Abstract
Protein N-glycosylation on human milk proteins assists in protecting an infant’s health and functions among others as competitive inhibitors of pathogen binding and immunomodulators. Due to the individual uniqueness of each mother’s milk and the overall complexity and temporal changes of protein N-glycosylation, analysis of the human milk N-glycoproteome requires longitudinal personalized approaches, providing protein- and N-site-specific quantitative information. Here, we describe an automated platform using hydrophilic-interaction chromatography (HILIC)-based cartridges enabling the proteome-wide monitoring of intact N-glycopeptides using just a digest of 150 μg of breast milk protein. We were able to map around 1700 glycopeptides from 110 glycoproteins covering 191 glycosites, of which 43 sites have not been previously reported with experimental evidence. We next quantified 287 of these glycopeptides originating from 50 glycoproteins using a targeted proteomics approach. Although each glycoprotein, N-glycosylation site, and attached glycan revealed distinct dynamic changes, we did observe a few general trends. For instance, fucosylation, especially terminal fucosylation, increased across the lactation period. Building on the improved glycoproteomics approach outlined above, future studies are warranted to reveal the potential impact of the observed glycosylation microheterogeneity on the healthy development of infants.
Keywords: human milk, glycoprotein, milk proteins, glycopeptides, HILIC-based platform, mass spectrometry, N-glycosylation, N-glycopeptide enrichment, targeted N-glycopeptide quantification, lactation
Introduction
Human milk is a remarkable multifunctional fluid of the mammary gland, providing varying amounts of nutritional and non-nutritional, bioactive components to satisfy the specific demand of a newborn.1 Glycans form a major component of human milk, occurring both in free form as lactose and oligosaccharides,2 as well as conjugated to glycoproteins primarily through N- and/or O-linked glycosylation.3 Protein glycosylation is of special interest in milk due to its widespread functional capacity. For instance, glycosylated proteins are relevant to proteolytic susceptibility,4 as competitive inhibitors of pathogen binding5,6 and immunomodulators,5 all together working to protect the infant’s health.7 Like other components in human milk, protein glycosylation is quite diverse among individual mothers, and it is thought to also vary quite dynamically across lactation.8 To understand the bioactive effects of the continuing changes of glycosylation in human milk, it is necessary that protein glycosylation analysis is done both in a personalized manner and longitudinally.
The characterization of human milk glycosylation is challenging due to the large dynamic range of human milk proteins9 and the complexity of protein glycosylation. The first challenge is posed by the co-occurrence of highly abundant non-glycosylated proteins in human milk, such as β-casein that accounts for ∼30% of the proteins by weight,9 which diminishes the detectability of glycopeptides in a standard proteomics approach. Furthermore, the microheterogeneity introduced by harboring different types of glycans on the same protein glycosylation site further confounds the complexity of the sample and, additionally, the specific analysis of glycopeptides.10,11
The glycopeptide-centric analysis of human milk protein glycosylation is still rather in its infancy. Thus, further developing this analysis is essential for obtaining information on site-specific glycosylation. Recent studies have mainly focused on the mapping of N-glycosylation sites of enzymatic deglycosylated peptides12−14 or on enzymatically released N-glycans.15,16 To name a few, a recent study from Cao et al.12 investigated human colostrum and mature milk samples, and reported 68 and 58 N-glycoproteins and 111 and 96 N-glycosites, respectively, using a lectin-enrichment approach and subsequent identification of the deglycosylated peptides. Picariello et al.14 used hydrophilic-interaction chromatography (HILIC) glycopeptide enrichment and applied this to a milk sample collected on day 7 after parturition, also merely analyzing the deglycosylated peptides, identifying 63 N-glycosylated sites originating from 32 glycoproteins. Released N-glycans have revealed that a specific feature of human milk glycosylation is the high degree of abundant fucosylation, when compared with bovine milk15 or healthy human serum.17 For instance, Nwosu et al.15 compared human and bovine milk and found substantial differences in their fucosylated N-glycans. They reported that 75% of the N-glycans in human milk correspond to fucosylated glycans, while this number was only 31% in bovine milk. Moreover, human milk exhibited more glycoforms harboring multiple fucoses when compared to bovine milk. About 60% of the fucosylated N-glycans in human milk were reported to be multi-fucosylated, whereas this number was just 30% in bovine milk. Also Dallas et al.16 quantified the abundance of N-glycans in human milk and found that 84% of the total were fucosylated N-glycans. They further reported that fucosylated N-glycans in human milk harbor either one (36%), two (32%), three (14%) or more (2%) fucose moieties. This high degree of fucosylation in human milk is in sharp contrast to what has been observed in human serum. For instance, Yoshida et al.17 reported that the majority of N-glycans in human serum were glycans without fucose (around 800 pmol/μL), followed by glycans with one fucose (around 300 pmol/μL) and glycans with two fucoses (only around 0.6 pmol/μL).
Here, we report on a high-throughput method to profile the human milk N-glycoproteome, taking a glycopeptide-centric approach, with the aim to monitor changes occurring over the lactation period in a single individual donor. Therefore, we collected human milk samples at nine time points covering the early, transitional, and mature lactational stages. We enriched for intact N-glycopeptides using an automated HILIC-based setup, and identified the N-glycopeptides by higher energy C-trap dissociation (HCD)-triggered electron-transfer/higher-energy collision dissociation (EThcD). We also quantified many N-glycopeptides using scheduled single ion monitoring (SIM) and parallel reaction monitoring (PRM). Overall, our dataset provides a comprehensive view of the human milk N-glycoproteome. We identify and profile 191 glycosites on 110 glycoproteins, of which 43 sites on 32 proteins can be considered novel. Moreover, these 191 sites harbor in total 1697 different glycans, i.e., on average 8 per site. We could also quantify 287 glycopeptides originating from 50 glycoproteins across the lactational period, which allowed us to observe distinctive site-specific glycosylation profiles. From this comprehensive profiling, a picture emerges that the human milk glycoproteome is rather complex and dynamic, likely reflecting the changing demands of the newborn’s growing and developing immune system and gut microbiota.
Experimental Procedures
Human Milk Sample Collection
Human milk samples were collected longitudinally from one healthy donor in weeks 1, 2, 3, 4, 6, 8, 10, 12, and 16 (for a total of nine time points). Written informed consent was obtained before the collection of any samples. All samples used were donated to Danone Nutricia Research in accordance to the Helsinki Declaration II. Milk samples were collected from one breast, as a complete breast expression, between the hours of 9 and 11 AM, taking the breast that had not been used for feeding within a 2 h window. Immediately after pumping, the samples were transferred to a 2 mL Eppendorf tube containing protease inhibitors (cOmplete Protease Inhibitor Cocktail Tablets from Roche) and were then frozen at −20 °C. The samples were transferred to the laboratory on dry ice and stored at −80 °C until thawed for analysis.
Experimental Design and Statistical Rationale
For all milk samples, three technical replicate analyses were performed. The mean ± standard deviation of the three technical replicates was calculated, and the proportion was calculated using the values of the mean of three technical replicates.
Chemicals and Materials
Unless otherwise specified, all chemicals and reagents were obtained from Sigma-Aldrich (Steinheim, Germany). Lys-C was obtained from Wako (Neuss, Germany). The Oasis PRiME HLB plate was purchased from Waters (Etten-Leur, the Netherlands). Formic acid (FA) was purchased from Merck (Darmstadt, Germany). Acetonitrile (ACN) was purchased from Biosolve (Valkenswaard, the Netherlands). The Pierce peptide retention time calibration (PRTC) mixture was obtained from Thermo Fisher Scientific (Bremen, Germany). Milli-Q was produced by an in-house system (Millipore, Billerica, MA).
Milk Serum Separation and Protein Digestion
For each time point, an aliquot of 200 μL of a thawed whole milk sample was centrifuged at 1000g for 1 h at 4 °C, whereafter the upper fat layer was discarded. The lower skimmed milk was further ultracentrifuged at 150 000g for 1 h at 4 °C to remove the insoluble pellet, and the supernatant milk serum was retained. The protein concentration was estimated by measuring the absorbance at 280 nm on a Nanodrop spectrophotometer (Nanodrop 2000, Thermo Scientific). Up to a final concentration of 1% w/v sodium deoxycholate (SDC), 100 mM Tris–HCl (pH 8.5), 5 mM tris(2-carboxyethyl)phosphine hydrochloride (TCEP), and 30 mM chloroacetamide (CAA) were added to the milk serum. Trypsin and Lys-C were added at ratios of 1:50 and 1:100 (w/w), respectively. Digestion was performed overnight at 37 °C. The next day, SDC was removed via acid precipitation (0.5% trifluoroacetic acid (TFA)), and the final peptide concentration was estimated by measuring the absorbance at 280 nm on a Nanodrop spectrophotometer (Nanodrop 2000, Thermo Scientific).18 The peptides were desalted using an Oasis PRiME HLB plate and then dried and stored at −80 °C.
Automated HILIC-Based Glycopeptide Enrichment
The HILIC-based glycopeptide enrichment was performed automatically with triplicates for each milk serum sample using an AssayMAP Bravo robot (Agilent technologies) coupled with HILIC-based cartridges (GlykoPrep APTS Cleanup Module, ProZyme, CA). The HILIC-based columns were first washed with 50 μL of 1% FA and equilibrated with 50 μL of loading buffer (80% ACN/0.5% TFA). The peptide digests (150 μg) were reconstituted with 50 μL of loading buffer and loaded onto the column. The cartridges were washed with 50 μL of loading buffer, and the glycopeptides were step-eluted with 75% ACN/0.5% TFA, 70% ACN/0.5% TFA, 65% ACN/0.5% TFA, and 0.5% FA. These samples were dried down and stored at −80 °C until subjected to liquid chromatography–tandem mass spectrometry (LC–MS/MS).
Full Proteome Analysis of Human Milk
To estimate protein abundances in the human milk proteome, 800 ng of the mixture of nonenriched tryptic peptides were analyzed using an Agilent 1290 Infinity high-performance liquid chromatography (HPLC) system (Agilent Technologies, Waldbronn, Germany) coupled on-line to a Q Exactive HF mass spectrometer (Thermo Fisher Scientific, Bremen, Germany). The peptides were first trapped using a 100 μm inner diameter 2 cm trap column (in-house packed with ReproSil-Pur C18-AQ, 3 μm) (Dr. Maisch GmbH, Ammerbuch-Entringen, Germany) coupled to a 50 m inner diameter 50 cm analytical column (in-house packed with Poroshell 120 EC-C18, 2.7 μm) (Agilent Technologies, Amstelveen, The Netherlands). The mobile-phase solvent A consisted of 0.1% FA in water, and the mobile-phase solvent B consisted of 0.1% FA in ACN. Trapping was performed at a flow rate of 5 μL/min for 5 min with 0% B, and peptides were eluted using a passively split flow of 300 nL/min for 170 min with 10–36% B over 155 min, 36–100% B over 3 min, 100% B for 1 min, 100–0% B over 1 min, and finally held at 0% B for 10 min. Peptides were ionized using a spray voltage of 1.9 kV and a heated capillary. The mass spectrometer was set to acquire full-scan MS spectra (375–1600 m/z) for a maximum injection time of 20 ms at a mass resolution of 60 000 and an automated gain control (AGC) target value of 3 × 106. Up to 15 of the most intense precursor ions were selected for tandem mass spectrometry (MS/MS). HCD MS/MS (200–2000 m/z) acquisition was performed in the HCD cell, with the readout in the Orbitrap mass analyzer at a resolution of 30 000 (isolation window of 1.4 Th) and an AGC target value of 1 × 105 or a maximum injection time of 50 ms with a normalized collision energy of 27%.
Raw shotgun LC–MS/MS data files were searched using a processing workflow in Proteome Discoverer (version 2.2, Thermo Scientific) using the Mascot search engine (version 2.6.1) against the Swiss-Prot database (release date: Feb 2017, 20 172 entries, taxonomy: Homo sapiens) using fixed Cys carbamidomethylation and variable Met oxidation of peptides as search variables. Trypsin was chosen for cleavage specificity with a maximum of two missed cleavages allowed. The searches were performed using a precursor mass tolerance of 10 ppm and a fragment mass tolerance of 0.05 Da, followed by data filtering using Percolator, resulting in a 1% false discovery rate (FDR). Only peptide-to-spectrum matches (PSMs) with Mascot score >20 were accepted. The full proteome search result was then used as a focused database for the identification of glycopeptides (1259 entries). For label-free quantification, the node called minora feature detector was used with high PSM confidence, a minimum of five nonzero points in a chromatographic trace, a minimum number of two isotopes, and a maximum retention time (RT) difference of 0.2 min for isotope peaks. The consensus workflow in Proteome Discoverer was used to open the search results and enable retention time (RT) alignment with a maximum RT shift of 5 min and mass tolerance of 10 ppm to match the precursor between runs. Label-free quantification of peptides was performed using the intensities of the extracted ion chromatogram (XIC). Protein intensity was determined by the average intensity of unique peptides. Protein abundance was estimated by taking the proportion of the protein intensity for each protein to the total protein intensity. To relatively compare the proportional change in each time point, the protein abundance in week 1 was defined as standard, giving an adjustment factor of 1. Other adjustment factors of proteins in other weeks were calculated using their relative abundances normalized to the measured value for that protein in week 1.
Identification of Human Milk Glycopeptides
All peptides from the automated HILIC-based glycopeptide enrichment were separated and analyzed using the same HPLC system as that used for the global proteome analysis, albeit now coupled on-line to an Orbitrap Fusion Lumos mass spectrometer (Thermo Fisher Scientific, Bremen, Germany) using a 90 min gradient, as follows: 0–5 min, 100% solvent A; 13–44% solvent B for 65 min; 44–100% solvent B for 5 min; 100% solvent B for 5 min; and 100% solvent A for 15 min. Peptides were ionized using a 2.0 kV spray voltage. For the MS scan, the mass range was set from 350 to 1800 m/z for a maximum injection time of 50 ms at a mass resolution of 60 000 and an AGC target value of 4 × 105 in the Orbitrap mass analyzer. The dynamic exclusion was set to 30 s for an exclusion window of 10 ppm with a cycle time of 3 s. Charge-state screening was enabled, and precursors with 2+ to 8+ charge states and intensities >1 × 105 were selected for tandem mass spectrometry (MS/MS). HCD MS/MS (120–2100 m/z) acquisition was performed in the HCD cell, with the readout in the Orbitrap mass analyzer at a resolution of 30 000 (isolation window of 1.6 Th) and an AGC target value of 5 × 104 or a maximum injection time of 75 ms with a normalized collision energy of 30%. If at least two out of three oxonium ions of glycopeptides (138.0545+, 204.0687+, or 366.1396+) were observed, EThcD MS/MS on the same precursor was triggered (isolation window of 1.6 Th) and fragment ions (120–2100 m/z) were analyzed in the Orbitrap mass analyzer at a resolution of 30 000, AGC target value of 2 × 105, or a maximum injection time of 250 ms with activation of electron-transfer dissociation (ETD) and supplemental activation with a normalized collision energy of 27%. To obtain MS/MS scans from more various precursors, we made reference samples with mixed glycopeptides from weeks 1, 4, 10 (ref (1)), weeks 2, 6, 12 (ref (2)), and weeks 3, 8, 16 (ref (3)) and measured each reference sample three times using the same LC–MS/MS method for glycopeptide identification, except that we applied various dynamic exclusion times, 30, 60, and 180 s, respectively.
All raw files obtained for the glycopeptide identification, including those of the reference samples, were processed in Proteome Discoverer (version 2.2, Thermo Scientific) using the Byonic node (Protein Metrics Inc., version 3.2.0) searching against a targeted milk protein database (1259 entries) based on our own data from the global milk proteome analysis using the following search parameters: trypsin digestion with a maximum of two missed cleavages, precursor ion mass tolerance, 10 ppm; fragmentation type, both HCD and EThcD; fragment mass tolerance, 20 ppm; carbamidomethylation of cysteines as a fixed modification; variable modifications: methionine oxidation. For glycan analysis, we used a Byonic database of 182 glycans with no multiple fucoses, whereby we added manually several reported glycan compositions with multiple fucoses (Table S1). The maximum number of precursors per scan was set to one and the FDR as 1%. Only PSMs matched to EThcD spectra with non-negligible error probabilities |log Prob| >4.0 and Byonic score ≥200 were accepted. The MS1 feature of high-confidence PSMs was captured by the node called minora feature detector, and the precursor was matched between runs by enabling retention time (RT) alignment with a maximum RT shift of 5 min and mass tolerance of 10 ppm.
Quantification of Human Milk Glycopeptides
First, we built a library in Skyline19 (Skyline-daily, version 4.2.1.18305) based on the m/z and retention time of each identified glycopeptide precursor and its HCD fragmentation pattern. Since the used version of Skyline was only able to accommodate b and y ions, we manually added oxonium ions and Y ions (peptide backbone ions carrying a glycan fragment from the glycosidic bond cleavage). The targeted peptides were selected based on their chromatographic trace and intensity. Due to the lack of appropriate stable isotope-labeled glycopeptide standards, we spiked PRTC peptides equally in all samples, which helped in monitoring the potential retention time shifts and variability between MS runs. Around 10% of the enriched glycopeptides and 50 fmol of the PRTC mixture were analyzed by LC–MS/MS with the same HPLC system and mass spectrometer as described above for the identification of the glycopeptides, except that MS and MS/MS scans were acquired in SIM and PRM modes, respectively. For MS scans, the m/z range was set from 350 to 2000 with a resolution of 30 000 using an AGC setting of 5 × 104, maximum IT of 54 ms, one microscan, a 1.6 Th isolation window, 27% normalized collision energy, and a 3 min retention time window. For MS/MS scans, the m/z range was set from 120 to 3000 with a resolution of 30 000 using an AGC setting of 5 × 104, maximum IT of 54 ms, one microscan, a 1.6 Th isolation window, 30% normalized collision energy, and a 3 min retention time window.
Further data analysis was done in Skyline as well. Skyline initially scanned for up to five isotopes of the precursors and all b and y fragment ions, oxonium ions, and Y ions for each peptide. Each peak was manually assessed, whereafter ions with interferences were removed. The retention time and rank of fragment ions were checked. If the fragment ions were co-eluted and the rank were the same as recorded in the above-mentioned library, the chromatographic peak of the precursor was undoubtedly pinpointed and the abundance of the glycopeptides was represented by the max intensity of the top three isotopes of the precursors. The abundances of the PRTC peptides were extracted to determine the coefficient of variation (CV). To relatively quantify the abundance of each glycopeptide over the lactation period, the abundance was normalized using the glycoprotein abundances obtained in our global human milk proteome analysis of the same samples.
Results and Discussion
Human Milk Sample Collection and Experimental Workflow
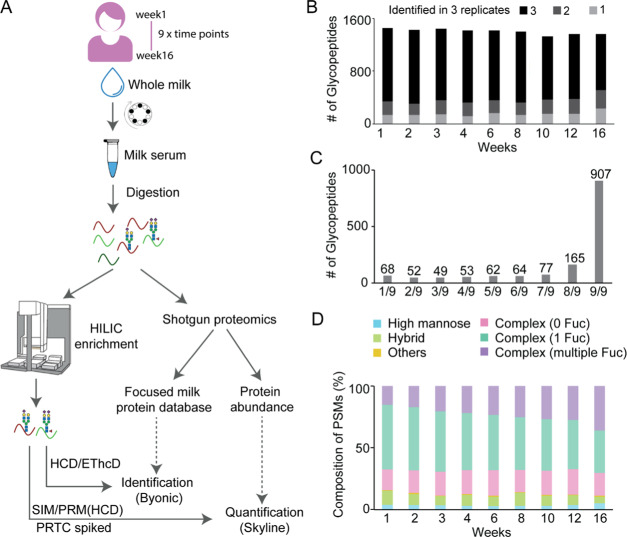
Here, we aimed to extensively characterize the temporal changes in the N-glycoproteome of a single mother’s breast milk during the course of a lactation period. The overall experimental workflow is depicted in Figure 1A and further detailed in the Experimental Procedures. In brief, we collected milk samples longitudinally from the same mother across early, transitional, and mature lactational stages, i.e., at weeks 1, 2, 3, 4, 6, 8, 10, 12, and 16. Following ultracentrifugation, the obtained milk serum was digested with LysC and trypsin, whereafter the total peptide concentration was estimated. First, abundances of the human milk proteins across the lactation period were determined by label-free standard shotgun proteomics approaches. In parallel, N-glycopeptides present in human milk were selectively enriched using HILIC-based cartridges implemented on an automated 96-well plate AssayMAP Bravo robot. The resulting glycopeptides were subsequently analyzed by LC–MS/MS, using HCD-triggered EThcD as the fragmentation method, whereafter the spectra were searched and identified using the Byonic node in Proteome Discoverer. A dedicated human milk protein database was made based on the 1259 proteins identified in the shotgun proteomics experiments. A selected set of human milk N-glycopeptides were quantified in more detail using a targeted assay with scheduled SIM and PRM, using a spiked-in PRTC peptide standard, to correct for the variability between runs and potential LC retention time shifts. These data were further quantitatively analyzed using Skyline. The intensity of each glycopeptide was normalized to the intensity of its protein of origin, as measured in the shotgun quantification of the same human milk sample.
Figure 1.
Experimental overview of the human milk glycopeptide-centric proteome analysis. (A) Overview of experimental steps taken. Human milk samples were collected longitudinally from the same individual across the early, transitional, and mature lactational stages, at weeks 1, 2, 3, 4, 6, 8, 10, 12, and 16. By centrifugation and ultracentrifugation, the separated milk serum was subsequently tryptic-digested, whereafter the total peptide concentration was determined. Milk proteins and their abundances across lactation were measured by standard shotgun proteomics approaches. N-Glycopeptides present in human milk were enriched using HILIC implemented on an automated AssayMAP Bravo platform. The resulting glycopeptides were then analyzed in HCD-triggered EThcD and searched by Byonic. A dedicated human milk protein database was used based on the proteomics data from the shotgun identification. N-Glycopeptides were next quantified by a targeted assay with scheduled SIM and PRM with spiked PRTC peptides to supervise the variability between runs and retention time shifts. These data were quantitatively analyzed using Skyline. The intensity of the glycopeptide was normalized to the intensity of its protein of origin, as measured in the shotgun quantification. (B) Total numbers of N-glycopeptides found in one, two, or three technical replicates. About 90% of the glycopeptides were identified in at least two replicates at all time points. Week 16 revealed somewhat lower reproducibility, albeit still 83%. (C) Total number of N-glycopeptides detected in at least two out of three technical replicates (1497 glycopeptides) found at all nine time points. Only 4.5% (68) were solely found at one time point, and the majority 60.6% (907) were detected at all time points, indicating the ability to reproducibly enrich from the different milk samples. (D) General composition of the observed glycoforms, based on the detected glycopeptide PSMs. The observed glycoforms were grouped into high mannose, hybrid, and complex types, whereby the complex type was further divided into subgroups without fucose, with one fucose, and with multiple fucoses, and others. See Table S5 for details about the glycan identifiers. The composition of complex with one fucose was observed to gradually decrease, while the composition of complex with multiple fucoses was found to relatively increase from early to mature lactation.
Mapping the Human Milk N-Glycoproteome across Lactation
To retain only the most confident glycopeptide identifications, we only accepted PSMs matched to EThcD spectra with strict thresholds, namely, a non-negligible error probability |log Prob| >4.0 and a Byonic score ≥200. In general, a high Bionic score improves accuracy but at the expense of lowering the number of IDs. We selected cutoff filters based on previous work by Lee et al.20 who investigated the relationship between the Byonic score and the accuracy of the IDs, whereby it was concluded that a Byonic score >200 was a stringent but good cutoff. For the log probability cutoff, we selected a value >4, as a value of >1 results in an estimated 0.33% FDR for glyco PSMs.21 After applying this stringent cutoff, in total, 22 614 PSMs could be identified (Table S2), resulting in 1697 unique intact N-glycopeptides (Table S3) originating from 110 different human milk glycoproteins. The identified glycoproteins spanned a range in abundance very similar to those observed in the full human milk proteome analysis (Figure S1). As a quality check, we next assessed the reproducibility in the automated enrichment of glycopeptides among all technical replicates (Figure 1B). About 90% of all glycopeptides were identified in at least two out of three technical replicates at all time points, whereby only week 16 revealed a somewhat lower reproducibility, albeit still around 83%, revealing the robustness and reproducibility of our automated glycopeptide enrichment.
We next inspected the 1497 glycopeptides, which were found in at least two out of three technical replicates among the nine different time points (Figure 1C). Only 4.5% (68) of these were uniquely found at one time point, and the majority 61% (907) were detected at all time points. Thus, qualitatively, the glycopeptide variety is rather constant over the lactation period. Following this, we evaluated the nature of the glycans identified based on the detected PSMs (Figure 1D). The observed glycoforms were grouped into high mannose, hybrid, and complex types, whereby the complex type was further divided into subgroups without fucose, with one fucose, and with multiple fucoses. The detailed compositions of the glycans can be found in Table S5. The majority of the identified glycans (67–70% of the PSMs) at each time point consisted of complex N-glycans with none, one, or multiple fucoses. This is in line with previous results for human milk analysis focused on released N-glycans.15,16 Among the complex N-glycans, we noticed that the glycans carrying one fucose gradually decreased, while those harboring multiple fucose moieties increased in time from early to mature lactation, again consistent with previous findings based on studies focused on released N-glycans.22
Having established a global picture of the glycan distribution as occurring on glycoproteins in human milk, we used these data to make a rough comparison to the global distribution of glycans in human serum, taking serum data reported earlier23 (Figure S2). This comparison revealed, not surprisingly, that global glycoproteome compositions in human milk and serum are quite distinct. In particular, hybrid structures and complex glycans harboring multiple fucoses seem to be relatively more abundant in human milk versus serum.
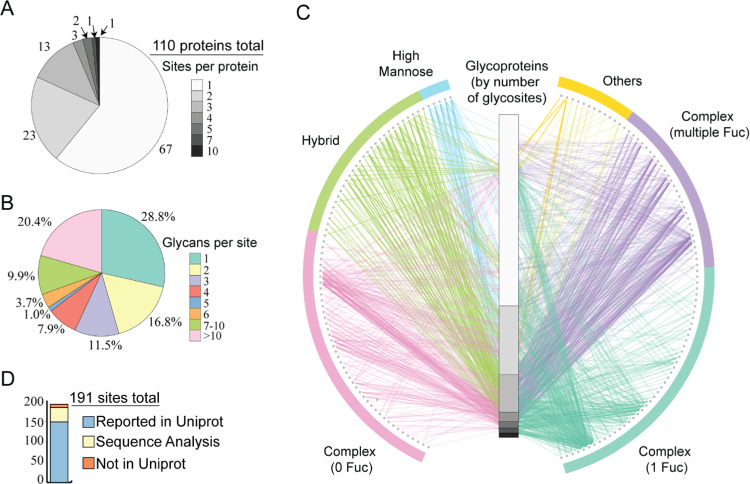
Since we detected the majority of glycopeptides at all investigated time points, we combined all data to get a comprehensive picture of the human milk N-glycoproteome. We first counted how many glycosites we identified for each glycoprotein (Figure 2A). The number of glycosites per glycoprotein varied from 1 to 10 (Table S4). In total, 191 glycosites were identified on cumulatively 110 glycoproteins. These proteins together harbor 546 N-glycosylation sites annotated in Uniprot by either experimental evidence or sequon prediction. Of all glycoproteins, 67 were detected with 1 glycosite, 23 with 2 glycosites, 13 with 3 glycosites, 3 with 4 glycosites, 2 with 5 glycosites, 1 with 7 glycosites, and 1 with 10 glycosites. The last one is the glycoprotein tenascin (TNC), which is known to be involved in the protection against viral infections, signifying the importance of glycans in host defense.24 We next investigated the degree of glycan microheterogeneity observed on each of the 191 glycosites (Figure 2B and Table S4). Overall, 72% of the glycosites were identified with more than 1 glycoform, and 20% of the glycosites were even harboring more than 10 glycoforms. This clearly indicates the high degree of glycan microheterogeneity observed on human milk glycoproteins. To further visualize the data, we adapted a glycoprotein–glycan network diagram21 (Figure 2C), in which the variety of glycans (outer nodes, 160 in total, Table S5) are connected to their glycoproteins of origin (inner column, organized by the number of glycosites, 110 in total). Again, the most eye-catching feature is the high prevalence of complex glycans harboring multiple fucoses. This network diagram also indicates that the complex glycans are found on nearly all glycoproteins, while the majority of high mannose and hybrid glycans occur mostly on glycoproteins harboring multiple glycosylation sites.
Figure 2.
Characteristics of the glycoproteins, glycopeptides, and glycosites observed in the human milk N-glycoproteome. (A) Distribution of the number of glycosites observed on each of the 110 identified glycoproteins. (B) Distribution of the number of different glycans observed on each glycosite. (C) Glycoprotein–glycan network map displaying what sort of glycans (outer circle, 160 total) modify which proteins (inner bar, 110 total). Glycoproteins are sorted as in (A), by the number of observed glycosites (note the same scale as the distribution in (A)). The observed glycans were, therefore, organized using a standard classification, and edges are colored by the glycan node from which they originate. See Table S5 for more details about the glycan identifiers. (D) Approximately 22.5% (43) of the identified glycosites are not yet annotated in UniProt. Of these, 35 could be hypothesized to be genuine N-glycosylation sites based on the presence of the N-X-S/T sequon, whereas 8 were not annotated at all.
Identification of Novel N-Glycosylation Sites
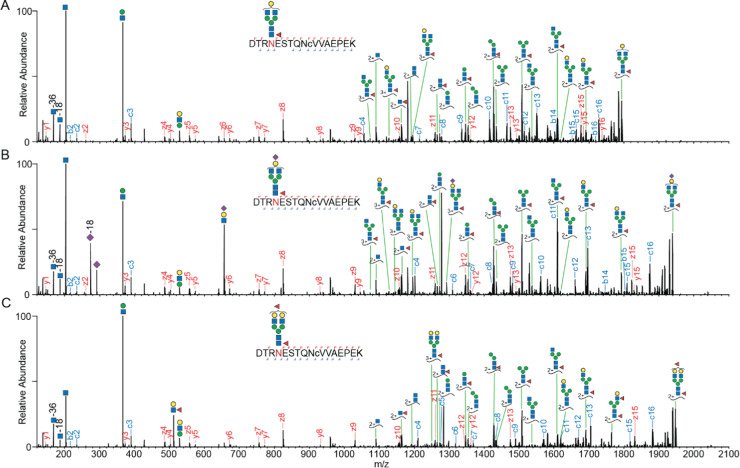
We evaluated the N-glycosylation sites found in our data versus those annotated in UniProt (Figure 2D). Most of the sites we report here (149 out of 191 glycosites) are annotated with experimental evidence in Uniprot; however, we detected 43 potentially novel glycosites, all displaying the well-known N-X-S/T sequon. In UniProt, these 43 were annotated as putative sites without any experimental evidence (35 glycosites) or not annotated at all (8 glycosites). In particular for these putative novel sites, we not only took the stringent cutoff in Byonic but also manually validated the MS/MS spectra using the following procedure. (1) All of these spectra should contain at least two Y ions (peptide backbone ion carrying a glycan fragment from the glycosidic bond cleavage) and two oxonium ions, which strongly indicate that the spectra originated from intact glycopeptides. (2) The EThcD fragmentation ions should contain the c or z ion pairs with a difference in mass of the intact glycan and corresponding amino acids, which is essential for an unambiguous site localization. Of all novel sites identified, the most surprising one was Asn69 (sequon: NES) in α-S1-casein, since this protein is quite abundant and a very well studied highly phosphorylated milk protein.25 We generated very strong evidence for Asn69 N-glycosylation, observing many different glycopeptides harboring this site. Due to a variable number of miscleavages, we identified EKQTDEIKDTRN69ESTQNCVVAEPEK (58–82), QTDEIKDTRN69ESTQNCVVAEPEK (60–82), DTRN69ESTQNCVVAEPEK (66–82), DTRN69ESTQNCVVAEPEKMESSISSSSEEMSLSK (66–98), and N69ESTQNCVVAEPEK (69–82). Among these, the backbone sequence of DTRN69ESTQNCVVAEPEK (66–82) was found to be detected with most PSMs and glycoform variants. The corresponding EThcD spectra showed both glycan and backbone fragmentation, enabling confident assignments of the glycoforms and glycosylation site (Figure 3). Moreover, the glycopeptides with two fucoses could be assigned as one core fucosylation and one terminal fucosylation by the Y1f ion (peptide backbone ion carrying a glycan fragment HexNAc(1)Fuc(1) from the glycosidic bond cleavage) and HexNAc(1)Hex(1)Fuc(1) oxonium ion (m/z 512.197+).
Figure 3.
Illustrative EThcD spectra of glycopeptides derived from α-S1-casein on a novel N-glycosylation site. Three examples of the same glycopeptide; DTRNESTQNCVVAEPEK of α-S1-casein (66–82), harboring various glycoforms. The EThcD fragmentation of these glycopeptides resulted in full sequence coverage, and the fragment ion (Y1f) of the amino acid backbone with HexNAc(1)Fuc(1) indicated one core fucose. (A) EThcD fragmentation spectra of the glycopeptide harboring a HexNAc(4)Hex(4)Fuc(1). (B) EThcD fragmentation spectra of the glycopeptide harboring a HexNAc(4)Hex(4)Fuc(1)Sia(1). (C) EThcD fragmentation spectra of the glycopeptide harboring a HexNAc(4)Hex(5)Fuc(2). Lower case c in the peptide sequence indicates a carbamylated cysteine.
Additionally, we investigated the site occupancy of this site by direct monitoring in the milk serum shotgun experiments (i.e., without any enrichment or depletion) for the DTRNESTQNCVVAEPEK peptide in its unmodified form and for the same peptide harboring the glycoform HexNAc(4)Hex(5)Fuc(1)Sia(1). We selected this glycopeptide as it was, on average, about 15 times more intense than all others (Figure S3A). The peak area of extracted ion chromatograms of the unmodified peptide at different time points showed a gradual decrease over lactation and was almost not detected in week 16, while that of the glycopeptide harboring the glycoform HexNAc(4)Hex(5)Fuc(1)Sia(1) decreased to a lesser extent (Figure S3B). By comparing the ion intensities of the most abundant glycopeptide and the corresponding unmodified peptide, we extracted that the site occupancy of Asn69 in α-S1-casein ranged from 20% at the early stages to almost fully occupied (98%) at later stages of lactation (Figure S3C). This substantial change in occupancy may indicate that the site has a functional relevance. However, at later lactational stages, we almost did not detect the unmodified peptide anymore, opening up the question of whether possibly other modifications may play a role as well. α-S1-casein is best known as a highly phosphorylated protein harboring close to a dozen phosphorylation sites. Among these phosphorylation sites, Ser71, which has recently been reported to be phosphorylated in breast milk,26 happens to be in the sequon of the N-glycosylation we found (NES) and, thus, on the peptide stretch we monitored. This opens the question of whether a potential positive or negative crosstalk between N-glycosylation and phosphorylation may occur. In humans, α-S1-casein has four isoforms produced by alternative splicing27 (Figure S4). Interestingly, in isoform 3, both Asn69 and Ser71 are deleted. Moreover, aligning α-S1-casein protein sequences across different species (Figure S4) revealed that only the human sequence carries this N-glycosylation site.
Targeted Monitoring of Intact Glycopeptides across Lactation
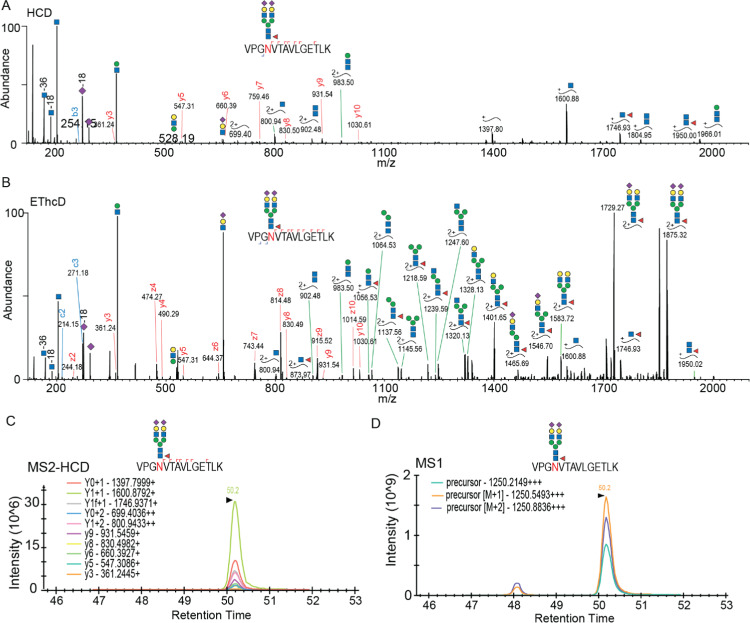
Observing that the glycan occupancy of certain N-glycopeptides varies substantially during the lactation period, we decided to monitor a subset of observed glycopeptides by a targeted proteomics assay. Therefore, we first built a library with relevant m/z and retention times of selected intact glycopeptide precursors and their corresponding HCD fragmentation pattern. The glycopeptide identification based on the HCD spectrum (Figure 4A) was confirmed by their matching EThcD spectra (Figure 4B). We then scheduled, for all selected glycopeptides, the precursor and its fragmentation ions to be measured. In a 90 min LC–MS gradient, we did target and measure 287 glycopeptides originating from 50 glycoproteins by selected ion monitoring (SIM) and parallel reaction monitoring (PRM), respectively, catching at least 10 data points in their chromatographic peaks (Figure S5A,B). In the MS/MS-HCD spectra, Y ions are normally quite abundant.28 We could use these ions in addition to the b and y ions to check whether they were co-eluted and compare the rank to that recorded in the above-mentioned library (Figure 4C). If so, we could undoubtedly pinpoint the chromatographic peak of the precursor and use the maximum intensity of the top three isotopes to represent the abundance of the glycopeptides (Figure 4D). We also noticed the separation of glycopeptide isoforms in MS1 XICs; however, we could often not directly distinguish their exact form. Therefore, in our analysis, we took the most intense MS1 XIC of the detected isoforms. For some glycopeptides containing Met, we noticed variable oxidation occurring; thus, we summed the intensities of the oxidation (±) forms. Considering the lack of commercial 13C-labeled glycopeptide standards, we spiked the PRTC mixture equally in all samples to monitor potential LC retention time shifts and variability between MS runs. We could calculate the CV of the intensities of PRTC peptides as a measure of variation. Of all PRTC peptides, 70% had CVs less than 20%; another 30% had CVs less than 40% (Figure S5C). Based on this observation, we concluded that the variability between runs was within a reasonable range. Based on our in parallel acquired proteome data, we noticed that the abundances of certain proteins also showed changes across the lactation period. Thus, to assess the total quantitative change in specific N-glycoform abundances, the raw intensity obtained from the targeted monitoring was normalized to the in parallel measured protein abundances, using the protein abundances from week 1 as the baseline (Table S6).
Figure 4.
Targeted quantitative monitoring of glycopeptides. For illustrative purposes, a glycopeptide of the polymeric immunoglobulin receptor (pIgR), VPGNVTAVLGETLK, is taken, harboring a HexNAc(4)Hex(5)Fuc(1)Sia(2) moiety. (A) HCD spectrum of the glycopeptide precursor (m/z 1250.2149+++). (B) EThcD spectrum of the same precursor, which improved the identification and site assignment. (C) Elution profile of Y and y ions, which were co-eluted and whose rank was matched with the HCD spectrum in (A). (D) The chromatographic peak of the precursor could be pinpointed by the MS2-HCD elution profile, whereby the max intensity of the top three isotopes was used to determine the abundance of the glycopeptide of interest.
Visual Summary for Each Glycoprotein and Each Glycosite
To better illustrate the dynamics observed in the site-specific glycosylation of all detected glycoproteins, we mapped on the UniProt sequence of each glycoprotein the detected glycans per site with their relative abundances over the lactation period (using log 10 of average normalized intensities), as shown in Figure S6 (Uniprot Accession). The diversity of glycan microheterogeneity can be exemplified by comparing different glycoproteins. Some proteins have only one glycosite that harbors a great variety of glycans, e.g., α-S1-casein (Figure S6, P47710), and immunoglobulin J chain (Figure S6, P01591). However, other glycoproteins have several glycosylation sites, with limited variability in the glycans modifying them, e.g., hemopexin (Figure S6, P02790). Also some glycosites exhibit higher variability in glycans than other sites on the same protein, e.g., lactoferrin (Figure S6, P02788). Interestingly, for lactoferrin, the glycosite with lower glycan heterogeneity (Asn642) is the site that is unique to human lactoferrin; other sites (Asn156, Asn497) are more conserved glycosylation sites.
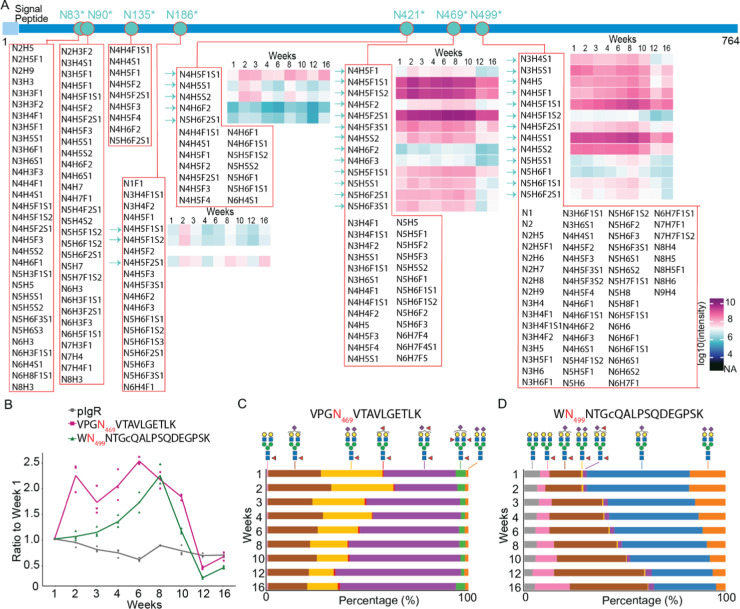
We choose to further demonstrate the variability of glycans and glycosites by focusing on one example, namely, the polymeric immunoglobulin receptor (pIgR) protein, which has seven potential N-glycosylation sites. The protein pIgR in human milk is quite abundant29 and plays an essential role in the formation and secretion of secretory immunoglobulin A (sIgA), which is the dominant immunoglobulin in human milk and essential for infant health.30 Not only is pIgR essential for the secretion of immunoglobulin A (IgA), but it has known antimicrobial functionality and can also be secreted on its own, providing protection to sIgA from degradation.31 We identified a stunning number of 201 glycopeptides in total, covering all seven potential glycosylation sites of pIgR, whereby each site displayed a striking different degree of heterogeneity (Figure 5A). We quantified 34 of the glycopeptides over nine time points, further elucidating quantitative differences. Among all detected glycosites, Asn469 and Asn499 accounted for the most PSMs and the highest degree of heterogeneity. We zoomed into the variation in occupancy of these two sites by different glycans, especially biantennary complex glycans with or without fucose moieties across lactation. First, as shown in Figure 5B, the pIgR protein does not change that much in abundance over the monitored lactation period, although it does show a gradual decrease by up to about 20% at the latest time point. After normalization for protein level variation, the glycopeptides harboring Asn469 and Asn499 in pIgR did reveal much more variability and site-specific changes over the lactation period. We next compared the longitudinal variations in the different glycans occupying these two sites. In Figure 5C, it is shown that for VPGN469VTAVLGETLK, glycoforms harboring one fucose decreased in abundance, while glycoforms with multiple fucoses increased, especially HexNAc(4)Hex(5)Fuc(2)NeuAc(1), from 36 to 58%. For WN499NTGCQALPSQDEGPSK (Figure 5D), the glycoforms harboring one fucose increased, while those without fucose decreased. Notably, glycan HexNAc(4)Hex(5)Fuc(1) increased from 5 to 18% and HexNAc(4)Hex(5)Fuc(1)NeuAc(1) from 16 to 39%, whereas HexNAc(4)Hex(5)NeuAc(1) dropped from 51 to 31% and HexNAc(4)Hex(5)NeuAc(2) from 18 to 5%. Our data are also supported by earlier studies that reported that fucosylation increases over lactation.32,33 For instance, Landberg et al.32 observed that with the progression of lactation, the levels of fucosylation on N-glycans of the bile-salt-stimulated lipase protein increased. Related to our work, Barboza et al.33 analyzed the released N-glycans of human milk lactoferrin from day 1 to 72 postpartum and found decreased levels of mono-fucosylation and increased levels of multiple fucosylation. They also showed that enzymatic removal of the fucose moieties from lactoferrin affected the ability of bacteria to bind to epithelial cells, whereas lactoferrin with fucose attached significantly inhibited pathogen adhesion. In our data, we observed similar trends for the glycopeptides of lactoferrin (Figure S6, P02788).
Figure 5.
Site-specific glycosylation varies across stages of lactation, as illustrated for the protein polymeric immunoglobulin receptor (pIgR). The temporal changes in abundance of pIgR and the glycans it harbors are depicted. (A) Overview of the pIgR protein sequence, the identified glycosites, the glycan compositions, and the abundances over the nine measured time points. The protein sequence chains are color-coded, wherein light blue and blue represent the signal peptide and the mature protein, respectively. Indigo dots inserted in the protein sequence with an asterisk indicate the UniProt-annotated N-glycosylation sites with evidence. A red circle indicates that the glycosite is identified in this study. (B) Taking the value in week 1 as the reference point, the abundance of the protein pIgR gradually decreases marginally over the 16 weeks of monitoring its abundance. In contrast, the glycan distribution on N469 and N499 (taken by the summed intensities of biantennary glycopeptides VPGN469VTAVLGETLK, WN499NTGCQALPSQDEGPSK, respectively) shows a diverging behavior with a 2–2.5-fold increase in abundance after 6–8 weeks, which decreases back to the abundance observed at 1 week at week 16. Microheterogeneity in glycan composition monitored over the lactation period as observed for (B) VPGN469VTAVLGETLK and (C) WN499NTGCQALPSQDEGPSK. For pIgR, the glycoforms harboring fucose moieties displayed a relative increase over the lactation period. In general, the site-specific glycosylation patterns were quite distinctive for each glycoprotein and each N-glycosite.
In mammals, core fucosylation, which is fucosylation on the GlcNAc linked to asparagine in the core of N-glycans, is the most common type of fucose modification.34 Thus, mono-fucosylation tends to be core fucosylation, while additional fucosylation of N-glycans is likely terminal fucosylation, for which we also find evidence in our EThcD spectra. The functions of multiple fucosylation in the course of lactation, especially the site-specific effects, are still understudied. Nevertheless, the feature of having terminal fucosylation on glycoproteins represents a structural homology to human milk oligosaccharides (HMOs), which may lead to a similar function of the fucosylated HMOs. There is evidence from studies investigating HMOs that have shown that HMOs increase in complexity and diversity, especially regarding fucosylation, throughout lactation to meet the needs of the increasing diversity of the gut microbiota.35−37 This could be an explanation for the changes observed in our study regarding the glycoproteome, especially with regard to increasing fucosylation observed from several different glycoproteins and varying glycosites across lactation. Potentially, then these observed differences are related to the changing needs of the infant to meet functional demands for growth and development and of diversifying gut microbiome. N-Glycans could be effectively released by the bacterium-derived glycohydrolases, e.g., endo-β-N-acetylglucosaminidases,38 and serve as the selective growth substrates for infant-associated gut microbes.39,40 Additionally, the attached N-glycans could slow down the proteolytic digestion in infants, which is conceivably supported by evidence that many glycoproteins such as lactoferrin41 and sIgA42 are not fully absorbed in the small intestine but rather can be found back as intact protein in infant stool samples, indicating that these proteins serve many important roles in pathogen defense and regulation of cellular proliferation and differentiation.5
Conclusions
In this study, we characterized the human milk N-glycoproteome of a single donor over nine time points during the lactation period, characterizing 191 glycosites on cumulatively 110 human milk glycoproteins. In total, we were able to assign 1697 glycopeptides, indicating that many sites were occupied by a wide variety of glycans. In more quantitative detail, we performed targeted proteomics on 287 glycopeptides originating from 50 glycoproteins monitoring their abundance over the lactational period, through which we observed distinctive site-specific changes in glycosylation. Compared to previous studies on the human milk glycoproteome, we took a more integral approach by focusing on intact glycopeptides and were able to reach considerably more depth. Other studies either focused on the released N-glycans15,16 or deglycosylated peptides.12−14 We consider the improvement in depth in our study to originate mainly from two factors: (1) the application of the robust and reproducible automated HILIC-based enrichment protocol, and (2) the high-end high-resolution mass spectrometry approach using HCD-triggered EThcD-based glycopeptide identification. The automated HILIC-based enrichment on the AssayMAP Bravo robot has the capacity to allow for the enrichment of 96 digested samples in parallel within 1 h, offering the potential for application on studies with numerous samples, such as the longitudinal time series presented here. We demonstrate that it is now possible to monitor personalized milk glycoproteomes, i.e., per individual donor. By doing this, our data revealed a fascinating variability in glycans and their site occupancy, whereby each human milk glycoprotein, each N-glycosylation site, and each glycan attached exhibit their own distinctive quantitative pattern across the monitored lactation period. All of these observed variations are very likely related to the changing needs of the growing and developing infant, and are hallmarked by the changing functional demands of the immune system and diversifying gut microbiome.
Acknowledgments
We acknowledge support from the Netherlands Organization for Scientific Research (NWO) funding the large-scale proteomics facility Proteins@Work (project 184.032.201) embedded in the Netherlands Proteomics Centre. A.J.R.H. acknowledges further support by the NWO TOP-Punt Grant 718.015.003 and the EU Horizon 2020 program INFRAIA project Epic-XS (Project 823839). J.Z. acknowledges support from the Chinese Scholarship Council (CSC). Additional support for this research was provided by Danone Nutricia Research. We thank Vojtech Franc and Karli Reiding (Utrecht University) for proofreading the manuscript.
Glossary
Abbreviations
- ACN
acetonitrile
- AGC
automated gain control
- CAA
chloroacetamide
- EThcD
electron-transfer/higher-energy collision dissociation
- FA
formic acid
- FDR
false discovery rate
- HCD
higher energy C-trap dissociation
- HILIC
hydrophilic-interaction chromatography
- HMOs
human milk oligosaccharides
- MS
mass spectrometry
- MS/MS
tandem mass spectrometry
- pIgR
polymeric immunoglobulin receptor
- PRM
parallel reaction monitoring
- PRTC
pierce peptide retention time calibration
- PSM
peptide-to-spectrum matches
- PTM
post-translational modification
- RT
retention time
- SDC
sodium deoxycholate
- sIgA
secretory immunoglobulin A
- SIM
selective ion monitoring
- TCEP
tris(2-carboxyethyl)phosphine hydrochloride
- TFA
trifluoroacetic acid
- XIC
extracted ion chromatogram
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.jproteome.9b00753.
Dynamic range of human milk proteome and identified glycoproteins (Figure S1); global comparison of the glycan composition of N-glycopeptides in human milk vs human serum (Figure S2); represented site occupancy of Asn69 from α-S1-casein by one major glycopeptide and unmodified peptide (Figure S3); alignment of protein sequences of α-S1-casein across different species (Figure S4); experimental parameters used in the scheduled targeted assay for glycopeptide quantification (Figure S5) (PDF)
Visual summary for each identified glycoprotein and schematic drawing of the sequences of the identified glycoproteins with identified sites and identified different glycan compositions (Figure S6) (PDF)
Two hundred possible N-glycan compositions in humans without sodium and with multiple fucoses (Table S1); peptide-to-spectrum matches for EThcD spectra (Table S2); identified N-glycopeptides in human milk across lactation (Table S3); glycan compositions at identified N-glycosites (Table S4); the identities of the outer nodes in the glycoprotein–glycan network diagram (Figure 2C) (Table S5); the normalized intensity of glycopeptides across lactation in a single donor (Table S6) (XLSX)
Author Contributions
⊥ J.Z. and Y.-H.L. contributed equally to this work.
The authors declare the following competing financial interest(s): M.M and B.S are employees of Danone Nutricia Research. J.Z and K.A.D were enrolled as PhD students at Utrecht University during this study and received partial financial support from Danone Nutricia Research. None of the authors have further conflicts of interest with regard to the content of this manuscript.
Notes
The mass spectrometry data have been deposited to the ProteomeXchange Consortium via the PRIDE43 partner repository with the dataset identifier PXD013764 and 10.6019/PXD013764.
Notes
Skyline data have been deposited to the ProteomeXchange Consortium via Panorama44 with the access URL (https://panoramaweb.org/BBgb9c.url).
Supplementary Material
References
- Hennet T.; Borsig L. Breastfed at Tiffany’s. Trends Biochem. Sci. 2016, 41, 508–518. 10.1016/j.tibs.2016.02.008. [DOI] [PubMed] [Google Scholar]
- Newburg D. S. Glycobiology of human milk. Biochemistry 2013, 78, 771–785. 10.1134/S0006297913070092. [DOI] [PubMed] [Google Scholar]
- Lis-Kuberka J.; Orczyk-Pawilowicz M. Sialylated Oligosaccharides and Glycoconjugates of Human Milk. The Impact on Infant and Newborn Protection, Development and Well-Being. Nutrients 2019, 11, 306 10.3390/nu11020306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi H.; Yamashita Y. Role of N-glycosylation in cell surface expression and protection against proteolysis of the intestinal anion exchanger SLC26A3. Am. J. Physiol.: Cell Physiol. 2012, 302, C781–C795. 10.1152/ajpcell.00165.2011. [DOI] [PubMed] [Google Scholar]
- Liu B.; Newburg D. S. Human milk glycoproteins protect infants against human pathogens. Breastfeed. Med. 2013, 8, 354–362. 10.1089/bfm.2013.0016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon D. L. The Role of Human Milk Immunomodulators in Protecting Against Viral Bronchiolitis and Development of Chronic Wheezing Illness. Children 2015, 2, 289–304. 10.3390/children2030289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgi G.; Bartke N.; Wiens F.; Stahl B. Functional glycans and glycoconjugates in human milk. Am. J. Clin. Nutr. 2013, 98, 578S–585S. 10.3945/ajcn.112.039065. [DOI] [PubMed] [Google Scholar]
- Fields D. A.; Schneider C. R.; Pavela G. A narrative review of the associations between six bioactive components in breast milk and infant adiposity. Obesity 2016, 24, 1213–1221. 10.1002/oby.21519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J.; Garrigues L.; Van den Toorn H.; Stahl B.; Heck A. J. R. Discovery and quantification of nonhuman proteins in human milk. J. Proteome Res. 2018, 18, 225–238. 10.1021/acs.jproteome.8b00550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Čaval T.; Tian W.; Yang Z.; Clausen H.; Heck A. J. R. Direct quality control of glycoengineered erythropoietin variants. Nat. Commun. 2018, 9, 3342 10.1038/s41467-018-05536-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caval T.; Zhu J.; Heck A. J. R. Simply Extending the Mass Range in Electron Transfer Higher Energy Collisional Dissociation Increases Confidence in N-Glycopeptide Identification. Anal. Chem. 2019, 91, 10401–10406. 10.1021/acs.analchem.9b02125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X.; Yang M.; Yang N.; Liang X.; Tao D.; Liu B.; Wu J.; Yue X. Characterization and comparison of whey N-glycoproteomes from human and bovine colostrum and mature milk. Food Chem. 2019, 276, 266–273. 10.1016/j.foodchem.2018.09.174. [DOI] [PubMed] [Google Scholar]
- Cao X.; Kang S.; Yang M.; Li W.; Wu S.; Han H.; Meng L.; Wu R.; Yue X. Quantitative N-glycoproteomics of milk fat globule membrane in human colostrum and mature milk reveals changes in protein glycosylation during lactation. Food Funct. 2018, 9, 1163–1172. 10.1039/C7FO01796K. [DOI] [PubMed] [Google Scholar]
- Picariello G.; Ferranti P.; Mamone G.; Roepstorff P.; Addeo F. Identification of N-linked glycoproteins in human milk by hydrophilic interaction liquid chromatography and mass spectrometry. Proteomics 2008, 8, 3833–3847. 10.1002/pmic.200701057. [DOI] [PubMed] [Google Scholar]
- Nwosu C. C.; Aldredge D. L.; Lee H.; Lerno L. A.; Zivkovic A. M.; German J. B.; Lebrilla C. B. Comparison of the human and bovine milk N-glycome via high-performance microfluidic chip liquid chromatography and tandem mass spectrometry. J. Proteome Res. 2012, 11, 2912–2924. 10.1021/pr300008u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dallas D. C.; Martin W. F.; Strum J. S.; Zivkovic A. M.; Smilowitz J. T.; Underwood M. A.; Affolter M.; Lebrilla C. B.; German J. B. N-linked glycan profiling of mature human milk by high-performance microfluidic chip liquid chromatography time-of-flight tandem mass spectrometry. J. Agric. Food Chem. 2011, 59, 4255–4263. 10.1021/jf104681p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida Y.; Furukawa J. I.; Naito S.; Higashino K.; Numata Y.; Shinohara Y. Quantitative analysis of total serum glycome in human and mouse. Proteomics 2016, 16, 2747–2758. 10.1002/pmic.201500550. [DOI] [PubMed] [Google Scholar]
- Bekker-Jensen D. B.; Kelstrup C. D.; Batth T. S.; Larsen S. C.; Haldrup C.; Bramsen J. B.; Sorensen K. D.; Hoyer S.; Orntoft T. F.; Andersen C. L.; Nielsen M. L.; Olsen J. V. An Optimized Shotgun Strategy for the Rapid Generation of Comprehensive Human Proteomes. Cell Syst. 2017, 4, 587.e4–599.e4. 10.1016/j.cels.2017.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLean B.; Tomazela D. M.; Shulman N.; Chambers M.; Finney G. L.; Frewen B.; Kern R.; Tabb D. L.; Liebler D. C.; MacCoss M. J. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics 2010, 26, 966–968. 10.1093/bioinformatics/btq054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee L. Y.; Moh E. S.; Parker B. L.; Bern M.; Packer N. H.; Thaysen-Andersen M. Toward Automated N-Glycopeptide Identification in Glycoproteomics. J. Proteome Res. 2016, 15, 3904–3915. 10.1021/acs.jproteome.6b00438. [DOI] [PubMed] [Google Scholar]
- Riley N. M.; Hebert A. S.; Westphall M. S.; Coon J. J. Capturing site-specific heterogeneity with large-scale N-glycoproteome analysis. Nat. Commun. 2019, 10, 1311 10.1038/s41467-019-09222-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai Y.; Tao J.; Zhou J.; Fan Q.; Liu M.; Hu Y.; Xu Y.; Zhang L.; Yuan J.; Li W.; Ze X.; Malard P.; Guo Z.; Yan J.; Li M. Fucosylated Human Milk Oligosaccharides and N-Glycans in the Milk of Chinese Mothers Regulate the Gut Microbiome of Their Breast-Fed Infants during Different Lactation Stages. mSystems 2018, 3, e00206-18 10.1128/mSystems.00206-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S.; Hu Y.; Jia L.; Eshghi S. T.; Liu Y.; Shah P.; Zhang H. Site-Specific Profiling of Serum Glycoproteins Using N-Linked Glycan and Glycosite Analysis Revealing Atypical N-Glycosylation Sites on Albumin and alpha-1B-Glycoprotein. Anal. Chem. 2018, 90, 6292–6299. 10.1021/acs.analchem.8b01051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouda G. G.; Jaeger F. H.; Amos J. D.; Ho C.; Kunz E. L.; Anasti K.; Stamper L. W.; Liebl B. E.; Barbas K. H.; Ohashi T.; et al. Tenascin-C is an innate broad-spectrum, HIV-1–neutralizing protein in breast milk. Proc. Natl. Acad. Sci. U.S.A. 2013, 110, 18220–18225. 10.1073/pnas.1307336110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y.; Weber D.; Xu W.; Durbin-Johnson B. P.; Phinney B. S.; Lonnerdal B. Absolute Quantification of Human Milk Caseins and the Whey/Casein Ratio during the First Year of Lactation. J. Proteome Res. 2017, 16, 4113–4121. 10.1021/acs.jproteome.7b00486. [DOI] [PubMed] [Google Scholar]
- Konig S.; Altendorfer I.; Saenger T.; Bleck E.; Vordenbaumen S.; Schneider M.; Jose J. Ser71 of alphaS1-Casein is Phosphorylated in Breast Milk-Evidence from Targeted Mass Analysis. Mol. Nutr. Food Res. 2017, 61, 1700496 10.1002/mnfr.201700496. [DOI] [PubMed] [Google Scholar]
- Johnsen L. B.; Rasmussen L. K.; Petersen T. E.; Berglund L. Characterization of three types of human alpha s1-casein mRNA transcripts. Biochem. J. 1995, 309, 237–242. 10.1042/bj3090237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng W.-F.; Liu M.-Q.; Zhang Y.; Wu J.-Q.; Fang P.; Peng C.; Nie A.; Yan G.; Cao W.; Liu C.; Chi H.; Sun R.-X.; Wong C. C. L.; He S.-M.; Yang P. pGlyco: a pipeline for the identification of intact N-glycopeptides by using HCD- and CID-MS/MS and MS3. Sci. Rep. 2016, 6, 25102 10.1038/srep25102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elwakiel M.; Boeren S.; Hageman J. A.; Szeto I. M.; Schols H. A.; Hettinga K. A. Variability of Serum Proteins in Chinese and Dutch Human Milk during Lactation. Nutrients 2019, 11, 499 10.3390/nu11030499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmeira P.; Carneiro-Sampaio M. Immunology of breast milk. Rev. Assoc. Med. Bras. 2016, 62, 584–593. 10.1590/1806-9282.62.06.584. [DOI] [PubMed] [Google Scholar]
- Phalipon A.; Corthesy B. Novel functions of the polymeric Ig receptor: well beyond transport of immunoglobulins. Trends Immunol. 2003, 24, 55–58. 10.1016/S1471-4906(02)00031-5. [DOI] [PubMed] [Google Scholar]
- Landberg E.; Huang Y.; Stromqvist M.; Mechref Y.; Hansson L.; Lundblad A.; Novotny M. V.; Pahlsson P. Changes in glycosylation of human bile-salt-stimulated lipase during lactation. Arch. Biochem. Biophys. 2000, 377, 246–254. 10.1006/abbi.2000.1778. [DOI] [PubMed] [Google Scholar]
- Barboza M.; Pinzon J.; Wickramasinghe S.; Froehlich J. W.; Moeller I.; Smilowitz J. T.; Ruhaak L. R.; Huang J.; Lonnerdal B.; German J. B.; Medrano J. F.; Weimer B. C.; Lebrilla C. B. Glycosylation of human milk lactoferrin exhibits dynamic changes during early lactation enhancing its role in pathogenic bacteria-host interactions. Mol. Cell. Proteomics 2012, 11, M111.015248 10.1074/mcp.M111.015248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider M.; Al-Shareffi E.; Haltiwanger R. S. Biological functions of fucose in mammals. Glycobiology 2017, 27, 601–618. 10.1093/glycob/cwx034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doherty A. M.; Lodge C. J.; Dharmage S. C.; Dai X.; Bode L.; Lowe A. J. Human Milk Oligosaccharides and Associations With Immune-Mediated Disease and Infection in Childhood: A Systematic Review. Front. Pediatr. 2018, 6, 91 10.3389/fped.2018.00091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donovan S. M.; Comstock S. S. Human Milk Oligosaccharides Influence Neonatal Mucosal and Systemic Immunity. Ann. Nutr. Metab. 2016, 69, 42–51. 10.1159/000452818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mezoff E. A.; Hawkins J. A.; Ollberding N. J.; Karns R.; Morrow A. L.; Helmrath M. A. The human milk oligosaccharide 2′-fucosyllactose augments the adaptive response to extensive intestinal. Am. J. Physiol.: Gastrointest. Liver Physiol. 2016, 310, G427–G438. 10.1152/ajpgi.00305.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrido D.; Nwosu C.; Ruiz-Moyano S.; Aldredge D.; German J. B.; Lebrilla C. B.; Mills D. A. Endo-beta-N-acetylglucosaminidases from Infant Gut-associated Bifidobacteria Release Complex N-glycans from Human Milk Glycoproteins. Mol. Cell. Proteomics 2012, 11, 775–785. 10.1074/mcp.M112.018119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M.; Bai Y.; Zhou J.; Huang W.; Yan J.; Tao J.; Fan Q.; Liu Y.; Mei D.; Yan Q.; Yuan J.; Malard P.; Wang Z.; Gu J.; Tanigchi N.; Li W. Core Fucosylation of Maternal Milk N-Glycan Evokes B Cell Activation by Selectively Promoting the l-Fucose Metabolism of Gut Bifidobacterium spp. and Lactobacillus spp. mBio 2019, 10, e00128-19 10.1128/mBio.00128-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karav S.; Le Parc A.; Bell J. M. L. N. D.; Frese S. A.; Kirmiz N.; Block D. E.; Barile D.; Mills D. A. Oligosaccharides Released from Milk Glycoproteins Are Selective Growth Substrates for Infant-Associated Bifidobacteria. Appl. Environ. Microbiol. 2016, 82, 3622–3630. 10.1128/AEM.00547-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lönnerdal B. Bioactive Proteins in Human Milk: Health, Nutrition, and Implications for Infant Formulas. J. Pediatr. 2016, 173, S4–S9. 10.1016/j.jpeds.2016.02.070. [DOI] [PubMed] [Google Scholar]
- Maruyama K.; Hida M.; Kohgo T.; Fukunaga Y. Changes in salivary and fecal secretory IgA in infants under different feeding regimens.. Pediatr. Int. 2009, 51, 342–345. 10.1111/j.1442-200X.2008.02748.x. [DOI] [PubMed] [Google Scholar]
- Vizcaíno J. A.; Csordas A.; Del-Toro N.; Dianes J. A.; Griss J.; Lavidas I.; Mayer G.; Perez-Riverol Y.; Reisinger F.; Ternent T.; Xu Q. W.; Wang R.; Hermjakob H. 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 2016, 44, 11033. 10.1093/nar/gkw880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma V.; Eckels J.; Taylor G. K.; Shulman N. J.; Stergachis A. B.; Joyner S. A.; Yan P.; Whiteaker J. R.; Halusa G. N.; Schilling B.; Gibson B. W.; Colangelo C. M.; Paulovich A. G.; Carr S. A.; Jaffe J. D.; MacCoss M. J.; MacLean B. Panorama: a targeted proteomics knowledge base. J. Proteome Res. 2014, 13, 4205–4210. 10.1021/pr5006636. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.