β stimulate Akt phosphorylation, and (5) why does TGF-β inhibit Akt phosphorylation at higher concentrations? Additional work needs to be done to elucidate these aspects of the HPSE–Akt pathway.
HPSE–VEGF signaling
Vascular endothelial growth factors are a family of proteins, which regulate vascular development, cell survival, and angiogenesis [62]. There are five known growth factors in the VEGF family: VEGF-A, VEGF-B, VEGF-C, VEGF-D, and the placenta growth factor P1GF [62]. VEGF is created by most parenchymal cells and plays key roles in both autocrine and paracrine signaling [62]. There are three known receptor tyrosine kinases (RTKs) through which VEGFs bind: vascular endothelial growth factor receptor 1 (VEGFR1), VEGFR2, and VEGFR3 (also known as Flt1, Flk1, and Flt4, respectively). Once a VEGF binds to a VEGFR, it induces a conformational change in the receptor, which phosphorylates the tyrosine residues on the receptor, activating the kinase for phosphorylation of downstream proteins, including PI3K–AKT, p38 MAPK, and ERK1/2 [62]. VEGF-A is one of the growth factors that can be bound and sequestered by HS [63].
VEGF upregulation has been observed in many different viral infections [64–66]. SARS-CoV-2-infected patients display higher VEGF expression [67], which may be linked to increased endothelial cell permeabilization and contribute to the development of noncardiogenic pulmonary edema in acute respiratory distress syndrome [68]. Uniquely, the Orf virus, a member of the poxvirus family, encodes a VEGF homolog which stimulates proangiogenic responses during infection [66, 69]. Certain viruses, such as EBV, KSHV, and the hepatitis viruses, are associated with oncogenic infections, and the increase in VEGF stimulates growth and angiogenesis [70]. Non-oncogenic viruses, such as HSV-1 and DENV, also increase VEGF expression during infection, which is associated with severe complications. Upregulation of VEGF during an ocular HSV-1 infection can lead to corneal neovascularization, and elevated VEGF levels during DENV infections contribute to dengue hemorrhagic fever [71, 72]. Inhibiting VEGF reduces the severity of viral infection and tumor growth [73–75].
Like its relationship with Akt, HPSE may be a key regulator of VEGF signaling. HPSE has been shown to significantly increase VEGF expression in a variety of cell lines, particularly VEGF-C [76]. The mechanism by which HPSE up-regulates VEGF is independent of its enzymatic activity, but is instead mediated by the activation of a SFK [77]. A sixfold increase in phospho-SFK was found in HPSE-transfected cells [78, 79]. Inhibition of SFK by PP2 abrogated VEGF stimulation upon HPSE addition [77]. Because PP2 and the antibody used were not specific for the Src protein, it may be the case that another member of the Src family was mediating the HPSE–VEGF mechanism [77]. This SFK may be responsible for mediating FAK activation in the HPSE–Akt pathway discussed before [36]. In addition, only the secreted forms of HPSE were able to up-regulate VEGF [77]. These results suggest that HPSE acts on the exterior of the cell to increase VEGF expression in an SFK-dependent mechanism.
HPSE may be able to directly release the VEGF bound to HS chains located on perlecan proteoglycans by cleaving HS [79, 80]. HPSE-induced VEGF-C upregulation enhanced tumor lymphangiogenesis in mouse models and was correlated with lymphatic vessel density in human patients [76]. Thus, HPSE may possess enzymatic and non-enzymatic means of increasing VEGF signaling. HPSE also indirectly impacts VEGFR2 via its role in syndecan shedding. In brief, enzymatically active HPSE stimulates the ERK signaling pathway which up-regulates matrix metalloproteinase 9 (MMP-9) [81]. MMP-9 then cleaves the syndecan-1 core protein from the cell surface [81]. When syndecan-1 sheds from the membrane, it can couple the integrin α4β1 (also known as VLA-4) and VEGFR2 together, forming a complex of three proteins [82]. The complex activates VEGFR2 which proceeds to enhance cell invasion, formation of endothelial tubes, and other tumorigenic responses [82]. This mechanism is inhibited by synstatins, proteins which can competitively inhibit the binding of either α4β1 or VEGFR2 to shed syndecan-1 [82, 83].
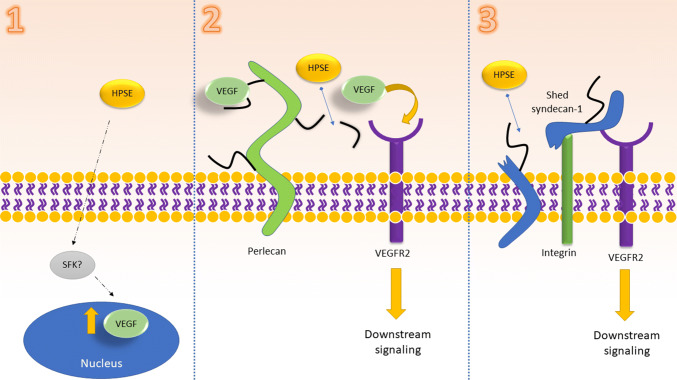
Thus, HPSE demonstrates three ways in which it activates the VEGF pathway (Fig. 2). Once secreted from the cell, it can induce SFK-mediated VEGF upregulation in an enzymatically independent manner. HPSE can also enzymatically cleave HS chains, liberating bound VEGF. It may also activate VEGFR2 via its role in syndecan-1 shedding. Further studies must be done to investigate precisely if and how HPSE activates SFKs. Additionally, the mechanism by which an SFK up-regulates VEGF needs to be explored.
Fig. 2.
Proposed mechanism by which HPSE activates VEGFR signaling. Proteins shown in gray have roles that are not fully understood. (1) Extracellular HPSE increases VEGF transcription in an SFK-dependent manner. (2) HPSE cleaves HS chains from perlecan, liberating VEGF to bind to VEGFR2 and stimulate downstream signaling. (3) HPSE activation leads to syndecan-1 shedding from the cell surface. The syndecan protein complexes with α4β1 and VEGFR2, activating the latter
In case of an HSV-1 infection, its immediate–early protein ICP4 acts as a transcription factor that up-regulates VEGF expression [84]. During the later stages of infection, HSV-1 up-regulates HPSE in an ICP34.5-dependent mechanism, which results in the cleavage of HS moieties from the cell surface [61]. Cleavage of HS releases VEGF which may then bind to VEGFR. Additionally, HSV-1 induces syndecan-1 shedding, which activates VEGFR, as well [85]. Activation of VEGFR stimulates downstream ERK signaling and the associated induction of inflammatory cytokines [86]. These cytokines trigger immune cell infiltration, and, in the case of ocular infections, can lead to neovascularization of the cornea [86]. Thus, while HSV-1 stimulates VEGF transcription directly, HPSE plays an important role in liberating extracellular VEGF to further bolster angiogenic responses.
HPSE–ERK signaling
The extracellular signal-regulated kinase (ERK) signaling pathway modulates the proliferation, survival, and differentiation of cells [87]. The process begins with the binding of ligands to a variety of membrane receptors: G-protein coupled receptors (GPCRs), RTKs, and ion channels among others [88]. For example, one method by which the pathway begins involves the binding of epidermal growth factor and transforming growth factor α (TGFα) to the epidermal growth factor receptor (EGFR), a tyrosine kinase [88]. EGFR dimerizes upon ligand binding and starts the signal transduction pathway, which sequentially forms a complex of the following proteins: ShC–Grb2, SOS, and the GTPase Ras [88]. The ShC–Grb2–SOS–Ras complex activates Ras which marks the start of the MAPK pathway [88]. The MAPK pathway consists of the sequential phosphorylation of the three kinases: MAPK 3-kinase (MAPKKK), MAPK 2 kinase (MAPKK), and MAPK [89]. In mammalian cells, these kinases are known as Raf, MEK1/2, and finally ERK1/2 [90]. Activated ERK then proceeds to translocate to the nucleus where it phosphorylates transcription factors that regulate activities such as proliferation, continued survival, and angiogenesis [88].
For similar reasons as the Akt and VEGF pathways, it is advantageous for viruses to exert control over the ERK signaling pathway. Promoting survival and delaying apoptosis allow viruses time to replicate and produce infectious progeny. Notably, all three classes of herpesviruses hijack the ERK pathway for their own benefit. The alphaherpesvirus HSV-1 activates ERK, which travels to the cell membrane and promotes the remodeling of the actin cortex [91]. This promotes filopodia formation and subsequent viral surfing, which aids HSV-1 find a suitable attachment point on the cell membrane for fusion [91]. The betaherpesvirus HCMV uses the ERK pathway to remodel its chromatin sites, an essential step in reactivation from latency [92]. The gamma herpesvirus KHSV activates ERK signaling to up-regulate the transcription factor Fos and stimulate the transcription of genes necessary for replication [93]. Additional DNA viruses that have been shown to impact ERK signaling include HPV, HBV, and vaccinia virus [94–96]. Certain RNA viruses hijack the pathway as well. Yellow fever virus, influenza, and HCV require an intact ERK pathway for successful replication [97–99]. The ERK pathway affects Ebola and Dengue viruses, as well, the former needing to repress ERK activity and the latter needing to stimulate it [99, 100]. Overall, a wide range of viruses have been demonstrated to regulate the ERK pathway for their own uses.
Again, HPSE influences the ERK pathway, and the mechanisms by which this occurs are discussed below. Enzymatically active HPSE has been shown to increase levels of ERK phosphorylation in macrophages, myeloma, and pituitary adenoma cells [81, 101]. HPSE-induced ERK activation then enhances the expression of matrix metallopeptidase 9 (MMP-9), which is a syndecan-1 sheddase, along with urokinase-type plasminogen activator (uPA) and uPA receptor (uPAR) levels. It has been shown that uPA and uPAR lead to the downstream activation of the proenzyme form of MMP-9 (pro-MMP-9) [102]. Activation of uPA, uPAR, and pro-MMP-9 results in the formation of active MMP-9. Another study discovered that syndecan-1 shedding was greatly inhibited when the activation of MMP-9, uPA, or uPAR was blocked, further supporting the relationship of these three proteins in stimulating syndecan-1 shedding [81]. However, it should be noted that in breast cancer cells, HPSE led to a decrease in MMP-9 expression, which suggests that the effect of HPSE may be cell-type dependent and elicit different pathways which impact MMP-9 expression [103].
The shed syndecan-1 can then bind to VEGF (also up-regulated by HPSE), and the complex attaches integrins to VEGFR2 [104]. They synergistically enhance ERK phosphorylation and invasion of the endothelial layer in myeloma cells [104]. The same study demonstrated that VEGF alone could facilitate a lesser degree of endothelial invasion, while syndecan-1 alone had no effect [104]. Thus, instead of independently promoting invasive behavior, the shed syndecan-1 may be further enhancing the effects of VEGF. The cause of this synergistic effect has been hypothesized to be due to the syndecan-1 protecting VEGF from degradation or aiding the binding of VEGF to receptors on the cell surface, but it is currently unknown [104]. The HPSE–VEGF–ERK pathway is thought to be taken advantage of during ocular HSV-1 infection to promote corneal neovascularization [86].
Because only enzymatically active variants of HPSE were shown to activate ERK phosphorylation in myeloma cells [81], the mechanism by which HPSE leads to ERK activation likely involves a downstream signal resulting from HS degradation. One candidate for this signal has been suggested by Purushothaman et al. to be hepatocyte growth factor (HGF) as it has been shown to bind to the HSPG syndecan-1 via its HS moieties in myeloma cells and stimulate Met signaling via the c-Met receptor, which, in turn, activates the ERK pathway [105, 106]. Although primarily made in mesenchymal cells, HGF is also produced in myeloma cells [107]. Enzymatically inactive HPSE has been shown to up-regulate HGF expression and secretion from the cell [108]. Thus, the enzymatic activity of HPSE stimulates syndecan-1 shedding which enhances the effects of the HGF that was already up-regulated by a non-enzymatic domain of HPSE. This may explain why non-enzymatically active HPSE is insufficient to stimulate ERK signaling. In addition, there is mixed evidence to support that HGF induces Akt phosphorylation via Met signaling in myeloma cells [105, 106]. If true, this would supplement the pro-survival signal response from the ERK pathway via the PI3K/Akt pathway.
Another facet of the HPSE–ERK axis includes the insulin receptor (IR). Upregulation of enzymatically active HPSE has been shown to increase the phosphorylation levels of the IR, a tyrosine kinase, in myeloma cells [87]. Antibodies targeting the IR inhibit ERK activation [87]. After HPSE activates the insulin receptor, the insulin receptor phosphorylates the insulin receptor substrate 1 (IRS-1) [87]. Additionally, HPSE was demonstrated to activate protein kinase C(PKC) which up-regulates IRS-1 [87]. Thus, HPSE enhances pIRS-1 levels by increasing both the production of IRS-1 and its phosphorylation. In other words, HPSE is responsible for increasing IRS-1 levels via PKC and for increasing phospho-IRS-1 levels via the insulin receptor. High levels of pIRS-1 then induce ERK activation [87].
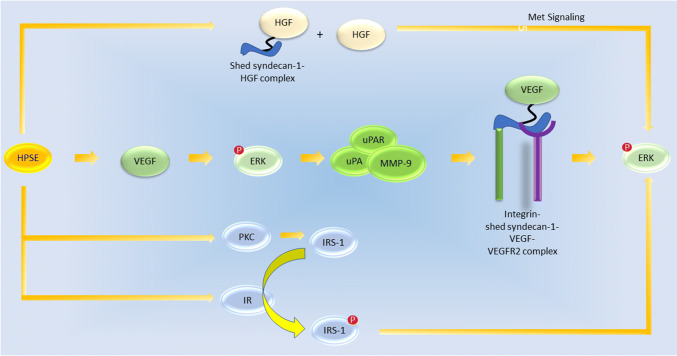
The results of these studies suggest that, in myeloma cells, HPSE increases VEGF which activates ERK (Fig. 3). ERK then stimulates the expression of MMP-9 and uPA/UPAR. This increases syndecan-1 shedding which has been shown to increase angiogenesis and endothelial invasion by myeloma cells [104]. In addition, the shed syndecan-1 can form a complex with VEGF to further enhance ERK signaling which may create a positive feedback loop. Removal of syndecan-1 from cells expressing high levels of HPSE reduced levels of ERK to baseline levels, highlighting the importance of syndecan-1 in magnifying the effects of VEGF and the larger effects on the ERK-signaling pathway [104]. In addition, HPSE-induced HGF activation along with the resultant HGF–syndecan-1 complex may also contribute to the activation of ERK signaling. Finally, HPSE can stimulate ERK activation via the insulin receptor pathway described previously. Thus, HPSE appears to exert its stimulatory effects on ERK phosphorylation from at least three mechanisms. Future work must be done exploring the role of the HPSE–HGF relationship in ERK signaling and whether the proposed mechanisms are present in cells other than myeloma or other cancerous cell lines.
Fig. 3.
Proposed mechanism by which HPSE activates ERK signaling. HPSE may induce syndecan-1 shedding in an enzymatically dependent manner and HGF upregulation in an enzymatically independent manner. Downstream Met signaling results in p-ERK formation. Another mechanism involves the HPSE-mediated VEGF activation which can lead to p-ERK activation. p-ERK can stimulate uPA, uPAR, and pro-MMP9 expression which leads to syndecan-1 shedding. Shed syndecan-1 forms a complex with VEGF as described before, and the downstream VEGF signaling can activate ERK. Finally, enzymatically active HPSE can activate PKC, which up-regulates IRS-1, and the IR, which phosphorylates IRS-1. p-IRS-1 can then stimulate ERK activation
HPSE–EGFR signaling
The epidermal growth factor receptor (EGFR) pathway is a key regulator of cell growth, proliferation, and survival [109]. EGFRs are a subset of RTKs and consist of four different types: EGFR, HER2, HER3, and HER4 [110]. EGFRs currently have 13 known ligands, including EGF, heparin-binding EGF-like growth factor (HB-EGF), and multiple neuregulins [111]. When a ligand binds to an EGFR, it induces either homo- or heterodimerization of the receptor and activates it [112]. Activation of EGFR leads to its clathrin-mediated endocytosis [113]. Inside the cell, EGFR can activate numerous, critical pathways for growth, survival, and migration such as the PI3K–Akt, MAPK–ERK, and signal transducers and activators of transcription (STAT) pathways [109, 111].
Certain viruses utilize EGFR signaling during infection. EGFR signaling causes the development of pulmonary fibrosis through regulation of wound-healing genes during coronavirus infections [114]. In an HBV infection, EGFR is required for the interaction of the virus with the host receptor NTCP and its subsequent internalization into the cell [115]. Loss of EGFR results in an attenuation of HBV entry despite the virus still binding to NTCP [115]. Similar results have been found with HCV, except the virus interacts with CD81 instead of NTCP [116]. Binding of CD81 activates EGFR, which induces the endocytosis of the HCV–CD81–EGFR complex [116]. HCMV is yet another virus which stimulates EGFR, this time by directly binding to it with its gB protein [117]. Activation of EGFR is required for the entry of HCMV into host monocytes [117]. The monocytes then migrate to peripheral tissues, further spreading the viral infection [117]. EGFR-dependent virus internalization is also seen in the Influenza A virus [118, 119]. The viral protein hemagglutinin binds to sialic acid residues on the host cell surface [119]. This leads to a re-organization of lipid rafts in the cell membrane which stimulates nearby EGFRs [119]. Following the EGFR activation, the Influenza A virus and the EGFR are endocytosed in a clathrin-dependent mechanism [119]. Depletion of sialic acid, lipid rafts, EGFR, or clathrin severely inhibits viral infection [118, 119]. Other respiratory viruses that have been shown to activate EGFR include rhinovirus (RV) and respiratory syncytial virus (RSV) [120, 121]. Additionally, Influenza A, RV, and RSV all have demonstrated the ability to stimulate the production of IL-8 via EGFR activation [122]. IL-8 inhibits the transcription of IRF1, and suppression of IRF1 leads to a reduction in CXCL10 levels, key cytokine which stimulates the migration of NK cells and T lymphocytes to the site of infection [122]. Further discussion of the interplay between viruses and EGFR signaling is covered in an excellent review by Zheng et al. [113].
HPSE expression increases EGFR phosphorylation levels in multiple tumor cell lines [39, 123]. In addition, HPSE was correlated with HER2 levels in human brain metastatic breast cancer cells [124]. This mechanism appears to be mediated by the activation of Src and occurs even when the enzymatic activity of HPSE is abolished [6, 39, 101]. It should be noted that the Src activity seen in these studies may be stemming from a SFK, not necessarily the Src protein. Notably, activation of Akt and ERK was not observed in pancreatic cancer cells upon addition of HPSE, suggesting that the effects of HPSE on signaling pathways may be highly sensitive to the cell type being used [123]. HPSE-induced EGFR activation resulted in increased proliferation and invasiveness in vitro [39, 123]. In brain metastatic breast cancer cells, it was reported that EGF activated HER2 and EGFR, which caused HPSE to translocate to the nucleolus in a dose- and time-dependent manner [124]. Once in the nucleolus, enzymatically active HPSE activates Topoisomerase I (Topo 1), which enhances cell proliferation [124]. Interestingly enough, Topo1 has been reported to be inhibited by HS, suggesting that HPSE may cleave HS bound to Topo1 [124]. This mechanism is completely arrested when both HER2 and EGFR are inhibited and partially arrested when only one of the receptors is inhibited [124]. Thus, in different cell lines, the sequence of activation regarding HPSE and EGFR varies. HPSE may induce the phosphorylation of EGFR, or EGFR may stimulate HPSE activity.
In addition to the EGFR proteins, HPSE was reported to increase the phosphorylation levels of the STAT3 and STAT5b proteins [6]. These two STAT proteins have been previously shown to be directly phosphorylated by Src [125, 126]. In particular, increased p-STAT3 in the cytoplasm correlated with the progression of tumors in a cohort of patients with head and neck squamous cell carcinoma [6]. In another study, administration of an HPSE inhibitor markedly reduced the levels of pSTAT3 in pancreatic cells, further supporting the link between HPSE and STAT3 [127]. Additionally, only cytoplasmic HPSE was able to induce the downstream phosphorylation of STAT3 via an SFK and EGFR [6]. STAT3 induction then drives tumorigenesis and inflammatory responses [40].
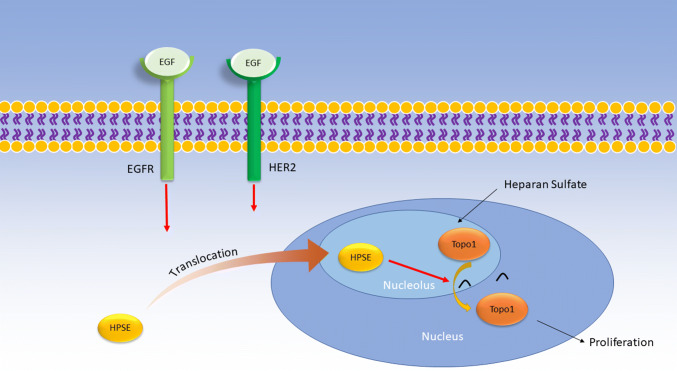
The results of these studies suggest the following mechanism: in certain cells, HPSE activates an SFK in an enzymatically independent manner which proceeds to stimulate EGFR. EGFR can then activate downstream pathways such as PI3K–Akt and MAPK–ERK. It is unknown whether the SFK or downstream EGFR signaling is responsible for phosphorylating STAT3, as inhibitors of either SFK or EGFR abrogated STAT3 activation. Activated STAT3 then drives tumorigenic responses. In brain metastatic breast cancer cells, EGF activates both EGFR and HER2 (Fig. 4). The downstream signaling cascade causes HPSE to translocate to the nucleolus and activate Topo1, an enzyme that drives cell proliferation. Since enzymatically inactive HPSE could not stimulate Topo1, HPSE may activate Topo1 by cleaving HS bound to it (Fig. 4).
Fig. 4.
Proposed role of HPSE in EGFR signaling in brain metastatic breast cancer cells. Extracellular EGF binds to both EGFR and HER2 receptors on the cell membrane. Activation of these receptors results in the nucleolar translocation of HPSE, where it can activate Topo1. HPSE may cleave HS bound to Topo1, but this is yet to be confirmed. Topo1 proceeds to drive cell proliferation
Again, the precise mechanism by which HPSE activates Src or an SFK is unknown and may be the subject of future studies. Elucidating the nature of the HPSE–SFK relationship could shed light not only on the EGFR signaling pathway but also on the other pathways discussed before. Additionally, it has not been confirmed that HPSE-mediated Topo1 activation is due to the cleavage of HS on Topo1, though it is likely.
HPSE in the immune cell response
Role of HPSE in macrophage activation
Macrophages are immune cells which detect and phagocytose foreign invaders that enter the body [128]. They are present in nearly all human tissues [128]. During a viral infection, macrophages engulf infected cells, degrade them in lysosomes, and present the processed antigens on MHC class II molecules on the cell membrane [129]. T cells use T-cell receptors (TCRs) to bind to the MHC class II molecules which facilitates their activation [129]. As professional antigen-presenting cells (APCs), macrophages play a key role in both the innate and adaptive immune response [129].
HPSE was demonstrated to be necessary for the activation and proper functioning of macrophages including their ability to secrete cytokines, to move to the site of infection, and to phagocytose foreign material [130]. Non-enzymatically active HPSE was able to maintain cytokine expression to normal levels, suggesting that the HS cleavage is not responsible for the effects of HPSE on macrophages [130]. Macrophages are activated upon expression of the cytokine macrophage inflammatory protein-2 (MIP-2), but this process is impaired in macrophages from HPSE-KO cells [130]. Interestingly, certain chemotherapeutics can stimulate macrophage activation and polarization into an M2 (pro-tumorigenic) phenotype in an HPSE-dependent mechanism [131].
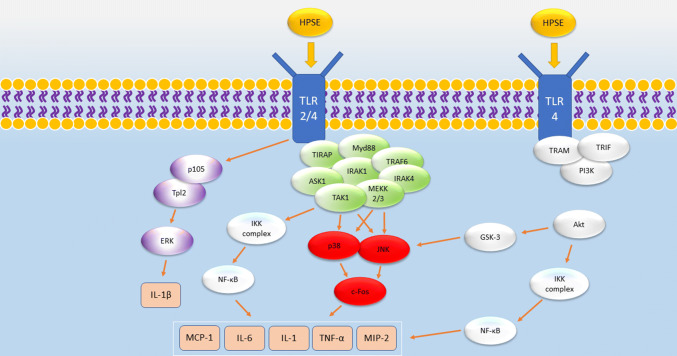
The inhibition of each of the three subfamilies of MAPK signaling (ERK, JNK, and p38) reduced cytokine expression in macrophages. JNK signaling and, to a lesser extent, ERK phosphorylation were responsible for the expression of TNF-α, IL6, and MIP-2 [130]. However, IL6 expression stemmed from ERK and p38 signaling [130]. In addition, the results of a transcription factor array revealed that c-Fos was downregulated in HPSE-KO macrophages, an effect which was replicated upon the addition of JNK and p38 inhibitors to HPSE-WT macrophages [130]. Because Toll-like receptors (TLRs) have been shown to be upstream of the HPSE-induced cytokine response [132], the authors proposed that HPSE activates membrane TLRs through an unknown mechanism. The results of this study indicate that HPSE upregulation activates TLRs which stimulate ERK, p38, and JNK signaling. This trio of proteins proceed to activate c-Fos which finally induces the cytokines needed for macrophage function [130].
Another study of HPSE and macrophage activation reported similar results with enzymatically inactive HPSE inducing the expression of the following cytokines: TNF-α, IL-1, MCP-1, and MMP-9 [38]. In the same study, the authors showed that mechanism corresponds to the activation of PI3K/Akt and MAPK/ERK pathways [38]. TLR2, TLR4, and NF-κB signaling were necessary for HPSE-induced cytokine production [38]. It appears that HPSE, by an unknown mechanism which does not involve enzymatic function or even the binding of HPSGs, activates TLR-2 and TLR-4. This stimulates NF-κB, MAPK, and PI3K signaling which promotes the downstream induction of cytokines that the macrophages require for activation and proper functioning. The mechanism by which TLRs activate NF-κB and JNK signaling is well understood and elucidated in great detail in a review by Takeda and Akira [133].
Yet, another study using mice with colitis reported a positive regulatory which shows HPSE-mediated macrophage activation [40]. HPSE-induced NF-κB signaling produced the following cytokines: IL-1, IL-6, MIP-1, MIP-2, and TNF-α. Enzymatically active HPSE also sensitized macrophages to LPS-induced activation of TLR-4 [40]. Because HS is a known inhibitor of TLR4 and HPSE has been noted to significantly reduce the HS located on the macrophage exterior, HPSE-mediated HS cleavage may clear space on the macrophage membrane and allow more LPS to bind to the macrophage TLR [40].
The activated macrophages then secret TNFα, which up-regulated HPSE expression in the epithelial cells [40]. Since TNFα and the early growth response 1 (EGR1) have both been shown to up-regulate HPSE [134, 135] and TNFα stimulates EGR1 [136], the authors hypothesized that the macrophage-secreted TNFα directly up-regulates HPSE via EGFR1 [40]. Activated macrophages also secrete cathepsin L in the extracellular space, which cleaves and activates the latent HPSE there [40]. The cycle continues, promoting a hyper inflammatory environment that aids tumorigenesis via an increase in vascularization and the chemokineIL-6 which promotes STAT3 signaling [40].
We propose the following mechanism based on the results of these studies: HPSE activates TLR2 and TLR4 through an unknown mechanism, but one that may involve hyaluronic acid. Downstream signaling from TLR2 and TLR4 activates p38, ERK, and JNK. p38 and ERK both function in IL1-β production involving the transcription factor c-Fos. The mechanism by which TLRs activate JNK is well understood and progresses through proteins such as IRAK-1, IRAK-4, TRAF6, and eventually TAK1. TAK1 then activates JNK and the IKK complex, the latter stimulating NF-κB activation. NF-κB translocates to the nucleus, binds to the specific sequences on the genome, and induces the transcription of cytokines, such as IL-1, MCP-1, interferon, and TNF-α, along with MMP-9. JNK signaling stimulates c-Fos which leads to the production of TNF-α, IL6, and MIP2. Syntheses of these cytokines stimulate the activation of macrophages (Fig. 5).
Fig. 5.
Proposed mechanism of the role in HPSE in stimulating the production of cytokines necessary for macrophage activation. By binding to TLR2 or TLR4, HPSE can stimulate the inhibition of p105, causing the release of Tpl2 and downstream ERK activation. ERK signaling results in IL-1β production. Activation of TLR2/4 can stimulate p38, JNK, and NF-κB signaling via an Myd88-dependent formation of a protein complex. Downstream signaling results in the synthesis of MCP-1, IL-6, IL-1, TNF-α, and MIP-2. Alternatively, the binding of HPSE to TLR4 in a TRAM- and TRIF-dependent mechanism which results in activation of PI3K and subsequently Akt. Akt stimulates GSK-3, further bolstering JNK signaling, and the IKK complex, activating NF-κB signaling
TLR4 can activate PI3K via a Myd88-independent mechanism utilizing the TRAM and TRIF proteins. PI3K continues the cascade through Akt which also activates NF-κB via phosphorylation and degradation of IκB. Akt also phosphorylates GSK-3β which activates JNK. Thus, TLRs may activate JNK and NF-κB signaling via two different pathways. More studies need to be done to elucidate the mechanism by which HPSE activates TLRs to stimulate cytokine production and how the TLRs may activate downstream pathways such as p38 and ERK.
Function of HPSE in T-cell adhesion
Active but not resting T cells are reported to produce HPSE [137]. Upon activation, EGR1-dependent HPSE expression level is induced in CD4+ antigen-specific T lymphocytes infiltrating the central nervous system during clinical disease [138]. HPSE has been demonstrated to bind to HS and adhere T cells to the ECM [139, 140]. The adhesive function of HPSE in binding T cells to the ECM was noted in lymphoma cells and cancerous lymphocytes [141, 142]. The mechanism for these adhesive properties was reported to be reliant on the presence of β1 integrin on T cells and vascular cell adhesion molecule (VCAM-1) on endothelial cells [140, 141]. HPSE alone was shown to adhere to T cells under shear flow conditions. The effect was enhanced with treatment of the chemokine SDF-1. Treatment with an α4β1 integrin antibody partially inhibited the adhesion of T cells [140]. Pre-treatment with HPSE to cleave HSPGs significantly reduced the capability of a later addition of HPSE to mediate T-cell adhesion to the ECM. Thus, the authors concluded that HPSE could augment the integrin-mediated adhesion of T cells [140].
HPSE binding to the T cells leads to upregulation of ERK and Pyk-2 kinases [140]. ERK phosphorylation may arise due to HPSE directly as described before. However, HPSE also reportedly augmented the effects of the chemokine SDF-1α on Pyk-2 and ERK phosphorylation, suggesting that it could play both a direct role and a indirect role in the activation of these kinases [140].
HPSE in NK-cell activation, migration, and invasion
Natural killer (NK) cells are innate lymphocytes which can kill infected cells and tumor cells. Heparanase has been implicated in natural killer cell activation, migration, and invasion. NK cells express natural cytotoxicity receptors (NCRs) which are known to induce cytokine production and thereby cytolytic activity in response to contact with specific ligands on the surfaces of infected cells [143]. HS acts as a co-ligand for NCR binding. In case of tumor cells, higher secretion of HPSE by tumor cells cleaves HS from NK cells and impedes NCR binding which impedes the activation of NK cells. On the other side, NK cells lacking HPSE expression also lack tumor-invasive properties. Therefore, the level of HS-NCR interactions regulates NK-cell activation, suggesting that maintaining optimal levels of HPSE is important for NK cell activation [143, 144].
Other roles of HPSE in viral infections
Cellular entry
HS is well established as an attachment receptor for HSV-1 [145]. Its glycoproteins gB and/or gC must bind to HS before entry can progress [145]. Treatment of single-cell neurons with heparanase significantly reduced subsequent HSV-1 entry compared to mock-treated controls [146]. This study supports the idea that HSV-1 must only up-regulate HPSE once entry has concluded. Accordingly, the γ34.5 gene of HSV-1 was found to activate HPSE transcription and is expressed during the later stages of infection [61]. Alongside HSV-1, other viruses which use HS as attachment receptors are HSV-2, HPV, KSHV, and DENV [147]. Pre-treatment of cells with HPSE prior to infection would be expected to hinder entry for these viruses. The respiratory syncytial virus (RSV) appears to require HS moieties on the surface of certain cells to initiate a productive infection [148]. Exogenous addition of bacterial heparinase reduces the susceptibility of HEK-293 and Hep-2 cells to RSV infection [148]. However, heparinase could not confer protection from RSV to HeLa cells [148].
The novel coronavirus SARS-CoV-2 is a member of the β-coronavirus subfamily alongside SARS-CoV and MERS-CoV [149]. Like SARS-CoV, SARS-Cov-2 encodes a spike glycoprotein used to bind to the angiotensin-converting enzyme 2 (ACE2) receptor of target cells during entry [150]. One study reported that α-coronavirus human coronavirus NL63 (HCoV-NL63) requires cell surface HS for attachment and entry. Addition of soluble HS competitively binds to HCov-NL63 and reduces viral adhesion in a dose-dependent manner regardless of the presence of ACE2 [151]. Another study reported that HSPGs are essential for SARS-CoV entry [152]. Prophylactic treatment of bacterial heparinase I or heparin to cells reduced SARS-CoV infectivity [152]. Thus, either loss of HS or competitive inhibition confers some protection to cells from SARS-CoV. Given the structural similarities between SARS-CoV and the novel SARS-CoV-2, it would be interesting to study the effects of HS removal on SARS-CoV-2 infections. As there are multiple ways to reduce viral contact with cell surface HS (HPSE, heparinase, heparin, soluble HS, and MMPs), investigating that this connection may shed light on SARS-CoV-2 entry and possible therapeutics.
Enhancing disease severity
During HSV-1 infections, transfection with a constitutively active HPSE protein (GS3-HPSE) worsened the symptoms of herpetic eye disease in vivo [61]. GS3-HPSE-transfected mice infected with HSV-1 exhibited increases in ocular discharge, ulcer formation, periorbital edema, and corneal vascularization [61]. GS3-HPSE transfection resulted in a delayed wound-healing response in vivo and in vitro [61]. Furthermore, overexpression of WT-HPSE inhibited the induction of the type I interferon response which stimulates antiviral signaling pathways [61].
The human papillomavirus (HPV) E6 gene induces upregulation of HPSE during infection in squamous cell carcinoma cells [153]. It is thought that the E6 protein inhibits p53 which normally limits HPSE transcription [153]. Subsequent cleavage of HS by HPSE results in a loss of ECM integrity and facilitates tumor invasion and metastasis. Furthermore, the release of growth factors from the HS fragments such as VEGF and HGF enhance tumor aggressiveness [153]. A pyranoside analog given to cervical cancer cells increased p53 activity, reducing HPSE transcription, and cell proliferation [154].
While still under investigation, a pair of studies has outlined a connection between HPSE and KSHV. One study found that the cellular miRNA 1258 (hsa-miR-1258) inhibits reactivation of KSHV from latency [155]. Hsa-miR-1258 also downregulates HPSE protein levels and restricts BMBC invasion [156]. Finally, expression of hsa-miR-1258 declines in KSHV-infected cells, suggesting that the virus seeks to remove this impediment to reactivation during infection [155]. It is possible that KSHV benefits from the increase in HPSE expression from inhibiting hsa-miR-1258, possibly through the induction of cell-survival signaling.
RSV up-regulates HPSE during infection in vivo [157]. In RSV infections, HPSE expression correlated with proteinuria severity [157].
Cellular egress
Multiple viruses utilize HPSE to facilitate egress from the host cell. HSV-1 up-regulates HPSE in the later stages of infection which goes on to cleave HS from the cell surface. While HS is an attachment receptor for the virus, its presence may restrict the release of mature virions. Cleavage of HS allows for viral egress to occur more efficiently. Similar loss of HS via HPSE upregulation was found during HSV-2 infections as well [158]. Additionally, HPSE has recently been shown to mediate syndecan-1 shedding from the cell surface during HSV-1 infection [85]. HPSE up-regulates MMP-3 and MMP-7 which translocate to the cell surface and cleave syndecan-1. Via a similar chain of reasoning, syndecan-1 shedding amplifies the loss of HS from the cell surface and promotes egress. Overexpression of HPSE results in increased viral release both in vitro and in vivo [159]. Conversely, knockdown of HPSE reduces HSV-1 and HSV-2 infectivity [158, 159]. Through a similar pathway involving the upregulation of HPSE and loss of HS, porcine reproductive and respiratory syndrome virus (PRRSV) utilizes HPSE to facilitate release from cells [160]. PRRSV activates NF-ĸB signaling which up-regulates HPSE transcription [160].
HPV virions escape from the host cell by binding to HS moieties and growth factors on HSPGs. As the HSPGs are naturally shed from the cell, the virus can spread to neighboring cells. Exogenous HPSE promotes HPV release and infectivity via cleavage of HS, and MMPs likewise remove syndecan-1 proteins from the cell surface [161]. Pharmacological inhibition of HPSE inhibits viral release to the ECM as well [161].
Conclusion
HPSE plays important enzymatic and non-enzymatic roles in the regulation of intracellular signaling pathways and viral infections. The enzymatic activity of HPSE is utilized in the turnover of HSPGs on the cell surface. Cleavage of HS helps to remodel the ECM and release signaling molecules that function in autophagy, exosome synthesis, and cell motility [8]. The non-enzymatic activity of HPSE also stimulates numerous cell-survival pathways as discussed in this review. Due to the variety of roles which it plays, its activity must be tightly regulated. Overexpression of HPSE transforms cell motility to invasiveness or platelet adhesion to thrombogenesis [162, 163]. Depending on the context and timing, HPSE can exacerbate or protect against viral infections.
Viruses often influence major cell-survival signaling pathways at various points, not just the initiation. However, viral activation of HPSE may sustain the activation of these pathways through the mechanisms proposed in this review. As approximately 20% of cancers are associated with viruses [164], sustained pro-survival signaling would contribute to tumor proliferation and metastasis. Both tumors and host cells can release HPSE in the tumor microenvironment, allowing HPSE to stimulate Akt, VEGFR, EGFR, ERK, and other pathways that aid in tumor progression. Thus, oncogenic viruses such as HPV may have an intimate relationship with HPSE-mediated signaling.
In this review, we have discussed the complex interplay between HPSE, viruses, and cell-signaling pathways. It is very clear that HPSE is intricately intertwined with a variety of signaling pathways important for viral pathogenesis. It possesses remarkable flexibility, influencing known cell signaling via enzymatic and non-enzymatic mechanisms, and key regulatory molecules including PI3K–Akt, ERK, VEGFR, and EGFR. Since viruses utilize the pathways regulated by these molecules during infection to promote cell survival or other aspects of viral lifecycle, they stand to gain from influencing HPSE activity, as well. Along the same lines, HPSE plays an important role in fine-tuning of innate and adaptive immune responses against viral pathogens. While a large body of work has previously been done to elucidate the role of HPSE in cell signaling, many key details of the mechanisms are still unknown. Future studies will be needed to develop a more precise understanding of HPSE as a major regulator of infection. For now, it does appear to regulate certain viral infections and interact with major pro-survival signaling pathways. If confirmed in the future, insights gleaned from this molecule could pave the way for new and broad-spectrum therapies for many persistent and acute viral infections.
Author contributions
Visualization—RK and RS; writing—original draft preparation, RK, RS, and DS; writing—review and editing. RK, RS, and DS. All authors have read and approved the manuscript.
Funding
This work was supported by the National Institutes of Health RO1 Grants EY029426, AI139768, EY024710 (to DS) and an NEI core Grant (EY001792).
Compliance with ethical standards
Conflict of interest
The authors declare that they have no competing interests.
Ethics approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
The authors give their consent to publish this review article.
Availability of data and material
Not applicable.
Code availability
Not applicable.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Rivara S, Milazzo F, Giannini G. Heparanase: a rainbow pharmacological target associated to multiple pathologies including rare diseases. Future Med Chem. 2016;8:647–680. doi: 10.4155/fmc-2016-0012. [DOI] [PubMed] [Google Scholar]
- 2.Masola V, Bellin G, Gambaro G, Onisto M. Heparanase: a multitasking protein involved in extracellular matrix (ECM) remodeling and intracellular events. Cells. 2018;7:236. doi: 10.3390/cells7120236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vlodavsky I, Ilan N, Naggi A, Casu B. Heparanase: structure, biological functions, and inhibition by heparin-derived mimetics of heparan sulfate. Curr Pharm Des. 2007;13:2057–2073. doi: 10.2174/138161207781039742. [DOI] [PubMed] [Google Scholar]
- 4.Abboud-Jarrous G, Atzmon R, Peretz T, Palermo C, Gadea B, Joyce J, Vlodavsky I. Cathepsin L is responsible for processing and activation of proheparanase through multiple cleavages of a linker segment. J Biol Chem. 2008;283:18167–18176. doi: 10.1074/jbc.M801327200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vlodavsky I, Singh P, Boyango I, Gutter-Kapon L, Elkin M, Sanderson R, Ilan N. Heparanase: from basic research to therapeutic applications in cancer and inflammation. Drug Resist Updates. 2016;29:54–75. doi: 10.1016/j.drup.2016.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cohen-Kaplan V, Jrbashyan J, Yanir Y, Naroditsky I, Ben-Izhak O, Ilan N, Doweck I, Vlodavsky I. Heparanase induces signal transducer and activator of transcription (STAT) protein phosphorylation: preclinical and clinical significance in head and neck cancer. J Biol Chem. 2012;287:6668–6678. doi: 10.1074/jbc.M111.271346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ilan N, Elkin M, Vlodavsky I. Regulation, function and clinical significance of heparanase in cancer metastasis and angiogenesis. Int J Biochem Cell Biol. 2006;38:2018–2039. doi: 10.1016/j.biocel.2006.06.004. [DOI] [PubMed] [Google Scholar]
- 8.Sanderson R, Elkin M, Rapraeger A, Ilan N, Vlodavsky I. Heparanase regulation of cancer, autophagy and inflammation: new mechanisms and targets for therapy. FEBS J. 2017;284:42–55. doi: 10.1111/febs.13932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thakkar N, Yadavalli T, Jaishankar D, Shukla D. Emerging roles of heparanase in viral pathogenesis. Pathogens. 2017;6:43. doi: 10.3390/pathogens6030043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Song G, Ouyang G, Bao S. The activation of Akt/PKB signaling pathway and cell survival. J Cell Mol Med. 2005;9:59–71. doi: 10.1111/j.1582-4934.2005.tb00337.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang L, Dan H, Sun M, Liu Q, Sun X, Feldman R, Hamilton A, Polokoff M, Nicosia S, Herlyn M, Sebti S, Cheng J. Akt/protein kinase B signaling inhibitor-2, a selective small molecule inhibitor of Akt signaling with antitumor activity in cancer cells overexpressing Akt. Cancer Res. 2004;64:4394–4399. doi: 10.1158/0008-5472.CAN-04-0343. [DOI] [PubMed] [Google Scholar]
- 12.Alessi D, Andjelkovic M, Caudwell B, Cron P, Morrice N, Cohen P, Hemmings B. Mechanism of activation of protein kinase B by insulin and IGF-1. EMBO J. 1996;15:6541–6551. doi: 10.1002/j.1460-2075.1996.tb01045.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Datta S, Brunet A, Greenberg M. Cellular survival: a play in three Akts. Genes Dev. 1999;13:2905–2927. doi: 10.1101/gad.13.22.2905. [DOI] [PubMed] [Google Scholar]
- 14.Gingis-Velitski S, Zetser A, Flugelman M, Vlodavsky I, Ilan N. Heparanase induces endothelial cell migration via protein kinase B/Akt activation. J Biol Chem. 2004;279:23536–23541. doi: 10.1074/jbc.M400554200. [DOI] [PubMed] [Google Scholar]
- 15.Rodriguez-Viciana P, Warne P, Dhand R, Vanhaesebroeck B, Gout I, Fry M, Waterfield M, Downward J. Phosphatidylinositol-3-OH kinase as a direct target of Ras. Nature. 1994;370:527–532. doi: 10.1038/370527a0. [DOI] [PubMed] [Google Scholar]
- 16.Hart J, Vogt P. Phosphorylation of AKT: a mutational analysis. Oncotarget. 2011;2:467–476. doi: 10.18632/oncotarget.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bozulic L, Hemmings B. PIKKing on PKB: regulation of PKB activity by phosphorylation. Curr Opin Cell Biol. 2009;21:256–261. doi: 10.1016/j.ceb.2009.02.002. [DOI] [PubMed] [Google Scholar]
- 18.Sarbassov D, Guertin D, Ali S, Sabatini D. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science. 2005;307:1098–1101. doi: 10.1126/science.1106148. [DOI] [PubMed] [Google Scholar]
- 19.Dunn E, Connor J. HijAkt: the PI3K/Akt pathway in virus replication and pathogenesis. Prog Mol Biol Transl Sci. 2012;106:223–250. doi: 10.1016/B978-0-12-396456-4.00002-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Manning B, Toker A. AKT/PKB signaling: navigating the network. Cell. 2017;169:381–405. doi: 10.1016/j.cell.2017.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Buchkovich N, Zampieri C, Yu Y, Alwine J. The TORrid affairs of viruses: effects of mammalian DNA viruses on the PI3K–Akt–mTOR signalling pathway. Nat Rev Microbiol. 2008;6:266–275. doi: 10.1038/nrmicro1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li J, Yen C, Liaw D, Podsypanina K, Bose S, Wang S, Puc J, Miliaresis C, Rodgers L, McCombie R, Bigner S, Giovanella B, Ittmann M, Tycko B, Hibshoosh H, Wigler M, Parsons R. PTEN, a putative protein tyrosine phosphatase gene mutated in human brain, breast, and prostate cancer. Science. 1997;275:1943–1947. doi: 10.1126/science.275.5308.1943. [DOI] [PubMed] [Google Scholar]
- 23.Kuo Y, Huang K, Yang C, Yang Y, Lee W, Chiang C. Regulation of phosphorylation of Thr-308 of Akt, cell proliferation, and survival by the B55alpha regulatory subunit targeting of the protein phosphatase 2A holoenzyme to Akt. J Biol Chem. 2008;283:1882–1892. doi: 10.1074/jbc.M709585200. [DOI] [PubMed] [Google Scholar]
- 24.Ji W, Liu H. PI3K–Akt signaling and viral infection. Recent Pat Biotechnol. 2008;2:218–226. doi: 10.2174/187220808786241042. [DOI] [PubMed] [Google Scholar]
- 25.Pasquereau S, Kumar A, Abbas W, Herbein G. Counteracting akt activation by HIV protease inhibitors in monocytes/macrophages. Viruses. 2018;10:190. doi: 10.3390/v10040190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Werden S, McFadden G. Pharmacological manipulation of the akt signaling pathway regulates myxoma virus replication and tropism in human cancer cells. J Virol. 2010;84:3287–3302. doi: 10.1128/JVI.02020-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Norman K, Sarnow P. Herpes simplex virus is Akt-ing in translational control. Genes Dev. 2010;24:2583–2586. doi: 10.1101/gad.2004510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moorman N, Cristea I, Terhune S, Rout M, Chait B, Shenk T. Human cytomegalovirus protein UL38 inhibits host cell stress responses by antagonizing the tuberous sclerosis protein complex. Cell Host Microbe. 2008;3:253–262. doi: 10.1016/j.chom.2008.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sodhi A, Chaisuparat R, Hu J, Ramsdell A, Manning B, Sausville E, Sawai E, Molinolo A, Gutkind J, Montaner S. The TSC2/mTOR pathway drives endothelial cell transformation induced by the Kaposi's sarcoma-associated herpesvirus G protein-coupled receptor. Cancer Cell. 2006;10:133–143. doi: 10.1016/j.ccr.2006.05.026. [DOI] [PubMed] [Google Scholar]
- 30.Cooray S. The pivotal role of phosphatidylinositol 3-kinase–Akt signal transduction in virus survival. J Gen Virol. 2004;85:1065–1076. doi: 10.1099/vir.0.19771-0. [DOI] [PubMed] [Google Scholar]
- 31.Benetti L, Roizman B. Protein kinase B/Akt is present in activated form throughout the entire replicative cycle of deltaU(S)3 mutant virus but only at early times after infection with wild-type herpes simplex virus 1. J Virol. 2006;80:3341–3348. doi: 10.1128/JVI.80.7.3341-3348.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chuluunbaatar U, Roller R, Feldman M, Brown S, Shokat K, Mohr I. Constitutive mTORC1 activation by a herpesvirus Akt surrogate stimulates mRNA translation and viral replication. Genes Dev. 2010;24:2627–2639. doi: 10.1101/gad.1978310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mizutani T, Fukushi S, Iizuka D, Inanami O, Kuwabara M, Takashima H, Yanagawa H, Saijo M, Kurane I, Morikawa S. Inhibition of cell proliferation by SARS-CoV infection in Vero E6 cells. FEMS Immunol Med Microbiol. 2006;46:236–243. doi: 10.1111/j.1574-695X.2005.00028.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chan C, Ma C, Chan W, Chan H. The SARS-coronavirus membrane protein induces apoptosis through modulating the Akt survival pathway. Arch Biochem Biophys. 2007;459:197–207. doi: 10.1016/j.abb.2007.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ben-Zaken O, Gingis-Velitski S, Vlodavsky I, Ilan N. Heparanase induces Akt phosphorylation via a lipid raft receptor. Biochem Biophys Res Commun. 2007;361:829–834. doi: 10.1016/j.bbrc.2007.06.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Riaz A, Ilan N, Vlodavsky I, Li J, Johansson S. Characterization of heparanase-induced phosphatidylinositol 3-kinase–AKT activation and its integrin dependence. J Biol Chem. 2013;288:12366–12375. doi: 10.1074/jbc.M112.435172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ramani V, Vlodavsky I, Ng M, Zhang Y, Barbieri P, Noseda A, Sanderson R. Chemotherapy induces expression and release of heparanase leading to changes associated with an aggressive tumor phenotype. Matrix Biol. 2016;55:22–34. doi: 10.1016/j.matbio.2016.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Blich M, Golan A, Arvatz G, Sebbag A, Shafat I, Sabo E, Cohen-Kaplan V, Petcherski S, Avniel-Polak S, Eitan A, Hammerman H, Aronson D, Axelman E, Ilan N, Nussbaum G, Vlodavsky I. Macrophage activation by heparanase is mediated by TLR-2 and TLR-4 and associates with plaque progression. Arterioscler, Thromb Vasc Biol. 2013;33:56–65. doi: 10.1161/ATVBAHA.112.254961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cohen-Kaplan V, Doweck I, Naroditsky I, Vlodavsky I, Ilan N. Heparanase augments EGF-receptor phosphorylation: correlation with head and neck tumor progression. Cancer Res. 2008;68:10077–10085. doi: 10.1158/0008-5472.CAN-08-2910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lerner I, Hermano E, Zacharia E, Rodkin D, Bulvik R, Doviner V, Rubinstein A, Ishai-Michaeli R, Atzmon R, Sherman Y, Meirovitz A, Peretz T, Vlodavsky I, Elkin M. Heparanase powers a chronic inflammatory circuit that promotes colitis-associated tumorigenesis in mice. J Clin Invest. 2011;121:1709–1721. doi: 10.1172/JCI43792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fux L, Feibish N, Cohen-Kaplan V, Gingis-Velitski S, Feld S, Geffen C, Vlodavsky I, Ilan N. Structure-function approach identifies a COOH-terminal domain that mediates heparanase signaling. Cancer Res. 2009;69:1758–1767. doi: 10.1158/0008-5472.CAN-08-1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Velling T, Nilsson S, Stefansson A, Johansson S. ß1-integrins induce phosphorylation of Akt on serine 473 independently of focal adhesion kinase and Src family kinases. EMBO Rep. 2004;5:901–905. doi: 10.1038/sj.embor.7400234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Velling T, Stefansson A, Johansson S. EGFR and β1 integrins utilize different signaling pathways to activate Akt. Exp Cell Res. 2008;314:309–316. doi: 10.1016/j.yexcr.2007.08.018. [DOI] [PubMed] [Google Scholar]
- 44.Fux L, Ilan N, Sanderson R, Vlodavsky I. Heparanase: busy at the cell surface. Trends Biochem Sci. 2009;34:511–519. doi: 10.1016/j.tibs.2009.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Parsons S, Parsons J. Src family kinases, key regulators of signal transduction. Oncogene. 2004;23:7906–7909. doi: 10.1038/sj.onc.1208160. [DOI] [PubMed] [Google Scholar]
- 46.Kükenshöner T, Schmit N, Bouda E, Sha F, Pojer F, Koide A, Seeliger M, Koide S, Hantschel O. Selective targeting of SH2 domain-phosphotyrosine interactions of Src family tyrosine kinases with monobodies. J Mol Biol. 2017;429:1364–1380. doi: 10.1016/j.jmb.2017.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Boyango I, Barash U, Naroditsky I, Li J, Hammond E, Ilan N, Vlodavsky I. Heparanase co-operates with Ras to drive breast and skin tumorigenesis. Cancer Res. 2014;74:4504–4514. doi: 10.1158/0008-5472.CAN-13-2962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Batool T, Fang J, Barash U, Moustakas A, Vlodavsky I, Li J. Overexpression of heparanase attenuated TGF-β-stimulated signaling in tumor cells. FEBS Open Bio. 2017;7:405–413. doi: 10.1002/2211-5463.12190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Leitinger B, Hogg N. The involvement of lipid rafts in the regulation of integrin function. J Cell Sci. 2002;115:963–972. doi: 10.1242/jcs.115.5.963. [DOI] [PubMed] [Google Scholar]
- 50.Xia H, Nho R, Kahm J, Kleidon J, Henke C. Focal adhesion kinase is upstream of phosphatidylinositol 3-kinase/Akt in regulating fibroblast survival in response to contraction of type I collagen matrices via a β1 integrin viability signaling pathway. J Biol Chem. 2004;279:33024–33034. doi: 10.1074/jbc.M313265200. [DOI] [PubMed] [Google Scholar]
- 51.Astier A, Avraham H, Manie S, Groopman J, Canty T, Avraham S, Freedman A. The related adhesion focal tyrosine kinase is tyrosine-phosphorylated after beta1-integrin stimulation in B cells and binds to p130cas. J Biol Chem. 1997;272:228–232. doi: 10.1074/jbc.272.1.228. [DOI] [PubMed] [Google Scholar]
- 52.Salazar E, Rozengurt E. Src family kinases are required for integrin-mediated but not for G protein-coupled receptor stimulation of focal adhesion kinase autophosphorylation at Tyr-397. J Biol Chem. 2001;276:17788–17795. doi: 10.1074/jbc.M100984200. [DOI] [PubMed] [Google Scholar]
- 53.Chan T, Rittenhouse S, Tsichlis P. AKT/PKB and other D3 phosphoinositide-regulated kinases: kinase activation by phosphoinositide-dependent phosphorylation. Annu Rev Biochem. 1999;68:965–1014. doi: 10.1146/annurev.biochem.68.1.965. [DOI] [PubMed] [Google Scholar]
- 54.Naser R, Aldehaiman A, Díaz-Galicia E, Arold S. Endogenous control mechanisms of FAK and PYK2 and their relevance to cancer development. Cancers. 2018;10:196. doi: 10.3390/cancers10060196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Beinke S, Phee H, Clingan J, Schlessinger J, Matloubian M, Weiss A. Proline-rich tyrosine kinase-2 is critical for CD8 T-cell short-lived effector fate. Proc Natl Acad Sci. 2010;107:16234–16239. doi: 10.1073/pnas.1011556107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gan X, Wang J, Su B, Wu D. Evidence for direct activation of mTORC2 kinase activity by phosphatidylinositol 3,4,5-trisphosphate. J Biol Chem. 2011;286:10998–11002. doi: 10.1074/jbc.M110.195016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zeller K, Idevall-Hagren O, Stefansson A, Velling T, Jackson S, Downward J, Tengholm A, Johansson S. PI3-kinase p110α mediates β1 integrin-induced Akt activation and membrane protrusion during cell attachment and initial spreading. Cell Signal. 2010;22:1838–1848. doi: 10.1016/j.cellsig.2010.07.011. [DOI] [PubMed] [Google Scholar]
- 58.Hemmings B, Restuccia D. PI3K-PKB/Akt Pathway. Cold Spring Harb Perspect Biol. 2012;4:a011189. doi: 10.1101/cshperspect.a011189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu X, Cohen J. The role of PI3K/Akt in human herpesvirus infection: From the bench to the bedside. Virology. 2015;479–480:568–577. doi: 10.1016/j.virol.2015.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dan H, Cooper M, Cogswell P, Duncan J, Ting J, Baldwin A. Akt-dependent regulation of NF-κB is controlled by mTOR and Raptor in association with IKK. Genes Dev. 2008;22:1490–1500. doi: 10.1101/gad.1662308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Agelidis A, Hadigal S, Jaishankar D, Shukla D. Viral activation of heparanase drives pathogenesis of Herpes Simplex Virus-1. Cell Rep. 2017;20:439–450. doi: 10.1016/j.celrep.2017.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Koch S, Claesson-Welsh L. Signal transduction by vascular endothelial growth factor receptors. Cold Spring Harb Perspect Med. 2012;2:a006502. doi: 10.1101/cshperspect.a006502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Elkin M, Ilan N, Ishai-Michaeli R, Friedmann Y, Papo O, Pecker I, Vlodavsky I. Heparanase as mediator of angiogenesis: mode of action. FASEB J. 2001;15:1661–1663. doi: 10.1096/fj.00-0895fje. [DOI] [PubMed] [Google Scholar]
- 64.Kalita J, Chauhan P, Mani V, Bhoi S, Misra U. VEGF and its receptors in dengue virus infection. J Med Virol. 2015;87:1449–1455. doi: 10.1002/jmv.24205. [DOI] [PubMed] [Google Scholar]
- 65.Li F, Cui J. Human telomerase reverse transcriptase regulates vascular endothelial growth factor expression via human papillomavirus oncogene E7 in HPV-18-positive cervical cancer cells. Med Oncol. 2015;32:199. doi: 10.1007/s12032-015-0649-0. [DOI] [PubMed] [Google Scholar]
- 66.Fleming S, Wise L, Mercer A. Molecular genetic analysis of orf virus: a poxvirus that has adapted to skin. Viruses. 2015;7:1505–1539. doi: 10.3390/v7031505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, Zhang L, Fan G, Xu J, Gu X, Cheng Z, Yu T, Xia J, Wei Y, Wu W, Xie X, Yin W, Li H, Liu M, Xiao Y, Gao H, Guo L, Xie J, Wang G, Jiang R, Gao Z, Jin Q, Wang J, Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Thickett D, Armstrong L, Christie S, Millar A. Vascular endothelial growth factor may contribute to increased vascular permeability in acute respiratory distress syndrome. Am J Respir Crit Care Med. 2001;164:1601–1605. doi: 10.1164/ajrccm.164.9.2011071. [DOI] [PubMed] [Google Scholar]
- 69.Lyttle D, Fraser K, Fleming S, Mercer A, Robinson A. Homologs of vascular endothelial growth factor are encoded by the poxvirus orf virus. J Virol. 1994;68:84–92. doi: 10.1128/jvi.68.1.84-92.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Alkharsah K. VEGF upregulation in viral infections and its possible therapeutic implications. Int J Mol Sci. 2018;19:1642. doi: 10.3390/ijms19061642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wuest T, Carr D. VEGF-A expression by HSV-1-infected cells drives corneal lymphangiogenesis. J Exp Med. 2010;207:101–115. doi: 10.1084/jem.20091385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Del Moral-Hernández O, Martínez-Hernández N, Mosso-Pani M, Hernández-Sotelo D, Illades-Aguiar B, Flores-Alfaro E, Antonio-Vejar V, Leyva-Vázquez M. Association DENV1 and DENV2 infection with high serum levels of soluble thrombomodulin and VEGF in patients with dengue fever and dengue hemorrhagic fever. Int J Clin Exp Med. 2014;7:370–378. [PMC free article] [PubMed] [Google Scholar]
- 73.Kalogeropoulos D, Geka A, Malamos K, Kanari M, Kalogeropoulos C. New therapeutic perceptions in a patient with complicated Herpes Simplex Virus 1 keratitis: a case report and review of the literature. Am J Case Rep. 2017;18:1382–1389. doi: 10.12659/ajcr.906506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Baghban Rahimi S, Mohebbi A, Vakilzadeh G, Biglari P, Razeghi Jahromi S, Mohebi S, Shirian S, Gorji A, Ghaemi A. Enhancement of therapeutic DNA vaccine potency by melatonin through inhibiting VEGF expression and induction of antitumor immunity mediated by CD8+ T cells. Arch Virol. 2018;163:587–597. doi: 10.1007/s00705-017-3647-z. [DOI] [PubMed] [Google Scholar]
- 75.Argiris A, Bauman J, Ohr J, Gooding W, Heron D, Duvvuri U, Kubicek G, Posluszny D, Vassilakopoulou M, Kim S, Grandis J, Johnson J, Gibson M, Clump D, Flaherty J, Chiosea S, Branstetter B, Ferris R. Phase II randomized trial of radiation therapy, cetuximab, and pemetrexed with or without bevacizumab in patients with locally advanced head and neck cancer. Ann Oncol. 2016;27:1594–1600. doi: 10.1093/annonc/mdw204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cohen-Kaplan V, Naroditsky I, Zetser A, Ilan N, Vlodavsky I, Doweck I. Heparanase induces VEGF C and facilitates tumor lymphangiogenesis. Int J Cancer. 2008;123:2566–2573. doi: 10.1002/ijc.23898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zetser A, Bashenko Y, Edovitsky E, Levy-Adam F, Vlodavsky I, Ilan N. Heparanase induces vascular endothelial growth factor expression: correlation with p38 phosphorylation levels and Src activation. Cancer Res. 2006;66:1455–1463. doi: 10.1158/0008-5472.CAN-05-1811. [DOI] [PubMed] [Google Scholar]
- 78.Thomas S, Brugge J. Cellular functions regulated by Src family kinases. Annu Rev Cell Dev Biol. 1997;13:513–609. doi: 10.1146/annurev.cellbio.13.1.513. [DOI] [PubMed] [Google Scholar]
- 79.Iozzo R, Sanderson R. Proteoglycans in cancer biology, tumour microenvironment and angiogenesis. J Cell Mol Med. 2011;15:1013–1031. doi: 10.1111/j.1582-4934.2010.01236.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kadenhe-Chiweshe A, Papa J, McCrudden K, Frischer J, Bae J, Huang J, Fisher J, Lefkowitch J, Feirt N, Rudge J, Holash J, Yancopoulos G, Kandel J, Yamashiro D. Sustained VEGF blockade results in microenvironmental sequestration of VEGF by tumors and persistent VEGF receptor-2 activation. Mol Cancer Res. 2008;6:1–9. doi: 10.1158/1541-7786.MCR-07-0101. [DOI] [PubMed] [Google Scholar]
- 81.Purushothaman A, Chen L, Yang Y, Sanderson R. Heparanase stimulation of protease expression implicates it as a master regulator of the aggressive tumor phenotype in myeloma. J Biol Chem. 2008;283:32628–32636. doi: 10.1074/jbc.M806266200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Jung O, Trapp-Stamborski V, Purushothaman A, Jin H, Wang H, Sanderson R, Rapraeger A. Heparanase-induced shedding of syndecan-1/CD138 in myeloma and endothelial cells activates VEGFR2 and an invasive phenotype: prevention by novel synstatins. Oncogenesis. 2016;5:e202. doi: 10.1038/oncsis.2016.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Rapraeger A. Synstatin: a selective inhibitor of the syndecan-1-coupled IGF1R–αvβ3 integrin complex in tumorigenesis and angiogenesis. FEBS J. 2013;280:2207–2215. doi: 10.1111/febs.12160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wuest T, Zheng M, Efstathiou S, Halford W, Carr D. The herpes simplex virus-1 transactivator infected cell protein-4 drives VEGF-A dependent neovascularization. PLoS Pathog. 2011;7:e1002278. doi: 10.1371/journal.ppat.1002278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hadigal S, Koganti R, Yadavalli T, Agelidis A, Suryawanshi R, Shukla D. Heparanase-regulated syndecan-1 shedding facilitates Herpes Simplex Virus 1 egress. J Virol. 2020;94:e01672–e1719. doi: 10.1128/JVI.01672-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Koujah L, Suryawanshi R, Shukla D. Pathological processes activated by herpes simplex virus-1 (HSV-1) infection in the cornea. Cell Mol Life Sci. 2019;76:405–419. doi: 10.1007/s00018-018-2938-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Purushothaman A, Babitz S, Sanderson R. Heparanase enhances the insulin receptor signaling pathway to activate extracellular signal-regulated kinase in multiple myeloma. J Biol Chem. 2012;287:41288–41296. doi: 10.1074/jbc.M112.391417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Arkun Y, Yasemi M. Dynamics and control of the ERK signaling pathway: sensitivity, bistability, and oscillations. PLoS ONE. 2018;13:e0195513. doi: 10.1371/journal.pone.0195513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kolch W. Meaningful relationships: the regulation of the Ras/Raf/MEK/ERK pathway by protein interactions. Biochem J. 2000;351(pt 2):289–305. doi: 10.1042/0264-6021:3510289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wennerberg K, Rossman K, Der C. The Ras superfamily at a glance. J Cell Sci. 2005;118:843–846. doi: 10.1242/jcs.01660. [DOI] [PubMed] [Google Scholar]
- 91.Zheng K, Xiang Y, Wang X, Wang Q, Zhong M, Wang S, Wang X, Fan J, Kitazato K, Wang Y (2014) Epidermal growth factor receptor-PI3K signaling controls cofilin activity to facilitate Herpes Simplex Virus 1 entry into neuronal cells. mBio 5:e00958–e00913. 10.1128/mBio.00958-13 [DOI] [PMC free article] [PubMed]
- 92.Kew V, Yuan J, Meier J, Reeves M. Mitogen and stress activated kinases act co-operatively with CREB during the induction of Human Cytomegalovirus immediate-early gene expression from latency. PLoS Pathog. 2014;10:e1004195. doi: 10.1371/journal.ppat.1004195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Li X, Du S, Avey D, Li Y, Zhu F, Kuanga E. ORF45-mediated prolonged c-Fos accumulation accelerates viral transcription during the late stage of lytic replication of Kaposi's sarcoma-associated herpesvirus. J Virol. 2015;89:6895–6906. doi: 10.1128/JVI.00274-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bowser B, Alam S, Meyers C. Treatment of a human papillomavirus type 31b-positive cell line with benzo[a]pyrene increases viral titer through activation of the Erk1/2 signaling pathway. J Virol. 2011;85:4982–4992. doi: 10.1128/JVI.00133-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Xia L, Huang W, Tian D, Zhu H, Zhang Y, Hu H, Fan D, Nie Y, Wu K. Up-regulated FoxM1 expression induced by hepatitis B virus X protein promotes tumor metastasis and indicates poor prognosis in hepatitis B virus-related hepatocellular carcinoma. J Hepatol. 2012;57:600–612. doi: 10.1016/j.jhep.2012.04.020. [DOI] [PubMed] [Google Scholar]
- 96.Bonjardim C. Viral exploitation of the MEK/ERK pathway—a tale of vaccinia virus and other viruses. Virology. 2017;507:267–275. doi: 10.1016/j.virol.2016.12.011. [DOI] [PubMed] [Google Scholar]
- 97.Ludwig S, Wolff T, Ehrhardt C, Wurzer W, Reinhardt J, Planz O, Pleschka S. MEK inhibition impairs influenza B virus propagation without emergence of resistant variants. FEBS Lett. 2004;561:37–43. doi: 10.1016/S0014-5793(04)00108-5. [DOI] [PubMed] [Google Scholar]
- 98.Menzel N, Fischl W, Hueging K, Bankwitz D, Frentzen A, Haid S, Gentzsch J, Kaderali L, Bartenschlager R, Pietschmann T. MAP-Kinase regulated cytosolic phospholipase A2 activity is essential for production of infectious Hepatitis C Virus particles. PLoS Pathog. 2012;8:e1002829. doi: 10.1371/journal.ppat.1002829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Albarnaz J, De Oliveira L, Torres A, Palhares R, Casteluber M, Rodrigues C, Cardozo P, De Souza A, Pacca C, Ferreira P, Kroon E, Nogueira M, Bonjardim C. MEK/ERK activation plays a decisive role in yellow fever virus replication: implication as an antiviral therapeutic target. Antiviral Res. 2014;111:82–92. doi: 10.1016/j.antiviral.2014.09.004. [DOI] [PubMed] [Google Scholar]
- 100.Smith J, Stein D, Shum D, Fischer M, Radu C, Bhinder B, Djaballah H, Nelson J, Früh K, Hirsch A. Inhibition of dengue virus replication by a class of small-molecule compounds that antagonize dopamine receptor d4 and downstream mitogen-activated protein kinase signaling. J Virol. 2014;88:5533–5542. doi: 10.1128/JVI.00365-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Rubinfeld H, Cohen-Kaplan V, Nass D, Ilan N, Meisel S, Cohen Z, Hadani M, Vlodavsky I, Shimon I. Heparanase is highly expressed and regulates proliferation in GH-secreting pituitary tumor cells. Endocrinology. 2011;152:4562–4570. doi: 10.1210/en.2011-0273. [DOI] [PubMed] [Google Scholar]
- 102.Ramos-DeSimone N, Hahn-Dantona E, Sipley J, Nagase H, French D, Quigley J. Activation of matrix metalloproteinase-9 (MMP-9) via a converging plasmin/stromelysin-1 cascade enhances tumor cell invasion. J Biol Chem. 1999;274:13066–13076. doi: 10.1074/jbc.274.19.13066. [DOI] [PubMed] [Google Scholar]
- 103.Bendre M, Gaddy-Kurten D, Mon-Foote T, Akel N, Skinner R, Nicholas R, Suva L. Expression of interleukin 8 and not parathyroid hormone-related protein by human breast cancer cells correlates with bone metastasis in vivo. Cancer Res. 2002;62:5571–5579. [PubMed] [Google Scholar]
- 104.Purushothaman A, Uyama T, Kobayashi F, Yamada S, Sugahara K, Rapraeger A, Sanderson R. Heparanase-enhanced shedding of syndecan-1 by myeloma cells promotes endothelial invasion and angiogenesis. Blood. 2010;115:2449–2457. doi: 10.1182/blood-2009-07-234757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Derksen P, Keehnen R, Evers L, van Oers M, Spaargaren M, Pals S. Cell surface proteoglycan syndecan-1 mediates hepatocyte growth factor binding and promotes Met signaling in multiple myeloma. Blood. 2002;99:1405–1410. doi: 10.1182/blood.v99.4.1405. [DOI] [PubMed] [Google Scholar]
- 106.Holt R, Fagerli U, Baykov V, Rø T, Hov H, Waage A, Sundan A, Børset M. Hepatocyte growth factor promotes migration of human myeloma cells. Haematologica. 2008;93:619–622. doi: 10.3324/haematol.11867. [DOI] [PubMed] [Google Scholar]
- 107.Börset M, Hjorth-Hansen H, Seidel C, Sundan A, Waage A. Hepatocyte growth factor and its receptor c-met in multiple myeloma. Blood. 1996;88:3998–4004. doi: 10.1182/blood.v88.10.3998.bloodjournal88103998. [DOI] [PubMed] [Google Scholar]
- 108.Ramani V, Yang Y, Ren Y, Nan L, Sanderson R. Heparanase plays a dual role in driving hepatocyte growth factor (HGF) signaling by enhancing HGF expression and activity. J Biol Chem. 2011;286:6490–6499. doi: 10.1074/jbc.M110.183277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kitano H, Oda K, Funahashi A, Matsuoka Y. A comprehensive pathway map of epidermal growth factor receptor signaling. Mol Syst Biol. 2005;1(2005):0010. doi: 10.1038/msb4100014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Seshacharyulu P, Ponnusamy M, Haridas D, Jain M, Ganti A, Batra S. Targeting the EGFR signaling pathway in cancer therapy. Expert Opin Ther Targets. 2012;16:15–31. doi: 10.1517/14728222.2011.648617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Yarden Y, Citri A. EGF–ERBB signalling: towards the systems level. Nat Rev Mol Cell Biol. 2006;7:505–516. doi: 10.1038/nrm1962. [DOI] [PubMed] [Google Scholar]
- 112.Capuani F, Conte A, Argenzio E, Marchetti L, Priami C, Polo S, Di Fiore P, Sigismund S, Ciliberto A. Quantitative analysis reveals how EGFR activation and down regulation are coupled in normal but not in cancer cells. Nat Commun. 2015;6:7999. doi: 10.1038/ncomms8999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zheng K, Kitazato K, Wang Y. Viruses exploit the function of epidermal growth factor receptor. Rev Med Virol. 2014;24:274–286. doi: 10.1002/rmv.1796. [DOI] [PubMed] [Google Scholar]
- 114.Venkataraman T, Frieman M. The role of epidermal growth factor receptor (EGFR) signaling in SARS coronavirus-induced pulmonary fibrosis. Antiviral Res. 2017;143:142–150. doi: 10.1016/j.antiviral.2017.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Iwamoto M, Saso W, Sugiyama R, Ishii K, Ohki M, Nagamori S, Suzuki R, Aizaki H, Ryo A, Yun J, Park S, Ohtani N, Muramatsu M, Iwami S, Tanaka Y, Sureau C, Wakita T, Watashi K. Epidermal growth factor receptor is a host-entry cofactor triggering hepatitis B virus internalization. PNAS. 2019;116:8487–8492. doi: 10.1073/pnas.1811064116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Diao J, Pantua H, Ngu H, Komuves L, Diehl L, Schaefer G, Kapadia S. Hepatitis C Virus induces epidermal growth factor receptor activation via CD81 binding for viral internalization and entry. J Virol. 2012;86:10935–10949. doi: 10.1128/JVI.00750-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Chan G, Nogalski M, Yurochko A. Activation of EGFR on monocytes is required for human cytomegalovirus entry and mediates cellular motility. PNAS. 2009;106:22369–22374. doi: 10.1073/pnas.0908787106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Wang W, Wu J, Zhang X, Hao C, Zhao X, Jiao G, Shan X, Tai W, Yu G. Inhibition of Influenza A Virus infection by fucoidan targeting viral neuraminidase and cellular EGFR pathway. Sci Rep. 2017;7:40760. doi: 10.1038/srep40760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Eierhoff T, Hrincius E, Rescher U, Ludwig S, Ehrhardt C. The epidermal growth factor receptor (EGFR) promotes uptake of influenza A viruses (IAV) into host cells. PLoS Pathog. 2010;6:e1001099. doi: 10.1371/journal.ppat.1001099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Liu K, Gualano R, Hibbs M, Anderson G, Bozinovski S. Epidermal growth factor receptor signaling to Erk1/2 and STATs control the intensity of the epithelial inflammatory responses to rhinovirus infection. J Biol Chem. 2008;283:9977–9985. doi: 10.1074/jbc.M710257200. [DOI] [PubMed] [Google Scholar]
- 121.Monick M, Cameron K, Staber J, Powers L, Yarovinsky T, Koland J, Hunninghake G. Activation of the epidermal growth factor receptor by respiratory syncytial virus results in increased inflammation and delayed apoptosis. J Biol Chem. 2005;280:2147–2158. doi: 10.1074/jbc.M408745200. [DOI] [PubMed] [Google Scholar]
- 122.Kalinowski A, Ueki I, Min-Oo G, Ballon-Landa E, Knoff D, Galen B, Lanier L, Nadel J, Koff J. EGFR activation suppresses respiratory virus-induced IRF1-dependent CXCL10 production. Am J Physiol Lung Cell Mol Physiol. 2014;307:L186–L196. doi: 10.1152/ajplung.00368.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Song J, Tan Y, Li S, Zhang S, Wan L, Ji S, Zhou H, Zhou Z, Gong F. Gemcitabine-induced heparanase promotes aggressiveness of pancreatic cancer cells via activating EGFR signaling. Oncotarget. 2017;8:58417–58429. doi: 10.18632/oncotarget.16911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Zhang L, Sullivan P, Suyama J, Marchetti D. Epidermal growth factor-induced heparanase nucleolar localization augments DNA topoisomerase I activity in brain metastatic breast cancer. Mol Cancer Res. 2010;8:278–290. doi: 10.1158/1541-7786.MCR-09-0375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Frank D. STAT3 as a central mediator of neoplastic cellular transformation. Cancer Lett. 2007;251:199–210. doi: 10.1016/j.canlet.2006.10.017. [DOI] [PubMed] [Google Scholar]
- 126.Pardoll D, Jove R, Yu H. STATs in cancer inflammation and immunity: a leading role for STAT3. Nat Rev Cancer. 2009;9:798–809. doi: 10.1038/nrc2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Khamaysi I, Singh P, Nasser S, Awad H, Chowers Y, Sabo E, Hammond E, Gralnek I, Minkov I, Noseda A, Ilan N, Vlodavsky I, Abassi Z. The role of heparanase in the pathogenesis of acute pancreatitis: a potential therapeutic target. Sci Rep. 2017;7:715. doi: 10.1038/s41598-017-00715-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Wynn T, Chawla A, Pollard J. Macrophage biology in development, homeostasis and disease. Nature. 2013;496:445–455. doi: 10.1038/nature12034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Blumenreich M. The white blood cell and differential count. In: Walker HK, Hall WD, Hurst JW, editors. Clinical methods: the history, physical, and laboratory examinations. 3. Boston: Butterworths; 1990. [PubMed] [Google Scholar]
- 130.Gutter-Kapon L, Alishekevitz D, Shaked Y, Li J, Aronheim A, Ilan N, Vlodavsky I. Heparanase is required for activation and function of macrophages. PNAS. 2016;113:E7808–E7817. doi: 10.1073/pnas.1611380113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Bhattacharya U, Gutter-Kapon L, Kan T, Boyango I, Barash U, Yang S, Liu J, Gross-Cohen M, Sanderson R, Shaked Y, Ilan N, Vlodavsky I. Heparanase and chemotherapy synergize to drive macrophage activation and enhance tumor growth. Cancer Res. 2020;80:1–12. doi: 10.1158/0008-5472.CAN-19-1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Bode J, Ehlting C, Häussinger D. The macrophage response towards LPS and its control through the p38MAPK–STAT3 axis. Cell Signal. 2012;24:1185–1194. doi: 10.1016/j.cellsig.2012.01.018. [DOI] [PubMed] [Google Scholar]
- 133.Takeda K, Akira S. TLR signaling pathways. Semin Immunol. 2004;16:3–9. doi: 10.1016/j.smim.2003.10.003. [DOI] [PubMed] [Google Scholar]
- 134.Chen G, Wang D, Vikramadithyan R, Yagyu H, Saxena U, Pillarisetti S, Goldberg I. Inflammatory cytokines and fatty acids regulate endothelial cell heparanase expression. Biochem. 2004;43:4971–4977. doi: 10.1021/bi0356552. [DOI] [PubMed] [Google Scholar]
- 135.de Mestre A, Rao S, Hornby J, Soe-Htwe T, Khachigian L, Hulett M. Early growth response gene 1 (EGR1) regulates heparanase gene transcription in tumor cells. J Biol Chem. 2005;280:35136–35147. doi: 10.1074/jbc.M503414200. [DOI] [PubMed] [Google Scholar]
- 136.Subbaramaiah K, Yoshimatsu K, Scherl E, Das K, Glazier K, Golijanin D, Soslow R, Tanabe T, Naraba H, Dannenberg A. Microsomal prostaglandin E synthase-1 is overexpressed in inflammatory bowel disease. Evidence for involvement of the transcription factor Egr-1. J Biol Chem. 2004;279:12647–12658. doi: 10.1074/jbc.M312972200. [DOI] [PubMed] [Google Scholar]
- 137.Naparstek Y, Cohen I, Fuks Z, Vlodavsky I. Activated T lymphocytes produce a matrix-degrading heparan sulphate endoglycosidase. Nature. 1984;310:241–244. doi: 10.1038/310241a0. [DOI] [PubMed] [Google Scholar]
- 138.de Mestre A, Staykova M, Hornby J, Willenborg D, Hulett M. Expression of the heparan sulfate-degrading enzyme heparanase is induced in infiltrating CD4+ T cells in experimental autoimmune encephalomyelitis and regulated at the level of transcription by early growth response gene 1. J Leukoc Biol. 2007;82:1289–1300. doi: 10.1189/jlb.0507315. [DOI] [PubMed] [Google Scholar]
- 139.Gilat D, Hershkoviz R, Goldkorn I, Cahalon L, Korner G, Vlodavsky I, Lider O. Molecular behavior adapts to context: heparanase functions as an extracellular matrix-degrading enzyme or as a T cell adhesion molecule, depending on the local pH. J Exp Med. 1995;181:1929–1934. doi: 10.1084/jem.181.5.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Sotnikov I, Hershkoviz R, Grabovsky V, Ilan N, Cahalon L, Vlodavsky I, Alon R, Lider O. Enzymatically quiescent heparanase augments T cell interactions with VCAM-1 and extracellular matrix components under versatile dynamic contexts. J Immunol. 2004;172:5185–5193. doi: 10.4049/jimmunol.172.9.5185. [DOI] [PubMed] [Google Scholar]
- 141.Goldshmidt O, Zcharia E, Cohen M, Aingorn H, Cohen I, Nadav L, Katz B, Geiger B, Vlodavsky I. Heparanase mediates cell adhesion independent of its enzymatic activity. FASEB J. 2003;17:1015–1025. doi: 10.1096/fj.02-0773com. [DOI] [PubMed] [Google Scholar]
- 142.Matasar M, Zelenetz A. Overview of lymphoma diagnosis and management. Radiol Clin North Am. 2008;46:175–198. doi: 10.1016/j.rcl.2008.03.005. [DOI] [PubMed] [Google Scholar]
- 143.Münz C, Chijioke O (2017) Natural killer cells in herpesvirus infections. F1000Res 6:F1000 Faculty Rev-1231. 10.12688/f1000research.11197.1. [DOI] [PMC free article] [PubMed]
- 144.Mayes K, Elsayed Z, Alhazmi A, Waters M, Alkhatib S, Roberts M, Song C, Peterson K, Chan V, Ailaney N, Malapati P, Blevins T, Lisnić B, Dumur C, Landry J. BPTF inhibits NK cell activity and the abundance of natural cytotoxicity receptor co-ligands. Oncotarget. 2017;8:64344–64357. doi: 10.18632/oncotarget.17834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Koganti R, Yadavalli T, Shukla D. Current and emerging therapies for ocular Herpes Simplex Virus type-1 infections. Microorganisms. 2019;7:429. doi: 10.3390/microorganisms7100429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Sharthiya H, Seng C, Kuppevelt T, Tiwari V, Fornaro M. HSV-1 interaction to 3-O-sulfated heparan sulfate in mouse-derived DRG explant and profiles of inflammatory markers during virus infection. J Neurovirol. 2017;23:483–491. doi: 10.1007/s13365-017-0521-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Cagno V, Tseligka E, Jones S, Tapparel C. Heparan sulfate proteoglycans and viral attachment: true receptors or adaptation bias? Viruses. 2019;11:E596. doi: 10.3390/v11070596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Dong L, Wang X, Guo Y, Wu J, Li S, Yu P, Wang Z. HS N-sulfation and iduronic acids play an important role in the infection of respiratory syncytial virus in vitro. Eur Rev Med Pharmacol Sci. 2013;17:1864–1868. [PubMed] [Google Scholar]
- 149.Wassenaar T, Zou Y. 2019_nCoV/SARS-CoV-2: rapid classification of betacoronaviruses and identification of traditional Chinese medicine as potential origin of zoonotic coronaviruses. Lett Appl Microbiol. 2020;70:342–348. doi: 10.1111/lam.13285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Hoffmann M, Kleine-Weber H, Schroeder S, Krüger N, Herrler T, Erichsen S, Schiergens T, Herrler G, Wu N, Nitsche A, Müller M, Drosten C, Pöhlmann S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Milewska A, Zarebski M, Nowak P, Stozek K, Potempa J, Pyrc K. Human Coronavirus NL63 utilizes heparan sulfate proteoglycans for attachment to target cells. J Virol. 2014;88:13221–13230. doi: 10.1128/JVI.02078-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Lang J, Yang N, Deng J, Liu K, Yang P, Zhang G, Jiang C. Inhibition of SARS Pseudovirus cell entry by lactoferrin binding to heparan sulfate proteoglycans. PLoS ONE. 2011;6:e23710. doi: 10.1371/journal.pone.0023710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Hirshoren N, Bulvik R, Neuman T, Rubinstein A, Meirovitz A, Elkin M. Induction of heparanase by HPV E6 oncogene in head and neck squamous cell carcinoma. J Cell Mol Med. 2014;18:181–186. doi: 10.1111/jcmm.12179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Song Y, Hu B, Qu H, Wang L, Zhang Y, Tao J, Cui J. Novel 1, 3-N, O-Spiroheterocyclic compounds inhibit heparanase activity and enhance nedaplatin-induced cytotoxicity in cervical cancer cells. Oncotarget. 2016;7:36154–36167. doi: 10.18632/oncotarget.8959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Yan Q, Ma X, Shen C, Cao X, Feng N, Qin D, Zeng Y, Zhu J, Gao S, Lu C. Inhibition of Kaposi's sarcoma-associated herpesvirus lytic replication by HIV-1 Nef and cellular microRNA hsa-miR-1258. J Virol. 2014;88:4987–5000. doi: 10.1128/JVI.00025-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Zhang L, Sullivan P, Goodman J, Gunaratne P, Marchetti D. MicroRNA-1258 suppresses breast cancer brain metastasis by targeting heparanase. Cancer Res. 2011;71:645–654. doi: 10.1158/0008-5472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Tao Y, Wang Z, Zhou Y. Expression of heparanase in kidney of rats with respiratory syncytial virus nephropathy and its relationship with proteinurina. Sichuan Da Xue Xue Bao Yi Xue. 2014;45(212–215):224. [PubMed] [Google Scholar]
- 158.Hopkins J, Yadavalli T, Agelidis A, Shukla D. Host enzymes heparanase and cathepsin L promote Herpes Simplex Virus 2 release from cells. J Virol. 2018;92:e01179–e1218. doi: 10.1128/JVI.01179-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Hadigal S, Agelidis A, Karasneh G, Antoine T, Yakoub A, Ramani V, Djalilian A, Sanderson R, Shukla D. Heparanase is a host enzyme required for herpes simplex virus-1 release from cells. Nat Commun. 2015 doi: 10.1038/ncomms7985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Guo C, Zhu Z, Guo Y, Wang X, Yu P, Xiao S, Chen Y, Cao Y, Liu X. Heparanase upregulation contributes to porcine reproductive and respiratory syndrome virus release. J Virol. 2017 doi: 10.1128/JVI.00625-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Surviladze Z, Sterkand R, Ozbun M. Interaction of human papillomavirus type 16 particles with heparan sulfate and syndecan-1 molecules in the keratinocyte extracellular matrix plays an active role in infection. J Gen Virol. 2015;96:2232–2241. doi: 10.1099/vir.0.000147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Cui H, Tan Y, Österholm C, Zhang X, Hedin U, Vlodavsky I, Li J. Heparanase expression up-regulates platelet adhesion activity and thrombogenicity. Oncotarget. 2016;7:39486–39496. doi: 10.18632/oncotarget.8960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Parish C, Hulett FC, M Heparanase: a key enzyme involved in cell invasion. BBA Rev Cancer. 2001;1471:M99–M108. doi: 10.1016/S0304-419X(01)00017-8. [DOI] [PubMed] [Google Scholar]
- 164.McLaughlin-Drubin M, Munger K. Viruses associated with human cancer. Biochim Biophys Acta. 2008;1782:127–150. doi: 10.1016/j.bbadis.2007.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]