E-cigarettes increase risk for oral diseases by shifting health-compatible ecosystems to virulence-rich communities.
Abstract
Six percent of Americans, including 3 million high schoolers, use e-cigarettes, which contain potentially toxic substances, volatile organic compounds, and metals. We present the first human study on the effects of e-cigarette exposure in the oral cavity. By interrogating both immunoinflammatory responses and microbial functional dynamics, we discovered pathogen overrepresentation, higher virulence signatures, and a brisk proinflammatory signal in clinically healthy e-cigarette users, equivalent to patients with severe periodontitis. Using RNA sequencing and confocal and electron microscopy to validate these findings, we demonstrate that the carbon-rich glycol/glycerol vehicle is an important catalyst in transforming biofilm architecture within 24 hours of exposure. Last, a machine-learning classifier trained on the metagenomic signatures of e-cigarettes identified as e-cigarette users both those individuals who used e-cigarettes to quit smoking, and those who use both e-cigarettes and cigarettes. The present study questions the safety of e-cigarettes and the harm reduction narrative promoted by advertising campaigns.
INTRODUCTION
A central characteristic of any ecosystem is that it responds to environmental perturbations with alterations in community structure, membership, and function. The oral cavity hosts an open microbial ecosystem with over 700 species of bacteria (1). It is now recognized that sustained oral health is predicated upon maintaining a stable, health-compatible microbial ecosystem and that dysbiosis within these communities triggers a florid inflammatory host response that results in disease (2). Ecosystem stability can be affected by two types of disturbances: long-term influences or “presses” and short-term effects or “pulses” (3). Anthropogenic stressors, due to their repetitive nature and addictive potential, take the form of a press perturbation. We have previously shown the effects one such anthropogenic pressor, tobacco smoking, on the oral microbiome and the impact of this destabilization on increasing the risk for periodontitis (4–6).
In 2003, a Chinese researcher patented an Electronic Nicotine Delivery System (known severally as ENDS, electronic cigarettes, e-cigarettes, or e-cigs), a battery-operated device that delivers a heated aerosol containing a mixture of nicotine, glycerol (and/or propylene glycol), and flavor with each puff. Nine years after their introduction in the United States, 20.4 million individuals (including 2.5 million high schoolers) are using this product (7–9). They are legal, touted in the media as safer than cigarettes, and claimed by consumers and the industry as smoking cessation aids. They also are popularly used in places where smoking is not permitted.
While the effects of ENDS on the respiratory system have received much attention (10), the oral cavity is not as well studied (11, 12). Oral bacterial communities have first access to these aerosols and, thus, have the greatest potential to be affected by them. The aim of this study was to investigate the effects of e-cigarettes on the subgingival microbiome using complementary approaches to achieve comprehensive insights into community assembly, dynamics, and function, as well as the impact of this community on the host’s immunoinflammatory response.
METHODS
Subject and site selection
We obtained approval for this study from the Office of Responsible Research Practices at The Ohio State University [IRB (Institutional Review Board) protocol number 2014H0062 and e-IBC protocol number 2015R00000005], and the study was conducted in accordance with approved guidelines. We recruited 123 systemically [ASA I (American Society of Anaesthesiologists Physical Status Classification I)] and periodontally healthy individuals [attachment loss ≤ 1; less than three sites with 4 mm of probe depths (PD); bleeding index (BOP) ≤ 20%] following informed consent and clinical and radiographic examination to each of five groups: (i) smokers (25), (ii) nonsmokers (25), (iii) e-cigarette users (20), (iv) former smokers currently using e-cigarettes (25), and (v) concomitant cigarette and e-cigarette users (28). Current smokers were those who had at least a five pack-year history and had no prior history of e-cigarette use. Never smokers were those who had smoked less than 100 cigarettes in their lifetime and none in the past year, and e-cigarette users were those who used e-cigarettes daily for at least 3 months, with at least one cartridge per day or 1 ml of liquid per day. Former smokers were those who had quit smoking for at least 1 year. Exclusion criteria for all groups included controlled or uncontrolled diabetes, HIV infection, use of immunosuppressant medications, bisphosphonates, or steroids, antibiotic therapy or oral prophylactic procedures within the preceding 3 months, and fewer than 20 teeth in the dentition. Sample size was estimated on the basis of the probability of least an 80% chance of detecting clades of bacterial genes that differed in abundance by more than 1%.
Sample collection
We collected and pooled subgingival plaque samples from 15 sites on six maxillary and mandibular anterior teeth using sterile endodontic paper points (Caulk-Dentsply, Milford, DE, USA). We also collected gingival crevicular fluid (GCF) by placing filter paper strips (Periopaper, Oraflow, Plainview, NY, USA) into the same sites. The fluid volume was measured with a calibrated device (Periotron 8000, Oraflow, Plainview, NY, USA).
DNA isolation, metagenomic sequencing, and analysis
We isolated bacterial DNA from paper points, using a Qiagen DNA MiniAmp kit (Qiagen, Valencia, CA, USA) after a 90-min incubation in lysozyme (2 mg/ml) (Thermo Fisher Scientific) and then quantified the DNA using a Qubit fluorometer. Library generation was completed. We used an Illumina TruSeq kit according to the manufacturer’s instructions for library generation, quantified and pooled the libraries and clustered them on the Illumina HiSeq 4000 (Illumina Inc., San Diego, CA) for 150–base pair (bp) paired-end sequencing. Sequences are deposited in the Sequence Read Archives under the project IDs PRJNA548383, PRJNA544061, and PRJNA508385. Trimmed and filtered sequences were uploaded to the MG-RAST metagenomics analysis pipeline (version 3.3.6) (13) (Argonne National Laboratory) for quality processing and basic functional analysis. The MG-RAST API and the custom Python library we have developed to access it and analyze/visualize results were used throughout the analysis process to download relevant data and pipeline results (available for download at http://github.com/smdabdoub/PyMGRAST). We used Nonpareil (14) to estimate coverage per sample. Comparisons of functional potential between groups were made in the context of the KEGG (Kyoto Encyclopedia of Genes and Genomes) (15), and the SEED subsystem (16) ontological hierarchies and statistical analysis of differential functional potential were performed using the DESeq2 package for R (17).
We used Kraken v1.1 (18) to determine the phylogenetic profile of each subject with a database constructed from a list of complete genomes from the Human Oral Microbiome Database, as of 19 September 2017. We computed alpha (within-group) and beta (between-group) diversity using PhyloToAST v1.4 and QIIME v1.9, respectively. The Shannon diversity index and Abundance Coverage Estimator (ACE) were used as estimators of species diversity and richness, and Bray-Curtis and Jaccard metrics were used to estimate beta diversity. We used principal coordinate analysis (PCoA) for dimensionality reduction and interrogated the significance of group-wise clustering using a permutational multivariate analysis of variance (MANOVA) (adonis function, vegan package for R). PCoA plots were generated with PhyloToAST (PCoA.py). We inducted CSS (cumulative sum scaling)–normalized species-level operational taxonomic unit (sOTU) counts into linear discriminant analysis (LDA) using scikit-learn v0.18.0 (19). Plots were visualized using PhyloToAST, and MANOVA/Wilks’ lambda was used to test for the significance of LDA clustering. We used SparCC (20) to test for differences in co-occurrence patterns between microbial communities from different ecosystems. Gephi (21) v0.9.1 was used to visualize the resultant networks. We used a machine learning algorithm (randomForest package in R) to test the ability of genes to discriminate between groups. Two-thirds of the dataset was used to train the algorithm, which was tested on the remaining data. This was iterated 10 times, and the mean “importance” was computed for each marker gene. The robustness of the classifier was evaluated using ROC (receiver operating characteristic) curves (ROCR package in R).
Quality control
All samples were sequenced in two runs; and to minimize batch effects, samples were randomly assigned to each run. Replicate sequencing was carried out for two samples in each batch, and the replicates showed good reliability across the five batches, with coefficient of variability (SD/mean) ranging from 0.26 to 1.3% for alpha diversity of taxonomy and 3.4 to 6.3% for predominant functions (carbohydrate metabolism, respiration, and virulence, disease, and defense).
Cytokine assay
We eluted GCF from the paper strips by adding 200 μl of phosphate-buffered saline and vortexing for 15 min. Levels of interferon-γ, interleukin-2 (IL-2), IL-4, IL-6, IL-8, IL-10, granulocyte-macrophage colony-stimulating factor (GM-CSF), and tumor necrosis factor–α (TNF-α) in GCF were determined using the Bio-Plex Pro Human Cytokine 8-plex assay (Bio-Rad Corporation, CA), according to the manufacturer’s instructions.
In vitro validation studies
Artificial saliva was made following the Marshall Group Research protocol for artificial saliva. SHI medium was prepared according to the protocol described by Tian et al. (22). The developed biofilms were developed using the modifications (23) from the protocol established by Guggenheim et al. (24). Briefly, sterilized, sintered hydroxyapatite (HA) disks (Clarkson Chromatography Products, South Williamsport, PA) were incubated in artificial saliva for 24 hours to establish a pellicle coat, following which multispecies commensal primary biofilms were generated by seeding six pioneer species [Streptococcus oralis (ATCC 35037), S. sanguis (10556), S. mitis (49456), Actinomyces naeslundii (12104), Neisseria mucosa (25997), and Veillonella parvula (17745)] and incubating under aerobic conditions in a 1:1(v/v) mixture of SHI media and artificial saliva. Pathogen-rich biofilms were created by further seeding the commensal biofilms with an intermediate bridging colonizer [Fusobacterium nucleatum (10953), secondary biofilm] followed 24 hours later by Porphyromonas gingivalis (33277), Filifactor alocis (35896), Selenomonas sputigena (35185), S. noxia (43541), Campylobacter gracilis (33236), Prevotella intermedia (25611), Parvimonas micra (33270), and Tannerella forsythia (43037) and incubating under anaerobic conditions for a further 24 hours (tertiary biofilms). Research grade cigarettes (IR4F) with a filter were obtained from the Kentucky Tobacco Research and Development Center (Lexington, KY). We prepared cigarette smoke extract (CSE) by bubbling smoke from two cigarettes into 5 ml of artificial saliva. To replicate our e-cigarette cohort’s usage, we used a moderate-sized e-cigarette vape pen, vaped by pressing “on” for 5 s and “off” for 30 s in between and repeated for 15 to 20 min. We prepared electronic cigarette vapor (ECV) by vaping two cartridges of nonflavored e-cigarette containing either 6 or 0 ml of nicotine through clean Pasteur pipettes into 5 ml of artificial saliva, until the whole cartridge was exhausted. CSE and ECV were prepared immediately before use. To maintain the consistency of CSE and ECV between experiments, an optical density of 0.65 at 600 nm represented 100% (25). CSE and ECV were diluted to 1%, and the biofilms were conditioned with these solutions for 24 hours. We isolated total RNA from the biofilms using the mirVana miRNA isolation kit (Applied Biosystems). Ribosomal RNA was depleted, and mRNA was enriched by modified capture hybridization approach (MICROBExpress mRNA enrichment kit, Thermo Fisher Scientific). Enriched mRNA served as a template for the polyadenylation reaction and complementary DNA synthesis. Microbial libraries were clustered on the Illumina HiSeq 4000 platform, and 150-bp paired-end sequencing was performed. The Illumina base-calling pipeline was used to process the raw fluorescence images and call sequences. Raw reads with >10% unknown nucleotides or with >50% low-quality nucleotides (quality value, <20) were discarded. Microbial transcripts were quality-filtered using Sickle v1.33 (default parameters) and aligned against the RefSeq nonredundant proteins database using DIAMOND v0.8.3.65 (26). Aligned sequences were annotated to the KEGG database using MEGAN 6 (27). We washed the HA disks with biofilms with glucose buffer, fixed, and dried them with ethanol and HMDS (hexamethyldisilazane). The dried specimens were mounted on stubs and then coated with a 10-nm layer of gold in a Leica ACE600 Sputter Coater. Images were collected at the Center for Electron Microscopy and Analysis (The Ohio State University) with staff assistance on a Thermo Fisher Scientific Apreo LoVac field emission scanning electron microscope (SEM) using the in-column detectors at specific magnifications and resolutions. Plain HA disks and HA disks conditioned with artificial saliva and SHI media were used as negative controls. To enable confocal microscopic imaging, we stained the biofilms using the BacLight kit (Life Technologies, NY) according to the manufacturer’s instructions. Briefly, the biofilms were incubated in 1.5 ml of 0.3% SYTO 9 and propidium iodide, and the fluorescence was measured at 486 and 520 nm using a Spectral FlowView confocal microscope at 10× magnification. The ratio of green to red fluorescence was computed, and Z-stack images were obtained. A minimum of eight images per group was obtained to generate volume and area graphs. Total surface area and volume were determined using Imaris v9 (http://bitplane.com) from the constructed three-dimensional images. Boxplot comparisons of areas and volumes were visualized using Seaborn v0.9.0, and the significance of pairwise differences was determined using Tukey’s post hoc test (JMP statistical software v13.0).
RESULTS
The e-cigarette–influenced periodontal ecosystem
To investigate the effects of e-cigarette use on the oral microbiome, we first created a catalog of bacterial genes found in the subgingival microbiome of e-cigarette users. To do this, we collected subgingival plaque samples from anterior teeth of 20 systemically (ASA I) and periodontally healthy e-cigarette users using paper points and curettes. Individuals were between 21 and 35 years of age, predominantly (90%) Caucasian, reported using e-cigarettes containing 6 to 18 mg of nicotine for 4 to 12 months, and did not report any other tobacco product use (table S1). Whole-genome shotgun sequencing generated 37 million paired-end sequences (see detailed methods). These sequences represented 9730 functionally annotated microbial genes (based on SEED subsystem classification). COGs (clusters of orthologous groups) that did not exceed a relative abundance of 0.01% were excluded from the analysis as an unsupervised feature reduction technique, yielding a total of 7035 COGs, of which 4907 were present in ≥80% of the sample population. To gain insights into the sources of variability in the e-cigarette–influenced microbiome, we used nonmetric multidimensional scaling (NMDS) on variance-stabilized abundances of genes and taxa. NMDS revealed the duration of e-cigarette use (<6 months versus >10 months) as the strongest source of variation. Nicotine concentration and type of flavoring agent did not emerge as discriminators of the biome (fig. S1).
The preponderant functionality in the e-cigarette–influenced microbiome was carbohydrate metabolism, representing 12 to 36% of the gene abundance in each individual. Within this pathway, genes encoding for kinases (glycerol, glycerate, and fructose), lysine and butyrate fermentation, and ATP-binding cassette (ABC) transporters demonstrated the greatest abundance (Fig. 1A). The other predominant pathways were protein and amino acid metabolism, together accounting for 3 to 12% of each individual metagenome. Genes involved in biofilm formation (cell wall and capsule synthesis, peptidoglycan, membrane transport, flagellar synthesis, etc.) demonstrated a relative abundance of 4 to 7% in all 20 individuals.
Fig. 1. Phylogenetic, functional, and immune characteristics of the subgingival microbiome of e-cigarette users.
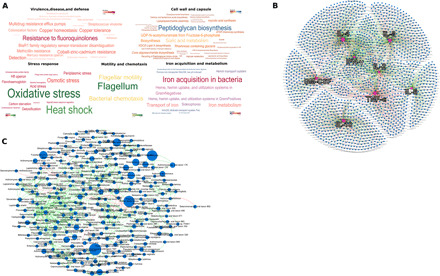
(A) Word map of the predominant microbial functions identified in e-cigarette users. The functions are colored by relative abundances (as indicated by the color map scale within each word map). (B) Network plot between eight immune mediators and the microbial metagenome, with the cytokines at the center of each hub. Interspecies networks are shown in (C). Each network graph contains nodes (circles sized by relative abundance per group) and edges (lines). Nodes represent cytokines and microbial genes in (B) and species-level OTUs in (C) and edges represent Spearman’s rho. In both network graphs, green edges indicate positive correlations and red edges indicate negative correlations. Only edges with correlation coefficient (Spearman’s rho) ≥ 0.80 and P < 0.05 are shown. The data supporting this figure can be found in table S1.
While 340 ± 37 (means ± SD) species were identified in each individual, only 59 species accounted for 75% of the microbial abundance, indicating that many of the species were rare taxa. However, we also noted that 73% of all species were shared by 80% of e-cigarette users, and 40% were found in the entire cohort (table S2). Network analysis revealed a highly interconnected topology, with multiple small tightly clustered hubs. The largest hub was anchored by 203 species belonging to the genera Actinomyces, Capnocytophaga, Filifactor, Fusobacterium, Treponema, Tannerella, Prevotella, Selenomonas, and Streptococcus (Fig. 1B). These species were part of the core microbial community and ranked high in betweenness centrality and node degree, indicating their potential in supervising or influencing the flow of resources and information in this ecosystem. Thus, both phylogenetically and functionally, the oral microbiome of e-cigarette users demonstrated remarkable homogeneity, suggesting that e-cigarettes might exert a selection pressure on the subgingival ecosystem. E-cigarette users also demonstrated significant correlations between proinflammatory cytokines and genes encoding for stress response, environmental response regulation, and transport of heavy metals (Fig. 1C).
The e-cigarette–influenced microbiome—Not a “Lucky Strike”
We investigated the magnitude of difference between the microbiomes of e-cigarette users, cigarette smokers, and never smokers by comparing exclusive e-cigarette users to 25 systemically (ASA 1) and periodontally healthy (attachment loss ≤ 1; less than three sites with 4 mm of PD; BOP ≤ 20%) never smokers (those who had smoked less than 100 cigarettes in their lifetime and none in the past year) and 25 current smokers (those who had smoked at least 10 cigarettes a day for the last 5 years and had no prior history of e-cigarette use). The groups were frequency matched for age, gender, ethnicity, and clinical periodontal parameters (table S1). One billion sequences were obtained from 70 individuals: 20 e-cigarette users, 25 cigarette smokers, and 25 never smokers (controls). Average coverage per sample ranged from 50 to 85% based on Nonpareil (14) and was not significantly different between groups (Tukey post hoc test, P > 0.05). Taxonomic identities were assigned with Kraken (18) and used to evaluate community composition and estimates of diversity.
Irrespective of exposure type, 30% of the metagenome was shared among all individuals, corroborating evidence from other human microbiome studies (28, 29). However, multivariate analysis demonstrated statistically significant group separation between smokers, e-cigarette users, and controls based on both functional and taxonomic profiles [P < 0.05, analysis of similarities (ANOSIM); Fig. 2, A and D]. Even within the shared metagenome, e-cigarette users demonstrated enrichment of 284 genes when compared to both smokers and controls [P < 0.05, false discovery rate (FDR)–adjusted Wald test; table S3]. These genes related to the following processes: alanine and arginine biosynthesis, polyamine metabolism, central carbohydrate metabolism, one-carbon metabolism, mono-, di-, and oligosaccharide metabolism, fermentation, and cell division and cell cycle, indicating that although central metabolic processes are present in all three groups, specific pathways contributing to these processes were different in each.
Fig. 2. Differences in microbial community structure and function between electronic cigarette users, smokers, and nonsmokers.
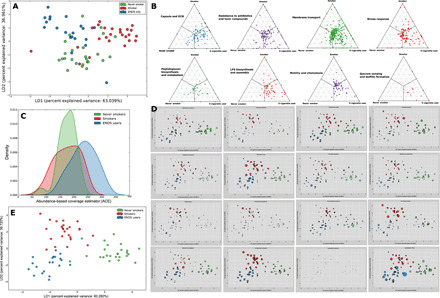
LDA of relative abundances of functional genes in periodontally and systemically healthy nonsmokers (green), smokers (red), and ENDS users (blue) is shown in (A). The microbial profiles of subjects clustered by exposure type, creating three statistically significant clusters (P = 0.008, MANOVA/Wilks). Barycentric plots of significantly different virulence functions in the three groups (P < 0.05, FDR-adjusted Wald test) are shown in (B). Each dot represents a gene. The three groups (smoker, control, or e-cigarette user) are used as vertices. Within each plot, the coordinates of each gene are determined by the weighted average of the coordinates of all genes, and the weights are given by the relative abundance of the gene in that group (smoker, control, or e-cigarette user). Density curves of alpha diversity (ACE) are shown in (C). The peak indicates the median values for each group, and the x axis shows the data range. E-cigarette users demonstrated significantly greater alpha diversity than the other two groups (P < 0.0001, Kruskal-Wallis). LDA of relative abundances of sOTUs is shown in (D), while the relative abundances of selected species in each subject are shown in (E).
When compared to smokers and controls, the metagenome of e-cigarette users demonstrated greater abundances of genes encoding for ABC transporters and RNA processing and modification systems, as well as virulence factors such as cell wall and capsular polysaccharides, peptidoglycan and lipopolysaccharide biosynthesis, stress response, quorum sensing and biofilm formation, resistance to antibiotics and toxic compounds, flagellar motility, and siderophores [P < 0.05, FDR-adjusted Wald test (DESeq2); Fig. 2B]. Taxonomically, alpha diversity (as measured by Chao1 and ACE indices) was significantly greater in e-cigarette users (P < 0.0001, Kruskal-Wallis; Fig. 2C). E-cigarette use was also associated with higher levels of Gram-negative facultatives while smoking selectively enriched for Gram-negative anaerobes. Candidatus Saccharibacteria oral taxon TM7x and species belonging to the genera Abiotrophia, Aggregatibacter, Eikenella, Granulicatella, Cardiobacterium, Hemophilus, Johnsenella, Kingella, Lachoanaerobaculum, Lautropia, Leptotrichia, Mogibacterium, Ottowia, Parvimonas, Peptostreptococcus, Rothia, Rhodobacter, Selenomonas, and Veillonella demonstrated significantly greater abundances [P < 0.05, FDR-adjusted Wald test (DESeq2); Fig. 2E] in ENDS users when compared to both never smokers and smokers.
Seventy percent of the metagenome in e-cigarette users was shared by more than 80% of subjects (table S3). By contrast, 80% of smokers and nonsmokers shared only 40 and 50% of the metagenomes, respectively. Moreover, the core microbiome of e-cigarette users contained 96% (130 of 136) of species that demonstrated significantly higher abundances when compared to the other two groups. The presence of a robust common core microbiome that differed significantly from those of smokers and nonsmokers suggests that e-cigarette aerosol affects the oral microbiome differently than cigarette smoke.
We then interrogated the ability of the metagenome to discriminate between e-cigarette aerosol and other exposures using a Random Forest Classifier (randomForest package in R). The robustness of the classifier was evaluated using ROC curves (ROCR package in R). Two-thirds of the dataset was used to train the classifier, which was tested on the remaining data (1000 trees/10-fold cross-validation). The mean importance was computed for each parameter and used to select robust parameters based on the methodology of Díaz-Uriarte (30). For all iterations of the test, a “confusion table” was created for each of the exposures based on the number of correctly classified and misclassified samples, and these data were used to compute sensitivity and specificity. The classifier was able to predict ENDS users with 90% sensitivity and 97% specificity, followed by controls (88% sensitivity and 92% specificity) and cigarette-smokers (84% sensitivity and 95% specificity). Forty-nine genes encoding for Gram-positive and Gram-negative cell wall, peptidoglycan biosynthesis, and enzymes within the protein and amino acid metabolism pathway were identified as discriminators of the e-cigarette aerosol-influenced microbiome (table S4).
Higher signals of harm found in e-cigarette users than in smokers and controls
Since the metagenome of e-cigarette users demonstrated greater virulence signatures than controls, we investigated their impact on the inflammatory burden in these cohorts. To do this, we quantified the levels of eight cytokines representing the T helper cell 1 (TH1) and TH2 types of inflammatory responses and correlated them to the metagenome using graph theory. To reduce bias induced by sparse data, we used core genes in each group to compute correlations. E-cigarette users demonstrated significantly higher levels of the proinflammatory cytokines IL-2, IL-6, GM-CSF, TNF-α, and INF-γ and lower levels of the antiinfammatory cytokine IL-10 when compared to the never smokers, while smokers demonstrated higher levels of IL-2, IL-6, IL-8, TNF-α, and INF-γ and lower levels of IL-10 when compared to the never smokers (P < 0.05, Dunn’s test), suggesting that both e-cigarettes and conventional cigarettes are associated with greater inflammation; however, they are mediated through different pathways (Fig. 3A).
Fig. 3. Inflammatory burden imposed by the e-cigarette–influenced microbiome.
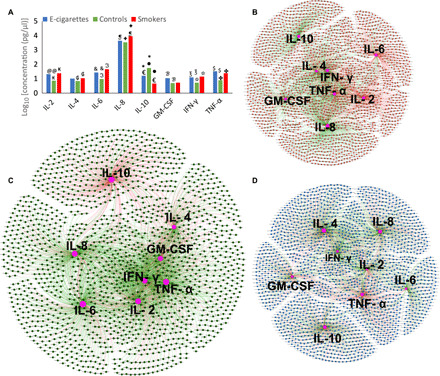
Levels of selected immune mediators in periodontally and systemically healthy nonsmokers (green), smokers (red), and e-cigarette users (blue) is shown in (A). The y axis represents log10-transformed concentrations. Bars with the same symbol are significantly different (P < 0.05, Dunn’s test). Co-occurrence networks between cytokines and microbial genes in each group are shown in (B to D). Smokers are shown in (B), nonsmokers in (C), and e-cigarette users in (D). Each network graph contains nodes (circles) and edges (lines). Nodes represent cytokines and KEGG-annotated genes, and edges represent Spearman’s rho. Edges are colored green for positive correlation and red for negative correlation. Only significant correlations (P < 0.05, t test) with a coefficient of at least 0.80 are shown.
We used graph theory to investigate whether the cytokine response was attributable to bacterial shifts. Strong positive correlations were observed between cytokines and genes encoding bacterial stress response and biofilm formation (table S5). Although IL-4 levels were not different between groups, network analysis revealed a robust IL-4–anchored hub (with higher betweenness and degree centrality) in e-cigarette users but not in smokers or controls. The majority of these correlations were negative. In contrast, while IL-6 emerged as a pivotal node in control samples, it did not demonstrate as high of a relevance in e-cigarette users or smokers. The higher concentrations of selected proinflammatory cytokines taken together with the dense metagenome-cytokine network topology point to a higher inflammatory burden imposed by the e-cigarette–influenced microbiome when compared to controls (Fig. 3, B to D).
Effects of e-cigarettes are different from the effects of smoking on the microbiome
From their inception, e-cigarettes have been positioned as a tobacco harm reduction strategy. Hence, we sought to quantify the magnitude of microbial shift when smokers partially or totally replaced conventional cigarettes with e-cigarettes. To do this, we investigated the microbiomes of 25 periodontally healthy, ASA 1 individuals who use conventional cigarettes and e-cigarettes concurrently (dual users) and 25 individuals who switched from conventional cigarettes to e-cigarettes (hitherto referred to as former smokers) (recruitment criteria explained in Methods). Both groups reported using e-cigarettes for 6 to 12 months, and former smokers reported quitting between 3 months and 1 year before study. We used PCoA to examine group separation between e-cigarette users, dual users, and former smokers and ANOSIM to assess statistical significance of group separation. There was no significant separation between the groups functionally or phylogenetically (P values, 0.27 and 0.35, respectively, ANOSIM). We then performed pairwise comparisons of the functional and phylogenetic profiles of the five groups (controls, e-cigarette users, dual users, former smokers, and current smokers) using the Bray-Curtis similarity index. Both phylogenetically and functionally, the microbiomes of dual users and former smokers exhibited significantly greater similarity to e-cigarette users than to never smokers and smokers (P < 0.00001, Tukey post hoc test of Bray-Curtis similarity index; Fig. 4, A and B).
Fig. 4. The synergistic effects of smoking and e-cigarettes on the microbiome.
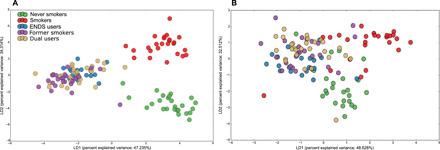
LDA of Bray-Curtis dissimilarity indices based on relative abundances of species level OTUs (A) and functional genes (B) in periodontally and systemically healthy individuals who exclusively use ENDS (blue) or cigarettes (red), dual users (who use cigarettes and ENDS concomitantly, in tan color), former smokers who currently use ENDS (purple) and nonsmokers (green) are shown. Three statistically significant clusters were observed, with dual smokers and former smokers clustering along with exclusive ENDS users (P = 0.003, MANOVA/Wilks).
To further assess the robustness of this effect, we trained a Random Forest machine learning classifier on the microbiomes of e-cigarette users and smokers (discovery set) and used the dual users and former smokers as the validation set. The algorithm identified dual users as e-cigarette users with 68% sensitivity and 72% specificity and former smokers with 76% sensitivity and 83% specificity. Thus, there is reason to infer that any potential harm reduction from e-cigarettes is not mediated through shifts in the microbiome.
E-cigarettes sculpt the subgingival microbial landscape
Since the network analysis revealed a dense multispecies hub in e-cigarette users, we interrogated the metagenome for potential biological underpinnings for this clustering. We computed the “minimal e-cigarette genome” as a set of functions expected to be present in most or all species constituting the hub, identifying three types of functions (table S6). The first was a cluster of housekeeping functions that included main metabolic pathways (e.g., carbohydrate metabolism and amino acid synthesis) and protein complexes such as RNA- and DNA-related proteins and general secretory apparatus. The second group was a common core of functions related to cell wall, cytoskeleton, and exopolysaccharide synthesis. In addition, we identified a group of genes with poorly characterized function and unknown contributions to metabolic pathways.
Simultaneously, we investigated aspects of the metagenome that best characterize the microbiome of e-cigarette users using a modification of Dufrene and Legrange’s indicator species analysis (31). This index is a composite of gene abundance (defined as specificity) and frequency (defined as fidelity). A Monte Carlo test of significance (P < 0.05) yielded 230 indicator genes, all of which belonged to a single functional family: cell wall and capsule (table S6).
Since genes encoding cell wall and capsule were (i) part of the minimal e-cigarette genome, (ii) indicators of e-cigarette exposure, (iii) significantly overrepresented in e-cigarette users when compared to controls, (iv) members of the common core metagenome of e-cigarette users, and (v) demonstrated significant positive correlations with proinflammatory mediators, we sought to investigate whether this enriched gene set had downstream effects on biofilm architecture. To do this, we used a validated in vitro biofilm model constructed with defined species, using metatranscriptomics to examine gene expression and confocal and SEM to quantify biofilm architecture.
To recapitulate the dynamics of oral biofilm colonization in humans, we sequentially seeded pioneer species, bridging organisms and tertiary colonizers onto saliva-conditioned HA disks in an e-cigarette aerosol–rich environment (see detailed methods). The aerosol consisted of propylene glycol and glycerol (PG/VG:50/50) and either 0 or 6 mg/ml of nicotine. Clean air–exposed biofilms were used as controls.
mRNA from the biofilms was sequenced, and the cell wall and capsule subsystem of the SEED database were used for gene calling. COGs (78%) were identified both in the e-cigarette–associated human metagenome and the in vitro metatranscriptome, suggesting that (i) the e-cigarette–associated human oral microbiome is transcriptionally active and (ii) the in vitro biofilm functionally recapitulates the in vivo microbiome. E-cigarette aerosol exposure of pioneer species induced synthesis of capsular polysaccharide, legionaminic acid, neuraminate, and lipoteichoic acid (table S6). In addition, we found exposure resulted in up-regulated lipid A, lipopolysaccharide, peptidoglycan and glycosaminoglycan biosynthesis, sialic acid metabolism, and rhamnose containing glycans processes by 4 to 134-fold. A concomitant down-regulation of colanic acid synthesis was observed. In an aerosol-rich environment, introduction of F. nucleatum (secondary biofilm) followed by tertiary colonizers (tertiary biofilm) led to further amplification of cell wall and capsule functionality, with induction of new genes and overexpression of previously up-regulated genes. However, certain other genes, notably those that participate in synthesis of glycosaminoglycans and polymixin resistance, were down-regulated in secondary and tertiary biofilms.
Knowing that cell wall and capsular polysaccharides play an important role in biofilm formation, we next explored the effects of e-cigarette aerosol on biofilm formation by surveying the metatranscriptome for associated genes in the KEGG classification (table S6). The greatest response to e-cigarette exposure was observed in 24-hour biofilms colonized by pioneer species; following introduction of F. nucleatum and tertiary colonizers, these functional amplifications were sustained or further up-regulated. Not unexpectedly, e-cigarettes induced or up-regulated two-component response systems, particularly, histidine kinase sensors. Quorum sensing (through the LuxR system) was also found to be induced, as well as expression of genes encoding for pellicle proteins.
We next studied the relative contributions of nicotine and aerosol to this transcriptional profile by comparing nicotine-free aerosol, aerosol with 6 mg of nicotine and 6 mg of nicotine without aerosol (liquid nicotine). Differential abundance analysis (DESeq2) of the global metatranscriptome revealed that, in primary biofilms, only 310 of 7120 genes were different between nicotine-free and nicotine-containing aerosol, while 2190 genes were different between nicotine-containing aerosol and aerosol-free nicotine. Moreover, there were no significant differences between nicotine-free and nicotine-containing e-cigarette aerosol in cell wall or biofilm pathways, while aerosol-free nicotine significantly down-regulated 158 genes coding for cell wall and capsular synthesis when compared to nicotine-containing aerosol.
We then used SEM to investigate whether this transcriptional activity was reflected in biofilm topography. While clean air-exposed primary biofilms demonstrated multiple microcolonies with well-defined cell outlines, e-cigarette–conditioned biofilms were enveloped in a dense matrix within 24 hours, making the cellular morphotypes indistinct (Fig. 5A). Therefore, we used confocal laser scanning microscopy to visualize the biofilms and computed surface area and volume with IMARIS. Exposure to aerosol without nicotine led to statistically significant increases in total surface area (37899.03 versus 24470 μm2; P = 0.0069, Dunn’s test with joint ranks) and volume (13346.25 versus 8357.28 μm3; P = 0.0121, Dunn’s test with joint ranks) in primary biofilms when compared to the clean air–exposed controls (Fig. 5B, i and ii). Nicotine containing aerosol led to a significant increase in area and volume when F. nucleatum was added (Fig. 5B, iii and iv), while nicotine-free aerosol created biofilms with greater surface area and volume when the biofilms were seeded with tertiary colonizers (Fig. 5B, v and vi). E-cigarette exposure did not significantly change the ratios of live:dead bacteria in primary, secondary, or tertiary biofilms (P > 0.05, Dunn’s test with joint ranks). Together, the metatranscriptomic and microscopic data suggest that (i) the sugar alcohol (glycerol and glycol) in the e-cigarette aerosol is a nutrient source and hence is a key catalyst in altering the topography of oral biofilms; (ii) this occurs within 24 hours of biofilm colonization; and (iii) although genes contributing to biofilm formation are differentially expressed during each stage of bacterial colonization, there is a continual increase in biofilm mass upon exposure to e-cigarettes.
Fig. 5. Effect of e-cigarettes on sculpting the biofilm landscape.
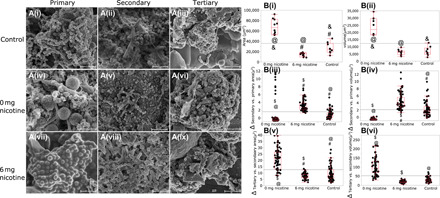
(A) SEM images of primary biofilms consisting of S. oralis, S. sanguis, S. mitis, A. naeslundii, N. mucosa, and V. parvula, secondary biofilms [following addition of an intermediate colonizer (F. nucleatum) to the primary biofilm] and tertiary biofilms (secondary biofilms seeded with P. gingivalis, F. alocis, Selenomas sputigena, S. noxia, C. gracilis, P. intermedia, P. micra, and T. forsythia) exposed to e-cigarette vapor with nicotine (6 or 0 mg/ml) and clean air controls. (B, i and ii) The effects of e-cigarette aerosol on area and volume of primary biofilms. (B, iii and iv) Changes in area and volume following addition of F. nucleatum. (B, v and vi) Changes in area and volume following addition of tertiary colonizers. Biofilms were visualized using confocal laser scanning microscopy in (B), and surface area and volume were computed with IMARIS. In all figures, groups connected by the same symbol are significantly different (P < 0.001, Dunn’s test with joint ranks).
DISCUSSION
Although e-cigarettes have been available in the U.S. market since 2009, we know little regarding their effects on the oral ecosystem. Therefore, we combined comparative metagenomics with a rigorous case-control human study and validated our findings using metatranscriptomics and confocal microscopy on an in vitro longitudinal model of biofilm colonization. To the best of our knowledge, this investigation provides some of the earliest experimental evidence documenting the effects of e-cigarette use on the oral microbial ecosystem; and it begins to explore the potential mechanisms that underlie this shift.
E-cigarettes stress the subgingival environment, as evidenced by significantly greater abundances of genes regulating stress response in the human metagenome, a brisk proinflammatory response, and robust, positive correlations between these cytokines and putative virulence factors. The in vitro model demonstrated that the greatest up-regulation of stress response occurs in the primary biofilm, with a down-regulation of stress in the secondary and tertiary biofilms. This is different from what we have previously observed with cigarette smoke, where commensal transcriptional activity was significantly lower following exposure to cigarette smoke (23). Thus, while the climax communities in both smokers and e-cigarette users are pathogen rich, the evidence begins to point to potentially different mechanisms of actions of these two tobacco products on the oral microbiome.
The most notable effect of e-cigarettes on the oral microbiome is in altering biofilm architecture. Network analysis of the climax community in e-cigarette users revealed a hub anchored by prolific biofilm formers, and genes participating in cell wall and capsule emerged as the single most influential characteristic of e-cigarette users. Microbial sialic acid metabolism has now been firmly established as a virulence determinant in a range of infectious diseases (32). We found that exposure of S. oralis, S. mitis, V. parvula, N. mucosa, A. naeslundii, and S. sanguis to e-cigarette aerosol led to an upsurge in transcription of several sialic acids, notably legionaminic acid and neuraminic acid, as well as the LuxR system. Our findings are supported by evidence in the literature that bacteria produce exopolysaccharides in response to stress and that this response is mediated through quorum sensing (33). Several factors are known to increase biofilm mass, e.g., temperature, alkaline pH, reactive oxygen species, and cytokines (34). It is possible that e-cigarettes mediate their effects through one or more of these mechanisms, warranting further exploration since alterations in the spatial architecture of biofilms have important implications for both dental caries and periodontitis.
Several aspects of our data suggest, albeit indirectly, that glycerol and propylene glycol might be drivers of the microbial shifts in e-cigarette users. The first indication came from the human data where the microbiomes did not segregate on the basis of flavoring agents or nicotine concentrations. This was initially unexpected, since studies on smokers have previously demonstrated dose-response to nicotine exposure (35). However, the validation experiments using a different sequencing approach (RNA instead of DNA) also demonstrated a high similarity between primary biofilms exposed to nicotine-containing or nicotine-free aerosol but not to aerosol-free nicotine. Together, the two experiments point to the existence of a factor in e-cigarettes that outweighs the effects of nicotine. The third indication came from amplification of biofilm pathways and up-regulation of histidine kinase receptors in response to e-cigarette exposure in both the human metagenome and the in vitro metatranscriptome. Biofilm formation demands large investments in energy and, hence, is carbon expensive (36). Bacteria are known to conscript substrate carbon, notably glycerol, to facilitate the synthesis of extracellular matrix (37). Glycerol is an essential precursor for the synthesis of lipids and for the building of (lipo)teichoic acids in many Gram-positive bacteria. The role of glycerol in intracellular growth of pathogenic bacteria is also well established. Moreover, bacteria sense the presence of glycerol through the histidine kinase signaling pathway (38). Thus, when our data are examined against the backdrop of what we know about biofilm formation, it strongly implicates the vehicle in e-cigarettes as a driver of community assembly.
One notable observation was that when smokers use e-cigarettes either to reduce or to quit smoking, the microbiome resembles that of e-cigarettes. Moreover, the smokers in our study (single use, dual use, and quitters) reported smoking for at least 5 years, while the e-cigarette users reported an average of 7 months of vaping history. While we acknowledge that the cross-sectional nature of this study does not permit us to make time-to-event type inferences, there is some indication that the effects of e-cigarettes on the oral microbiome might be evident much earlier than with smoking. E-cigarettes are being promoted for their harm reduction potential, and some evidence suggests that they might be effective smoking cessation aids (39). However, as our data demonstrate, the risk-for-harm from e-cigarettes is different from, but not less than, conventional cigarettes.
As holobionts, it behooves humans to protect and sustain our coexisting microbial ecosystems. Our data demonstrate that e-cigarettes exert a powerful, detrimental effect on the subgingival ecosystem, altering the immunotolerance of the host. Note that none of the 123 individuals in our study had periodontal disease, yet the functional signatures bore remarkable resemblance to those of individuals with periodontitis (28, 40), attesting to the risk for harm posed by e-cigarettes to the oral ecosystem. Maintaining equilibrium between the host immune system and resident biofilms is a determinant of health. Current smokers are known to have a fourfold increase in odds of having periodontitis, and we have previously demonstrated that dysbiosis involving pathogen-rich, commensal poor communities persists well before the onset of clinical disease (5). Periodontitis is a chronic disease, sometimes taking over a decade to manifest as clinically measurable change; hence, intermediate biomarker analysis is our only “window into the future.” When we examine the present data in the context of what we already know about bacterial virulence mechanisms and host inflammatory response, our intermediate biomarker analysis strongly suggests that e-cigarettes have the potential to shift the host-microbiome equilibrium, posing a significant risk for future disease, and that the pathogenetic mechanisms might not be similar to what we have learned from studying disease in smokers. Further, longitudinal studies, preferably in humans or animal models using an oral mode of exposure to e-cigarette vapor, are urgently needed to understand the manifold effects of these drug delivery devices on human health.
Supplementary Material
Acknowledgments
Funding: This work was supported by NIH R01DE027857 to P.S.K. and by U.S. Food and Drug Administration GRT00009123 to P.S.K. Author contributions: S.M.G.: Subject recruitment, sample collection, data analysis (instead of sequence analysis) analysis, MS preparation, and imaging. S.M.D.: Data analysis and MS preparation. H.N.N.: Data analysis and MS preparation. M.L.S.: Metatranscriptomic analysis, imaging, and MS preparation. S.P.: Clinical examination, sample collection, and processing. M.L.B., P.G.S., and M.E.W.: MS preparation. P.S.K.: Study design, clinical examination, sample collection, data analysis, and MS preparation. Competing interests: The authors declare that they have no competing interests. Data and Materials Availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors. Sequences generated from this study are deposited in the Sequence Read Archives of the NCBI.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/22/eaaz0108/DC1
REFERENCES AND NOTES
- 1.Socransky S. S., Haffajee A. D., Periodontal microbial ecology. Periodontol. 2000 38, 135–187 (2005). [DOI] [PubMed] [Google Scholar]
- 2.Darveau R. P., Tanner A., Page R. C., The microbial challenge in periodontitis. Periodontol. 2000 14, 12–32 (1997). [DOI] [PubMed] [Google Scholar]
- 3.K. C. Weathers, D. L. Strayer, G. E. Likens, Fundamentals of Ecosystem Science (Elsevier/AP, 2013), pp. 312. [Google Scholar]
- 4.Joshi V., Matthews C., Aspiras M., de Jager M., Ward M., Kumar P., Smoking decreases structural and functional resilience in the subgingival ecosystem. J. Clin. Periodontol. 41, 1037–1047 (2014). [DOI] [PubMed] [Google Scholar]
- 5.Kumar P., Matthews C. R., Joshi V., de Jager M., Aspiras M., Tobacco smoking affects bacterial acquisition and colonization in oral biofilms. Infect. Immun. 79, 4730–4738 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mason M. R., Preshaw P. M., Nagaraja H. N., Dabdoub S. M., Rahman A., Kumar P. S., The subgingival microbiome of clinically healthy current and never smokers. ISME J. 9, 268–272 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Centers for Disease Control and Prevention , Notes from the field: Electronic cigarette use among middle and high school students—United States, 2011–2012. MMWR Morb. Mortal. Wkly. Rep. 62, 729–730 (2013). [PMC free article] [PubMed] [Google Scholar]
- 8.Centers for Disease Control and Prevention, E-cigarette use triples among middle and high school students in just one year. CDC Online Newsroom, (2015).
- 9.Dutra L. M., Glantz S. A., Electronic cigarettes and conventional cigarette use among U.S. adolescents: A cross-sectional study. JAMA Pediatr. 168, 610–617 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lappas A. S., Tzortzi A. S., Konstantinidi E. M., Teloniatis S. I., Tzavara C. K., Gennimata S. A., Koulouris N. G., Behrakis P. K., Short-term respiratory effects of e-cigarettes in healthy individuals and smokers with asthma. Respirology 23, 291–297 (2018). [DOI] [PubMed] [Google Scholar]
- 11.Cuadra G. A., Smith M. T., Nelson J. M., Loh E. K., Palazzolo D. L., A comparison of flavorless electronic cigarette-generated aerosol and conventional cigarette smoke on the survival and growth of common oral commensal Streptococci. Int. J. Environ. Res. Public Health 16, E1669 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stewart C. J., Auchtung T. A., Ajami N. J., Velasquez K., Smith D. P., De La Garza R. II, Salas R., Petrosino J. F., Effects of tobacco smoke and electronic cigarette vapor exposure on the oral and gut microbiota in humans: A pilot study. PeerJ 6, e4693 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wilke A., Bischof J., Gerlach W., Glass E., Harrison T., Keegan K. P., Paczian T., Trimble W. L., Bagchi S., Grama A., Chaterji S., Meyer F., The MG-RAST metagenomics database and portal in 2015. Nucleic Acids Res. 44, D590–D594 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rodriguez-R R. L., Konstantinidis K. T., Nonpareil: A redundancy-based approach to assess the level of coverage in metagenomic datasets. Bioinformatics 30, 629–635 (2014). [DOI] [PubMed] [Google Scholar]
- 15.Kanehisa M., Goto S., KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Overbeek R., Olson R., Pusch G. D., Olsen G. J., Davis J. J., Disz T., Edwards R. A., Gerdes S., Parrello B., Shukla M., Vonstein V., Wattam A. R., Xia F., Stevens R., The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST). Nucleic Acids Res. 42, D206–D214 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wood D. E., Salzberg S. L., Kraken: Ultrafast metagenomic sequence classification using exact alignments. Genome Biol. 15, R46 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pedregosa F., Varoquaux G., Gramfort A., Michel V., Thirion B., Grisel O., Blondel M., Prettenhofer P., Weiss R., Dubourg V., Vanderplas J., Passos A., Cournapeau D., Brucher M., Perrot M., Duchesnay É., Scikit-learn: Machine learning in python. J. Mach. Learn. Res. 12, 2825–2830 (2011). [Google Scholar]
- 20.Friedman J., Alm E. J., Inferring correlation networks from genomic survey data. PLOS Comput. Biol. 8, e1002687 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bastian M., Heymann S., Jacomy M., Gephi: An open source software for exploring and manipulating networks. ICWSM 8, 361–362 (2009). [Google Scholar]
- 22.Tian Y., He X., Torralba M., Yooseph S., Nelson K. E., Lux R., McLean J. S., Yu G., Shi W., Using DGGE profiling to develop a novel culture medium suitable for oral microbial communities. Mol. Oral Microbiol. 25, 357–367 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shah S. A., Ganesan S. M., Varadharaj S., Dabdoub S. M., Walters J. D., Kumar P. S., The making of a miscreant: Tobacco smoke and the creation of pathogen-rich biofilms. NPJ Biofilms Microbiomes 3, 26 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guggenheim B., Gmür R., Galicia J. C., Stathopoulou P. G., Benakanakere M. R., Meier A., Thurnheer T., Kinane D. F., In vitro modeling of host-parasite interactions: The ‘subgingival’ biofilm challenge of primary human epithelial cells. BMC Microbiol. 9, 280 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baglole C. J., Maggirwar S. B., Gasiewicz T. A., Thatcher T. H., Phipps R. P., Sime P. J., The aryl hydrocarbon receptor attenuates tobacco smoke-induced cyclooxygenase-2 and prostaglandin production in lung fibroblasts through regulation of the NF-κB family member RelB. J. Biol. Chem. 283, 28944–28957 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Buchfink B., Xie C., Huson D. H., Fast and sensitive protein alignment using DIAMOND. Nat. Methods 12, 59–60 (2015). [DOI] [PubMed] [Google Scholar]
- 27.Huson D. H., Auch A. F., Qi J., Schuster S. C., MEGAN analysis of metagenomic data. Genome Res. 17, 377–386 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dabdoub S. M., Ganesan S. M., Kumar P. S., Comparative metagenomics reveals taxonomically idiosyncratic yet functionally congruent communities in periodontitis. Sci. Rep. 6, 38993 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Human Microbiome Project Consortium , Structure, function and diversity of the healthy human microbiome. Nature 486, 207–214 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Díaz-Uriarte R., Alvarez de Andrés S., Gene selection and classification of microarray data using random forest. BMC Bioinformatics 7, 1 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dufrêne M., Legendre P., Species assemblages and indicator species: The need for a flexible asymmetrical approach. Ecol. Monogr. 67, 345–366 (1997). [Google Scholar]
- 32.Vimr E. R., Kalivoda K. A., Deszo E. L., Steenbergen S. M., Diversity of microbial sialic acid metabolism. Microbiol. Mol. Biol. Rev. 68, 132–153 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Serra D. O., Hengge R., Stress responses go three dimensional – the spatial order of physiological differentiation in bacterial macrocolony biofilms. Environ. Microbiol. 16, 1455–1471 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pöllänen M. T., Paino A., Ihalin R., Environmental stimuli shape biofilm formation and the virulence of periodontal pathogens. Int. J. Mol. Sci. 14, 17221–17237 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lim M. Y., Yoon H. S., Rho M., Sung J., Song Y.-M., Lee K., Ko G. P., Analysis of the association between host genetics, smoking, and sputum microbiota in healthy humans. Sci. Rep. 6, 23745 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pang C. M., Liu W.-T., Biological filtration limits carbon availability and affects downstream biofilm formation and community structure. Appl. Environ. Microbiol. 72, 5702–5712 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kalai Chelvam K., Yap K. P., Chai L. C., Thong K. L., Variable responses to carbon utilization between planktonic and biofilm cells of a human carrier strain of Salmonella enterica serovar typhi. PLOS ONE 10, e0126207 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shemesh M., Chai Y., A combination of glycerol and manganese promotes biofilm formation in Bacillus subtilis via histidine kinase KinD signaling. J. Bacteriol. 195, 2747–2754 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hajek P., Phillips-Waller A., Przulj D., Pesola F., Myers Smith K., Bisal N., Li J., Parrott S., Sasieni P., Dawkins L., Ross L., Goniewicz M., Wu Q., McRobbie H. J., A randomized trial of e-cigarettes versus nicotine-replacement therapy. N. Engl. J. Med. 380, 629–637 (2019). [DOI] [PubMed] [Google Scholar]
- 40.Duran-Pinedo A. E., Chen T., Teles R., Starr J. R., Wang X., Krishnan K., Frias-Lopez J., Community-wide transcriptome of the oral microbiome in subjects with and without periodontitis. ISME J. 8, 1659–1672 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/22/eaaz0108/DC1


