Abstract
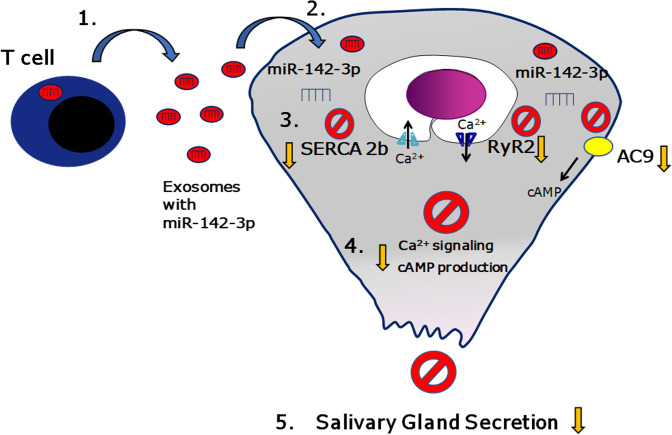
Sjögren’s syndrome (SS) is a systemic autoimmune disease that mainly affects exocrine salivary and lacrimal glands. Local inflammation in the glands is thought to trigger glandular dysfunction and symptoms of dryness. However, the mechanisms underlying these processes are incompletely understood. Our work suggests T cell exosome–derived miR-142-3p as a pathogenic driver of immunopathology in SS. We first document miR-142-3p expression in the salivary glands of patients with SS, both in epithelial gland cells and within T cells of the inflammatory infiltrate, but not in healthy volunteers. Next, we show that activated T cells secreted exosomes containing miR-142-3p, which transferred into glandular cells. Finally, we uncover a functional role of miR-142-3p–containing exosomes in glandular cell dysfunction. We find that miR-142-3p targets key elements of intracellular Ca2+ signaling and cAMP production — sarco(endo)plasmic reticulum Ca2+ ATPase 2b (SERCA2B), ryanodine receptor 2 (RyR2), and adenylate cyclase 9 (AC9) — leading to restricted cAMP production, altered calcium signaling, and decreased protein production from salivary gland cells. Our work provides evidence for a functional role of the miR-142-3p in SS pathogenesis and promotes the concept that T cell activation may directly impair epithelial cell function through secretion of miRNA-containing exosomes.
Keywords: Autoimmunity
Keywords: Autoimmune diseases
Introduction
Sjögren’s syndrome (SS) is a systemic autoimmune disease that affects exocrine glands, primarily salivary and lacrimal (1, 2). Mechanisms of SS pathogenesis are not fully understood, and disease triggers are not conclusively identified. However, it is thought that local inflammation in glandular tissues mediates exocrine gland dysfunction and leads to the primary disease manifestations of xerostomia (dry mouth) and xeropthalmia (dry eyes) in SS patients’ glands (1, 2). Yet, how inflammation triggers salivary gland dysfunction is not clearly delineated.
Previous work in our laboratory has focused on the role of miRNAs in the pathogenesis of salivary gland dysfunction in SS (3). miRNAs are endogenous small noncoding RNA molecules that regulate the expression of target genes through translational repression and/or degradation of mRNAs. miRNAs are involved in virtually every cellular process and are essential in cell development, differentiation, and physiology (4). Deregulation of miRNA transcription and function has been associated thus far with a variety of human pathological conditions (4). Dysregulation of miRNA expression has been also documented in SS and hypothesized to play a role in disease pathogenesis (3, 5).
We hypothesized that miRNA dysregulation may be involved in the pathogenesis of salivary gland dysfunction observed in SS. We therefore investigated miRNAs dysregulated in SS that could target elements/pathways involved in saliva secretion and exocrine gland physiology. Saliva secretion is a tightly regulated and coordinated process (6). Studies over the past 4 decades have demonstrated that Ca2+ and cAMP pathways are the primary signaling systems in secretory epithelia that control virtually all secretory gland functions (7). Our previous screening of miRNAs upregulated in Sjögren’s salivary glands revealed miR-142-3p as a candidate miRNA involved in the secretory dysfunction observed in Sjögren’s (3). Based on target prediction algorithms, miR-142-3p could target critical components of the Ca2+ and cAMP pathways. The adenylate cyclase 9 (AC9), the sarco(endo)plasmic reticulum Ca2+ ATPase 2b (SERCA2B) pump, and the ryanodine receptor 2 (RyR2) are all predicted targets of miR-142-3p and play a critical role in salivary gland secretion (7).
However, miR-142-3p has been previously documented to be expressed primarily in the immune compartment (8) and to play important roles in T cell functions (9). We therefore hypothesized that miR-142-3p may be secreted by inflammatory cells through exosomes in the lesions of SS, may target glandular cells, and may affect their function. Indeed, it is well recognized that miRNAs are transported through exosomes (10). These small noncoding RNAs can be transferred from their cells of origin into the recipient cells and alter their function (10). The transfer of miRNAs through exosomes has been shown to play important roles in the regulation of physiological immune responses but also in the development of autoimmune and carcinogenic diseases (11, 12).
Our current work demonstrates that miRNA-containing exosomes from activated T cells impair the function of salivary gland epithelial cells. We show that T cell–derived exosomes impair mechanisms linked to epithelial secretory function, including Ca2+ flux, cAMP production, and protein secretion. We identify miR-142-3p as a candidate regulator of glandular cell function in SS. Indeed, we document that miR-142-3p affects expression and function of SERCA2B and RyR2 that impairs intracellular Ca2+ signaling. Finally, we confirm the upregulation of miR-142-3p in lesions of patients with SS and correlate its expression with the downregulation of its targets SERCA2B and RyR2 in lesions of disease.
Results
miR-142-3p is overexpressed in salivary gland lesions of SS patients.
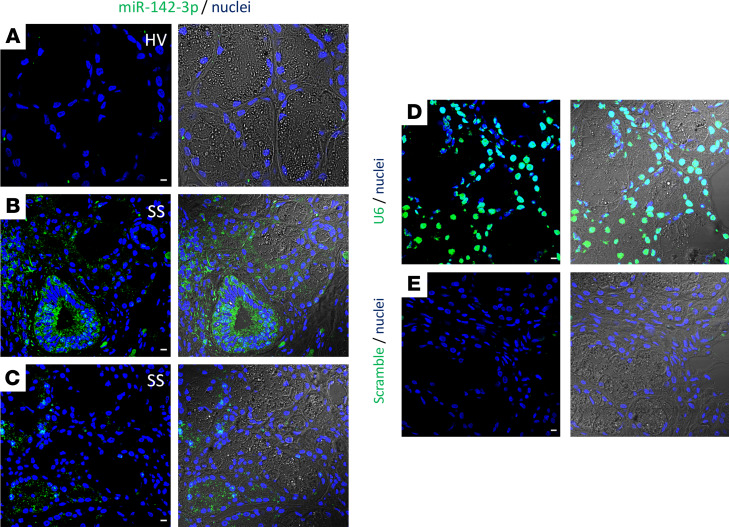
We first investigated miRNA overexpression in salivary gland lesions of patients with SS. miR-142-3p was expressed within salivary gland tissues of patients with SS (≈5.7-fold upregulation in patients with severe inflammation; Supplemental Figure 1A; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.133497DS1) (3). Furthermore, miR-142-3p was predicted to target molecules known to be involved in the secretory function of epithelial cells (SERCA2B, RyR2, AC9) (Supplemental Figure 1B). We first examined expression of miR-142-3p in labial salivary glands of healthy volunteers and patients with SS. To evaluate expression of miR-142-3p in tissues, we established an in situ hybridization (ISH) staining assay (Figure 1, A–E). We found that miR-142-3p was minimally detected or undetected in salivary glands of healthy volunteers (Figure 1A). However, in salivary glands of patients with SS, miR-142-3p was abundantly detected both within the inflammatory cell infiltrate (Figure 1B) and within the acinar/secretory cells and ductal cells of the gland (Figure 1C). These results demonstrated that miR-142-3p is highly expressed throughout SS salivary glands and led us to hypothesize that miR-142-3p may be involved in secretory dysfunction in SS disease.
Figure 1. miR-142-3p expression in minor salivary gland tissues.
(A–C) ISH of miR-142-3p in human minor salivary gland biopsies of healthy volunteers (HVs) and patients with SS. (D and E) ISH of U6 (positive control) and scrambled miRNA (negative control). (A–E) Left panels are ISH of miR-142-3p, U6, and negative control. Right panels represent transmission light combined with ISH for miR-142-3p, U6, and scramble RNA negative control. miRNA and U6 were stained by green, and cell nuclei were stained by blue. Scale bar: 10 μm. HVs; n = 3, SS patients; n = 4.
Overexpression of miR-142-3p targets SERCA2B, RyR2, and AC9 expression in salivary epithelial cells.
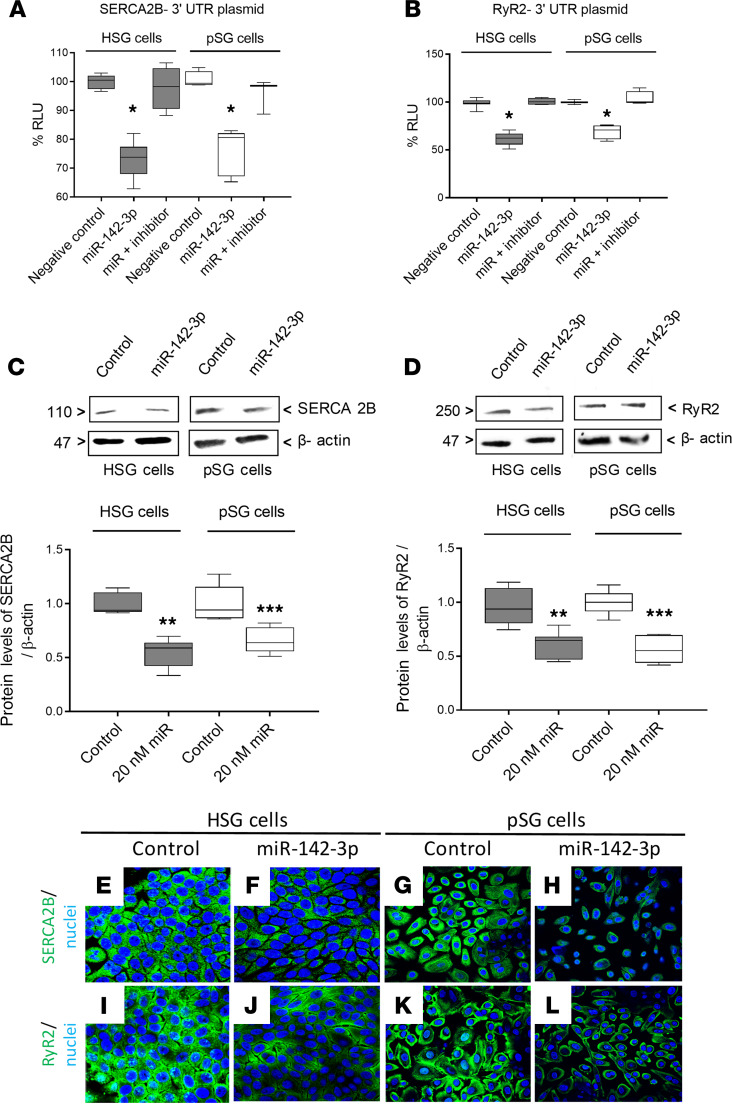
We next investigated the ability of miR-142-3p to target SERCA2B and RyR2 in a human submandibular gland cell line (HSG) system and in human-derived primary salivary gland (pSG) epithelial cells. To evaluate whether miR-142-3p targets the 3′-UTR of SERCA2B and RyR2, we cotransfected cells with miR-142-3p mimic and luciferase reporter, constructs containing either SERCA2B 3′-UTR or RyR2 3′-UTR. In these assays, luciferase activity indicated the expression of SERCA2B or RyR2, and reduced luciferase activity reflected inhibition due to binding of the miRNA to the UTR of the respective genes. Transfection with miR-142-3p mimic resulted in significant downregulation of luciferase activity for the SERCA2B 3′-UTR reporter in both HSG and pSG cells (Figure 2A). A significant downregulation in the luciferase activity of RyR2 3′-UTR reporter was also observed in miR-142-3p mimic–transfected HSG and pSG cells (Figure 2B). Cotransfection with a miR-142-3p hairpin inhibitor reversed the effect of miR-142-3p mimic for both SERCA2B 3′-UTR (Figure 2A) and RyR2 3′-UTR (Figure 2B). Overexpression of miR-142-3p mimic also led to significant downregulation of endogenous SERCA2B (Figure 2C) and RyR2 (Figure 2D) protein levels in both HSG and pSG cells (this effect was concentration dependent; Supplemental Figure 2). Decrease in protein levels of SERCA2B and RyR2 was supported by immunofluorescence staining of SERCA2B (Figure 2, E–H) and RyR2 (Figure 2, I–L) in miR-142-3p mimic–transfected HSG cells and pSG cells. miR-142-3p also targeted AC9 in epithelial cells (Supplemental Figure 3). AC9 is a validated target of miR-142-3p in T cells (13). Overexpression of miR-142-3p mimic led to decreased AC9 protein levels in HSG in salivary gland epithelial cells (as shown by both Western blot and immunofluorescence analysis; Supplemental Figure 3, A–C). These data thus validate SERCA2B, RyR2, and AC9 as targets for miR-142-3p.
Figure 2. SERCA2B and RyR2 are both targets of miR-142-3p in HSG and pSG cells.
(A and B) Dual luciferase reporter assays in HSG and pSG. Cells were cotransfected with plasmid 3′-UTR SERCA2B or 3′-UTR RyR2 and miR-142-3p mimic or miRNA hairpin inhibitor. Luciferase activity was measured in relative light units (RLU) (n = 4, median, maximum, and minimum shown). Statistical significance was determined by Mann-Whitney nonparametric test; *P < 0.05. (C and D) Protein levels of SERCA2B and RyR2 in HSG and pSG transfected with or without miR-142-3p mimic. (n = 5, median, maximum, and minimum shown; **P < 0.01, and ***P < 0.001 determined by Mann-Whitney nonparametric test.) The box plots depict the minimum and maximum values (whiskers), the upper and lower quartiles, and the median. The length of the box represents the interquartile range. (E–L) Immunofluorescence staining for SERCA2B and RyR2 (both green) in HSG and pSG transfected with or without miR-142-3p mimic. Cell nuclei were stained DAPI (blue). Scale bar: 10 μm. (n = 3 experiments per condition, 3 fields of view evaluated per experiment.)
Calcium signaling and cAMP production are downregulated by miR-142-3p in epithelial cells.
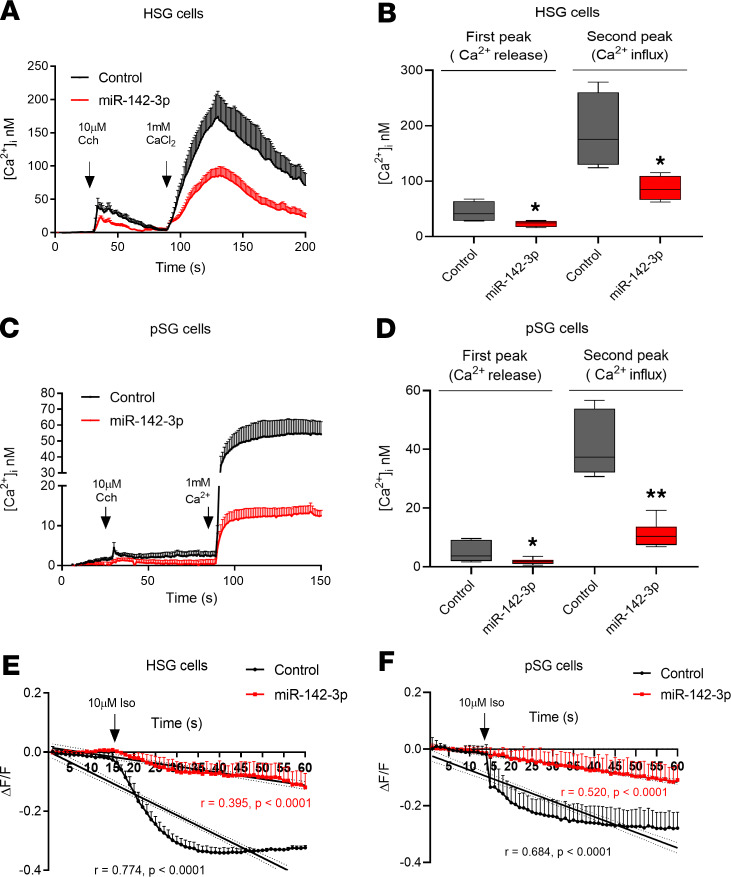
Because SERCA2B, RyR2, and AC9 are considered key elements for Ca2+ signaling and cAMP production, we hypothesized that overexpression of miR-142-3p would result in functional consequences mimicking impaired Ca2+ signaling and cAMP production in salivary gland epithelial cells. We therefore analyzed the functional effects of miR-142-3p overexpression on epithelial cells. For these experiments, HSG and pSG cells were transfected with miR-142-3p mimic or control mimic for 48 hours before loading with Fluo-4 acetoxymethyl ester calcium indicator. Intracellular calcium signaling was induced by carbachol (Cch) stimulation, and thereafter Ca2+ influx was further triggered by adding external 1 mM Ca2+ in solution. Cch 10 μM induced a rapid and transient elevation of [Ca2+]i in HSG cells (Figure 3, A and B, black first peak). However, HSG cells transfected with miR-142-3p mimic displayed significantly reduced levels of [Ca2+]i compared with control transfected HSG cells (44.46 ± 9.44 nM vs. 23.37 ± 2.84 nM) (Figure 3, A and B, first peak). Addition of external Ca2+ solution further increased [Ca2+]i levels in control HSG cells, while this increase was significantly blunted in HSG cells transfected with miR-142-3p mimic (188.4 ± 34.54 nM vs. 86.76 ± 11.1 nM) (Figure 3, A and B, second peak). Similar results were obtained in pSG cells (first peak: 4.97 ± 1.42 nM vs. 1.82 ± 0.40 nM and second peak: 41.86 ± 5.08 nM vs. 11.08 ± 1.67 nM) (Figure 3, C and D).
Figure 3. Ca2+ signaling and cAMP production are disrupted in miR-142-3p–transfected HSG and pSG cells.
(A and C) HSG and pSG cells loaded with Fluo-4-AM were stimulated with carbachol (Cch) (concentration, 10 μM) with or without 1 mM Ca2+ externally. Time course of [Ca2+]i in HSG and pSG cells. (B and D) Quantification of calcium release and influx peaks in HSG and pSG cells that were transfected with (red) and without (black) miR-142-3p mimic (n = 4, median, maximum, and minimum shown). Statistical significance was determined by Mann-Whitney nonparametric test; *P < 0.05, and **P < 0.01 (20 cells per condition, n = 4 experiments). The box plots depict the minimum and maximum values (whiskers), the upper and lower quartiles, and the median. The length of the box represents the interquartile range. (E and F) Time course of cAMP production. cADD, a cAMP biosensor, loaded in HSG and pSG cells that were stimulated with isoproterenol (Iso) (concentration, 10 μM). Linear regression analysis of cAMP production versus time in control salivary epithelial cells (black) and miR-142-3p mimic–transfected salivary epithelial cells (red). Averages of 4 independent experiments are shown with their standard deviation (20 cells per condition, n = 4 experiments).
Next, we evaluated the effect of miR-142-3p mimic overexpression on cAMP production. Both in HSG (Figure 3E) and in pSG cells (Figure 3F), cAMP production was significantly reduced in epithelial cells transfected by miR-142-3p mimic (shown in red) as compared with control epithelial cells (shown in black). Linear regression analysis of cAMP production versus time in control versus miR-142-3p mimic–transfected salivary epithelial cells was performed. Regression slopes were significantly different between control and miR-142-3p mimic–transfected cells (Figure 3, E and F; and Supplemental Figure 4, A and B). These data demonstrate that overexpression of miR-142-3p mimic leads to functional consequences, including decreased Ca2+ signaling and cAMP production in salivary epithelial cells.
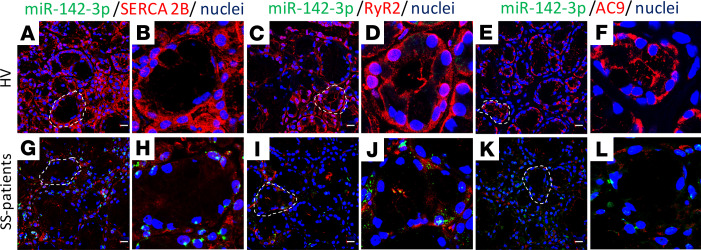
SERCA2B, RyR2, and AC9 expression is decreased in salivary gland tissues of Sjögren’s patients.
Given the overexpression of miR-142-3p in SS, we inquired whether the targets of miR-142-3p are downregulated in SS salivary glands (SGs). We thus investigated the presence of miR-142-3p targets SERCA2B, RYR2, and AC9 in minor SGs from SS patients and healthy volunteers. Double ISH/immunofluorescence staining for miR-142-3p and its target proteins in minor SG biopsies showed that SERCA2B, RyR2, and AC9 protein levels were higher in healthy volunteer biopsies (Figure 4, A–F) compared with tissues from SS patients (Figure 4, G–L). It is noteworthy that low protein levels of SERCA2B, RyR2, and AC9 were detected in the epithelial cells of SS patients where miR-142-3p was highly expressed (Figure 4, H, J, and L). These results suggest that the presence of miR-142-3p is associated with a decrease in SERCA2B, RyR2, and AC9 expression within SS tissue epithelial cells and reinforce the notion that miR-142-3p is a negative regulator of SERCA2B, RyR2, and AC9 protein expression.
Figure 4. Expression of SERCA2B, RyR2, and AC9 is reduced in SGs of patients with SS.
(A–L) Double ISH/immunofluorescence staining in paraffin-embedded sections from minor SG biopsies of healthy volunteers and patients with SS. B, D, F, H, J, and L are higher magnification (×60) images of indicated (outlined) areas in A, C, E, G, I, and K. (A and G) miR-142-3p expression (green) and SERCA2B (red), (C and I) miR-142-3p (green) and RyR2 (red), and (E and K) miR-142-3p (green) and AC9 (red). Cell nuclei were stained with DAPI (blue). Scale bar: 10 μm. Representative of healthy volunteers, n = 3; SS patients, n = 4.
T cell–derived exosomes contain miR-142-3p and can transfer into epithelial cells.
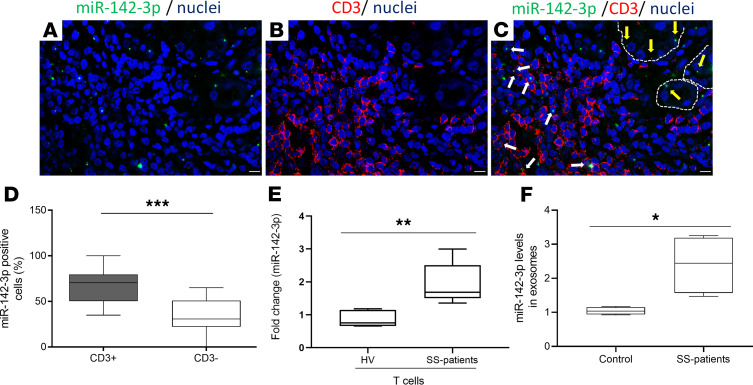
miR-142-3p is reported to be primarily expressed in the hematopoietic compartment and particularly in T cells (Supplemental Figure 5) (14). Indeed, miR-142-3p is contained in exosomes in sera of SS patients significantly more than sera of healthy volunteers (Figure 5F). Importantly, peripheral T cells from SS patients express significantly higher levels of miR-142-3p compared with T cells from healthy volunteers (Figure 5E and Supplemental Figure 6). We therefore hypothesized that T cells in SGs of patients with SS express miR-142-3p and can transfer it through exosomes to epithelial cells to disrupt salivary function. We first assessed miR-142-3p expression within the T cell infiltrate of SS SG biopsies and observed that miR-142-3p was present within CD3+ T cells in the lesion (white arrows in Figure 5, A–C). In fact, the majority of miR-142-3p–expressing inflammatory cells were CD3+ T cells (Figure 5D). miR-142-3p–positive epithelial cells were also evident (yellow arrows in Figure 5C).
Figure 5. miR-142-3p is upregulated in salivary gland lesions and within secreted T cell exosomes from patients with SS.
(A–C) ISH for miR-142-3p (green) and immunofluorescence staining for CD3+ T cells (red) were performed on paraffin-embedded sections from minor SG biopsies of patients with SS. Merged image shows several CD3+ T cell/miR-142-3p–coexpressing cells. White arrows point to miR-142-3p expressed by CD3+ T cells. Yellow arrows point to miR-142-3p expressed by epithelial cells. Cell nuclei were stained with DAPI (blue). Scale bar: 10 μm. (Representative images n = 4 SS patients.) (D) Fluorescence intensity quantification of miR-142-3p in T cells (CD3+) and non-T cells (CD3–) from minor SG biopsies of SS patients. (SS patients, n = 4.) (n = 12, median, maximum, and minimum shown.) Statistical significance was determined by Mann-Whitney nonparametric test; ***P < 0.001. (E) Expression of miR-142-3p in CD3+ T cells from PBMCs of healthy volunteers and SS patients (healthy volunteers, n = 4; SS patients, n = 4) (n = 4, median, maximum, and minimum shown). Statistical significance was determined by Mann-Whitney nonparametric test; **P < 0.01. (F) Expression of miR-142-3p in serum exosomes from healthy volunteers and SS patients (healthy volunteers, n = 4; SS patients, n = 4) (n = 4, median, maximum, and minimum shown). Statistical significance was determined by Mann-Whitney nonparametric test; *P < 0.05. The box plots depict the minimum and maximum values (whiskers), the upper and lower quartiles, and the median. The length of the box represents the interquartile range.
Our next aim was to investigate whether miR-142-3p secreted in exosomes from CD3+ T cells can be transferred into epithelial cells. We therefore activated T cells by anti-CD3/CD28 antibody stimulation and isolated exosomes from the culture supernatant. We detected upregulation of miR-142-3p expression in activated T cells and within secreted exosomes (with minimal presence of miR-142-3p in macrovesicles), compared with naive T cells, despite comparable quantities of total exosomes (Supplemental Figure 7, A–C).
We next transferred T cell exosomes onto SG cells in vitro and accessed intracellular miR-142-3p expression. We observed that treatment with T cell exosomes led to a robust and significant increase in the levels of miR-142-3p in both HSG and pSG cells, compared with treatment with exosome-depleted media (Supplemental Figure 7D). These results suggest transfer of miR-142-3p through exosomes into SG epithelial cells as a plausible mechanism.
Treatment with T cell–derived exosomes targets SERCA2B, RyR2, and AC9 in SG cells.
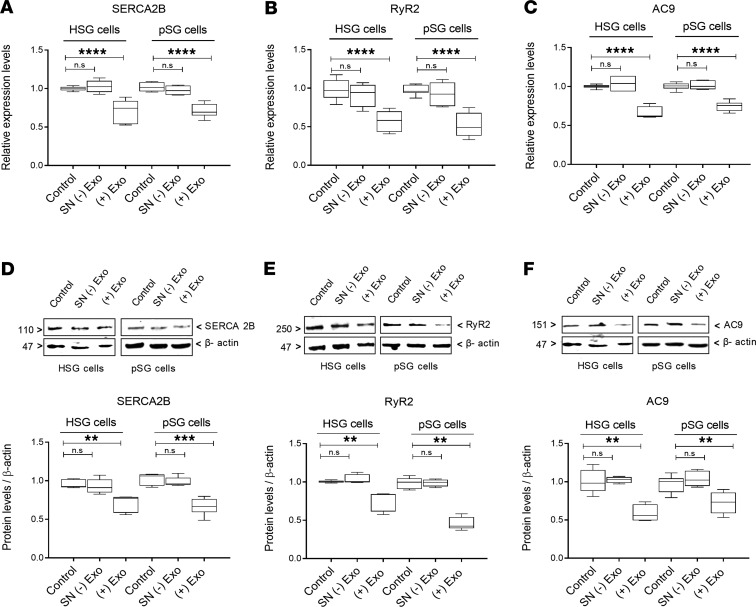
Having observed that treatment with T cell–derived exosomes establishes transfer of miR-142-3p into salivary epithelial cells, we investigated whether T cell–derived exosome treatment leads to downregulation of target proteins SERCA2B, RyR2, and AC9. We documented that treatment with T cell exosomes led to significantly reduced mRNA levels for targets SERCA2B, RyR2, and AC9 (Figure 6, A–C) in both HSG cells and pSG cells. Unlike treatment with exosomes, treatment with culture supernatants (depleted of exosomes) did not affect mRNA levels for the targets. Exosome-mediated inhibition of SERCA2B, RyR2, and AC9 expression in epithelial cells was dose dependent (Supplemental Figure 8). Protein levels for SERCA2B, RyR2, and AC9 were also significantly reduced in both HSG cells and pSG cells following treatment with T cell exosomes (Figure 6, D–F). These results indicate that release of activated T cell exosomes can affect miR-142-3p targets SERCA2B, RyR2, and AC9, which are key proteins involved in SG secretion.
Figure 6. SERCA2B, RyR2, and AC9 expression in epithelial cells is altered by T cell exosomes.
(A–C) mRNA expression for SERCA2B, RyR2, and AC9 in HSG and pSG cells. (D–F) Representative Western blots and relative protein levels of SERCA2B, RyR2, and AC9 in HSG and pSG cells obtained by densitometric analysis. HSG and pSG cells were treated with complete epithelial cell medium (control), exosome-depleted supernatant [SN (-) Exo], or pure exosomes isolated from activated T cells [(+) Exo]. Figure 6, D and F, are derived from the same blots and thus share the same loading control. The box plots depict the minimum and maximum values (whiskers), the upper and lower quartiles, and the median. The length of the box represents the interquartile range. (n = 5, median, maximum, and minimum shown; **P < 0.01, ***P < 0.001, and ****P < 0.0001, determined by Mann-Whitney nonparametric test.) n.s, not significant.
Exosomes from activated T cells disrupt Ca2+ signaling, cAMP production, and amylase secretion in 3D cultures of HSG acini.
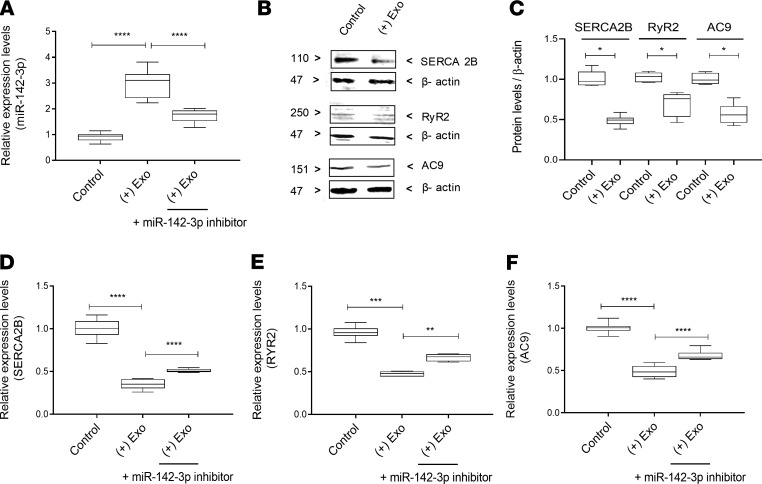
We next wanted to evaluate whether treatment with T cell–derived exosomes will impair key aspects of epithelial SG cell function, including Ca2+ signaling, cAMP production, and protein secretion (amylase secretion). For these experiments, we used a 3D culture model of HSG cells to recapitulate the function of SG epithelium. Indeed, HSG 3D acinar cell cultures provide an optimal system to study salivary protein secretion (15). We first confirmed that treatment with T cell–derived exosomes will transfer miR-142-3p to acinar cells in 3D culture (Figure 7A). We also used a miR-142-3p inhibitor that would partially inhibit miR-142-3p to assess specificity of miR-142-3p function in our experiments (Figure 7A).
Figure 7. SERCA2B, RyR2, and AC9 expression is altered by T cell exosomes in 3D cultures of HSG acini.
(A) miR-142-3p expression in 3D HSG acini that were treated with complete epithelial cell medium (control), pure exosomes isolated from activated T cells, or pure exosomes isolated from activated T cells and miR-142-3p hairpin inhibitor. (B) Western blots of SERCA2B, RyR2, and AC9 in 3D HSG acini that were treated with complete epithelial cell medium (control) and pure exosomes isolated from activated T cells. (C) Graph shows relative protein levels of SERCA2B, RyR2, and AC9 in 3D HSG acini obtained by densitometric analysis. (D–F) Relative mRNA levels of SERCA2B, RyR2, and AC9 in 3D HSG acini that were treated with complete epithelial cell medium (control), pure exosomes isolated from activated T cells, or pure exosomes isolated from activated T cells and miR-142-3p hairpin inhibitor. (n = 5, median, maximum, and minimum shown; *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001, determined by Mann-Whitney nonparametric test.) The box plots depict the minimum and maximum values (whiskers), the upper and lower quartiles, and the median. The length of the box represents the interquartile range.
We found that treatment with T cell–derived exosomes would target SERCA2B, RyR2, and AC9 protein (Figure 7, B and C) and mRNA expression (Figure 7, D–F). Addition of a miR-142-3p inhibitor hairpin in these experiments partially inhibited the effect of exosomes, confirming the role of miR-142-3p in target downregulation and suggesting that additional T cell exosome factors might be targeting these genes. SERCA2B, RyR2, and AC9 protein levels were also decreased in 3D HSG acini treated with exosomes from activated T cells (Figure 7C).
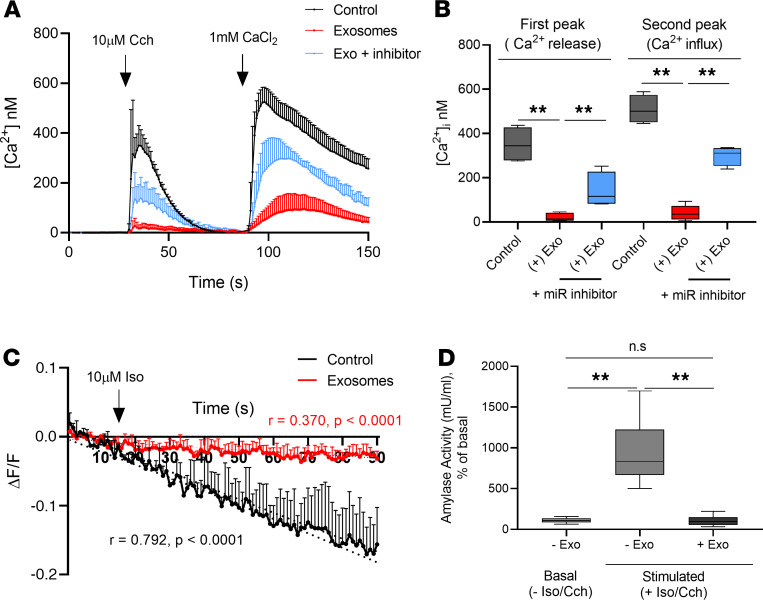
Next, we evaluated the effect of exosome treatment on SG epithelial function in the 3D HSG model. We found that addition of T cell–derived exosomes significantly reduced ER Ca2+ release. Ca2+ influx was also significantly reduced in 3D HSG acini treated with exosomes (Figure 8, A and B). Addition of a miR-142-3p inhibitor hairpin in these experiments partially inhibited the effect of exosomes. Furthermore, cAMP production from 3D HSG acini was significantly reduced upon treatment with exosomes (red line showing treated, compared with control acini shown with black line; Figure 8C and Supplemental Figure 4C). We documented that the slopes of cAMP production were significantly different between exosome-treated and untreated cells. Finally, given the well-established central role of Ca2+ and cAMP in promoting salivary protein secretion, we also examined downstream functional effects by assessing the secretion of the salivary protein amylase. Analysis of Cch/Iso-mediated amylase secretion in the 3D HSG acini model revealed significantly decreased amylase production in the 3D HSG acini treated with T cell–derived exosomes compared with those treated with complete epithelial cell medium (Figure 8D). Taken together, these results demonstrate that T cell–derived exosomes target key proteins involved in SG epithelial cell function and impair Ca2+ signaling, cAMP production, and amylase secretion.
Figure 8. T cell–derived exosomes affect Ca2+ signaling, cAMP production, and amylase secretion in 3D cultures of HSG acini.
(A and B) Internal Ca2+ concentration was measured in 3D HSG acini. 3D HSG acini were treated with complete epithelial cell medium (black), exosomes isolated from activated T cells (red), or exosomes and miR-142-3p inhibitor (light blue). 3D HSG acini were loaded with Fluo-4-AM. Time course of [Ca2+]i induced by stimulation of 3D HSG acini with 10 μM Cch in the absence or presence of external CaCl2(A) and quantification of calcium release entry peaks (B) (n = 4, median, maximum, and minimum shown). Statistical significance was determined by Mann-Whitney nonparametric test; **P < 0.01. (There were 15 cells per condition, n = 4 experiments.) (C) Time course of cAMP production in cADDis cAMP biosensor–loaded 3D acini that were stimulated with Iso (10 μM). Linear regression analysis of cAMP production versus time in 3D acini control (black) and 3D acini treated with pure exosomes isolated from activated T cells (red). (There were 15 cells per condition, n = 3 experiments.) (D) Summary for the percentage of amylase activity with respect to basal condition under 10 μM Cch and 10 μM Iso stimulation (n = 6, median, maximum, and minimum shown). Statistical significance was determined by Mann-Whitney nonparametric test; **P < 0.01. The box plots depict the minimum and maximum values (whiskers), the upper and lower quartiles, and the median. The length of the box represents the interquartile range.
Discussion
SS is a common autoimmune disease of unknown etiology. To date, the pathophysiological mechanisms that drive disease pathogenesis are not fully elucidated. The hallmark of the disease from both a clinical and pathognomonic standpoint is exocrine dysfunction, including dysfunction of salivary and lacrimal glands. Thus, the primary clinical manifestations of SS are xerostomia and xeropthalmia, which lead to morbidity and dramatic impairment of quality of life for patients (1, 16). Treatment for the disease remains largely palliative, with a focus of stimulating remaining exocrine gland secretion or replacing exocrine fluids (saliva and tears). However, available treatments are largely unsuccessful in improving symptomatology and disease progression (17, 18). Therefore, further elucidation of mechanisms leading to exocrinopathy in SS may not only further the understanding of disease mechanisms but also can potentially reveal targets for therapeutic intervention.
In our current study we identify a novel mechanism linked to impaired salivary glandular function in SS. We show that inflammation in the gland, and specifically, activated T cells may release miRNA-containing exosomes, which transfer into salivary epithelial cells to impair their function. We show that a specific microRNA, miR-142-3p, contained in T cell exosomes can target key proteins (SERCA2B, RyR2, AC9) involved in intracellular Ca2+ signaling, cAMP production, and protein secretion in SG epithelial cells.
Ca2+ and cAMP pathways are the primary signaling systems in secretory epithelia that control virtually all secretory gland functions (7). Ca2+ signals are critical for the control of fluid and enzyme secretion from exocrine glands (19). Synergistic function of Ca2+-mediated signaling and cAMP production are linked to protein secretion in acinar cells of exocrine glands (20). Importantly, the targets of miR-142-3p (SERCA2B, RyR2, AC9) are all involved in key events of Ca2+ signaling and cAMP-mediated secretion. SERCA2B is a Ca2+ ATPase that transfers Ca2+ from the cytosol to the lumen of the sarco/endoplasmic reticulum. In SGs, SERCA2B has been shown to be expressed in the luminal pole of acinar and ductal cells and to be critical for initiation and propagation of Ca2+ waves (21). The ryanodine receptor (RyR) is a calcium channel that mediates Ca2+ release from the ER and uptake by mitochondria. This uptake is critical for functional Ca2+ signaling in SG acinar cells (22, 23). The expression of functional RyRs in salivary acinar cells has been previously reported (24, 25). The RyRs are gated by elevations in cytoplasmic Ca2+ via a mechanism termed calcium-induced calcium release and contribute to the spread of [Ca2+]i increase in acinar cells following stimulation (24). Finally, AC9 is a membrane-bound enzyme that catalyzes the formation of cAMP from ATP. Adenylyl cyclases have recognized roles in exocrine function (26). SERCA2B and RyR2 have been previously shown to be downregulated in epithelial cells of patients with SS (15, 25).
Recent studies have identified that SG dysfunction both in the setting of radiation-induced SG damage and in the setting of SS is linked to defects in critical Ca2+-signaling mechanisms (27). Indeed, our group has previously reported that agonist-regulated intracellular Ca2+ release, critically required for Ca2+ entry and fluid secretion, is defective in acini from patients with SS. Similar decreases in Cch-stimulated Ca2+ elevation were detected in acinar cells from lymphotoxin-α–transgenic mice, a murine model for SS (28). However, the exact mechanism(s) causing the loss of fluid secretion, in either condition, are not clearly understood. We show that miR-142-3p targets key elements of Ca2+ signaling to impair exocrine secretion. Until now, miR-142-3p has been shown to play immunomodulatory roles in various pathological conditions by regulating several target genes. Particularly, it appears to have strong effects on immune responses related to various physiological and disease settings (14). Previous work has also established the presence of miR-142-3p in exosomes. Studies have demonstrated that miR-142-3p can survive in different body fluids due to being enclosed in exosomes (14, 29). Furthermore, in vitro studies have shown that miR-142-3p released by activated T cells can be taken up by endothelial cells (30). More recently, a study demonstrated that miR-142-3p released from activated T cells can be transferred to pancreatic β cells and impair their function. In that context, miR-142-3p affected exocrine cell function in another organ (pancreas) through a different mechanism involving triggering of chemokine expression and induction of apoptosis in β cells (30).
Involvement of miRNAs in the pathophysiology of SS has been hypothesized for a few years now (5, 31, 32). The concept that miRNAs transferred through exosomes will participate in mechanisms of disease has also been previously presented (12, 33, 34). Finally, previous work in our laboratory has shown that another miRNA, ebv-miR-BART13-3p, could be transferred via exosomes from B cells into salivary epithelial cells to regulate epithelial gland secretory function (35). Given that T cells are a feature of early Sjögren’s lesions while B cells accumulate in later stages of disease (36), we hypothesize that T cell–derived exosomes may be more critical in early initiation steps of disease and SG dysfunction and could conceivably represent more optimal targets for early-stage therapy. Indeed, miRNAs have recently emerged both as critical biomarkers in various diseases and also as functional drivers of disease pathology and are now increasingly considered therapeutic targets (37).
Beyond the potential of clinical targeting of miRNA-containing exosomes in SS, our current study introduces a potentially novel concept in its pathogenesis. We propose that activated T cells release exosomes with miRNAs that can impair SG function. Our data connect the inflammatory pathways observed in early lesions of Sjögren’s with the outcome of exocrinopathy and tie together aspects of autoimmunity with SG dysfunction.
Methods
Human subjects.
The study group included 16 patients with SS and 12 healthy volunteers (Supplemental Tables 1 and 2). Healthy volunteers (HVs) were free of sicca symptoms, chronic medical illnesses, or pregnancy and did not have HIV or hepatitis B or C infections. Importantly, HVs did not fulfill primary SS classification criteria, including an absence of serum autoantibodies (negative for ANA, SSA, and SSB autoantibodies) and did not exhibit focal lymphocytic sialadenitis (focus scores = 0) in their minor SG biopsies. The subjects with SS fulfilled the American-European criteria for SS classification and did not present with any other concomitant autoimmune disease.
Cell lines, cell cultures, and transfection.
The HSG cell line, originally established from an irradiated HSG (38), was cultured in DMEM F-12 (MilliporeSigma) supplemented with 5% FBS (Hyclon) and antibiotics (100 U/mL penicillin and 100 μg/mL streptomycin; Gibco, Thermo Fisher Scientific). Human-derived pSG epithelial cells were cultured in Keratinocyte Growth Medium (LONZA) (39). HSG and pSG cells were maintained under conditions previously described and were plated on 10-cm plastic culture dishes. Cells were transfected for 6 hours with 20 nM of miR-142-3p mimic (Dharmacon) using the HiPerfect Transfection reagent (QIAGEN) according to the manufacturer’s instructions. Six hours posttransfection, the transfection medium was changed to DMEM F-12; supplemented with 5% FBS, 100 U/mL penicillin, and 100 μg/mL streptomycin; and maintained for 24 hours at 37°C in a humidified 5% CO2 incubator. HSG and pSG cells were seeded (count of 4 × 105 and 2 × 105 cells/well, respectively) in a collagen-coated 6-well plate (Corning).
3D HSG acini.
HSG were seeded on Cultrex Reduced Growth Factor Basement Membrane Extract (Trevigen), as previously described (15).
T cell activation, microvesicles, and exosome isolation.
Peripheral blood mononuclear cells (PBMCs) were obtained by leukapheresis of HVs (Department of Transfusion Medicine, NIH) and T cells purified by counterflow centrifugal elutriation. Human T cells obtained from HVs were suspended in RPMI 1640 medium with 25 mM HEPES, 2 mM glutamine (Gibco, Thermo Fisher Scientific), 10% fetal calf serum (FCS) (Gemini Bio Products), 100 IU/mL penicillin, and 50 μg/mL streptomycin (P/S). To activate T cells, 125 μL (25 μL/mL) of ImmunoCult Human CD3/CD28 T Cell Activator (STEMCELL Technologies) was added to the cell suspension (2 × 106 cells/mL). Human T cells were incubated for 24 hours at 37°C in a humidified 5% CO2 incubator. The cell suspension then was centrifugated at 600 g for 10 minutes to remove cell debris, and microvesicles contained in the resulting supernatant were isolated by ultracentrifugation at 10,000 g and 4°C for 30 minutes. To isolate exosomes, the supernatant fraction–free microvesicles were collected, and EXOQuick-TC (SBI) was used according to the manufacturer’s instructions. Both exosomes and culture medium without exosomes were collected.
Sorting of single T cells.
Single-cell suspensions of PBMCs from HVs and SS subjects were incubated with mouse serum (Jackson ImmunoResearch Laboratories) and fluorochrome-conjugated antibodies against surface markers (CD3-FITC; Thermo Fisher Scientific SK7) in FACS buffer (PBS with 2.5% FBS) for 20 minutes at 4°C in the dark, then washed with FACS buffer twice. Dead cells were excluded with Live/Dead fixable dye (DAPI; Thermo Fisher Scientific). Cells were purified by FACS sorting using BD FACSAria ΙΙ and FACSDiva software (BD Biosciences). CD3+ cells were isolated as T cells.
Stimulation of 3D HSG acini and epithelial cells.
HSG, pSG, and 3D HSG acini were cultured under conditions described previously (15, 39). Cells were stimulated for 24 hours at 37°C with exosomes obtained from naive T cells or activated T cells. RPMI 1640 medium with 10% FCS and P/S or media without exosomes were used as controls for the T cell exosome stimulation.
miRNA target predictions.
Prediction analysis of the miR-142-3p target was performed using mirDIP 4.1: integrative database of human microRNA target predictions (http://ophid.utoronto.ca/mirDIP/index.jsp#r ) (40).
Luciferase reporter assays.
HSG and pSG cells were seeded in 24-well plates. One day after seeding, cells were cotransfected with 1 μg pEZX-MT06-3′-UTR luciferase reporter and 20 nM hsa-miR-142-3p mimic or 200 nM miRNA hairpin inhibitor (Dharmacon). Scrambled miRNA was used as a negative control. Luciferase reporter, miR-142-3p mimic, scrambled miRNA, and miRNA hairpin were transfected using the Attractene Transfection Reagent (QIAGEN) according to the manufacturer’s instructions. Twenty-four hours posttransfection, cells were lysed, and extracts were measured with the Luc-Pair Duo-Luciferase HS Assay Kit according to the manufacturer’s instructions (GeneCopoeia). The expression of Renilla luciferase (included in pEZX-MT06-3′-UTR plasmid; GeneCopoeia) was used to monitor transfection efficiency. Results are represented as the ratio of Firefly to Renilla luciferase activity.
ISH and immunofluorescent double staining.
To investigate specific distribution of miR-142-3p in human SG samples, ISH was performed on paraffin-embedded sections from HVs’ and SS patients’ minor SG biopsies. The 5′- and 3′- digoxigenin-labeled locked nucleic acid (LNA) probe complementary to the mature miR-142-3p (Exiqon, Woburn, Massachusetts, USA) was used to perform ISH. The LNA scramble and LNA U6 probes (Exiqon, Vedbaek, Denmark) were used as negative and positive controls, respectively. Tissue sections of 5 μm were deparaffinized in xylene and ethanol solutions and placed in PBS. Antigen was recovered by incubating the slides in 0.01 M citrate buffer (pH 6.0) for 60 minutes at 99°C. After 20 minutes of cooling, the slides were washed in PBS 3 times for 5 minutes each. Sections were then incubated in 3% H2O2/methanol for 30 minutes. A prehybridization step was performed by incubating the slides in ISH buffer (50% deionized formamide, 0.25% SDS, 200 μg/mL yeast tRNA, 1X Denhardt’s, 600 mM NaCl, 1 mM EDTA, 10% dextran sulfate) for 1 hour at 37°C. The 5′- and 3′- digoxigenin-labeled LNA probe, LNA scramble, and LNA U6 probes were denatured at 90°C for 5 minutes and diluted to a concentration of 100 nM in ISH buffer. Tissue sections were hybridized with probe diluted in ISH buffer overnight at 42°C and then were washed for 15 minutes at 42°C in 2X SSC (3 washes) and 0.2X SSC (2 washes). The sections then were washed in PBS 3 times for 5 minutes at room temperature (RT), incubated with blocking buffer (3% BSA in TBS buffer, pH 7.4) at RT for 1 hour, and incubated overnight at 4°C with a mouse anti-digoxigenin unconjugated antibody (Abcam ab420; dilution 1:100). Briefly, sections were then washed in PBS 3 times for 5 minutes at RT and the miRNA signal was detected with the Alexa Fluor 488 Tyramide SuperBoost Kit (Invitrogen, Thermo Fisher Scientific) following the manufacturer’s directions. To perform double ISH/immunofluorescence staining in sections from minor SG biopsies, ISH was performed as described, and SERCA2B, RyR2, AC9, and CD3 were then detected using goat anti-SERCA2B antibody (LSBio B10343), rabbit anti-RyR2 antibody (LSBio C330906), rabbit anti-AC9 antibody (Abcam ab191423), and rabbit anti-CD3 antibody (Abcam ab5690) with incubation overnight at 4°C (dilution 1:100). After 3 washes in PBS the slides were incubated for 1 hour at RT with Alexa Fluor 633–conjugated goat anti-rabbit antibody (Invitrogen, Thermo Fisher Scientific, A21070) and Alexa Fluor 594–conjugated donkey anti-goat antibody (Invitrogen, Thermo Fisher Scientific, A11058) diluted 1:200 in blocking buffer. The slides were mounted with ProLong Gold Antifade Mountant with DAPI (Invitrogen, Thermo Fisher Scientific). The slides were evaluated using a confocal microscope (Olympus Fluoview MP). Images were taken using the same laser intensity levels for all the samples analyzed. The images were analyzed with ImageJ (NIH) software, and to visualize the localization of signals and measure colocalization, a plug-in for ImageJ called EzColocalization was used (41).
Immunofluorescence staining.
Immunofluorescence of SERCA2B, RyR2, and AC9 was performed in HSG and pSG cells grown on glass-bottom culture dishes (MatTek) and transfected with hsa-miR-142-3p mimic. Twenty-four hours posttransfection, epithelial cells were washed with PBS, fixed with 4% paraformaldehyde, and blocked with 3% BSA in PBS for 1 hour at RT. Primary antibodies were then added and incubated at 4°C overnight. Cells were washed with PBS and incubated with Alexa Fluor 488–conjugated secondary antibody (Invitrogen, Thermo Fisher Scientific) for 1 hour at RT, washed, and mounted using ProLong Gold Antifade Mountant with DAPI (Invitrogen, Thermo Fisher Scientific). The slides were imaged using a confocal microscope (Olympus Fluoview MP).
Quantitative PCR.
Total RNA was collected from transfected and nontransfected cells or cells stimulated with exosomes from activated T cells using RNeasy Plus Mini Kit (QIAGEN) according to the manufacturer’s instructions. A total of 1 μg of total RNA was reverse-transcribed using High Capacity cDNA Reverse Transcription Kit (Thermo Fisher Scientific). Quantitative PCR was performed with TaqMan probes of genes of interest and TaqMan Fast Universal PCR Master Mix in triplicate using QuantStudio 6 Flex Real-Time PCR System (Thermo Fisher Scientific). Expression of mRNA was normalized using β-actin (ACTB). TaqMan probes for SERCA2B (ATP2A2), RyR2, AC9 (ADCY9), and ACTB were used (Thermo Fisher Scientific HS00544877_m1, HS00181461_m1, HS00181599_m1, and HS01060665_g1) according to the protocol provided by the manufacturer. For miR-142-3p quantification, 500 ng of the same RNA was reverse-transcribed with TaqMan MicroRNA Reverse Transcription Kit (Thermo Fisher Scientific). TaqMan probes for miR-142-3p and U6snRNA (as endogenous control; Thermo Fisher Scientific 000464 and 001973). The relative quantification of miRNA and mRNA was performed using the comparative cycle threshold (2-ΔCt) method. For analysis of miR-142-3p expression in supernatant and exosomes from T cells, total RNA was extracted with miRCURY RNA Isolation Kit (Exiqon), and SYBR Green–based real-time quantitative PCR reactions were performed according to the manufacturer’s protocol. The miRCURY LNA PCR primer set was used for miR-142-3p, and miRCURY LNA RNA Spike-in Kit (QIAGEN) was used to normalize miR-142-3p values. Quantification of miR-142-3p expression in CD3+ T cells from PBMCs was performed using TaqMan MicroRNA Reverse Transcription Kit, TaqMan PreAmp Master Mix, and TaqMan Fast Universal PCR Master Mix (Thermo Fisher Scientific) (42).
Protein extraction and Western blot.
HSG acini in 3D culture and HSG and pSG 2D cell cultures were homogenized in RIPA buffer in the presence of Complete Protease Inhibitor Cocktail EDTA-free Mini Tablets (Roche) at 4°C. The protein concentration of extracts was determined using DC protein assay kits (Bio-Rad). Before protein extraction of 3D acini, the isolated 3D acini were detached from the Matrigel. The isolated 3D acini were made with a Cultrex 3D Culture Cell Harvesting kit (Trevigen) according to the manufacturer’s instructions. A total of 30 μg of total HSG cells’ protein extract was loaded and resolved in 4–12% NuPAGE gels (Invitrogen, Thermo Fisher Scientific). Goat anti-SERCA2B antibody (LSBio B10343), rabbit anti-RyR2 antibody (LSBio C330906), rabbit anti-AC9 antibody (Abcam 191423), and anti–β-actin antibody (MilliporeSigma A2228) were used at 1:1000, 1:500, 1:1000, and 1:1000 dilutions, respectively. Protein bands were detected by ODYSSEY Fc System (LI-COR, Biosciences). The intensity of immunoreactive bands was quantified using ImageJ. Protein levels were normalized to β-actin.
Measurement of [Ca2+]i.
HSG and pSG cells were seeded on glass-bottom culture dishes (MatTek) and were transfected with miR-142-3p mimic using the HiPerfect Transfection reagent according to the manufacturer’s instructions. For 3D HSG acini calcium signaling, 3D acini were stimulated with exosomes from activated T cells. Twenty-four hours posttransfection or stimulation, cells were loaded with 5 μM Fluo-4 acetoxymethyl ester (Invitrogen, Thermo Fisher Scientific) in saline buffer (140 mM NaCl, 1 mM MgCl2, 5 mM KCl, 1 mM CaCl2, 5 mM glucose, and 10 mM HEPES, pH: 7.4) for 30 minutes at 37°C. After loading, the cells were washed with saline buffer for 5 minutes. For measure of [Ca2+]i, calcium signaling was stimulated by 10 μM Cch in the absence of external Ca2+ (Ca2+ release) and in the presence of external calcium by addition of 1 mM MgCl2 (Ca2+ influx). Confocal fluorescent images were obtained with a confocal microscope (Olympus Fluoview MP). Fluo-4 excitation at 488 nm was delivered from an argon laser, and the collected emission was high-pass filtered at 515 nm. Regions of interest (X by Y microns) from the image obtained with a ×60 oil immersion objective were captured every 10 seconds. For converting fluorescence data into [Ca2+]i, we used the following equation: [Ca2+]i = KD × (F – Fmin)/(Fmax – F) (43). The images were stored for later analysis with ImageJ software (44).
cAMP assay.
3D acini were stimulated with exosomes from activated T cells, and epithelial cells were transfected with miR-142-3p mimic. Twenty-four hours after transfection stimulation, cAMP intracellular accumulation was measured using cADDis cAMP Assay (Montana Molecular) according to the protocol provided by the manufacturer. Twenty-four hours posttransduction with complete medium containing 60 μL BacMam sensor (green downward cAMP difference detector in situ), cells were incubated for 24 hours at 37°C with complete medium containing sodium butyrate. To measure cAMP intracellular accumulation, cAMP production was stimulated by 10 μM Iso. Confocal fluorescence images were obtained with a confocal microscope (Olympus Fluoview MP). Regions of interest (X by Y microns) from the image obtained with a ×60 oil immersion objective were captured every 10 seconds.
Amylase assay.
After 24 hours of stimulation with 10 μM Cch and 10 μM Iso, the level of amylase secreted in the supernatant (50 μL) was assayed using the Amylase Activity Assay Kit (LSBio) according to the manufacturer’s protocol. The results were expressed as the amylase activity (mU/mL = nmol/min/mL).
Statistics.
Two-tailed unpaired t test or Mann-Whitney test was used when comparing 2 groups. P values less than 0.05 were considered statistically significant. Data were analyzed using Prism 7 (GraphPad Software).
Study approval.
Sera, PBMC, and labial minor SG biopsies were obtained from patients who provided informed consent to IRB-approved clinical protocols (NCT01425892 and NCT02327884) at the NIH. All subjects signed the informed consent document before any procedures.
Author contributions
IA and JCT conceived the research study. IA, JCT, and NMM designed experiments. JCT and PP performed experiments. JCT, IA, NMM, PP, SIJ, BMW, and TI analyzed data. PP, JH, SIJ, BMW, and TI did technical support and consultations. IA and BMW collected clinical samples. JCT and NMM wrote the manuscript. JCT, NM, IA, PP, JH, TGW, and TI edited the manuscript.
Supplementary Material
Acknowledgments
This research was supported by the Intramural Program of the NIH, NIDCR. We would like to also thank the NIDCR Imaging Core (ZIC DE000750-01) for technical assistance with imaging, NIDCR Combined Technical Research Core for technical assistance with cell sorting, and Mayank Tandon for computational advice and comments on the manuscript.
Version 1. 05/07/2020
Electronic publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2020, American Society for Clinical Investigation.
Reference information: JCI Insight. 2020;5(9):e133497.https://doi.org/10.1172/jci.insight.133497.
Contributor Information
Juan Cortes-Troncoso, Email: juan.cortestroncoso@nih.gov.
Shyh-Ing Jang, Email: jangs@mail.nih.gov.
Paola Perez, Email: pperezriv@gmail.com.
Tomoko Ikeuchi, Email: tomoko.ikeuchi@nih.gov.
Blake M. Warner, Email: blake.warner@nih.gov.
Ilias Alevizos, Email: alevizos@gmail.com.
References
- 1.Mavragani CP, Moutsopoulos HM. Sjögren’s syndrome. Annu Rev Pathol. 2014;9:273–285. doi: 10.1146/annurev-pathol-012513-104728. [DOI] [PubMed] [Google Scholar]
- 2.Brito-Zerón P, et al. Influence of geolocation and ethnicity on the phenotypic expression of primary Sjögren’s syndrome at diagnosis in 8310 patients: a cross-sectional study from the Big Data Sjögren Project Consortium. Ann Rheum Dis. 2017;76(6):1042–1050. doi: 10.1136/annrheumdis-2016-209952. [DOI] [PubMed] [Google Scholar]
- 3.Alevizos I, Alexander S, Turner RJ, Illei GG. MicroRNA expression profiles as biomarkers of minor salivary gland inflammation and dysfunction in Sjögren’s syndrome. Arthritis Rheum. 2011;63(2):535–544. doi: 10.1002/art.30131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gebert LFR, MacRae IJ. Regulation of microRNA function in animals. Nat Rev Mol Cell Biol. 2019;20(1):21–37. doi: 10.1038/s41580-018-0045-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kapsogeorgou EK, Gourzi VC, Manoussakis MN, Moutsopoulos HM, Tzioufas AG. Cellular microRNAs (miRNAs) and Sjögren’s syndrome: candidate regulators of autoimmune response and autoantigen expression. J Autoimmun. 2011;37(2):129–135. doi: 10.1016/j.jaut.2011.05.003. [DOI] [PubMed] [Google Scholar]
- 6.Catalán MA, Nakamoto T, Melvin JE. The salivary gland fluid secretion mechanism. J Med Invest. 2009;56(suppl):192–196. doi: 10.2152/jmi.56.192. [DOI] [PubMed] [Google Scholar]
- 7.Ahuja M, Jha A, Maléth J, Park S, Muallem S. cAMP and Ca2+ signaling in secretory epithelia: crosstalk and synergism. Cell Calcium. 2014;55(6):385–393. doi: 10.1016/j.ceca.2014.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lu X, et al. miR-142-3p regulates the formation and differentiation of hematopoietic stem cells in vertebrates. Cell Res. 2013;23(12):1356–1368. doi: 10.1038/cr.2013.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sun Y, et al. Mature T cell responses are controlled by microRNA-142. J Clin Invest. 2015;125(7):2825–2840. doi: 10.1172/JCI78753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Valadi H, Ekström K, Bossios A, Sjöstrand M, Lee JJ, Lötvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9(6):654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 11.Théry C, Ostrowski M, Segura E. Membrane vesicles as conveyors of immune responses. Nat Rev Immunol. 2009;9(8):581–593. doi: 10.1038/nri2567. [DOI] [PubMed] [Google Scholar]
- 12.Katsiougiannis S. Extracellular Vesicles: Evolving Contributors in Autoimmunity. For Immunopathol Dis Therap. 2015;6(3-4):163–170. doi: 10.1615/ForumImmunDisTher.2016016491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang B, et al. miR-142-3p restricts cAMP production in CD4+CD25- T cells and CD4+CD25+ TREG cells by targeting AC9 mRNA. EMBO Rep. 2009;10(2):180–185. doi: 10.1038/embor.2008.224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sharma S. Immunomodulation: A definitive role of microRNA-142. Dev Comp Immunol. 2017;77:150–156. doi: 10.1016/j.dci.2017.08.001. [DOI] [PubMed] [Google Scholar]
- 15.Cortés J, et al. Synaptotagmin-1 overexpression under inflammatory conditions affects secretion in salivary glands from Sjögren’s syndrome patients. J Autoimmun. 2019;97:88–99. doi: 10.1016/j.jaut.2018.10.019. [DOI] [PubMed] [Google Scholar]
- 16.Ramos-Casals M, Tzioufas AG, Font J. Primary Sjögren’s syndrome: new clinical and therapeutic concepts. Ann Rheum Dis. 2005;64(3):347–354. doi: 10.1136/ard.2004.025676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mavragani CP, Moutsopoulos NM, Moutsopoulos HM. The management of Sjögren’s syndrome. Nat Clin Pract Rheumatol. 2006;2(5):252–261. doi: 10.1038/ncprheum0165. [DOI] [PubMed] [Google Scholar]
- 18.Saraux A, Pers JO, Devauchelle-Pensec V. Treatment of primary Sjögren syndrome. Nat Rev Rheumatol. 2016;12(8):456–471. doi: 10.1038/nrrheum.2016.100. [DOI] [PubMed] [Google Scholar]
- 19.Petersen OH, Tepikin AV. Polarized calcium signaling in exocrine gland cells. Annu Rev Physiol. 2008;70:273–299. doi: 10.1146/annurev.physiol.70.113006.100618. [DOI] [PubMed] [Google Scholar]
- 20.Tang S, Beharry S, Kent G, Durie PR. Synergistic effects of cAMP- and calcium-mediated amylase secretion in isolated pancreatic acini from cystic fibrosis mice. Pediatr Res. 1999;45(4 pt 1):482–488. doi: 10.1203/00006450-199904010-00005. [DOI] [PubMed] [Google Scholar]
- 21.Lee MG, et al. Polarized expression of Ca2+ pumps in pancreatic and salivary gland cells. Role in initiation and propagation of [Ca2+]i waves. J Biol Chem. 1997;272(25):15771–15776. doi: 10.1074/jbc.272.25.15771. [DOI] [PubMed] [Google Scholar]
- 22.Zhang X, et al. Ryanodine and inositol trisphosphate receptors are differentially distributed and expressed in rat parotid gland. Biochem J. 1999;340(pt 2):519–527. [PMC free article] [PubMed] [Google Scholar]
- 23.Kopach O, Kruglikov I, Pivneva T, Voitenko N, Fedirko N. Functional coupling between ryanodine receptors, mitochondria and Ca(2+) ATPases in rat submandibular acinar cells. Cell Calcium. 2008;43(5):469–481. doi: 10.1016/j.ceca.2007.08.001. [DOI] [PubMed] [Google Scholar]
- 24.Ambudkar IS. Ca2+ signaling and regulation of fluid secretion in salivary gland acinar cells. Cell Calcium. 2014;55(6):297–305. doi: 10.1016/j.ceca.2014.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Melvin JE, Yule D, Shuttleworth T, Begenisich T. Regulation of fluid and electrolyte secretion in salivary gland acinar cells. Annu Rev Physiol. 2005;67:445–469. doi: 10.1146/annurev.physiol.67.041703.084745. [DOI] [PubMed] [Google Scholar]
- 26.Sabbatini ME, Gorelick F, Glaser S. Adenylyl cyclases in the digestive system. Cell Signal. 2014;26(6):1173–1181. doi: 10.1016/j.cellsig.2014.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ambudkar I. Calcium signaling defects underlying salivary gland dysfunction. Biochim Biophys Acta Mol Cell Res. 2018;1865(11 pt B):1771–1777. doi: 10.1016/j.bbamcr.2018.07.002. [DOI] [PubMed] [Google Scholar]
- 28.Teos LY, et al. IP3R deficit underlies loss of salivary fluid secretion in Sjögren’s Syndrome. Sci Rep. 2015;5:13953. doi: 10.1038/srep13953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sukma Dewi I, et al. Exosomal miR-142-3p is increased during cardiac allograft rejection and augments vascular permeability through down-regulation of endothelial RAB11FIP2 expression. Cardiovasc Res. 2017;113(5):440–452. doi: 10.1093/cvr/cvw244. [DOI] [PubMed] [Google Scholar]
- 30.Guay C, et al. Lymphocyte-derived exosomal micrornas promote pancreatic β cell death and may contribute to type 1 diabetes development. Cell Metab. 2019;29(2):348–361.e6. doi: 10.1016/j.cmet.2018.09.011. [DOI] [PubMed] [Google Scholar]
- 31.Alevizos I, Illei GG. MicroRNAs in Sjögren’s syndrome as a prototypic autoimmune disease. Autoimmun Rev. 2010;9(9):618–621. doi: 10.1016/j.autrev.2010.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reale M, D’Angelo C, Costantini E, Laus M, Moretti A, Croce A. MicroRNA in Sjögren’s syndrome: their potential roles in pathogenesis and diagnosis. J Immunol Res. 2018;2018:7510174. doi: 10.1155/2018/7510174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kapsogeorgou EK, Abu-Helu RF, Moutsopoulos HM, Manoussakis MN. Salivary gland epithelial cell exosomes: a source of autoantigenic ribonucleoproteins. Arthritis Rheum. 2005;52(5):1517–1521. doi: 10.1002/art.21005. [DOI] [PubMed] [Google Scholar]
- 34.Michael A, et al. Exosomes from human saliva as a source of microRNA biomarkers. Oral Dis. 2010;16(1):34–38. doi: 10.1111/j.1601-0825.2009.01604.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gallo A, et al. Targeting the Ca(2+) sensor STIM1 by exosomal transfer of Ebv-miR-BART13-3p is associated with Sjögren’s syndrome. EBioMedicine. 2016;10:216–226. doi: 10.1016/j.ebiom.2016.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Christodoulou MI, Kapsogeorgou EK, Moutsopoulos HM. Characteristics of the minor salivary gland infiltrates in Sjögren’s syndrome. J Autoimmun. 2010;34(4):400–407. doi: 10.1016/j.jaut.2009.10.004. [DOI] [PubMed] [Google Scholar]
- 37.Rupaimoole R, Slack FJ. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov. 2017;16(3):203–222. doi: 10.1038/nrd.2016.246. [DOI] [PubMed] [Google Scholar]
- 38.Shirasuna K, Sato M, Miyazaki T. A neoplastic epithelial duct cell line established from an irradiated human salivary gland. Cancer. 1981;48(3):745–752. doi: 10.1002/1097-0142(19810801)48:3<745::AID-CNCR2820480314>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 39.Jang SI, Ong HL, Gallo A, Liu X, Illei G, Alevizos I. Establishment of functional acinar-like cultures from human salivary glands. J Dent Res. 2015;94(2):304–311. doi: 10.1177/0022034514559251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tokar T, et al. mirDIP 4.1-integrative database of human microRNA target predictions. Nucleic Acids Res. 2018;46(D1):D360–D370. doi: 10.1093/nar/gkx1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stauffer W, Sheng H, Lim HN. EzColocalization: an ImageJ plugin for visualizing and measuring colocalization in cells and organisms. Sci Rep. 2018;8(1):15764. doi: 10.1038/s41598-018-33592-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Le Carré J, Lamon S, Léger B. Validation of a multiplex reverse transcription and pre-amplification method using TaqMan® MicroRNA assays. Front Genet. 2014;5:413. doi: 10.3389/fgene.2014.00413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Paredes RM, Etzler JC, Watts LT, Zheng W, Lechleiter JD. Chemical calcium indicators. Methods. 2008;46(3):143–151. doi: 10.1016/j.ymeth.2008.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9(7):671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.