Abstract
Anucleate platelets, long viewed as merely cell fragments with a limited repertoire of rapid-acting hemostatic functions, are now recognized to have a complex and dynamic transcriptome mirroring that of many nucleated cells. The field of megakaryocyte and platelet transcriptomics has been rapidly growing, particularly with the advent of newer technologies such as next-generation RNA-sequencing. Studies interrogating the megakaryocyte and platelet transcriptome have led to a number of key insights into human health and disease. In this brief focused review, we will discuss some of the recent discoveries made through transcriptome analysis of megakaryocytes and platelets. We will also highlight the utility of integrating ribosome footprint analysis to augment discoveries. Both bulk and single-cell sequencing approaches will be reviewed, along with comparative studies between human and murine platelets under basal healthy settings and during acute systemic inflammatory diseases.
Keywords: gene expression profiling, hemostasis, megakaryocytes, ribosomes, transcriptome
Highlights.
Megakaryocyte and platelet transcriptomics enable discoveries in human health and disease.
While exquisitely durable in health, the platelet transcriptomic is dynamically altered in disease states.
Single-cell RNA sequencing of megakaryocytes enables insights into megakaryopoiesis and thrombopoiesis.
Platelets are abundant, circulating blood cells canonically known for their roles in hemostasis and thrombosis. Emerging studies highlight that platelets have key functions spanning immune, inflammatory, and thrombotic continuums.1–3 Although anucleate, platelets have a rich and complex transcriptome of mRNA, miRNA, long noncoding RNA, pre-mRNA, and circular RNA.4–7 It has been shown that platelets are capable of processing pre-mRNAs in signal dependent fashion to generate mRNA.8,9 mRNAs in platelets can be translated into proteins that influence platelet functional responses. In platelets, miRNAs have been shown to not only regulate direct platelet functions but also participate in cell-cell interactions and host responses.10
It is estimated that the majority of transcripts in platelets are acquired from their parental cell, the megakaryocyte, at the time of platelet formation (thrombopoiesis) while a smaller proportion may be acquired through cell-cell transfer while platelets are circulating11 (Figure 1). Platelets also harbor alternative structural features of RNA that diversify the platelet transcriptome and proteome and are known to alter platelet function, including alternative start and stop sites, exon skipping, and intron retention. Moreover, emerging and established data highlight that the platelet transcriptome is not fixed. Rather, in response to inflammatory signals, invading pathogens, cancer, or other stressors, the platelet transcriptome dynamically changes. Platelets are also capable of de novo protein synthesis, both basally and upon activation.6,8,9,12–14 As platelets lack a nucleus, this diverse and dynamic transcriptome enables platelets to synthesize new proteins and modulate their functions to participate in host thrombo-inflammatory responses.
Figure 1.
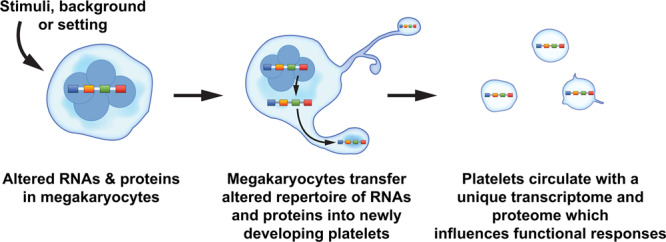
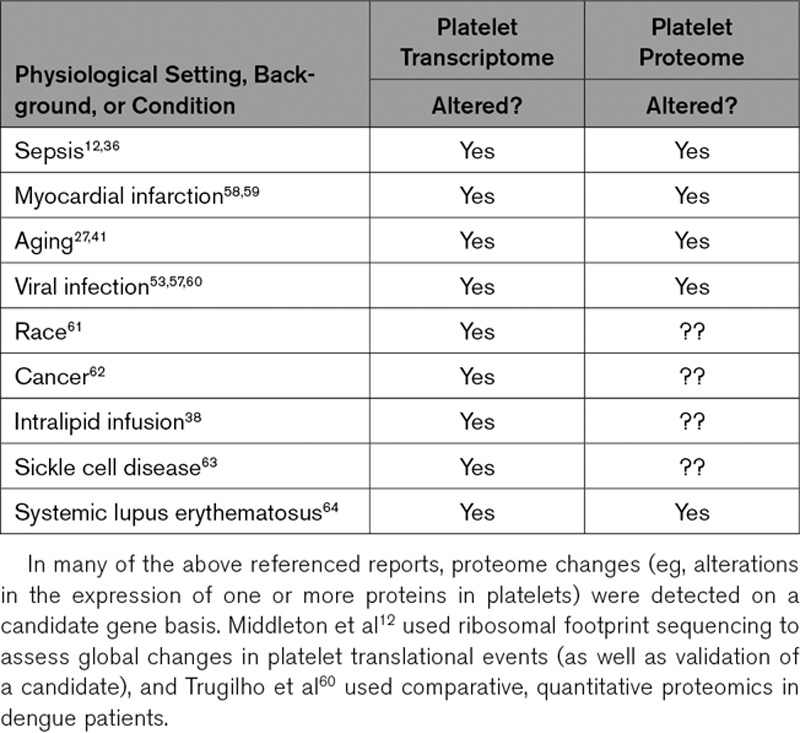
Alterations in the megakaryocyte transcriptome and proteome may be passed on to newly developing platelets. The overarching theme depicted in this schematic is that physiological stimuli, diseases, and differences in racial background may alter the portfolio of RNAs and proteins in megakaryocytes. Megakaryocytes may then invest newly developing platelets with an altered set of RNAs (eg, transcriptome) and proteins (eg, proteome) that influences cellular functions and host responses. The accompanying Table 1 in the article lists some examples of settings where the platelet transcriptome and proteome are altered. Of note, many studies referenced in the text interrogated changes in the platelet proteome for a limited subset of genes, rather than performing global analyses of the proteome.
Table.
Examples of Settings Where Published Studies Demonstrate That the Platelet Transcriptome and Proteome of Humans Is Altered
Seminal work, spanning >70 years, demonstrating that platelets contain mRNA and possess the machinery to translate mRNAs9 has opened the door to diverse RNA-based investigations that have significantly advanced the field of megakaryocyte and platelet biology.
With the development of techniques such as next-generation RNA-sequencing (RNA-seq), transcriptome analyses of platelets and megakaryocytes are increasingly used for discoveries on novel aspects of platelet biology, as diagnostic and prognostic markers, and for therapeutic development efforts. For example, platelet RNA-seq (gene expression profiling) led to the identification of the causative gene for gray platelet syndrome.15 Moreover, in recent years, the concept of tumor-educated platelets has emerged as an innovative way to detect and track the progression of certain solid tumors. In these patients, platelet RNA-seq can reveal specific transcriptional signatures of lung, brain, and breast tumors.16,17
In this focused review, we will highlight selected recent studies leveraging platelet and megakaryocyte transcriptomics for genetic discoveries, for intriguing insights into disease pathology, and for diagnostics and prognostics efforts.
Platelet Transcriptomics for Comparisons Between Mice and Humans
Next-generation RNA sequencing allows for comparative analyses of human and murine platelet transcriptomes. In general, transcripts in human platelets overlap with those in C57bl/6 murine platelets and for the most part, the most abundant transcripts in human platelets are also abundant in mouse platelets and vice versa.18 However, some transcripts have only been detected in human platelets (eg, PAFR and PAR1) while others have only been detected in murine platelets (eg, PAR3 and F5) under baseline conditions. The divergence in platelet function caused by this notable difference in gene expression for each of these genes has been well established. Thus, RNA-seq in mouse platelets can be used to understand functional differences between human and mouse platelets. It may also be valuable for identifying functional differences in mouse models of disease and inferring their relevance to their human disease counterpart.
Recently, comparative analyses between the human and murine platelet transcriptome were extended to disease settings. Middleton et al12 employed parallel strategies of RNA-seq and ribosomal footprinting in isolated platelets from septic patients and mice subjected to the cecal ligation and puncture model of sepsis. Ribosomal footprint profiling allows for preservation and subsequent sequencing of RNAs with ribosomes attached, which can be indicative of specific mRNAs undergoing active translation.19 When RNA-seq and ribosomal footprint profiling are done in parallel on the same isolated platelet samples, data generated allow one to simultaneously evaluate both transcriptional and translational changes under the same conditions. The application of this strategy to human and murine platelets during sepsis (and, for comparison, healthy or basal conditions) enabled the discovery that not only are the platelet transcriptional and translational landscapes significantly altered in sepsis, but also that for hundreds of genes, sepsis-induced changes are similar between humans and mice. Correlations between clinical and experimental sepsis held for both changes in total RNA as well as for changes in ribosome-protected mRNAs. This suggests that for many genes, transcription and translation are similarly altered in platelets during sepsis.
For example, of the many differentially regulated genes at the transcriptional (RNA-seq) and translational (ribosomal footprint) levels in mice and humans with sepsis, ITGA2B (encoding for αIIb protein) was substantially elevated. Further validation showed that platelets from mice and humans with sepsis exhibit higher amounts of αIIb that directly correlate with mortality during clinical and experimental sepsis. These sequencing data are publicly available (https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA521077) and may serve to help identify when murine models are relevant for studies of platelet gene expression in sepsis, and when they are not.
Longitudinal Assessment of Platelet Transcriptomes
Although disease can markedly alter the platelet transcriptome, recent longitudinal studies demonstrate that in the absence of disease, platelet gene expression is remarkably stable within individuals over time.20 Platelets were isolated repeatedly up to 4 years from 2 independent cohorts of healthy individuals, and stability of gene expression and alternative splicing RNA-seq assessed. Only a limited number of transcripts varied substantially within individuals over time suggesting a minor contribution of acute environmental cues to gene expression variation in healthy individuals. One of these environmental triggers that affects gene expression in self-reported healthy individuals appears to be inflammation. The most variable transcripts within individuals in both cohorts were enriched for genes in the inflammation gene ontology.
As expected, gene expression varied more between individuals than within individuals. Between individual variation accounted for most of the variation for the majority (>50%) of transcripts. As previously identified,21,22 sex and race accounted for some differences between individuals, but only for a small number of genes. Genetic markers associated with gene expression called expression quantitative trait loci played a more prominent role in gene expression differences between individuals. Genes that varied the most between individuals, yet were relatively stable within individuals (ie, were highly repeatable) were enriched for genes with known expression quantitative trait loci signal, and several novel expression quantitative trait loci genes were found among the most repeatable genes.
Beyond gene expression, several exon skipping events were identified in platelets. Like gene expression, the level of splicing was relatively stable over time. Among the most repeatable exon skipping events was exon 14 of SELP (P-selectin). This exon, which codes for the transmembrane portion of SELP and affects the ratio of surface to soluble SELP protein, was only retained in 38% of transcripts in platelets from some individuals and 92% in platelets from other individuals. A single nucleotide polymorphism, rs6128, within exon 14 accounted for the difference. Intriguingly, this platelet splice site quantitative trait loci is associated with race with the a/a allele found predominantly in blacks and African Americans, whereas white almost exclusively harbor the g/g allele. Cell culture experiments confirmed that this variant is causative for SELP exon 14 splicing. So far, rs6128 is the only site quantitative trait loci that has been identified for platelets. However, other exon skipping events beside SELP were also highly repeatable and are likely regulated by site quantitative trait loci. Figure 2 highlights some of the discoveries spanning from this recent work.
Figure 2.
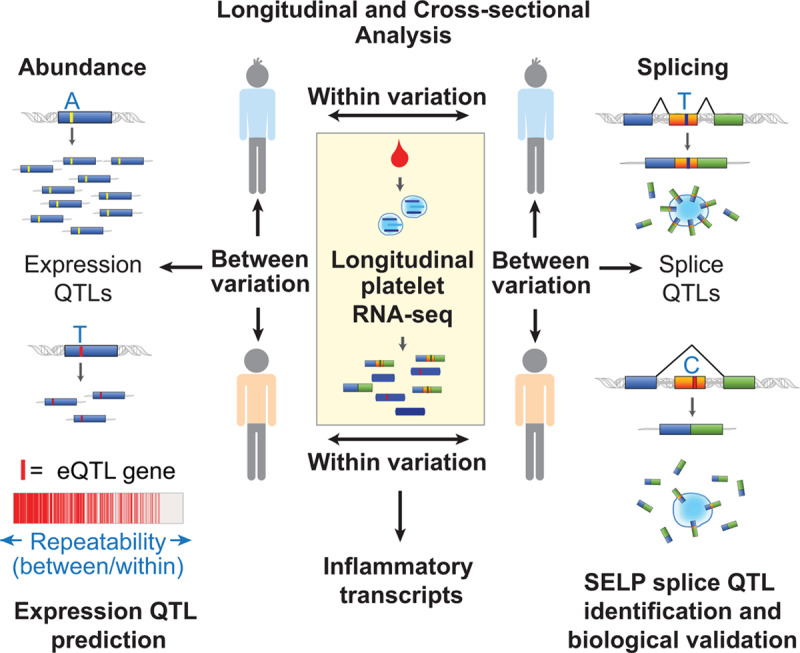
Insights into the platelet transcriptome from longitudinal and cross-sectional analysis of gene expression and splicing. In a report by Rondina et al,20 platelets were sampled repeatedly from 38 individuals for up to 4 y. Variation in gene expression and transcript splicing (exon skipping) between individuals and within individuals over time was assessed by RNA-sequencing (RNA-seq). Transcripts that varied the most within individuals were enriched for those related to inflammation. Transcripts that varied the most between individuals, but were relatively stable within individuals over time (ie, were repeatable), were predictive of identifiable cis-expression quantitative trait loci (eQTL) signal. Repeatable splicing events were also identified, including SELP (P-selectin) exon 14 skipping. Platelet SELP exon 14 skipping was associated with a single nucleotide genetic polymorphism in exon 14. In vitro experiments in cell lines using mini-gene constructs that varied by this single-nucleotide demonstrated increased exon skipping for the C/C compared with T/T variant. This resulted in reduced transmembrane domain inclusion, decreased surface, and increased soluble P-selectin.
Single-Cell Sequencing of Megakaryocytes
Bulk analysis of cultured human or murine megakaryocytes remains a valuable analytical tool to study endogenous and experimental aspects of megakaryocytes. This approach has been used to identify gene pathways involved in megakaryocyte maturation and proplatelet formation.23–25 Other complementary approaches, such as cell-sorting by flow cytometry, are suitable to isolate cultured or native megakaryocytes (eg, directly isolated from bone marrow or liver). This method requires considerable megakaryocyte enrichment of the samples before the cell sorting. In addition, it is necessary to employ larger sorting nozzles (ideally 200 μm) and low-pressure conditions which increase the sorting time required for acquisition of an adequate number of viable cells.26,27
With the introduction of single-cell RNA-seq (scRNA-seq), which uses barcode labeling of each cell present within the population of interest, the potential for de novo discovery within heterogenous cellular populations has increased. For detailed methodological descriptions of the different scRNA-seq platforms available, aspects of sample preparation and workflow, and bioinformatics analyses, the reader is referred to the following resources.28–32 One consideration when choosing scRNA-seq platforms relates to the sequencing coverage. Some systems may provide full-length transcript sequencing, while others may only sequence the 3′-end transcripts. Full length sequencing allows higher-level studies such as alternative splicing analysis, but typically is more expensive and requires more complex bioinformatic analyses.
Several groups have recently used scRNA-seq to gain a deeper understanding of the gene pathways influencing how hematopoietic precursors develop into megakaryocyte-erythroid progenitors (MEPs), erythrocytes, and megakaryocytes. For example, Psaila et al33 performed scRNA-seq on flow cytometry-sorted human MEPs and discovered that immunophenotypically defined MEPs encompass a transcriptionally heterogeneous population of cells with distinctive potentials for differentiation into megakaryocytes and erythrocytes. Intriguingly, some cells retained the potential for both erythroid and megakaryocytic output. Similarly, Lu et al34 employed scRNA-seq to compare the transcriptome of MEPs, common myeloid progenitors, erythroid, and megakaryocyte progenitors. This analysis led them to identify key genes involved in fate determination of MEPs and demonstrated that differential modulation of cell cycle speed dictates whether MEPs differentiate into erythroid progenitors or megakaryocyte progenitors. These and other studies have advanced the field by establishing novel and valuable methodological approaches to study the development of MEPs, erythroid progenitors, and megakaryocyte progenitors. Data sets generated also provide a roadmap of the transcriptomic landscape of these complex and heterogeneous cellular populations. Current platforms and protocols for scRNA-seq are not yet able to capture and sequence platelets given the lower amount of RNA per platelet compared with a nucleated cell (estimated to be ≈2.2 fg/platelet17,35). Once developed and validated, however, scRNA-seq of platelets is anticipated to offer substantial new insights.
We recently employed scRNA-seq to analyze and compare the transcriptome of native, freshly isolated, bone marrow megakaryocytes from young and aged mice as a way to begin to understand the mechanisms underlying aging-associated platelet hyperreactivity.27 We enriched for megakaryocytes using a BSA density gradient followed by CD61 positive selection with magnetic beads. This process raised the percentage of megakaryocytes from ≈0.1% to ≈2% of bone marrow cells. Megakaryocytes were transcriptionally defined by identifying cells with the highest expression of megakaryocytes specific genes (eg, Vwf, Pf4, Gata-1, Selp, Gp6, and Gp1ba). Our bioinformatics pipeline led us to identify 7 distinctive clusters of transcriptionally identified megakaryocytes with progressively increasing expression of megakaryocytes maturation markers (ie, increased expression of the β3 integrin as depicted in Figure 3). Enriched pathways in megakaryocytes from aged mice included mitochondrial dysfunction, oxidative phosphorylation, and inflammation. Numerous transcriptional differences identified in megakaryocytes were functionally evident in circulating platelets from aged mice. We used these transcriptional insights to elucidate how increased TNF-α (tumor necrosis factor α) in aging promoted platelet hyperreactivity and thrombosis. Ongoing efforts by our groups and others are using these approaches to delineate the transcriptome of human megakaryocytes during megakaryopoiesis and thrombopoiesis.
Figure 3.
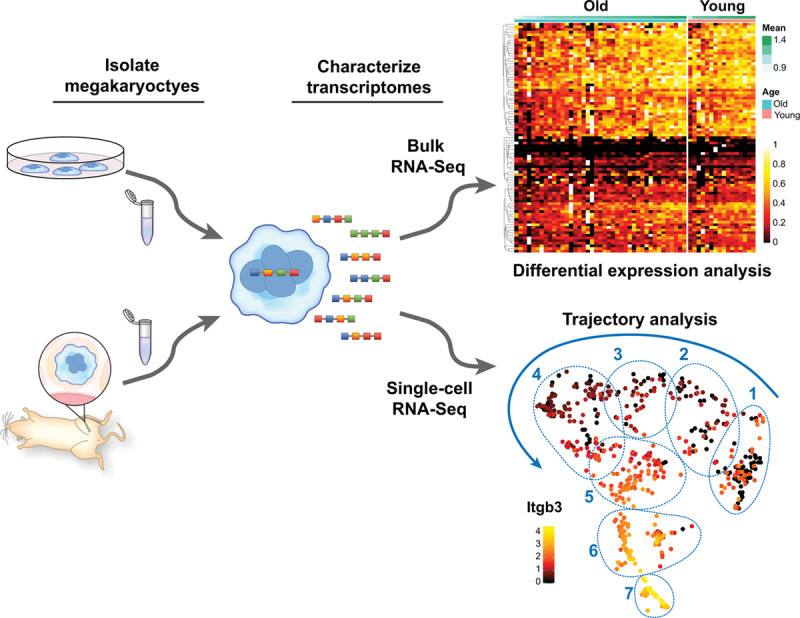
Transcriptomic analyses of megakaryocytes (MKs). Primary or cultured MKs can be subjected to different experimental conditions in vitro or in vivo before isolation for RNA sequencing. RNA-seq can be performed in bulk fashion (top right), allowing comparative analysis of MK populations under specific experimental conditions. Shown is a representative heat map of comparative, bulk, bone marrow MK transcriptomic analyses from older and younger mice. Recently, we optimized a protocol to perform single-cell RNA (scRNA-seq) sequencing of freshly isolated MKs from mice. Similar to bulk RNA-seq, scRNA-seq can be employed to compare large populations of MKs as a whole, or between clearly defined subpopulations of MKs (regions 1–7). However, scRNA-seq also affords the possibility to identify, track, and predict gene pathways involved in megakaryocyte a development and maturation. Shown is a representative trajectory analysis of 7 unique MK clusters at different maturation stages as evidenced by the expression levels of Itgb3 (β3 integrin)
Platelet Transcriptomics in Health and Disease
A number of recent studies have leveraged RNA-seq to globally examine the platelet transcriptome in healthy donors across the aging spectrum, during disease, and in comparative analyses between aged and newly released platelets in the circulation (Figure 1 and Table). More recent selected studies are highlighted in greater detail below, and we also refer the reader to other studies we, unfortunately, do not have the room to discuss here.36–40
Human aging is generally associated with an increased risk of thrombo-inflammatory diseases. Yet, mechanisms underpinning this heightened risk remain incompletely understood. Earlier, microarray-based profiling of the platelet transcriptome in 154 healthy donors aged 18 to 46 years old identified a number of age-related, differentially expressed (DE) mRNAs (n=129) and miRNAs (n=15). Intriguingly, GO enrichment analyses of mRNAs predicted to be targeted by the 15 miRNAs that were DE by age suggested categories relevant to platelet activation and function.21 Of note, the authors of this study created an interactive, public web-based tool allowing for dataset queries (www.plateletomics.com).
More recently, we used RNA-seq to perform comparative analyses of the platelet transcriptome between older (age ≥65 years old) and younger (age <45 years old) apparently healthy donors. Consistent with prior work, we identified numerous (n=514) transcripts DE in older adults. Most of these DE transcripts (n=455/514; 89%) were upregulated in older adults, as compared with younger adults.41 While we focused in detail in this study on dissection of one DE mRNA (eg, Granzyme A), the platelet RNA-seq data set is publicly available (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA397446).
In contrast to older adults, aging at the opposite end of the spectrum (eg, neonates) is associated with platelet hyporeactivity. Clinically, this may translate into a higher risk of bleeding in neonates (particularly in settings of prematurity or illness). Recently, Caparros-Perez et al used microarray to perform the first comparative analyses of platelet transcriptomes from full-term neonates and healthy adult human donors. Importantly, these investigators independently replicated analyses in 2 separate laboratories, thus providing increased rigor to differential-expression analyses. They identified 201 transcripts DE in newborn platelets. Similar to studies in older adults, the majority (162/201; 81%) were upregulated. Interestingly, among the significantly upregulated transcripts in newborn platelets, the most prevalent were those related to protein synthesis, trafficking, and degradation. Additionally, DE genes in newborn platelets were also implicated in platelet adhesion, activation, and aggregation responses—among others. Validation studies performed by these investigators confirmed that both ADRA2A (adrenoreceptor alpha 2A) and GNAZ (G-protein subunit alpha Z) were downregulated in newborn’s platelets and may account for prior observations that neonatal platelets exhibit decreased responses to epinephrine.42,43
Aging can also occur in platelets as they circulate in the blood. Reticulated platelets (RPs) represent a subset of circulating platelets that are thought to be newly released. RPs are generally larger and contain more RNA compared with mature platelets, and in some settings are hyperreactive.44–47 Clinically, RPs are associated with cardiovascular events48,49 and may also predict adverse outcomes in sepsis,50 where thrombosis risk is elevated.51 By flow cytometry, investigators separated RPs (defined as platelets with the highest thiazole orange staining) from mature platelets.52 Cell populations were then sequenced and compared. As has been previously reported, isolated RPs were hyperreactive compared with mature platelets. Consistent with this cellular phenotype, there were a large number of DE transcripts in RPs (n=1744) and biological processes related to platelet activation and hemostasis were enriched in RPs. Small-RNA sequencing identified only a small number of DE miRNAs in RPs (n=9). However, all 9 miRNAs were downregulated in RPs.
Genes upregulated in RPs and involved in platelet activation and hemostasis included the thrombin receptor PAR4, the thromboxane A2 receptor, and number integrin and adhesion molecules, including ITGA2B (CD41, or αIIb protein). This transcriptional-based study, therefore, provides new insights into prior observations that RPs are hyperreactive and supports identified associations between RPs and thrombotic events. As with other publications highlighted in this review, these investigators have made their sequencing data publicly available (https://www.ncbi.nlm.nih.gov/geo/GSE126448).
Platelet transcriptional profiling can also provide unique insights into (patho)physiological processes occurring during human disease. We highlight 2 recent examples where RNA-seq led to new biological discoveries during inflammatory diseases. In the first example, investigators studied the platelet transcriptome in persons living with HIV (all of which were on anti-retroviral therapies and had HIV RNA viral loads <200 copies/mL) and healthy donors.53 Clinical studies have demonstrated that persons living with HIV have an increased risk of cardiovascular disease, even when virally suppressed.54 Yet the mechanisms driving this increased cardiovascular disease risk remain incompletely understood.
Similar to other reports,55,56 platelets from persons living with HIV were hyperreactive—in this case, evidenced by increased adhesion and signaling to endothelial cells in vitro. Platelets from persons living with HIV also demonstrated a number (n=73) of significantly, DE transcripts. Notably, the expression of genes involved in leukocyte activation were increased in platelet from persons living with HIV. The most upregulated gene encoded for ABCC4 (ATP-binding cassette subfamily C), which regulates platelet activation. The expression of ABCC4 mRNA in platelets was significantly and positively associated with platelet surface SELP levels (r=0.72, P=0.046) and integrin αIIbβ3 activation (r=0.77, P=0.025).
The investigators of this study then elegantly pursued the cellular location, function, and impact of increased ABCC4 expression in platelets from persons living with HIV. They localized ABCC4 in platelets to the membrane of dense granules, which facilitates ADP export. Accordingly, ABCC overexpression in HIV was associated with reduced phosphorylation of VASP (vasodilator-stimulated phosphoprotein), a preferential cAMP (cyclic AMP)-dependent protein kinase phosphorylation site. Platelet ABCC4 impaired cAMP homeostasis, thereby promoting platelet activation in persons living with HIV. Further, the investigators were then able to rescue platelet-driven, enhanced endothelial cell and monocyte activation in HIV by inhibiting ABCC4. Collectively, these data demonstrate new biology for platelet ABCC4 in HIV in mediating direct platelet responses, as well as heterotypic platelet interactions with other cells.
As a second example, our group recently used RNA-seq to elucidate new biology of megakaryocytes and platelets during acute viral infections. Hypothesizing that anti-viral immune genes would be upregulated in platelets and megakaryocytes in response to invading viral pathogens, we performed RNA-seq on patients acutely infected with either dengue or influenza virus. In both cohorts, the interferon-sensitive gene IFITM3 (interferon-inducible transmembrane protein 3) was markedly (≈40- to 60-fold) upregulated. IFITM3 protein, normally undetectable in platelets from healthy donors, was also turned on. Functionally, IFITM3 in megakaryocytes served to limit viral infection, not only in megakaryocytes but also in stem cells through immune bystander effects.57
Conclusions
In conclusion, platelet RNA-seq and ribosomal footprinting analyses offer a global view to transcriptional and translation landscapes of platelets and megakaryocytes. The combination of these 2 analytical tools enhances the discovery potential as evidence by our recent work. In addition to comprehensive data sets, these toolsets enable focused discovery efforts at the single gene level, as well as for gene networks and interactions. As technologies continue to advance, there is the tangible potential that current roadblocks, such as single-cell sequencing of platelets, will be overcome. This is indeed an exciting time in the field of platelet transcriptomics, and we look forward to future insights into new roles for platelets and megakaryocytes during health and disease.
Acknowledgments
We appreciate the excellent editorial assistance of Antoinette Blair and the superb creative assistance with graphics by Diana Lim.
Sources of Funding
This work was supported in part by Merit Review Award Number I01 CX001696 to M.T. Rondina from the US Department of Veterans Affairs Clinical Sciences R&D (CSRD) Service. This material is the result of work supported with resources and the use of facilities at the George E. Wahlen VA Medical Center, Salt Lake City, Utah. This work was also supported by the National Institute on Aging (AG048022 and AG059877 to M.T. Rondina), the National Heart Lung and Blood Institute (HL142804 and HL141783 to M.T. Rondina and HL144957 to J.W. Rowley), and a Maternal and Child Health Bureau Grant H30MC24049 (to P. Davizon-Castillo).
Disclosures
None.
Nonstandard Abbreviations and Acronyms
- ABCC4
- ATP-binding cassette subfamily C
- DE
- differentially expressed
- MEPs
- megakaryocyte-erythroid progenitors
- RNA-seq
- RNA-sequencing
- RPs
- reticulated platelets
- scRNA-seq
- single-cell RNA-seq
- SELP
- P-selectin
For Sources of Funding and Disclosures, see page 1438.
References
- 1.Guo L, Rondina MT. The era of thromboinflammation: platelets are dynamic sensors and effector cells during infectious diseases. Front Immunol. 2019;10:2204. doi: 10.3389/fimmu.2019.02204. doi: 10.3389/fimmu.2019.02204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Deppermann C, Kubes P. Start a fire, kill the bug: the role of platelets in inflammation and infection. Innate Immun. 2018;24:335–348. doi: 10.1177/1753425918789255. doi: 10.1177/1753425918789255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McDonald B, Dunbar M. Platelets and intravascular immunity: guardians of the vascular space during bloodstream infections and sepsis. Front Immunol. 2019;10:2400. doi: 10.3389/fimmu.2019.02400. doi: 10.3389/fimmu.2019.02400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bray PF, McKenzie SE, Edelstein LC, Nagalla S, Delgrosso K, Ertel A, Kupper J, Jing Y, Londin E, Loher P, et al. The complex transcriptional landscape of the anucleate human platelet. BMC Genomics. 2013;14:1. doi: 10.1186/1471-2164-14-1. doi: 10.1186/1471-2164-14-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schubert S, Weyrich AS, Rowley JW. A tour through the transcriptional landscape of platelets. Blood. 2014;124:493–502. doi: 10.1182/blood-2014-04-512756. doi: 10.1182/blood-2014-04-512756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nassa G, Giurato G, Cimmino G, Rizzo F, Ravo M, Salvati A, Nyman TA, Zhu Y, Vesterlund M, Lehtiö J, et al. Splicing of platelet resident pre-mRNAs upon activation by physiological stimuli results in functionally relevant proteome modifications. Sci Rep. 2018;8:498. doi: 10.1038/s41598-017-18985-5. doi: 10.1038/s41598-017-18985-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nilsson RJ, Balaj L, Hulleman E, van Rijn S, Pegtel DM, Walraven M, Widmark A, Gerritsen WR, Verheul HM, Vandertop WP, et al. Blood platelets contain tumor-derived RNA biomarkers. Blood. 2011;118:3680–3683. doi: 10.1182/blood-2011-03-344408. doi: 10.1182/blood-2011-03-344408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rondina MT, Schwertz H, Harris ES, Kraemer BF, Campbell RA, Mackman N, Grissom CK, Weyrich AS, Zimmerman GA. The septic milieu triggers expression of spliced tissue factor mRNA in human platelets. J Thromb Haemost. 2011;9:748–758. doi: 10.1111/j.1538-7836.2011.04208.x. doi: 10.1111/j.1538-7836.2011.04208.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Denis MM, Tolley ND, Bunting M, Schwertz H, Jiang H, Lindemann S, Yost CC, Rubner FJ, Albertine KH, Swoboda KJ, et al. Escaping the nuclear confines: signal-dependent pre-mRNA splicing in anucleate platelets. Cell. 2005;122:379–391. doi: 10.1016/j.cell.2005.06.015. doi: 10.1016/j.cell.2005.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Edelstein LC, McKenzie SE, Shaw C, Holinstat MA, Kunapuli SP, Bray PF. MicroRNAs in platelet production and activation. J Thromb Haemost. 2013;11(suppl 1):340–350. doi: 10.1111/jth.12214. doi: 10.1111/jth.12214. [DOI] [PubMed] [Google Scholar]
- 11.Best MG, Wesseling P, Wurdinger T. Tumor-educated platelets as a noninvasive biomarker source for cancer detection and progression monitoring. Cancer Res. 2018;78:3407–3412. doi: 10.1158/0008-5472.CAN-18-0887. doi: 10.1158/0008-5472.CAN-18-0887. [DOI] [PubMed] [Google Scholar]
- 12.Middleton EA, Rowley JW, Campbell RA, Grissom CK, Brown SM, Beesley SJ, Schwertz H, Kosaka Y, Manne BK, Krauel K, et al. Sepsis alters the transcriptional and translational landscape of human and murine platelets. Blood. 2019;134:911–923. doi: 10.1182/blood.2019000067. doi: 10.1182/blood.2019000067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schwertz H, Rowley JW, Zimmerman GA, Weyrich AS, Rondina MT. Retinoic acid receptor-α regulates synthetic events in human platelets. J Thromb Haemost. 2017;15:2408–2418. doi: 10.1111/jth.13861. doi: 10.1111/jth.13861. [DOI] [PubMed] [Google Scholar]
- 14.Schwertz H, Köster S, Kahr WH, Michetti N, Kraemer BF, Weitz DA, Blaylock RC, Kraiss LW, Greinacher A, Zimmerman GA, et al. Anucleate platelets generate progeny. Blood. 2010;115:3801–3809. doi: 10.1182/blood-2009-08-239558. doi: 10.1182/blood-2009-08-239558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kahr WH, Hinckley J, Li L, Schwertz H, Christensen H, Rowley JW, Pluthero FG, Urban D, Fabbro S, Nixon B, et al. Mutations in NBEAL2, encoding a BEACH protein, cause gray platelet syndrome. Nat Genet. 2011;43:738–740. doi: 10.1038/ng.884. doi: 10.1038/ng.884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sol N, Wurdinger T. Platelet RNA signatures for the detection of cancer. Cancer Metastasis Rev. 2017;36:263–272. doi: 10.1007/s10555-017-9674-0. doi: 10.1007/s10555-017-9674-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Best MG, In ‘t Veld SGJG, Sol N, Wurdinger T. RNA sequencing and swarm intelligence-enhanced classification algorithm development for blood-based disease diagnostics using spliced blood platelet RNA. Nat Protoc. 2019;14:1206–1234. doi: 10.1038/s41596-019-0139-5. doi: 10.1038/s41596-019-0139-5. [DOI] [PubMed] [Google Scholar]
- 18.Rowley JW, Oler AJ, Tolley ND, Hunter BN, Low EN, Nix DA, Yost CC, Zimmerman GA, Weyrich AS. Genome-wide RNA-seq analysis of human and mouse platelet transcriptomes. Blood. 2011;118:e101–e111. doi: 10.1182/blood-2011-03-339705. doi: 10.1182/blood-2011-03-339705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ingolia NT. Ribosome footprint profiling of translation throughout the genome. Cell. 2016;165:22–33. doi: 10.1016/j.cell.2016.02.066. doi: 10.1016/j.cell.2016.02.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rondina MT, Voora D, Simon LM, Schwertz H, Harper JF, Lee O, Bhatlekar SC, Li Q, Eustes AS, Montenont E, et al. Longitudinal RNA-seq analysis of the repeatability of gene expression and splicing in human platelets identifies a platelet SELP splice QTL. Circ Res. 2020;126:501–516. doi: 10.1161/CIRCRESAHA.119.315215. doi: 10.1161/CIRCRESAHA.119.315215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Simon LM, Edelstein LC, Nagalla S, Woodley AB, Chen ES, Kong X, Ma L, Fortina P, Kunapuli S, Holinstat M, et al. Human platelet microRNA-mRNA networks associated with age and gender revealed by integrated plateletomics. Blood. 2014;123:e37–e45. doi: 10.1182/blood-2013-12-544692. doi: 10.1182/blood-2013-12-544692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Faraday N, Goldschmidt-Clermont PJ, Bray PF. Gender differences in platelet GPIIb-IIIa activation. Thromb Haemost. 1997;77:748–754. [PubMed] [Google Scholar]
- 23.Cecchetti L, Tolley ND, Michetti N, Bury L, Weyrich AS, Gresele P. Megakaryocytes differentially sort mRNAs for matrix metalloproteinases and their inhibitors into platelets: a mechanism for regulating synthetic events. Blood. 2011;118:1903–1911. doi: 10.1182/blood-2010-12-324517. doi: 10.1182/blood-2010-12-324517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang F, He J, Zhu FM, Liu JH, Qin F, Chen S, Xu G, Lü XJ, Yan LX. [Analysis of mRNA expression profiles of megakaryocytes from human cord blood CD34+ cells ex vivo expanded using Solexa sequencing]. Zhongguo Yi Xue Ke Xue Yuan Xue Bao. 2011;33:529–532. [PubMed] [Google Scholar]
- 25.Opalinska JB, Bersenev A, Zhang Z, Schmaier AA, Choi J, Yao Y, D’Souza J, Tong W, Weiss MJ. MicroRNA expression in maturing murine megakaryocytes. Blood. 2010;116:e128–e138. doi: 10.1182/blood-2010-06-292920. doi: 10.1182/blood-2010-06-292920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heazlewood SY, Williams B, Storan MJ, Nilsson SK. The prospective isolation of viable, high ploidy megakaryocytes from adult murine bone marrow by fluorescence activated cell sorting. Methods Mol Biol. 2013;1035:121–133. doi: 10.1007/978-1-62703-508-8_10. doi: 10.1007/978-1-62703-508-8_10. [DOI] [PubMed] [Google Scholar]
- 27.Davizon-Castillo P, McMahon B, Aguila S, Bark D, Ashworth K, Allawzi A, Campbell RA, Montenont E, Nemkov T, D’Alessandro A, et al. TNF-α-driven inflammation and mitochondrial dysfunction define the platelet hyperreactivity of aging. Blood. 2019;134:727–740. doi: 10.1182/blood.2019000200. doi: 10.1182/blood.2019000200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lafzi A, Moutinho C, Picelli S, Heyn H. Tutorial: guidelines for the experimental design of single-cell RNA sequencing studies. Nat Protoc. 2018;13:2742–2757. doi: 10.1038/s41596-018-0073-y. doi: 10.1038/s41596-018-0073-y. [DOI] [PubMed] [Google Scholar]
- 29.Haque A, Engel J, Teichmann SA, Lönnberg T. A practical guide to single-cell RNA-sequencing for biomedical research and clinical applications. Genome Med. 2017;9:75. doi: 10.1186/s13073-017-0467-4. doi: 10.1186/s13073-017-0467-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pellin D, Loperfido M, Baricordi C, Wolock SL, Montepeloso A, Weinberg OK, Biffi A, Klein AM, Biasco L. A comprehensive single cell transcriptional landscape of human hematopoietic progenitors. Nat Commun. 2019;10:2395. doi: 10.1038/s41467-019-10291-0. doi: 10.1038/s41467-019-10291-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vieth B, Parekh S, Ziegenhain C, Enard W, Hellmann I. A systematic evaluation of single cell RNA-seq analysis pipelines. Nat Commun. 2019;10:4667. doi: 10.1038/s41467-019-12266-7. doi: 10.1038/s41467-019-12266-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen G, Ning B, Shi T. Single-cell RNA-seq technologies and related computational data analysis. Front Genet. 2019;10:317. doi: 10.3389/fgene.2019.00317. doi: 10.3389/fgene.2019.00317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Psaila B, Barkas N, Iskander D, Roy A, Anderson S, Ashley N, Caputo VS, Lichtenberg J, Loaiza S, Bodine DM, et al. Single-cell profiling of human megakaryocyte-erythroid progenitors identifies distinct megakaryocyte and erythroid differentiation pathways. Genome Biol. 2016;17:83. doi: 10.1186/s13059-016-0939-7. doi: 10.1186/s13059-016-0939-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lu YC, Sanada C, Xavier-Ferrucio J, Wang L, Zhang PX, Grimes HL, Venkatasubramanian M, Chetal K, Aronow B, Salomonis N, et al. The molecular signature of megakaryocyte-erythroid progenitors reveals a role for the cell cycle in fate specification. Cell Rep. 2018;25:3229. doi: 10.1016/j.celrep.2018.11.075. doi: 10.1016/j.celrep.2018.11.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Teruel-Montoya R, Kong X, Abraham S, Ma L, Kunapuli SP, Holinstat M, Shaw CA, McKenzie SE, Edelstein LC, Bray PF. MicroRNA expression differences in human hematopoietic cell lineages enable regulated transgene expression. PLoS One. 2014;9:e102259. doi: 10.1371/journal.pone.0102259. doi: 10.1371/journal.pone.0102259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Freishtat RJ, Natale J, Benton AS, Cohen J, Sharron M, Wiles AA, Ngor WM, Mojgani B, Bradbury M, Degnan A, et al. Sepsis alters the megakaryocyte-platelet transcriptional axis resulting in granzyme B-mediated lymphotoxicity. Am J Respir Crit Care Med. 2009;179:467–473. doi: 10.1164/rccm.200807-1085OC. doi: 10.1164/rccm.200807-1085OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rose JJ, Voora D, Cyr DD, Lucas JE, Zaas AK, Woods CW, Newby LK, Kraus WE, Ginsburg GS. Gene expression profiles link respiratory viral infection, platelet response to aspirin, and acute myocardial infarction. PLoS One. 2015;10:e0132259. doi: 10.1371/journal.pone.0132259. doi: 10.1371/journal.pone.0132259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Beaulieu LM, Vitseva O, Tanriverdi K, Kucukural A, Mick E, Hamburg N, Vita J, Freedman JE. Platelet functional and transcriptional changes induced by intralipid infusion. Thromb Haemost. 2016;115:1147–1156. doi: 10.1160/TH15-09-0739. doi: 10.1160/TH15-09-0739. [DOI] [PubMed] [Google Scholar]
- 39.Clancy L, Beaulieu LM, Tanriverdi K, Freedman JE. The role of RNA uptake in platelet heterogeneity. Thromb Haemost. 2017;117:948–961. doi: 10.1160/TH16-11-0873. doi: 10.1160/TH16-11-0873. [DOI] [PubMed] [Google Scholar]
- 40.Koupenova M, Mick E, Corkrey HA, Singh A, Tanriverdi SE, Vitseva O, Levy D, Keeler AM, Ezzaty Mirhashemi M, ElMallah MK, et al. Pollen-derived RNAs are found in the human circulation. iScience. 2019;19:916–926. doi: 10.1016/j.isci.2019.08.035. doi: 10.1016/j.isci.2019.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Campbell RA, Franks Z, Bhatnagar A, Rowley JW, Manne BK, Supiano MA, Schwertz H, Weyrich AS, Rondina MT. Granzyme A in human platelets regulates the synthesis of proinflammatory cytokines by monocytes in aging. J Immunol. 2018;200:295–304. doi: 10.4049/jimmunol.1700885. doi: 10.4049/jimmunol.1700885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Corby DG, O’Barr TP. Decreased alpha-adrenergic receptors in newborn platelets: cause of abnormal response to epinephrine. Dev Pharmacol Ther. 1981;2:215–225. [PubMed] [Google Scholar]
- 43.Tweeten KA, Bulla LA, Jr, Consigli RA. Characterization of an alkaline protease associated with a granulosis virus of Plodia interpunctella. J Virol. 1978;26:703–711. [PMC free article] [PubMed] [Google Scholar]
- 44.Bernlochner I, Goedel A, Plischke C, Schüpke S, Haller B, Schulz C, Mayer K, Morath T, Braun S, Schunkert H, et al. Impact of immature platelets on platelet response to ticagrelor and prasugrel in patients with acute coronary syndrome. Eur Heart J. 2015;36:3202–3210. doi: 10.1093/eurheartj/ehv326. doi: 10.1093/eurheartj/ehv326. [DOI] [PubMed] [Google Scholar]
- 45.Armstrong PC, Hoefer T, Knowles RB, Tucker AT, Hayman MA, Ferreira PM, Chan MV, Warner TD. Newly formed reticulated platelets undermine pharmacokinetically short-lived antiplatelet therapies. Arterioscler Thromb Vasc Biol. 2017;37:949–956. doi: 10.1161/ATVBAHA.116.308763. doi: 10.1161/ATVBAHA.116.308763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hannawi B, Hannawi Y, Kleiman NS. Reticulated platelets: changing focus from basics to outcomes. Thromb Haemost. 2018;118:1517–1527. doi: 10.1055/s-0038-1667338. doi: 10.1055/s-0038-1667338. [DOI] [PubMed] [Google Scholar]
- 47.Hilt ZT, Ture SK, Mohan A, Arne A, Morrell CN. Platelet-derived β2m regulates age related monocyte/macrophage functions. Aging (Albany NY) 2019;11:11955–11974. doi: 10.18632/aging.102520. doi: 10.18632/aging.102520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cesari F, Marcucci R, Gori AM, Caporale R, Fanelli A, Casola G, Balzi D, Barchielli A, Valente S, Giglioli C, et al. Reticulated platelets predict cardiovascular death in acute coronary syndrome patients. Insights from the AMI-florence 2 study. Thromb Haemost. 2013;109:846–853. doi: 10.1160/TH12-09-0709. doi: 10.1160/TH12-09-0709. [DOI] [PubMed] [Google Scholar]
- 49.Ibrahim H, Schutt RC, Hannawi B, DeLao T, Barker CM, Kleiman NS. Association of immature platelets with adverse cardiovascular outcomes. J Am Coll Cardiol. 2014;64:2122–2129. doi: 10.1016/j.jacc.2014.06.1210. doi: 10.1016/j.jacc.2014.06.1210. [DOI] [PubMed] [Google Scholar]
- 50.Muronoi T, Koyama K, Nunomiya S, Lefor AK, Wada M, Koinuma T, Shima J, Suzukawa M. Immature platelet fraction predicts coagulopathy-related platelet consumption and mortality in patients with sepsis. Thromb Res. 2016;144:169–175. doi: 10.1016/j.thromres.2016.06.002. doi: 10.1016/j.thromres.2016.06.002. [DOI] [PubMed] [Google Scholar]
- 51.Kaplan D, Casper TC, Elliott CG, Men S, Pendleton RC, Kraiss LW, Weyrich AS, Grissom CK, Zimmerman GA, Rondina MT. VTE incidence and risk factors in patients with severe sepsis and septic shock. Chest. 2015;148:1224–1230. doi: 10.1378/chest.15-0287. doi: 10.1378/chest.15-0287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bongiovanni D, Santamaria G, Klug M, Santovito D, Felicetta A, Hristov M, von Scheidt M, Aslani M, Cibella J, Weber C, et al. Transcriptome analysis of reticulated platelets reveals a prothrombotic profile. Thromb Haemost. 2019;119:1795–1806. doi: 10.1055/s-0039-1695009. doi: 10.1055/s-0039-1695009. [DOI] [PubMed] [Google Scholar]
- 53.Marcantoni E, Allen N, Cambria MR, Dann R, Cammer M, Lhakhang T, O’Brien MP, Kim B, Worgall T, Heguy A, et al. Platelet transcriptome profiling in HIV and ATP-binding cassette subfamily C member 4 (ABCC4) as a mediator of platelet activity. JACC Basic Transl Sci. 2018;3:9–22. doi: 10.1016/j.jacbts.2017.10.005. doi: 10.1016/j.jacbts.2017.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Torriani FJ, Komarow L, Parker RA, Cotter BR, Currier JS, Dubé MP, Fichtenbaum CJ, Gerschenson M, Mitchell CK, Murphy RL, et al. ACTG 5152s Study Team. Endothelial function in human immunodeficiency virus-infected antiretroviral-naive subjects before and after starting potent antiretroviral therapy: the ACTG (AIDS Clinical Trials Group) study 5152s. J Am Coll Cardiol. 2008;52:569–576. doi: 10.1016/j.jacc.2008.04.049. doi: 10.1016/j.jacc.2008.04.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mesquita EC, Hottz ED, Amancio RT, Carneiro AB, Palhinha L, Coelho LE, Grinsztejn B, Zimmerman GA, Rondina MT, Weyrich AS, et al. Persistent platelet activation and apoptosis in virologically suppressed HIV-infected individuals. Sci Rep. 2018;8:14999. doi: 10.1038/s41598-018-33403-0. doi: 10.1038/s41598-018-33403-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.O’Brien M, Montenont E, Hu L, Nardi MA, Valdes V, Merolla M, Gettenberg G, Cavanagh K, Aberg JA, Bhardwaj N, et al. Aspirin attenuates platelet activation and immune activation in HIV-1-infected subjects on antiretroviral therapy: a pilot study. J Acquir Immune Defic Syndr. 2013;63:280–288. doi: 10.1097/QAI.0b013e31828a292c. doi: 10.1097/QAI.0b013e31828a292c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Campbell RA, Schwertz H, Hottz ED, Rowley JW, Manne BK, Washington AV, Hunter-Mellado R, Tolley ND, Christensen M, Eustes AS, et al. Human megakaryocytes possess intrinsic antiviral immunity through regulated induction of IFITM3. Blood. 2019;133:2013–2026. doi: 10.1182/blood-2018-09-873984. doi: 10.1182/blood-2018-09-873984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Eicher JD, Wakabayashi Y, Vitseva O, Esa N, Yang Y, Zhu J, Freedman JE, McManus DD, Johnson AD. Characterization of the platelet transcriptome by RNA sequencing in patients with acute myocardial infarction. Platelets. 2016;27:230–239. doi: 10.3109/09537104.2015.1083543. doi: 10.3109/09537104.2015.1083543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schmidt RA, Morrell CN, Ling FS, Simlote P, Fernandez G, Rich DQ, Adler D, Gervase J, Cameron SJ. The platelet phenotype in patients with ST-segment elevation myocardial infarction is different from non-ST-segment elevation myocardial infarction. Transl Res. 2018;195:1–12. doi: 10.1016/j.trsl.2017.11.006. doi: 10.1016/j.trsl.2017.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Trugilho MRO, Hottz ED, Brunoro GVF, Teixeira-Ferreira A, Carvalho PC, Salazar GA, Zimmerman GA, Bozza FA, Bozza PT, Perales J. Platelet proteome reveals novel pathways of platelet activation and platelet-mediated immunoregulation in dengue. PLoS Pathog. 2017;13:e1006385. doi: 10.1371/journal.ppat.1006385. doi: 10.1371/journal.ppat.1006385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Edelstein LC, Simon LM, Montoya RT, Holinstat M, Chen ES, Bergeron A, Kong X, Nagalla S, Mohandas N, Cohen DE, et al. Racial differences in human platelet PAR4 reactivity reflect expression of PCTP and miR-376c. Nat Med. 2013;19:1609–1616. doi: 10.1038/nm.3385. doi: 10.1038/nm.3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Best MG, Sol N, Kooi I, Tannous J, Westerman BA, Rustenburg F, Schellen P, Verschueren H, Post E, Koster J, et al. RNA-seq of tumor-educated platelets enables blood-based pan-cancer, multiclass, and molecular pathway cancer diagnostics. Cancer Cell. 2015;28:666–676. doi: 10.1016/j.ccell.2015.09.018. doi: 10.1016/j.ccell.2015.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Raghavachari N, Xu X, Harris A, Villagra J, Logun C, Barb J, Solomon MA, Suffredini AF, Danner RL, Kato G, et al. Amplified expression profiling of platelet transcriptome reveals changes in arginine metabolic pathways in patients with sickle cell disease. Circulation. 2007;115:1551–1562. doi: 10.1161/CIRCULATIONAHA.106.658641. doi: 10.1161/CIRCULATIONAHA.106.658641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lood C, Amisten S, Gullstrand B, Jönsen A, Allhorn M, Truedsson L, Sturfelt G, Erlinge D, Bengtsson AA. Platelet transcriptional profile and protein expression in patients with systemic lupus erythematosus: up-regulation of the type I interferon system is strongly associated with vascular disease. Blood. 2010;116:1951–1957. doi: 10.1182/blood-2010-03-274605. doi: 10.1182/blood-2010-03-274605. [DOI] [PubMed] [Google Scholar]


