Abstract
Evidence accumulates that the functional plasticity of insulin and insulin-like growth factor signaling in insects could spring, among others, from the multiplicity of insulin receptors (InRs). Their multiple variants may be implemented in the control of insect polyphenism, such as wing or caste polyphenism. Here, we present a comprehensive phylogenetic analysis of insect InR sequences in 118 species from 23 orders and investigate the role of three InRs identified in the linden bug, Pyrrhocoris apterus, in wing polymorphism control. We identified two gene clusters (Clusters I and II) resulting from an ancestral duplication in a late ancestor of winged insects, which remained conserved in most lineages, only in some of them being subject to further duplications or losses. One remarkable yet neglected feature of InR evolution is the loss of the tyrosine kinase catalytic domain, giving rise to decoys of InR in both clusters. Within the Cluster I, we confirmed the presence of the secreted decoy of insulin receptor in all studied Muscomorpha. More importantly, we described a new tyrosine kinase-less gene (DR2) in the Cluster II, conserved in apical Holometabola for ∼300 My. We differentially silenced the three P. apterus InRs and confirmed their participation in wing polymorphism control. We observed a pattern of Cluster I and Cluster II InRs impact on wing development, which differed from that postulated in planthoppers, suggesting an independent establishment of insulin/insulin-like growth factor signaling control over wing development, leading to idiosyncrasies in the co-option of multiple InRs in polyphenism control in different taxa.
Keywords: insulin signaling, insulin receptor, decoy of insulin receptor, wing polyphenism, gene structure, insects
Introduction
The insulin/insulin-like growth factor signaling (IIS) is a conserved regulatory pathway ubiquitous in Metazoa (Vitali et al. 2018), activated by the interaction of insulin-like peptides (ILPs) with insulin receptors (InRs). IIS is the key element in the nutrient-sensing cascade responsible for redirecting of resources into various, often competing metabolic and developmental processes. Large body of evidence on the multiple functions of IIS pathway has been accumulated in insects. IIS is known to be implicated directly, or through a crosstalk with other major regulatory pathways (TOR and FOXO pathways, juvenile hormone, ecdysteroids), in postembryonic development, nutrition-based phenotypic plasticity, and body size control (Chen et al. 1996; Brogiolo et al. 2001; Junger et al. 2003; Emlen et al. 2012; Snell-Rood and Moczek 2012; Hattori et al. 2013; Koyama et al. 2013; Ding et al. 2017; Lin et al. 2018; Pan et al. 2018), reproduction (Badisco et al. 2013; Abrisqueta et al. 2014), and, last but not least, the diapause, circadian rhythmicity (Sim and Denlinger 2008, 2013; Vafopoulou and Steel 2014), and behavior (Erion and Sehgal 2013). Due to its central role in metabolism, growth, and reproduction, IIS also acts as a key regulator of insect lifespan (Clancy et al. 2001; Tatar et al. 2001; Broughton et al. 2005; Giannakou and Partridge 2007; Bai et al. 2012; Kannan and Fridell 2013).
Interaction of ILPs with InRs is an upstream step in a highly conserved, though complex and versatile IIS transduction cascade. Canonical insulin receptor consists of a dimer of >1,000 amino acid long transmembrane InR proteins. The binding of the ligand to its extracellular domain induces structural changes in the receptor, leading to autophosphorylation of tyrosine residues within the intracellular tyrosine kinase (TK) domain, which in turn phosphorylates the insulin receptor substrate (Maruyama 2014). The downstream intracellular action then proceeds via activation of specific kinases with further targets Akt/protein kinase B, TOR, and FOXO. An alternative system, sensitive to some ILPs and acting through leucine-rich repeat-containing G-protein coupled receptor, has been described in Drosophila (Colombani et al. 2015).
The complexity of IIS effects is not only due to its interactions with multiple downstream targets but springs also from the multiplicity of ILPs. The numbers of identified ILPs vary remarkably among insects. Although in some of them, such as the locusts, no more than a single ILP was identified (Hetru et al. 1991; Badisco et al. 2008), 8 ilp genes are known in Drosophila, and as many as 38 ilp genes were detected in the silk moth Bombyx mori (Kondo et al. 1996; Wu and Brown 2006). Differential functions, expression, and release dynamics of different ILPs were the best characterized in Drosophila (Kannan and Fridell 2013; Nassel et al. 2015), but addressed also in other models, such as mosquitoes (Wen et al. 2010), honey bees (Wang et al. 2013), or beetles (Okada et al. 2019).
Yet another level of IIS versatility may result from the multiplicity of InRs. In contrast to traditional models, that is, Drosophila and Caenorhabditis elegans, which only have a single InR gene, a list of insect taxa with two or even three InR copies has been growing over the past decade. First noticed in social Hymenoptera (Wheeler et al. 2006; Ament et al. 2008; de Azevedo and Hartfelder 2008; Lu and Pietrantonio 2011), two InRs were later detected in some beetles (Sang et al. 2016), but also in more basal clades of Hemiptera (Xu et al. 2015; Guo et al. 2016; Sang et al. 2016; Ding et al. 2017) and Blattodea (including termites) (Terrapon et al. 2014; Ding et al. 2017; Harrison et al. 2018), suggesting that all these cases may result from an ancient InR duplication. It has first been estimated to take place prior to the differentiation of Hemiptera (Sang et al. 2016), later analyses situated this duplication deeper in the evolutionary history by including two InR homologs (and an additional one arising from a more recent duplication) found in the genome of the termite Zootermopsis nevadensis (Xu and Zhang 2017). Two recent studies, concurrent with our research, proposed that the ancient InR duplication in fact took place prior to the evolution of flight (Armisén et al. 2018; Kremer et al. 2018). All current scenarios propose multiple secondary losses of one InR paralog in individual lineages, including the large crown group containing Diptera and Lepidoptera, which is in line with previous observations of a single InR in the genomes of mosquitoes and flies (Vogel et al. 2013), but also some beetles (Lavine et al. 2013) or Hymenoptera (Nasonia vitripennis) (Ding et al. 2017).
The presence of more than one InR gene in some lineages prompts the hypothesis that the InR multiplicity may have functional implications. Differential expression patterns and distinct roles in the development and reproduction have been described for the two InRs in the red flour beetle, Tribolium castaneum (Sang et al. 2016). By contrast, exclusive functions were not clearly identified for two InR genes in the aphid Aphis citricidus (Ding et al. 2017). Eventual specificity of individual InRs may be particularly relevant for the understanding of insect polyphenism. A pioneering study assigned antagonistic roles to two InRs in the control of wing polymorphism in planthoppers (Xu et al. 2015; Xu and Zhang 2017). On the contrary, differential functions of the two InRs were not found in the control of the male ornaments in the dung beetle Onthophagus taurus (Casasa and Moczek 2018), in contrast to previous results, showing the critical role of IIS in the regulation of exaggerated male traits in the rhinoceros beetle Trypoxylus dichotomus (Emlen et al. 2012). These controversies suggest that the pattern of insect polyphenism control by multiple InRs may not be universal.
Yet another case of insect polyphenism, which is likely to be controlled by multiple InRs, is the caste polyphenism of social insects. Strong evidences were accumulated for the implication of IIS in the caste determination of social Hymenoptera (Corona et al. 2016; Nijhout and McKenna 2018). Even though the eventual differential role of the two InRs is not fully understood, it is considered as likely, at least based on their differential expression in queen-destined and worker-destined honey bee larvae (de Azevedo and Hartfelder 2008), and caste- and tissue-specific expressions in the ant Solenopsis invicta (Lu and Pietrantonio 2011) and the bumblebee Bombus terrestris (Jedlička et al. 2016). Prior to the description of multiple InRs in the genomes of termites (Terrapon et al. 2014; Harrison et al. 2018; Kremer et al. 2018), the role of at the time single-known InR has been demonstrated in morphogenesis of termite soldiers (Hattori et al. 2013). A recent study by Kremer et al. (2018) underlines an exclusive occurrence of three InRs in cockroaches and termites and draws appealing conclusions that the evolution of the third InR may have been a prerequisite for the caste polyphenism in termites (Kremer et al. 2018).
The objectives of the present study are 2-fold. First, the apparent complexity in InR distribution across Insecta begets a comprehensive reconstruction of InR evolution, because the previous phylogenies only covered a maximum of nine insect orders (Armisén et al. 2018). To this goal, we systematically explored the available genomic and transcriptomic information (including two de novo assembled genomes and four transcriptomes) and manually annotated putative InR protein sequences. We selected a subset of taxa showing sufficient quality sequences with emphasis on a maximum representation of insect diversity, reconstructed InR evolution, and mapped the InR duplications and losses in 98 species across 23 insect orders. To track in detail important evolutionary events, we performed an additional series of taxon-specific analyses with 20 more insect species. To avoid erroneous interpretations of false negative results, InR losses were only considered if independently confirmed from genomes and/or transcriptomes of multiple representatives of a monophyletic clade. To draw a complete picture of InR evolution, we did not restrict our analyses only to InR genes coding for all functional domains of a canonical insulin receptor protein, but also focused on their homologs arising through the loss of the TK catalytic domain, the so-called “decoys of insulin receptor.” Though lacking the TK domain, the Drosophila secreted decoy of insulin receptor (SDR) has been confirmed as a functional protein able to bind the ILPs and having antagonistic function to the canonical InR (Okamoto et al. 2013). Transformation of InRs into decoys of insulin receptors thus may be viewed as an adaptive event rather than a simple loss of InRs, and as such it deserves attention in phylogenetic interpretations and functional studies. Therefore, we mapped the genes coding for SDRs and another, previously undescribed class of TK-less decoy of insulin receptor, across insects.
And second, we addressed one of the most prominent functions assigned to InR multiplicity, that is, its participation in wing polyphenism control. IIS has been recently postulated as major regulator of wing polyphenism in planthoppers and the opposite roles of their two InRs deemed to be related to structural features characteristic for each of the two receptor proteins (Xu et al. 2015; Xu and Zhang 2017). Our aim was to verify the contribution of IIS to wing elongation/shortening in an unrelated insect model and depict eventual commonalities and differences in wing polymorphism control by multiple InRs and causalities related to their protein structures. To this goal, we applied reverse genetic approach to investigate the roles of the three InRs, characterized in this study, in the wing-polymorphic linden bug, Pyrrhocoris apterus (Heteroptera).
Results
Ancestral InR Duplication Coincides with Wing Evolution and Remains Conserved in Most Winged Insects
As multiple InRs were reported in various animals including Platyhelminthes (You et al. 2010), Vertebrata (Hernandez-Sanchez et al. 2008), Crustacea (Boucher et al. 2010), and Insecta, we have first compared InR protein sequences in key animal phyla (supplementary fig. 1, Supplementary Material online). We observed independent InR multiplications in Porifera, Platyhelminthes, Nematoda, vertebrates, and crustaceans. In vertebrates, the InRs branch together with the insulin-like growth factor receptor and the insulin receptor-related receptor. By contrast, only one InR has been found in springtails, hexapod relatives of insects, suggesting that the InR duplicity observed in many insects resulted from duplication in their late hexapod ancestor or within basal Insecta.
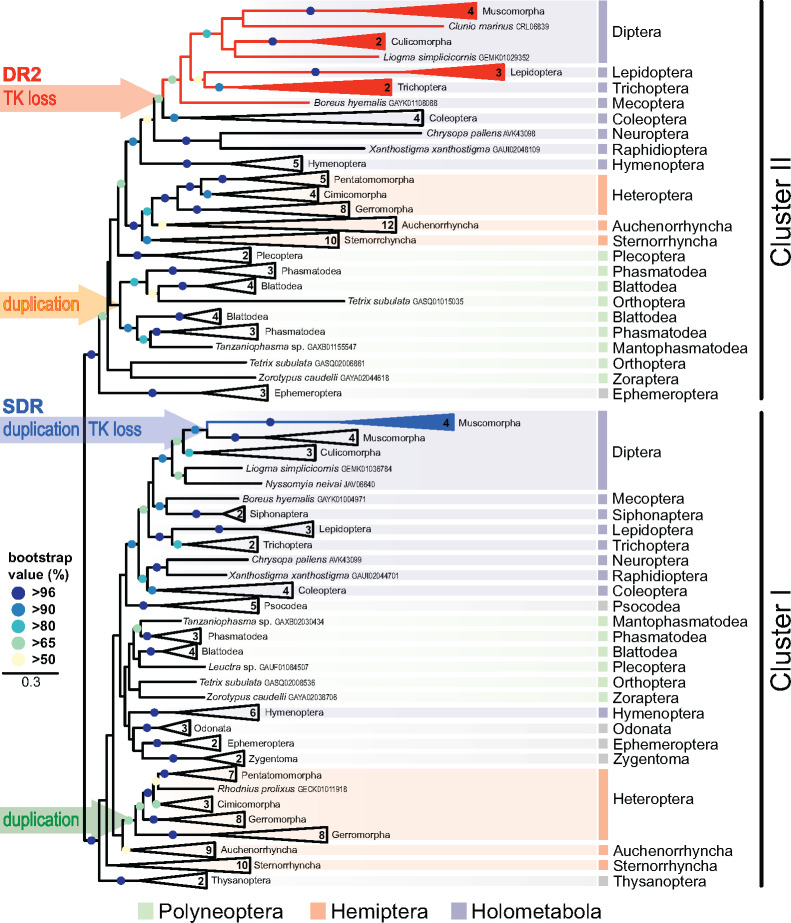
The main phylogenetic analysis of 98 insect species from 23 orders is presented in the simplified maximum likelihood (ML) tree (fig. 1), supplementary figures 2‒4 (Supplementary Material online) show detailed views of this tree and a phylogeny obtained using Bayesian inference. In all reconstructions, InR protein sequences were split into two well-supported clades. In both clades, majority of studied lineages was represented, with only a few exceptions indicating secondary losses of one InR paralog. Because previous studies are largely inconsistent in the nomenclature of insect InRs (see supplementary figs. 2 and 3, Supplementary Material online), we avoid the assignments InR1, InR2, or others. Instead, we define two clusters, Clusters I and II, and throughout the text, tables, and figures, we provide accession numbers to individual sequences.
Fig. 1.
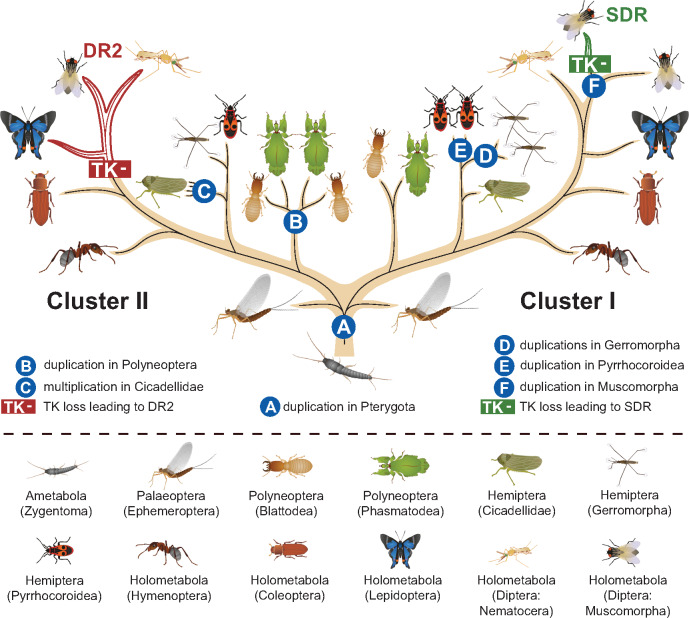
Phylogenetic tree of insect insulin receptors and decoy of insulin receptors identified in 98 species from 23 orders. The numbers in condensed branches indicate the numbers of species studied. Orange arrow marks the Cluster II duplication within Polyneoptera. Red arrow (DR2) indicates the loss of the tyrosine kinase domain in advanced Holometabola, giving rise to the decoy of insulin receptor gene DR2 (red). Green arrow marks the Cluster I duplication in Gerromorpha, blue arrow (SDR) marks the loss of the tyrosine kinase domain in Muscomorpha, giving rise to the secreted decoy of insulin receptor gene SDR (blue). The topology and branching supports were inferred using RAxML maximum likelihood algorithm with WAG + Γ model (−ln = 285839.374747). The bootstrap values calculated from 500 replicates are shown for branches represented in >50% of trees. Full version of the tree, together with the taxon coverage and nomenclature used in previous studies, is given in the supplementary figures 2 and 3 (Supplementary Material online).
Despite a systematic effort, we were able to retrieve only the InR genes belonging to Cluster I in the two studied wingless insects (Zygentoma). By contrast, in the basal-winged insects, Palaeoptera, we identified the Cluster II gene in all three studied mayflies (Ephemeroptera), in addition to the Cluster I gene found in two of them. Interestingly, only the Cluster I homologs were retrieved in the three studied dragonflies (Odonata), the sister order to Ephemeroptera within Palaeoptera. Although more genomes/transcriptomes from basal insects are needed for accurate reconstruction of the ancestral duplication, the most likely scenario is that the Cluster I and Cluster II InRs diversification closely coincides with the evolution of wings in Pterygota.
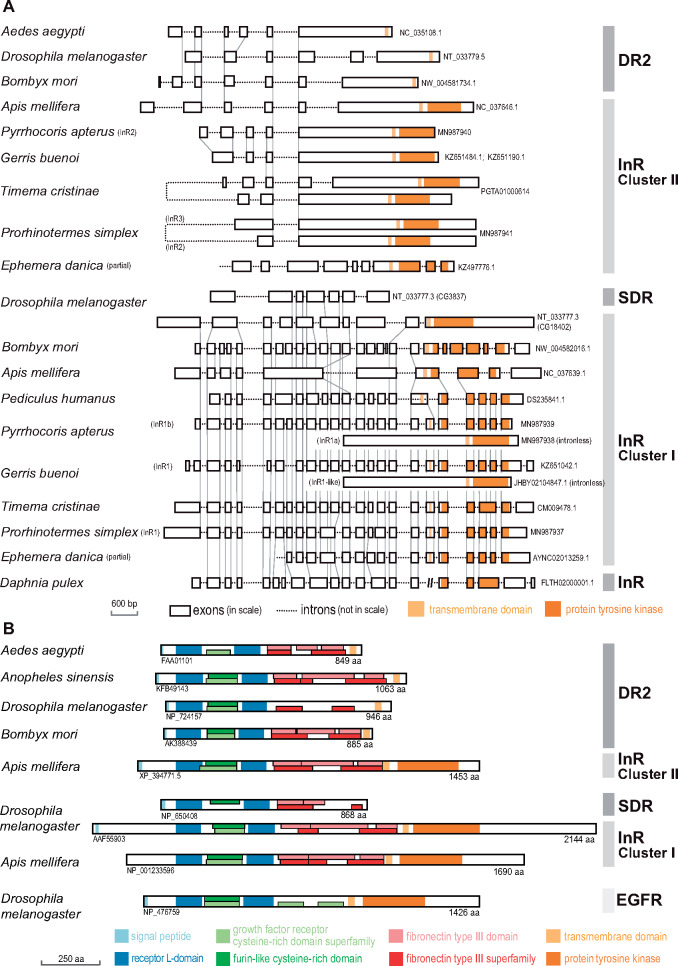
To elucidate the origin of Cluster I and Cluster II InR genes, we compared the gene structures in ten representative insects and one outgroup, the crustacean Daphnia pulex (fig. 2). Even a visual inspection revealed a different pattern of exon–intron structures between the Cluster I genes, consisting of larger number of exons (up to 26), when compared with Cluster II genes with a maximum of 9 exons. High similarity of the Cluster I gene structures and exon–intron boundaries in D. pulex and all Hemimetabola unambiguously identified Cluster I as the ancestral insect InR. Cluster I genes showed a conspicuous reduction of intron numbers in the honey bee and the fruit fly, resulting in a drop of gene structure similarity with both, hemimetabolous insects and other Holometabola (e.g., B. mori). General comparison of the two InR clusters with respect to evolutionary rates using the averaged ML distances indicated that Cluster I clade is more conserved (average ML distance = 1.31197, SD = 0.61146) compared with the faster evolving Cluster II (1.98940, 0.73031). In addition, the relative number of sequences, whose base compositions did not correspond to the employed WAG model, was significantly lower in the Cluster I (4%) than in the Cluster II (21%), whereas the relative number of parsimony-informative sites, a proxy of the phylogenetic signal, remained comparable in the two clades (86% and 91%).
Fig. 2.
Gene structures and protein domains of InRs and decoys of insect insulin receptors. (A) Intron and exon structures and homologous intron–exon boundaries (gray vertical lines) in Cluster I and Cluster II InR, DR2, and SDR genes in selected insect representatives, including the taxa, in which important duplications were detected (stick insects, termites, semiaquatic bugs, and linden bug) and compared with InR gene structure in the sister group of insects, the crustacean Daphnia pulex. (B) Protein domains recognized in the protein sequences reconstructed from the transcripts of Cluster I and Cluster II InRs, DR2, and SDR, and compared with EGFR protein.
In both clusters, the main phylogenetic analysis retrieved all studied insect orders as monophyletic. While in the Cluster II, most relationships among individual orders corresponded to current phylogenetic hypotheses, the internal topology of Cluster I diverged substantially more from the general phylogeny of Pterygota. Hymenoptera clustered outside Holometabola, Thysanoptera, and Sterrnorhyncha were relatively remote from other insects, and Zygentoma, Palaeoptera, and Polyneoptera were not retrieved as basal to the Cluster I tree. Important evolutionary events spotted in Clusters I and II (fig. 1 andsupplementary figs. 2‒4, Supplementary Material online) are individually presented below.
InR Duplication in Polyneoptera
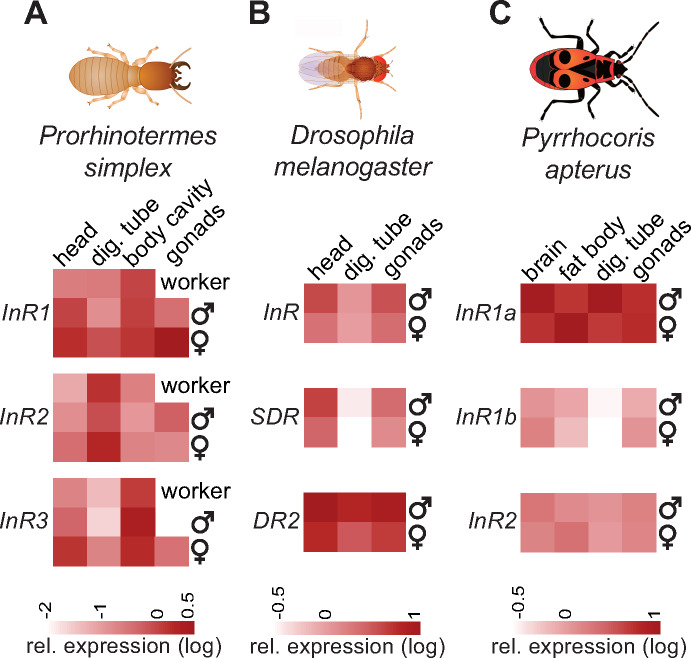
The main analysis indicated the presence of two Cluster II paralogs in all four studied species of Blattodea (one cockroach and three termites species), and all three stick insects. This observation prompted us to conduct a more detailed analysis of these advanced polyneopteran clades (supplementary fig. 5, Supplementary Material online). We confirmed the same pattern in four species of termites, four cockroaches from both, Blaberoidea and Blattoidea (including Cryptocercus as the closest relative to termites), and in all five studied genera of Phasmatodea, with strong support to both Cluster II subclades. This suggests the origin of the duplication in an ancestor of the crown polyneopteran lineage. The two Cluster II InRs show a high level of mutual similarity also at the level of gene structure, when both copies are inversely situated on one locus (fig. 2). Expression analysis of the three polyneopteran InRs in the termite Pro. simplex revealed a strong tissue-specific and in some cases also caste-specific InR expression profiles, which, at the same time, showed clearly distinct and independent trends among the three InR paralogs (fig. 3A). Detailed expression analysis and statistical comparisons are given in supplementary figure 8 (Supplementary Material online).
Fig. 3.
Relative expression of insulin receptor genes and decoy of insulin receptor genes in different tissues of termite workers and 10-day-old male and female neotenic reproductive (A), adult male and female fruit flies (B), and adult male and female linden bugs (C). The heatmaps represent mean expression values from two to four replicates per phenotype and tissue, relative to the control gene rp49 and shown on a log scale. Detailed comparisons are given in the supplementary figures 12, 14, and 15 (see Supplementary Material online).
To spot an eventual Cluster II duplication even earlier in the polyneopteran evolution, we next focused in detail on more basal Polyneoptera, especially Orthoptera (supplementary figs. 6 and 7, Supplementary Material online). With the exception of crickets (Laupala and Gryllus), all the basal polyneopterans possessed at least one Cluster II paralog (supplementary fig. 6, Supplementary Material online). All these observations indicate that Cluster II InR duplication might have taken place already in basal Polyneoptera, because paralogs belonging either to one or to the other Cluster II subclades are found in basal groups, and that subsequent losses of one or both Cluster II InRs occurred in individual lineages. Nevertheless, the low support of some InR assignments, evidenced also in the supplementary figure 7 (Supplementary Material online), calls for a more cautious interpretation of the exact dating of the Cluster II duplication in basal Polyneoptera.
InR Duplications in Hemiptera
In hemipteran orders, we identified a complex pattern of InR duplications. First, as shown in figure 1, all eight analyzed Gerromorpha contained a Cluster I paralog, manifested as a sister branch to the other Cluster I copy present in all Heteroptera. This paralog is an intronless gene, exclusively present in semiaquatic bugs (figs. 1 and 2;supplementary fig. 3, Supplementary Material online).
Another independent Cluster I duplication was recorded in two other Heteroptera, P. apterus and Largus californicus (both Pentatomorpha: Pyrrhocoroidea). As evidenced from the analysis of our genomic and transcriptomic data on P. apterus, both paralogs are very similar at the protein level and contain all characteristic domains (supplementary figs. 9 and 10, Supplementary Material online), but differ in the gene structure (fig. 2). Although the likely ancestral paralog (KX087104, hereafter InR1b) is coded by a gene containing at least 20 exons, the InR1a gene (KX087103) is intronless and probably originated through reverse transcription of InR1b.
A detailed view on the Cluster II revealed duplication events within Auchenorrhyncha, leading to InR multiplication in Cicadellidae (supplementary fig. 2, Supplementary Material online). Two Cluster II genes were found in the leafhoppers Graminella nigrifrons, Graphocephala atropunctata, and two species of Homalodisca, and as many as four in Euscelidius variegatus, suggesting up to three InR duplications in this clade.
Secondary Losses of Cluster II InR in Chalcid Wasps, Thrips, Fleas, and Psocodea
In five of the six studied Hymenoptera included in the main analysis, we recovered paralogs of both InR clusters (fig. 1), except for the solitary chalcid wasp N. vitripennis, lacking the Cluster II paralog in the genome and transcriptome. We then examined in more detail 3 genera of Symphyta and 12 Apocrita, including 5 Chalcidoidea. We retrieved Cluster I and Cluster II paralogs in all included species, with the exception of all five Chalcidoidea, in which the Cluster II gene was missing (supplementary fig. 11, Supplementary Material online). This supports the hypothesis on the secondary loss of Cluster II InR in Chalcidoidea.
Similarly, we did not detect any Cluster II InR in Psocodea (five transcriptomes and one genome analyzed), Thysanoptera (transcriptomes of two species), and Siphonaptera (transcriptomes of two species), suggesting secondary losses of the gene also in these orders (fig. 1 andsupplementary figs. 2‒4, Supplementary Material online).
Decoys of Insulin Receptor—InR-Like Genes without TK Domain
During our quest for InRs in Holometabola, we systematically encountered the genes and transcripts showing in approximately three-quarters of their translated sequence (from N-terminus) a high similarity with insulin receptors and clear separation from epidermal growth factor receptor (fig. 2). They consisted of most sequence features typical for InRs, that is, the signal peptide, two receptor L-domains surrounding one furin-like cysteine-rich domain, followed by several fibronectin type III domains, and in some cases a transmembrane region (TM) at the C-terminus. All of them were lacking the TK domain critical for signal transduction in canonical InR signaling cascade (fig. 2 andsupplementary figs. 9 and 10, Supplementary Material online). Phylogenetic analysis separated these proteins into two distinct groups. The first one was exclusive to flies and homologous to SDR (Okamoto et al. 2013). SDRs were clearly monophyletic and branched together within Diptera as a sister group to canonical InRs of flies inside the Cluster I (fig. 1 andsupplementary figs. 3 and 4, Supplementary Material online). The second monophylum lacking TK domain consisted of genes from Diptera, Lepidoptera, Trichoptera, and Mecoptera, and branched as a sister clade to Cluster II InRs of beetles (fig. 1 andsupplementary figs. 2 and 4, Supplementary Material online). Hereafter, we refer to this newly identified TK-less gene as decoy of insulin receptor 2 (DR2). The affiliation of DR2 protein sequences into the Cluster II is supported also by approximately unbiased test, which rejected the enforced monophyly of DR2 and Cluster I (P = 0.027).
To elucidate the origin of DR2 and SDR independently of the phylogenetic analysis, we compared the positions of exon–intron boundaries in selected dipteran and lepidopteran genes with ancestral InRs (fig. 2). The position of two introns is conserved between Apis mellifera Cluster II InR and all studied DR2. Vice versa, the origin of SDR from Cluster I InR is supported by two exon–intron boundaries conserved between D. melanogaster InR and SDR. Structural differences between the two receptor decoy clades can be found in certain sequence motifs, especially in the furin-like domain and in the presence of C-terminal TM region in all DR2s, contrasting with its absence in SDRs (fig. 2 andsupplementary figs. 9 and 10, Supplementary Material online).
We compared the expression of InR, SDR, and DR2 in somatic organs and gonads of adult D. melanogaster flies. The results, summarized in figure 3B, show that all three genes were expressed in all studied tissues of both sexes and had rather similar expression patterns with low values in the digestive tube and higher expression in the head and gonads, DR2 having the highest transcript abundance of the three genes. Detailed expression analysis and statistical comparisons are given in supplementary figure 12 (Supplementary Material online).
InR Multiplicity and Wing-Morph Determination in P. apterus
Having identified three InRs in the heteropteran model, the linden bug P. apterus (Heteroptera: Pyrrhocoridae) (supplementary figs. 2 and 3, Supplementary Material online), we decided to study experimentally their role. We first confirmed that all three InRs are expressed in the adult bugs and that the expression patterns differ among tissues, among the three InRs and in some cases also between the two sexes (fig. 3C). Detailed expression analysis and statistical comparisons are given in supplementary figure 13 (Supplementary Material online). We next explored the possible participation of the three InR variants and three identified ILPs in wing morph determination using a series of RNA interference (RNAi) experiments.
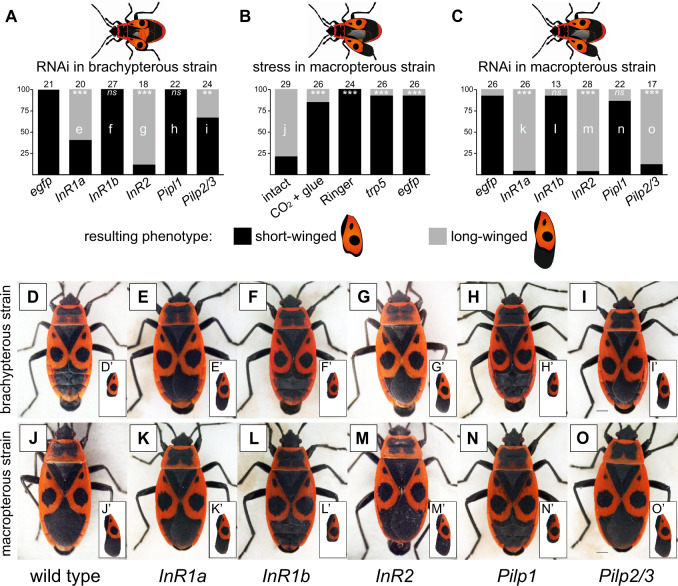
Pyrrhocoris apterus is a flightless species with usually miniature hind wings, short front wings, and underdeveloped flight muscles (brachypterous morph, short-winged phenotype, SW), although long-winged (LW) adults (macropterous morph) occasionally occur. In wild populations, the percentage of LW morphs depends on abiotic and biotic factors, such as temperature and population density (Socha 1993; Honěk 1995). Although flightless, both LW sexes show higher dispersal ability (Socha and Zemek 2003) and delayed female reproduction with lower number of egg batches laid (Socha 2013), altogether strongly resembling the migratory or dispersal morphs of other insects. We took advantage of two established inbred laboratory lines with opposite wing phenotypes, the brachypterous and the macropterous strain, and tested the antagonistic roles of Cluster I and Cluster II InRs in wing length regulation, postulated in planthoppers (Xu et al. 2015). The results of RNAi assays are given in figure 4. Injection of InR2 dsRNA to the fourth instar larvae of the brachypterous strain resulted in 89% of LW adults, whereas the control animals (nontreated or egfp dsRNA injected) were all SW. Similar trend was observed after knockdown of InR1a, where 60% animals became LW. By contrast, injection of InR1b dsRNA did not affect the resulting wing morphology when compared with controls. Thus, in spite of the close sequence identity of InR1a and InR1b paralogs (>75%), we succeeded in obtaining sufficient RNAi specificity for each gene (see supplementary fig. 14, Supplementary Material online) and observed a different phenotypic effect after their silencing.
Fig. 4.
Interaction of insulin signaling pathway, stress, and wing polymorphism in Pyrrhocoris apterus studied by means of RNAi experiments. One-day-old fourth instar P. apterus larvae of brachypterous or macropterous strain were subjected to the treatment, raised to adults, and the proportion of obtained short- and long-winged adult phenotypes was scored. (A) Brachypterous strain larvae injected with InR1a, InR1b, InR2, Pilp1 or Pilp2/3 dsRNA, or negative control egfp dsRNA. (B) Macropterous strain larvae subjected to stress stimuli, that is, handling (intact), CO2 anesthesia and immobilization (CO2 + glue), buffer injection (Ringer), injection of control dsRNA (trp5), and negative control dsRNA (egfp). (C) Macropterous strain larvae injected with InR1a, InR1b, InR2, Pilp1 or Pilp2/3 dsRNA, or negative control egfp dsRNA. (D‒O) Photographs of dominant phenotypes observed after different treatments for each of the two strains, that is, wild-type adult males (D and J), individuals injected with InR1a dsRNA (E and K), InR1b dsRNA (F and L), InR2 dsRNA (G and M), Pilp1 dsRNA (H and N), and Pilp2/3 dsRNA (I and O). Numbers above each column indicate numbers of evaluated adults in each treatment. Letters inside the columns (e‒o) refer to the photographs of representative phenotypes (E‒O). For each set of experiments (A, B, and C), the obtained proportions of long- versus short-winged adult phenotypes were compared with the control treatment (first column) using an equivalent of Dunnett’s test adjusted for proportion data (Zar 1999). *, **, and *** denote significant differences at P < 0.05, P < 0.01, and P < 0.001, respectively. Scale bars for all photographs indicate 1 mm. Insets show separate views on forewings of the given phenotype.
To further explore the role of IIS in wing development, we knocked down the expression of three P. apterus insulin-like peptides (Pilp’s) in fourth instar larvae of the brachypterous strain. Although the silencing of Pilp1 did not promote the development of LW phenotypes, the knock down of Pilp2/3 (Pilp2 and Pilp3 are 89% identical) induced 33% of LW adults (see supplementary fig. 15 for RNAi efficiency, Supplementary Material online).
Similar set of experiments was then performed with the macropterous strain, usually containing 85–90% LW adults (Honěk 1995). To our surprise, we observed a strong phenotypic effect of experimental manipulation on the wing morphology. The injection of nonspecific (egfp) or P. apterus-specific (trp5) control dsRNAs significantly decreased the proportion of LW adults, when compared with intact animals. Similar effect was observed for other stress-inducing manipulations, that is, the injection of Ringer’s buffer or even the CO2 anesthesia itself; both treatments strongly reduced the number of LW adults (fig. 4). Nevertheless, the RNAi silencing of InRs and Pilp’s successfully overrode the impact of stress and shifted the SW/LW proportions in similar manner as for the brachypterous strain, when compared with control injection treatments (egfp, trp5, Ringer): InR2, InR1a, and Pilp2/3 significantly increased the LW proportions, whereas InR1b and Pilp1 injection did not change the SW/LW ratio (fig. 4).
Discussion
Evolution of Insulin Receptors and Decoy of Insulin Receptors in Insects
Phylogenetic analysis of InR sequences in key animal phyla revealed that the ancestral InR duplication, leading to the presence of two InR genes in most insect taxa, took place at earliest in a late hexapod ancestor of Insecta, or within basal Insecta, as recently suggested by Kremer et al. (2018). Detailed analysis of 98 insects from 23 orders showed the presence of only Cluster I InR in wingless insects. At the same time, we recorded in the most basal-winged insects (Palaeoptera) the first occurrence of InRs from the Cluster II in three species of mayflies, including Ephemera danica, in which the two InRs were signaled previously (Armisén et al. 2018; Kremer et al. 2018). This leads us to propose that Cluster II InR originated from Cluster I InR in a late common ancestor of the extant Pterygota.
Gene structure comparison independently confirmed that Cluster II InR is a derived paralog from the original arthropod InR, because the intron-rich Cluster I InR in most studied insect taxa shares multiple structural features with the single InR identified in crustaceans, unlike the intron-poor Cluster II InR. Within the Cluster I, we observed a conspicuous reduction in intron number in the fruit fly and the honey bee, contrasting both, with basal insects and other, more advanced Holometabola. Mutation accumulation during the intron losses through double-strand break repair (Farlow et al. 2011; Rodgers and McVey 2016) thus may eventually explain the noncanonical branching of Hymenoptera in our phylogenetic reconstructions of Cluster I InRs.
Within winged insects, we highlighted several important evolutionary events in the two InR clusters, as summarized in figure 5. We identified duplication in the Cluster II in all termites, cockroaches, and Phasmatodea. This duplication was previously deemed as exclusive to Blattodea (termites and cockroaches) (Xu and Zhang 2017; Armisén et al. 2018; Kremer et al. 2018), and the arising InR multiplicity proposed to be co-opted for the control of termite polyphenism (Kremer et al. 2018). Although this function is likely, as can also be concluded from differential expression within and among the three InRs, which we recorded in different castes and tissues of termites (fig. 3A), our data challenge the dating of the Cluster II duplication. The presence of two Cluster II paralogs, and their comparable gene structure with two inverted gene repeats in all Phasmatodea, suggests that the duplication is roughly by 130 My older than Blattodea (Evangelista et al. 2019). Our data suggest that the Cluster II duplication might have occurred even earlier, because InRs from opposite subclades are found in Zoraptera versus Plecoptera as well as in some Orthoptera. According to the current phylogeny (Wipfler et al. 2019), the Cluster II duplication would thus be situated to very basal Polyneoptera. Yet, this assumption suffers a low sampling and branching support; additional data, especially for Dermaptera, may provide conclusive proof for the ancestral polyneopteran origin of the duplication.
Fig. 5.
Evolution of insect insulin receptor genes and decoy of insulin receptor genes. The tree represents InR gene duplication events and losses of the tyrosine kinase domain leading to decoy of insulin receptors, mapped on the simplified phylogenetic tree of insects according to Misof et al. (2014). The tree does not display the losses of complete InR genes, nor the duplications detected at the level of individual species, and it only shows selected taxa and lineages important for the understanding of InR evolution.
We recorded complex InR pattern in hemipteran orders. All representatives of semiaquatic bugs (Gerromorpha) showed a Cluster I duplication exclusive to this basal clade of Heteroptera. This duplication has been recently reported by Armisén et al. (2018); as shown by the authors, the newly arising InR gene is intronless, which indicates its likely origin as an RNA-based retroposed gene. Interestingly, the independent Cluster I duplication that we detected in two genera of another heteropteran lineage (Pyrrhocoroidea), also probably results from reverse transposition, giving rise to an intronless gene.
In another hemipteran lineage, Cicadellidae, we spotted one to two Cluster II duplications in several genera, leading to up to four InR copies. Thus, it appears that hemipteran orders are particularly prone to InR duplications, which may contribute to their phenotypic plasticity, including frequent cases of wing polyphenism (Armisén et al. 2018).
In apical lineages of Holometabola, we identified in both InR clusters the losses of TK domains, giving rise to decoys of insulin receptors. Within the Cluster I, we recorded an InR paralog lacking both the TK and the TM domains, previously described as SDR and only known in Drosophila (Okamoto et al. 2013), in all four included flies (Muscomorpha). Its high similarity with genuine Cluster I InR, also present in flies, suggests that InR duplication preceded the loss of the functional domains. In the Cluster II, another TK-less gene occurs in all representatives of the crown group Mecopterida (Diptera + Lepidoptera + Trichoptera + Mecoptera). We labeled this new gene as DR2 and unlike in SDR, we observed that it always possesses the TM domain. Both genes remained conserved for long evolutionary time, and especially the presence of DR2 spans over at least 300 My (Wiegmann et al. 2009; Misof et al. 2014). This suggests that these TK-less genes retained or gained functions in IIS pathway. Indeed, a functional significance has been attributed to SDR in Drosophila as a protein constantly secreted to the hemolymph and interacting with ILPs independently of insulin binding protein Imp-L2, acting thus as an antagonist of IIS signaling (Okamoto et al. 2013). By contrast, the putative function of the newly identified DR2 gene is elusive and remains to be addressed.
The ubiquitous nonnegligible expression and indications of tissue- and sex-specific regulation (fig. 3B) suggest that DR2 indeed has a functional role. Its origin and high-sequence similarity with Cluster II InRs indicate that just like SDR also DR2 might be able to bind insulin peptides. By contrast, due to the presence of the TM domain consisting of a hydrophobic α-helix at the C-terminal region, different mode of action might be expected for both proteins, because SDR lacks this domain. It would be interesting to see whether DR2 can eventually dimerize together with either InRs or SDR in vivo.
In addition to SDR and DR2, we also spotted a few individual cases of TK-less sequences, supplementing the Cluster II InRs (e.g., the beetle T. castaneum, supplementary fig. 16, Supplementary Material online). However, because these observations represent isolated cases at low taxonomic levels, it is difficult to discriminate them from assembly artifacts or pseudogenes. More importantly, decoy of InR may be, in principle, coded by full receptor gene and produced through alternative RNA splicing or other posttranscriptional modifications, such as RNA editing creating a premature stop codon. Notably, systematic search in mammalian cells identified 31 dominant-negative secreted variant isoforms of receptor TKs that are produced by activation of intronic poly(A) sites (Vorlova et al. 2011). It is interesting to note that TK-less InR paralog was documented also among the four InR genes in the genome of the crustacean D. pulex (Boucher et al. 2010).
Beside the duplications and transformations of InR genes into decoy receptors, discussed above and depicted in figure 5, several InR secondary losses have been recorded at higher taxonomic levels. Cluster II InR has not been retrieved in lice and other Psocodea, in the parasitoid chalcid wasps, thrips, and fleas. Even though the InR absence must be taken with caution (namely in the latter two taxa where only two genera were analyzed and no genomes were explored), it invites to speculate about the significance of eventual InR losses in the context of life strategies of these often parasitic and often wingless taxa.
Role of Insulin Receptor Multiplicity in Wing Polymorphism
Wing polymorphism is found in various insect taxa and drives the differential seasonal, population-specific, and nutrition-dependent dispersal modalities of these insects. Wing polymorphism can be determined exclusively at the genetic level (genetic polymorphism), result entirely from environmental variations (polyphenism), or, which is the most common situation, be controlled by a combination of both environmental and genetic factors (Harrison 1980; Roff 1986). Experimental evidence indicates that the mechanistic control of wing polymorphism does not have to be conserved even between related taxa. Within Hemiptera, ecdysone signaling is deemed to be the main wing polyphenism regulator in aphids, whereas the principal role of IIS was proposed in planthoppers (reviewed in Zhang et al. 2019). Remarkably, opposing effects of InR1 and InR2 activation in the development of SW and LW phenotypes was reported in three planthopper species, Nilaparvata lugens, Sogatella furcifera, and Laodelphax striatellus (Xu et al. 2015).
As we successfully used RNAi in P. apterus to study the metamorphosis (Smykal, Daimon, et al. 2014), reproduction (Smykal, Bajgar, et al. 2014; Urbanova et al. 2016), and reproductive diapause (Kotwica-Rolinska et al. 2017), we decided to use the reverse genetic techniques to elucidate the role of the three identified InRs in the control of wing polyphenism (figs. 4 and 6). Our experiments confirmed the role of IIS in wing morph determination, previously postulated in planthoppers (Xu et al. 2015; Xu and Zhang 2017). However, although the effect of Cluster II InRs (InR2) silencing in LW suppression appears as similar in P. apterus and Nil. lugens, the analogy is less clear for Cluster I orthologs (InR1) (fig. 6). Although the InR1 gene in Nil. lugens plays a perfectly antagonistic role to that of InR2, InR1a in P. apterus seems to have a role similar to InR2. Moreover, structurally close InR1b, from which InR1a likely evolved through reverse transcription, does not affect the wing development. This observation challenges the argument by Xu et al. (2015) and Xu and Zhang (2017) that the distinct actions of InR1 and InR2 proteins are defined by sequence motifs in the furin-like cysteine-rich domain specific to Cluster I and Cluster II InRs, respectively. In fact, these cluster-specific motifs are conserved and clearly distinguishable in all insect InRs and are even further conserved in SDR and DR2 (fig. 2 andsupplementary fig. 9, Supplementary Material online). However, in case of P. apterus, InRs representing both clusters of InR evolution have analogous effects on LW growth (InR2 and InR1a).
Fig. 6.
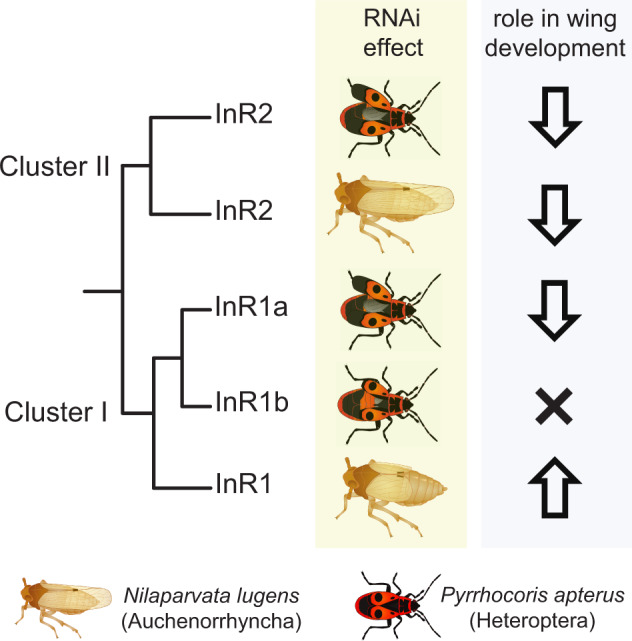
Scheme summarizing the effects of RNAi-mediated gene silencing of insulin receptor genes and inferred roles of individual paralogs in the control of wing polyphenism in the linden bug Pyrrhocoris apterus, compared with the observations by Xu et al. (2015) in the planthopper Nilaparvata lugens.
At the same time, our study points at an evolutionarily conserved role of stress in wing polyphenism. Even mere manipulation, including CO2 anesthesia necessary during injections of P. apterus nymphs, leads to SW morphology. The resulting experimental limitations are in line with observations that wounding during nymphal stages increases the SW ratio in Nil. lugens (Lin et al. 2016) and indicate the underlying complexity of wing development interactions with environmental cues, reported in various insect species. These cues include, among others, the food quality, as shown for planthoppers, in which the rising glucose concentrations increase the proportions of LW phenotypes synergistically with population density (Lin et al. 2018).
In conclusion, our results highlight the versatile function of InR multiplicity, which is to a large extent independent of phylogenetic constraints and is manifested by idiosyncrasies in IIS use in individual taxa. This raises questions on the possible role of multiple (up to five genes in some Cicadellidae) InRs in the control of other remarkable cases of polyphenism and developmental peculiarities, such as the extreme variability of anatomical novelties in treehoppers, or extremely long, yet tightly regulated development in “periodical cicadas” (Cicadellidae: Magicicada), where populations with either 13- or 17-year life cycle coexist within three species groups (Du et al. 2019).
Materials and Methods
Selection of Taxa and Data Sets for the Reconstruction of InRs and Decoy of InRs Evolution
We conducted multiple phylogenetic analyses to respond a series of different questions. First, to verify the monophyletic origin of insect InRs, we compared the InR sequences in a subset of 48 insect species and 19 noninsect representatives across Animalia.
Second, the main phylogenetic analysis of InRs and decoys of InRs within Insecta was designed so as to include a maximum of insect orders. As the taxonomic coverage of genomic and transcriptomic data is largely uneven across insect phylogeny (Li et al. 2019), we performed taxon-specific searches of InR sequences, followed by their manual curation. Instead of using a universal inclusion criteria based on overall genome or transcriptome parameters, we included the sequencing projects in which the detected InR sequence(s) covered the desired alignment span and coded for “diagnostic” protein domains (see below) and retrieved the candidate species to balance the coverage of individual insect orders and avoid multiple entries of congeneric species. The resulting data set of 98 species from 23 insect orders was used for the main analysis.
And third, subsequent more detailed analyses of selected insect lineages of particular interest included additional 20 species of Orthoptera, Phasmatodea, Blattodea, and Hymenoptera, including those for which only partial sequences were available. The analyzed taxa, together with accession numbers for identified InR and decoy of InR genes, are listed in supplementary tables 1 and 2 (Supplementary Material online). Full sequences and alignments are available in Dryad data repository under 10.5061/dryad.4xgxd255n.
Data Origin, Identification of InR, and Decoy of InR Genes
GenBank was BLAST examined for InR and InR homologs in taxon-specific searches through NCBI web page interface. In addition, genome drafts of Blatella germanica, E. danica, and Homalodisca vitripennis were downloaded from the database of I5k genome sequencing initiative (http://i5k.github.io/arthropod_genomes_at_ncbi) and prospected using BLAST algorithm in Geneious program (Biomatters, Ltd.). In case of Lepisma saccharina, de novo assembly of transcriptomes was performed using Trinity algorithm from sequence read archive available at NCBI (https://www.ncbi.nlm.nih.gov/sra) and the resulting database was subsequently explored with BLAST in Geneious. Embiratermes neotenicus, Pro. simplex, and P. apterus InRs were retrieved from in-house transcriptomic databases assembled using Illumina reads with Trinity algorithm, verified, and completed by Sanger sequencing and PCR using primers specific to each individual paralog (supplementary table 3, Supplementary Material online). In the latter two species, InR multiplicity and sequence accuracy were independently confirmed and the gene structures reconstructed from draft genome assemblies based on Oxford Nanopore sequencing of genomic DNA.
All retrieved protein sequences were explored for the presence of characteristic InR features (receptor L-domain, furin-like cysteine-rich domain, fibronectin type III domain, and TK domain) using InterProScan (http://www.ebi.ac.uk/interpro/search/sequence-search), transmembrane domains were predicted on TMHMM Server v. 2.0 (http://www.cbs.dtu.dk/services/TMHMM/) and corresponding protein schemes were depicted in Adobe Illustrator. Fast tree and RAxML phylogenetic analyses of Clustal W alignments (both in Geneious) and manual scoring for the presence of characteristic domain modules were used to classify a protein as a candidate for InR or decoy of insulin receptor, which differ from InR only by absence of TK domain.
Sequence Alignment and Phylogenetic Analysis of InRs and Decoy of InR Genes
In the first analysis, aiming at verification of monophyletic origin of insect InRs, 67 animal InR protein sequences were aligned using MAFFT v7.308 (Katoh and Standley 2013) with the E-INS-i multiple alignment method and BLOSUM80 scoring matrix. Poorly aligned regions were identified visually and removed. The resulting alignment (available as InR_AllPhyla-SI-alignment in the Dryad data repository under 10.5061/dryad.4xgxd255n) was used for ML analysis in RAxML ver. 8.2.12 (Stamatakis 2014) with 500 bootstrap replicates under the best fitting WAG + Γ model.
For the main phylogenetic analysis of InRs and decoy of InRs evolution within Insecta, 205 protein sequences were aligned (MAFFT v7.308, E-INS-i multiple alignment method, BLOSUM80). Spurious and poorly aligned regions were removed from the alignment by trimAl (http://trimal.cgenomics.org/) using “gappyout” parameter (available as 205einsi.gappy—length1301 in the Dryad data repository under 10.5061/dryad.4xgxd255n). ML analyses were performed in RAxML (WAG + Γ model, 500 bootstrap replicates). Bayesian analysis was performed in PhyloBayes v4.1 (Lartillot et al. 2009) using the CAT+GTR model of evolution (Lartillot and Philippe 2004). Two intertwined chains were run in parallel and the run stopped once all discrepancies were ≤0.2 and all effective sizes >100. Burn-in period was equal to one fifth of the total chain length. To test the robustness of the affiliation of DR2 into the Cluster II, we constrained the monophyly of DR2 and Cluster I InR and reoptimized the topology in RAxML under the parameters described earlier. Per-site log likelihood values were then estimated for both original and alternative topologies in RAxML. Statistical significance of observed differences was then tested using the approximately unbiased test (Shimodaira 2004) as implemented in CONSEL (Shimodaira and Hasegawa 2001; Shimodaira 2004).
In the subsequent detailed analyses of selected insect lineages, sequences were aligned (MAFFT v7.308, E-INS-i multiple alignment method, BLOSUM80), poorly aligned regions identified visually and removed (alignments accessible in the Dryad repository under 10.5061/dryad.4xgxd255n). ML analyses were performed in RAxML (WAG + Γ model, 500 bootstrap replicates). For analysis of Orthopteran InRs, Bayesian analysis was conducted in MrBayes v3.0 (Ronquist and Huelsenbeck 2003) using WAG + Γ model of evolution. Posterior probabilities were estimated over 1,000,000 generations via two independent runs of four simultaneous Markov chain Monte Carlo calculations with every 100th tree saved. Tracer v1.6 (Rambaut and Drummond 2007) was used to set the length of the burn-in period (20%).
Expression of InRs in Termites, Fruit Flies, and Linden Bug
Expression profiles of three InRs in different castes and tissues of the termite Pro. simplex, one InR and two decoy of insulin receptors (SDR and DR2) in different organs of adult males and females in the fruit fly D. melanogaster, and three InRs of adult male and female linden bugs P. apterus were deciphered using qPCR as described in detail in supplementary materials and methods (Supplementary Material online).
Role of Multiple InRs in Wing Polyphenism of the Linden Bug, P. apterus
Pyrrhocoris apterus forewing has two melanized spots on each side, a small one and a more distal large one. The latter one is separated by the red part of the corium from the membrane. The SW phenotype is defined as having the membrane length equal or shorter than the red “separating” corium. The LW or macropterous phenotype is characteristic by the forewing membrane reaching the tip of the abdomen and representing about one-third of the wing length. In our experiments, we classified the adults as LW if at least one forewing membrane was more than twice as long as the red corium between the large spot and the membrane. All other adults were scored as SW.
Two P. apterus strains were studied. Brachypterous strain Oldrichovec (Pivarciova et al. 2016) and Czech Macropterous Line strain (Honěk 1995; Socha 2013). Typically, 99% adults are SW in Oldrichovec strain and 85–90% adults of Czech Macropterous Line strain are LW. Bugs were kept in 0.5 l glass jars at density of 30–40 individuals per jar with linden seeds and water ad libitum in long photoperiod (18 h light:6 h dark) at 25 °C.
The three InR genes and three Pilp’s genes identified in P. apterus were functionally tested by RNAi with respect to their impact on wing development. Technical details of the RNAi experiment design and statistics are given in supplementary materials and methods (Supplementary Material online).
Supplementary Material
Acknowledgments
We thank James Davies for critical reading of the article and Martina Hajdušková (www.biographix.cz) for insect schemes used in figures 3‒6. We thank Helena Štěrbová for the P. apterus MLC strain and Marek Jindra for the injection facility. This work was supported by the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation program (Grant Agreement No. 726049). R.H., O.L., and P.J. were supported by the Institute of Organic Chemistry and Biochemistry, Czech Academy of Sciences (RVO 61388963), and by the Czech Science Foundation (18-21200S). O.B. and J.P were supported by the Czech Science Foundation (17-01003S and 15-23681S, respectively).
Author Contributions
D.D. designed the study. M.P., J.P., O.L., A.H., and I.F. performed the phylogenetic analysis and sequence assemblies. V.S. performed P. apterus functional experiments with help from H.V. P.J. and O.B. determined expression levels by qRT PCR. D.D. and R.H. wrote the article.
Full sequences and alignments used for phylogenetic reconstructions are available in Dryad data repository under 10.5061/dryad.4xgxd255n. InR sequences newly generated from transcriptomes or genomes of Pyrrhocoris apterus, Prorhinotermes simplex, and Embiratermes neotenicus were deposited in GenBank (for accession numbers, see fig. 2 and Supplementary Material online); complete assembled transcriptomes and genomes will be published elsewhere.
References
- Abrisqueta M, Suren-Castillo S, Maestro JL.. 2014. Insulin receptor-mediated nutritional signalling regulates juvenile hormone biosynthesis and vitellogenin production in the German cockroach. Insect Biochem Mol Biol. 49:14–23. [DOI] [PubMed] [Google Scholar]
- Ament SA, Corona M, Pollock HS, Robinson GE.. 2008. Insulin signaling is involved in the regulation of worker division of labor in honey bee colonies. Proc Natl Acad Sci U S A. 105(11):4226–4231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armisén D, Rajakumar R, Friedrich M, Benoit JB, Robertson HM, Panfilio KA, Ahn S-J, Poelchau MF, Chao H, Dinh H, et al. 2018. The genome of the water strider Gerris buenoi reveals expansions of gene repertoires associated with adaptations to life on the water. BMC Genomics 19(1):832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badisco L, Claeys I, Van Hiel M, Clynen E, Huybrechts J, Vandersmissen T, Van Soest S, Vanden Bosch L, Simonet G, Vanden Broeck J.. 2008. Purification and characterization of an insulin-related peptide in the desert locust, Schistocerca gregaria: immunolocalization, cDNA cloning, transcript profiling and interaction with neuroparsin. J Mol Endocrinol. 40(3):137–150. [DOI] [PubMed] [Google Scholar]
- Badisco L, Van Wielendaele P, Vanden Broeck J.. 2013. Eat to reproduce: a key role for the insulin signaling pathway in adult insects. Front Physiol. 4:202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai H, Kang P, Tatar M.. 2012. Drosophila insulin-like peptide-6 (dilp6) expression from fat body extends lifespan and represses secretion of Drosophila insulin-like peptide-2 from the brain. Aging Cell 11(6):978–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boucher P, Ditlecadet D, Dube C, Dufresne F.. 2010. Unusual duplication of the insulin-like receptor in the crustacean Daphnia pulex. BMC Evol Biol. 10(1):305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brogiolo W, Stocker H, Ikeya T, Rintelen F, Fernandez R, Hafen E.. 2001. An evolutionarily conserved function of the Drosophila insulin receptor and insulin-like peptides in growth control. Curr Biol. 11(4):213–221. [DOI] [PubMed] [Google Scholar]
- Broughton SJ, Piper MDW, Ikeya T, Bass TM, Jacobson J, Driege Y, Martinez P, Hafen E, Withers DJ, Leevers SJ, et al. 2005. Longer lifespan, altered metabolism, and stress resistance in Drosophila from ablation of cells making insulin-like ligands. Proc Natl Acad Sci U S A. 102(8):3105–3110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casasa S, Moczek AP.. 2018. Insulin signalling’s role in mediating tissue-specific nutritional plasticity and robustness in the horn-polyphenic beetle Onthophagus taurus. Proc R Soc B. 285(1893):20181631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Jack J, Garofalo RS.. 1996. The Drosophila insulin receptor is required for normal growth. Endocrinology 137(3):846–856. [DOI] [PubMed] [Google Scholar]
- Clancy DJ, Gems D, Harshman LG, Oldham S, Stocker H, Hafen E, Leevers SJ, Partridge L.. 2001. Extension of life-span by loss of CHICO, a Drosophila insulin receptor substrate protein. Science 292(5514):104–106. [DOI] [PubMed] [Google Scholar]
- Colombani J, Andersen DS, Boulan L, Boone E, Romero N, Virolle V, Texada M, Leopold P.. 2015. Drosophila Lgr3 couples organ growth with maturation and ensures developmental stability. Curr Biol. 25(20):2723–2729. [DOI] [PubMed] [Google Scholar]
- Corona M, Libbrecht R, Wheeler DE.. 2016. Molecular mechanisms of phenotypic plasticity in social insects. Curr Opin Insect Sci. 13:55–60. [DOI] [PubMed] [Google Scholar]
- de Azevedo SV, Hartfelder K.. 2008. The insulin signaling pathway in honey bee (Apis mellifera) caste development – differential expression of insulin-like peptides and insulin receptors in queen and worker larvae. J Insect Physiol. 54(6):1064–1071. [DOI] [PubMed] [Google Scholar]
- Ding B-Y, Shang F, Zhang Q, Xiong Y, Yang Q, Niu J-Z, Smagghe G, Wang J-J.. 2017. Silencing of two insulin receptor genes disrupts nymph-adult transition of alate brown citrus aphid. Int J Mol Sci. 18(2):357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Z, Hasegawa H, Cooley JR, Simon C, Yoshimura J, Cai W, Sota T, Li H.. 2019. Mitochondrial genomics reveals shared phylogeographic patterns and demographic history among three periodical cicada species groups. Mol Biol Evol. 36(6):1187–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emlen DJ, Warren IA, Johns A, Dworkin I, Lavine LC.. 2012. A mechanism of extreme growth and reliable signaling in sexually selected ornaments and weapons. Science 337(6096):860–864. [DOI] [PubMed] [Google Scholar]
- Erion R, Sehgal A.. 2013. Regulation of insect behavior via the insulin-signaling pathway. Front Physiol. 4:353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evangelista DA, Wipfler B, Béthoux O, Donath A, Fujita M, Kohli MK, Legendre F, Liu S, Machida R, Misof B, et al. 2019. An integrative phylogenomic approach illuminates the evolutionary history of cockroaches and termites (Blattodea). Proc R Soc B. 286(1895):20182076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farlow A, Meduri E, Schlötterer C.. 2011. DNA double-strand break repair and the evolution of intron density. Trends Genet. 27(1):1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannakou ME, Partridge L.. 2007. Role of insulin-like signalling in Drosophila lifespan. Trends Biochem Sci. 32(4):180–188. [DOI] [PubMed] [Google Scholar]
- Guo S-S, Zhang M, Liu T-X.. 2016. Insulin-related peptide 5 is involved in regulating embryo development and biochemical composition in pea aphid with wing polyphenism. Front Physiol. 7:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison MC, Jongepier E, Robertson HM, Arning N, Bitard-Feildel T, Chao H, Childers CP, Dinh H, Doddapaneni H, Dugan S, et al. 2018. Hemimetabolous genomes reveal molecular basis of termite eusociality. Nat Ecol Evol. 2(3):557–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison RG. 1980. Dispersal polymorphisms in insects. Annu Rev Ecol Syst. 11(1):95–118. [Google Scholar]
- Hattori A, Sugime Y, Sasa C, Miyakawa H, Ishikawa Y, Miyazaki S, Okada Y, Cornette R, Lavine LC, Emlen DJ, et al. 2013. Soldier morphogenesis in the damp-wood termite is regulated by the insulin signaling pathway. J Exp Zool Mol Dev Evol. 320(5):295–306. [DOI] [PubMed] [Google Scholar]
- Hernandez-Sanchez C, Mansilla A, de Pablo F, Zardoya R.. 2008. Evolution of the insulin receptor family and receptor isoform expression in vertebrates. Mol Biol Evol. 25(6):1043–1053. [DOI] [PubMed] [Google Scholar]
- Hetru C, Li KW, Bulet P, Lagueux M, Hoffmann JA.. 1991. Isolation and structural characterization of an insulin-related molecule, a predominant neuropeptide from Locusta migratoria. Eur J Biochem. 201(2):495–499. [DOI] [PubMed] [Google Scholar]
- Honěk A. 1995. Factors and consequences of a nonfunctional alary polymorphism in Pyrrhocoris apterus (Heteroptera, Pyrrhocoridae). Res Popul Ecol. 37(1):111–118. [Google Scholar]
- Jedlička P, Ernst UR, Votavová A, Hanus R, Valterová I.. 2016. Gene expression dynamics in major endocrine regulatory pathways along the transition from solitary to social life in a bumblebee. Front Physiol. 7:574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junger MA, Rintelen F, Stocker H, Wasserman JD, Vegh M, Radimerski T, Greenberg ME, Hafen E.. 2003. The Drosophila forkhead transcription factor FOXO mediates the reduction in cell number associated with reduced insulin signaling. J Biol. 2:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannan K, Fridell Y-W.. 2013. Functional implications of Drosophila insulin-like peptides in metabolism, aging, and dietary restriction. Front Physiol. 4:288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM.. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo H, Ino M, Suzuki A, Ishizaki H, Iwami M.. 1996. Multiple gene copies for bombyxin, an insulin-related peptide of the silkmoth Bombyx mori: structural signs for gene rearrangement and duplication responsible for generation of multiple molecular forms of bombyxin. J Mol Biol. 259(5):926–937. [DOI] [PubMed] [Google Scholar]
- Kotwica-Rolinska J, Pivarciova L, Vaneckova H, Dolezel D.. 2017. The role of circadian clock genes in the photoperiodic timer of the linden bug Pyrrhocoris apterus during the nymphal stage. Physiol Entomol. 42(3):266–273. [Google Scholar]
- Koyama T, Mendes C, Mirth C.. 2013. Mechanisms regulating nutrition-dependent developmental plasticity through organ-specific effects in insects. Front Physiol. 4: 263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremer LPM, Korb J, Bornberg-Bauer E.. 2018. Reconstructed evolution of insulin receptors in insects reveals duplications in early insects and cockroaches. J Exp Zool Mol Dev Evol. 330(5):305–311. [DOI] [PubMed] [Google Scholar]
- Lartillot N, Lepage T, Blanquart S.. 2009. PhyloBayes 3: a Bayesian software package for phylogenetic reconstruction and molecular dating. Bioinformatics 25(17):2286–2288. [DOI] [PubMed] [Google Scholar]
- Lartillot N, Philippe H.. 2004. A Bayesian mixture model for across-site heterogeneities in the amino-acid replacement process. Mol Biol Evol. 21(6):1095–1109. [DOI] [PubMed] [Google Scholar]
- Lavine LC, Hahn LL, Warren IA, Garczynski SF, Dworkin I, Emlen DJ.. 2013. Cloning and characterization of an mRNA encoding an insulin receptor from the horned scarab beetle Onthophagus nigriventris (Coleoptera: Scarabaeidae). Arch Insect Biochem Physiol. 82(1):43–57. [DOI] [PubMed] [Google Scholar]
- Li F, Zhao X, Li M, He K, Huang C, Zhou Y, Li Z, Walters JR.. 2019. Insect genomes: progress and challenges. Insect Mol Biol. 28(6):739–758. [DOI] [PubMed] [Google Scholar]
- Lin X, Xu Y, Jiang J, Lavine M, Lavine LC.. 2018. Host quality induces phenotypic plasticity in a wing polyphenic insect. Proc Natl Acad Sci U S A. 115(29):7563–7568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X, Yao Y, Wang B, Lavine MD, Lavine LC.. 2016. FOXO links wing form polyphenism and wound healing in the brown planthopper, Nilaparvata lugens. Insect Biochem Mol Biol. 70:24–31. [DOI] [PubMed] [Google Scholar]
- Lu H-L, Pietrantonio PV.. 2011. Insect insulin receptors: insights from sequence and caste expression analyses of two cloned hymenopteran insulin receptor cDNAs from the fire ant. Insect Mol Biol. 20(5):637–649. [DOI] [PubMed] [Google Scholar]
- Maruyama IN. 2014. Mechanisms of activation of receptor tyrosine kinases: monomers or dimers. Cells 3(2):304–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misof B, Liu S, Meusemann K, Peters RS, Donath A, Mayer C, Frandsen PB, Ware J, Flouri T, Beutel RG, et al. 2014. Phylogenomics resolves the timing and pattern of insect evolution. Science 346(6210):763–767. [DOI] [PubMed] [Google Scholar]
- Nassel DR, Liu Y, Luo J.. 2015. Insulin/IGF signaling and its regulation in Drosophila. Gen Comp Endocrinol. 221:255–266. [DOI] [PubMed] [Google Scholar]
- Nijhout HF, McKenna KZ.. 2018. The distinct roles of insulin signaling in polyphenic development. Curr Opin Insect Sci. 25:58–64. [DOI] [PubMed] [Google Scholar]
- Okada Y, Katsuki M, Okamoto N, Fujioka H, Okada K.. 2019. A specific type of insulin-like peptide regulates the conditional growth of a beetle weapon. PLoS Biol. 17(11):e3000541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto N, Nakamori R, Murai T, Yamauchi Y, Masuda A, Nishimura T.. 2013. A secreted decoy of InR antagonizes insulin/IGF signaling to restrict body growth in Drosophila. Genes Dev. 27(1):87–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan J, Di YQ, Li YB, Chen CH, Wang JX, Zhao XF.. 2018. Insulin and 20-hydroxyecdysone oppose each other in the regulation of phosphoinositide-dependent kinase-1 expression during insect pupation. J Biol Chem. 293(48):18613–18623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pivarciova L, Vaneckova H, Provaznik J, Wu BC, Pivarci M, Peckova O, Bazalova O, Cada S, Kment P, Kotwica-Rolinska J, et al. 2016. Unexpected geographic variability of the free running period in the linden bug Pyrrhocoris apterus. J Biol Rhythms. 31(6):568–576. [DOI] [PubMed] [Google Scholar]
- Rambaut A, Drummond AJ.. 2007. Tracer v1.6. Available from: http://beast.bio.ed.ac.uk/Tracer.
- Rodgers K, McVey M.. 2016. Error-prone repair of DNA double-strand breaks. J Cell Physiol. 231(1):15–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roff DA. 1986. The evolution of wing dimorphism in insects. Evolution 40(5):1009–1020. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP.. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19(12):1572–1574. [DOI] [PubMed] [Google Scholar]
- Sang M, Li C, Wu W, Li B.. 2016. Identification and evolution of two insulin receptor genes involved in Tribolium castaneum development and reproduction. Gene 585(2):196–204. [DOI] [PubMed] [Google Scholar]
- Shimodaira H. 2004. Approximately unbiased tests of regions using multistep-multiscale bootstrap resampling. Ann Stat. 32(6):2616–2641. [Google Scholar]
- Shimodaira H, Hasegawa M.. 2001. CONSEL: for assessing the confidence of phylogenetic tree selection. Bioinformatics 17(12):1246–1247. [DOI] [PubMed] [Google Scholar]
- Sim C, Denlinger D.. 2013. Insulin signaling and the regulation of insect diapause. Front Physiol. 4:189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sim C, Denlinger DL.. 2008. Insulin signaling and FOXO regulate the overwintering diapause of the mosquito Culex pipiens. Proc Natl Acad Sci U S A. 105(18):6777–6781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smykal V, Bajgar A, Provaznik J, Fexova S, Buricova M, Takaki K, Hodkova M, Jindra M, Dolezel D.. 2014. Juvenile hormone signaling during reproduction and development of the linden bug, Pyrrhocoris apterus. Insect Biochem Mol Biol. 45:69–76. [DOI] [PubMed] [Google Scholar]
- Smykal V, Daimon T, Kayukawa T, Takaki K, Shinoda T, Jindra M.. 2014. Importance of juvenile hormone signaling arises with competence of insect larvae to metamorphose. Dev Biol. 390(2):221–230. [DOI] [PubMed] [Google Scholar]
- Snell-Rood EC, Moczek AP.. 2012. Insulin signaling as a mechanism underlying developmental plasticity: the role of FOXO in a nutritional polyphenism. PLoS One 7(4):e34857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Socha R. 1993. Pyrrhocoris apterus (Heteroptera) – an experimental model species: a review. Eur J Entomol. 90:241–286. [Google Scholar]
- Socha R. 2013. Do long- and short-winged adult females of the bug Pyrrhocoris apterus (Heteroptera: Pyrrhocoridae) differ in lifespan and reproductive capacity? Eur J Entomol. 110(1):115–121. [Google Scholar]
- Socha R, Zemek R.. 2003. Wing morph-related differences in the walking pattern and dispersal in a flightless bug, Pyrrhocoris apterus (Heteroptera). Oikos 100(1):35–42. [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30(9):1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatar M, Kopelman A, Epstein D, Tu MP, Yin CM, Garofalo RS.. 2001. A mutant Drosophila insulin receptor homolog that extends life-span and impairs neuroendocrine function. Science 292(5514):107–110. [DOI] [PubMed] [Google Scholar]
- Terrapon N, Li C, Robertson HM, Ji L, Meng X, Booth W, Chen Z, Childers CP, Glastad KM, Gokhale K, et al. 2014. Molecular traces of alternative social organization in a termite genome. Nat Commun. 5(1):3636. [DOI] [PubMed] [Google Scholar]
- Urbanova V, Bazalova O, Vaneckova H, Dolezel D.. 2016. Photoperiod regulates growth of male accessory glands through juvenile hormone signaling in the linden bug, Pyrrhocoris apterus. Insect Biochem Mol Biol. 70:184–190. [DOI] [PubMed] [Google Scholar]
- Vafopoulou X, Steel C.. 2014. Synergistic induction of the clock protein PERIOD by insulin-like peptide and prothoracicotropic hormone in Rhodnius prolixus (Hemiptera): implications for convergence of hormone signaling pathways. Front Physiol. 5:41–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitali V, Horn F, Catania F.. 2018. Insulin-like signaling within and beyond metazoans. Biol Chem. 399(8):851–857. [DOI] [PubMed] [Google Scholar]
- Vogel KJ, Brown MR, Strand MR.. 2013. Phylogenetic investigation of Peptide hormone and growth factor receptors in five dipteran genomes. Front Endocrinol. 4:193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vorlova S, Rocco G, Lefave CV, Jodelka FM, Hess K, Hastings ML, Henke E, Cartegni L.. 2011. Induction of antagonistic soluble decoy receptor tyrosine kinases by intronic polyA activation. Mol Cell. 43:927–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Azevedo SV, Hartfelder K, Amdam GV.. 2013. Insulin-like peptides (AmILP1 and AmILP2) differentially affect female caste development in the honey bee (Apis mellifera L.). J Exp Biol. 216(23):4347–4357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen Z, Gulia M, Clark KD, Dhara A, Crim JW, Strand MR, Brown MR.. 2010. Two insulin-like peptide family members from the mosquito Aedes aegypti exhibit differential biological and receptor binding activities. Mol Cell Endocrinol. 328(1–2):47–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler DE, Buck N, Evans JD.. 2006. Expression of insulin pathway genes during the period of caste determination in the honey bee, Apis mellifera. Insect Mol Biol. 15(5):597–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiegmann BM, Trautwein MD, Kim J-W, Cassel BK, Bertone MA, Winterton SL, Yeates DK.. 2009. Single-copy nuclear genes resolve the phylogeny of the holometabolous insects. BMC Biol. 7(1):34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wipfler B, Letsch H, Frandsen PB, Kapli P, Mayer C, Bartel D, Buckley TR, Donath A, Edgerly-Rooks JS, Fujita M, et al. 2019. Evolutionary history of Polyneoptera and its implications for our understanding of early winged insects. Proc Natl Acad Sci U S A. 116(8):3024–3029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Q, Brown MR.. 2006. Signaling and function of insulin-like peptides in insects. Annu Rev Entomol. 51(1):1–24. [DOI] [PubMed] [Google Scholar]
- Xu HJ, Xue J, Lu B, Zhang XC, Zhuo JC, He SF, Ma XF, Jiang YQ, Fan HW, Xu JY, et al. 2015. Two insulin receptors determine alternative wing morphs in planthoppers. Nature 519(7544):464–467. [DOI] [PubMed] [Google Scholar]
- Xu HJ, Zhang CX.. 2017. Insulin receptors and wing dimorphism in rice planthoppers. Philos Trans R Soc B. 372(1713):20150489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You H, Zhang W, Jones MK, Gobert GN, Mulvenna J, Rees G, Spanevello M, Blair D, Duke M, Brehm K, et al. 2010. Cloning and characterisation of Schistosoma japonicum insulin receptors. PLoS One 5(3):e9868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zar JH. 1999. Biostatistical analysis. New Jersey: Prentice Hall, Inc. [Google Scholar]
- Zhang CX, Brisson JA, Xu HJ.. 2019. Molecular mechanisms of wing polymorphism in insects. Annu Rev Entomol. 64(1):297–314. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.