Abstract
Objective
To define the uniqueness of chest CT infiltrative features associated with COVID-19 image characteristics as potential diagnostic biomarkers.
Methods
We retrospectively collected chest CT exams including n = 498 on 151 unique patients RT-PCR positive for COVID-19 and n = 497 unique patients with community-acquired pneumonia (CAP). Both COVID-19 and CAP image sets were partitioned into three groups for training, validation, and testing respectively. In an attempt to discriminate COVID-19 from CAP, we developed several classifiers based on three-dimensional (3D) convolutional neural networks (CNNs). We also asked two experienced radiologists to visually interpret the testing set and discriminate COVID-19 from CAP. The classification performance of the computer algorithms and the radiologists was assessed using the receiver operating characteristic (ROC) analysis, and the nonparametric approaches with multiplicity adjustments when necessary.
Results
One of the considered models showed non-trivial, but moderate diagnostic ability overall (AUC of 0.70 with 99% CI 0.56–0.85). This model allowed for the identification of 8–50% of CAP patients with only 2% of COVID-19 patients.
Conclusions
Professional or automated interpretation of CT exams has a moderately low ability to distinguish between COVID-19 and CAP cases. However, the automated image analysis is promising for targeted decision-making due to being able to accurately identify a sizable subsect of non-COVID-19 cases.
Key Points
• Both human experts and artificial intelligent models were used to classify the CT scans.
• ROC analysis and the nonparametric approaches were used to analyze the performance of the radiologists and computer algorithms.
• Unique image features or patterns may not exist for reliably distinguishing all COVID-19 from CAP; however, there may be imaging markers that can identify a sizable subset of non-COVID-19 cases.
Keywords: COVID-19, Biomarkers, Pneumonia, Neural network
Introduction
The novel coronavirus disease (COVID-19) affects a large portion of the world population and has been associated with over one hundred thousand deaths worldwide. The limited understanding of this disease has contributed to the failure to contain its impact. Recent studies have described the characteristics of COVID-19 on chest CT images. The primary findings of lung infiltrates present such patterns as peripheral distribution, ground-glass opacity, vascular thickness, and pleural effusion [1–7]. Chest CT scan has demonstrated high sensitivity (~ 98%) [8] in diagnosing COVID-19 in a screening setting; however, the specificity has been low (~ 25% [9]) in distinguishing COVID-19 infiltrates from other diseases associated with lung infiltrates. Therefore, the effort to discover image characteristics or biomarkers that can improve the CT scan specificity would significantly contribute to the clinical management of patients with or under suspicion for COVID-19.
Bai et al [10] investigated the performance of radiologists to differentiate COVID-19 (n = 219) from viral pneumonia (n = 205) on chest CT scans. They reported that the radiologists had moderate sensitivity in distinguishing COVID-19 from viral pneumonia. Several reports found that COVID-19 pneumonia was more likely to have a peripheral distribution (80% vs. 57%, p < 0.001), ground-glass opacity (91% vs. 68%, p < 0.001), fine reticular opacity (56% vs. 22%, p < 0.001), and vascular thickening (59% vs. 22%, p < 0.001) [5, 7, 11–13] as compared with non-COVID-19 pneumonia [10]. These observations suggest that the frequencies of specific infiltrative patterns of COVID-19 are similar to non-COVID-19 pneumonia, but the existence of specific image patterns uniquely associated with COVID-19 has not been established. A preliminary analysis by Li et al [14] supported a potential role for deep learning in discriminating COVID-19 from community-acquired pneumonia (CAP).
To determine whether unique image biomarkers can distinguish COVID-19 infiltrates from other types of pneumonia, we developed several three-dimensional (3D) convolutional neural network (CNN) models to classify CT scans from subjects with COVID-19 and CAP. We leveraged the availability of the source code developed by Li et al [14, 15] and applied it to our independent test set. Additionally, two experienced radiologists were asked to visually interpret the same images to distinguish patients with COVID-19. We compared the performance of the deep learning solution with the two radiologists’ interpretations.
Materials and methods
Image datasets
We retrospectively collected a dataset consisting of 498 CT scans acquired on 151 subjects positive for COVID-19 by RT-PCR and chest CT imaging findings (Table 1). All subjects had close contact with individuals from Wuhan or had a travel history to Wuhan. Namely, the collected cases were either imported cases or secondary infection cases. Most of the subjects had multiple CT scans acquired at different time points (every 3~10 days) to assess disease progression including change in infiltrative pattern. We also retrospectively collected a dataset consisting of 497 CT examinations acquired on different subjects with other types of pneumonia (Table 1). The majority of the collected CAP cases were caused by influenza (flu) A and B viruses, human parainfluenza virus (I, II, and III), human rhinovirus, and adenovirus pneumonia. The CT scans in the COVID-19 and non-COVID-19 datasets were split into three groups at the patient level: (1) training, (2) internal validation, and (3) independent test (Table 1). All data used in this study were de-identified with all protected health information removed. This study was approved by both the Ethics Committee at the Xian Jiaotong University The First Affiliated Hospital (XJTU1AF2020LSK-012) and the University of Pittsburgh Institutional Review Boards (IRB) (# STUDY20020171).
Table 1.
The summary of the collected dataset
| COVID-19 | Non-COVID-19 | |||||||
|---|---|---|---|---|---|---|---|---|
| Training | Validation | Testing | Total | Training | Validation | Testing | Total | |
| Patient | 97 | 27 | 27 | 151 | 393 | 55 | 50 | 498 |
| Mild or moderate | 85 | 25 | 25 | 135 | 366 | 52 | 47 | 465 |
| Severe | 8 | 1 | 1 | 10 | 21 | 2 | 3 | 26 |
| Critical | 4 | 1 | 1 | 6 | 6 | 1 | 0 | 7 |
| CT exams | 301 | 55 | 50 | 497 | 393 | 55 | 50 | 498 |
| Age | 49.4 ± 15.4 | 40.7 ± 13.7 | 53.2 ± 17.4 | 45.7 ± 15.7 | 46.9 ± 17.9 | 40.0 ± 17.4 | 45.5 ± 15.3 | 45.9 ± 17.7 |
| Male (%) | 52 (53.6) | 15 (55.6) | 16 (59.3) | 83 (55.0) | 206 (52.4) | 30 (54.5) | 27 (54) | 263 (52.8) |
Classification of the CT examinations using 3D CNNs
In the past years, the deep learning technology, namely, convolutional neural network (CNN), has been emerging a novel solution for a variety of medical image analysis problems, including classification, detection, segmentation, and registrations, and demonstrated remarkable performance. In architecture, a CNN is typically formed by several building blocks, including convolution layers, activation functions, pooling layers, batch normalization layers, flatten layers, and fully connected (FC) layers. Organizing these blocks in different ways results in different types of CNN architectures that may present different performance. The strength of CNN lies in the ability to automatically learn specific features (or feature map) by repeatedly applying the convolutional layers to an image.
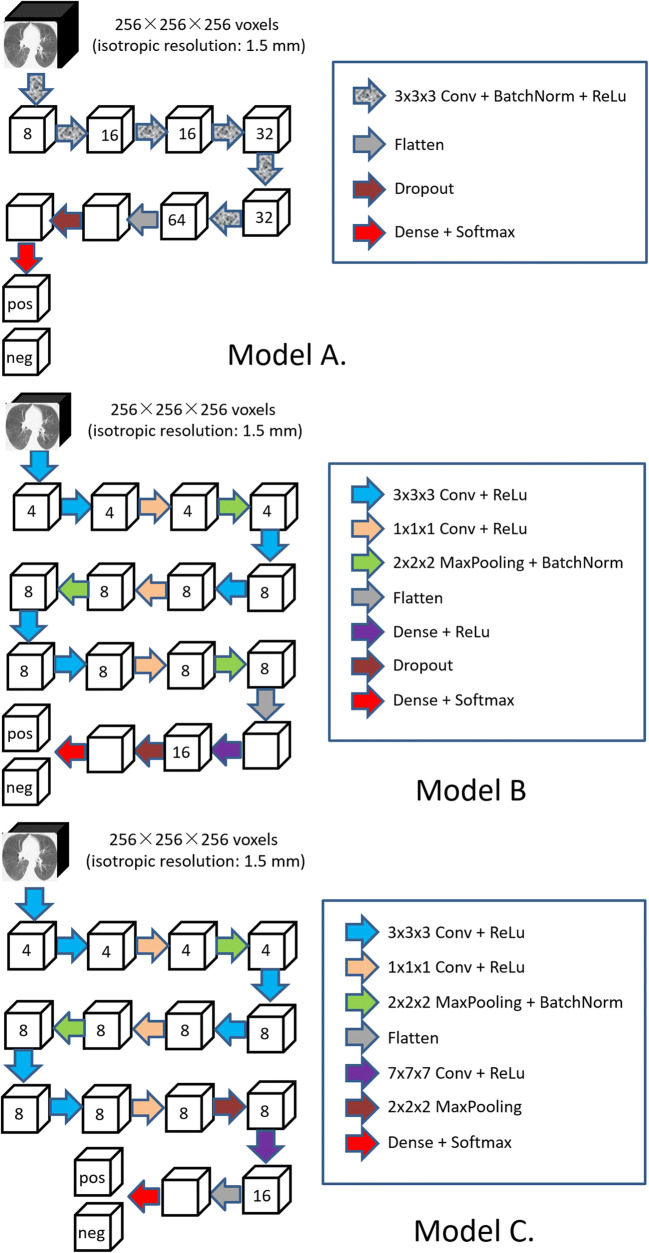
We implemented three classifiers based on 3D CNNs in an attempt to discriminate CT scans that originated from COVID-19 and CAP (Fig. 1). With consideration of the memory limit of the graphics processing unit (GPU), we resized and padded the CT images at 256 × 256 × 256 voxels with an isotropic resolution of 1.5 mm in order to send an entire CT scan to the network for training and inference. The CNN architectures used different numbers of filters at different layers. Batch normalization and rectified linear unit (ReLu) activation were used. A dropout regularization between the fully connected (FC) layers with a probability of 0.5 was used in Models A and B. In these models, the last FC layers, which were activated by the Softmax function, output the prediction probabilities of being COVID-19. The binary cross-entropy (BCE) loss function, as defined by (1), was minimized to obtain optimal classification performances.
| 1 |
where N is the number of classes (i.e., N = 14), yi is the ground truth, and is the predicted probability.
Fig. 1.
The architectures of the developed image classifiers based on 3D CNN
In the implementation, the maximum pooling layers are used to compute the maximum over a region of a feature map and thus is a feature map with the most prominent features of the previous feature map but reduced dimensionality. This dimensionality reduction can reduce the number of extracted features to avoid overfitting. If there is no activation function, the neural network will work like a linear regression model, which will prevent the machine from learning complex patterns from the data. As a non-linear activation function, rectified linear units (ReLU) is often used as a way to characterize complicated shapes or domains. ReLU does not activate all the neurons at the same time but those whose outputs are above zero. This activation strategy can significantly improve the efficiency of the computation as compared with other activation functions (e.g., sigmoid and tanh function). The batch normalization is used to improve the stability of a neural network by normalizing the input layer. The data augmentation increases the diversity of the training dataset and enables the learning of the same features from various versions of the images. This learning strategy can alleviate the impact of noise on the performance and improve the robustness of the trained models. The neurons in an FC layer have full connections to all activations in the previous layers. The goal is to determine specific global configurations of the image features identified by the previous convolutional layers for classification purposes. Hence, an FC layer is typically at the end of the convolutional layers but before the classification output of a CNN. Softmax function outputs a vector that represents the probability distributions of a list of potential outcomes.
When training these models, we used the training and internal validation subgroups listed in Table 1. The batch size was set at 8 for the models. To improve the data diversity and the reliability of the models, we augmented the 3D images via geometric and intensity transformations, such as rotation, translation, vertical/horizontal flips, Hounsfield Unit (HU) shift [− 25 HU, 25 HU], smoothing (blurring) operation, and Gaussian noises. The initial learning rate was set as 0.001 and would be reduced by a factor of 0.2 if the validation performance did not increase in three epochs. Adam optimizer was used, and the training procedure would stop when the validation performance of the current epoch did not improve compared with the previous ten epochs.
The CNN architecture developed by Li et al [14, 15] was trained and tested on our datasets to compare with the performance of our three models.
Subjective classification of the CT examinations
Two radiologists with 11 and 25 years of experience, including experience visualizing > 200 COVID positive images from a different image set, who were blind to the diagnosis, independently viewed and rated the 100 CT scans (50 COVID-19 and 50 non-COVID-19) in the testing dataset. They rated the CT scans as COVID-19 positive or negative. During the interpretations, these scans were randomly presented to each reader. The readers were allowed to adjust parameters, such as the window levels and the image dimensions (e.g., zoom in and out), to facilitate their interpretations.
Statistical analyses
The testing set, including 50 COVID-19 CT exams s (D = 1) and 50 CAP CT exams (D = 0), were used for diagnostic performance evaluation. The accuracy of radiologists’s decisions (Trad = 1 as “positive” or Trad = 1 = 0 as “negative” for COVID-19) were characterized with empirical estimates “sensitivity,” Se = Pr(Trad = 1|D = 1), and “specificity,” Sp = Pr(Trad = 0|D = 0) [16]. Kappa coefficient was used to quantify the agreement on the suspected COVID-19 positivity, separately for patients without and with verified COVID-19 pneumonia. Statistical inferences were based on the two-sided 95% confidence intervals (CI) estimated using the empirical cluster bootstrap approach [17, 18] with patient as a sampling unit. Models’ predicted probabilities (scores Tmod∈(0, 1)) were used to estimate receiver operating characteristic (ROC) curves (with Se(ξ) = Pr(Tmod > ξ|D = 1) as a function of 1-Sp(ξ) = Pr(Tmod > ξ|D = 0)) and the related parameters [19]. The area under the ROC curve (AUC) was used to evaluate the overall predictive ability of a model, using the empirical cluster bootstrap CIs with 99% coverage for multiplicity-adjusted testing. Model’s usefulness for accurate identification of a sizable subset of non-COVID-19 patients was quantified using the estimated specificity-at-98%-sensitivity [19], Sp(Se−1(0.95)), with the two-sided 95% CI being used to identify the size of corresponding the low-risk subset.
Results
Both radiologists demonstrated moderate diagnostic accuracy with sensitivity levels of 42/50 (84%, CI 72–93%) and 40/50 (80%, CI 62–96%) and specificity levels of 28/50 (56%, CI 42–70%) and 31/50 (62%, CI 48–76%). While there were no significant differences in radiologists’ sensitivity or specificity levels (p > 0.48), the radiologists demonstrated only slight agreement with κ = 0.22 (CI − 0.04 to 0.50) and κ = 0.19 (CI − 0.29 to 0.53) for patients without and with verified COVID-19 respectively.
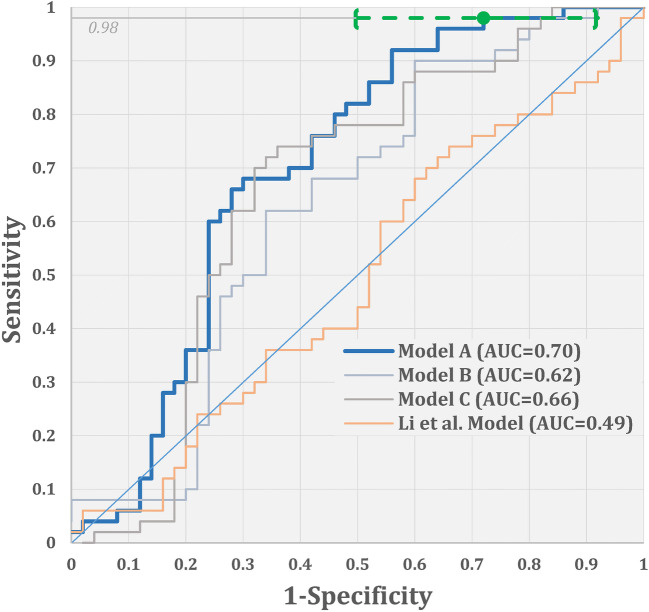
The empirical ROC curves for the three models and Li et al’s model are illustrated in Fig. 2. Under the multiplicity adjustment, only model A demonstrated statistically significant improvement over the chance performance (AUC of 0.70 with 99%, CI 0.56–0.85). For discriminating between the COVID-19 and CAP cases in our data, Li et al’s model had an accuracy level (AUC of 0.49, 99% CI 0.34–0.66) which is significantly lower than Model A’s accuracy level (p = 0.008).
Fig. 2.
Empirical ROC curves and the corresponding AUCs for the three models and Li et al model [14] (green dot and bracketed line indicate an ROC point corresponding to the “low-risk” threshold, the estimate of the specificity-at-98%-sensitivity, and the corresponding two-sided 95% CI)
The shape of Model A’s ROC curve indicates the potential usefulness of the model for accurate identification of a sizable subset of non-COVID-19 cases. In particular, the model allows for identification of 8–50% of non-COVID-19 cases with less than 2% of false negatives (namely, specificity-at-98%-sensitivity of 28%, CI 8–50%). In our sample, the corresponding low-risk subset of cases (corresponding to the model’s score of less than 0.3022) included 14 CT scans from non-COVID-19 cases and 1 CT from a case with verified COVID-19 (all from different subjects). The only COVID-19 case that was included in the low-risk subset was missed by both radiologists. At the same time, 14 non-COVID-19 cases in the low-risk subset were challenging, with one radiologist identifying 50% of these (7/14) as COVID-19-positive, and the other identifying 29% (4/14) as COVID-19-positive.
Discussion
In an attempt to distinguish COVID-19 from CAP, we developed and tested several CNN models. Although deep learning works as a black-box and cannot report which features were associated with COVID-19, the performance of the classifications based on deep learning can at least tell us whether there are features implied in the images that can distinguish CT scans from COVID-19 and CAP patients. Unfortunately, our experimental results did not identify imaging markers with high diagnostic ability to distinguish COVID-19 from simply CAP. However, our imaging marker identified a sizable subset of non-COVID-19 cases and a very low fraction of COVID-19 cases. We note that this was observed for the model optimized based on the general objective function (Eq. (1)); the targeted model building (optimized using a task-oriented objective function) would lead to further improvement in accuracy identifying a low-risk subset of cases.
The performance of the radiologists in our study was similar to Bai et al’s study [10]. Seven radiologists in their study reviewed and rated CT scans from COVID-19 (n = 219) and viral pneumonia (n = 205) subjects. The sensitivity of the seven radiologists ranged from 67 to 97%, while the specificity ranged from 7 and 100%. They concluded that the radiologists had moderate sensitivity in distinguishing COVID-19 from viral pneumonia.
Our CNN models did not perform as well as the CNN model reported in the article by Li et al [14]. They reported an AUC of 0.96 for discriminating CT scans from subjects with COVID-19 and CAP. We applied their publicly available algorithm to our CT scan dataset. However, the training procedure did not converge, suggesting that it had difficulty in accurately distinguishing CT scans from COVID-19 and CAP in our dataset. Although Li et al [14] used the class activation map (CAM) to visualize the features, the features identified did not appear to be unique to COVID-19 based on the figures presented in their publication [14].
To perform this study, we collected 498 CT scans acquired on 151 COVID-19 patients and (positive by RT-PCR) and 497 CT scans on different subjects with CAP. The data collection protocols for COVID-19 and CAP were not the same. First, we can access a large number of cases with CAP in practice but a limited number of cases with COVID-19. Second, due to the limited understanding of COVID-19, a series of CT scans were performed by some medical institutes to monitor the progress of COVID-19. In contrast, very few follow-up CT scans were performed to monitor community-acquired pneumonia. Third, the COVID-19 progressed very rapidly and there were obvious differences across the series of CT scans acquired at different time points. The utilization of the multiple CT scans acquired on the same subjects enables excellent data augmentation. In addition, most of these COVID-19 cases had mild and moderate severity of the disease. Only a few had severe COVID-19. This is primarily because the COVID-19 cases were collected from Shaanxi Province, China, which is about 500 miles away from the epidemic center (Wuhan City, Hubei Province, China). The COVID-19 cases were either imported or secondary infection cases. Although there are few severe cases with COVID-19, we believe our dataset is relatively representative and will not affect the conclusion in this study, because most of the COVID-19 subjects in clinical practices typically have mild or moderate severity.
We are aware that we used a relatively small dataset; however, we did not expect that the scale of our dataset significantly affected the conclusions. The images were augmented to improve the diversity of the dataset as a way to improve the reliability of the training. Hence, additional effort from a third party would further clarify this issue. Also, our dataset did not include subjects without pneumonia because the primary purpose of this study was to differentiate COVID-19 from other types of pneumonia.
Conclusion
We used deep learning technology and high-resolution CT images to investigate whether there are any unique image features associated with COVID-19. Our results suggest that unique image features or patterns may not exist to reliably distinguish all COVID-19 from CAP. However, our results demonstrate the potential of the imaging markers to assist in identifying a sizable subset of non-COVID-19 cases.
Acknowledgments
This work is supported by National Institutes of Health (NIH) (Grant No. R01CA237277 and R01HL096613).
Abbreviations
- AUC
Area under the curve
- BCE
Binary cross-entropy
- CAP
Community-acquired pneumonia
- CI
Confidencial interval
- CNN
Convolutional neural network
- COVID-19
Novel coronavirus
- CT
Computed tomography
- FC
Fully connected
- GPU
Graphics processing unit
- IRB
Institutional review boards
- ROC
Receiver operating characteristic
Authors’ contributions
JP, FS, and DW designed the study. JL, AB, and CJ performed the analyses. JS, PD, CF, SK, and YG collected and reviewed the images. JY, BY, and JP developed deep learning models. JP, JL, and JB wrote the articles.
Compliance with ethical standards
Guarantor
The scientific guarantor of this publication are Chenwang Jin, M.D., Ph.D. Professor of Radiology, Xi’an Jiaotong University The First Affiliated Hospital, (jin1115@mail.xjtu.edu.cn) and Andriy Bandos, Ph.D. Associate Professor of Biostatistics, University of Pittsburgh, (anb61@pitt.edu).
Conflicts of interest
The authors declare that there is no conflict of interest.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Jiantao Pu, Email: puj@upmc.edu.
Chenwang Jin, Email: jin1115@mail.xjtu.edu.cn.
References
- 1.Guan CS, Lv ZB, Yan S et al (2020) Imaging features of coronavirus disease 2019 (COVID-19): evaluation on thin-section CT. Acad Radiol. 10.1016/j.acra.2020.03.002 [DOI] [PMC free article] [PubMed]
- 2.Hope MD, Raptis CA, Shah A, Hammer MM, Henry TS; six signatories (2020) A role for CT in COVID-19? What data really tell us so far. Lancet. 10.1016/S0140-6736(20)30728-5 [DOI] [PMC free article] [PubMed]
- 3.Ufuk F (2020) 3D CT of novel coronavirus (COVID-19) pneumonia. Radiology. 201183. 10.1148/radiol.2020201183
- 4.Caruso D, Zerunian M, Polici M et al (2020) Chest CT features of COVID-19 in Rome, Italy. Radiology 201237. 10.1148/radiol.2020201237 [DOI] [PMC free article] [PubMed]
- 5.Xiong Y, Sun D, Liu Y et al (2020) Clinical and high-resolution CT features of the COVID-19 infection: comparison of the initial and follow-up changes. Invest Radiol. 10.1097/RLI.0000000000000674 [DOI] [PMC free article] [PubMed]
- 6.Lee EYP, Ng MY, Khong PL (2020) COVID-19 pneumonia: what has CT taught us? Lancet Infect Dis. 10.1016/S1473-3099(20)30134-1 [DOI] [PMC free article] [PubMed]
- 7.Zhou S, Wang Y, Zhu T, Xia L (2020) CT Features of Coronavirus Disease 2019 (COVID-19) pneumonia in 62 patients in Wuhan, China. AJR Am J Roentgenol 2020:1–8. 10.2214/AJR.20.22975 [DOI] [PubMed]
- 8.Fang Y, Zhang H, Xie J et al (2020) Sensitivity of chest CT for COVID-19: comparison to RT-PCR. Radiology. 200432. 10.1148/radiol.2020200432 [DOI] [PMC free article] [PubMed]
- 9.Ai T, Yang Z, Hou H et al (2020) Correlation of chest CT and RT-PCR testing in coronavirus disease 2019 (COVID-19) in China: a report of 1014 cases. Radiology. 200642. 10.1148/radiol.2020200642 [DOI] [PMC free article] [PubMed]
- 10.Bai HX, Hsieh B, Xiong Z et al (2020) Performance of radiologists in differentiating COVID-19 from viral pneumonia on chest CT. Radiology. 200823. 10.1148/radiol.2020200823 [DOI] [PMC free article] [PubMed]
- 11.Chua F, Armstrong-James D, Desai SR et al (2020) The role of CT in case ascertainment and management of COVID-19 pneumonia in the UK: insights from high-incidence regions. Lancet Respir Med. 10.1016/S2213-2600(20)30132-6 [DOI] [PMC free article] [PubMed]
- 12.Liu J, Yu H, Zhang S (2020) The indispensable role of chest CT in the detection of coronavirus disease 2019 (COVID-19). Eur J Nucl Med Mol Imaging. 10.1007/s00259-020-04795-x [DOI] [PMC free article] [PubMed]
- 13.Shi H, Han X, Jiang N et al (2020) Radiological findings from 81 patients with COVID-19 pneumonia in Wuhan, China: a descriptive study. Lancet Infect Dis. 10.1016/S1473-3099(20)30086-4 [DOI] [PMC free article] [PubMed]
- 14.Li L, Qin L, Xu Z et al (2020) Artificial intelligence distinguishes COVID-19 from community acquired pneumonia on chest CT. Radiology. 200905. 10.1148/radiol.2020200905
- 15.COVNet. Available from: https://github.com/bkong999/COVNet
- 16.Pepe MS (2003) The statistical evaluation of medical test for classification and prediction. Oxford University Press
- 17.Field CA, Welsh AH. Bootstrapping clustered data. J R Stat Soc Series B Stat Methodology. 2007;69(3):369–390. doi: 10.1111/j.1467-9868.2007.00593.x. [DOI] [Google Scholar]
- 18.Davison AC, Hinkley DV (1997) Bootstrap methods and their application. Cambridge University Press
- 19.Zhou XH, Obuchowski NA, McClish DK (2011) Statistical methods in diagnostic medicine. Wiley