Gut microbiota diversity has become the subject of extensive research in human and nonhuman animals, linking diversity and composition to gut function and host health. Because wild primates are good indicators of tropical ecosystem health, we developed the idea that they are a suitable model to observe the consequences of advancing global change (e.g., habitat degradation) on gut microbiota. So far, most of the studies focus mainly on gut bacteria; however, they are not the only component of the gut: fungi also serve essential functions in gut homeostasis. Here, for the first time, we explore and measure diversity and composition of both bacterial and fungal microbiota components of two tropical primate species living in highly different habitat types (intact versus degraded forests). Results on their microbiota diversity and composition are discussed in light of conservation issues and potential applications.
KEYWORDS: Tanzania, Udzungwa, bacteria, conservation, fungi, gut microbiota, habitat degradation, primates, red colobus, yellow baboon
ABSTRACT
Human exploitation and destruction of tropical resources are currently threatening innumerable wild animal species, altering natural ecosystems and thus, food resources, with profound effects on gut microbiota. Given their conservation status and the importance to tropical ecosystems, wild nonhuman primates make excellent models to investigate the effect of human disturbance on the diversity of host-associated microbiota. Previous investigations have revealed a loss of fecal bacterial diversity in primates living in degraded compared to intact forests. However, these data are available for a limited number of species, and very limited information is available on the fungal taxa hosted by the gut. Here, we estimated the diversity and composition of gut bacterial and fungal communities in two primates living sympatrically in both human-modified and pristine forests in the Udzungwa Mountains of Tanzania. Noninvasively collected fecal samples of 12 groups of the Udzungwa red colobus (Procolobus gordonorum) (n = 89), a native and endangered primate (arboreal and predominantly leaf-eating), and five groups of the yellow baboon (Papio cynocephalus) (n = 69), a common species of least concern (ground-feeding and omnivorous), were analyzed by the V1-V3 region of the 16S rRNA gene (bacterial) and ITS1-ITS2 (fungal) sequencing. Gut bacterial diversities were associated with habitat in both species, most likely depending on their ecological niches and associated digestive physiology, dietary strategies, and locomotor behavior. In addition, fungal communities also show distinctive traits across hosts and habitat type, highlighting the importance of investigating this relatively unexplored gut component.
IMPORTANCE Gut microbiota diversity has become the subject of extensive research in human and nonhuman animals, linking diversity and composition to gut function and host health. Because wild primates are good indicators of tropical ecosystem health, we developed the idea that they are a suitable model to observe the consequences of advancing global change (e.g., habitat degradation) on gut microbiota. So far, most of the studies focus mainly on gut bacteria; however, they are not the only component of the gut: fungi also serve essential functions in gut homeostasis. Here, for the first time, we explore and measure diversity and composition of both bacterial and fungal microbiota components of two tropical primate species living in highly different habitat types (intact versus degraded forests). Results on their microbiota diversity and composition are discussed in light of conservation issues and potential applications.
INTRODUCTION
Human exploitation and destruction of natural ecosystems are causing the extinction of wild animal species on a global scale (1–3). Recent research also suggests that disturbing the habitat of wild animal populations may trigger the loss of “microbiodiversity,” the array of microorganisms hosted by various body niches, such as the skin and gut (4–6). Given that this microbiota, especially the bacterial component, is known to play a crucial role in animal development, immunity, and nutrition (7–9), loss of this microbiodiversity could have an effect on an individual’s behavior, health, and ultimately on its lifetime fitness (10, 11). In vulnerable animal species, a decrease in individual health and reproductive output could accelerate population decline and species extinction (12, 13).
Tropical nonhuman primates have been a particular focus of microbiota research, given their phylogenetic affinity to humans (14), but also for their contribution to forest regeneration and ecosystem health (15) and conservation status (11), with half of the species being classified as endangered or critically endangered (16, 17). In primates, gut bacterial communities are known to be species specific (18), but they vary widely with age (19), social structure (20), and diet (21, 22) with significant losses of bacterial diversity in captivity (23, 24). Gut microbiota composition and diversity have also been found to vary between contrasting habitat type (4, 5, 19, 25, 26). For example, populations living in fragments of both the black howler monkey (Alouatta pigra) (5) and Udzungwa red colobus (Procolobus gordonorum) (4) have been shown to have a lower gut bacterial diversity in fragmented habitats compared to intact habitats (but see reference 27 for Ugandan red colobus [Procolobus rufomitratus] and black and white colobus [Colobus guereza] and reference 26 for Chlorocebus monkeys). In red colobus, bacterial diversity loss is potentially linked to a decreased ability to digest toxic plant compounds (4), while the gut microbiota of black howler monkeys from secondary forests were more enriched by potentially pathogenic bacterial families, associated with malnutrition and disease (25). Similarly, ring-tailed lemurs (Lemur catta) living in marginal areas were found to be overrepresented by Coprococcus probably as a result of increased contact with human and domestic animal waste (19).
Until now, microbiota research has focused on the bacterial component of the gut ecosystem. Fungi are another relevant, yet relatively neglected taxa contributing to host immunity and gut health, and are possibly associated with disease susceptibility (for a review, see reference 28). In humans, specific fungal communities (“mycobiota”) have also been associated with obesity (29). The mycobiota in wild vertebrate populations are less explored (30, 31), but the composition and diversity of plant-degrading fungi in the gut appear to be linked with phylogeny in herbivorous mammals (32–38). Gut fungi have also recently been shown to vary with sex, age, and season in a group of macaques (39). However, the effect of habitat fragmentation and host lifestyle on gut fungi is unknown, as are the interactions between fungal and bacterial microbiota components.
The present research uses wild primates to investigate the impact of human disturbance and habitat fragmentation on bacterial and fungal gut communities. In this study, we characterize and compare the bacterial and fungal components of the gut microbiota in two nonhuman primate species, one arboreal (the Udzungwa red colobus [Procolobus gordonorum]), and one ground-feeding (the yellow baboon [Papio cynocephalus]) living in both protected and fragmented habitats using metataxonomics of noninvasive fecal samples.
Habitat quality is expected to be crucial in determining the diet ingested by the host, which in turn is one of the most plausible drivers of gut microbiota composition and diversity (4, 40–42); animals feeding in disturbed and fragmented forest tend to have less diverse food sources than in more pristine and continuous forests (43); forests surrounded by cultivations and livestock are also more likely affected by treatments of antimycotics, antibiotics, and pesticides. Thus, we tested whether aspects of primate ecology (diet and behavior) were associated with gut microbiota diversities, asking specifically the three following questions. (i) Are gut microbiota bacterial and fungal diversities higher in a leaf-eating, arboreal (the red colobus) host compared to an omnivorous, terrestrial (yellow baboon) host, as predicted (44, 45)? (ii) Are gut microbiota bacterial and fungal diversities higher in hosts inhabiting protected forests than in those living in fragmented and degraded ones? (iii) Can microbiota composition be used to predict sample origin?
RESULTS
A total of 2,112,896 high-quality reads were obtained from 69 yellow baboon and 89 red colobus fecal samples. Among these reads, 973,226 reads (mean ± standard deviation [SD], 6,160 ± 2,042 reads/sample) belonged to bacteria, while 1,139,670 reads (7,213 ± 4,564 reads/sample) were fungal sequences. After rarefaction and classification, a total of 4,620 bacterial sequence variants (SVs) were recruited from all 158 samples, and 1,419 fungal SVs were recovered from 142 samples.
Are gut microbiota bacterial and fungal diversities higher in a leaf-eating arboreal host (the red colobus) compared to an omnivorous, terrestrial (yellow baboon) host? (i) Bacteria: taxonomy, abundance, and diversity across hosts.
Of the 11 bacterial phyla detected, seven were identified in both primate species (Actinobacteria, Bacteroidetes, Cyanobacteria/chloroplast, Firmicutes, Verrucomicrobia, Spirochaetes, and Proteobacteria), while Fusobacteria, Elusimicrobia, Tenericutes, Lentisphaerae, and Fibrobacteres were found only in yellow baboons (Fig. 1a; see also Data Set S1 in the supplemental material [sheet 1] [no red colobus phyla were unique]).
FIG 1.
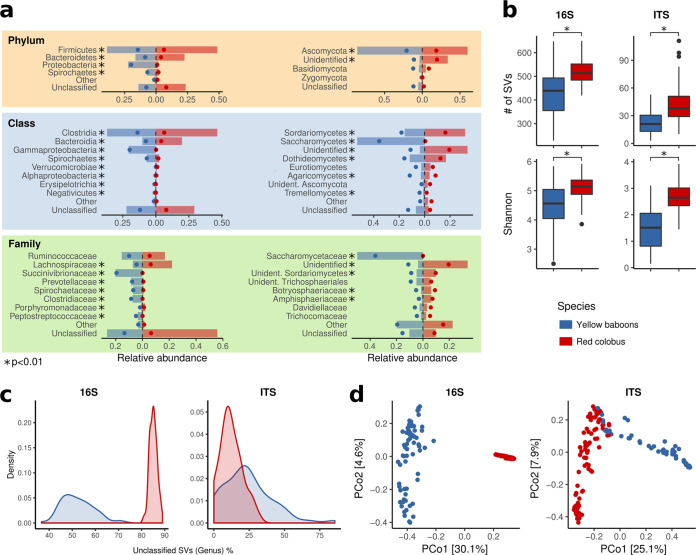
Taxonomic analyses of fecal bacteria (16S) and fungi (ITS) for red colobus (red in all four panels) and yellow baboon (blue). (a) Dominant phyla, classes, and families. The bars show the mean relative abundances expressed as percentages, while the dots show standard deviations. Unidentified refers to SVs present in the reference database; thus, SVs classified as kingdom Fungi for which no lower classification was possible. (b) Bacterial and fungal diversity indices. The number of observed SVs (richness) and Shannon entropy are depicted. (c) Distribution of percentage unclassified SVs at the genus level. (d) Principal coordinate analysis (PCoA) of Bray-Curtis dissimilarities. *, P < 0.01.
(Sheet 1) Median, first quartile (Q1), third quartile (Q3), mean and standard deviation (SD) of relative abundances of bacterial (16S) SVs at the phylum level in yellow baboons (Papio cynocephalus) and red colobus (Procolobus gordonorum). (Sheet 2) Median, first quartile (Q1), third quartile (Q3), mean and standard deviation (SD) of relative abundances of bacterial (16S) SVs at the class level in yellow baboons (Papio cynocephalus) and red colobus (Procolobus gordonorum). (Sheet 3) Wilcoxon rank sum test between yellow baboons (Papio cynocephalus) and red colobus (Procolobus gordonorum) of the relative abundances of 16S SVs at the phylum, class, and family levels. For each taxon, we report the median (“estimate” column), the 95% nonparametric confidence interval (“conf.low” and “conf.high” columns) of the relative abundances difference between the two species, the test statistic, the P value, and the FDR-corrected P value (“p.value.adj” column). (Sheet 4) Median, first quartile (Q1), third quartile (Q3), mean and standard deviation (SD) of relative abundances of bacterial (16S) SVs at the family level in yellow baboons (Papio cynocephalus) and red colobus (Procolobus gordonorum). (Sheet 5) Median, first quartile (Q1), third quartile (Q3), mean and standard deviation (SD) of relative abundances of fungal (ITS) SVs at the family level in yellow baboons (Papio cynocephalus) and red colobus (Procolobus gordonorum). (Sheet 6) Median, first quartile (Q1), third quartile (Q3), mean and standard deviation (SD) of relative abundances of fungal (ITS) SVs at the phylum level in yellow baboons (Papio cynocephalus) and red colobus (Procolobus gordonorum). (Sheet 7) Median, first quartile (Q1), third quartile (Q3), mean and standard deviation (SD) of relative abundances of fungal (ITS) SVs at the class level in yellow baboons (Papio cynocephalus) and red colobus (Procolobus gordonorum). (Sheet 8) Wilcoxon rank sum test between yellow baboons (Papio cynocephalus) and red colobus (Procolobus gordonorum) of the relative abundances of ITS SVs at the phylum, class, and family levels. For each SV, we report the median (“estimate” column), the 95% nonparametric confidence interval (“conf.low” and “conf.high” columns) of the difference in relative abundances between the two species, the test statistic, the P value, and the FDR-corrected P value (“p.value.adj” column). (Sheet 9) Median, first quartile (Q1), third quartile (Q3), mean and standard deviation (SD) of relative abundances of unclassified 16S and ITS SVs at the genus level. (Sheet 10) CCREPE results for yellow baboon (Papio cynocephalus). Significant correlations between 16S and ITS SVs calculated on each forest (Mwanihana, a PF, and Magombera, a FF). (Sheet 11) CCREPE results for red colobus (Procolobus gordonorum). Significant correlation between 16S and ITS SVs calculated on each forest (Mwanihana, a PF, and Magombera, a FF). (Sheet 12) MICTools results (yellow baboons). TICe (total information coefficient) P values (TICePVal), Pearson correlation coefficients (PearsonR), Spearman’s rank correlation coefficients (SpearmanRho), and maximal information coefficient (MICe) for each significant relationship between 16S SVs across forests. (Sheet 13) MICTools results (red colobus). TICe P values (TICePVal), Pearson correlation coefficients (PearsonR), Spearman’s rank correlation coefficients (SpearmanRho), and maximal information coefficient (MICe) for each significant relationship between 16S SVs across forests. (Sheet 14) MICTools results (yellow baboons). TICe P values (TICePVal), Pearson correlation coefficients (PearsonR), Spearman’s rank correlation coefficients (SpearmanRho), and maximal information coefficient (MICe) for each significant relationship between ITS SVs across forests. (Sheet 15) MICTools results (red colobus). TICe P values (TICePVal), Pearson correlation coefficients (PearsonR), Spearman’s rank correlation coefficients (SpearmanRho), and maximal information coefficient (MICe) for each significant relationship between ITS SVs across forests. (Sheet 16) Results of DeSeq2 analysis showing significantly different abundances of 16S SVs between forests (protected versus fragmented) across species. The table reports the species, SV identifier, mean of normalized counts (baseMean), base 2 logarithm of the fold change (log2FoldChange), standard errors of the log2FoldChange (lfcSE), test statistics (stat), P values (pval), corrected P values (padj), and predicted taxonomy. (Sheet 17) Results of DeSeq2 analysis showing significantly different abundances of ITS SVs between forests (protected versus fragmented) across species. (Sheet 18) Mean and standard deviation (SD) of 16S SV importances estimated by the “habitat prediction” RF models trained on yellow baboons (Papio cynocephalus) and red colobus (Procolobus gordonorum) samples (Species column). (Sheet 19) Mean and standard deviation (SD) of ITS SV importances estimated by the “habitat prediction” RF models trained on yellow baboons (Papio cynocephalus) and red colobus (Procolobus gordonorum) samples (Species column). Download Data Set S1, XLSX file, 0.7 MB (750KB, xlsx) .
Copyright © 2020 Barelli et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The four most abundant phyla accounted for 84.6% and 75.3% of total bacterial SVs for baboons and red colobus, respectively, and the mean percentages of each differed between species. Due to the plausible implications of the Firmicutes/Bacteroidetes (F/B) ratio to obesity (46), this ratio has been suggested, despite limitations (47, 48), as an index of the health gut microbiome. Thus, we calculated F/B ratio between species and across habitats. The F/B ratio was significantly higher in yellow baboons than in red colobus (F/B ratio = 2.60 and 2.17, respectively; Wilcoxon rank sum test, P = 0.038), but within species, there was no difference between fragmented forest (FF) and protected forest (PF) (yellow baboons, P = 0.326; red colobus, P = 0.565; see Fig. S1 in the supplemental material).
Violin plots indicating the distribution of the Firmicutes/Bacteroidetes ratio across species and forests. From the bottom to the top, the horizontal lines indicate the first, second (i.e., median), and third quartiles. Download FIG S1, PDF file, 0.03 MB (27.2KB, pdf) .
Copyright © 2020 Barelli et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Among the 21 bacterial classes identified, 19 were detected in baboons, and 13 were detected in red colobus (Data Set S1, sheet 2), with Clostridia (mean relative abundances of 37% and 46.6% in baboons and red colobus, respectively; P = 1.11 × 10−6, Data Set S1, sheet 3) and Bacteroidia (10.3% and 19.8%, respectively; P = 7.06 × 10−15) being the two most abundant classes in both primate species. Gammaproteobacteria were highly represented in baboons (20.6% ± 19.9%), but not in red colobus (0.04% ± 0.2%; P = 5.39 × 10−26). At the family level, 36 microbial families were identified for baboons versus 21 for red colobus, with the two most abundant being the Succinivibrionaceae (20.2%) and the Ruminococcaceae (15.5%) in baboons and the Lachnospiraceae (22.1%) and Ruminococcaceae (16.8%) in red colobus (Data Set S1, sheet 4). The two primate species had very different distributions of unclassified bacteria: the fraction of SVs that could not be classified at the genus level reached a mean of 85.1% in red colobus samples but only 52.2% in yellow baboons (Fig. 1c).
Bacterial alpha diversity differed significantly across the two primate species. Both the Shannon index and number of observed SVs showed that red colobus have significantly higher diversity than yellow baboons (P = 2.3 × 10−8 and P = 7.2 × 10−11 for Shannon index and number of observed SVs, respectively; Wilcoxon rank sum test, Fig. 1b). Bray-Curtis dissimilarities also confirmed that bacterial compositions of the two host species were significantly different from each other, as is clear in Fig. 1d (permutational multivariate analysis of variance [PERMANOVA], R2= 0.296, P ≤ 0.0001).
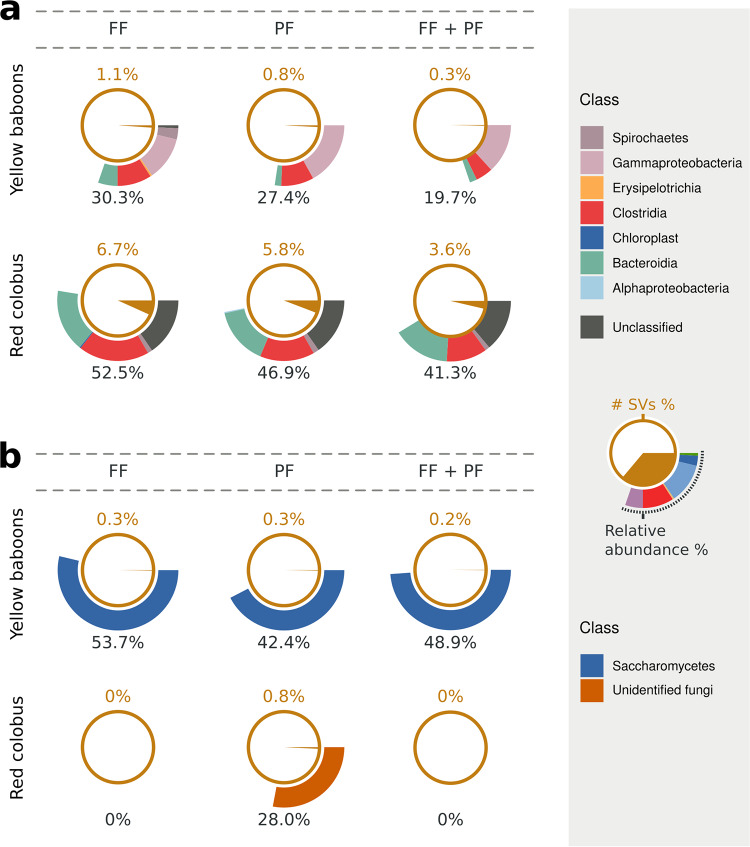
Given a primate species and forest, a very low percentage of bacterial SVs were shared between individuals, even lower when results from the two habitats per species were combined (Fig. 2, inner pie charts). However, among the SVs present in at least 75% of the samples (bacterial “core communities”), those that were shared belonged to the classes representing 20 to 50% of the total relative abundances in each sample (except for red colobus fungal SVs; Fig. 2, outer rings). Yellow baboons shared only about 1% or less of SVs, in both single forests and when they were combined (Fig. 2a). Instead, red colobus shared about 3 to 7% in all combinations of habitats (Fig. 2a).
FIG 2.
Core bacterial (a) and fungal (b) microbiota calculated for each forest separately (fragmented forest [FF] or protected forest [PF]) or combined (FF + PF) for yellow baboons (top pie charts) and red colobus (bottom pie charts). The core microbiota is expressed as the fraction of the number of SVs (inner brown pie charts and top brown numbers) and as mean relative abundance (separated at class level, outer rings and bottom black numbers).
(ii) Fungi: taxonomy, abundance, and diversity across hosts.
A total of 123 fungal families (79 in yellow baboons and 101 in red colobus) were identified (Data Set S1, sheet 5). These families derived primarily from three phyla: Ascomycota, Basidiomycota, and Zygomycota (Data Set S1, sheets 6 and 7). At the class level, significant differences between primate species were found in Sordariomycetes (P = 4.55 × 10−8), Saccharomycetes (P = 6.53 × 10−22), Dothideomycetes (P = 1.67 × 10−3), Agaricomycetes (P = 9.29 × 10−4), and Tremellomycetes (P = 1.15 × 10−3; Data Set S1, sheet 8). In contrast to bacterial SVs, the fraction of unclassified fungal SVs at the genus level was much lower, although higher in yellow baboons than in red colobus (23.7% and 12.3%, respectively; Fig. 1c and Data Set S1, sheet 9). Like bacteria, the fungal alpha diversity differed across host species with red colobus having significantly higher fungal richness than yellow baboons (Fig. 1b) regardless of the index (number of SVs, P = 1.1 × 10−9; Shannon entropy index, P = 6.7 × 10−15). Moreover, the internal transcribed spacer (ITS) profiles between species differed significantly (R2 = 0.182, P ≤ 10−4) in terms of Bray-Curtis dissimilarity (Fig. 1d).
As for bacteria, very few fungal SVs were shared between individuals of each species (Fig. 2). However, the core fungal composition of baboon fecal samples was dominated by a few SVs of the Saccharomycetes class that represented on average 50% of the relative abundances in both forests (Fig. 2b). Red colobus individuals shared virtually no fungal SVs, and those that they did share could not be identified at taxonomic levels lower than domain (Fig. 2b).
(iii) Correlation between and within bacterial and fungal microbiota.
For each primate species, we also explored correlations between diversity indices between bacteria and fungi and within each domain. Only red colobus showed negatively correlated Shannon indices between domains (Pearson r = −0.301, P = 0.007; Fig. S2), but the same pattern was not noted in this species for other alpha diversity indices such as the number of observed SVs or Chao1. No correlation of alpha diversities between bacteria and fungi was detected in yellow baboons. Compositionality corrected by renormalization and permutation (CCREPE) analysis of relative abundances between domains in each species also indicated significant correlations between bacteria and fungi, mainly involving Clostridia and Bacteroidia from the bacterial classes and Sordariomycetes and Dothideomycetes among the fungal classes. Again, a greater number of correlations were found in red colobus (24 in Mwanihana; eight in Magombera; Data Set S1, sheet 10) compared to yellow baboons (only one correlation in Mwanihana; Data Set S1, sheet 11).
MICTools analysis. Significant relationships between the relative abundances of SVs across species and habitats for both 16S (a) and ITS (b) data. SVs are grouped by the predicted taxonomy; the size of the circle represents the number of significant SV relationships for each pair of classes. Download FIG S2, PDF file, 0.01 MB (12.9KB, pdf) .
Copyright © 2020 Barelli et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Considering relative abundances within each domain, we found significant positive and negative correlations in each primate species (MICtools analysis, see details in Data Set S1, sheets 12 to 15). Specifically, the numbers of correlations for fungi were similar between primate species, while many more correlations were noted for bacteria in baboons (Fig. S3). Again, the majority of correlations included unidentified fungi and Clostridia and Bacteroidia bacteria.
Results of MICTools analysis, showing significant relationships between the relative abundances of SVs across species and habitats for both 16S (a) and ITS (b) data. SVs are grouped by predicted taxonomy; the size of the circle represents the number of significant SV relationships for each pair of classes. Download FIG S3, PDF file, 0.02 MB (26.2KB, pdf) .
Copyright © 2020 Barelli et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Are gut microbiota bacterial and fungal diversities higher in hosts inhabiting protected forests than in those living in fragmented and degraded ones? (i) Bacteria.
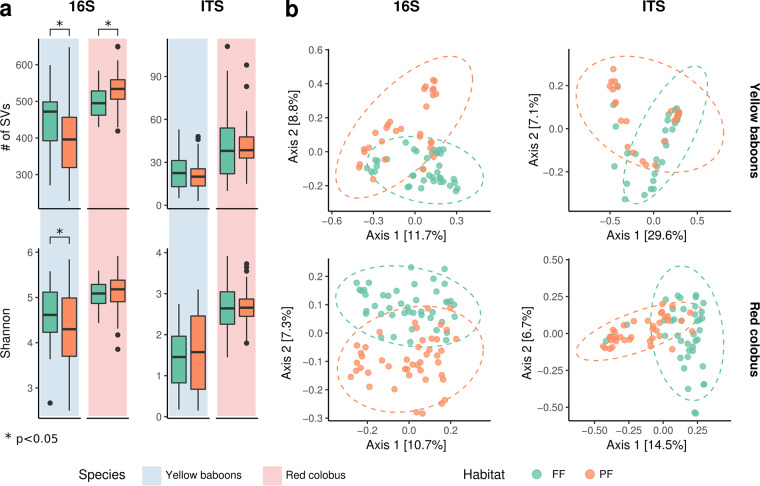
Red colobus had a significantly higher number of observed bacterial SVs in PF than in FF (P = 0.036, generalized linear model), but not a higher Shannon entropy (P = 0.659; Fig. 3a). The opposite was found in yellow baboons, with individuals from FF having a significantly higher bacterial alpha diversity than those from PF for both indices (number of observed SVs and Shannon index: P = 0.002 and P = 0.003, respectively; Fig. 3a).
FIG 3.
Diversity of bacterial and fungal gut communities in red colobus and yellow baboons in different forest types. (a) Alpha diversity of bacterial (16S) and fungal (ITS) communities estimated as number of observed SVs (top panel) and Shannon entropy (bottom panel) in yellow baboons (blue background) and red colobus (red background) in fragmented forest (FF) (green bars) and protected forest (PF) (orange bars). *, P < 0.05. (b) Principal coordinate analysis (PCoA) of Bray-Curtis dissimilarities of bacterial (16S) and fungal (ITS) data (see also Data Set S1, sheet 7, in the supplemental material) within species and across habitats. Dashed ellipses represent 95% confidence interval, while the numbers in square brackets refer to the variance explained by the PCoA axes.
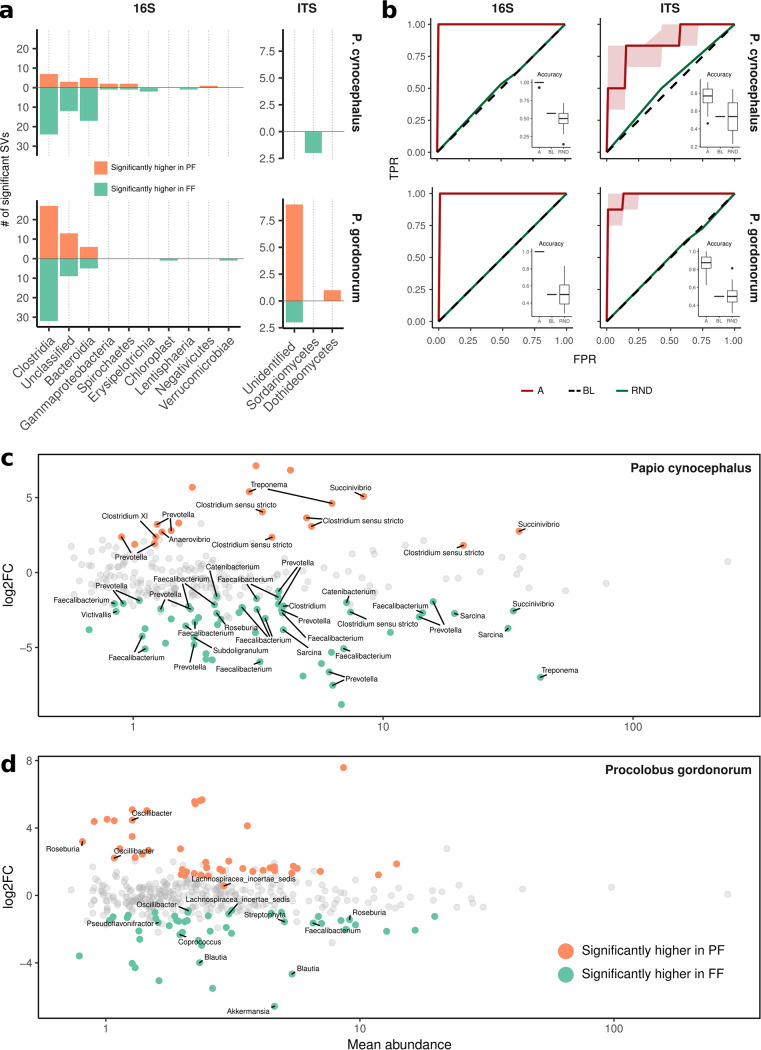
DESeq2 was used to identify changes in the relative abundance of taxa between primate species and habitat types. DESeq2 identified differentially abundant bacterial SVs between forests, independently for each primate species (false discovery rate [FDR] corrected using the Benjamini-Hochberg procedure, P < 0.01; Fig. 4a, left panel), highlighting that among the 31 Clostridia SVs identified as differentially abundant in yellow baboons, 24 were more abundant in FF and 7 were more abundant in PF. Similarly, for 22 Bacteroidia differentially abundant in yellow baboons, 17 were more abundant in FF and five in PF. In addition, the classes Erysipelotrichia and Lentisphaeria were enriched only in FF and Negativicutes only in PF (Fig. 4a). For red colobus, 32 Clostridia SVs were differentially abundant in FF and 27 in PF; five Bacteroidia SVs were differentially abundant in FF and six in PF (Fig. 4a). DESeq2 analyses also found that yellow baboon samples from FF were enriched in 10 genera; for example, 16 Faecalibacterium and 12 Prevotella (Fig. 4c), while the same species from PF was enriched with six other genera (Fig. 4c). Conversely, although most bacterial genera in red colobus were unclassified, nine were differentially abundant in FF and three in PF (Fig. 4c, bottom portion). Among those that could be classified, Faecalibacterium, Coprococcus, Pseudoflavonifractor, Streptophyta, Blautia, and Akkermansia were found only in FF (Fig. 4c, bottom portion; see also Data Set S1, sheet 16).
FIG 4.
Significant differentially abundant SVs (P < 0.01) grouped by class (a) and genus (c) level. The green bars in panel a show SVs that are significantly higher in fragmented forest (FF), while the orange bars show SVs that are significantly higher in protected forest [PF]), estimated from the number of SVs. (b) Random Forest (RF) predictive performances (receiver operating characteristic [ROC] curves and accuracy [insets] are shown for both bacterial [left panel] and fungal [right panel] domains and across primate species [top and bottom plots]). Abbreviations: TPR, true positive rate; FPR, false-positive rate; A, actual; BL, baseline classifier; RND, random classifier. (c) Logarithm-transformed fold change (log2FC) of each SV against its mean relative abundance for yellow baboons (top panel) and red colobus (bottom panel). Significant SVs (colored points) without the taxonomic annotation are “Unclassified” SVs at the genus level.
(ii) Fungi.
Fungal diversity was not statistically different between habitats for either primate species (yellow baboons, P = 0.506, P = 0.395; red colobus, P = 0.741, P = 0.785; Fig. 3a).
Instead, comparison of beta diversity revealed that the composition of bacterial and fungal communities was unique for each species across habitats (Bray-Curtis dissimilarities and weighted UniFrac for 16S in yellow baboons, R2 = 0.067, P < 0.001 and R2 = 0.077, P < 0.001; in red colobus, R2 = 0.059, P < 0.001 and R2 = 0.042, P < 0.001; for ITS in yellow baboons, R2 = 0.036, P = 0.017 and R2 = 0.023, P = 0.17; in red colobus, R2 = 0.075, P = 0.001 and R2 = 0.080, P = 0.001; Fig. 3b; see also Table S1 in the supplemental material).
When considering the ratio between the number of 16S and ITS SVs, we did not find any significant differences (Wilcoxon rank sum test, P = 0.397) between individuals living in different habitats. However, yellow baboons had a higher ratio than red colobus did (P = 1.29 × 10−4; yellow baboons, median = 19.7, interquartile range [IQR] = 21.0; red colobus, median = 13.1, IQR = 8.94).
Fungal communities presented a smaller number of differentially abundant SVs than bacterial communities did (Fig. 4a, right panels): only yellow baboon samples from FF were enriched in two Sordariomycetes classes. Red colobus from FF had two differentially abundant unidentified classes, while those from PF were enriched in Dothideomycetes and nine unidentified classes (Fig. 4a; see also Data Set S1, sheet 17).
Can microbiota composition be used to predict habitat origin?
Machine learning algorithms were used to test whether the origin of each sample from the composition of its bacterial and fungal microbiota can be predicted. The random forest classification algorithm revealed that sample origin could be predicted on the basis of bacterial composition for both red colobus and yellow baboons (for both species accuracy and Matthews correlation coefficient [MCC], median = 1, IQR = 0; out-of-bag [OOB] error [mean ± SD], yellow baboons, 0.03 ± 0.02; red colobus, 0.001 ± 0.004; Fig. 4b, left panels). The most relevant SVs explaining the habitat prediction model were distinct for red colobus and yellow baboons, with SVs belonging to the Ruminococcaceae and Lachnospiraceae families in red colobus and to the Prevotellaceae and Clostridiaceae in yellow baboons (Data Set S1, sheet 18; Fig. S4). Thus, the taxa involved in explaining the model were highly different between primate species, confirming the results found for the DESeq2 analysis (Fig. 4c).
Top 15 16S SVs in terms of feature importance identified by the habitat prediction RF models trained on yellow baboons (Papio cynocephalus) and red colobus (Procolobus gordonorum) samples. The thick colored bars and intervals represent, respectively, the means and 2 standard deviations of the normalized Gini importances calculated over the 50 cross-validation iterations. Download FIG S4, PDF file, 0.02 MB (21.4KB, pdf) .
Copyright © 2020 Barelli et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Similarly, fungal composition also predicted sample origin (Fig. 4b, right panels) but with less accuracy (for yellow baboons, accuracy, median = 0.77, IQR = 0.15; MCC, median = 0.55, IQR = 0.32; for red colobus, accuracy, median = 0.88, IQR = 0.13; MCC, median = 0.76, IQR = 0.21; OOB error [mean ± SD] for yellow baboons, 0.28 ± 0.05; for red colobus, 0.14 ± 0.03). Although the most relevant SVs involved in explaining the model were “Unidentified” at the family level for red colobus, those SVs associated with habitat were again very different between species (Data Set S1, sheet 19; Fig. S5).
Top 15 ITS SVs in terms of feature importance identified by the habitat prediction RF models trained on yellow baboons (Papio cynocephalus) and red colobus (Procolobus gordonorum) samples. The thick colored bars and intervals represent, respectively, the means and 2 standard deviations of the normalized Gini importances calculated over the 50 cross-validation iterations. Download FIG S5, PDF file, 0.02 MB (21.4KB, pdf) .
Copyright © 2020 Barelli et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
DISCUSSION
Here, we test whether the bacterial and fungal components of the gut microbiota differ between two species of wild primates with different ecologies living in both protected and fragmented forests in a tropical African biodiversity hotspot. Because seasonal shifts may drive variation in dietary patterns and, consequently, in gut microbiota compositions (e.g., see references 21, 49, and 50), individual fecal sample collection was performed in a very narrow temporal window to avoid any potential seasonal effects. Thus, any intraspecific differences should reflect the effects of habitat type. Our findings revealed several patterns: bacterial and fungal microbiota composition of arboreal, leaf-eating red colobus monkeys was very different from that of ground-dwelling omnivorous yellow baboons, with red colobus having higher bacterial and fungal gut diversities than yellow baboons, as predicted from differences in dietary strategies. However, while red colobus living in protected forests had higher levels of diversity than those in unprotected forests, as predicted and noted in previous studies (4), unexpectedly, yellow baboons showed the opposite correlation, probably due to their ecologically different niches represented by their omnivorous diet and terrestrial habits. In addition, some correlations between bacterial and fungal diversity and composition were detected, and we showed that using machine learning models, it is possible to identify habitat of origin from fecal sample microbiota diversity. These results are discussed with regards to conservation issues and potential applications.
Gut microbiota diversities are higher in the leaf-eating red colobus than in the omnivorous yellow baboons.
Despite the lack of explicit data on the diet ingested by the hosts in our results, we do refer to the extensive data on dietary habit and intake available from the literature of these and other red colobus and baboon populations. Bacterial richness (alpha diversity) was significantly higher in leaf-eating red colobus than in omnivorous yellow baboons, highlighting the differences in dietary strategies and digestive physiology of the two species. Although these two species live sympatrically and both belong to the Cercopithecidae, they possess ecologically different adaptations. Yellow baboons have an anatomically simple digestive tract, are hindgut fermenters and omnivorous, consuming a variety of food items, such as fruits, leaves, seeds, insects, roots, and small vertebrates (51) with 6% to 20% of fiber intake (52). Udzungwa red colobus, like other red colobus, have a specialized digestive system similar to ruminants, are foregut fermenters and rely almost exclusively on a folivorous diet, with up to 75% of fiber intake (53, 54). Although baboons occasionally include leaves and seeds in their diet, it is unlikely that their diet contains fiber contents comparable to that of red colobus. Thus, our results are in line with those revealing that greater fiber intake is associated with higher bacterial richness (41, 42, 55, 56). Similar findings have been reported for other primate species (20) and humans under strictly vegetarian diets compared to omnivorous ones (44). The two host species also showed highly different fungal richness, resembling the pattern found in bacteria but at a much lower scale (150-fold lower). Leaf-eating red colobus monkeys again presented higher fungal richness than the omnivorous baboons, consistent with previous findings (38) indicating that hosts with high gut fungal richness might be more efficient in breaking down plant fibers (57). Another interesting difference between the two host species concerns the surprisingly low interindividual variation of bacterial composition in red colobus compared to yellow baboons. Such stable and subtle interindividual variation likely indicates that red colobus gut bacteria are more phylogenetically similar because they have similar and highly specialized functions (i.e., degrading complex plant material such as cellulose, hemicellulose, and xylan [4]), compared to the gut bacteria of yellow baboons, which show greater dietary plasticity and greater ability to inhabit novel environments (i.e., human settings).
Gut microbiota diversities are higher in protected forests for red colobus but not yellow baboons.
Ecological theory as well as empirical data have long shown that large expanses of natural forest habitat usually host a high autochthonous tree diversity, offering their wild animal inhabitants a more diverse diet than smaller tracts of degraded and fragmented habitat (58). Furthermore, it has been suggested that greater dietary variation promotes higher bacterial diversity in the host gut (59). Thus, if we consider the two host species living in intact and degraded habitats, we expected that higher bacterial alpha diversity should be found in the species residing in their natural, intact habitat. Our results strongly support this hypothesis for the red colobus, confirming our previous report noting associations between higher bacterial richness and habitat integrity (4), which are corroborated in other species as well (black howler monkey [Alouatta pigra] [5]; Andean bears [Tremarctos ornatus] [60]). However, baboons had an unexpectedly high gut bacterial diversity in FF compared to PF. This may be because fragmentation or disturbance of vegetation may stimulate a higher number of tolerant tree species in secondary-growth forest (61), and/or provoke modification in both diet composition and quality, allowing for a greater diversity of food consumption (62) in generalist feeders. Therefore, the higher bacterial diversity found in the omnivorous baboons could be due to a more diverse diet of baboons in these habitats. However, these differences in gut diversity could also be due to differences in behavior. Because of the rapid conversion of tropical forest to human-modified croplands, baboons have altered their socioecological behavior, especially in the neighboring areas where human density is the highest (like FF compared to PF), spending more time on the ground feeding on human food and food waste in villages, as well as raiding cultivations (i.e., vegetables and sugarcane). In this way, baboons increase their nutrient intake (63–65), but they also come into more frequent contact with soil, livestock, and humans, as well as their associated microbiota, either directly or through fecal-oral transmission. Higher gut flora diversity in mammals living on human-derived diets have been found in olive baboons [Papio anubis] (66), polar bears [Ursus maritimus] feeding on bone piles (67), and cattle straying into suburban areas (68), as well as in some species in captivity (including rhinoceros and red-crown crane [Grus japonensis] [69, 70] but see references 71 and 72). More recently, Grieneisen and colleagues have suggested that soil properties also alter soil microbiota and the microbiota of terrestrial animal species (73). Together, these results suggest that disturbed habitats may promote a higher diet diversity rather than a lower diet diversity and/or modify behavior, both of which contribute to the biodiversity of host gut communities. Since bacterial richness has been suggested as an indicator of “gut health,” either animals in proximity to human settlements and consuming human-derived food are healthier than those living in PF or microbiota richness is not in fact an indicator of gut health. For example, in the polar bear, the higher gut diversity of bears feeding on bone piles is also associated with a higher number of potential pathogens (67). Moreover, in light of new results (73), we cannot exclude the possibility that the soil microbiota of the two forests may directly shape microbial communities in this baboon population, regardless of diet and behavior.
Gut microbiota composition and function.
A closer examination of more specific components of the bacterial and fungal microbiota confirms the relevance of the diverse dietary strategies of the two primate species in different habitats (41, 74). For example, leaf-eating red colobus monkeys shared, as expected, clades of the Bacteroidia and Clostridia classes most likely associated with the digestion of cellulose (75). In fact, the bacterial families Ruminococcaceae and Lachnospiraceae dominated the red colobus guts. Because those families are associated with degradation of complex plant material like cellulose and lignin (75, 76), the overrepresentation in red colobus may reflect their folivorous diet, as already found in most of the leaf-eating primates and ruminants (20, 77, 78). Our findings that baboon guts were predominantly associated with Gammaproteobacteria which are linked to starch digestion and carbohydrate fermentation confirm previous results (79, 80) and support their higher consumption of sugar-rich items. More specifically, baboon guts were dominated by Succinivibrionaceae (20.2%), which is a starch-digesting bacterial family that assists in the fermentation of carbohydrates to succinate and acetate (81). For fungi, the guts of yellow baboons were mainly dominated by Ascomycota (the largest phylum of the kingdom fungi), a few Basidiomycota and Zygomycota, consistent with previous findings in humans and nonhuman primates (30, 39, 82), whereas the guts of red colobus, beside considerable Ascomycota and a few Basidiomycota and Zygomycota, showed many unidentified fungi. An examination of core SVs follows this pattern: there were no shared bacterial and fungal SVs across species; however, within host species, a few core SVs accounted for about half of the microbiota composition.
Bacterial and fungal community composition also differed considerably across protected (PF) and fragmented (FF) habitats that just 60 years ago were naturally connected, before human settlements and intensive agriculture separated them (83). In fact, each species in each habitat had a unique gut microbiota, for both bacteria and fungi, again most likely linked to differences in diet discussed above. For example, among the classified SVs, only baboons of Magombera (FF), but not those of PF, were enriched with the bacterial genera Sarcina, Prevotella, and Faecalibacterium. Sarcina are synthesizers of microbial cellulose and linked to chitinolytic or protein-degrading functions, consistent with arthropod consumption, as reported for the white-faced capuchin monkeys (49). Since arthropods are a regular part of the diet of this and other baboons regardless of habitat (51, 84), the enrichment of Sarcina in the FF forest may be associated with feeding on a high-protein human-derived food resource, such as human waste. Indeed, Prevotella are frequently associated with consumption of fiber- and sugar-rich diets (40), consistent with the baboon diet in FF which includes sugarcane; similarly, Prevotella was also enriched in captive douc langurs consuming sugar-rich fruits (22). However, factors other than fiber and sugar ingestion contribute to high diversity and abundance; for example, Sarcina and Prevotella diversity are also linked with chronic gut inflammatory conditions and are implicated in causing gut diseases (85–87). Interestingly, in contrast to the potential negative associations of these genera, the butyrate-producing Roseburia and the Faecalibacterium genus were also overrepresented in baboons living in FF, two taxa generally associated with promoting intestinal barrier function and thus, metabolic benefits. On the other hand, baboons living in PF had a relatively high abundance of Clostridium sensu stricto, another group of bacteria providing protective mechanisms involved in the resistance to infection (88). More-detailed investigations on the role of such taxa are necessary to assess these interactions and gut homeostasis.
Considering the central role that fungi have in maintaining intestinal homeostasis and systemic immunity (89), variation in their diversity may impair host health and gut homeostasis. Thus, investigating either the direct interaction with the host and the competitive association between bacterial and fungal microorganisms in the gut become crucial for the conservation of animal species, especially for those that face constant threat due to habitat modification such as primate hosts. For fungi, we found that 10 different classes of unidentified fungi and one class of the Dothideomycetes were significantly enriched in PF in red colobus, presumably in response to a diet more rich in fiber. Baboons from FF were instead significantly enriched in two SVs belonging to the Sordariomycetes class. Although still poorly understood, the different dietary habits may drive diversification of fungal communities across species. However, given the lack of genomic databases and knowledge on the metabolic functions of fungi (compared to those of bacteria), at present, we can only note the differences and that patterns are similar to those found for bacteria, but we are unable to conjecture about potential explanations for these patterns.
The positive correlations between fungal and bacterial communities in the leaf-eating monkeys (especially those living in protected forests) and between bacterial taxa of baboons are intriguing, although their biological significance is still unclear, given the rarity of the taxa involved. Further investigation into these relationships are needed to unravel the significance of these patterns.
The Udzungwa hotspot is associated with a high proportion of unclassified SVs.
About 10 to 20% of the total SVs could not be assigned even to a phylum as noted previously for bacteria (4). We contend that the high level of native vertebrate and invertebrate species reported thus far from the Udzungwa hotspot (90) is also reflected in primate gut microbiota, resulting in a high number of unclassifiable bacterial and fungal taxa unlisted in any database, and therefore, new to science, as also suggested for lemurs (91), Verreaux’s sifakas (50), and human hunter-gatherers (55). Interestingly, only about half of the yellow baboon gut bacteria are unidentified at the genus level compared to 85% of red colobus gut bacteria (Fig. 1c), possibly reflecting the higher contact between baboons and human residents, their omnivorous diet and similar digestive system, and therefore, possibly sources of gut microbiota (which is much better known for humans). However, further investigations of these unknown microrganisms are ongoing.
Rethinking gut microbiota diversity as a tool for conservation.
Since greater taxonomic diversity might mirror greater functional diversity (9, 92), it is plausible that higher gut diversity might have an effect on “gut health” (93), and therefore, on overall host health (94, 95). Therefore, it has been suggested that gut microbiota could be used as a tool for assessing individual health of threatened taxa, especially in the case of noninvasive sampling (96). As mentioned above, the leaf-eating red colobus had higher bacterial and fungal richness than omnivorous baboons, indicating that their fiber-rich diet may promote a healthier gut; however, unexpectedly, baboons in FF had a higher gut diversity than baboons in PF. Thus, determining general patterns of associations between microbiota diversity and habitat characteristics remains elusive. However, we believe that species-specific ecological adaptations may mask the understanding of the general patterns (97). Based on gut microbiota diversity and composition, some species might be less resilient than others and respond differently to environmental variation. In this context, specialist leaf-eating primates, dependent on forest integrity, are more likely to show a greater impact to habitat degradation also in terms of gut microbiota diversity and composition. Conversely, species not dependent on a specialized diet, such as the omnivorous baboons that adapt better and show higher tolerance to human disturbance, may widen their dietary choices and potentially increase their gut microbiota. Although the possibility that omnivorous primates overcome the effects of habitat degradation by consuming human food and may be in better shape than those that are less unable to adapt like the red colobus is not excluded, we suggest that standard microbiota indices per se might not be a universal indicator of a “healthy gut.” Thus, deeper investigations into the functional analysis of each gut component are needed, as well as more appropriate data sets to better test ecological theory (98) for understanding which processes shape diversity in the host-associated microbial ecosystem. Moreover, to evaluate the association between gut health and diversity, and its value as a conservation tool, the present experimental design should be repeated using additional species, or even entire communities, with alternative lifestyles.
Besides the increasing impact of machine learning models on health care (99), to our knowledge, this is the first use of machine learning to investigate whether gut microbiota composition is associated with the habitat of origin in wildlife. Our analysis showed that almost all samples were correctly assigned to the forest where they were collected on the basis of their bacterial and fungal compositions. Therefore, the gut bacterial microbiota of each Udzungwa primate population is a distinctive signature of the forested area from which it belongs, and could potentially be used as a biomarker of their habitat of origin. Additionally, abiotic factors, such as soil properties and microbial communities of the surrounding environment, should be included to better predict host microbial diversity. Moreover, if gut microbiota accurately reflects the specificity of their habitat of origin, continued longitudinal monitoring of changes in this composition could indicate changes in habitat and/or individual health and species conservation status, as well as how quickly these are occurring. Further analyses of the data set to identify additional biomarkers for conservation and management purposes are also ongoing.
MATERIALS AND METHODS
Study site.
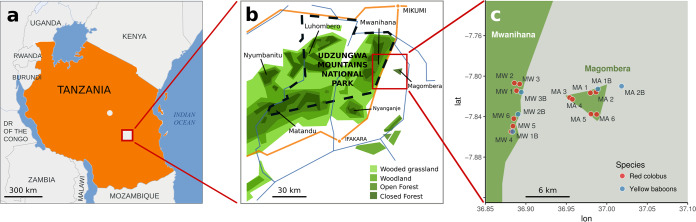
The Udzungwa Mountains of Tanzania (Fig. 5) form part of an internationally recognized biodiversity hotspot (www.conservation.org); however, these relatively pristine forests are threatened by intensive agriculture (100), and primates living outside protected areas are particularly vulnerable to hunting and other human disturbance (101). The omnivorous yellow baboon is terrestrial, foraging for arthropods, seeds, fruit, and leaves, although they opportunistically also raid crops; this species is categorized as “least concern” by the International Union for the Conservation of Nature and Natural Resources (IUCN) (16). In contrast, the diet of the arboreal Udzungwa red colobus mainly consists of leaves and stems; it is native and endangered (16). Both these species live in Mwanihana (in a protected forest [PF]), an intact semideciduous to submontane and montane evergreen forest located within the boundaries of the Udzungwa Mountains National Park since 1992 (covering an area of 150 km2; altitude, 351 to 2,263 meters above sea level [ASL]), and Magombera (a fragmented forest [FF]) a flat, groundwater forest fragment surrounded by villages and sugarcane plantations approximately 6 km east of Mwanihana (covering an area of 12 km2; altitude, 269 to 302 meters ASL).
FIG 5.
Map of the Udzungwa Mountains in Tanzania (a). Enlargement indicating the two forest blocks, Mwanihana and Magombera (b), and sample sites for the 12 social groups of Udzungwa red colobus and 5 social groups of yellow baboons (c). The dashed line in panel b indicates the border of the Udzungwa Mountain National Park. Colors identify species (i.e., red and blue for red colobus and yellow baboons, respectively).
Noninvasive fecal sample collection.
C. Barelli and two trained local field assistants followed the primate social groups (average group size, approximately 25 individuals) unobtrusively from the ground and collected fecal samples deposited by a total of 69 yellow baboons (belonging to five different social groups) and 89 red colobus (from 12 social groups; Fig. 5). The data collection was conducted over 4 weeks between June and July 2016 to avoid seasonal variation in diet (53) following already established procedures (4). Three social groups of yellow baboons were sampled from PF and two from FF. Six of the sampled groups of red colobus inhabited PF, while the remaining six groups were in FF. Samples were preserved in 96% ethanol, kept at 4°C, transported to the Fondazione E. Mach in Italy, and kept at –80°C until shipment to the University of Illinois, USA, for analysis. Sample collection complied with laws governing wildlife research in Tanzania under research permit 2016-267-ER-2009-49 obtained from the Tanzania Commission for Science and Technology (COSTECH), Tanzania Wildlife Research Institute (TAWIRI) and Tanzania National Parks (TANAPA).
DNA extraction and Illumina sequencing.
Whole DNA was extracted from 0.25 g of each fecal sample using the QIAamp PowerFecal DNA kit (Qiagen Group, Hilden, Germany), following manufacturer’s instructions, quantified on a Qubit (Life Technologies) using the High Sensitivity DNA kit, and amplified by the DNA Services Laboratory, Roy J. Carver Biotechnology Center, University of Illinois, Urbana-Champaign, IL, USA. Library preparation for Illumina MiSeq sequencing of the V1-V3 regions of the 16S rRNA gene was carried out using the primer pair 28F and 519R for the PCR amplification of bacteria, while the 5′-GCATCGATGAAGAACGCAGC (forward) and 5′-TCCTCCGCTTATTGATATGC (reverse) primers were used to amplify the ITS1-ITS2 region of fungi (protocols found in references 121 and 122; further information in Text S1 in the supplemental material). Sterile water and human fecal DNA samples were also included as negative and positive controls during each step.
Detailed information of bacterial and fungal extraction methods, PCR amplification, and sequencing. Additional information on software used for data processing for both bacterial and fungal sequences and statistical analyses. Detailed scripts used to analyze bacterial (16S) and fungal (ITS) sequences for the Micca pipeline. Additional information on the intradomain relationships between SVs relative abundances, specifically, the correlations across classes of bacteria and fungi among the two primate hosts. Download Text S1, DOCX file, 0.02 MB (25.2KB, docx) .
Copyright © 2020 Barelli et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Data processing and statistical analyses.
Raw bacterial 16S rRNA gene sequences were processed using the open-source MICCA (v1.7.0) software (102). After primer trimming, forward 16S reads shorter than 225 bp and with an expected error rate (103) of higher than 0.5% were discarded. Filtered sequences were denoised using the UNOISE (104) algorithm. Denoising methods (105–107) were chosen to correct sequencing errors and determine true biological sequences at the single nucleotide resolution by generating amplicon sequence variants (SVs) rather than operational taxonomic units (OTUs) defined using fixed similarity threshold (e.g., 97%) (108). Bacterial SVs were taxonomically classified using the Ribosomal Database Project (RDP) Classifier v2.11 (109). Multiple sequence alignments (MSA) were performed on the denoised reads applying the Nearest Alignment Space Termination (110) (NAST) algorithm, and the phylogenetic tree was inferred using FastTree v2.1.8 (110, 111).
Raw overlapping internal transcribed spacer (ITS) paired-end reads were assembled using the procedure described in reference 103. Paired-end reads with an overlap length smaller than 50 and with more than 15 mismatches were discarded. After forward and reverse primer trimming, merged reads shorter than 225 bp and with an expected error rate higher than 0.5% were removed. Filtered sequences were denoised as described above, and SVs were classified using the RDP Classifier v2.11 and the UNITE (112) database. Multiple sequence alignment was performed on SVs using MUSCLE v3.8.31 (113), and a phylogenetic tree was inferred using FastTree. Finally, SVs with less than 75% similarity to the sequences present in the UNITE database (clustered at 85%; release 2017/12/01) were discarded using VSEARCH v2.3.4 (114). The complete list of commands is reported in Text S1 in the supplemental material.
Downstream analyses were performed using R v3.4.2 with the phyloseq v1.25.3 and vegan v2.5-2 packages (115). 16S and ITS data from 158 individuals were rarefied (without replacement) at 2,400 reads per sample resulting in 16 ITS samples being discarded, as well as two 16S and 49 ITS SVs. Wilcoxon rank sum tests were performed to detect significant differences in relative abundances between the red colobus and the yellow baboon samples at the phylum, class, and family levels. P values were corrected using the Benjamini-Hochberg procedure. For each taxon, we estimated the median (and the 95% nonparametric confidence interval) of the relative abundance difference between the two species. Alpha diversity indices (observed number of SVs and Shannon entropy) for each fecal sample were estimated using the phyloseq library. The comparison of alpha diversity indices between the primate species was performed using the Wilcoxon rank sum test implemented in the R package stats.
To determine whether the core microbiota of each primate species was influenced by habitat, the number of SVs shared across individuals within each species was estimated overall and for each forest type. Core taxa were established by considering SVs present at least one time (on the rarefied SV matrix) and in at least 75% of samples, and between-forest richness analysis was performed using generalized linear models (GLM) available in the R package stats, with quasi-Poisson (log link function) and Gaussian families for the number of observed SVs and Shannon entropy, respectively. Permutational multivariate analyses of variance were performed by the function adonis (available in the R package vegan) on Bray-Curtis dissimilarities and weighted UniFrac distances (with 9,999 permutations). Differential abundance testing was carried out removing the 16 ITS samples with less than 2,400 reads and using the R package DESeq2 (116) using the nonrarefied data as suggested in reference 117. Only SVs with at least five counts in more than 10 samples were considered in this analysis. P values were false discovery rate corrected using the Benjamini-Hochberg procedure implemented in DESeq2.
To infer whether bacterial and fungal profiles could predict the habitat of sample origin, random forest predictive classification models were built using the Python package scikit-learn (118). Generalization accuracy, Matthews correlation coefficient (MCC), out-of-bag (OOB) errors, and receiver operating characteristic (ROC) curves were estimated by a stratified random subsampling cross-validation (CV) scheme (50 splits) using the rarefied data. For each cross-validation split, the model was fitted using 90% of samples, and the classification accuracy was computed predicting the remaining 10% (blind set). Models were trained on rarefied SVs in order to avoid biases due to different sequencing depths. The number of estimators in the RF classifier was set at 500, and no model selection was performed. The RF feature importances were computed as the average of the normalized Gini importances calculated in each decision tree. Baseline accuracy values were calculated by a “dummy” classifier, which predicts the most frequent label in the training set. The “random” accuracy values were calculated generating random predictions according to the class distribution of the training set.
By combining available bacterial and fungal sequencing data from the same fecal samples, intradomain correlation analyses were performed using MICtools (119). All comparisons of relationships (i.e., bacterium-bacterium, fungus-fungus, fungus-bacterium within host species and forest) were assessed based on the relative abundance of bacterial and fungal SVs. Only SVs with a relative abundance higher than 0.1% in at least 30 samples were included in this analysis. To detect significant interactions between bacteria and fungi SVs, we additionally use the CCREPE R library (http://huttenhower.sph.harvard.edu/ccrepe) version 1.20.0. The number of bootstrap and permutation iterations was set at 100, and P values were FDR corrected using the qvalue (120) R package.
Data availability.
DNA sequences have been deposited in the European Nucleotide Archive (ENA) under study accession number PRJEB37770. 16S and ITS raw sequences generated for this study and metadata are publicly available at https://doi.org/10.5281/zenodo.3725526. The machine learning algorithm is available at the repository link: https://github.com/compmetagen/barelli_et_al_msystems_2020.
Permutational multivariate analysis of variance (PERMANOVA) independently applied for each marker (16S and ITS) and species, using Bray-Curtis dissimilarity and weighted UniFrac distance. The table reports the F statistics (F), partial R-squared (R2), and uncorrected P values [Pr(>F)]. Download Table S1, DOCX file, 0.01 MB (15.7KB, docx) .
Copyright © 2020 Barelli et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
ACKNOWLEDGMENTS
We are grateful to the Tanzania Commission for Science and Technology (COSTECH), Tanzania Wildlife Research Institute (TAWIRI) and Tanzania National Parks (TANAPA) for granting us permissions to conduct the study (COSTECH permit 2016-267-ER-2009-49). We also thank the ranger staff of Udzungwa Mountains National Park and A. Mwakisoma and R. Laizzer for providing invaluable field assistance.
Data collection and analyses were funded by the European Union’s Horizon 2020 research and innovation program under the Marie Sklodowska-Curie grant agreement 752399 (project WILDGUT).
REFERENCES
- 1.Cardinale BJ, Duffy JE, Gonzalez A, Hooper DU, Perrings C, Venail P, Narwani A, Mace GM, Tilman D, Wardle DA, Kinzig AP, Daily GC, Loreau M, Grace JB, Larigauderie A, Srivastava DS, Naeem S. 2012. Biodiversity loss and its impact on humanity. Nature 486:59–67. doi: 10.1038/nature11148. [DOI] [PubMed] [Google Scholar]
- 2.Ripple WJ, Chapron G, López-Bao JV, Durant SM, Macdonald DW, Lindsey PA, Bennett EL, Beschta RL, Bruskotter JT, Campos-Arceiz A, Corlett RT, Darimont CT, Dickman AJ, Dirzo R, Dublin HT, Estes JA, Everatt KT, Galetti M, Goswami VR, Hayward MW, Hedges S, Hoffmann M, Hunter LTB, Kerley GIH, Letnic M, Levi T, Maisels F, Morrison JC, Nelson MP, Newsome TM, Painter L, Pringle RM, Sandom CJ, Terborgh J, Treves A, Van Valkenburgh B, Vucetich JA, Wirsing AJ, Wallach AD, Wolf C, Woodroffe R, Young H, Zhang L. 2016. Saving the world’s terrestrial megafauna. Bioscience 66:807–812. doi: 10.1093/biosci/biw092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Davis M, Faurby S, Svenning J-C. 2018. Mammal diversity will take millions of years to recover from the current biodiversity crisis. Proc Natl Acad Sci U S A 115:11262–11267. doi: 10.1073/pnas.1804906115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barelli C, Albanese D, Donati C, Pindo M, Dallago C, Rovero F, Cavalieri D, Tuohy KM, Hauffe HC, De Filippo C. 2015. Habitat fragmentation is associated to gut microbiota diversity of an endangered primate: implications for conservation. Sci Rep 5:14862. doi: 10.1038/srep14862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Amato KR, Yeoman CJ, Kent A, Righini N, Carbonero F, Estrada A, Gaskins HR, Stumpf RM, Yildirim S, Torralba M, Gillis M, Wilson BA, Nelson KE, White BA, Leigh SR. 2013. Habitat degradation impacts black howler monkey (Alouatta pigra) gastrointestinal microbiomes. ISME J 7:1344–1353. doi: 10.1038/ismej.2013.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang B-H, Chang C-W, Huang C-W, Gao J, Liao P-C. 2017. Composition and functional specialists of the gut microbiota of frogs reflect habitat differences and agricultural activity. Front Microbiol 8:2670. doi: 10.3389/fmicb.2017.02670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tremaroli V, Bäckhed F. 2012. Functional interactions between the gut microbiota and host metabolism. Nature 489:242–249. doi: 10.1038/nature11552. [DOI] [PubMed] [Google Scholar]
- 8.Hooper LV, Littman DR, Macpherson AJ. 2012. Interactions between the microbiota and the immune system. Science 336:1268–1273. doi: 10.1126/science.1223490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lozupone CA, Stombaugh JI, Gordon JI, Jansson JK, Knight R. 2012. Diversity, stability and resilience of the human gut microbiota. Nature 489:220–230. doi: 10.1038/nature11550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Montiel-Castro AJ, González-Cervantes RM, Bravo-Ruiseco G, Pacheco-López G. 2013. The microbiota-gut-brain axis: neurobehavioral correlates, health and sociality. Front Integr Neurosci 7:70. doi: 10.3389/fnint.2013.00070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stumpf RM, Gomez A, Amato KR, Yeoman CJ, Polk JD, Wilson BA, Nelson KE, White BA, Leigh SR. 2016. Microbiomes, metagenomics, and primate conservation: new strategies, tools, and applications. Biol Conserv 199:56–66. doi: 10.1016/j.biocon.2016.03.035. [DOI] [Google Scholar]
- 12.Hauffe HC, Barelli C. 2019. Conserve the germs: the gut microbiota and adaptive potential. Conserv Genet 20:19–27. doi: 10.1007/s10592-019-01150-y. [DOI] [Google Scholar]
- 13.West AG, Waite DW, Deines P, Bourne DG, Digby A, McKenzie VJ, Taylor MW. 2019. The microbiome in threatened species conservation. Biol Conserv 229:85–98. doi: 10.1016/j.biocon.2018.11.016. [DOI] [Google Scholar]
- 14.Amato KR. 2019. Missing links: the role of primates in understanding the human microbiome. mSystems 4:e00165-19. doi: 10.1128/mSystems.00165-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cowlishaw G, Dunbar R. 2000. Primate conservation biology. University of Chicago Press, Chicago, IL. [Google Scholar]
- 16.International Union for the Conservation of Nature and Natural Resources (IUCN). 2019. IUCN red list of threatened species version 2019-2. International Union for the Conservation of Nature and Natural Resources, Gland, Switzerland. http://www.iucnredlist.org. Accessed 27 November 2019.
- 17.Estrada A, Garber PA, Rylands AB, Roos C, Fernandez-Duque E, Di Fiore A, Nekaris K-I, Nijman V, Heymann EW, Lambert JE, Rovero F, Barelli C, Setchell JM, Gillespie TR, Mittermeier RA, Arregoitia LV, de Guinea M, Gouveia S, Dobrovolski R, Shanee S, Shanee N, Boyle SA, Fuentes A, MacKinnon KC, Amato KR, Meyer ALS, Wich S, Sussman RW, Pan R, Kone I, Li B. 2017. Impending extinction crisis of the world’s primates: why primates matter. Sci Adv 3:e1600946. doi: 10.1126/sciadv.1600946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yildirim S, Yeoman CJ, Sipos M, Torralba M, Wilson BA, Goldberg TL, Stumpf RM, Leigh SR, White BA, Nelson KE. 2010. Characterization of the fecal microbiome from non-human wild primates reveals species specific microbial communities. PLoS One 5:e13963. doi: 10.1371/journal.pone.0013963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bennett G, Malone M, Sauther ML, Cuozzo FP, White B, Nelson KE, Stumpf RM, Knight R, Leigh SR, Amato KR. 2016. Host age, social group, and habitat type influence the gut microbiota of wild ring‐tailed lemurs (Lemur catta). Am J Primatol 78:883–892. doi: 10.1002/ajp.22555. [DOI] [PubMed] [Google Scholar]
- 20.Trosvik P, de Muinck EJ, Rueness EK, Fashing PJ, Beierschmitt EC, Callingham KR, Kraus JB, Trew TH, Moges A, Mekonnen A, Venkataraman VV, Nguyen N. 2018. Multilevel social structure and diet shape the gut microbiota of the gelada monkey, the only grazing primate. Microbiome 6:84. doi: 10.1186/s40168-018-0468-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Amato KR, Leigh SR, Kent A, Mackie RI, Yeoman CJ, Stumpf RM, Wilson BA, Nelson KE, White BA, Garber PA. 2015. The gut microbiota appears to compensate for seasonal diet variation in the wild black howler monkey (Alouatta pigra). Microb Ecol 69:434–443. doi: 10.1007/s00248-014-0554-7. [DOI] [PubMed] [Google Scholar]
- 22.Clayton JB, Al-Ghalith GA, Long HT, Van Tuan B, Cabana F, Huang H, Vangay P, Ward T, Van Minh V, Tam NA, Dat NT, Travis DA, Murtaugh MP, Covert H, Glander KE, Nadler T, Toddes B, Sha JCM, Singer R, Knights D, Johnson TJ. 2018. Associations between nutrition, gut microbiome, and health in a novel nonhuman primate model. Sci Rep 8:11159. doi: 10.1038/s41598-018-29277-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Clayton JB, Vangay P, Huang H, Ward T, Hillmann BM, Al-Ghalith GA, Travis DA, Long HT, Van Tuan B, Van Minh V, Cabana F, Nadler T, Toddes B, Murphy T, Glander KE, Johnson TJ, Knights D. 2016. Captivity humanizes the primate microbiome. Proc Natl Acad Sci U S A 113:10376–10381. doi: 10.1073/pnas.1521835113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lugano SD, Nyerere KA, Kariuki WK, Samuel K, Joseph K, Apondi OJ. 2018. Gastrointestinal microbial flora in wild and captive olive baboons (Papio anubis). Am J Infect Dis Microbiol 6:30–37. doi: 10.12691/ajidm-6-1-5. [DOI] [Google Scholar]
- 25.Amato KR, Martinez-Mota R, Righini N, Raguet-Schofield M, Corcione FP, Marini E, Humphrey G, Gogul G, Gaffney J, Lovelace E, Williams L, Luong A, Dominguez-Bello MG, Stumpf RM, White B, Nelson KE, Knight R, Leigh SR. 2016. Phylogenetic and ecological factors impact the gut microbiota of two Neotropical primate species. Oecologia 180:717–733. doi: 10.1007/s00442-015-3507-z. [DOI] [PubMed] [Google Scholar]
- 26.Trosvik P, Rueness EK, de Muinck EJ, Moges A, Mekonnen A. 2018. Ecological plasticity in the gastrointestinal microbiomes of Ethiopian Chlorocebus monkeys. Sci Rep 8:20. doi: 10.1038/s41598-017-18435-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McCord AI, Chapman CA, Weny G, Tumukunde A, Hyeroba D, Klotz K, Koblings AS, Mbira DNM, Craggier M, White BA, Leigh SR, Goldberg TL. 2014. Fecal microbiomes of non-human primates in Western Uganda reveal species-specific communities largely resistant to habitat perturbation. Am J Primatol 76:347–354. doi: 10.1002/ajp.22238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Enaud R, Vandenborght L-E, Coron N, Bazin T, Prevel R, Schaeverbeke T, Berger P, Fayon M, Lamireau T, Delhaes L. 2018. The mycobiome: a neglected component in the microbiota-gut-brain axis. Microorganisms 6:22. doi: 10.3390/microorganisms6010022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mar Rodríguez M, Pérez D, Javier Chaves F, Esteve E, Marin-Garcia P, Xifra G, Vendrell J, Jové M, Pamplona R, Ricart W, Portero-Otin M, Chacón MR, Fernández Real JM. 2015. Obesity changes the human gut mycobiome. Sci Rep 5:14600. doi: 10.1038/srep14600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Strati F, Di Paola M, Stefanini I, Albanese D, Rizzetto L, Lionetti P, Calabrò A, Jousson O, Donati C, Cavalieri D, De Filippo C. 2016. Age and gender affect the composition of fungal population of the human gastrointestinal tract. Front Microbiol 7:1227. doi: 10.3389/fmicb.2016.01227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nam Y-D, Chang H-W, Kim K-H, Roh SW, Kim M-S, Jung M-J, Lee S-W, Kim J-Y, Yoon J-H, Bae J-W. 2008. Bacterial, archaeal, and eukaryal diversity in the intestines of Korean people. J Microbiol 46:491–501. doi: 10.1007/s12275-008-0199-7. [DOI] [PubMed] [Google Scholar]
- 32.Hager CL, Ghannoum MA. 2017. The mycobiome: role in health and disease, and as a potential probiotic target in gastrointestinal disease. Dig Liver Dis 49:1171–1176. doi: 10.1016/j.dld.2017.08.025. [DOI] [PubMed] [Google Scholar]
- 33.Underhill DM, Iliev ID. 2014. The mycobiota: interactions between commensal fungi and the host immune system. Nat Rev Immunol 14:405–416. doi: 10.1038/nri3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huffnagle GB, Noverr MC. 2013. The emerging world of the fungal microbiome. Trends Microbiol 21:334–341. doi: 10.1016/j.tim.2013.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sokol H, Leducq V, Aschard H, Pham H-P, Jegou S, Landman C, Cohen D, Liguori G, Bourrier A, Nion-Larmurier I, Cosnes J, Seksik P, Langella P, Skurnik D, Richard ML, Beaugerie L. 2017. Fungal microbiota dysbiosis in IBD. Gut 66:1039–1048. doi: 10.1136/gutjnl-2015-310746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mukherjee PK, Sendid B, Hoarau G, Colombel J-F, Poulain D, Ghannoum MA. 2015. Mycobiota in gastrointestinal diseases. Nat Rev Gastroenterol Hepatol 12:77–87. doi: 10.1038/nrgastro.2014.188. [DOI] [PubMed] [Google Scholar]
- 37.Oever JT, ten Oever J, Netea MG. 2014. The bacteriome-mycobiome interaction and antifungal host defense. Eur J Immunol 44:3182–3191. doi: 10.1002/eji.201344405. [DOI] [PubMed] [Google Scholar]
- 38.Liggenstoffer AS, Youssef NH, Couger MB, Elshahed MS. 2010. Phylogenetic diversity and community structure of anaerobic gut fungi (phylum Neocallimastigomycota) in ruminant and non-ruminant herbivores. ISME J 4:1225–1235. doi: 10.1038/ismej.2010.49. [DOI] [PubMed] [Google Scholar]
- 39.Sun B, Gu Z, Wang X, Huffman MA, Garber PA, Sheeran LK, Zhang D, Zhu Y, Xia D-P, Li J-H. 2018. Season, age, and sex affect the fecal mycobiota of free-ranging Tibetan macaques (Macaca thibetana). Am J Primatol 80:e22880. doi: 10.1002/ajp.22880. [DOI] [PubMed] [Google Scholar]
- 40.Gomez A, Rothman JM, Petrzelkova K, Yeoman CJ, Vlckova K, Umaña JD, Carr M, Modry D, Todd A, Torralba M, Nelson KE, Stumpf RM, Wilson BA, Blekhman R, White BA, Leigh SR. 2016. Temporal variation selects for diet-microbe co-metabolic traits in the gut of Gorilla spp. ISME J 10:532. doi: 10.1038/ismej.2015.252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.De Filippo C, Cavalieri D, Di Paola M, Ramazzotti M, Poullet JB, Massart S, Collini S, Pieraccini G, Lionetti P. 2010. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc Natl Acad Sci U S A 107:14691–14696. doi: 10.1073/pnas.1005963107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Clemente JC, Pehrsson EC, Blaser MJ, Sandhu K, Gao Z, Wang B, Magris M, Hidalgo G, Contreras M, Noya-Alarcón Ó, Lander O, McDonald J, Cox M, Walter J, Oh PL, Ruiz JF, Rodriguez S, Shen N, Song SJ, Metcalf J, Knight R, Dantas G, Dominguez-Bello MG. 2015. The microbiome of uncontacted Amerindians. Sci Adv 1:e1500183. doi: 10.1126/sciadv.1500183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Marshall AR. 2008. Ecological report on Magombera forest. University of York, York, United Kingdom. [Google Scholar]
- 44.Losasso C, Eckert EM, Mastrorilli E, Villiger J, Mancin M, Patuzzi I, Di Cesare A, Cibin V, Barrucci F, Pernthaler J, Corno G, Ricci A. 2018. Assessing the influence of vegan, vegetarian and omnivore oriented Westernized dietary styles on human gut microbiota: a cross sectional study. Front Microbiol 9:317. doi: 10.3389/fmicb.2018.00317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kohl KD, Miller AW, Marvin JE, Mackie R, Dearing MD. 2014. Herbivorous rodents (Neotoma spp.) harbour abundant and active foregut microbiota. Environ Microbiol 16:2869–2878. doi: 10.1111/1462-2920.12376. [DOI] [PubMed] [Google Scholar]
- 46.Koliada A, Syzenko G, Moseiko V, Budovska L, Puchkov K, Perederiy V, Gavalko Y, Dorofeyev A, Romanenko M, Tkach S, Sineok L, Lushchak O, Vaiserman A. 2017. Association between body mass index and Firmicutes/Bacteroidetes ratio in an adult Ukrainian population. BMC Microbiol 17:120. doi: 10.1186/s12866-017-1027-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mariat D, Firmesse O, Levenez F, Guimarăes VD, Sokol H, Doré J, Corthier G, Furet JP. 2009. The Firmicutes/Bacteroidetes ratio of the human microbiota changes with age. BMC Microbiol 9:123. doi: 10.1186/1471-2180-9-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Indiani C, Rizzardi KF, Castelo PM, Ferraz LFC, Darrieux M, Parisotto TM. 2018. Childhood obesity and Firmicutes/Bacteroidetes ratio in the gut microbiota: a systematic review. Child Obes 14:501–509. doi: 10.1089/chi.2018.0040. [DOI] [PubMed] [Google Scholar]
- 49.Orkin JD, Campos FA, Myers MS, Cheves Hernandez SE, Guadamuz A, Melin AD. 2019. Seasonality of the gut microbiota of free-ranging white-faced capuchins in a tropical dry forest. ISME J 13:183–196. doi: 10.1038/s41396-018-0256-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Springer A, Fichtel C, Al-Ghalith GA, Koch F, Amato KR, Clayton JB, Knights D, Kappeler PM. 2017. Patterns of seasonality and group membership characterize the gut microbiota in a longitudinal study of wild Verreaux’s sifakas (Propithecus verreauxi). Ecol Evol 7:5732–5745. doi: 10.1002/ece3.3148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Norton GW, Rhine RJ, Wynn GW, Wynn RD. 1987. Baboon diet: a five-year study of stability and variability in the plant feeding and habitat of the yellow baboons (Papio cynocephalus) of Mikumi National Park, Tanzania. Folia Primatol (Basel) 48:78–120. doi: 10.1159/000156287. [DOI] [PubMed] [Google Scholar]
- 52.Wasser SK, Thomas R, Nair PP, Guidry C, Southers J, Lucas J, Wildt DE, Monfort SL. 1993. Effects of dietary fibre on faecal steroid measurements in baboons (Papio cynocephalus cynocephalus). Reproduction 97:569–574. doi: 10.1530/jrf.0.0970569. [DOI] [PubMed] [Google Scholar]
- 53.Steel RI. 2012. The effects of habitat parameters on the behavior, ecology, and conservation of the Udzungwa red colobus monkey (Procolobus gordonorum). Ph.D. thesis. Duke University, Durham, NC.
- 54.Danish L, Chapman CA, Hall MB, Rode KD, Worman C. 2012. The role of sugar in diet selection in redtail and red colobus monkeys, p 473 In Hohmann G, Robbins MM, Boesch C (ed), Feeding ecology in apes and other primates. Cambridge Studies in Biological and Evolutionary Anthropology, Book 48. Cambridge University Press, Cambridge, United Kingdom. [Google Scholar]
- 55.Schnorr SL, Candela M, Rampelli S, Centanni M, Consolandi C, Basaglia G, Turroni S, Biagi E, Peano C, Severgnini M, Fiori J, Gotti R, De Bellis G, Luiselli D, Brigidi P, Mabulla A, Marlowe F, Henry AG, Crittenden AN. 2014. Gut microbiome of the Hadza hunter-gatherers. Nat Commun 5:3654. doi: 10.1038/ncomms4654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tap J, Furet J-P, Bensaada M, Philippe C, Roth H, Rabot S, Lakhdari O, Lombard V, Henrissat B, Corthier G, Fontaine E, Doré J, Leclerc M. 2015. Gut microbiota richness promotes its stability upon increased dietary fibre intake in healthy adults. Environ Microbiol 17:4954–4964. doi: 10.1111/1462-2920.13006. [DOI] [PubMed] [Google Scholar]
- 57.Akin DE, Gordon GL, Hogan JP. 1983. Rumen bacterial and fungal degradation of Digitaria pentzii grown with or without sulfur. Appl Environ Microbiol 46:738–748. doi: 10.1128/AEM.46.3.738-748.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Corlett RT. 2016. Plant diversity in a changing world: status, trends, and conservation needs. Plant Divers 38:10–16. doi: 10.1016/j.pld.2016.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Clarke SF, Murphy EF, O’Sullivan O, Lucey AJ, Humphreys M, Hogan A, Hayes P, O’Reilly M, Jeffery IB, Wood-Martin R, Kerins DM, Quigley E, Ross RP, O’Toole PW, Molloy MG, Falvey E, Shanahan F, Cotter PD. 2014. Exercise and associated dietary extremes impact on gut microbial diversity. Gut 63:1913–1920. doi: 10.1136/gutjnl-2013-306541. [DOI] [PubMed] [Google Scholar]
- 60.Borbón-García A, Reyes A, Vives-Flórez M, Caballero S. 2017. Captivity shapes the gut microbiota of Andean bears: insights into health surveillance. Front Microbiol 8:1316. doi: 10.3389/fmicb.2017.01316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hill JL, Curran PJ. 2001. Species composition in fragmented forests: conservation implications of changing forest area. Appl Geogr 21:157–174. doi: 10.1016/S0143-6228(01)00002-9. [DOI] [Google Scholar]
- 62.Abbas F, Morellet N, Hewison AM, Merlet J, Cargnelutti B, Lourtet B, Angibault J-M, Daufresne T, Aulagnier S, Verheyden H. 2011. Landscape fragmentation generates spatial variation of diet composition and quality in a generalist herbivore. Oecologia 167:401–411. doi: 10.1007/s00442-011-1994-0. [DOI] [PubMed] [Google Scholar]
- 63.Higham JP, Warren Y, Adanu J, Umaru BN, MacLarnon AM, Sommer V, Ross C. 2009. Living on the edge: life-history of olive baboons at Gashaka-Gumti National Park, Nigeria. Am J Primatol 71:293–304. doi: 10.1002/ajp.20651. [DOI] [PubMed] [Google Scholar]
- 64.McLennan MR, Ganzhorn JU. 2017. Nutritional characteristics of wild and cultivated foods for chimpanzees (Pan troglodytes) in agricultural landscapes. Int J Primatol 38:122–150. doi: 10.1007/s10764-016-9940-y. [DOI] [Google Scholar]
- 65.Riley EP, Tolbert B, Farida WR. 2013. Nutritional content explains the attractiveness of cacao to crop raiding Tonkean macaques. Curr Zool 59:160–169. doi: 10.1093/czoolo/59.2.160. [DOI] [Google Scholar]
- 66.Tsukayama P, Boolchandani M, Patel S, Pehrsson EC, Gibson MK, Chiou KL, Jolly CJ, Rogers J, Phillips-Conroy JE, Dantas G. 2018. Characterization of wild and captive baboon gut microbiota and their antibiotic resistomes. mSystems 3:e00016-18. doi: 10.1128/mSystems.00016-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Watson SE, Hauffe HC, Bull M, Atwood TA, McKinney MA, Pindo M, Perkins SE. 2019. Global change-driven use of onshore habitat impacts polar bear faecal microbiota. ISME J 13:2916–2926. doi: 10.1038/s41396-019-0480-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lau SKP, Teng JLL, Chiu TH, Chan E, Tsang AKL, Panagiotou G, Zhai S-L, Woo P. 2018. Differential microbial communities of omnivorous and herbivorous cattle in southern China. Comput Struct Biotechnol J 16:54–60. doi: 10.1016/j.csbj.2018.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.McKenzie VJ, Song SJ, Delsuc F, Prest TL, Oliverio AM, Korpita TM, Alexiev A, Amato KR, Metcalf JL, Kowalewski M, Avenant NL, Link A, Di Fiore A, Seguin-Orlando A, Feh C, Orlando L, Mendelson JR, Sanders J, Knight R. 2017. The effects of captivity on the mammalian gut microbiome. Integr Comp Biol 57:690–704. doi: 10.1093/icb/icx090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Xie Y, Xia P, Wang H, Yu H, Giesy JP, Zhang Y, Mora MA, Zhang X. 2016. Effects of captivity and artificial breeding on microbiota in feces of the red-crowned crane (Grus japonensis). Sci Rep 6:33350. doi: 10.1038/srep33350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cheng Y, Fox S, Pemberton D, Hogg C, Papenfuss AT, Belov K. 2015. The Tasmanian devil microbiome—implications for conservation and management. Microbiome 3:76. doi: 10.1186/s40168-015-0143-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kong F, Zhao J, Han S, Zeng B, Yang J, Si X, Yang B, Yang M, Xu H, Li Y. 2014. Characterization of the gut microbiota in the red panda (Ailurus fulgens). PLoS One 9:e87885. doi: 10.1371/journal.pone.0087885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Grieneisen LE, Charpentier MJ, Alberts SC, Blekhman R, Bradburd G, Tung J, Archie EA. 2019. Genes, geology and germs: gut microbiota across a primate hybrid zone are explained by site soil properties, not host species. Proc Biol Sci 286:20190431. doi: 10.1098/rspb.2019.0431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yang J, Yu J. 2018. The association of diet, gut microbiota and colorectal cancer: what we eat may imply what we get. Protein Cell 9:474–487. doi: 10.1007/s13238-018-0543-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Brulc JM, Antonopoulos DA, Miller MEB, Wilson MK, Yannarell AC, Dinsdale EA, Edwards RE, Frank ED, Emerson JB, Wacklin P, Coutinho PM, Henrissat B, Nelson KE, White BA. 2009. Gene-centric metagenomics of the fiber-adherent bovine rumen microbiome reveals forage specific glycoside hydrolases. Proc Natl Acad Sci U S A 106:1948–1953. doi: 10.1073/pnas.0806191105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Biddle A, Stewart L, Blanchard J, Leschine S. 2013. Untangling the genetic basis of fibrolytic specialization by Lachnospiraceae and Ruminococcaceae in diverse gut communities. Diversity 5:627–640. doi: 10.3390/d5030627. [DOI] [Google Scholar]
- 77.Hale VL, Tan CL, Niu K, Yang Y, Knight R, Zhang Q, Cui D, Amato KR. 2018. Diet versus phylogeny: a comparison of gut microbiota in captive colobine monkey species. Microb Ecol 75:515–527. doi: 10.1007/s00248-017-1041-8. [DOI] [PubMed] [Google Scholar]
- 78.Amato KR, Leigh SR, Kent A, Mackie RI, Yeoman CJ, Stumpf RM, Wilson BA, Nelson KE, White BA, Garber PA. 2014. The role of gut microbes in satisfying the nutritional demands of adult and juvenile wild, black howler monkeys (Alouatta pigra). Am J Phys Anthropol 155:652–664. doi: 10.1002/ajpa.22621. [DOI] [PubMed] [Google Scholar]
- 79.Ren T, Grieneisen LE, Alberts SC, Archie EA, Wu M. 2016. Development, diet and dynamism: longitudinal and cross-sectional predictors of gut microbial communities in wild baboons. Environ Microbiol 18:1312–1325. doi: 10.1111/1462-2920.12852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tung J, Zhou X, Alberts SC, Stephens M, Gilad Y. 2015. The genetic architecture of gene expression levels in wild baboons. Elife 4:e04729. doi: 10.7554/eLife.04729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Stackebrandt E, Hespell RB. 2006. The family Succinivibrionaceae, p 419–429. In Dworkin M, Falkow S, Rosenberg E, Schleifer K-H, Stackebrandt E (ed), The prokaryotes: a handbook on the biology of bacteria, 3rd ed, vol 3. Springer, New York, NY. [Google Scholar]
- 82.Suhr MJ, Banjara N, Hallen-Adams HE. 2016. Sequence-based methods for detecting and evaluating the human gut mycobiome. Lett Appl Microbiol 62:209–215. doi: 10.1111/lam.12539. [DOI] [PubMed] [Google Scholar]
- 83.Struhsaker TT, Marshall AR, Detwiler K, Siex K, Ehardt C, Lisbjerg DD, Butynski TM. 2004. Demographic variation among Udzungwa red colobus in relation to gross ecological and sociological parameters. Int J Primatol 25:615–658. doi: 10.1023/B:IJOP.0000023578.08343.4e. [DOI] [Google Scholar]
- 84.Rovero F, Marshall AR, Jones T, Perkin A. 2009. The primates of the Udzungwa Mountains: diversity, ecology and conservation. J Anthropol Sci 87:93–126. [PubMed] [Google Scholar]
- 85.Dillon SM, Lee EJ, Kotter CV, Austin GL, Gianella S, Siewe B, Smith DM, Landay AL, McManus MC, Robertson CE, Frank DN, McCarter MD, Wilson CC. 2016. Gut dendritic cell activation links an altered colonic microbiome to mucosal and systemic T-cell activation in untreated HIV-1 infection. Mucosal Immunol 9:24–37. doi: 10.1038/mi.2015.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ley RE. 2016. Gut microbiota in 2015: Prevotella in the gut: choose carefully. Nat Rev Gastroenterol Hepatol 13:69–70. doi: 10.1038/nrgastro.2016.4. [DOI] [PubMed] [Google Scholar]
- 87.Ushida K, Segawa T, Tsuchida S, Murata K. 2016. Cecal bacterial communities in wild Japanese rock ptarmigans and captive Svalbard rock ptarmigans. J Vet Med Sci 78:251–257. doi: 10.1292/jvms.15-0313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lopetuso LR, Scaldaferri F, Petito V, Gasbarrini A. 2013. Commensal Clostridia: leading players in the maintenance of gut homeostasis. Gut Pathog 5:23. doi: 10.1186/1757-4749-5-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Iliev ID, Leonardi I. 2017. Fungal dysbiosis: immunity and interactions at mucosal barriers. Nat Rev Immunol 17:635–646. doi: 10.1038/nri.2017.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Mittermeier RA, Turner WR, Larsen FW, Brooks TM, Gascon C. 2011. Global biodiversity conservation: the critical role of hotspots, p 3–22. In Zachos FE, Habel JC (ed), Biodiversity hotspots: distribution and protection of conservation priority areas. Springer-Verlag, Berlin, Germany. [Google Scholar]
- 91.Fogel AT. 2015. The gut microbiome of wild lemurs: a comparison of sympatric Lemur catta and Propithecus verreauxi. Folia Primatol (Basel) 86:85–95. doi: 10.1159/000369971. [DOI] [PubMed] [Google Scholar]
- 92.Cho I, Blaser MJ. 2012. The human microbiome: at the interface of health and disease. Nat Rev Genet 13:260–270. doi: 10.1038/nrg3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Heiman ML, Greenway FL. 2016. A healthy gastrointestinal microbiome is dependent on dietary diversity. Mol Metab 5:317–320. doi: 10.1016/j.molmet.2016.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Clemente JC, Ursell LK, Parfrey LW, Knight R. 2012. The impact of the gut microbiota on human health: an integrative view. Cell 148:1258–1270. doi: 10.1016/j.cell.2012.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Marchesi JR, Adams DH, Fava F, Hermes GDA, Hirschfield GM, Hold G, Quraishi MN, Kinross J, Smidt H, Tuohy KM, Thomas LV, Zoetendal EG, Hart A. 2016. The gut microbiota and host health: a new clinical frontier. Gut 65:330–339. doi: 10.1136/gutjnl-2015-309990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bahrndorff S, Alemu T, Alemneh T, Lund Nielsen J. 2016. The microbiome of animals: implications for conservation biology. Int J Genomics 2016:5304028. doi: 10.1155/2016/5304028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Amato KR, Mallott EK, McDonald D, Dominy NJ, Goldberg T, Lambert JE, Swedell L, Metcalf JL, Gomez A, Britton GAO, Stumpf RM, Leigh SR, Knight R. 2019. Convergence of human and Old World monkey gut microbiomes demonstrates the importance of human ecology over phylogeny. Genome Biol 20:201. doi: 10.1186/s13059-019-1807-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Costello EK, Stagaman K, Dethlefsen L, Bohannan BJ, Relman DA. 2012. The application of ecological theory toward an understanding of the human microbiome. Science 336:1255–1262. doi: 10.1126/science.1224203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Reynolds RJ, Day SM. 2018. The growing role of machine learning and artificial intelligence in developmental medicine. Dev Med Child Neurol 60:858–859. doi: 10.1111/dmcn.13917. [DOI] [PubMed] [Google Scholar]
- 100.Laurance WF, Useche DC, Rendeiro J, Kalka M, Bradshaw CJA, Sloan SP, Laurance SG, Campbell M, Abernethy K, Alvarez P, Arroyo-Rodriguez V, Ashton P, Benítez-Malvido J, Blom A, Bobo KS, Cannon CH, Cao M, Carroll R, Chapman C, Coates R, Cords M, Danielsen F, De Dijn B, Dinerstein E, Donnelly MA, Edwards D, Edwards F, Farwig N, Fashing P, Forget P-M, Foster M, Gale G, Harris D, Harrison R, Hart J, Karpanty S, Kress WJ, Krishnaswamy J, Logsdon W, Lovett J, Magnusson W, Maisels F, Marshall AR, McClearn D, Mudappa D, Nielsen MR, Pearson R, Pitman N, van der Ploeg J, Plumptre A, Poulsen J, et al. 2012. Averting biodiversity collapse in tropical forest protected areas. Nature 489:290–294. doi: 10.1038/nature11318. [DOI] [PubMed] [Google Scholar]
- 101.Rovero F, Mtui A, Kitegile A, Jacob P, Araldi A, Tenan S. 2015. Primates decline rapidly in unprotected forests: evidence from a monitoring program with data constraints. PLoS One 10:e0118330. doi: 10.1371/journal.pone.0118330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Albanese D, Fontana P, De Filippo C, Cavalieri D, Donati C. 2015. MICCA: a complete and accurate software for taxonomic profiling of metagenomic data. Sci Rep 5:9743. doi: 10.1038/srep09743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Edgar RC, Flyvbjerg H. 2015. Error filtering, pair assembly and error correction for next-generation sequencing reads. Bioinformatics 31:3476–3482. doi: 10.1093/bioinformatics/btv401. [DOI] [PubMed] [Google Scholar]
- 104.Edgar RC. 2016. UNOISE2: improved error-correction for Illumina 16S and ITS amplicon sequencing. bioRxiv 10.1101/081257. [DOI]
- 105.Amir A, McDonald D, Navas-Molina JA, Kopylova E, Morton JT, Xu ZZ, Kightley EP, Thompson LR, Hyde ER, Gonzalez A, Knight R. 2017. Deblur rapidly resolves single-nucleotide community sequence patterns. mSystems 2:e00191-16. doi: 10.1128/mSystems.00191-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, Holmes SP. 2016. DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods 13:581–583. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Nearing JT, Douglas GM, Comeau AM, Langille M. 2018. Denoising the Denoisers: an independent evaluation of microbiome sequence error-correction approaches. PeerJ 6:e5364. doi: 10.7717/peerj.5364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Callahan BJ, McMurdie PJ, Holmes SP. 2017. Exact sequence variants should replace operational taxonomic units in marker-gene data analysis. ISME J 11:2639–2643. doi: 10.1038/ismej.2017.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wang Q, Garrity GM, Tiedje JM, Cole JR. 2007. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.DeSantis TZ, Hugenholtz P, Keller K, Brodie EL, Larsen N, Piceno YM, Phan R, Andersen GL. 2006. NAST: a multiple sequence alignment server for comparative analysis of 16S rRNA genes. Nucleic Acids Res 34:W394–W399. doi: 10.1093/nar/gkl244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Price MN, Dehal PS, Arkin AP. 2010. FastTree 2 – approximately maximum-likelihood trees for large alignments. PLoS One 5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kõljalg U, Larsson K-H, Abarenkov K, Nilsson RH, Alexander IJ, Eberhardt U, Erland S, Høiland K, Kjøller R, Larsson E, Pennanen T, Sen R, Taylor AFS, Tedersoo L, Vrålstad T, Ursing BM. 2005. UNITE: a database providing web-based methods for the molecular identification of ectomycorrhizal fungi. New Phytol 166:1063–1068. doi: 10.1111/j.1469-8137.2005.01376.x. [DOI] [PubMed] [Google Scholar]
- 113.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Rognes T, Flouri T, Nichols B, Quince C, Mahé F. 2016. VSEARCH: a versatile open source tool for metagenomics. PeerJ 4:e2584. doi: 10.7717/peerj.2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.McMurdie PJ, Holmes S. 2013. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 8:e61217. doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.McMurdie PJ, Holmes S. 2014. Waste not, want not: why rarefying microbiome data is inadmissible. PLoS Comput Biol 10:e1003531. doi: 10.1371/journal.pcbi.1003531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, Blondel M, Prettenhofer P, Weiss R, Dubourg V. 2011. Scikit-learn: machine learning in Python. J Mach Learn Res 12:2825–−2830. [Google Scholar]
- 119.Albanese D, Riccadonna S, Donati C, Franceschi P. 2018. A practical tool for maximal information coefficient analysis. Gigascience 7:giy032. doi: 10.1093/gigascience/giy032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Storey JD, Tibshirani R. 2003. Statistical significance for genomewide studies. Proc Natl Acad Sci U S A 100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Mallott EK, Garber PA, Malhi RS. 2017. Integrating feeding behavior, ecological data, and DNA barcoding to identify developmental differences in invertebrate foraging strategies in wild white-faced capuchins (Cebus capucinus). Am J Phys Anthropol 162:241–254. doi: 10.1002/ajpa.23113. [DOI] [PubMed] [Google Scholar]
- 122.Mallott EK, Malhi RS, Garber PA. 2015. High-throughput sequencing of fecal DNA to identify insects consumed by wild Weddell’s saddleback tamarins (Saguinus weddelli, Cebidae, Primates) in Bolivia. Am J Phys Anthropol 156:474–481. doi: 10.1002/ajpa.22654. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(Sheet 1) Median, first quartile (Q1), third quartile (Q3), mean and standard deviation (SD) of relative abundances of bacterial (16S) SVs at the phylum level in yellow baboons (Papio cynocephalus) and red colobus (Procolobus gordonorum). (Sheet 2) Median, first quartile (Q1), third quartile (Q3), mean and standard deviation (SD) of relative abundances of bacterial (16S) SVs at the class level in yellow baboons (Papio cynocephalus) and red colobus (Procolobus gordonorum). (Sheet 3) Wilcoxon rank sum test between yellow baboons (Papio cynocephalus) and red colobus (Procolobus gordonorum) of the relative abundances of 16S SVs at the phylum, class, and family levels. For each taxon, we report the median (“estimate” column), the 95% nonparametric confidence interval (“conf.low” and “conf.high” columns) of the relative abundances difference between the two species, the test statistic, the P value, and the FDR-corrected P value (“p.value.adj” column). (Sheet 4) Median, first quartile (Q1), third quartile (Q3), mean and standard deviation (SD) of relative abundances of bacterial (16S) SVs at the family level in yellow baboons (Papio cynocephalus) and red colobus (Procolobus gordonorum). (Sheet 5) Median, first quartile (Q1), third quartile (Q3), mean and standard deviation (SD) of relative abundances of fungal (ITS) SVs at the family level in yellow baboons (Papio cynocephalus) and red colobus (Procolobus gordonorum). (Sheet 6) Median, first quartile (Q1), third quartile (Q3), mean and standard deviation (SD) of relative abundances of fungal (ITS) SVs at the phylum level in yellow baboons (Papio cynocephalus) and red colobus (Procolobus gordonorum). (Sheet 7) Median, first quartile (Q1), third quartile (Q3), mean and standard deviation (SD) of relative abundances of fungal (ITS) SVs at the class level in yellow baboons (Papio cynocephalus) and red colobus (Procolobus gordonorum). (Sheet 8) Wilcoxon rank sum test between yellow baboons (Papio cynocephalus) and red colobus (Procolobus gordonorum) of the relative abundances of ITS SVs at the phylum, class, and family levels. For each SV, we report the median (“estimate” column), the 95% nonparametric confidence interval (“conf.low” and “conf.high” columns) of the difference in relative abundances between the two species, the test statistic, the P value, and the FDR-corrected P value (“p.value.adj” column). (Sheet 9) Median, first quartile (Q1), third quartile (Q3), mean and standard deviation (SD) of relative abundances of unclassified 16S and ITS SVs at the genus level. (Sheet 10) CCREPE results for yellow baboon (Papio cynocephalus). Significant correlations between 16S and ITS SVs calculated on each forest (Mwanihana, a PF, and Magombera, a FF). (Sheet 11) CCREPE results for red colobus (Procolobus gordonorum). Significant correlation between 16S and ITS SVs calculated on each forest (Mwanihana, a PF, and Magombera, a FF). (Sheet 12) MICTools results (yellow baboons). TICe (total information coefficient) P values (TICePVal), Pearson correlation coefficients (PearsonR), Spearman’s rank correlation coefficients (SpearmanRho), and maximal information coefficient (MICe) for each significant relationship between 16S SVs across forests. (Sheet 13) MICTools results (red colobus). TICe P values (TICePVal), Pearson correlation coefficients (PearsonR), Spearman’s rank correlation coefficients (SpearmanRho), and maximal information coefficient (MICe) for each significant relationship between 16S SVs across forests. (Sheet 14) MICTools results (yellow baboons). TICe P values (TICePVal), Pearson correlation coefficients (PearsonR), Spearman’s rank correlation coefficients (SpearmanRho), and maximal information coefficient (MICe) for each significant relationship between ITS SVs across forests. (Sheet 15) MICTools results (red colobus). TICe P values (TICePVal), Pearson correlation coefficients (PearsonR), Spearman’s rank correlation coefficients (SpearmanRho), and maximal information coefficient (MICe) for each significant relationship between ITS SVs across forests. (Sheet 16) Results of DeSeq2 analysis showing significantly different abundances of 16S SVs between forests (protected versus fragmented) across species. The table reports the species, SV identifier, mean of normalized counts (baseMean), base 2 logarithm of the fold change (log2FoldChange), standard errors of the log2FoldChange (lfcSE), test statistics (stat), P values (pval), corrected P values (padj), and predicted taxonomy. (Sheet 17) Results of DeSeq2 analysis showing significantly different abundances of ITS SVs between forests (protected versus fragmented) across species. (Sheet 18) Mean and standard deviation (SD) of 16S SV importances estimated by the “habitat prediction” RF models trained on yellow baboons (Papio cynocephalus) and red colobus (Procolobus gordonorum) samples (Species column). (Sheet 19) Mean and standard deviation (SD) of ITS SV importances estimated by the “habitat prediction” RF models trained on yellow baboons (Papio cynocephalus) and red colobus (Procolobus gordonorum) samples (Species column). Download Data Set S1, XLSX file, 0.7 MB (750KB, xlsx) .
Copyright © 2020 Barelli et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Violin plots indicating the distribution of the Firmicutes/Bacteroidetes ratio across species and forests. From the bottom to the top, the horizontal lines indicate the first, second (i.e., median), and third quartiles. Download FIG S1, PDF file, 0.03 MB (27.2KB, pdf) .
Copyright © 2020 Barelli et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
MICTools analysis. Significant relationships between the relative abundances of SVs across species and habitats for both 16S (a) and ITS (b) data. SVs are grouped by the predicted taxonomy; the size of the circle represents the number of significant SV relationships for each pair of classes. Download FIG S2, PDF file, 0.01 MB (12.9KB, pdf) .
Copyright © 2020 Barelli et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Results of MICTools analysis, showing significant relationships between the relative abundances of SVs across species and habitats for both 16S (a) and ITS (b) data. SVs are grouped by predicted taxonomy; the size of the circle represents the number of significant SV relationships for each pair of classes. Download FIG S3, PDF file, 0.02 MB (26.2KB, pdf) .
Copyright © 2020 Barelli et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Top 15 16S SVs in terms of feature importance identified by the habitat prediction RF models trained on yellow baboons (Papio cynocephalus) and red colobus (Procolobus gordonorum) samples. The thick colored bars and intervals represent, respectively, the means and 2 standard deviations of the normalized Gini importances calculated over the 50 cross-validation iterations. Download FIG S4, PDF file, 0.02 MB (21.4KB, pdf) .
Copyright © 2020 Barelli et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Top 15 ITS SVs in terms of feature importance identified by the habitat prediction RF models trained on yellow baboons (Papio cynocephalus) and red colobus (Procolobus gordonorum) samples. The thick colored bars and intervals represent, respectively, the means and 2 standard deviations of the normalized Gini importances calculated over the 50 cross-validation iterations. Download FIG S5, PDF file, 0.02 MB (21.4KB, pdf) .
Copyright © 2020 Barelli et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Detailed information of bacterial and fungal extraction methods, PCR amplification, and sequencing. Additional information on software used for data processing for both bacterial and fungal sequences and statistical analyses. Detailed scripts used to analyze bacterial (16S) and fungal (ITS) sequences for the Micca pipeline. Additional information on the intradomain relationships between SVs relative abundances, specifically, the correlations across classes of bacteria and fungi among the two primate hosts. Download Text S1, DOCX file, 0.02 MB (25.2KB, docx) .
Copyright © 2020 Barelli et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Permutational multivariate analysis of variance (PERMANOVA) independently applied for each marker (16S and ITS) and species, using Bray-Curtis dissimilarity and weighted UniFrac distance. The table reports the F statistics (F), partial R-squared (R2), and uncorrected P values [Pr(>F)]. Download Table S1, DOCX file, 0.01 MB (15.7KB, docx) .
Copyright © 2020 Barelli et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Data Availability Statement
DNA sequences have been deposited in the European Nucleotide Archive (ENA) under study accession number PRJEB37770. 16S and ITS raw sequences generated for this study and metadata are publicly available at https://doi.org/10.5281/zenodo.3725526. The machine learning algorithm is available at the repository link: https://github.com/compmetagen/barelli_et_al_msystems_2020.
Permutational multivariate analysis of variance (PERMANOVA) independently applied for each marker (16S and ITS) and species, using Bray-Curtis dissimilarity and weighted UniFrac distance. The table reports the F statistics (F), partial R-squared (R2), and uncorrected P values [Pr(>F)]. Download Table S1, DOCX file, 0.01 MB (15.7KB, docx) .
Copyright © 2020 Barelli et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.