Abstract
Background Early detection and efficient management of sepsis are important for improving health care quality, effectiveness, and costs. Due to its high cost and prevalence, sepsis is a major focus area across institutions and many studies have emerged over the past years with different models or novel machine learning techniques in early detection of sepsis or potential mortality associated with sepsis.
Objective To understand predictive analytics solutions for sepsis patients, either in early detection of onset or mortality.
Methods and Results We performed a systematized narrative review and identified common and unique characteristics between their approaches and results in studies that used predictive analytics solutions for sepsis patients. After reviewing 148 retrieved papers, a total of 31 qualifying papers were analyzed with variances in model, including linear regression ( n = 2), logistic regression ( n = 5), support vector machines ( n = 4), and Markov models ( n = 4), as well as population (range: 24–198,833) and feature size (range: 2–285). Many of the studies used local data sets of varying sizes and locations while others used the publicly available Medical Information Mart for Intensive Care data. Additionally, vital signs or laboratory test results were commonly used as features for training and testing purposes; however, a few used more unique features including gene expression data from blood plasma and unstructured text and data from clinician notes.
Conclusion Overall, we found variation in the domain of predictive analytics tools for septic patients, from feature and population size to choice of method or algorithm. There are still limitations in transferability and generalizability of the algorithms or methods used. However, it is evident that implementing predictive analytics tools are beneficial in the early detection of sepsis or death related to sepsis. Since most of these studies were retrospective, the translational value in the real-world setting in different wards should be further investigated.
Keywords: sepsis, predictive analytics, machine learning, algorithms, data modeling
Background and Significance
Sepsis is a severe complication stemmed from an infection in the body and can lead to potential tissue damage, organ failure, or even death. More than 1.7 million individuals are diagnosed with sepsis annually in the United States and has a one in three mortality rate. 1 Sepsis is a strain on the hospitals and health care system as it is a disease of high prevalence and cost. In 2013, almost $24 billion was spent on care for sepsis patients, making it the most expensive condition to treat in U.S. hospitals. 2 Unfortunately, sepsis can stem from a vast array of initial infections, such as pneumonia or a urinary tract infection. Despite the high occurrence and prevalence, detection and diagnosis of sepsis remain a challenge due to its nondescript early-onset symptoms, such as high heart rate and clammy skin. 3 However, as it can quickly progress to a life-threatening stage, it is crucial to treat sepsis patients earlier and more efficiently to increase survival outcomes. Furthermore, patients diagnosed with sepsis tend to remain in the hospital for a significantly longer period of time when compared with those without the condition; thus, using more resources and hampering the ability to move patients out of the emergency department (ED) and into beds efficiently.
Currently, there are various metrics in use to define and identify sepsis in the clinical setting. In 1991, the Sepsis-1 definition of sepsis, severe sepsis, and septic shock was released. Sepsis was then described as a systemic inflammatory response syndrome (SIRS) due to a present infection, with at least two of the following criteria: (1) temperature > 38°C or < 36°C, (2) heart rate > 90 beats per minute, (3) respiratory rate > 20 or PaCO 2 < 32 mm Hg, or (4) white blood cell > 12,000/mm 3 , < 4,000/mm 3 , or > 10% bands; severe sepsis was having sepsis resulting in organ dysfunction while septic shock was the occurrence of sepsis-induced hypotension. 4 In 2001, an update resulted in the introduction of the Sepsis-2 definition, which added confirmed or suspected infection to the sepsis definition. 4 However, in 2016, Sepsis-3 was created and sepsis is now described as a life-threatening organ dysfunction caused by a dysregulated host to infection. 5
While the definitions of sepsis have evolved, so has data collection in the clinical setting. In 2010, the United States government established a three-stage incentive program, aptly titled “Meaningful Use,” which established the requirement to use electronic health records (EHRs). With Meaningful Use stage one, EHRs were widely adopted and now streams of patient data are constantly being collected. Many researchers and clinicians are now trying to leverage and integrate the data to create tools that aid in early detection of sepsis. Many of these tools and predictive solutions use machine learning (ML) techniques or hazards model to assist in predicting sepsis onset or mortality. ML is the application of artificial intelligence to aid with automatic learning, detection, or classification, without being explicitly programmed, and can potentially be useful with medical data. 6 Additionally, the type and size of the feature set is important for the efficacy and interpretability of ML techniques as irrelevant features may lower the effectiveness. 7
Because there are many ML models and feature sets that can be used for sepsis predictive analytics, we systematically identified various studies to understand the current state of sepsis prediction tools. Furthermore, we sought to determine how predictive analytics are being implemented for septic patients and to see if there are any optimal solutions for sepsis detection or mortality associated with sepsis currently being explored.
Methods
We performed a literature search on PubMed in November 2018 to identify current practices and studies that have used predictive analytics for septic patients, aiding in both early detection of onset or mortality, using the following query: (“sepsis” OR “septicemia” OR “septic” OR “septic shock” OR “severe sepsis”) AND (“prediction” OR “predict” OR “analytics”) AND (“machine learning” OR “big data” OR “AI” OR “NLP” OR “neural network” OR “algorithm”). We included common synonyms and popular phrases for sepsis and predictive analytics to cast a greater net when searching. After querying the database, we identified and categorized relevant articles by reviewing the article titles and abstracts to ensure a ML technique or model was being used for prediction or detection of sepsis. Our search was not limited to the intensive care unit (ICU) as sepsis does not solely occur in such a setting. Furthermore, this allowed a larger scope to explore novel techniques being developed in other clinical settings. Our overall initial inclusion criteria are detailed below:
Study was published in a peer-reviewed journal or conference.
Study was published in English.
Study was published after 2008.
Study used at least one ML or model technique.
Study identified the features and data set used.
Study presented their summary statistics and/or compared with previous studies.
Studies that did not meet these criteria were automatically excluded. However, to increase the breadth of our search and to include seminal studies, we added studies based on their titles and abstracts that were commonly found in the references, regardless of publication year. We did not restrict our search to a specific population age for similar reasons, but most of the reviewed studies centered around the general adult population aged 18 to 65. After curating our set of relevant articles, we identified and categorized the characteristics of each study by analyzing the common themes and differences between them to better understand the issues in applying predictive analytics to sepsis detection.
Results
Fig. 1 depicts the article selection process. Our PubMed search resulted in 148 articles, where 31 articles were immediately excluded due to publication date. After reviewing the abstracts of the remaining articles, 95 articles were further excluded as they were not relevant to our question. A total of 22 full-text articles were extracted from our PubMed search. From these articles, a total of 11 cited references were added based on their titles and abstracts as they were commonly cited papers between many of the articles that met our inclusion criteria. However, two articles were then removed as there were no summary statistics presented. In total, 31 papers were analyzed. Full results are summarized in Table 1 , detailing the algorithm or model, population and feature size, “gold standard” definitions, and summary statistics.
Fig. 1.
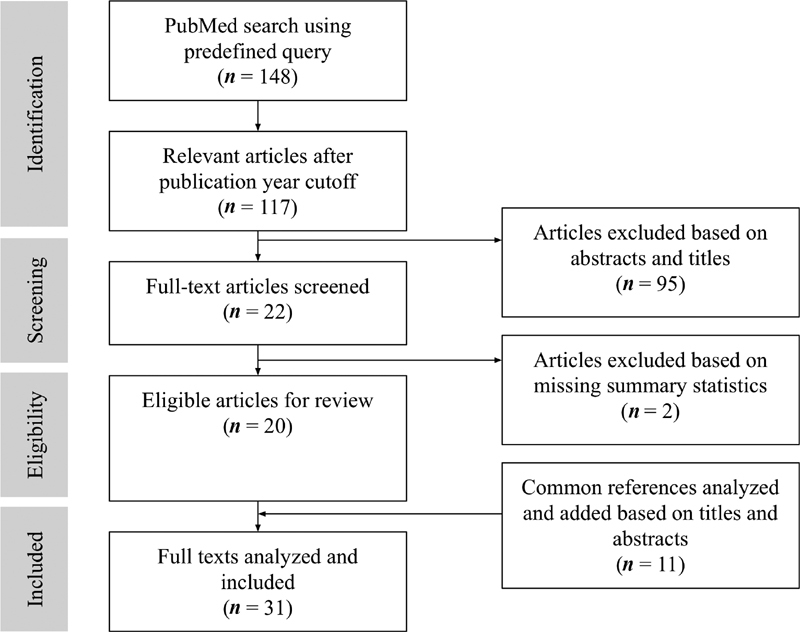
Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) adapted diagram.
Table 1. Overview of unique characteristics and differences between methods.
| Author | Year | Goal | Population location and size | Feature set size | “Gold standard” definition | Model and performance metrics |
|---|---|---|---|---|---|---|
| Carrara et al 8 | 2015 | Mortality prediction in septic shock patients | MIMIC II (ICU) 30,000+ patients |
30 variables | Septic shock: 1991 SIRS criteria, ICD-9 code for septic shock, abnormal interval must exceed 5 h for each feature, SIRS 2 + , SIRS with low SBP despite adequate fluid resuscitation | Multivariate linear regression with Shrinkage Techniques model Mean square error (MSE): 0.03 |
| Danner et al 9 | 2017 | Assess the value of HR-to-systolic ratio in the accuracy of sepsis prediction after ED presentation | Local (ED) 53,313 patients |
9 vitals/variables | Sepsis: Discharge diagnosis of sepsis, evaluated vitals, demographics, chief complaints | Multivariate linear regression model - Accuracy: 0.74 - HR to systolic ratio accounted for 69% of overall predictive ability |
| Capp et al 10 | 2015 | Describe key patient characteristics present within 4 h of ED arrival that are associated with developing septic shock between 4 and 48 h of ED arrival | Local (ED) 1,316 patients |
5 risk factors | Sepsis: manual chart review with SIRS 2 + , evidence of infection (excluded if gastrointestinal bleed) Septic shock: SBP > 90 mm Hg despite appropriate fluid hydration of 30 mL/kg with presence of hypotension for at least 2 h after |
Multivariable logistic regression model Found risk factors associated with progression of sepsis to septic shock between 4 and 48 h of ED arrival: - Female: 1.59 odds ratio (OR) - Nonpersistent hypotension: 6.24 OR - Lactate > 4 mmol/L: 5.30 OR - Bandemia > 10%: 2.60 OR - Past medical of coronary heart disease: 2.01 OR |
| Faisal et al 11 | 2018 | To develop a logistic regression model to predict the risk of sepsis following emergency admission using the patient's first electronically recorded vital signs and blood test results and to validate this novel computer-aided risk of sepsis model, using data from another hospital | Local (ED) 57,243 patients |
12 vitals/variables | Sepsis: ICD-10 codes without organ failure Severe sepsis: ICD-10 codes with 1+ organ failure or septic shock |
Logistic regression models All area under the receiver operator curve (AUROC): 0.79 Sepsis AUROC: 0.70 Severe sepsis AUROC: 0.81 |
| Ho et al 12 | 2012 | Investigate how different imputation methods can overcome the handicap of missing information | MIMIC II (ICU) Sample size not stated |
6 vitals | Sepsis: ICD-9 Septic shock: examined clinical chart records |
- Sepsis: Multivariate logistic regression models - Septic shock: multivariate logistic regression, linear kernel SVM, and regression trees H: Clinical history feature set P: initial physiological state feature set Sepsis AUROC (imputed mean and matrix factorization-based approaches) All H: 0.791 (0.792) Stepwise H: 0.790 (0.791) All H ∪ P: 0.821 (0.822) Stepwise H ∪ P: 0.823 (0.823) Septic shock AUROC: 0.773–0.786 |
| Langley et al 13 | 2013 | Examine clinical features, plasma metabolome, and proteome of patients to predict patient survival of sepsis | CAPSOD (ED) 1,152 individuals with suspected, community-acquired sepsis; Discovery set of 150 patients |
4 vitals/variables | Acute infection + 2+ SIRS | Logistic regression (sepsis prediction) and SVM model (survival and death prediction) Logistic regression AUROC: 0.847 Logistic regression accuracy: 0.851 *best stats occurred at enrollment SVM AUROC: 0.740 SVM accuracy: 0.746 |
| Sutherland et al 14 a | 2011 | Use gene expression biomarkers to prospectively distinguish patients with sepsis from those who experience systemic inflammation from healing of surgery | Local (ICU) 85 patients |
42 biomarkers | Likely enter sepsis cohort if met ACCP/SCCM consensus statement and clinical suspicion of systemic infection Confirmation performed retrospectively |
Classifier: Recursive partitioning, LASSO, logistic regression. Individual genes examined via Bayes-adjusted linear model. MT-PCR diagnostic classifier generated using a LogitBoost ML algorithm (tree-based) PCR Accuracy: 92% AUROC: 0.86–0.92 |
| Gultepe et al 15 | 2014 | Develop a decision support system to identify patients with hyperlactatemia and to predict mortality from sepsis using predicted lactate levels | Local (ED) 741 patients |
7 vitals/laboratories | Sepsis: determined from EHR diagnosis and SIRS criteria | SVM classifier Accuracy: 0.73 AUROC: 0.73 |
| Horng et al 16 | 2017 | To demonstrate the incremental benefit of using free text data in addition to vital sign and demographic data to identify patients with suspected infection in the emergency department | Local (ED) 198,833 control 32,103 cases |
12 vitals/variables | ED ICD-9-CM code | Linear SVM and free text models Bag of words AUROC: 0.86 Bag of words sensitivity: 0.78 Bag of words specificity: 0.79 Topic model AUROC: 0.85 Topic model sensitivity: 0.80 Topic model specificity: 0.75 |
| Thottakkara et al 17 | 2016 | To compare performance of risk prediction models for forecasting postoperative sepsis and acute kidney injury | Local (in-patient) 50,318 patients |
285 variables | Forecast postop sepsis and acute kidney injury AHRQ definition of “post-op sepsis” and organ failure associated with sepsis was identified by ICD-9-CM code for acute organ dysfunction |
Comparison of models that used logistic regression, generalized additive models (GAM), naive Bayes, SVM Naive Bayes performed the worst in the comparison; GAMs and SVMs had good performance; PCA feature extraction (reduced to 5 features) improved predictive performance for all models Severe sepsis AUROC: 0.76–0.91 |
| Vieira et al 18 | 2013 | Proposed a modified binary particle swarm optimization method for feature selection to predict mortality in septic patients | MEDAN (ICU) 382 patients |
Model chooses custom number of features (2–7) | MEDAN data set prelabeled patients for abdominal septic shock | Support vector machine for mortality prediction Modified binary particle swarm optimization (MBPSO): feature selection MBPSO 12 (28) features: No-FS Accuracy: 72.6% (89%) Accuracy: 76.5% (94.4%) |
| Ghosh et al 19 | 2017 | Predict septic shock for ICU patients using noninvasive waveform measurements | MIMIC II (ICU) 1,519 patients |
3 vitals/laboratories | Sepsis: ICD-9 Septic shock: examining clinical chart records |
Coupled hidden Markov models (CHMM) with varying gap interval and observation window sizes CHMM average: 0.85 Multichannel patterns (MCP)-CHMM average: 0.86 |
| Peelen et al 20 | 2010 | Develop a set of complex Markov models based on clinical data to extract meaningful clinical patterns and to provide prediction for sepsis and other diseases | Local (ICU) 2,271 patients |
6 variables | Sever sepsis: SIRS 2+ within 24 h of ICU admission and 1+ dysfunctioning organ system (SOFA) | 3 Markov models (amount of organ failure, type of organ failure, differences between development and persistence of organ failure) ICU death the error rates were 17.7%, 18.1%, and 17.8% and the AUCs were 0.79, 0.79, and 0.80 for models I, II, and III |
| Stanculescu et al 21 | 2014 | Demonstrate that by adding a higher-level discrete variable with semantics sepsis/nonsepsis, can detect changes in the physiological factors that signal the presence of sepsis | Local (NICU) 24 neonates |
Bradycardia, desaturation | Laboratory result of blood culture for neonatal sepsis | Hierarchical switching linear dynamical system (HSLDS) Autoregressive (AR)-HMM AUROC: 0.72 HSLDS deep learning AUROC: 0.69 HSLDS known factors AUROC: 0.62 |
| Stanculescu et al 22 | 2014 | Detect and identify sepsis in neonates before a blood sample is drawn. Furthermore, they wanted to identify which physiological event would contribute most for detecting sepsis | Local (NICU) 24 neonates |
6 vitals/variables | Positive cultures as pathogens: proven sepsis Positive cultures as mixed growth/skin commensal: “suspected sepsis” |
AR-HMM AUROC: 0.74–0.75 AUROC with missing data: 0.72–0.73 AUROC with bradycardia and minibradycardia: 0.79–0.80 AUROC with desaturation: 0.76–0.78 AUROC with all states: 0.79–0.80 |
| Gultepe et al 23 | 2012 | Use a Bayesian network to detect sepsis early | Local (ICU) 1,492 patients |
BN1: 5 variables BN2: 7 variables |
“Sepsis occurrence” | Bayesian network (BN) models BN-1 (vitals) goodness of fit: 15.4 BN-2 (vitals + MAP) goodness of fit: 19.9 Found that lactate is a driver in both models and maybe an important feature for early sepsis detection |
| Nachimuthu and Haug 24 | 2012 | Detect sepsis right after patients are admitted to the ED | Local (ED) 3,100 patients |
11 vitals/variables | Clinician determined “sepsis” during retrospective chart review | Dynamic Bayesian network 3 h after admission AUROC: 0.911 6 h after admission AUROC: 0.915 12 h after admission AUROC: 0.937 24 h after admission AUROC: 0.944 |
| Calvert et al 25 | 2016 | Detect and predict the onset of septic shock for alcohol-use disorder patients in the ICU | MIMIC III (ICU) 1,394 patients |
9 vitals/variables | Septic shock: SIRS 2 + , ICD-9, organ dysfunction, SBP < 90 mm Hg for 1 h, total fluid replacement ≥ 1,200 mL or 20 mL/kg for 24 h |
InSight
Sensitivity: 0.93 Specificity: 0.91 Accuracy: 0.91 F1 score: 0.161 |
| Calvert et al 26 | 2016 | To develop high-performance early sepsis prediction technology for the “general patient population” | MIMIC II (ICU) 29,083 patients |
10 vitals/variables | Sepsis: ICD-9 code, 1991 SIRS for 5 h |
InSight
Sensitivity: 0.90 Specificity: 0.81 AUROC: 0.92 Accuracy: 0.83 |
| Desautels et al 27 | 2016 | To validate InSight with the new Sep-3 definition and make predictions using minimal set of variables | MIMIC III (ICU) 22,583 patients |
8 vitals/laboratories | Sepsis: Sep-3 definition, suspicion of infection equated with an order of culture laboratory draw and dose of antibiotics |
InSight
AUROC: 0.88 APR: 0.60 |
| Mao et al 28 | 2018 | Validate the InSight algorithm for detection and prediction of sepsis and septic shock | MIMIC III (ICU) Local (ED, general) 61,532 stays |
6 vitals/laboratories | Sepsis: ICD-9 + SIRS 2+ (995.91) Severe sepsis: ICD-9 (955.92), organ dysfunction, SIRS 2+ Septic shock: ICD-9 (785.52), SBP < 90 mm Hg (at least 30 min), resuscitated with ≥ 20 mL/kg over 24 h, ≥ 1,200mL in total fluids |
InSight
Detect sepsis AUROC: 0.92 Detect severe sepsis AUROC: 0.87 Detect 4 h before onset sepsis AUROC: 0.96 Detect 4 h before onset severe sepsis AUROC: 0.85 |
| McCoy and Das 29 | 2017 | Aimed to improve sepsis-related patient outcomes through a revised sepsis management approach | Local (ICU) 407 patients |
6 vitals/variables | Severe sepsis: SIRS 2 + , qSOFA score |
Dascena
Sep-3 AUROC: 0.91 Sep-3 sensitivity: 0.83 Sep-3 specificity: 0.96 Severe sepsis AUROC: 0.96 Severe sepsis sensitivity: 0.90 Severe sepsis specificity: 0.85 |
| Shimabukuro et al 30 a | 2017 | Randomized control trial to show lowered mortality and length of stay using a machine learning sepsis prediction algorithm | Local (ICU) 75 controls 67 cases |
7 vitals/laboratories | Severe sepsis: “organ dysfunction caused by sepsis” Random allocation sequence to put patients in groups |
InSight
AUROC: 0.952 Sensitivity: 0.9 Specificity: 0.9 Average length of stay decreased from 13 to 10.3 d In-hospital mortality decreased by 12.3% |
| Henry et al 32 | 2015 | Create and test a score that predicts which patients will develop septic shock | MIMIC II (ICU) 16,234 patients |
54 features | Suspicion of infection: ICD-9 or by presence of clinical note that mentioned sepsis or septic shock Sepsis: suspicion + SIRS Severe sepsis: sepsis + organ dysfunction |
TREWScore (Cox proportional hazards model using the time until the onset of septic shock as the supervisory signal) AUROC: 0.83 Specificity: 0.67 Sensitivity: 0.85 “Patients were identified a median of 28.2 h before shock onset” |
| Lukaszewski et al 31 a | 2008 | Detect and identify septic patients before displaying symptoms for ICU patients | Local (ICU) 92 patients |
7 cytokines | Admission diagnosis upon ICU entry | Neural networks using cytokine and chemokine data Sensitivity: 0.91 Specificity: 0.80 Accuracy: 0.95 |
| Nemati et al 35 | 2018 | Aimed to develop and validate an artificial intelligence sepsis algorithm for early prediction of sepsis | Local (ICU) 33,069 patients |
65 variables | Sepsis: Sepsis-3 | Modified Weibull–Cox proportional hazards model 4 h in advance AUROC: 0.85 |
| Pereira et al 33 | 2011 | Examined different approaches to predicting septic shock with missing data | MEDAN (ICU) 139 patients |
2 sets of 12 and 28 “selected features” | Septic shock: associated with abdominal causes (not clearly defined, data may be prelabeled) | Zero-Order-Hold (ZOH) Fuzzy c-means clustering based on partial distance calculation strategy (FCM-PDS) Performance improvements occur where up to 60% of the data are missing ZOH-FCM-PDS 12 (28) feature AUROC: 0.899 (0.649); FCM-PDS 12 (28) feature AUROC: 0.786 (0.631) |
| Ribas et al 36 | 2011 | Demonstrate that a SVM variant can provide automatic ranking of mortality predictor and have higher accuracy that current methods | Local (ICU) 354 patients |
4 vitals/laboratories | Severe sepsis: organ dysfunction (SOFA) | Relevance vector machine AUROC: 0.80 Error rate: 0.24 Sensitivity: 0.66 Specificity: 0.80 |
| Sawyer et al 38 a | 2011 | Evaluate if implementing an automated sepsis screening and alert system can facilitate in early interventions by identifying non-ICU patients at risk for developing sepsis | Local (Non-ICU) 270 patients |
9 vitals/variables | Intervention group: real-time sepsis alert generated from Clinical Desktop | Recursive partitioning regression tree analysis Within 12 h of sepsis alert, 70.8% of patients in the intervention group received treatment versus 55.8% in control |
| Shashikumar et al 37 | 2017 | Investigates the utility of high-resolution blood pressure and heart rate times series dynamics for the early prediction of sepsis | Local (ICU) 242 patients |
11 vitals/variables | Sepsis: Seymour (Sep-3) at some point during ICU stay | Elastic Net logistic classifier: 3 models: (1) entropy features, (2) EMR + sociodemographic-patient history features, (3) models 1 + 2 Model 1 AUROC (Accuracy): 0.67 (0.47) Model 2 AUROC (Acc): 0.70 (0.50) Model 3 AUROC (Acc): 0.78 (0.61) |
| Taylor et al 39 | 2016 | Compare a machine learning approach to existing clinical decision rules to predict sepsis in-hospital mortality | Local (ED) 4,676 patients |
20 variables | ICD-9 with AHRQ clinical classification software to obtain more exhaustive list of patients | Random forest model AUROC: 0.86 |
Abbreviations: AHRQ, Agency for Healthcare Research and Quality; ACCP/SCCM, American College of Chest Physicians/Society of Critical Care; ED, emergency department; EHR, electronic health record; HR, heart rate; ICD, International Classification of Disease; ICU, intensive care unit; MEDAN, Medical Data Warehousing and Analysis; MIMIC, Medical Information Mart for Intensive Care; ML, machine learning; MT-PCR, multiplex tandem-polymerase chain reaction; NICU, neonatal intensive care unit; PCA, principal component analysis; SBP, systolic blood pressure; SIRS, systemic inflammatory response syndrome; SOFA, sequential organ failure assessment; SVM, support vector machine.
Indicates prospective.
For the majority of the studies analyzed, a variety of metrics were used to report results, such as area under the receiver operator curve (AUROC) and accuracy. The reported metrics were dependent on the ML technique or model used, the features that were selected, and the size of the study data. From our review, we identified four key differences between the analyzed studies (1) variability in ML or modeling techniques, (2) variability in feature selection, (3) variability in data sample selection and size, and (4) variability in “gold standard” sepsis definitions.
Variability in Machine Learning or Modeling Techniques
A wide range of models and ML techniques were used to predict or detect sepsis onset, septic shock, severe sepsis, or mortality. The variety of methods used, summarized in Table 2 , added to the richness of this systematic scoping review. Common methods include linear regression ( n = 2), 8 9 logistic regression ( n = 5), 10 11 12 13 14 support vector machines ( n = 4), 15 16 17 18 Markov models ( n = 4), 19 20 21 22 and Bayesian networks ( n = 2). 23 24 Additionally, a few studies ( n = 6), 25 26 27 28 29 30 used an industry created tool, InSight (Dascena Inc.), to validate performance compared with the more commonly used methods. In particular, Mao et al, used InSight to test the predictive abilities of the industry-created sepsis detection algorithm on open source and local data sets, determining the transferability of the algorithm across varying data sets. 28 Similarly, a few studies, including Danner et al, Gultepe et al, and Thottakkara et al used multiple methods and algorithms for comparison purposes against their own developed solution. 9 15 17 Many of the general results concluded that utilizing predictive analytics were beneficial in the detection or prediction sepsis onset or mortality. Predictive performance measures for each study are detailed in Table 1 . Due to heterogeneity in methods and standards used, the predictive performance measure varies across the analyzed studies. Table 1 details the goals of the studies, the “gold standard” or definition used for sepsis, septic shock, or severe sepsis, the best performance markers, and summary statistics.
Table 2. Count of methods used among analyzed studies.
| Method | Retrospective count | Prospective count |
|---|---|---|
| InSight/Dascena | 5 | 1 |
| Regression models | 6 (4 logistic, 2 linear) | 1 (logistic) |
| Markov models | 4 (3 HMM, 1 MM) | – |
| Vector machine | 4 (3 SVM, 1 RVM) | – |
| Bayesian network | 2 | – |
| Hazard models | 2 | – |
| Neural network | – | 1 |
| Fuzzy c-means clustering | 1 | – |
| Regression tree | – | 1 |
| Net classifier | 1 | – |
| Linear dynamic | 1 | – |
| Random forest | 1 | – |
Abbreviations: HMM, hidden Markov model; MM, Markov model; RVM, relevance vector machine; SVM, support vector machine.
Variability in Feature Selection
A majority chose common vital signs, including heart rate, temperature, respiratory rate, and diastolic and systolic blood pressures, for predicting sepsis onset, septic shock, severe sepsis, or mortality. However, some studies went beyond these common vital signs and found that including biological data in tandem with these common features could potentially help enhance prediction and detection. Sutherland et al used blood cultured for gene expression analysis to help with their detection solution. 14 In addition to the common vital and test variables, they included 145 biomarkers to compare gene expression change from the Affymetrix GeneChip data and were able to conclude that gene expression biomarker test had the ability to detect early evidence of sepsis before the availability of microbiology results. Langley et al used blood and protein profiles to help curate individualized detection of sepsis. 13 Using these profiles, they found that patients with severe sepsis had more skewed distribution of metabolomic measurements and distinct metabolic differences between sepsis survivor and death groups. Afterwards, they created a solution that took clinical features and various metabolites to predict survival of patients with sepsis. Lukaszewski et al created neural network models that would predict which ICU patients would develop sepsis from two daily samples of blood. 31 They used various leukocytes and cytokines (IL-1B, IL-6, Il-8, IL-10, TNF-a, CCL-2, Fas-L) as features for model development. However, they mentioned that the model may also be identifying individuals who are more likely to develop sepsis from a genetic predisposition. Although a few studies included biological data to aid with sepsis prediction and detection, it may not always provide better results. Stanculescu et al used biological data from neonates for their real-time sepsis prediction tool. 22 They found that this addition was not statistically significant from their previous work using hidden Markov models and vital signs. From this, it is apparent that feature selection for ML techniques varies and there is not one set of features that is more ideal than another.
Variability in Data Sample Selection and Size
Many studies used publicly available data sets, such as Medical Information Mart for Intensive Care (MIMIC) ( n = 8), 8 12 19 25 26 27 28 32 or the less commonly used Medical Data Warehousing and Analysis (MEDAN) project ( n = 2), 18 33 to help train and validate their tools. The MIMIC-III data set contains 53,423 distinct hospital admissions for adult patients (aged 16 years or above) admitted to critical care units between 2001 and 2012, while the MEDAN data set contains data from 71 German ICUs from 1998 to 2002. 18 34 These data sets are extensive and provide researchers with real, de-identified data that can be used as testing, training, or validation sets when using predictive analytics. Additionally, many studies ( n = 22) used ICU data (either local 14 17 20 21 22 23 29 30 31 35 36 37 or MIMIC), while nine studies used ED 9 10 11 13 15 16 24 38 39 data. While local data varied greatly in size, ranging from 24 to 198,833, some used MIMIC in addition to their local data sets, which created a potentially more generalizable set of data to increase statistical significance and to increase the transfer of learning. Nemati et al used local data as the development cohort and MIMIC data as the validation cohort rather than a random split of local data for both the development and validation cohorts. 35 This allowed them to claim that their solution is more generalizable and has the potential to work sufficiently well across institutions. Similarly, Mao et al integrated both ICU and non-ICU using local mixed wards data to increase generalizability and MIMIC to increase transferability. 28
Additionally, in our review, most of the studies reviewed were retrospective as they used data that was previously collected to create predictive analytics solutions, but there were four prospective studies in which tools were created to assist in the real-time clinical setting. Sutherland et al prospectively predicted sepsis onset by using the American College of Chest Physicians/Society of Critical Care consensus statement and if the patient had suspected infection based on microbiological diagnosis. 14 Using recursive partitioning, LASSO, and logistic regression on microarray procedures, they examined individual genes via a Bayes-adjusted linear model and leave-one-out cross-validation. Later, they used 42 genes to generate a diagnostic classifier using a LogitBoost ML algorithm and applied the classifier to the validation set. Although the diagnosis of sepsis was unknown at the time of enrollment, confirmation was done retrospectively, and they found their real-time detection tool was able to perform before the availability of microbiology results.
Similarly, Lukaszewski et al prospectively monitored molecular changes to identify presymptomatic individuals with an admission diagnosis of “likely septic.” 31 They used real-time polymerase chain reaction to predict sepsis at an early stage of microbial infection, before overt clinical symptoms were to appear. Furthermore, they built five neural network classifiers, each with 30% of the data, to assess nonlinear patterns and used a chi-squared test to ascertain whether the neural network derived predictive accuracies that were statistically significant. Although their solution was able to predict sepsis before the comparative method using the SIRS criteria, they found that clinicians might have trouble understanding the results from the neural network tool. Sawyer et al pilot tested a real-time automated sepsis alert that would increase the rate of interventions within 12 hours of detection. 38 They found that their alert system resulted in an increase in early intervention for those who were identified to be at risk for sepsis.
Variability in “Gold Standard” Definitions
When implementing a retrospective predictive analytics solution, defining the outcome variable can greatly impact the performance. Among the analyzed studies, there was variability in defining sepsis. Some studies determined if a patient had sepsis by using the presence of an International Classification of Disease Code 9 or 10, while others opted for a more rule-based approach based on the Sepsis-2, Sepsis-3, SIRS criteria, and/or organ dysfunction presence. Detailed information regarding gold standard definitions can be seen in Table 1 . Additionally, a few studies relied on manual chart review for determining septic patients. On the other hand, prospective studies utilized a different approach. One utilized an admission diagnosis code upon ICU entry, 31 while another study determined septic patients based on a real-time sepsis alert generated from their clinical alert system. 38
Discussion
In this study, we systematically reviewed the literature to identify all relevant studies that used a predictive analytics solution, including ML and hazards models, to predict onset or mortality of sepsis in hospitalized patients. We identified 31 studies and detailed the various methods and models each study utilized. Because the studies selected were not homogeneous in nature, there are a few distinct differences that should be noted.
Most of the studies reviewed were retrospective, while a few were prospective. Although many of the results show improved accuracy and early detection of sepsis onset or mortality, it remains unknown how effective and efficient many of these predictive analytics solutions are in a real-time patient care setting. To fully understand the usability and accuracy of these solutions, they should be studied prospectively and observed in the health care setting. Similarly, Michael found that prospective cohort studies could potentially capture clinically relevant variables that are absent from retrospective data sets and they could also gather data in a more representative and accurate manner. 40
Furthermore, a lot of studies used ICU data, most likely due to data availability. The algorithms created using just ICU data may not be transferable to other departments due to the high variation in patient population, differences in scoring schemes, and possible missingness for features found in ICU data potentially being utilized in the predictive tool. For instance, often times organ dysfunction is measured using different scoring schemes in the ICU and ED environments. 41 Although many studies presented favorable predictive value, it is nearly impossible to conclude from the reviewed studies if one predictive analytics solution is more effective than another as there were differences among gold standards. These gold standard definitions could have resulted in definition-specific results and if modified could yield differing results. Furthermore, the population and data sizes used for each study were different and the features that were used ranged from solely vital signs all the way to including free text and administrative data. 42 The heterogeneous nature of all the reviewed studies shows that there are many approaches for solving the question of applying predictive analytics for sepsis.
A few studies used industry-created solutions that were sponsored by the respective company. These results can even be seen advertised on the company's Web sites themselves. 43 Because of this connection, there may have been publication bias present. It is best to be aware of this and interpret their respective results accordingly. 44
Limitations of Predictive Analytics Solutions Used
Because of the heterogeneity of the data used, there were many differences in types of predictive analytic solutions used. Most predictive analytics techniques used linear classifiers, such as Naive Bayes and linear or linear and logistic regressions. Cross-validation was also a common technique that was used among the analyzed studies. The use of cross-validation indicates that the selected sample sizes in some of these explored studies may not be large enough. By utilizing cross-validation, an artificial large sample size is created; however, by doing so, there is a risk of overfitting. Using k-fold cross-validation can help reduce the effects of overfitting but does not eliminate the risk. 45 With ML, larger and more representative data sets can result in more realistic outcomes and higher predictive power. Therefore, it is important to consider the effects of data size. However, some models, such as basic linear regression, may oversimplify a real-world scenario as features and response variables may not follow a linear relationship. Multivariate linear regression can produce a more complete model in understanding the independent impact of predictor variables on an outcome; similarly, multivariate logistic regression can only be used when the outcome variable is categorical, which may not always be the intended case. 46
Nonlinear models that were explored include neural networks and Markov models. Neural networks are easy to conceptualize, they are slower, do not have as great of performance metrics, requires tuning many parameters, and if a multilayer neural network is used, then it is even harder to train. 47 Hidden Markov models were also considered, which are memoryless, and make assumptions that the next event is only dependent on the current event and not the past event. Markov models are state machines with the state changes being probabilities. In a hidden Markov models, the probabilities are not known, but the outcome is known. However, implementing a neural network may be too much of a black box and may not be ideal in a health care setting where doctors and clinicians would most likely want to be aware of the computations and reasoning behind the outputs. 48
In addition to the common methods previously mentioned, there were a few unique methods that were used worth discussing. One study used Symbolic Gate Approximation which helped reduce the set of features necessary. Another study used Bayesian principal component analysis (PCA), which can be advantageous for small data sets in “high dimensions as it can avoid the singularities associated with maximum likelihood PCA by suppressing unwanted degrees of freedom in the model.” 49
Generalized additive models were also used. These can be powerful in that they allow us to fit a nonlinear function to each predictor potentially allowing for more accurate predictions when compared with a linear method. Furthermore, because the model is additive, the effect of each predictor can be analyzed when fixing the remaining predictors; however, this additive characteristic can also be hindering. 50 One study used a Weibull–Cox proportional hazards model, which is a good method for analyzing survival data, and is smoother than just a standard Cox model. Finally, random forests were also explored. It is a bagging technique for both classification and regression. The general concept is that you divide your data into several portions, use a relatively weak classifier/regressor to process, and then combine them. Random forest is flexible and can enhance the accuracy of the weak algorithm to a better extent at the expense of heavier computational resources required. 51 However, if the data are not meaningful to begin with, the end result will still not be meaningful. While these uniquely applied predictive analytics solutions are interesting, they were not specifically differentiated from the more common tools that were used. 50 Overall, there was no clear-cut best algorithm; however, when selecting a predictive analytics solution to implement, one must consider the bias–variance tradeoff and sample size of the data.
Limitations in Findings
There are some limitations in terms of how the systematic scoping review was conducted and designed. We chose our search query to be fairly broad to be able to capture the variety of predictive analytics solutions being created for septic patients. Many of these studies had a different objective in their approach to using predictive analytics making it is difficult to determine whether one approach was better than another. If we narrowed the objective of our review to solely include studies that aimed to decrease sepsis mortality or onset, the search would have brought in different studies and examples. Furthermore, the definition of sepsis is ever changing, and this solely marks, generally, what has been done until now.
The availability of data seems to deeply affect and influence potential research opportunity and scope. The most commonly used data were obtained from the ICU followed by the ED. This skew may be due to the availability of public open-access data, such as MIMIC and MEDAN. Therefore, we do not have good information in terms of whether or not predictive analytics tools are better applied in the ICU setting for better patient outcomes. As more data sets become available, we should be careful in interpreting where the application of these algorithms should be best assigned and used. The definition of sepsis that was used as well as target population contributed to the variation. Stanculescu et al looked at developing an alert system for neonates. Because the number of neonates that met their eligibility criteria was low, their study population size was thus limited. 21 22 Most of the studies included in our analysis attempted to detect sepsis or death related to sepsis earlier than what is currently available. Furthermore, most of the studies were retrospective, but there were a few prospective studies performed, and even a randomized control trial. When using ML to predict sepsis onset, many studies used vital signs or laboratory test results as their features to train and test their solutions. However, Sutherland et al show that waiting for the microbiology results could potentially be avoided if gene expression analysis from blood plasma were to be utilized instead. 14 They were able to show strong findings of detecting sepsis before the availability of microbiology results. Shimabukuro et al performed a randomized control trial by using an algorithm created by Dascena, Inc. and found that their predictor decreased the average length of stay and in-hospital mortality rate. 30 Nemati et al found that they can predict, accurately, sepsis onset 4 to 12 hours prior to clinical recognition through the use of their modified hazards model. 35 Horng et al found that utilizing the unstructured text data improved the accuracy of models that solely used the structured data. 16 However, since most of these studies were performed retrospectively, a prospective approach would be needed to determine the feasibility and clinical utility of these predictive analytics methods. For those that did use a prospective approach, there were varying results in accuracy metrics as some found that there was no major improvement in patient outcomes, while others found decreased in-hospital mortality and length of stay rates. Additionally, since the definitions of sepsis currently available rely on clinical features, bias in prediction models will be present as there will be an overlap in the feature set and outcome. Further research and exploration would be necessary in this area.
Limitations in Search Strategy
There are a few limitations in our search query. We used a simplistic and more accessible search phrase query without using search tags for our exploration purposes. Recent systematic or literature reviews that have been published in the last few months utilize more extensive queries and use a range of Boolean and search tags in a wider set of databases. Fleuren et al performed searches on not only PubMed, but also Embase and Scopus. 51 Peiffer-Smadja et al used a general search query to identify general infectious diseases on PubMed, Embase, Google Scholar, BioXiv, Acm Digital Library, arXiV, and IEEE. 52 Schinkel et al performed a review similar to ours by searching only PubMed, but they excluded studies that did not have an AUROC statistic. 53 No search query is perfect; in fact, Salvador-Olivan et al found that almost 93% of search strategies in systematic reviews contained at least one error in their respective search queries. 54
Conclusion
Overall, we found variation in the domain of predictive analytics tools for septic patients, from feature and population size to choice of method or algorithm. However, implementing predictive analytics tools may be beneficial in the early detection of sepsis or death related to sepsis. Since most of these studies were retrospective, the translational value in the real-world setting should be further investigated as other variables such as changes in workflow may also have an impact on outcome. Additionally, many solely used one data set, which is not generalizable across institutions, or even within departments. It will be interesting to see if a predictive analytics tool can be built on top of institutions that have implemented a common data model.
Clinical Relevance Statement
As the amount of data being collected by EHRs continue to increase, it is important to consider ways to harness the data to aid patients. Sepsis is a leading cause of death in hospitals and using retrospective data to create predictive solutions can have the potential in reducing the number of patients affected by sepsis. Prospective-based solutions using real-time vital signs and features can be used to identify septic patients early and potentially decrease morbidity and costs. Because it is important to understand the techniques being used in these solutions, we identified various studies and analyzed their variation in techniques and provide a review for those in the field to consider.
Multiple Choice Questions
-
Predictive analytics approaches using the MIMIC data set:
Have demonstrated poor success in retrospective clinical scenarios.
Have had high accuracy levels to help in the emergency department.
Have been a commonly used source to explore sepsis approaches.
Were not used in tandem with local data.
Correct Answer: The correct answer is option c. We discuss various sources of data that have been used in studies for predictive analytics for sepsis. Often times, open-source data sets, such as MIMIC, have been used as they are easily accessible online and would not require as stringent of human subjects' protections as local institution data. Therefore, these open data sets are a very common way to explore clinical data with machine learning, especially with sepsis.
-
Predictive analytics sepsis prediction approaches using local institution data have:
Used the same common set of standard features.
Used features specific to the study.
Coded features in SNOMED.
All of the above.
Correct Answer: The correct answer is option b. We discuss various feature sets that have been used across studies. Most common approaches were to use feature sets that complemented the data found at the local institution. For example, although most feature sets included common vitals such as heart rate and temperature, a few approaches using local data went a bit further by incorporating clinical text note information or blood tests. This is to highlight that although there are a few common features used among the studies, many still used a curated feature set for their own institution and goals.
Funding Statement
Funding This work was supported by the U.S. Department of Health and Human Services, National Library of Medicine Training Grant T15LM007442
Conflict of Interest None declared.
Protection of Human and Animal Subjects
Human and/or animal subjects were not included in this project.
References
- 1.Data & Reports | Sepsis | CDC. Centers for Disease Control and Prevention. Available at:https://www.cdc.gov/sepsis/datareports/index.html. Accessed August 11, 2019
- 2.O'Brien J. The Cost of Sepsis. Centers for Disease Control and Prevention. Published November 18,2016. Available at:https://blogs.cdc.gov/safehealthcare/the-cost-of-sepsis/. Accessed August 11, 2019
- 3.Sepsis. Mayo Clinic. Published November 16,2018. Available at:https://www.mayoclinic.org/diseases-conditions/sepsis/symptoms-causes/syc-20351214. Accessed August 11, 2019
- 4.Balk R A. Systemic inflammatory response syndrome (SIRS): where did it come from and is it still relevant today? Virulence. 2014;5(01):20–26. doi: 10.4161/viru.27135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Singer M, Deutschman C S, Seymour C W et al. The Third International Consensus Definitions for Sepsis and Septic Shock (Sepsis-3) JAMA. 2016;315(08):801–810. doi: 10.1001/jama.2016.0287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Foster K R, Koprowski R, Skufca J D. Machine learning, medical diagnosis, and biomedical engineering research - commentary. Biomed Eng Online. 2014;13(01):94. doi: 10.1186/1475-925X-13-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheng T-H, Wei C-P, Tseng V S.Feature Selection for Medical Data Mining: Comparisons of Expert Judgment and Automatic ApproachesIn:19th IEEE Symposium on Computer-Based Medical Systems (CBMS'06) Salt Lake City, UT: IEEE; 2006165–170. [Google Scholar]
- 8.Carrara M, Baselli G, Ferrario M.Mortality prediction in septic shock patients: towards new personalized models in critical careIn:2015 37th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC) Milan: IEEE; 20152792–2795. [DOI] [PubMed] [Google Scholar]
- 9.Danner O K, Hendren S, Santiago E, Nye B, Abraham P. Physiologically-based, predictive analytics using the heart-rate-to-systolic-ratio significantly improves the timeliness and accuracy of sepsis prediction compared to SIRS. Am J Surg. 2017;213(04):617–621. doi: 10.1016/j.amjsurg.2017.01.006. [DOI] [PubMed] [Google Scholar]
- 10.Capp R, Horton C L, Takhar S S et al. Predictors of patients who present to the emergency department with sepsis and progress to septic shock between 4 and 48 hours of emergency department arrival. Crit Care Med. 2015;43(05):983–988. doi: 10.1097/CCM.0000000000000861. [DOI] [PubMed] [Google Scholar]
- 11.Faisal M, Scally A, Richardson D et al. Development and external validation of an automated computer-aided risk score for predicting sepsis in emergency medical admissions using the patient's first electronically recorded vital signs and blood test results. Crit Care Med. 2018;46(04):612–618. doi: 10.1097/CCM.0000000000002967. [DOI] [PubMed] [Google Scholar]
- 12.Ho J C, Lee C H, Ghosh J.Imputation-enhanced prediction of septic shock in ICU patients categories and subject descriptorsHI-KDD 2012: ACM SIGKDD Workshop on Health Informatics;2012
- 13.Langley R J, Tsalik E L, van Velkinburgh J C et al. An integrated clinico-metabolomic model improves prediction of death in sepsis. Sci Transl Med. 2013;5(195):195ra95. doi: 10.1126/scitranslmed.3005893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sutherland A, Thomas M, Brandon R A et al. Development and validation of a novel molecular biomarker diagnostic test for the early detection of sepsis. Crit Care. 2011;15(03):R149. doi: 10.1186/cc10274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gultepe E, Green J P, Nguyen H, Adams J, Albertson T, Tagkopoulos I. From vital signs to clinical outcomes for patients with sepsis: a machine learning basis for a clinical decision support system. J Am Med Inform Assoc. 2014;21(02):315–325. doi: 10.1136/amiajnl-2013-001815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Horng S, Sontag D A, Halpern Y, Jernite Y, Shapiro N I, Nathanson L A. Creating an automated trigger for sepsis clinical decision support at emergency department triage using machine learning. PLoS One. 2017;12(04):e0174708. doi: 10.1371/journal.pone.0174708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thottakkara P, Ozrazgat-Baslanti T, Hupf B B et al. Application of machine learning techniques to high-dimensional clinical data to forecast postoperative complications. PLoS One. 2016;11(05):e0155705. doi: 10.1371/journal.pone.0155705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vieira S M, Mendonça L F, Farinha G J, Sousa J MC. Modified binary PSO for feature selection using SVM applied to mortality prediction of septic patients. Appl Soft Comput. 2013;13(08):3494–3504. [Google Scholar]
- 19.Ghosh S, Li J, Cao L, Ramamohanarao K. Septic shock prediction for ICU patients via coupled HMM walking on sequential contrast patterns. J Biomed Inform. 2017;66:19–31. doi: 10.1016/j.jbi.2016.12.010. [DOI] [PubMed] [Google Scholar]
- 20.Peelen L, de Keizer N F, Jonge Ed, Bosman R J, Abu-Hanna A, Peek N. Using hierarchical dynamic Bayesian networks to investigate dynamics of organ failure in patients in the intensive care unit. J Biomed Inform. 2010;43(02):273–286. doi: 10.1016/j.jbi.2009.10.002. [DOI] [PubMed] [Google Scholar]
- 21.Stanculescu I, Williams C KI, Freer Y.A hierarchical switching linear dynamical system applied to the detection of sepsis in neonatal condition monitoringUAI'14 Proceedings of the Thirtieth Conference on Uncertainty in Artificial Intelligence;2014752–761.
- 22.Stanculescu I, Williams C KI, Freer Y. Autoregressive hidden Markov models for the early detection of neonatal sepsis. IEEE J Biomed Health Inform. 2014;18(05):1560–1570. doi: 10.1109/JBHI.2013.2294692. [DOI] [PubMed] [Google Scholar]
- 23.Gultepe E, Nguyen H, Albertson T, Tagkopoulos I. A Bayesian network for early diagnosis of sepsis patients: a basis for a clinical decision support system. 2012 IEEE 2nd International Conference on Computational Advances in Bio and Medical Sciences, ICCABS 2012;20121–5.
- 24.Nachimuthu S K, Haug P J. Early detection of sepsis in the emergency department using Dynamic Bayesian Networks. AMIA Annu Symp Proc. 2012;2012:653–662. [PMC free article] [PubMed] [Google Scholar]
- 25.Calvert J, Desautels T, Chettipally U et al. High-performance detection and early prediction of septic shock for alcohol-use disorder patients. Ann Med Surg (Lond) 2016;8:50–55. doi: 10.1016/j.amsu.2016.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Calvert J S, Price D A, Chettipally U K et al. A computational approach to early sepsis detection. Comput Biol Med. 2016;74:69–73. doi: 10.1016/j.compbiomed.2016.05.003. [DOI] [PubMed] [Google Scholar]
- 27.Desautels T, Calvert J, Hoffman J et al. Prediction of sepsis in the intensive care unit with minimal electronic health record data: a machine learning approach. JMIR Med Inform. 2016;4(03):e28. doi: 10.2196/medinform.5909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mao Q, Jay M, Hoffman J L et al. Multicentre validation of a sepsis prediction algorithm using only vital sign data in the emergency department, general ward and ICU. BMJ Open. 2018;8(01):e017833. doi: 10.1136/bmjopen-2017-017833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McCoy A, Das R. Reducing patient mortality, length of stay and readmissions through machine learning-based sepsis prediction in the emergency department, intensive care unit and hospital floor units. BMJ Open Qual. 2017;6(02):e000158. doi: 10.1136/bmjoq-2017-000158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shimabukuro D W, Barton C W, Feldman M D, Mataraso S J, Das R. Effect of a machine learning-based severe sepsis prediction algorithm on patient survival and hospital length of stay: a randomised clinical trial. BMJ Open Respir Res. 2017;4(01):e000234. doi: 10.1136/bmjresp-2017-000234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lukaszewski R A, Yates A M, Jackson M Cet al. Presymptomatic prediction of sepsis in intensive care unit patients Clin Vaccine Immunol 200815071089–1094.18480235 [Google Scholar]
- 32.Henry K E, Hager D N, Pronovost P J, Saria S. A targeted real-time early warning score (TREWScore) for septic shock. Sci Transl Med. 2015;7(299):299ra122. doi: 10.1126/scitranslmed.aab3719. [DOI] [PubMed] [Google Scholar]
- 33.Pereira R DMA, Fialho A S, Cismondi F 2011. pp. 2507–2512.
- 34.Johnson A E, Pollard T J, Shen L et al. MIMIC-III, a freely accessible critical care database. Sci Data. 2016;3(01):160035. doi: 10.1038/sdata.2016.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nemati S, Holder A, Razmi F, Stanley M D, Clifford G D, Buchman T G. An interpretable machine learning model for accurate prediction of sepsis in the ICU. Crit Care Med. 2018;46(04):547–553. doi: 10.1097/CCM.0000000000002936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ribas V J, Lopez J C, Ruiz-Sanmartin A . Boston, MA: IEEE; 2011. In: 2011 Annual International Conference of the IEEE Engineering in Medicine and Biology Society; pp. 100–103. [DOI] [PubMed] [Google Scholar]
- 37.Shashikumar S P, Stanley M D, Sadiq I et al. Early sepsis detection in critical care patients using multiscale blood pressure and heart rate dynamics. J Electrocardiol. 2017;50(06):739–743. doi: 10.1016/j.jelectrocard.2017.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sawyer A M, Deal E N, Labelle A J et al. Implementation of a real-time computerized sepsis alert in nonintensive care unit patients. Crit Care Med. 2011;39(03):469–473. doi: 10.1097/CCM.0b013e318205df85. [DOI] [PubMed] [Google Scholar]
- 39.Taylor R A, Pare J R, Venkatesh A K et al. Prediction of in-hospital mortality in emergency department patients with sepsis: a local big data-driven, machine learning approach. Acad Emerg Med. 2016;23(03):269–278. doi: 10.1111/acem.12876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Clark M.Retrospective versus prospective cohort study designs for evaluating treatment of pressure ulcers: a comparison of 2 studies J Wound Ostomy Continence Nurs 20083504391–394., quiz 395–396 [DOI] [PubMed] [Google Scholar]
- 41.Raith E P, Udy A A, Bailey M et al. Prognostic accuracy of the SOFA score, SIRS criteria, and qSOFA score for in-hospital mortality among adults with suspected infection admitted to the intensive care unit. JAMA. 2017;317(03):290–300. doi: 10.1001/jama.2016.20328. [DOI] [PubMed] [Google Scholar]
- 42.Govindan S, Prescott H C, Chopra V, Iwashyna T J. Sample size implications of mortality definitions in sepsis: a retrospective cohort study. Trials. 2018;19(01):198. doi: 10.1186/s13063-018-2570-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.UCSF Case Study: Dascena. Available at:https://www.dascena.com/ucsf. Accessed August 11, 2019
- 44.Nassir Ghaemi S, Shirzadi A A, Filkowski M. Publication bias and the pharmaceutical industry: the case of lamotrigine in bipolar disorder. Medscape J Med. 2008;10(09):211. [PMC free article] [PubMed] [Google Scholar]
- 45.Lever J, Krzywinski M, Altman N. Points of significance: model selection and overfitting. Nat Methods. 2016;13(09):703–704. [Google Scholar]
- 46.Worster A, Fan J, Ismaila A. Understanding linear and logistic regression analyses. CJEM. 2007;9(02):111–113. doi: 10.1017/s1481803500014883. [DOI] [PubMed] [Google Scholar]
- 47.Miotto R, Wang F, Wang S, Jiang X, Dudley J T. Deep learning for healthcare: review, opportunities and challenges. Brief Bioinform. 2018;19(06):1236–1246. doi: 10.1093/bib/bbx044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang H, Wu Z, Xing E P. Removing confounding factors associated weights in deep neural networks improves the prediction accuracy for healthcare applications. Pac Symp Biocomput. 2019;24:54–65. [PMC free article] [PubMed] [Google Scholar]
- 49.Bishop C M, Bayesian P CA.MIT Press; 1999382–388.. Available at:http://papers.nips.cc/paper/1549-bayesian-pca.pdf. Accessed April 17, 2020 [Google Scholar]
- 50.Ravindra K, Rattan P, Mor S, Aggarwal A N. Generalized additive models: building evidence of air pollution, climate change and human health. Environ Int. 2019;132:104987. doi: 10.1016/j.envint.2019.104987. [DOI] [PubMed] [Google Scholar]
- 51.Schinkel M, Paranjape K, Nannan Panday R S, Skyttberg N, Nanayakkara P WB. Clinical applications of artificial intelligence in sepsis: a narrative review. Comput Biol Med. 2019;115:103488. doi: 10.1016/j.compbiomed.2019.103488. [DOI] [PubMed] [Google Scholar]
- 52.Fleuren L M, Klausch T LT, Zwager C L et al. Machine learning for the prediction of sepsis: a systematic review and meta-analysis of diagnostic test accuracy. Intensive Care Med. 2020;46(03):383–400. doi: 10.1007/s00134-019-05872-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Peiffer-Smadja N, Rawson T M, Ahmad Ret al. Machine learning for clinical decision support in infectious diseases: a narrative review of current applications Clin Microbiol Infect 2019(September):S1198-743X(19)30494-X. [DOI] [PubMed] [Google Scholar]
- 54.Salvador-Oliván J A, Marco-Cuenca G, Arquero-Avilés R. Errors in search strategies used in systematic reviews and their effects on information retrieval. J Med Libr Assoc. 2019;107(02):210–221. doi: 10.5195/jmla.2019.567. [DOI] [PMC free article] [PubMed] [Google Scholar]


