Abstract
Capicua (CIC), a member of the high mobility group–box (HMG-box) superfamily of transcriptional repressors, is frequently mutated in human oligodendrogliomas. However, its functions in brain development and tumorigenesis remain poorly understood. Here, we report that brain-specific deletion of Cic compromises developmental transition of neuroblasts to immature neurons in mouse hippocampus and compromises normal neuronal differentiation. Combined gene expression and ChIP-seq analyses identified VGF as an important CIC-repressed transcriptional surrogate involved in neuronal lineage regulation. Aberrant VGF expression promotes neural progenitor cell proliferation by suppressing their differentiation. Mechanistically, we demonstrated that CIC represses VGF expression by tethering SIN3-HDAC to form a transcriptional corepressor complex. Mass spectrometry analysis of CIC-interacting proteins further identified the BRG1-containing mSWI/SNF complex whose function is necessary for transcriptional repression by CIC. Together, this study uncovers a potentially novel regulatory pathway of CIC-dependent neuronal differentiation and may implicate these molecular mechanisms in CIC-dependent brain tumorigenesis.
Keywords: Oncology, Stem cells
Keywords: Brain cancer, Neuronal stem cells, Transcription
Dynamic transcriptional repression by CIC through neuronal differentiation is critical, and its dysregulation may drive the pathogenesis of oligodendrogliomas.
Introduction
Oligodendroglioma, a diffusely infiltrating primary malignant brain tumor in adults, is histologically characterized by its composition of neoplastic cells morphologically resembling oligodendroglial cells. Despite their generally better prognosis and response to early chemotherapy, most oligodendroglial tumors recur eventually, with many of them progressing to a higher-grade lesion (1). Genetically, oligodendrogliomas are defined by a combined loss of chromosome arms 1p and 19q (2). A more recent high-throughput sequencing approach has further identified the capicua (CIC; orthog of Drosophila capicua) gene, which is mapped to 19q13.2, as one of the most frequently mutated genes in oligodendrogliomas (3–5), pointing to a critical role of CIC in brain development and oligodendroglioma pathogenesis.
CIC is a transcriptional repressor with a SOX-like high mobility group (HMG) DNA binding domain (6). Studies in Drosophila revealed CIC as a key mediator of RAS/MAPK signaling that regulates embryonic patterning and intestinal stem cell proliferation (7, 8). In mammals, CIC has been shown to play a critical role in T cell development, adaptive immunity (9, 10), lung alveolarization, abdominal wall closure during the development (11, 12), and bile acid homeostasis (13). As to its function in CNS development, interestingly, a previous study of mouse brain–specific Cic deletion by glial fibrillary acidic protein promoter–driven cre recombinase (hGFAP-cre) reported no gross developmental abnormalities (12). By contrast, another study using Emx1-cre revealed that CIC was important for neurodevelopment (14). Loss of CIC disrupted organization and maintenance of upper-layer cortical neurons has led to mouse hyperactivity, with defective learning and memory loss. More recently, an additional study using Foxg1-cre revealed that mouse forebrain–specific deletion of Cic also caused abnormal increase of oligodendrocyte progenitor cells (OPC) and immature oligodendrocytes populations (15, 16), likely at the expense of neuronal propagation. Despite those efforts, the molecular functions of CIC in brain development and tumorigenesis remain poorly understood. Here, we describe a mouse model in which we applied embryonic neural progenitor cells (NPC) targeting Nestin-cre (Nes-cre) to broadly inactivate CIC in CNS. Our data show that the loss of CIC compromises the normal developmental transition of neuroblasts to neurons and, therefore, their differentiation. Through integrated expression and ChIP-seq analyses, we identified VGF as a CIC surrogate that mediates its function in regulating neuronal lineage differentiation. We further show that CIC transcriptionally represses its target gene expression by recruiting mammalian switch/sucrose nonfermentable (mSWI/SNF) and SIN3-HDAC repressor complexes. Our findings suggest CIC as a critical developmental transcriptional repressor that blocks tumorigenesis by facilitating neuronal maturation.
Results
Defective cerebral cortex development in CicKO mouse.
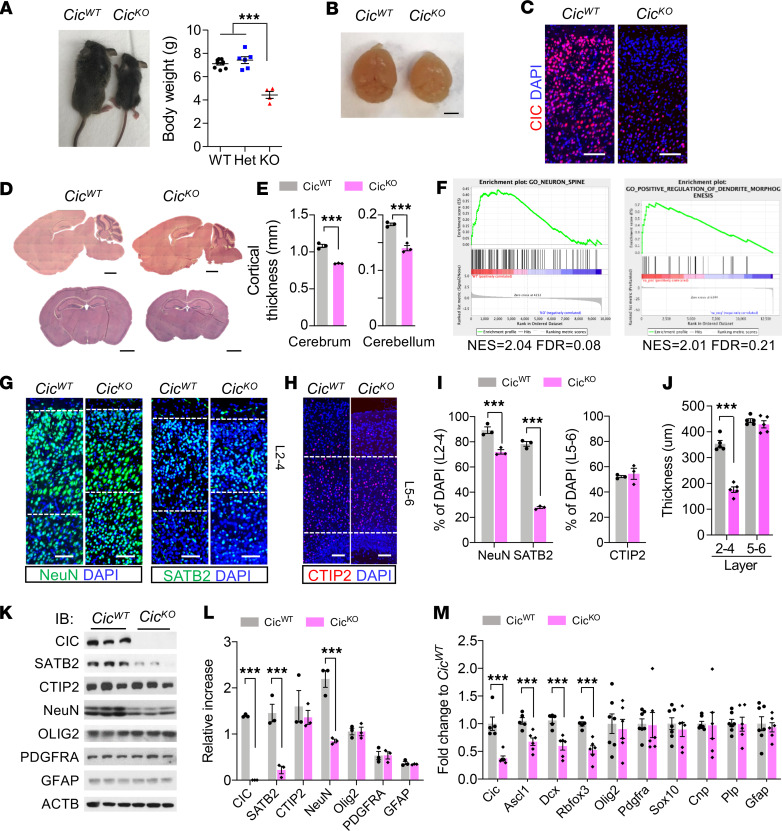
To investigate the role of CIC in neurogenesis, we engineered a conditional Cic floxed mouse allele and crossed it with Nes-cre animal to target Cic deletion in embryonic neural precursors and their progenies (Supplemental Figure 1A; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.135826DS1). Recombination of floxed Cic allele in brains of Nes-cre; Cicfl/fl (CicKO) animals was confirmed by PCR (Supplemental Figure 1, B–D). Developmentally, the CicKO mice displayed severe growth retardation and reduction of brain volume by P14 (Figure 1, A and B). Although largely indistinguishable from their littermate WT and heterozygous counterparts at birth, the growth retardation became evident in CicKO pups by P6. None of CicKO pups survived beyond P20–P22 (n = 21). The efficiency of CIC depletion was confirmed by immunofluorescence (IF) analysis (Figure 1C). Histologic examination of CicKO pups at P14 further revealed significant reductions of cerebral and cerebellar cortical thickness compared with CicWT littermates (n = 10) (Figure 1, D and E), indicating that CIC is required for early brain development.
Figure 1. Defective cerebral cortex developments in CicKO mouse.
(A) The picture (left) and the plot for body weights (right) at P14. Statistical significance was determined by 1-way ANOVA. Mean ± SEM of 4–10 experimental animals. ***P < 0.001. (B) The brains at P14. (C) IF analysis for CIC in the cerebral cortex. Scale bar: 100 μm. (D) H&E staining of CicWT and CicKO brains. Scale bar: 1 mm. For B–D, 3 brains per group were examined, and representative images are shown. (E) Quantification of the thickness cerebral cortex (left) and cerebellar cortex (right) are plotted. Mean ± SEM of 3 experimental animals. (F) GSEA in CicKO versus CicWT brains. Two brains per group were analyzed. (G and H) IF analysis for NeuN, SATB2, and CTIP2 in the cerebral cortex of P14 mouse. Scale bar: 100 μm. (I) Quantifications at layers 2–4 of NeuN+ or SATB2+ and layers 5–6 of CTIP2+ numbers are plotted. Mean ± SEM of 200 DAPI+ nuclei from 3 animals. (J) Measurement the thickness for SATB2+ layers 2–4 and CTIP2+ layers 5–6. Mean ± SEM of 3 images from 3 animals. (K) WB analysis for indicated protein expressions in cerebral cortex lysates. Samples were run on 3 gels, and the most representative ACTB blot is shown. (L) Densitometry analysis of multiple WB in J is plotted. Mean ± SEM of 3 experimental animals. (M) qPCR results of indicated genes in P14 CicKO versus CicWT brains. Mean ± SEM of 5–7 experimental animals. For I, J, L, and M, statistical significance was determined by unpaired t test. ***P < 0.001.
To uncover the underlying cause of the observed brain phenotype, we performed RNA sequencing (RNA-seq) analysis of P0.5 brains from control and CicKO animals. Gene set enrichment analysis (GSEA) (17, 18) revealed that downregulated genes in CicKO mouse were enriched in the gene sets related with neuronal development (Figure 1F). Indeed, IF analysis of P14 animals showed markedly reduced numbers of NeuN+ neurons in cortical layers 2–4 compared with those of CicWT controls. Interestingly, although the number of SATB2+ cortical neurons and the thickness of layers 2–4 were evidently reduced in P14 CicKO animals, we did not find significant differences in the number of CTIP2+ cortical neurons and the thickness of layers 5–6 when compared with those of P14 CicWT control pups (Figure 1, G–J). These observations were further corroborated by both Western blot (WB) and quantitative PCR (qPCR) analysis of brain cortices from corresponding animals (Figure 1, K–M), suggesting that CIC is necessary for the normal development of the cerebral cortex.
Notably, 2 previous studies (15, 16) reported that deletion of Cic caused increased number of either OLIG2+/PDGFRA+ OPC or GFAP+ astrocytes in the affected mouse brain. To test this in our setting, we next analyzed the expression of various lineage-specific genes for oligodendrocytes (e.g., Olig2, Pdgfra, Sox10, Cnp, and Plp), astrocytes (e.g., Gfap), and neurons (e.g., Ascl1, Dcx, and Rbfox3) in brain samples derived from P14 CicKO and their littermate CicWT animals. Surprisingly, although the expressions of neuronal genes were consistently reduced in CicKO compared with their CicWT control animals, there were no evident differences in either oligodendrocytic or astrocytic lineage marker expressions (Figure 1M). WB and GSEA analysis of P0.5 animals confirmed that neither downregulated nor upregulated genes in CicKO mouse were significantly enriched in gene sets related with oligodendrocyte or astrocyte differentiation (Supplemental Figure 2, A–D). Consistently, the immunohistochemistry (IHC) analysis did not show abnormal expansion of OLIG2+ or GFAP+ cells in P14 CicKO brains (Supplemental Figure 2, E and F). In addition, we did not observe reduction of myelin basic protein (MBP) staining in P14 CicKO brains (Supplemental Figure 2, E and F), suggesting that CIC is mainly involved in neuronal lineage differentiation regulation during brain development.
Loss of CIC compromises neuronal maturation.
Next, we determined CIC protein expression in different CNS cell types to understand its role in brain development. IF analysis of early postnatal and adult mouse brains revealed that CIC is highly expressed in NeuN+ neurons (Figure 2, A and B). But its expression in GFAP+ astrocytes and OLIG2+ oligodendrocytes is much weaker than that of neurons (Supplemental Figure 3, A–D). This is consistent with the finding that CIC is primarily involved in regulation of neurogenesis.
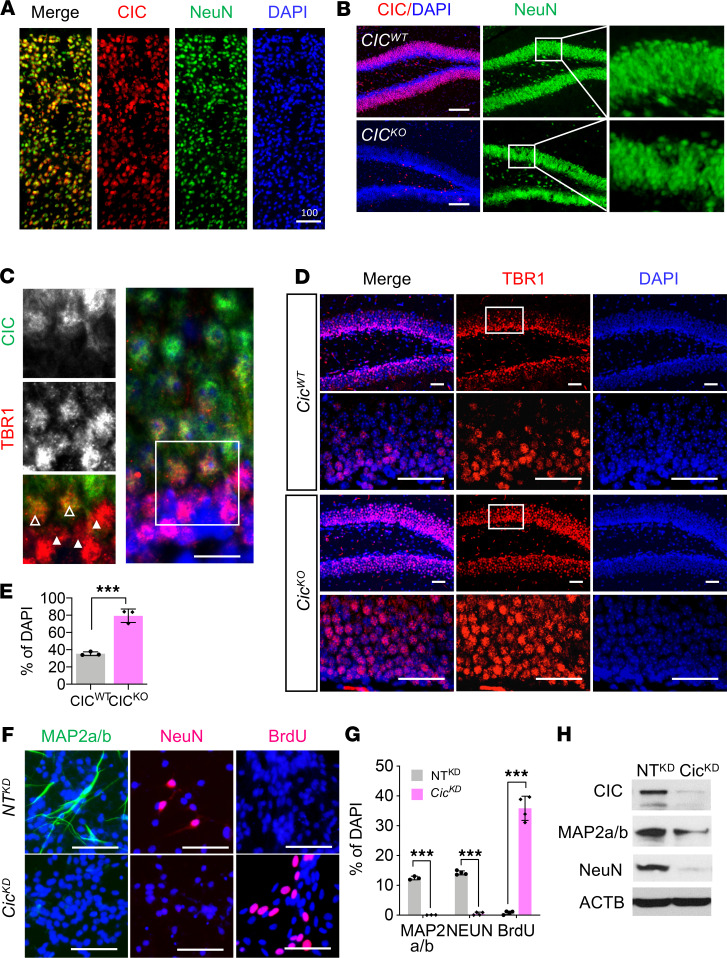
Figure 2. Loss of CIC compromises neuronal maturation.
(A and B) IF analysis for CIC and NeuN. Scale bar: 200 μm. Inset magnification: 400×. (C) Co-IF for TBR1 and CIC expression in hippocampus. Open arrowheads point to TBR1-low cells, and closed arrowheads are on TBR1-high cells. Inset magnification: 800×. (D) IF analysis for TBR1 in P14 CicWT and CicKO brains. Scale bar: 50 μm. Inset magnification: 400×. For A–D, 3 animals were examined, and representative images are shown. (E) TBR1-high cells of images in D are counted. Mean ± SEM of the measurement from 50–60 DAPI from 3 animals. (F) IF analysis for MAP2a/b, NeuN, and BrdU in NTKD NPC and CicKD NPC at 8 days of differentiation. Scale bar: 50 μm. (G) Quantitation of experiments in F. Mean ± SEM of the measurement from 100 DAPI+ nuclei from 3 independent experiments. For E and G, statistical significance was determined by unpaired t test. *** P < 0.001. (H) WB analysis for each specific marker at day 5 of differentiation. Three experiments were conducted, and a representative result is shown. Samples were run on 2 gels, and the representative ACTB blot is shown.
To examine a potential function of CIC along the neuronal differentiation process, we measured cellular CIC expression in the neurogenic subgranular zone (SGZ) and subventricular zone (SVZ) areas of adult mouse brains. Coimmunostaining with a panel of stage-specific markers showed a pattern of progressively increased CIC expression along the neuronal differentiation. Specifically, compared with the mature NeuN+ neurons that ubiquitously registered high levels of CIC expression (Figure 2, A and B), the Nestin+ (or SOX2+) type 1 (or type B) neural stem cells, TBR2+ type 2A, ASCL1+ type C transit-amplifying progenitor cells, or DCX+ neuroblasts all exhibited relatively low CIC expression (Supplemental Figure 4, A–C). Consistently, CIC expression was weak in TBR1-high neuroblast cells or immature neurons but became increasingly strong later in TBR1-low or NeuN+ mature neurons (Figure 2C). This pattern of dynamically regulated CIC expression along the process of neurogenesis supports the premise that it plays an important role during neuronal maturation. Indeed, the analysis of hippocampal granular layers of CicKO brains revealed abnormally expanded TBR1-high cells but reduced populations of TBR1-low cells compared with the CicWT controls (Figure 2, D and E).
To further explore the function of CIC in neuronal differentiation, we adopted a previously established in vitro differentiation system by introducing tet-inducible ASCL1 into immortalized NPC (19). Sixty percent of NPC highly expressed ASCL1 and were rapidly differentiated to DCX+ neuroblast upon treatment of doxycycline (Supplemental Figure 5, A and B). Similar to primary neural stem/progenitor cells, these cells could be further differentiated to NeuN+ neurons (Supplemental Figure 5, C and D) and cease to proliferate, as evidenced by lack of BrdU incorporation (Supplemental Figure 5E). To determine the role of CIC in neuronal differentiation, we generated Cic–knocked down and control NPC using short hairpin RNA (CicKD and NTKD, respectively). Importantly, NeuN+ mature neurons were not detected in CicKD NPC on day 8 of differentiation, unlike NTKD NPC (Figure 2, F–H). To further determine whether the loss of CIC sustains proliferation, we analyzed differentiated cells for BrdU incorporation. On day 8 of differentiation, about 20% of CicKD NPC continued to be BrdU+. These results suggest that CIC is necessary for complete exit from cell cycle and terminal neuronal differentiation.
CIC represses transcriptional targets in developing brains.
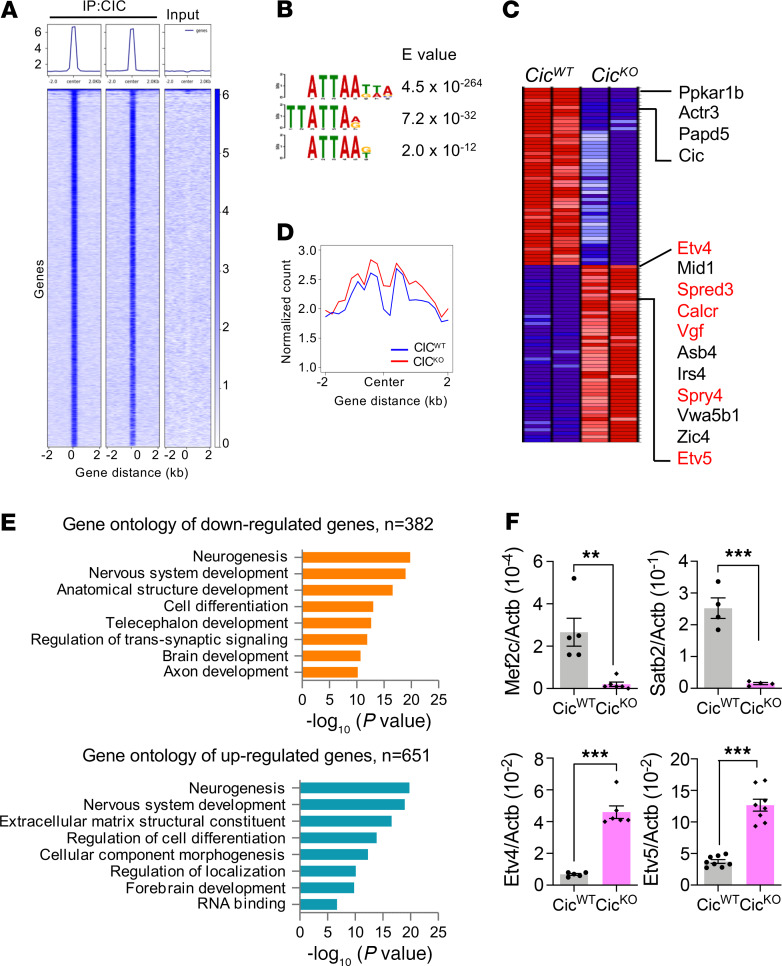
CIC is a member of the HMG-box superfamily of transcriptional repressors (6). To identify its direct transcriptional surrogates that may play a role in neurogenesis, we performed ChIP-seq concurrently with RNA-seq using P0.5 control and CicKO mouse brains that were undergoing neuronal maturation. Two biological replicates showed concordant genome-wide peaks that are absent in input control (Figure 3A), verifying quality of the data. De novo motif analysis further revealed that CIC bindings are strongly enriched at the AT-rich regions (Figure 3B). A significant portion of CIC peaks were resided within the promoter regions (5%) or introns (20%), whereas the rest were in intergenic regions (45%). In agreement with its presumed role as a transcriptional repressor, RNA-seq comparisons of P0.5 CicWT control and CicKO brain samples revealed that CIC depletion increased the expression of genes with CIC-interactive promoters. Out of 50 most differentially upregulated, 13 genes (Etv4, Spred3, Calcr, Vgf, Spry4, Etv5, Hcrtr1, Car10, Alk, Shc4, Maff, Dusp4, Fign) have CIC peaks in their promoter regions, suggesting direct transcriptional repression by CIC binding (Figure 3C and Supplemental Table 1). Alignment of CIC peaks with corresponding H3K27Ac ChIP-seq annotations confirmed that signal density of active promoter/enhancer–designated H3K27Ac peak centers was higher in CicKO samples than that of CicWT (Figure 3D). By contrast, the expression of genes with only intronic CIC peaks (e.g., Skap2, Lipa, and Cntnap2) did not show significant changes following CIC loss, suggesting that CIC may exert its transcriptional repression function mainly at promoter/enhancer regions.
Figure 3. CIC represses transcriptional targets in developing brains.
(A) Heatmaps at CIC peaks showing CIC (n = 2) and input. (B) Motif analysis. (C) Heatmap of the top 50 ranked differentially expressed genes in RNA-seq analysis of CicKO vs. CicWT. Genes with CIC ChIP peaks are marked red. (D) Average ChIP-seq signal of H3K27Ac at CIC peaks from P0.5 brains of CicWT and CicKO. (E) Gene ontology analysis with RNA-seq in P0.5 brains of CicWT and CicKO. The up- or downregulated genes were retrieved with P < 0.02. (F) qPCR results of Mef2c, Satb2, Etv4, and Etv5 mRNA expressions in P0.5 brains of CicWT and CicKO. Mean ± SEM of 5 experimental animals. Statistical significance was determined by unpaired t test. **P < 0.01, *** P < 0.001.
To explore underlying cellular and developmental mechanisms associated with CIC loss, we next performed the gene ontology comparison of gene expression of CicWT control and CicKO brain samples. Annotation of the 1033 differentially expressed genes (P < 0.02) indeed revealed neurogenesis as a highly dysregulated biological process following CIC depletion (Figure 3E), which is consistent with phenotypes observed in the CicKO brains. qPCR analysis further confirmed that expression of the neural transcriptional regulators Mef2c and Satb2 were significantly downregulated in CicKO brains (Figure 3F). In contrast, previously reported CIC transcriptional targets (e.g., Etv4, Etv5) were upregulated in CicKO brains.
Aberrant expression of VGF precludes development of mature neurons in CIC-deficient cells.
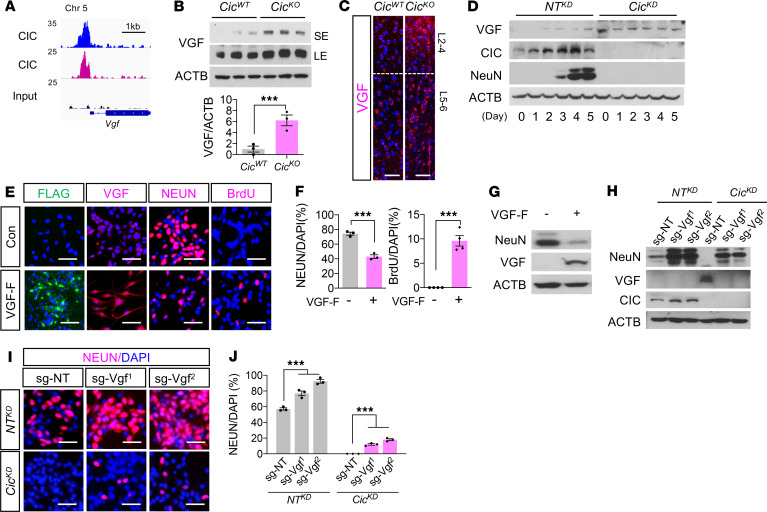
We next leveraged transcriptional and ChIP-seq profiles of the paired CicWT control and CicKO brains to identify CIC transcriptional targets that are potentially involved in regulation of neurogenesis. Among those that exhibited strong CIC binding in their promoter regions, the mRNA expression of Vgf (VGF nerve growth factor inducible) was consistently upregulated in CicKO brains (Figure 4A). VGF is a neuropeptide that is important for neuronal maturation during development (20) and also plays a crucial role promoting glioma stem cell survival and stemness in glioma pathogenesis (21). Consistent with its increased mRNA expression following CIC depletion, WB and IF staining showed that protein levels of VGF were markedly elevated in CicKO brain lysates and cortex in comparison with WT counterparts (Figure 4, B and C). Increased VGF protein expression was also observed in cultured NPC and their in vitro differentiated progenies following CicKD (Figure 4D). Importantly, enforced VGF expression compromised neuronal differentiation capacity of NPC and blocked its cell cycle exit under differentiation induction (Figure 4, E–G), recapitulating the phenotype observed in CicKD NPC. In addition, we generated VGF–knocked down cells utilizing CRISPR/Cas9 to further determine the role of VGF in neuronal differentiation. Two independent guide RNA sequences targeting VGF (single guide RNA for VGF; sg-Vgf) partially restored neuronal differentiation in CicKD NPC, confirming its role as a negative regulator of terminal neuronal differentiation (Figure 4, H–J). These findings establish VGF as a crucial mediator of CIC function during neurogenesis.
Figure 4. Aberrant expression of VGF compromises neurogenesis in CIC-deficient brains.
(A) Biological replicates of CIC ChIP-seq tracks at Vgf promoter regions. (B) WB analysis in P0.5 brains of CicWT and CicKO mouse. SE, short exposure; LE, long exposure. The blots were run on same gel. Mean ± SEM of 3 experimental animals. (C) IF analysis for VGF in the P14 brains of CicWT and CicKO. Scale bar: 100 μm. (D) WB analysis for VGF in NTKD NPC and CicKD NPC during differentiation. The blots were run on same gel. (E) IF analysis for Flag, VGF, NeuN, and BrdU in VGF-Flag–expressed NPC (VGF-F) and control NPC (con) at 7 days of differentiation. Scale bar: 50 μm. (F) Quantitations for NeuN+ or BrdU+ numbers are plotted. Mean ± SEM of 100 DAPI+ nuclei from 3 experiments. (G) WB analysis for indicated proteins in control NPC and VGF-F NPC at 5 days of differentiation. (H) WB analysis for indicated proteins in NTKD NPC and CicKD NPC at 4 days of differentiation. sg-NT, nontargeted control single-guide RNA. For G and H, samples were run on 2 gels, and the representative ACTB blot is shown. (I) IF analysis for NEUN in NTKD NPC and CicKD NPC at 7 days of differentiation. (J) Quantifications for images from I are plotted. Mean ± SEM of 100 DAPI+ nuclei from 3 experiments. Statistical significance was determined by unpaired t test for B and F and 1-way ANOVA for J. *** P < 0.001. For C, D, E, G, H, and I, 3 experiments were conducted, and each representative result is shown.
CIC transcriptionally represses VGF expression.
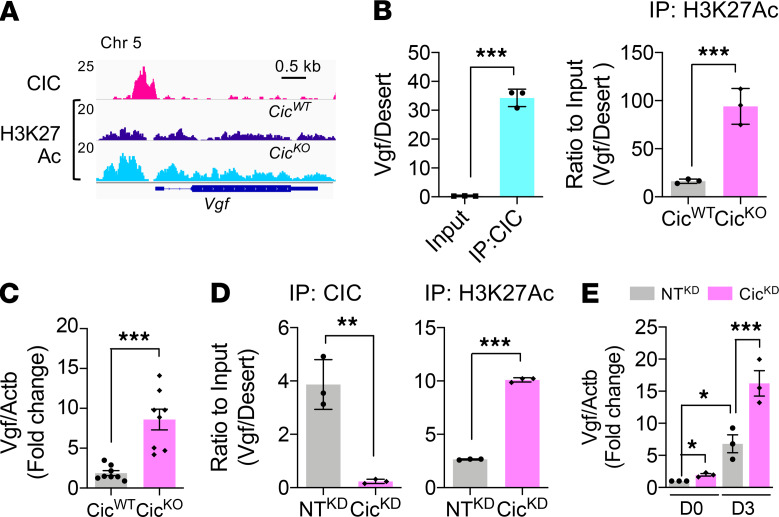
To confirm VGF as a direct CIC transcriptional target, we analyzed the ChIP-seq profile from paired P0.5 CicWT and CicKO brain samples. The CIC ChIP-seq track revealed an evident peak at the vicinity of the transcription start site (TSS) of the Vgf gene (Figure 5A). The Vgf promoter–specific CIC binding was verified by ChIP-qPCR results from 2 independent sets of P0.5 brain samples (Figure 5B). In agreement with its function as a member of the HMG-box superfamily of transcriptional repressors, ChIP and qPCR analysis from independent sets of paired CicWT and CicKO brain samples showed that brain-specific CIC depletion elicited significant increase of H3K27 acetylation within the Vgf promoter region and also elevation of Vgf mRNA expression (Figure 5, B and C). Similar enrichment of Vgf promoter–specific CIC occupancy, regional increase of H3K27Ac density, and correlated increase in its mRNA level following CIC depletion were also observed in cultured NPC and their differentiated progenies (Figure 5, D and E). Together, these findings indicate that VGF is a bona fide CIC transcriptional downstream target.
Figure 5. CIC transcriptionally represses VGF.
(A) Indicated ChIP-seq tracks at Vgf promoter region. (B) ChIP-qPCR results from P0.5 brains at Vgf promoter region. Mean ± SEM of 3 experimental animals. (C) qPCR results of Vgf mRNA expressions in P0.5 cerebral cortex of CicWT and CicKO. Mean ± SEM of 8 experimental animals. (D) ChIP-qPCR results from NTKD NPC and CicKD NPC at Vgf promoter region. Mean ± SEM of 3 experiments. (E) qPCR results of Vgf mRNA expression in the NTKD NPC and CicKD NPC at day 0 (D0) and D3 of differentiation. Mean ± SEM of 3 experiments. Statistical significance was determined by unpaired t test for B, C, and D and by 1-way ANOVA for E. *P < 0.05, **P < 0.01, *** P < 0.001.
CIC interacts with mSWI/SNF complex during neurogenesis.
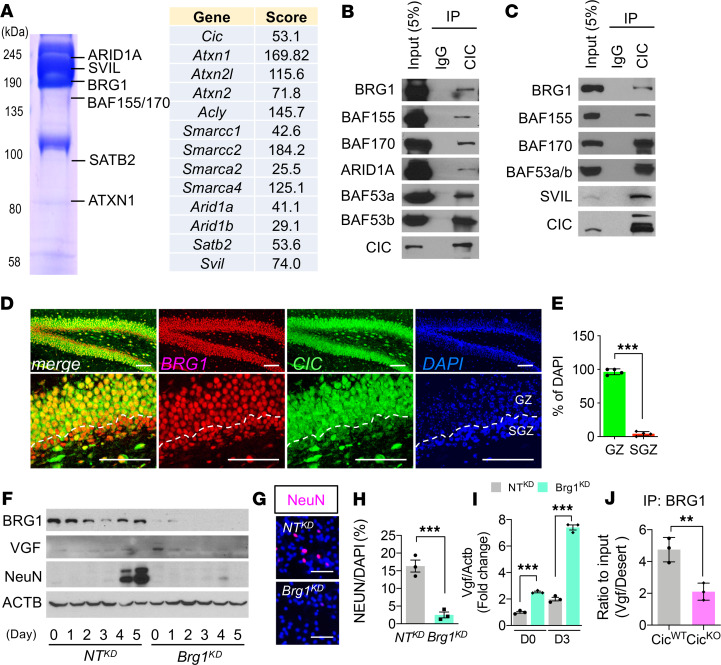
To explore the molecular mechanism underlying CIC-mediated transcriptional repression during neurogenesis, we performed CIC immunoprecipitation (IP) from P1 brain lysates followed by mass spectrometry (MS). Besides previously reported CIC-interacting proteins like ACLY (22) and ATXN1 (23), the IP-MS analysis uncovered many core components of mSWI/SNF complex, including ARID1A, ARID1B, SMARCC1 (BAF155), SMARCC2 (BAF170), SMARCA2 (BAF190B, BRM), and BRG1 (SMARCA4) (Figure 6A and Supplemental Table 2). The specific interaction of CIC with the mSWI/SNF complex was further verified by co-IP analysis of either CIC-transfected HEK293 cells (Figure 6B) or P0.5 CicWT mouse brains (Figure 6C).
Figure 6. mSWI/SNF complex is a molecular partner for CIC-dependent transcriptional regulation.
(A) The partial list of CIC-interacting proteins analyzed by mass spectrometry. (B) Co-IP analysis of CIC with components of the mSWI/SNF complex after expression in HEK293T cells. (C) Co-IP analysis of endogenous proteins from P0.5 brains. For B and C, a single experiment was conducted. (D) Co-IF analysis of CIC and BRG1 in P14 brains. Scale bar: 100 μm. (E) Ratio of CIC and BRG1 coexpressed cells to DAPI+ nuclei in D. GZ, granular zone. Mean ± SEM of the measurement from 50 DAPI+ nuclei from 3 animals. (F) WB analysis of differentiation time course of NTKD NPC and Brg1KD NPC. The blots were run on same gel. Three experiments were conducted, and a representative result is shown. (G) IF analysis for NeuN at day 7 of differentiation. Scale bar: 50 μm. (H) Quantitation of NeuN+ cells from G. Mean ± SEM of 100 DAPI+ nuclei from 3 experiments. (I) qPCR results for Vgf mRNA expression in NTKD NPC and Brg1KD NPC at day 0 (D0) and D3 of differentiation. Mean ± SEM of 3 experiments. (J) ChIP-qPCR results from IP of BRG1 from NTKD NPC and Brg1KD NPC at Vgf promoter regions. Mean ± SEM of 3 experiments. Statistical significance was determined by unpaired t test for E, H, and J and 1-way ANOVA for I. **P < 0.01, *** P < 0.001.
The mSWI/SNF, an ATP-dependent multisubunit chromatin remodeling complex, plays critical roles in the regulation of neural stem cell proliferation, neurogenesis, and neocortical development (24–26). Indeed, immunostaining found that BRG1, the core subunit of mSWI/SNF, was highly expressed in the granular cell layer of hippocampus, similar to the CIC expression pattern (Figure 6, D and E). To test whether disruption of the mSWI/SNF complex would recapitulate CIC loss–associated developmental and molecular phenotypes, we applied CRISPR/Cas9-based genomic editing to generate BRG1-depleted (Brg1KD) NPC (Figure 6F). Depletion of BRG1 not only compromised the differentiation capacity and neuronal marker expression during neuronally directed differentiation induction (Figure 6, G and H), but it also elevated VGF expression in the NPC and its differentiated progenies (Figure 6I). In addition, BRG1 ChIP-qPCR from P0.5 CicWT mouse brains showed enriched occupancy within the Vgf promoter region (Figure 6J). Importantly, this BRG1 binding was significantly reduced in CicKO brains, indicating that mSWI/SNF-mediated chromatin remodeling in regulation of VGF transcription requires CIC.
In addition to the mSWI/SNF complex, we also identified SVIL, an actin-binding protein encoded by supervillin gene, as a CIC- and mSWI/SNF complex–associated protein. IP of BRG1 followed by immunoblot analysis of P0.5 brain lysates confirmed the interaction of SVIL with mSWI/SNF complex (Supplemental Figure 6A). SVIL has been implicated in the regulation of neuronal differentiation (27). Interestingly, IF analysis of NPC revealed that SVIL was diffusively distributed throughout the cytoplasm, but it became mostly nuclear as the NPC underwent neuronal differentiation induction (Supplemental Figure 6B). To search for the motifs or amino acid residues critical for its localization switch, we performed MotifScan and identified S243 within its N-terminal nuclear localization signal region as a strong CDK5 phosphorylation site (Supplemental Figure 6C). Since CDK5 plays an important role in neuronal differentiation (28), we next tested whether its activity is required for differentiation-induced SVIL nuclear translocation. Indeed, treatment of NPC with CDK5 inhibitor roscovitine blocked the nuclear switch of SVIL when the cells were subjected to neuronal differentiation induction and compromised their differentiation (Supplemental Figure 6, D and E). Similarly, knockdown of SVIL (SvilKD) in NPC also suppressed their neuronal differentiation capacity, as evidenced by the lack of mature neuronal marker MAP2a/b and NeuN expressions. In addition, compared with the control NPC that fully underwent differentiation and exited cell cycle, approximately 5% of the SVIL-depleted cells continued to incorporate BrdU, even after 8 days of differentiation induction (Supplemental Figure 6, F–H). Together, these findings indicate that SVIL functions together with CIC-mSWI/SNF complex to facilitate neuronal differentiation.
CIC tethers SIN3-HDAC corepressor complex and mSWI/SNF complex to VGF promoter during neurogenesis.
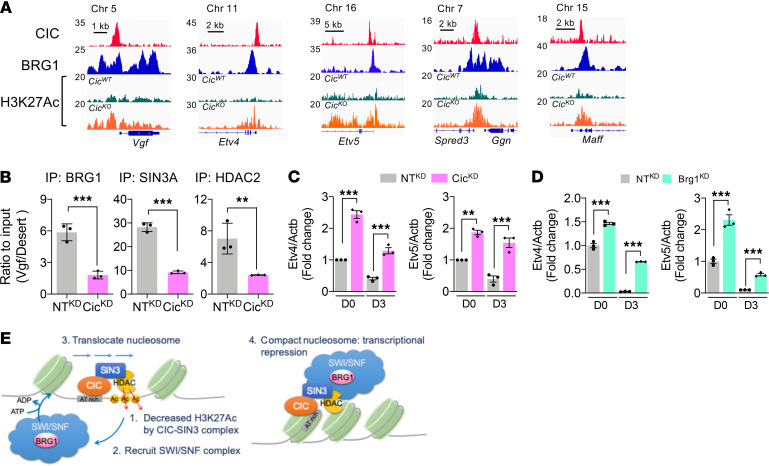
We found that CIC and BRG1 (29) showed largely overlapping peaks at the promoter regions of CIC target genes in developing mouse brains (Figure 7A). Consistent with the expression levels, H3K27Ac coverage in the CIC peak region was consistently higher on these genes in CicKO than CicWT brains, suggesting a HDAC-dependent transcriptional repression mechanism. SIN3 is a scaffold for the HDAC-associated transcriptional corepressor complex (30). Since SIN3 can interact with CIC and the mSWI/SNF complex through BRG1 (31, 32), we next tested whether the SIN3 corepressor complex is necessary for CIC-mSWI/SNF complex–mediated repression of VGF transcription. ChIP-qPCR analysis of NPC samples confirmed significantly enriched occupancies of SIN3A and neuronal SIN3 corepressor complex–associated HDAC2 within the Vgf promoter region, where CIC and BRG1 peaks overlap (33) (Figure 7B). Importantly, the regional enrichment of BRG1, SIN3A, and HDAC2 were all evidently reduced following CIC depletion, indicating that their recruitment to the Vgf promoter is dependent on the presence of CIC. Notably, CIC-dependent enrichment of BRG1/SIN3A/HDAC2 was not restricted to the Vgf promoter. Analysis of Etv4 and Etv5, known CIC targets, also showed coenrichment of CIC, BRG1, SIN3A, and HDAC2 within their promoter regions in control NPC (Supplemental Figure 7, A and B). Similar to the Vgf promoter, depletion of CIC reduced BRG1, SIN3A, and HDAC2 occupancy within Etv4 and Etv5 promoter regions, which led to increased H3K27Ac and mRNA expression. Concordantly, depletion of CIC or BRG1 was sufficient to upregulate the mRNA expression of Etv4 and Etv5 in differentiating NPC (Figure 7, C and D). Together, our results support a model that CIC functions as a neurogenic regulator by recruiting mSWI/SNF and SIN3-HDAC repressor complexes to transcriptionally regulate its target gene expression during neurogenesis (Figure 7E).
Figure 7. CIC represses VGF through tethering mSWI/SNF-SIN3 complex.
(A) ChIP tracks on CIC transcription targets. (B) ChIP-qPCR results from NTKD NPC and CicKD NPC at Vgf promoter region. Mean ± SEM of 3 experiments. (C) qPCR results of Etv4 and Etv5 mRNA expressions in NTKD NPC and CicKD NPC at day 0 (D0) and D3 of differentiation. Mean ± SEM of 3 experiments. (D) qPCR results of Etv4 and Etv5 mRNA expressions in NTKD NPC and Brg1KD NPC. Mean ± SEM of 3 experiments. Statistical significance was determined by unpaired t test for B and 1-way ANOVA for C and D. **P < 0.01, *** P < 0.001. (E) The model schema for transcriptional regulatory mechanism by CIC-mSWI/SNF-SIN3 complex.
Discussion
CIC is frequently mutated in oligodendrogliomas (34). This compelling genetic evidence argues for its role in neuro-oncogenesis. But its function in brain development and tumorigenesis remains poorly understood. Recent studies reported smaller cerebral size and behavioral deficit of forebrain in targeted Emx1-cre; Cicfl/fl mouse (14). These and the current study commonly found the reduction in number of neurons in cortical layers 2–4. Another study using Foxg1-cre–driven CicKO animals showed increased glia at the expense of neuronal differentiation (15). Considering the impaired neuronal maturation or terminal neural cell differentiation underlying neuro-oncogenesis, we hypothesized that CIC functions toward neuronal differentiation and development.
The reduced cortical thickness and hippocampal dentate gyrus neuronal density phenotype of broadly deleting Nes-cre–driven CicKO brain was consistent with that of forebrain-specific Emx1-cre–driven CicKO animals (14). We suspect loss of CIC during embryonic brain development impaired neuronal differentiation and maturation, as Cic-null P0.5 brains already showed decreased neuron-specific gene expression (Supplemental Figure 2, A and B). This is consistent with the report by Ahmad et al. (15). We also observed that reduced overall brain size became progressively pronounced postnatally, a finding consistent with the reported phenotype of Foxg1-cre:Cicfl/fl μ animals. The newly formed brain continues to grow by a rapid increase in number of axons, dendrites, synapses, and glial cells. Thus, postnatal brain growth problems of CIC-deficient brains may arise from disrupted connectivity due to defects in processes such as elaboration of axons and dendrites, spinogenesis and maturation, synaptogenesis and remodeling, gliogenesis, and myelination. We speculate that decreased maturation of neurons contributes to the phenotype, as the loss of CIC expression has a profound impact on the expression of dendrite and synaptogenesis genes based on RNA-seq analysis (Figure 1F). Gliogenesis and myelination changes appear less likely causes. Emx1-cre; Cicfl/fl (14) or Nes-cre; Cicfl/fl mice showed normal range of corpus callosum thickness (Supplemental Figure 2, E and F). In contrast, previous studies emphasized altered neural stem cell fate with expansion of OPC in CIC-deficient mouse brains. Foxg1-cre; Cicfl/fl animals at P2 displayed decreased corpus callosum and Mbp1/Cnp levels due to defective terminal differentiation of OPC (15). Similarly, increased OLIG2+ OPC in whole body CicKO at P28 and decreased oligodendrocytes differentiation of CicKO neural stem cells were noted (16). In our study, however, oligodendrocytes (OLIG2+), OPC (PDGFRA+), and astrocytes (GFAP+) were not expanded broadly in Cic-deleted postnatal brains. Also, MBP staining did not show appreciable differences, suggesting unaltered terminal differentiation of OPC into myelinating oligodendrocytes (Supplemental Figure 2, E and F). This lack of phenotype is unlikely due to the inefficient targeting of Cic in glia, given that the rate of Nes-cre driven recombination reaches nearly 100% in neural and glial progenitors during perinatal development (35). These phenotypes suggest that glial cell development was only marginally impacted in CIC-null brain.
Notably, Nes-cre–driven CicKO animals showed decreased postnatal growth, leading to mortality around weaning age. This is consistent with the lethality of previously reported telencephalon-targeted Foxg1-cre–driven CicKO animals (15). Based on the spectrum of different cre drivers, it is likely that Cic deletion in the basal ganglia may be the cause of lethality. The physiological role of CIC in keeping the viability of animals warrants further investigation.
Previous studies demonstrated that VGF-derived peptide — TLQP-62, the C-terminal 62 amino acid peptide — enhances neurogenesis of hippocampal cells in vitro and in vivo (36). Interestingly, VGF acted on early phases of neurogenesis by promoting neural progenitor proliferation while it inhibited terminal neuronal differentiation. It is noteworthy that CIC expression is dynamically upregulated through neuronal differentiation (Figure 2, A and C, and Figure 4D). In the hippocampal dentate gyrus neurogenesis, where CIC has a clear impact, we found that nuclear CIC is readily detected beginning in immature neurons with low TBR1 expression. The role of CIC lies at the transition from high TBR1 to low TBR1, as suggested by the expansion of high TBR1 cells upon the loss of CIC. These findings are concordant with the mechanism connecting CIC to VGF repression during neurogenic differentiation. We also noted that aberrantly increased VGF expression is not limited to phenotypically affected layers 2–4, but also present in layers 5–6 of the CicKO P14 cortex (Figure 4C). Derepressed VGF expression from the deep layer may have functions separate from CIC-mediated neuronal differentiation and maturation. Corroborating this possibility, a recent study demonstrated that VGF has a critical role in mood regulation (37, 38).
In our study, by combined ChIP-seq and RNA-seq analysis of P0.5 brains, we found previously identified bona fide transcriptional targets of CIC, including negative regulators of the RTK/RAS/MAPK pathway (e.g., Dusp4, Spred3, Spry4, and Nf1) and the PEA3 subfamily of ETS transcription factors (e.g., Etv4 and Etv5). These were all upregulated in Cic-deficient brains. Those repressed targets showed the CIC peak localized near TSS and largely overlapping with BRG1, in agreement with their molecular interaction. These CIC peaks cooccurred with H3K27Ac peaks, whose coverage is conspicuously increased upon the loss of CIC expression. This finding is consistent with a previous study from Weissman et al. (32) demonstrating SIN3-HDAC corepressor complex recruitment to target loci by CIC. Indeed, we found decreased SIN3A and HDAC2 on target promoters in Cic–knocked down cells, supporting our model (Figure 7E).
Interestingly, a subset of genes with intronic CIC peaks (e.g., Satb2, Mef2c, and Myt1l) showed downregulation in CicKO brains. These peaks, however, did not colocalize with H3K27Ac or BRG1 peaks, unlike genes that are repressed by CIC (Supplemental Figure 7C). This lack of overlap excludes its role on enhancer- or promoter-dependent regulation of those genes. Professional lineage-determining transcription factors like OLIG2 showed distinct mechanisms of target gene regulation engaging the BRG1-containing mSWI/SNF complex. OLIG2 prepatterns the recruitment of the mSWI/SNF complex to the enhancer of oligodendrocyte-specific genes (25). The mSWI/SNF complex recruited to distal lineage-specific enhancers interacts with p300 to modulate H3K27Ac (39). In contrast, CIC-mediated transcriptional regulations primarily occurred proximal to the TSS, unlike those lineage-determining transcription factors. This is likely due to the acute and dynamic regulation of CIC activity by the RAS/MAPK signaling pathway (32). CIC integrates extracellular signals to maintain the homeostasis. Its role is likely to facilitate the transition of differentiating cells by coordinating gene expression according to the extracellular cue. Whether and how intronic CIC peaks regulate the gene expression warrants further investigation.
In conclusion, these molecular mechanisms outlining how CIC regulates neuronal differentiation may help our understanding of CIC-deregulated brain tumorigenesis and facilitate the development of novel therapeutic strategies.
Methods
Generation of Cicfl/fl μ mouse.
Mice were maintained on a 12-hour light/dark cycle, and food and water were provided ad libitum. All mice were healthy, with no obvious behavioral phenotypes, and none of the experimental mice were immune compromised. For all mouse studies, mice of either sex were used, and mice were randomly allocated to experimental groups. Animals aged from P0.5–P14 were used. Specific end-point developmental ages used for each experiment are indicated in the figure legends. Cic targeting vector was acquired from the KOMP repository of KO mouse project (Vector, PG00139_Y_4_H08 – Cic, KOMP; https://www.komp.org/pdf.php?projectid=47159). The vector element is depicted in Supplemental Figure 1A. One hundred forty-four embryonic stem cell clones were screened for homologous recombination. Two clones were injected into blastocysts. A clone generated high-percentage chimeras and achieved germ-line transmission when crossed to C57BL/6 females. The heterozygous Cicneo–loxP/+ mouse was crossed with FLPe transgenic mouse (Jackson Laboratory, 003800) for recombination of FLPe recognition target (Frt) cassette. Cicfl/fl μ mice was crossed to Nes-cre transgenic mouse (Jackson Laboratory, 003771) for recombination of loxP sites. The recombination was confirmed by PCR. Mouse genotypes were determined by PCR using genomic DNA. The primer sets are described in Supplemental Table 3.
NPC cultures and differentiation into terminally differentiated neurons.
Ink/Arf–/– NPC (40) were transduced with viral particles of pInducer-ASCL1 (NPC). pDONR221-ASCL1 (DNASU, HsCD0004025) was gateway cloned into pInducer vector (Addgene, 44012). After selecting the culture with hygromycin 50 μg/mL (MilliporeSigma, 31282-04-9), the cells were divided and transduced with virus encoding shRNA for Cic (MilliporeSigma, TRCN0000302060 and TRCN0000082010), nontarget shRNA for control (MilliporeSigma, SHC016), pLu-VGF-Flag (in-house), or lentiCrisprV2-sgRNA (Addgene, 52961). After selected with puromycin (3 μg/mL, InvivoGen, anti–pr-1) or blasticidin (10 μg/mL, InvivoGen, anti–bl-1), all NPC are seeded on plates coated with fibronectin and poly-L-ornitine. On day 0, culture medium was replaced with N2 containing BDNF (10 ng/mL, PeproTech, 450-02), NT-3 (10 ng/mL, PeproTech, 450-03), B27 (Thermo Fisher Scientific, 1704044), and/or doxycycline (2 μg/mL, Research Products, D43020). On day 2, 0.5% FBS was added to the medium to support astrocyte survival, and the medium was changed every 2 days as we have done previously (19).
Productions of viruses.
A total of 1.5 × 107 293T cells were seeded in 150-mm tissue culture dishes. After 24 hours, the medium was replaced and cells were transfected with 4.5 μg of pMD2.G (Addgene, 12259), 9 μg of psPAX2 (Addgene, 12260), and 18 μg of target plasmid. After 48 hours and 72 hours of transfection, the medium containing viral particles was collected and was cleaned by filtration through a 0.45-μm cellulose acetate membrane. The viral particles were concentrated by ultracentrifugation for 2 hours at 70,000 × g and 4°C; then, the viral pellet was resuspended with 1 mL of Opti-MEM (Thermo Fisher Scientific) overnight at 4°C and stored in aliquots at –80°C.
Generation of knockdown cells with CRISPR-Cas9 strategy.
sgRNAs were cloned into lentiCrisprV2 as previously published (41). Viral particles were produced and infected to NPC, followed by puromycin (3 μg/mL) selection. Gene targeting was confirmed by IF and WB analysis. Target sequences for sgRNAs are listed in Supplemental Table 3.
IF analysis.
Cells were fixed with 4% paraformaldehyde (Thermo Fisher Scientific, 28906) for 15 minutes at room temperature, followed by permeabilization with 0.2% Triton X-100 (MilliporeSigma, T8787) in PBS (MilliporeSigma, P5493). The cells were subjected to IF staining with anti-CIC (1:500, in-house, ref. 23), anti-SATB2 (1:100, Abcam, ab51502/SATBA4B10-C-term), anti-CTIP2 (1:300, Abcam, ab18465/25B6), anti-BRG1 (1:1000, MilliporeSigma, 07-478), anti-VGF (1:300, Abcam, ab69989), anti-SVIL (1:1000, MilliporeSigma, S8695), anti-nestin (1:1000, DSHB, rat-401), anti-GFAP (1:2,00, Origene, UM5000005/UMAB5), anti-NeuN (1:300, Cell Signaling Technology, 24307), anti-MAP2a/b (1:100, Abcam, 36447, MT07), anti-DCX (1:300, Santa Cruz Biotechnology Inc., SC-8066/C-18), anti-TBR1 (1:100, Abcam, ab31940), anti-TBR2 (1:100, Abcam, ab23345), or anti-SOX2 (1:500, Abcam, ab97959) antibodies incubated overnight at 4°C and followed by labeling with Alexa 488–labeled anti-rabbit (Thermo Fisher Scientific, A21206) and Alexa 568–labeled anti-mouse secondary antibodies at room temperature for 1 hour. For BrdU IF staining, cells were incubated with 0.5 μg/mL of BrdU (MilliporeSigma, B5002) for 3 hours, followed by 1N HCl (MilliporeSigma, H1758) for 10 minutes on ice and 2N HCl at 37°C for 20 minutes and neutralization with 0.1M sodium borate buffer, pH 8.5, for 10 minutes at room temperature. IF staining was performed with anti-BrdU (1:500, Dako, M0744) antibody overnight at 4°C, followed by labeling with Alexa 568–labeled anti-mouse secondary antibody at room temperature for 1 hour. Images were acquired using EVOS FL Cell Auto imaging system (Thermo Fisher Scientific, AMAFD1000).
WB analysis.
Cells were lysed by RIPA buffer (Thermo Fisher Scientific, 89901) or Laemmli buffer (MilliporeSigma, 38733), followed by sonication (30 watt/5 sec/10 cycles). Protein concentration was determined by using Pierce BCA protein assay kit (Thermo Fisher Scientific, 23227). A total of 30 μg of proteins were fractionated by SDS-PAGE electrophoresis and transferred to PVDF membrane (Thermo Fisher Scientific, 88518) using a transfer apparatus (Bio-Rad, 1703930) following manufacturer instructions. After incubation with 5% skim milk in TBST (10 mM Tris [MilliporeSigma, 93362, pH 8.0], 150 mM NaCl (MilliporeSigma, S7653), 0.5% Tween 20 [MilliporeSigma, P7949]) for 1 hour, the membrane was incubated with antibodies against β-actin, CIC (1:1000), SATB2 (1:500), CTIP2 (1:2000), NeuN (1:1000), DCX (1:2,000, Cell Signaling Technology, 4604), BRG1 (1:3000), SVIL (1:1000), α-tubulin (1:10000, clone4A1, DSHB, AB_2732839), OLIG2 (1:5000, MilliporeSigma, AB9610), PDGFRA (1:1000, Cell Signaling Technology, 3174), CNPase (1:2000, Abcam, ab6319), GFAP (1:5000), or VGF (1:1000, made by Salton SR, Icahn School of Medicine, New York, New York, USA) (42) overnight at 4°C. Membranes were washed 3 times with TBST for 30 minutes and then incubated with HRP-conjugated anti-mouse (Thermo Fisher Scientific, 31430), anti-rabbit (Thermo Fisher Scientific, 31460), or anti–guinea pig (Thermo Fisher Scientific, A18769; 1:10,000) diluted in 3% skim milk for 1 hour. Blots were washed with TBST 3 times and developed with the SuperSignal West Pico Chemiluminescent substrate (Thermo Fisher Scientific, 34080) according to the manufacturer’s protocols.
IP analysis.
Cell or tissues were lysed using 1% TNT buffer (135 mM NaCl, 20 mM Tris-HCl, 1 mM EDTA (MilliporeSigma, E6758), and 1% Triton X-100) for 30 minutes on ice. After centrifugation for 15 minutes at 4°C (16,300 × g) to remove the debris, 500 μg of protein was incubated with 1–3 μg of antibody at 4°C for 16 hours. A total of 15 μL of Dynabeads Protein G (Thermo Fisher Scientific, 1009D) was added and incubated 4°C for 3 hours. The beads were washed 3 times with 1% TNT buffer and eluted the proteins with 2× Laemmli buffer. The protein interaction was determined by WB protocol.
qPCR analysis.
Total RNAs were extracted from cells or tissues by using NucleoSpin RNA kit (Macherey-Nagel, 740955.25). Reverse transcription was carried out on 500 ng of total RNA using RevertAid RT kit (Thermo Fisher Scientific, K1691). qPCR was performed on cDNA samples using the PowerUp SYBR Green Master Mix (Thermo Fisher Scientific, A25778) and was performed the qPCR on the 7500 Fast Real-time PCR system (Thermo Fisher Scientific, 4351106). Primer sequences are listed in Supplemental Table 3. Each sample was run as duplicates, and the mRNA level of each sample was normalized to that of ACTB mRNA. The relative mRNA level was presented as unit values of 2ΔCt.
MS analysis.
The analysis was performed at the Proteomics and Metabolomics Core facility of Weill Cornell Medicine. In-gel digestion was performed according to a previous published protocol (43). Briefly, gel pieces were excised and distained, followed by reduction, alkylation, and digestion with trypsin. The peptides were then extracted from the gels, desalted, and analyzed by liquid chromatography–tandem MS (LC-MS/MS). An EASY-nLC 1200 coupled on-line to a Fusion Lumos mass spectrometer was used for LC-MS/MS analysis. Buffer A (0.1% formic acid in water) and buffer B (0.1% formic acid and 80% acetonitrile in water) were used as mobile phases for gradient separation. A 75 μm × 15 cm chromatography column was packed in-house for peptide separation. Peptides were separated with a gradient of 5%–40% buffer B over 20 minutes and 40%–100% B over 5 minutes at a flow rate of 300 nL/min. The Fusion Lumos mass spectrometer was operated in data-dependent mode with 1-second cycle time. Survey scans were acquired in the Orbitrap mass analyzer over a range of 300–1500 m/z with resolution 120,000 at m/z 200. The most abundant precursors from the survey scan were selected with an isolation window of 1.6 Thomsons and fragmented by higher-energy collisional dissociation with normalized collision energy of 35. MS/MS scans were acquired in the ion trap mass analyzer with rapid scan rate. The automatic gain control target value was 1 × 106 for MS scans and 1 × 104 for MS/MS scans, and the maximum ion injection time was 60 ms for both. The raw files were processed using the MaxQuant computational proteomics platform (44) (version 1.5.5.1). The fragmentation spectra were searched against the UniProt mouse protein data base (contain 80,593 sequences) and allowed up to 2 missed tryptic cleavages. Oxidation of methionine and protein N-terminal acetylation were used as variable modifications for data base searching. Carbamidomethylation of cysteine was used as a fixed modification. The precursor and fragment mass tolerances were set to 7 ppm and 0.5 Da, respectively. Both peptide and protein identifications were filtered at 1% FDR.
RNA-seq and data analysis.
Total RNAs were isolated from P0.5 brains of CicWT and CicKO mouse and subjected to RNA-seq at the Genomics Resources Core facility of Weill Cornell Medicine. RNA-seq libraries were prepared using the Illumina TruSeq stranded mRNA library preparation kit and sequenced on HiSeq4000 sequencer (Illumina). RNA-seq data were aligned to the mm9 reference genome using STAR 2.3.0e (45). Raw counts of each transcript were measured by featureCounts v1.4.6-p5 (46). Lists of differentially expressed genes were generated by DESeq2-1.4.5 in R (47). GSEA in this manuscript were generated from a GSEA preranked model. The input of GSEA is the gene expression level logFC (CicKO mouse versus control). Pathway analysis (Figure 3E) was performed using DAVID 6.8 (48, 49).
ChIP-seq and qPCR analysis.
We used whole brain from P0.5 mouse or 1 × 107 of NPCs for each analysis. ChIP analysis was performed with modifications (50). In brief, tissues or cells were reacted with 5 mM EGS (Thermo Fisher Scientific, 21565) for 45 minutes at room temperature to cross-link protein-to-protein interaction. After centrifugation and removal of supernatant at 1700 × g for 5 minutes at 4°C, cross-linked for 10 minutes with 1% paraformaldehyde, and quenched with 120 mM glycine (MilliporeSigma, 50046) for 5 minutes at room temperature. After nucleus isolation, the chromatin was digested with 6000 gel units of micrococcal nuclease for 10 minutes at 37°C. The enzyme reaction was quenched by adding 20 mM EDTA. We resuspended the pellet in shearing buffer (50 mM Tris-HCl, 10 mM EDTA, and 0.1% SDS [MilliporeSigma, 71725]) and broke the nuclear membrane using the Covaris M220 Focused ultrasonicator according to the manufacturer’s instructions. IP was performed with 10 μg of anti-CIC, anti-BRG1 (Cell signaling Technology, 49360), anti-H3K27Ac (Abcam, 4729), anti-Sin3A (Abcam, ab3479), or anti-HDAC2 (Cell Signaling Technology, 57165) overnight at 4°C. A total of 30 μL of precleared Dynabeads Protein G was added and incubated for 3 hours at 4°C. We washed the beads with high salt buffer (50 mM HEPES, 500 mM NaCl, 1 mM EDTA, 0.1% SDS, 1% Triton X-100, and 0.1% sodium deoxycholate) and RIPA buffer (including LiCl) and eluted the chromatin by elution buffer (50 mM Tris-HCl, 10 mM EDTA, and 1% SDS). After being treated with RNase and Proteinase K, the DNA was incubated at 65°C overnight to reverse cross-linking. DNA was extracted using NucleoSpin Gel and a PCR clean-up DNA extraction kit (Macherey-Nagel, 740609.25) and carried out size-selection to obtain about 300 bp of DNA fragments using SPRIselect Reagent (Beckman Coulter, B23317). qPCR was performed using specific primers described in Supplemental Table 3.
Libraries were made using KAPA Hyper Prep kit (KAPA Biosystems, KR0961) following manufacturer’s instructions. Briefly, 30 ng from each immunoprecipitated or input DNA were end-repaired, phosphorylated, A-tailed, and ligated to adaptors. Ligated products were size selected with 0.8× SPRI beads to obtain 250–350 bp of DNA. After purification, 8-cycle PCR amplification reaction was performed. PCR product was cleaned by the use of 1× SPRI beads. The final product was resuspended in 30 μL of Tris buffer. Final yields were quantified in a Qubit 4.0 Fluorometer, and the quality of the library was assessed by running on a DNA1000 Bioanalyzer chip. Libraries were normalized to 2 nM and pooled at the desired plexity. Sequencing and postprocessing of the raw data were performed at the Epigenomics Core at Weill Cornell Medicine as follows. Libraries were clustered at 6 pM on single-read flow cell and sequenced for 50 cycles on an Illumina HiSeq 2500. Illumina’s CASAVA 1.8.2 software was used to perform image capture, base calling, and demultiplexing.
ChIP-seq data were aligned to the mm9 reference genomes using bowtie-0.12.9 with default parameters -n 2 and -best (51). Peak calling was performed by macs14 1.4.2 (52) with default parameters. BigWig files of ChIP-seq data track and analysis of read density in peak regions were generated using deeptools 3.1.3 (53). Read density of specific genomic regions were displayed using Integrative Genomics Viewer (IGV) 2.4.19 (54).
Data availability.
The RNA-seq and ChIP-seq data sets produced in this study are available at NCBI GEO (GSE131302; http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE131302).
Statistics.
We determined experimental sample sizes on the basis of preliminary data. All results are expressed as mean ± SEM. GraphPad Prism software was used for all statistical analyses. Normal distribution of the sample sets was determined before applying unpaired Student’s 2-tailed t test for 2 group comparisons. One-way ANOVA was used to assess the differences between multiple groups. The mean values of each group were compared by the Bonferroni’s post hoc procedure. Differences were considered significant when P < 0.05.
Study approval.
All animal use was approved by the IACUC of the Weill Cornell Medicine (no. 2011-0088).
Author contributions
Conception and design were contributed by IH and JP. Development of methodology was contributed by IH and HP. Acquisition of data was contributed by IH. Analysis and interpretation of data (e.g., statistical analysis, biostatistics, computational analysis) were contributed by IH, JY, HP, HZ, and JP. Writing of the manuscript was contributed by IH, HZ, and JP. Administrative, technical, or material support (e.g., reporting or organizing data, constructing data bases) was contributed by IH, OE, HZ, and JP. Study supervision was contributed by JP and OE.
Supplementary Material
Acknowledgments
Authors thank Stephen Salton for the VGF antibody; Natalia De Marco for the Emx1-cre mouse; Yoontae Lee for providing CIC expression plasmids; Huda Zhoghbi for CIC antisera, which was used for the initial optimization of CIC immunoblotting; and Angana Biswas for proofreading this manuscript. We thank Zhe Cheng and Guoan Zhang at the Proteomics and Metabolomics Core Facility and Tuo Zhang at the Genomics Core Facility of The Weill Cornell Medicine for MS and sequencing data analysis. JP is supported by the Irma T. Hirschl Award and NIH grant AG048284.
Version 1. 03/31/2020
In-Press Preview
Version 2. 05/07/2020
Electronic publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2020, American Society for Clinical Investigation.
Reference information: JCI Insight. 2020;5(9):e135826.https://doi.org/10.1172/jci.insight.135826.
Contributor Information
Inah Hwang,, Email: nahwang@gmail.com.
Heng Pan,, Email: hep2007@med.cornell.edu.
Jun Yao, Email: JYao1@mdanderson.org.
Olivier Elemento,, Email: ole2001@med.cornell.edu.
Hongwu Zheng,, Email: hoz4001@med.cornell.edu.
Jihye Paik, Email: jep2025@med.cornell.edu.
References
- 1.Cairncross G, et al. Phase III trial of chemoradiotherapy for anaplastic oligodendroglioma: long-term results of RTOG 9402. J Clin Oncol. 2013;31(3):337–343. doi: 10.1200/JCO.2012.43.2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cairncross JG, et al. Specific genetic predictors of chemotherapeutic response and survival in patients with anaplastic oligodendrogliomas. J Natl Cancer Inst. 1998;90(19):1473–1479. doi: 10.1093/jnci/90.19.1473. [DOI] [PubMed] [Google Scholar]
- 3.Bettegowda C, et al. Mutations in CIC and FUBP1 contribute to human oligodendroglioma. Science. 2011;333(6048):1453–1455. doi: 10.1126/science.1210557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jiao Y, et al. Frequent ATRX, CIC, FUBP1 and IDH1 mutations refine the classification of malignant gliomas. Oncotarget. 2012;3(7):709–722. doi: 10.18632/oncotarget.588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yip S, et al. Concurrent CIC mutations, IDH mutations, and 1p/19q loss distinguish oligodendrogliomas from other cancers. J Pathol. 2012;226(1):7–16. doi: 10.1002/path.2995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee CJ, et al. CIC, a member of a novel subfamily of the HMG-box superfamily, is transiently expressed in developing granule neurons. Brain Res Mol Brain Res. 2002;106(1-2):151–156. doi: 10.1016/S0169-328X(02)00439-4. [DOI] [PubMed] [Google Scholar]
- 7.Jiménez G, Guichet A, Ephrussi A, Casanova J. Relief of gene repression by torso RTK signaling: role of capicua in Drosophila terminal and dorsoventral patterning. Genes Dev. 2000;14(2):224–231. [PMC free article] [PubMed] [Google Scholar]
- 8.Jin Y, et al. EGFR/Ras Signaling Controls Drosophila Intestinal Stem Cell Proliferation via Capicua-Regulated Genes. PLoS Genet. 2015;11(12):e1005634. doi: 10.1371/journal.pgen.1005634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Park S, et al. Capicua deficiency induces autoimmunity and promotes follicular helper T cell differentiation via derepression of ETV5. Nat Commun. 2017;8:16037. doi: 10.1038/ncomms16037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tan Q, et al. Loss of Capicua alters early T cell development and predisposes mice to T cell lymphoblastic leukemia/lymphoma. Proc Natl Acad Sci USA. 2018;115(7):E1511–E1519. doi: 10.1073/pnas.1716452115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee Y, et al. ATXN1 protein family and CIC regulate extracellular matrix remodeling and lung alveolarization. Dev Cell. 2011;21(4):746–757. doi: 10.1016/j.devcel.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Simón-Carrasco L, et al. Inactivation of Capicua in adult mice causes T-cell lymphoblastic lymphoma. Genes Dev. 2017;31(14):1456–1468. doi: 10.1101/gad.300244.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim E, et al. Deficiency of Capicua disrupts bile acid homeostasis. Sci Rep. 2015;5:8272. doi: 10.1038/srep08272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lu HC, et al. Disruption of the ATXN1-CIC complex causes a spectrum of neurobehavioral phenotypes in mice and humans. Nat Genet. 2017;49(4):527–536. doi: 10.1038/ng.3808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ahmad ST, et al. Capicua regulates neural stem cell proliferation and lineage specification through control of Ets factors. Nat Commun. 2019;10(1):2000. doi: 10.1038/s41467-019-09949-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang R, et al. Cic Loss Promotes Gliomagenesis via Aberrant Neural Stem Cell Proliferation and Differentiation. Cancer Res. 2017;77(22):6097–6108. doi: 10.1158/0008-5472.CAN-17-1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mootha VK, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34(3):267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 18.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hwang I, et al. Far Upstream Element-Binding Protein 1 Regulates LSD1 Alternative Splicing to Promote Terminal Differentiation of Neural Progenitors. Stem Cell Reports. 2018;10(4):1208-1221. doi: 10.1016/j.stemcr.2018.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alder J, et al. Brain-derived neurotrophic factor-induced gene expression reveals novel actions of VGF in hippocampal synaptic plasticity. J Neurosci. 2003;23(34):10800–10808. doi: 10.1523/JNEUROSCI.23-34-10800.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang X, et al. Reciprocal Signaling between Glioblastoma Stem Cells and Differentiated Tumor Cells Promotes Malignant Progression. Cell Stem Cell. 2018;22(4):514–528.e5. doi: 10.1016/j.stem.2018.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chittaranjan S, et al. Mutations in CIC and IDH1 cooperatively regulate 2-hydroxyglutarate levels and cell clonogenicity. Oncotarget. 2014;5(17):7960–7979. doi: 10.18632/oncotarget.2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lam YC, et al. ATAXIN-1 interacts with the repressor Capicua in its native complex to cause SCA1 neuropathology. Cell. 2006;127(7):1335–1347. doi: 10.1016/j.cell.2006.11.038. [DOI] [PubMed] [Google Scholar]
- 24.Ninkovic J, et al. The BAF complex interacts with Pax6 in adult neural progenitors to establish a neurogenic cross-regulatory transcriptional network. Cell Stem Cell. 2013;13(4):403–418. doi: 10.1016/j.stem.2013.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yu Y, et al. Olig2 targets chromatin remodelers to enhancers to initiate oligodendrocyte differentiation. Cell. 2013;152(1-2):248–261. doi: 10.1016/j.cell.2012.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sokpor G, Xie Y, Rosenbusch J, Tuoc T. Chromatin Remodeling BAF (SWI/SNF) Complexes in Neural Development and Disorders. Front Mol Neurosci. 2017;10:243. doi: 10.3389/fnmol.2017.00243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Laurent B, et al. A specific LSD1/KDM1A isoform regulates neuronal differentiation through H3K9 demethylation. Mol Cell. 2015;57(6):957–970. doi: 10.1016/j.molcel.2015.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jessberger S, Gage FH, Eisch AJ, Lagace DC. Making a neuron: Cdk5 in embryonic and adult neurogenesis. Trends Neurosci. 2009;32(11):575–582. doi: 10.1016/j.tins.2009.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Attanasio C, et al. Tissue-specific SMARCA4 binding at active and repressed regulatory elements during embryogenesis. Genome Res. 2014;24(6):920–929. doi: 10.1101/gr.168930.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Laherty CD, Yang WM, Sun JM, Davie JR, Seto E, Eisenman RN. Histone deacetylases associated with the mSin3 corepressor mediate mad transcriptional repression. Cell. 1997;89(3):349–356. doi: 10.1016/S0092-8674(00)80215-9. [DOI] [PubMed] [Google Scholar]
- 31.Kuzmichev A, Zhang Y, Erdjument-Bromage H, Tempst P, Reinberg D. Role of the Sin3-histone deacetylase complex in growth regulation by the candidate tumor suppressor p33(ING1) Mol Cell Biol. 2002;22(3):835–848. doi: 10.1128/MCB.22.3.835-848.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weissmann S, Cloos PA, Sidoli S, Jensen ON, Pollard S, Helin K. The Tumor Suppressor CIC Directly Regulates MAPK Pathway Genes via Histone Deacetylation. Cancer Res. 2018;78(15):4114–4125. doi: 10.1158/0008-5472.CAN-18-0342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Baltan S, Bachleda A, Morrison RS, Murphy SP. Expression of histone deacetylases in cellular compartments of the mouse brain and the effects of ischemia. Transl Stroke Res. 2011;2(3):411–423. doi: 10.1007/s12975-011-0087-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cancer Genome Atlas Research Network, et al. Comprehensive, Integrative Genomic Analysis of Diffuse Lower-Grade Gliomas. N Engl J Med. 2015;372(26):2481–2498. doi: 10.1056/NEJMoa1402121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liang H, Hippenmeyer S, Ghashghaei HT. A Nestin-cre transgenic mouse is insufficient for recombination in early embryonic neural progenitors. Biol Open. 2012;1(12):1200–1203. doi: 10.1242/bio.20122287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thakker-Varia S, et al. VGF (TLQP-62)-induced neurogenesis targets early phase neural progenitor cells in the adult hippocampus and requires glutamate and BDNF signaling. Stem Cell Res. 2014;12(3):762–777. doi: 10.1016/j.scr.2014.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mizoguchi T, Hara H, Shimazawa M. VGF has Roles in the Pathogenesis of Major Depressive Disorder and Schizophrenia: Evidence from Transgenic Mouse Models. Cell Mol Neurobiol. 2019;39(6):721–727. doi: 10.1007/s10571-019-00681-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jiang C, et al. VGF and its C-terminal peptide TLQP-62 in ventromedial prefrontal cortex regulate depression-related behaviors and the response to ketamine. Neuropsychopharmacology. 2019;44(5):971–981. doi: 10.1038/s41386-018-0277-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Alver BH, et al. The SWI/SNF chromatin remodelling complex is required for maintenance of lineage specific enhancers. Nat Commun. 2017;8:14648. doi: 10.1038/ncomms14648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Khoo CM, Carrasco DR, Bosenberg MW, Paik JH, Depinho RA. Ink4a/Arf tumor suppressor does not modulate the degenerative conditions or tumor spectrum of the telomerase-deficient mouse. Proc Natl Acad Sci USA. 2007;104(10):3931–3936. doi: 10.1073/pnas.0700093104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sanjana NE, Shalem O, Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods. 2014;11(8):783–784. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chakraborty TR, Tkalych O, Nanno D, Garcia AL, Devi LA, Salton SR. Quantification of VGF- and pro-SAAS-derived peptides in endocrine tissues and the brain, and their regulation by diet and cold stress. Brain Res. 2006;1089(1):21–32. doi: 10.1016/j.brainres.2006.02.124. [DOI] [PubMed] [Google Scholar]
- 43.Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal Chem. 1996;68(5):850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- 44.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26(12):1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 45.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30(7):923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 47.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 49.Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37(1):1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee TI, Johnstone SE, Young RA. Chromatin immunoprecipitation and microarray-based analysis of protein location. Nat Protoc. 2006;1(2):729–748. doi: 10.1038/nprot.2006.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10(3):R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9(9):R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ramírez F, et al. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 2016;44(W1):W160–W165. doi: 10.1093/nar/gkw257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Robinson JT, et al. Integrative genomics viewer. Nat Biotechnol. 2011;29(1):24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-seq and ChIP-seq data sets produced in this study are available at NCBI GEO (GSE131302; http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE131302).