Abstract
Muscle progenitor cell fusion is required for the formation and regeneration of multinucleated skeletal muscle fibers. Chronic muscle regeneration in Duchenne muscular dystrophy (DMD) is characterized by ongoing fusion of satellite cell (SC) progeny, but the effects of fusion on disease and the mechanisms by which fusion is accomplished in this setting are not fully understood. Using the mdx mouse model of DMD, we deleted the fusogenic protein Myomaker in SCs or myofibers. Following deletion in SCs, mice displayed a complete lack of myocyte fusion, resulting in severe muscle loss, enhanced fibrosis, and significant functional decline. Reduction of Myomaker in mature myofibers in mdx mice, however, led to minimal alterations in fusion dynamics. Unexpectedly, myofiber-specific deletion of Myomaker resulted in improvement of disease phenotype, with enhanced function and decreased muscle damage. Our data indicate that Myomaker has divergent effects on dystrophic disease severity depending upon its compartment of expression. These findings show that myocyte fusion is absolutely required for effective regeneration in DMD, but persistent Myomaker expression in myofibers due to ongoing fusion may have unintended deleterious consequences for muscle integrity. Thus, sustained activation of a component of the myogenic program in dystrophic myofibers exacerbates disease.
Keywords: Muscle Biology, Stem cells
Keywords: Skeletal muscle
Myomaker-mediated fusion is necessary to maintain muscle during muscular dystrophy disease progression, but sustained expression of Myomaker in myofibers has deleterious consequences for membrane stability.
Introduction
Skeletal muscle fibers are formed from the fusion of mononucleated muscle progenitor cells generating multinucleated myofibers (1, 2). This process occurs in embryonic and postnatal development as well as during regeneration of damaged tissue (3). Following a muscle injury, satellite cells (SCs) become activated and differentiate, ultimately leading to fusion and formation of regenerated myofibers (4, 5). Regenerative fusion can occur following acute injury, such as traumatic insult, in which case muscle structure and function are completely restored upon the conclusion of regeneration. Alternatively, chronic muscle injury is characterized by cycles of degeneration and regeneration, and thus there is continuous activation of the SC population. Such is the case in the muscular dystrophies, which comprise a group of inherited diseases leading to progressive muscle loss and dysfunction (6). The most common of these, Duchenne muscular dystrophy (DMD), is caused by mutations in dystrophin, a critical component of the dystrophin-glycoprotein complex, which serves to stabilize the muscle membrane during contraction (7). The lack of a functional dystrophin-glycoprotein complex leads to muscle damage with consequent inflammation and fibrotic remodeling (8, 9). In contrast to the regulation of muscle progenitor fusion during embryogenesis and in response to acute injury, the importance and mechanisms of fusion in a chronic regenerative setting like dystrophic disease progression are not well understood.
There has been much interest in the role of SCs in DMD because their activity is an endogenous mechanism with the potential to mitigate dystrophic disease. It has been shown that dystrophic SCs possess cell-intrinsic deficits that hamper their ability to achieve optimal regeneration, and strategies to enhance or rejuvenate SC function, along with exogenous stem cell therapies, are among the leading approaches for therapeutic development in DMD (10–12). A number of studies have shown that augmenting SC expansion can improve disease phenotype, while limiting expansion or promoting SC pool exhaustion worsens pathology. Mdx mice with deletion of syndecan-3 show increased activation and proliferation of SCs, leading to enhanced regeneration (13). Transient inhibition of STAT3 in dystrophic mice leads to SC expansion and enhanced muscle repair, and genetic deletion of chicken ovalbumin upstream promoter–transcription factor II (COUP-TFII) has similar effects, while COUP-TFII overexpression limits SC proliferation and leads to more severe myopathy (14, 15). Additionally, dystrophic mice lacking telomerase activity show reduced SC regenerative capacity and exacerbated disease progression (16). However, early stem cell ablation studies in dystrophic mice suggested there may be histological improvements following irradiation, and more recently, it has been shown that delaying regeneration can potentially ameliorate DMD severity (17–19).
Despite the clear relevance of SCs to DMD pathophysiology, the direct contributions of SCs to dystrophic muscle, as well as the effects of their absence, has not been experimentally tested. Unlike in acute injury, the chronic regenerative state of DMD is characterized by the continual fusion of SC progeny over months or years. This system provides a unique setting to determine mechanistic underpinnings of fusion, such as the requirement for fusion factors on the cells that undergo fusion in dystrophic muscle. Fusion in DMD can occur symmetrically between multiple myoblasts to form de novo myofibers, or myoblasts can fuse asymmetrically to damaged existing myofibers, but it is not understood if the machinery is the same for each of these fusion events (20). Additionally, the chronic myocyte fusion in dystrophic muscle represents a distinct pathological situation, one that never occurs during the life span of a normal animal. Thus, the consequences of ongoing fusion and membrane remodeling on muscle integrity in DMD could have a profound impact on the course of the disease.
In vertebrate skeletal muscle, fusion is mediated through the activity of Myomaker, a skeletal muscle–specific transmembrane protein (21–23). Indeed, genetic deletion of Myomaker leads to a complete lack of fusion and blocks formation of multinucleated muscle fibers during embryogenesis and after acute injury in the adult (24). Myomaker is required on the cell membrane for the initial step of the fusion process (25). While the biochemical function of Myomaker is not known, current hypotheses include that it possesses an activity that promotes cell membrane merger (26). The expression of Myomaker is highly restricted to times of myogenesis, but it is downregulated after the fusion process is complete and thus is absent in healthy, unstimulated adult muscle (24, 27). In contrast, Myomaker is activated in skeletal muscle of adult mdx mice (24), but the dynamics and consequences of Myomaker’s fusogenic activity in a chronic disease setting are unknown.
In this study, we investigated the role for fusion in determining the disease course of DMD and interrogated the requirement of Myomaker for effective fusion. Specifically, we examined the consequences of Myomaker expression in each of 2 compartments in dystrophic skeletal muscle that undergo fusion: SCs and mature myofibers. We found that the activity of Myomaker in SCs is absolutely required for effective regeneration, and loss of fusogenicity in SC progeny leads to severely exacerbated pathology. Unexpectedly, we found that reduction of Myomaker in myofibers did not affect fusion dynamics but led to improved indices of muscle function and myofiber integrity. These findings suggest that Myomaker expression in myofibers is not needed for fusion with myocytes but indicate that Myomaker in myofibers has a deleterious effect on overall membrane integrity. Together, our data indicate that the fusogenic activity of Myomaker is required in SCs for chronic muscle regeneration but paradoxically contributes to the pathogenesis of DMD at the level of the myofiber.
Results
Fusion in muscular dystrophy is blocked by deletion of Myomaker in Pax7+ SCs.
Using a MymkLacZ allele in which one copy of the Myomaker gene is effectively replaced with a LacZ cassette, we previously showed that Myomaker is upregulated in skeletal muscle of mdx mice because of ongoing SC activation and regeneration (24). However, whether Myomaker is expressed in activated SCs or myofibers was not known. Using a similar approach, we found LacZ+ myofibers in dystrophic muscle, indicating that Myomaker transcription occurs in both activated SCs and myofibers in DMD (Supplemental Figure 1; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.136095DS1). To examine the impact of SC progeny fusion on the disease course of DMD, we used a MymkloxP/loxP mouse line and crossed this mouse with mdx4cv mice and introduced the tamoxifen-inducible Pax7CreERT2 allele. This mouse line (mdx4cv MymkloxP/loxP Pax7CreERT2) allowed conditional deletion of Myomaker in SCs of dystrophic mice after treatment with tamoxifen. We treated mdx4cv MymkloxP/loxP (mdx) and mdx4cv MymkloxP/loxP Pax7CreERT2 (mdx MymkscKO) mice with tamoxifen at 2 months of age and analyzed these mice at 7 months of age (Figure 1A).
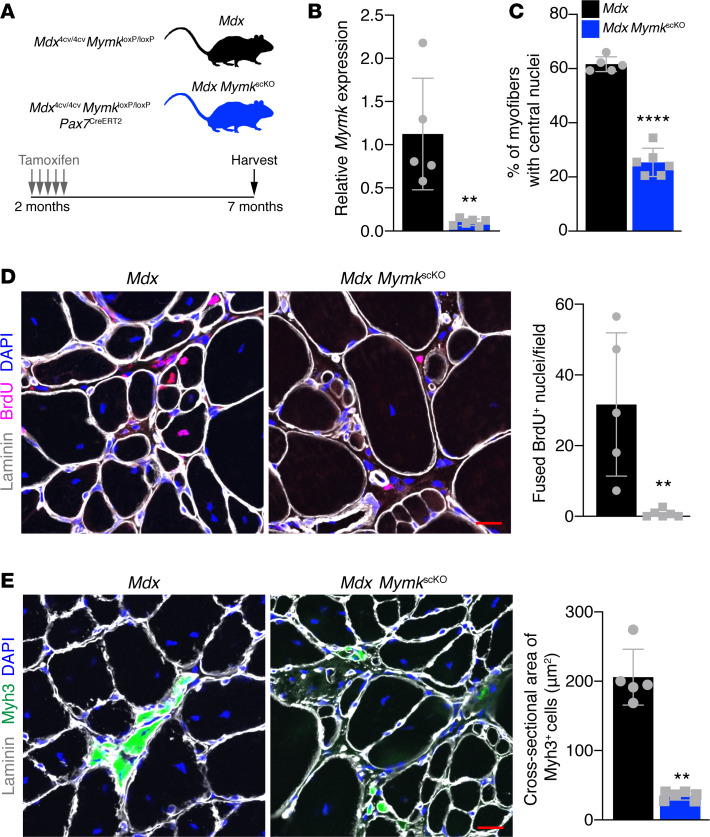
Figure 1. Genetic deletion of Myomaker in SCs of mdx mice leads to complete loss of fusion.
(A) Schematic showing the mouse model and timing of tamoxifen injections. (B) qPCR analysis of Myomaker from whole diaphragm indicates that Myomaker is significantly reduced in mdx MymkscKO muscle. (C) Percentage of myofibers with central nucleation in laminin and DAPI-stained tibialis anterior (TA) sections. (D) Mice were given intraperitoneal injections of BrdU for 7 days before sacrifice to label proliferating nuclei. BrdU+DAPI+ nuclei within the borders of laminin-labeled myofibers were defined as fused nuclei. Quantification shows loss of fusion in mdx MymkscKO mice. (E) Immunofluorescence staining for Myh3+ (embryonic myosin) myofibers reveals loss of de novo myofiber formation in mdx MymkscKO mice. Quantification of cross-sectional area of Myh3+ cells supports lack of true myofiber formation. Statistical analyses and data presentation: (B, D, and E) Mann-Whitney U test; **P < 0.01; (C) unpaired t test; ****P < 0.0001. Data are represented as mean ± SD. Scale bar: 20 μm. n = 5–6.
Quantitative PCR (qPCR) analysis revealed Myomaker mRNA transcripts were significantly downregulated in skeletal muscle of mdx MymkscKO mice (Figure 1B). Analysis of myofibers with central nuclei, a hallmark of regeneration and fusion, showed a significant reduction in mdx MymkscKO muscle (Figure 1C). These data suggest that ongoing fusion contributes heavily to central nucleation in dystrophic muscle. To more directly assay fusion, mice were treated with BrdU for 7 days before sacrifice to label proliferating nuclei, including myogenic progenitors that could then be tracked for their incorporation into myofibers. A fusion event was defined as a BrdU+ nucleus located within a myofiber labeled by laminin. BrdU+ nuclei within myofibers were observed in mdx mice, showing that fusion was occurring in this time frame, but mdx MymkscKO mice showed a complete lack of BrdU+ nuclei in myofibers (Figure 1D). Another hallmark of regeneration and fusion is the presence of de novo myofibers, which are marked by embryonic myosin (Myh3). Immunostaining with Myh3 antibodies showed the presence of large Myh3+ cells in mdx mice (Figure 1E). In contrast, only small Myh3+ cells were observed in mdx MymkscKO muscle (Figure 1E). Analysis of the cross-sectional area of Myh3+ cells showed that these cells in mdx MymkscKO were smaller than in controls (Figure 1E). These data indicate that SCs in mdx MymkscKO muscle can differentiate but that their progeny fail to fuse properly, resulting in stunted growth of de novo myofibers. Additionally, we found that the presence of Pax7+ and Myog+ cells in the muscle was unchanged in mdx MymkscKO muscle, indicating the persistence of a differentiated but nonfusogenic SC population following deletion of Myomaker (Supplemental Figure 2, A and B). These data show that Myomaker is required in the SC compartment for fusion between progenitors to form de novo myofibers and fusion between a myogenic progenitor and myofiber. Thus, we have established a dystrophic mouse model that maintains a reservoir of SCs but lacks any fusion of their progeny.
Myocyte fusion is required for regeneration of dystrophic muscle.
The body weights of mdx and mdx MymkscKO mice were monitored after induction of fusion incompetence, and mice lacking fusion displayed a dramatic decline in body weight (Figure 2A). At 5 months after tamoxifen administration, mdx MymkscKO mice were visibly smaller and showed exacerbated kyphosis of the spine, an established marker of disease severity in DMD (Figure 2B). Analysis of muscle weights revealed significantly reduced muscle mass in mdx MymkscKO mice compared with mdx controls (Figure 2C). Similarly, muscle function as assessed by forelimb grip strength was severely impaired in mdx MymkscKO mice (Figure 2D). Histological analysis of the TA also revealed overall smaller muscle area in mdx MymkscKO mice but the presence of large myofibers (Figure 2E). We also assessed the inflammatory state of the tissue after loss of Myomaker in SCs, which revealed an aggravated dystrophic phenotype. We observed an increased inflammatory response with elevated numbers of CD68+ macrophages in mdx MymkscKO mice (Figure 2F). Additionally, collagen deposition and fibrosis were significantly increased (Figure 2G). These data establish that fusion is absolutely required for regeneration and muscle function in DMD and that dystrophic muscle is unable to properly compensate for damage-induced degeneration without fusing SC progeny.
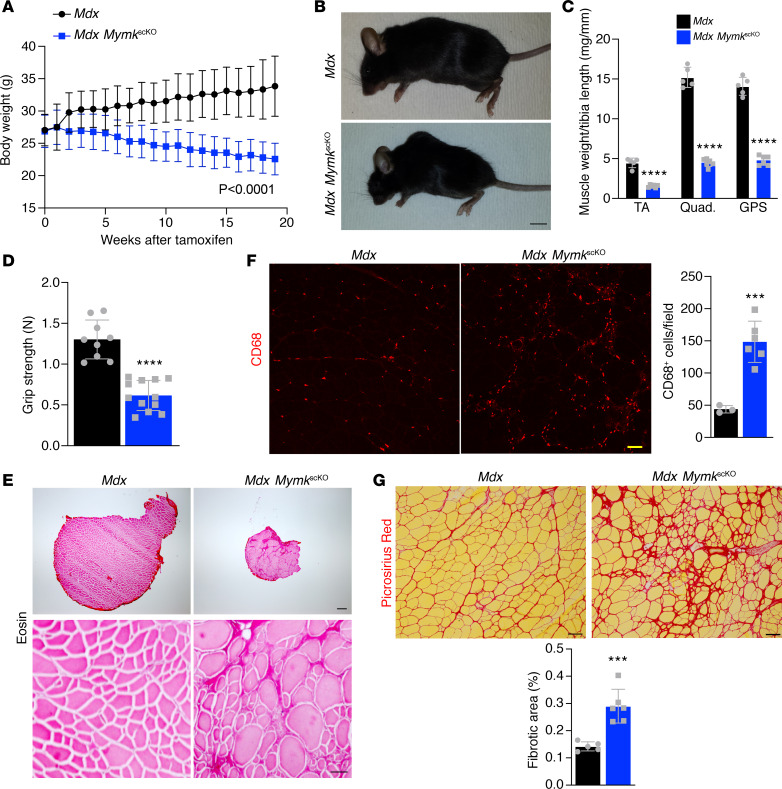
Figure 2. Deletion of Myomaker in SCs of dystrophic muscle leads to severe pathology.
(A) Body weight was assessed weekly and dropped steadily following loss of Myomaker in SCs (n = 9–12). (B) Mdx MymkscKO show loss of muscle mass and kyphotic spinal morphology by gross examination. (C) Dry weights of tibialis anterior (TA), quadriceps, and gastrocnemius/plantaris/soleus (GPS) following dissection. (D) Muscle function was assessed using a forelimb grip strength meter (n = 9–12). (E) Eosin stains on cross section of TA muscles show reduced muscle cross-sectional area and altered myofiber morphology. (F) CD68 immunofluorescence of TA sections shows enhanced inflammatory process in mdx MymkscKO muscle (n = 3–6). (G) Picrosirius red–stained sections of TA muscle demonstrate increased fibrosis in fusion-incompetent dystrophic muscle. Statistical analyses and data presentation: (A) linear regression with slopes comparison; (D, F, and G) unpaired t test; ***P < 0.001, and ****P < 0.0001. Data are represented as mean ± SD. Scale bars: 1 cm (B), 200 μm (top right) and 50 μm (bottom right) (E), 50 μm (F), 100 μm (G). n = 5–6 except where noted.
It is well established that mdx mice exhibit the most profound muscle degeneration early in life, with the onset of severe muscle damage occurring between 3 and 6 weeks of age (28). There is likely an important role of fusion during this early time frame, but whether fusion occurs throughout the dystrophic disease process is not known. We assessed fusion rates in mdx mice across the life span through our BrdU assay and showed that myocyte fusion rates are highest early in life, with a steep drop-off in levels of fusion by 6 months (Supplemental Figure 3A). This is despite the fact that SC levels are persistently elevated compared with WT controls (29). We next asked whether myocyte fusion is required for regeneration later in the disease course of mdx mice, when there is less ongoing fusion. We treated mdx MymkscKO mice with tamoxifen starting at 12 months of age and again followed up for 5 months (Supplemental Figure 3B). We also observed significant exacerbation of pathology with reduced overall body and muscle weights (Supplemental Figure 3, C–E) and a reduction in grip strength (Supplemental Figure 3F). The effects were more modest in older mice, likely reflecting reduced activity of SCs, but nonetheless indicate that fusion is required for effective regeneration in DMD throughout life.
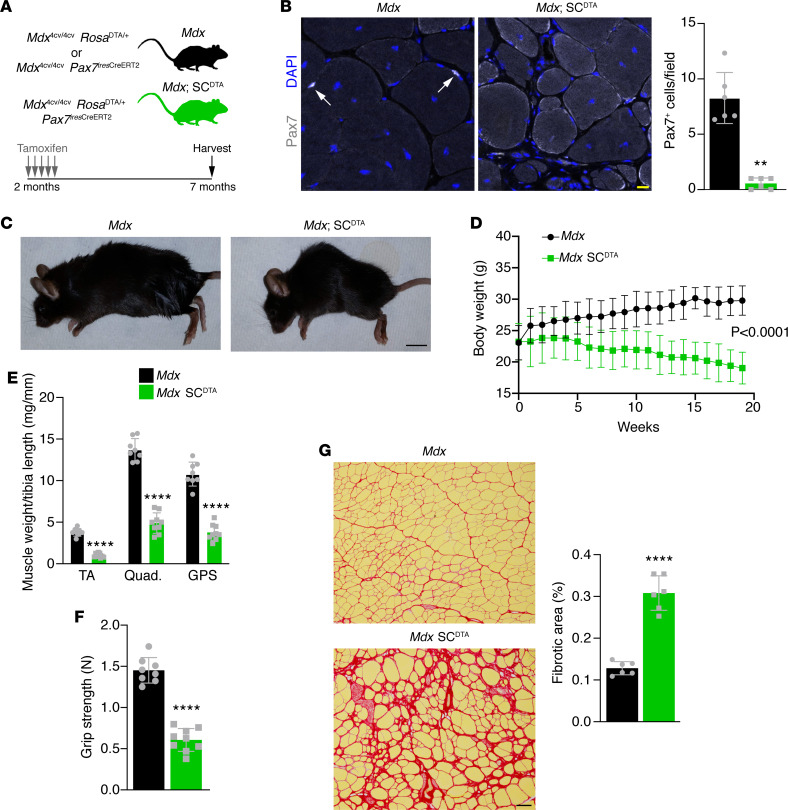
Our mdx MymkscKO model is notable for the continued presence of SCs and their activated progeny in a regenerative setting, while blocking the ability of those cells to fuse. This provided a unique opportunity to investigate whether SCs might have any impact on disease course beyond their role in fusion. Multiple studies have indicated that SCs may play a role in muscle physiology outside of their function in regeneration (30, 31). To test whether there may be a fusion-independent role for SCs in DMD, we generated a model in which SCs would be completely ablated using a Rosa26DTA/+ allele in combination with a tamoxifen-inducible Pax7CreERT2 in the mdx4cv background (mdx SCDTA). These mice were treated with tamoxifen at 2 months of age, similar to our experiment with mdx MymkscKO mice (Figure 3A). Five months after tamoxifen administration, we observed efficient ablation of Pax7+ SCs in muscles from these mice (Figure 3B). Mdx SCDTA mice exhibited an analogous pattern of decline to mdx MymkscKO mice when followed after SC ablation (Figure 3C). Body weight and muscle weights dropped dramatically (Figure 3, D and E), grip strength was significantly impaired (Figure 3F), and collagen deposition and fibrosis were also increased (Figure 3G). Direct comparisons of muscle weights, grip strength, and fibrosis between the mdx MymkscKO and mdx SCDTA groups revealed equivalent effect sizes, indicating the 2 models result in a similar exacerbation of pathology (Supplemental Figure 4, A–C). The phenotypic equivalence of the mdx MymkscKO and mdx SCDTA models is strong evidence that SCs have a profound beneficial effect on regeneration in muscular dystrophy and that they exert this effect primarily through the function of their fusogenic progeny to repair damaged muscle.
Figure 3. Ablation of SCs in mdx mice leads to loss of muscle mass and diminished function.
(A) Schematic showing the mdx SCDTA mouse model and timing of tamoxifen injections. (B) Pax7 immunofluorescence shows loss of SCs in mdx SCDTA tibialis anterior (TA) muscle. Quantification is of Pax7+DAPI+ cells. (C) Mdx mice lacking SCs show diminished size and exaggerated spinal kyphosis. (D) Body weight was assessed weekly and dropped steadily in SC-ablated mdx mice. (E) Individual dry muscle weights of TA, quadriceps, and gastrocnemius/plantaris/soleus (GPS) muscles. (F) Forelimb grip strength measurements taken 5 months after tamoxifen administration. (G) Picrosirius red staining of TA muscles reveals aggravated fibrosis in mdx SCDTA mice. Statistical analyses and data presentation: (D) linear regression with slopes comparison; (E–G) unpaired t test; ****P < 0.0001. Data are represented as mean ± SD. Scale bars: 10 μm (B), 100 μm (G). n = 6–9.
Myomaker expression in myofibers has minimal impact on fusion dynamics and regeneration.
Myomaker is known to be required bilaterally on both fusing membranes during myoblast fusion in vitro, but the symmetry and cell specificity of its activity in vivo have not been established (20, 25). Evidence from the synergist ablation model of muscle hypertrophy suggests that Myomaker is not strongly upregulated in adult myofibers even during periods of muscle growth and myocyte fusion (27). This raises the question of Myomaker’s requirement in and activity on myofibers in vivo. In DMD, fusion occurs chronically, both symmetrically between myoblasts as well as asymmetrically between myoblasts and damaged myofibers; thus, we reasoned that DMD is an appropriate model system in which to assess the role of Myomaker on mature myofibers and its impact on dystrophic pathology. We crossed the mdx MymkloxP/loxP mice with the myofiber-specific Acta1CreERT2 allele (mdx MymkfiberKO) and induced myofiber-specific deletion of Myomaker starting at 2 months of age (Figure 4A). Myomaker transcripts were significantly reduced in whole skeletal muscle of mdx MymkfiberKO mice upon harvesting 5 months later (Figure 4B). We next directly assessed myofiber-specific knockdown of Myomaker by isolating RNA from single myofibers from the extensor digitorum longus muscle of mdx and mdx MymkfiberKO after short-term tamoxifen treatment (Supplemental Figure 5A). We observed a significant decrease of Mymk transcripts in mdx MymkfiberKO myofibers by qPCR (Supplemental Figure 5B). We also determined that Mymk transcripts were not altered in the mononuclear compartment from mdx MymkfiberKO mice (Supplemental Figure 5C). Taken together, Mymk transcripts are significantly reduced in myofibers but not in muscle progenitors.
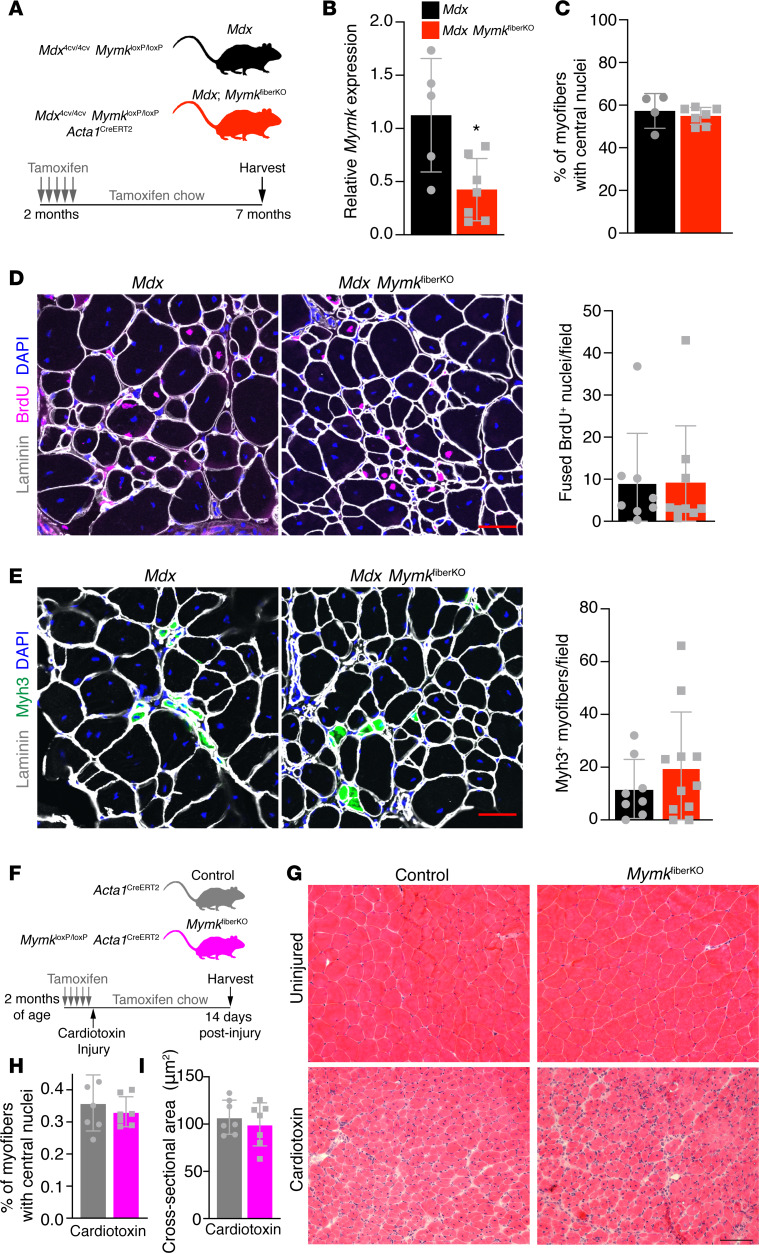
Figure 4. Deletion of Myomaker in mature myofibers does not affect fusion dynamics in WT or mdx mice.
(A) Schematic showing the mouse model and the timing of tamoxifen administration. (B) qPCR analysis from whole diaphragm muscle indicates reduced Myomaker expression in mdx MymkfiberKO mice (n = 5–7). (C) Quantification of centrally nucleated myofibers in tibialis anterior (TA) muscle revealed no change following deletion of Myomaker in myofibers (n = 4–7). (D) Fusion of BrdU+ nuclei is unaffected in mdx MymkfiberKO mice (n = 8–11). (E) Formation of de novo (Myh3+) myofibers is not significantly altered in mdx MymkfiberKO muscle (n = 8–11). (F) Schematic showing acute injury mouse model, timing of cardiotoxin injury, and tamoxifen regimen. (G) H&E stain of TA muscle 14 days postinjury. (H) Quantification of myofibers with central nuclei from laminin-stained sections (not shown) (n = 7). (I) Average cross-sectional area of TA myofibers was unchanged following Myomaker deletion in myofibers (n = 7). Statistical analysis and data presentation: (B) unpaired t test, *P < 0.05. Data are represented as mean ± SD. Scale bars: 50 μm (D and E), 100 μm (G).
Intriguingly, after 5 months of tamoxifen administration, we found no evidence of significantly altered fusion dynamics. First, quantification of centrally nucleated myofibers displayed no difference between mdx MymkfiberKO mice and controls, suggesting no long-term change in fusion as seen in the mdx MymkscKO model (Figure 4C). Total fusion rate was unchanged relative to control mice, as indicated by comparable levels of fused nuclei within myofibers following a 7-day BrdU regimen (Figure 4D). Moreover, de novo myofibers (Myh3+) were also detected at comparable levels in both groups (Figure 4E). Thus, Myomaker reduction in myofibers leads to minimal impact on fusion in dystrophic muscle in this model.
These results suggested that myofiber expression of Myomaker is dispensable for fusion in vivo. We sought to confirm these findings in a nondystrophic model of short-term regenerative fusion using cardiotoxin (CTX) injury of the TA muscle. We performed CTX injury of MymkloxP/loxP Acta1CreERT2 mice (MymkfiberKO, nondystrophic) and assessed muscle regeneration 14 days after injury (Figure 4F). We have previously performed this experiment using a MymkscKO mouse line and shown that in the absence of SC expression of Myomaker, muscle-regenerative capacity is completely lost and no myofibers are formed (24). In contrast, the MymkfiberKO muscles were able to regenerate effectively (Figure 4G). We assessed central nucleation and cross-sectional area of myofibers in the focus of injury and found no significant differences (Figure 4, H and I), indicating that the regenerated myofibers are comparable and that the fusion process is unaltered in the absence of myofiber-specific Myomaker expression. In combination with our findings in dystrophic mice, these results suggest that Myomaker is dispensable on the myofiber during regeneration in vivo and that Myomaker primarily drives membrane fusion through its activity in SC progeny.
Myomaker expression in myofibers contributes to membrane damage and degeneration.
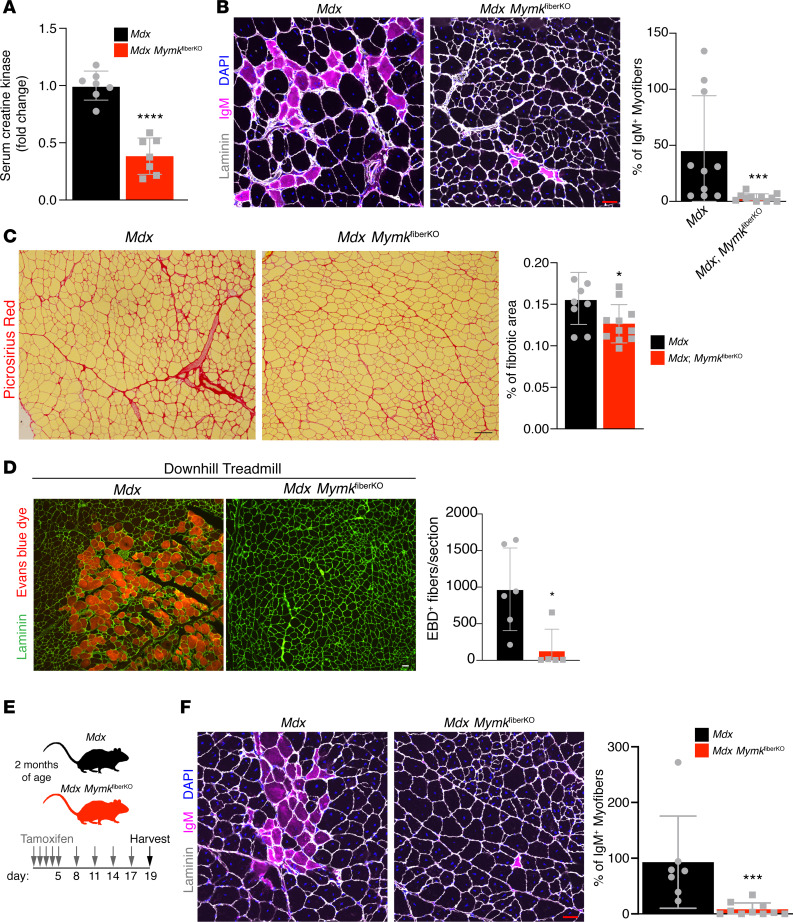
Although we found that reduction of Myomaker expression in myofibers exerts no detectable influence on fusion during regeneration, we nevertheless asked whether Myomaker in myofibers had any impact on dystrophic phenotype. Surprisingly, we found that serum creatine kinase (CK) was significantly reduced in mdx MymkfiberKO compared with controls (Figure 5A). Serum CK is a common clinical marker of DMD severity and is a reflection of systemic muscle damage. We therefore performed additional histological assays to assess muscle damage. One such assay is IgM immunofluorescence, which labels myofibers with unstable sarcolemmas that allow for infiltration of extracellular contents. Remarkably, mdx MymkfiberKO mice showed almost complete absence of IgM+ myofibers upon immunofluorescence, whereas groups of IgM+ myofibers were visible in controls (Figure 5B). Additionally, a significant reduction in fibrosis was also observed in mdx MymkfiberKO mice (Figure 5C), further indicating less overall muscle damage. We next performed a validated damage-inducing downhill treadmill protocol on mdx and mdx MymkfiberKO mice (32, 33), with injection of Evans blue dye (EBD), a vital dye that is used as a marker of sarcolemmal instability by its ability to penetrate compromised myofiber membranes. This experiment served as an independent assay for membrane integrity and assessed the protective effect of mdx MymkfiberKO mice under an acute damage stimulus. Mdx MymkfiberKO showed a significant reduction in EBD+ damaged myofibers following completion of the protocol (Figure 5D). Reduced damage after downhill exercise in mdx MymkfiberKO mice suggests the direct protective effect is due to the reduction of Mymk in myofibers.
Figure 5. Deletion of Myomaker in myofibers leads to reduced muscle damage in mdx mice.
(A) Serum CK is reduced in mdx MymkfiberKO mice (n = 7). (B) IgM immunostaining of quadriceps (rectus femoris) sections shows reduction of damaged myofibers following deletion of myofiber-specific Myomaker expression (n = 8–11). (C) Picrosirius red stain of tibialis anterior (TA) muscle sections indicates less fibrosis in mdx MymkfiberKO mice (n = 8–11). (D) Rectus femoris muscle from mice treated with Evans blue dye (EBD) and subjected to a damage-inducing forced treadmill running protocol, with EBD+ myofibers quantified (n = 5–6). (E) Schematic of short-term mdx MymkfiberKO experiment showing continued tamoxifen administration for 2 weeks following initial 5-day induction. (F) Following short-term deletion of Myomaker in myofibers, IgM+ myofibers are reduced in rectus femoris muscle (n = 7–10). Statistical analyses and data presentation: (A and C) unpaired t test; *P < 0.05, and ****P < 0.0001; (B, D, and F) Mann-Whitney U test; *P < 0.05, and ***P < 0.001. Data are represented as mean ± SD. Scale bars: 50 μm (B, D, and F), 100 μm (C).
To rule out the possibility that the improvement in muscle damage indices could be a secondary effect of an undetected fusion phenotype that had accumulated over months, we harvested muscles from an acute time point, 2 weeks after initial tamoxifen induction (Figure 5E). IgM+ myofibers were again nearly absent in mdx MymkfiberKO mice, while prevalent in control mdx muscles (Figure 5F). Additionally, mice treated with vehicle injections rather than tamoxifen showed no difference in IgM+ myofibers (Supplemental Figure 6, A and B). Finally, to rule out the further possibility that reduced muscle damage could be a side effect of the ongoing activation of Acta1CreERT2 when mice are on a tamoxifen diet, we generated an additional control mouse line with the Acta1CreERT2 alone crossed into the mdx background (Supplemental Figure 6C). No significant differences in IgM staining were observed when compared with mdx controls (Supplemental Figure 6D). Altogether, these data show that ablation of Myomaker expression in myofibers leads to a significant improvement in disease phenotype, indicating that the cell biological consequence of Myomaker expression in myofibers is its contribution to muscle damage.
Deletion of Myomaker in dystrophic myofibers leads to improved muscle function.
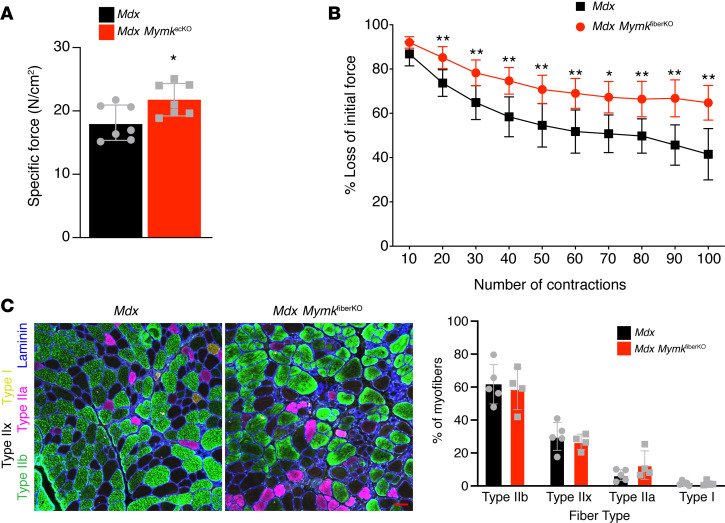
We next asked whether this reduction in muscle damage led to functional benefits for the muscle of mdx MymkfiberKO mice. We performed in situ muscle functional measurements to assess force production and fatigability of the TA muscle. We found that mdx MymkfiberKO mice displayed increased specific force relative to controls, an indication of greater intrinsic muscle quality (Figure 6A). In addition, mdx MymkfiberKO displayed significantly enhanced resistance to muscle fatigue upon repeated stimulation when compared with control mice (Figure 6B). Although improved resistance to fatigue can often be associated with a shift toward oxidative myofiber types, we found no change in myofiber type composition in the TA muscle of mdx MymkfiberKO mice (Figure 6C), suggesting that the improvement is directly related to reduced susceptibility to myofiber damage and overall improved muscle quality. In summary, our results demonstrate that reduction of Myomaker in mature dystrophic myofibers reduces myofiber damage and improves muscle function.
Figure 6. Muscle function is improved in mdx mice following myofiber-specific deletion of Myomaker.
(A) Specific force of tibialis anterior (TA) muscle upon in situ muscle functional measurement (n = 7). (B) Upon repeated nerve stimulation to assay fatigability, mdx MymkfiberKO mice showed greater resistance to muscle fatigue (n = 7). (C) Immunofluorescence staining for myofiber type–specific myosin heavy chains revealed no change in myofiber type distribution of mdx MymkfiberKO TA muscle (n = 4–5). Statistical analyses and data presentation: (A and B) unpaired t test; *P < 0.05, **P < 0.01. Data are represented as mean ± SD. Scale bar: 50 μm (C).
Myomaker expression in myofibers is dependent upon fusion of SC progeny.
A key remaining question concerned the origin of Myomaker expression in dystrophic muscle and specifically in myofibers. We sought to distinguish between 2 possibilities: first, that Mymk transcription is upregulated in resident myofiber nuclei in chronic regeneration, and second, that Mymk is primarily transcribed in activated SCs, which contribute Myomaker-expressing nuclei to nascent and existing myofibers through fusion. To distinguish these possibilities, we returned to the mdx MymkscKO model and used the MymkLacZ allele to assess active Mymk transcription in dystrophic muscle when the fusion of SC progeny has been blocked. In the MymkLacZ construct, the LacZ cassette is inserted in intron 1 of the Mymk locus, resulting in a null allele (24), which was combined with a floxed allele to achieve total inducible knockout of Myomaker in SCs (mdx Pax7CreERT2 MymkLacZ/loxP, which we designate as mdx MymkLacZ/scKO, with mdx MymkLacZ/loxP controls). The MymkLacZ allele will still result in active LacZ transcription in Myomaker-depleted SCs, resulting in the presence of LacZ+ mononuclear cells, but assessment of LacZ+ myofibers should reveal whether Myomaker expression in myofibers is SC dependent. We observed a negligible number of LacZ+ myofibers in mdx MymkLacZ/scKO muscle, which we quantified by measuring the cross-sectional area of all LacZ+ cells (Figure 7, A and B). We supported this finding with an independent assay, using single-molecule RNA-FISH (smRNA-FISH) to assess directly Mymk transcripts in control and mdx MymkscKO muscle. We found that Myomaker again localized to both small and large myofibers, and mononuclear cells in mdx muscle, whereas mdx MymkscKO samples showed a near absence of Mymk signal and no Mymk+ myofibers (Figure 7C). These experiments indicate that the ultimate reservoir of Mymk transcriptional capacity in dystrophic muscles are activated SCs, which contribute Mymk-expressing nuclei to myofibers through ongoing fusion.
Figure 7. Myomaker expression in myofibers is contributed through fusion of myocytes.
(A) X-gal staining of tibialis anterior (TA) muscle from mdx MymkLacZ/fl and mdx MymkLacZ/scKO mice, with cross-sectional area distribution of LacZ+ cells (n = 5–6). (B) X-gal staining of diaphragm with cross-sectional area distribution (n = 5–6). (C) smRNA-FISH for Mymk transcripts in quadriceps muscles of mdx MymkscKO mice and controls, with laminin immunofluorescence staining (n = 5). Quantification of Mymk+ cells per field with average cross-sectional area shown on right. Statistical analyses and data presentation: (A and B) Mann-Whitney U and unpaired t tests; *P < 0.05, **P <0.01, ***P < 0.001, and ****P < 0.0001; (C) unpaired t tests; **P <0.01. Data are represented as mean ± SD. Scale bars: 100 μm (A and B), 20 μm (C).
Discussion
In this study we evaluated the role of Myomaker in SCs and myofibers in muscular dystrophy. Dystrophic fusion occurs within a relatively intact tissue, with both SC progeny and myofibers participating in membrane fusion events. We found that Myomaker has a differential effect on the pathology dependent upon its compartment of expression. Expression of Myomaker in activated SCs is absolutely required for fusion and consequent regeneration of dystrophic muscle, whereas myofiber expression of Myomaker has minimal effect on fusion but is detrimental to membrane health and muscle function. Overall, we propose a paradigm whereby, in contrast to normal adult muscle adapting to increased workload (27), Myomaker is ectopically expressed in dystrophic myofibers. The source of this ectopic expression is continual activation and fusion of myocytes, which lead to persistent expression of fusogenic factors, such as Myomaker, in myofibers. While regenerative fusion is necessary for preserving muscle function, the chronic loading of Myomaker onto myofiber membranes contributes to myofiber instability and disease progression. Blocking fusion entirely, however, leads to significant loss of muscle mass and functional deterioration, which establishes that SCs are required for regeneration in muscular dystrophy. These data are consistent with many studies showing perturbation of SCs results in exacerbated disease (15, 16, 34, 35). Curiously, ablation of SCs entirely leads to an indistinguishable phenotype, which suggests that the ongoing presence of Pax7+ SCs and activated myoblasts in the mdx MymkscKO model has little impact on disease pathology. This supports a model in which the exclusive function of SCs in dystrophic muscle is fusion to drive de novo myofiber formation or repair damaged myofibers. We did not observe an accumulation of SCs or activated myoblasts in mdx MymkscKO muscle, which could indicate the possibility of death or fate change as activated SC progeny cannot fuse. Further experiments would be necessary to investigate these possibilities. Altogether, while there could be fusion-independent roles for SCs in this and other muscle contexts as suggested elsewhere (30, 31), the effects of those roles are not easily revealed when fusion is abrogated.
A central question raised in this study is the symmetry of Myomaker’s activity in vivo: is Myomaker required on both fusing membranes (myocyte and myofiber) for fusion to occur? Myomaker exhibits a bilateral requirement on myoblasts both in vivo during development and in vitro using cell lines, but these systems may not recapitulate the membrane dynamics of myocyte-to-myofiber fusion. One limitation of a definitive answer for the need for Myomaker on the myofiber for fusion concerns the syncytial nature of this cell type. Multiple nuclei within a shared cytoplasm could represent a challenge in achieving complete deletion in all myonuclei, and the continual addition of fusing SCs may also provide an ongoing source of Myomaker transcription. This challenge is reflected in our mdx MymkfiberKO mice, where loss of Mymk was not complete in either whole muscle or isolated myofibers. Analysis of mRNA expression from whole muscle does not distinguish between expression in muscle progenitors or myofibers, and Mymk transcription by activated SC progeny was unaffected in these mice. Similarly, isolated myofibers can be contaminated by transcripts from mononuclear cell populations and very recently fused nuclei, making interpretation of precise levels of remaining Mymk transcription in myonuclei difficult.
Despite technical hurdles, we have obtained multiple indirect results suggesting that Myomaker is not needed on the myofiber for fusion. It may be expected that if Myomaker were needed on the myofiber for fusion, then nuclei within myofibers would possess the capability to upregulate the transcript in the absence of SC fusion. However, in this study, we found that Myomaker expression in dystrophic myofibers is dependent upon fusion of SC progeny, indicating that myofiber nuclei cannot independently upregulate Myomaker and that Myomaker’s principal role in driving fusion is in the muscle progenitor. Moreover, we show here that reduction of Myomaker in myofibers (mdx MymkfiberKO mice) does not influence effective fusion. Finally, experiments performed in healthy mice undergoing muscle overload hypertrophy have also indicated that Myomaker is dispensable on myofiber membranes during muscle growth in a nondiseased setting (27). Taken together, several lines of experimentation now suggest that Myomaker expression in myofibers has minimal impact on fusion dynamics. In turn, these findings raise the questions of what factors are required on myofiber membranes for fusion and whether fusion competency is mechanistically distinct in myofibers compared with myocytes.
Directly related to the question of Myomaker’s activity on myofibers is the question of the mechanistic basis of the improvement seen in mdx MymkfiberKO mice, which can be attributed to one of 2 possible models. The first possibility is that Myomaker acts on dystrophic myofiber membranes to drive fusion of SCs to myofibers, thereby helping maintain damaged and degenerative myofibers. In this case, maintaining these “unhealthy” existing myofibers actually has a negative effect on overall muscle health, and shunting fusogenic SCs toward formation of de novo myofibers instead provides a net benefit to the tissue. The lack of IgM+ myofibers would then be explained by dropout of degenerating myofibers that do not receive fusing SCs. While an intriguing possibility, this model is unsupported by the lack of any significant change in fusion dynamics or de novo myofiber formation. Additionally, the preservation of membrane integrity following forced treadmill running in mdx MymkfiberKO mice, in comparison with widespread damage in controls, is essentially incompatible with the notion of myofiber dropout. The second model would be that Myomaker, as a transmembrane protein active in driving the membrane-remodeling steps of fusion, is a destabilizing factor on dystrophic myofiber membranes. It has been previously suggested that the process of membrane fusion may further exacerbate the sarcolemmal instability that is fundamental to DMD pathophysiology (17). Our findings suggest that fusion itself may not be deleterious, but the sustained presence of fusion factors in myofibers may underlie the effect on dystrophic membranes. The observed deleterious effects may be the consequence of a membrane-remodeling activity for Myomaker, which remains to be elucidated. However, Myomaker is required for hemifusion and may act as a driver of the lipid rearrangements necessary for outer membrane leaflet merger (26). Further experiments would be required to determine whether Myomaker directly contributes to membrane instability at the biochemical level. Additionally, the implication of membrane repair proteins, such as the ferlins and Ano5, in the myoblast fusion process may also suggest a link between fusion and membrane destabilization (36–38). In the context of fragile dystrophic membranes, the fusion machinery may actually injure the sarcolemma in a manner normally compensated for in healthy muscle. In this sense, the mdx MymkscKO and mdx SCDTA models provide important context for these considerations by establishing that the net effect of ongoing fusion is beneficial to dystrophic muscle.
In conclusion, we have shown that DMD is characterized by ongoing fusion of SC progeny that is fundamentally required for regeneration and maintenance of muscle mass, while Myomaker’s presence in myofibers simultaneously contributes to muscle tissue damage and membrane instability. These results likely explain the reason why Myomaker is downregulated after the fusion process ceases during postnatal development and adult regeneration (24). One possible explanation for the tight regulation of expression of the fusion machinery could be that the proteins responsible for fusion, although necessary for muscle growth and regeneration, also cause membrane destabilization that can be detrimental, especially in the case of an already-unstable membrane that occurs in DMD. In this case, persistent loading of the fusion machinery in dystrophic myofibers would represent a previously unappreciated aspect of DMD pathophysiology. We speculate that the concept proposed here — that expression of the myogenic development program in dystrophic myofibers can result in deleterious consequences — may extend beyond the expression of Myomaker and include other fundamental muscle differentiation factors, including MyoD, myogenin, and the second fusion factor Myomerger (39–41). In conclusion, these findings guide our understanding of the role of SCs during DMD and the cell biology of muscle cell fusion and overall may inform efforts to modulate cell therapy for therapeutic benefits.
Methods
Mice.
All mice used in this study were maintained on a C57BL/6 background. The MymkLacZ and MymkloxP/loxP strains have been described previously (21, 24). For the present study, MymkloxP/loxP mice were bred into the mdx4cv (JAX strain 002378) background. To achieve cell type–specific inducible deletion of Mymk, one of 2 Cre recombinase alleles was then introduced: the satellite cell–specific Pax7CreERT2 (exon 1 is replaced with CreERT2) (42) or myofiber-specific Acta1CreERT2 (43). For MymkscKO experiments, both floxed-only and vehicle-treated controls were used and combined. For MymkfiberKO experiments, floxed-only controls were used except where indicated. For SC ablation, we crossed the Rosa26-EGFP-DTA line (JAX strain 006331) with mdx4cv along with an inducible Pax7CreERT2 containing an iresCreERT2 cassette (44). Mdx Pax7CreERT2/+ and mdx R26DTA/+ controls were treated with tamoxifen and combined for subsequent analysis. All experiments were performed on sex- and age-matched cohorts, where both male and female mice were used, and at least 2 independent experiments were performed for all mouse models and time points.
Tamoxifen and BrdU treatment.
Tamoxifen (MilliporeSigma) was prepared in corn oil with 10% ethanol at a concentration of 25 mg/mL. Mice were given intraperitoneal injections of tamoxifen (0.075 mg/kg/d) for 5 days to induce recombination. For experiments with the Acta1CreERT2 allele, mice were then maintained on tamoxifen either by injections every third day or by tamoxifen chow (Envigo) for long-term experiments. To label proliferating nuclei and assess fusion of SCs, mice were given BrdU (Thermo Fisher Scientific) by intraperitoneal injection at 10 μL/g as indicated.
CTX injury.
CTX from Naja pallida (MilliporeSigma 217503) was aliquoted in sterile saline at a concentration of 10 μM. Mice were anesthetized using isoflurane for sterile injection of TA muscles. Mouse legs were shaved and 50 μL of CTX was injected using a 28-gauge needle.
Muscle collection and sample preparation.
Mouse hind limb muscles and diaphragm were dissected, dried, and weighed. Tibias were dissected and remaining tissue was digested with proteinase K (0.4 mg/mL) overnight at 55°C, after which tibia length was measured using digital calipers. Muscles were embedded in 10% tragacanth/PBS (MilliporeSigma) and frozen in 2-methylbutane cooled in liquid nitrogen. We used 10-μm sections for all histology. For RNA preparations, tissues were flash-frozen in liquid nitrogen immediately upon dissection. For RNA isolation from muscle fibers, whole extensor digitorum longus muscles were incubated in 0.2% type I collagenase (MilliporeSigma C0130) in DMEM at 37°C for 90 minutes. Following incubation, muscles were triturated in PBS to release individual myofibers, which were then collected and centrifuged at centrifuged at 10,000 g to form a pellet for RNA isolation with TRIzol (Invitrogen, Thermo Fisher Scientific).
To isolate mononuclear cell fractions from skeletal muscle, we performed enzymatic dissociation as previously described with modifications (25). Briefly, mouse limb muscles (gastrocnemius, TA, and quadriceps for MymkscKO experiments or gastrocnemius and triceps for MymkfiberKO experiments) were dissected and dissociated in gentleMACS C Tubes (Miltenyi Biotec) using a gentleMACS Dissociator. Muscles were then digested in 2% type I collagenase (Invitrogen, Thermo Fisher Scientific) in low-glucose DMEM (Invitrogen, Thermo Fisher Scientific) for 90 minutes at 37°C, at which time samples were triturated using 10-mL pipettes. Dispase (Roche) was added at 4.8 U/mL, and samples were incubated for an additional 30 minutes. Following trituration with a 20-gauge needle, samples were passed through a 40-μm filter, centrifuged, washed in PBS, and recentrifuged. The resulting pellet was then dissolved in TRIzol (Invitrogen, Thermo Fisher Scientific) for subsequent RNA isolation.
Histological analyses.
X-gal stain was performed as described previously (24). Sections were costained with 0.7% eosin and mounted with VectaMount (Vector Laboratories). Picrosirius red and H&E stains were performed on frozen sections using standard protocols. Immunohistochemical studies were performed as described previously with minor modifications (27). Briefly, sections were fixed in 1% PFA/PBS and permeabilized with 0.2% Triton X-100/PBS. Sections were blocked using 1% BSA, 1% heat-inactivated goat serum, and 0.025% Tween-20/PBS, and for mouse primary antibodies, we also included AffiniPure Fab fragment goat anti-mouse IgG (Jackson ImmunoResearch). Primary antibodies were incubated overnight at 4°C, and secondary Alexa Fluor antibodies (1:200) (Invitrogen, Thermo Fisher Scientific) were applied at room temperature for 1 hour. For anti-Pax7 (gift from Christoph Lepper, The Ohio State University, Columbus, Ohio, USA) and anti-myogenin (Developmental Studies Hybridoma Bank, DSHB; F5D) staining, an antigen retrieval step was included, boiling slides in 10× Antigen Retrieval Citra Plus Solution (Biogenex HK086-9K) for 30 minutes before blocking with M.O.M. mouse IgG blocking reagent (Vector Laboratories). For anti-BrdU (Roche 11170376001) staining, slides were fixed in 4% PFA and denatured with 2 M HCL in 0.5% Triton X-100/PBS, followed by neutralization with TBS (pH 8.4). Other antibodies used include anti-Myh3 (DSHB F1.652), anti-CD68 (Abcam ab53444), FITC-conjugated anti-mouse IgM (MilliporeSigma F9259), and anti-laminin (MilliporeSigma L9393). For myofiber-type–specific myosin immunofluorescence, the following antibodies were used from DSHB: Type IIb (BF-F3), Type IIa (SC-71), Type I (BA-D5), and Type IIx myofibers were identified by absence of fluorescent signal.
Muscle force measurements.
Forelimb grip strength was measured using a forelimb strength measurement meter (Columbus Instruments). In situ isometric muscle force measurement was performed as described previously (45). Briefly, under anesthesia via isoflurane inhalation, the distal tendon of the TA was surgically exposed, cut, and attached to the lever arm (Aurora Scientific, 305C) by silk suture. Two electrodes were placed under the sciatic nerve, and electrical stimulation (0.2-ms pulse at 50 mA) was applied to elicit peak isometric twitch force at optimal muscle length. To determine peak isometric tetanic forces (Po), electrical frequency was increased from 25 to 150 Hz for 350 ms every 2 minutes. After 5 minutes’ rest, fatigue resistance of the TA muscle was tested by repeated isometric tetanic contractions at 150 Hz for 350-ms duration every 10 seconds for 100 contractions.
Immediately after the force measurement, mice were euthanized, and TA muscle weight and length were measured. The physiological cross-sectional area (PCSA) of TA muscle was estimated by the following equation: muscle mass (g)/(1.06 × muscle length [cm] × 0.6) (46). Po was normalized to PCSA to calculate specific force. All data were collected and analyzed by Dynamic Muscle Control and Dynamic Muscle Analysis software (Aurora Scientific).
Forced downhill treadmill exercise and EBD uptake.
To assess the sarcolemmal stability of mice in response to exercise-induced damage, we performed a downhill treadmill exercise protocol with EBD injection as previously described (32, 33). Briefly, mice were run on a 6-lane treadmill set to a 15-degree downhill angle on 4 consecutive days to induce hind limb muscle damage. Mice were allowed 5 minutes’ acclimatization to the treadmill, after which speed was increased at 2 m/min increments to a maximum of 18 m/min. Animals were run to the completion of the protocol or exhaustion, which was defined as 10 consecutive seconds on the shock grid without attempt to continue running. Mice were injected intraperitoneally with EBD (10 mg/mL in sterile PBS at a dose of 0.1 mL per 10 g mouse body weight) at 24 hours before the final exercise session, after which they were immediately sacrificed.
RNA analysis.
Total RNA was isolated from muscle samples using established TRIzol protocols. cDNA was synthesized with the MultiScribe kit using random primers (Applied Biosystems, Thermo Fisher Scientific). Standard qPCR methods were used with PowerUp SYBR Green Master Mix (Applied Biosystems, Thermo Fisher Scientific), and the assay was performed on the Bio-Rad CFX96 Real-Time System with the following SYBR primers: Myomaker, 5′-ACCATGTTCTTTGTGGCGTTC-3′ (forward) and 5′-GTTCATCAAAGTCGGCCAGT-3′ (reverse); and GAPDH, 5′-TGCGACTTCAACAGCAACTC-3′ (forward) and 5′-GCCTCTCTTGCTCAGTGTCC-3′ (reverse).
Single-molecule RNA-FISH.
Single-molecule FISH experiments probing for Mymk mRNA were performed using the RNAscope system (ACDBio) according to the manufacturer’s protocols. Fresh-frozen, midbelly, 10-μm cross sections of quadriceps muscles were used. After the final wash step of the RNAscope protocol, we performed immunofluorescent laminin staining to visualize myofibers. Sections were blocked for 30 minutes using 1% BSA, 1% heat-inactivated goat serum, and 0.025% Tween-20/PBS. Samples were then incubated with anti-laminin (1:100; MilliporeSigma L9393) for 30 minutes at room temperature, followed by secondary Alexa Fluor antibodies (1:200; Invitrogen, Thermo Fisher Scientific) for 30 minutes at room temperature. Coverslips were mounted using VECTASHIELD with DAPI (Vector Laboratories).
Microscopy and image analysis.
Immunostained slides were imaged using a Nikon A1R confocal system. X-gal and H&E stains were visualized using an Olympus BX51 inverted bright-field microscope. For quantification of BrdU+ nuclei, centrally located myonuclei, and CD68+ cells, counts were made in ImageJ (NIH) for up to 4 separate fields per animal (dependent upon section size) and averaged. Quantification of fibrotic area from Picrosirius red sections was performed using a threshold intensity algorithm in NIS Elements. Myofiber cross-sectional area and myofiber number measurements were made using a binary thresholding algorithm in NIS Elements. All image analysis was performed in a blinded fashion.
Statistics.
All statistical analysis was performed using GraphPad Prism 8 software. Data are presented as mean ± SD. Groups were assessed for normality using a Shapiro-Wilk test and analyzed using either an unpaired 2-tailed t test or Mann-Whitney U test for single comparisons or 1-way ANOVA with post hoc Tukey’s for multiple comparisons. Specific statistical tests are noted in the figure legends. Body weight data were assessed using linear regression best-fit models, and slopes were tested for significant differences. Statistical significance throughout was set at P values less than 0.05.
Study approval.
All animal procedures were approved by Cincinnati Children’s Hospital Medical Center’s Institutional Animal Care and Use Committee (IACUC2017-0053).
Author contributions
MJP and DPM designed experiments. TS performed muscle function experiments in the laboratory of SS. All authors analyzed the data and contributed to interpretations. MJP and DPM wrote the manuscript with editing from all authors.
Supplementary Material
Acknowledgments
We thank Justin Boyer and Jeffery Molkentin at Cincinnati Children’s Hospital Medical Center for helpful discussions and Qingnian Goh for technical assistance. This work was supported by grants to DPM from the Cincinnati Children’s Hospital Research Foundation, National Institutes of Health (R01AR068286, R01AG059605), and Pew Charitable Trusts. MJP was supported by a training grant from the National Institutes of Health (National Heart, Lung, and Blood Institute T32HL007752). SS has received support from National Institutes of Health grants R01 HL130356, R56 HL139680, R01 AR067279, R01 HL105826 R01 HL143490, and R13 HL149313; American Heart Association 2019 Institutional Undergraduate Student (19UFEL34380251) and Transformational Project (19TPA34830084) awards; and MyoKardia, AstraZeneca, Merck, and Amgen. TS is supported by an American Heart Association Postdoctoral Fellowship (19POST34380448).
Version 1. 04/20/2020
In-Press Preview
Version 2. 05/07/2020
Electronic publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2020, American Society for Clinical Investigation.
Reference information: JCI Insight. 2020;5(9):e136095.https://doi.org/10.1172/jci.insight.136095.
Contributor Information
Michael J. Petrany, Email: petranml@mail.uc.edu.
Taejeong Song, Email: songtg@ucmail.uc.edu.
Sakthivel Sadayappan, Email: sadayasl@uc.edu.
Douglas P. Millay, Email: douglas.millay@cchmc.org.
References
- 1.Brukman NG, Uygur B, Podbilewicz B, Chernomordik LV. How cells fuse. J Cell Biol. 2019;218(5):1436–1451. doi: 10.1083/jcb.201901017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kim JH, Jin P, Duan R, Chen EH. Mechanisms of myoblast fusion during muscle development. Curr Opin Genet Dev. 2015;32:162–170. doi: 10.1016/j.gde.2015.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sampath SC, Sampath SC, Millay DP. Myoblast fusion confusion: the resolution begins. Skelet Muscle. 2018;8(1):3. doi: 10.1186/s13395-017-0149-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dumont NA, Bentzinger CF, Sincennes MC, Rudnicki MA. Satellite cells and skeletal muscle regeneration. Compr Physiol. 2015;5(3):1027–1059. doi: 10.1002/cphy.c140068. [DOI] [PubMed] [Google Scholar]
- 5.Yin H, Price F, Rudnicki MA. Satellite cells and the muscle stem cell niche. Physiol Rev. 2013;93(1):23–67. doi: 10.1152/physrev.00043.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mercuri E, Bönnemann CG, Muntoni F. Muscular dystrophies. Lancet. 2019;394(10213):2025–2038. doi: 10.1016/S0140-6736(19)32910-1. [DOI] [PubMed] [Google Scholar]
- 7.Rahimov F, Kunkel LM. The cell biology of disease: cellular and molecular mechanisms underlying muscular dystrophy. J Cell Biol. 2013;201(4):499–510. doi: 10.1083/jcb.201212142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gao QQ, McNally EM. The dystrophin complex: structure, function, and implications for therapy. Compr Physiol. 2015;5(3):1223–1239. doi: 10.1002/cphy.c140048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Serrano AL, Muñoz-Cánoves P. Fibrosis development in early-onset muscular dystrophies: mechanisms and translational implications. Semin Cell Dev Biol. 2017;64:181–190. doi: 10.1016/j.semcdb.2016.09.013. [DOI] [PubMed] [Google Scholar]
- 10.Dumont NA, et al. Dystrophin expression in muscle stem cells regulates their polarity and asymmetric division. Nat Med. 2015;21(12):1455–1463. doi: 10.1038/nm.3990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tedesco FS, Dellavalle A, Diaz-Manera J, Messina G, Cossu G. Repairing skeletal muscle: regenerative potential of skeletal muscle stem cells. J Clin Invest. 2010;120(1):11–19. doi: 10.1172/JCI40373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chang NC, Chevalier FP, Rudnicki MA. Satellite cells in muscular dystrophy - lost in polarity. Trends Mol Med. 2016;22(6):479–496. doi: 10.1016/j.molmed.2016.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pisconti A, et al. Loss of niche-satellite cell interactions in syndecan-3 null mice alters muscle progenitor cell homeostasis improving muscle regeneration. Skelet Muscle. 2016;6:34. doi: 10.1186/s13395-016-0104-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tierney MT, et al. STAT3 signaling controls satellite cell expansion and skeletal muscle repair. Nat Med. 2014;20(10):1182–1186. doi: 10.1038/nm.3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xie X, Tsai SY, Tsai MJ. COUP-TFII regulates satellite cell function and muscular dystrophy. J Clin Invest. 2016;126(10):3929–3941. doi: 10.1172/JCI87414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sacco A, et al. Short telomeres and stem cell exhaustion model Duchenne muscular dystrophy in mdx/mTR mice. Cell. 2010;143(7):1059–1071. doi: 10.1016/j.cell.2010.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Granata AL, Vecchi C, Graciotti L, Fulgenzi G, Maggi S, Corsi A. Gamma irradiation can reduce muscle damage in mdx dystrophic mice. Acta Neuropathol. 1998;96(6):564–568. doi: 10.1007/s004010050936. [DOI] [PubMed] [Google Scholar]
- 18.Wakeford S, Watt DJ, Partridge TA. X-irradiation improves mdx mouse muscle as a model of myofiber loss in DMD. Muscle Nerve. 1991;14(1):42–50. doi: 10.1002/mus.880140108. [DOI] [PubMed] [Google Scholar]
- 19.Rossi G, et al. Silencing Nfix rescues muscular dystrophy by delaying muscle regeneration. Nat Commun. 2017;8(1):1055. doi: 10.1038/s41467-017-01098-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Petrany MJ, Millay DP. Cell fusion: merging membranes and making muscle. Trends Cell Biol. 2019;29(12):964–973. doi: 10.1016/j.tcb.2019.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Millay DP, et al. Myomaker is a membrane activator of myoblast fusion and muscle formation. Nature. 2013;499(7458):301–305. doi: 10.1038/nature12343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Landemaine A, Rescan PY, Gabillard JC. Myomaker mediates fusion of fast myocytes in zebrafish embryos. Biochem Biophys Res Commun. 2014;451(4):480–484. doi: 10.1016/j.bbrc.2014.07.093. [DOI] [PubMed] [Google Scholar]
- 23.Luo W, Li E, Nie Q, Zhang X. Myomaker, regulated by MYOD, MYOG and miR-140-3p, promotes chicken myoblast fusion. Int J Mol Sci. 2015;16(11):26186–26201. doi: 10.3390/ijms161125946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Millay DP, Sutherland LB, Bassel-Duby R, Olson EN. Myomaker is essential for muscle regeneration. Genes Dev. 2014;28(15):1641–1646. doi: 10.1101/gad.247205.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Millay DP, et al. Structure-function analysis of myomaker domains required for myoblast fusion. Proc Natl Acad Sci U S A. 2016;113(8):2116–2121. doi: 10.1073/pnas.1600101113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leikina E, et al. Myomaker and myomerger work independently to control distinct steps of membrane remodeling during myoblast fusion. Dev Cell. 2018;46(6):767–780.e7. doi: 10.1016/j.devcel.2018.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goh Q, Millay DP. Requirement of myomaker-mediated stem cell fusion for skeletal muscle hypertrophy. Elife. 2017;6:e20007 [Google Scholar]
- 28.Duddy W, et al. Muscular dystrophy in the mdx mouse is a severe myopathy compounded by hypotrophy, hypertrophy and hyperplasia. Skelet Muscle. 2015;5:16. doi: 10.1186/s13395-015-0041-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jiang C, Wen Y, Kuroda K, Hannon K, Rudnicki MA, Kuang S. Notch signaling deficiency underlies age-dependent depletion of satellite cells in muscular dystrophy. Dis Model Mech. 2014;7(8):997–1004. doi: 10.1242/dmm.015917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fry CS, Kirby TJ, Kosmac K, McCarthy JJ, Peterson CA. Myogenic progenitor cells control extracellular matrix production by fibroblasts during skeletal muscle hypertrophy. Cell Stem Cell. 2017;20(1):56–69. doi: 10.1016/j.stem.2016.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alexakis C, Partridge T, Bou-Gharios G. Implication of the satellite cell in dystrophic muscle fibrosis: a self-perpetuating mechanism of collagen overproduction. Am J Physiol, Cell Physiol. 2007;293(2):C661–C669. doi: 10.1152/ajpcell.00061.2007. [DOI] [PubMed] [Google Scholar]
- 32.Goonasekera SA, et al. Mitigation of muscular dystrophy in mice by SERCA overexpression in skeletal muscle. J Clin Invest. 2011;121(3):1044–1052. doi: 10.1172/JCI43844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vanhoutte D, et al. Thrombospondin expression in myofibers stabilizes muscle membranes. Elife. 2016;5:e17589. doi: 10.7554/eLife.17589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gallot YS, Straughn AR, Bohnert KR, Xiong G, Hindi SM, Kumar A. MyD88 is required for satellite cell-mediated myofiber regeneration in dystrophin-deficient mdx mice. Hum Mol Genet. 2018;27(19):3449–3463. doi: 10.1093/hmg/ddy258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Megeney LA, Kablar B, Garrett K, Anderson JE, Rudnicki MA. MyoD is required for myogenic stem cell function in adult skeletal muscle. Genes Dev. 1996;10(10):1173–1183. doi: 10.1101/gad.10.10.1173. [DOI] [PubMed] [Google Scholar]
- 36.Griffin DA, et al. Defective membrane fusion and repair in Anoctamin5-deficient muscular dystrophy. Hum Mol Genet. 2016;25(10):1900–1911. doi: 10.1093/hmg/ddw063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Doherty KR, et al. Normal myoblast fusion requires myoferlin. Development. 2005;132(24):5565–5575. doi: 10.1242/dev.02155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Posey AD, Demonbreun A, McNally EM. Ferlin proteins in myoblast fusion and muscle growth. Curr Top Dev Biol. 2011;96:203–230. doi: 10.1016/B978-0-12-385940-2.00008-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Quinn ME, et al. Myomerger induces fusion of non-fusogenic cells and is required for skeletal muscle development. Nat Commun. 2017;8:15665. doi: 10.1038/ncomms15665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bi P, et al. Control of muscle formation by the fusogenic micropeptide myomixer. Science. 2017;356(6335):323–327. doi: 10.1126/science.aam9361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang Q, et al. The microprotein Minion controls cell fusion and muscle formation. Nat Commun. 2017;8:15664. doi: 10.1038/ncomms15664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lepper C, Conway SJ, Fan CM. Adult satellite cells and embryonic muscle progenitors have distinct genetic requirements. Nature. 2009;460(7255):627–631. doi: 10.1038/nature08209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schuler M, Ali F, Metzger E, Chambon P, Metzger D. Temporally controlled targeted somatic mutagenesis in skeletal muscles of the mouse. Genesis. 2005;41(4):165–170. doi: 10.1002/gene.20107. [DOI] [PubMed] [Google Scholar]
- 44.Murphy MM, Lawson JA, Mathew SJ, Hutcheson DA, Kardon G. Satellite cells, connective tissue fibroblasts and their interactions are crucial for muscle regeneration. Development. 2011;138(17):3625–3637. doi: 10.1242/dev.064162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Boyer JG, et al. ERK1/2 signaling induces skeletal muscle slow fiber-type switching and reduces muscular dystrophy disease severity. JCI Insight. 2019;5:127356. doi: 10.1172/jci.insight.127356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brooks SV, Faulkner JA. Contractile properties of skeletal muscles from young, adult and aged mice. J Physiol (Lond) 1988;404:71–82. doi: 10.1113/jphysiol.1988.sp017279. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.