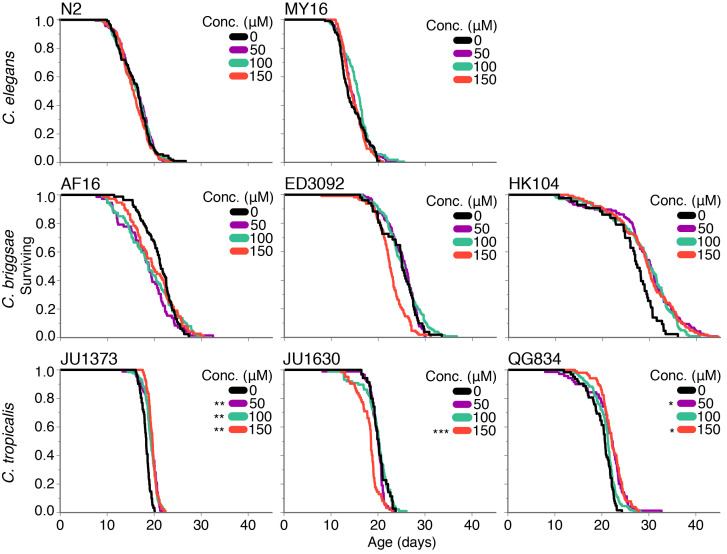
Figure 1.
Survival under adult obeticholic acid exposure: Survival curves for C. elegans strains N2 and MY16, C. briggsae strains AF16, ED3092 and HK104, and C. tropicalis strains JU1373, JU1630 and QG834 exposed to obeticholic acid at various concentrations starting on day one of adulthood. Each line represents lifespan data from multiple replicates. JU1373 lifespan differed significantly from control (mean=18.2 days) at 50 µM, 100 µM, and 150 µM (mean=19.2, 19.2 and 19.6, days; p=0.006, 0.008 and 0.001, respectively). JU1630 survival differed significantly from control (mean=20.2 days) at 150 µM (mean=18.1 days; p<0.001). QG834 differed significantly from control (mean=19.9 days) at 50 µM and 150 µM (mean=21.3 and 22.2 days; p=0.01 and 0.04, respectively). Statistical comparisons are from the Cox proportional hazards (CPH) model with mixed effects using the coxme package v2.2-10 in R (Therneau 2018; R Core Team 2019). Asterisks represent p-values from the CPH model such that ***p<0.001, **p<0.01, and *p<0.05.
Description
The Caenorhabditis Intervention Testing Program (CITP) is a multi-institutional, National Institute on Aging (NIA)-funded consortium. The goal of the program is to identify chemical compounds that extend lifespan robustly and reproducibly across genetically diverse Caenorhabditis strains (Lucanic et al. 2017). The CITP test compounds are selected if they are consistently highly ranked via computational prediction for lifespan or healthspan effects (Coleman-Hulbert et al. 2019), if they are predicted or known to interact with known lifespan-regulating pathways, or if they have previously been reported as extending lifespan or healthspan in laboratory animals. Obeticholic acid is an analog of the natural bile acid chenode oxycholic acid, which acts as an agonist of the farnesoid X receptor (FXR) (Neuschwander-Teri et al. 2015), a nuclear receptor (NR) closely involved with hepatic triglyceride homeostasis. Obeticholic acid is most commonly used to treat the autoimmune liver disease, primary biliary cholangitis. The most likely homolog of FXR in C. elegans is DAF-12, which can bind and be activated by human bile acids (Held et al. 2006; Zhi et al. 2011). DAF-12 modulation is of particular interest because it is closely linked to dauer formation, lifespan extension, and metabolism homeostasis (Antebi 2015).
We assayed lifespan in response to different concentrations of obeticholic acid exposure in three Caenorhabditis species using the flatbed scanner-based Automated Lifespan Machine (ALM) workflow previously published (Banse et al. 2019). To summarize, the worms were age synchronized by egg-lays on standard 60 mm diameter Nematode Growth Media (NGM) plates with lawns of Escherichia coli OP50-1, and transferred to compound-treated 38 mm NGM plates containing 51 μM 5-Fluoro-2′-deoxyuridine (FUdR) at a density of 50 worms per plate on day one of adulthood. For treatment plates, we used standardized protocols (Caenorhabditis Intervention Testing Program 2020); in short, obeticholic acid (Apexbio Technology) was dissolved in dimethyl sulfoxide (DMSO) and diluted appropriately such that addition of 7.5 µl of stock solution and 125 µl water for 35 mm diameter plates, and 17.5 µl of stock solution and 232.5 µl water for 50 mm diameter scanner plates would generate 50, 100, and 150 µM final obeticholic acid concentrations. For control plates, DMSO was added instead of stock solution using the same method. The worms were maintained at 20ºC and transferred to new treatment plates again on day two of adulthood. One week after age-synchronization (day five of adulthood for C. elegans and C. briggsae, day four for C. tropicalis), the worms were transferred to compound-treated scanner plates and loaded onto the ALM. At this point, automated survival monitoring began, and the scanner data was collected and analyzed using Lifespan Machine software (https://github.com/nstroustrup/lifespan; Stroustrup et al.. 2013).
Our results indicate that obeticholic acid does not have a consistent beneficial effect on lifespan in any of the C. elegans or C. briggsae strains tested at the concentrations used. Although we did see some significant differences from the control for some of the concentrations in the C. tropicalis strains, overall the difference was not robust. We actually saw a significant decrease in lifespan in C. tropicalis JU1630, a weakly significant increase in C. tropicalis QG834 at some concentrations, and a relatively significant increase in C. tropicalis JU1373, but with only a 5.7-7.9% change in mean survival from the control (Fig. 1). In summary, our results do not indicate a robust effect of obeticholic acid on Caenorhabditis lifespan. This conclusion is based upon two biological replicates at each concentration performed in one lab, resulting in an average of 104 individuals measured per strain and concentration, and should be considered preliminary. The effect on lifespan in this study may pertain to a lack of physiological relevance of obeticholic acid to Caenorhabditis. Obeticholic acid was of interest to the CITP because of its effect on the mammalian NR FXR. Although DAF-12 has been identified as a potential Caenorhabditis homolog of FXR and other bile acids have been shown to bind with DAF-12 (Zhi et al. 2011), it is possible that obeticholic acid was not able to bind with high affinity to the receptor, therefore eliciting little to no effect on lifespan. Alternatively, obeticholic acid may be rapidly metabolized in Caenorhabditis.
Acknowledgments
Acknowledgments
We acknowledge all members of the Lithgow, Driscoll, and Phillips labs for helpful discussions, Stephen A. Banse and Anna L. Coleman-Hulbert in particular. We thank the CITP Advisory Committee and Ronald Kohanski (National Institute on Aging) for extensive discussion. All strains were procured from the NIH Office of Research Infrastructure Programs-funded (P40 OD010440) Caenorhabditis Genetics Center (CGC).
Funding
This work was supported by National Institutes on Aging U01AG045829 and U24AG056052 to PCP, U01AG045844 to GJL, and U01AG045864 to MD.
References
- Antebi A. Nuclear receptor signal transduction in C. elegans. WormBook. 2015 Jun 01;:1–49. doi: 10.1895/wormbook.1.64.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banse SA, Lucanic M, Sedore CA, Coleman-Hulbert AL, Plummer WT, Chen E, Kish JL, Hall D, Onken B, Presley MP, Jones EG, Blue BW, Garrett T, Abbott M, Xue J, Guo S, Johnson E, Foulger AC, Chamoli M, Falkowski R, Melentijevic I, Harinath G, Huynh P, Patel S, Edgar D, Jarrett CM, Guo M, Kapahi P, Lithgow GJ, Driscoll M, Phillips PC. Automated lifespan determination across Caenorhabditis strains and species reveals assay-specific effects of chemical interventions. Geroscience. 2019 Dec 10;41(6):945–960. doi: 10.1007/s11357-019-00108-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caenorhabditis Intervention Testing Program (2020): Caenorhabditis Intervention Testing Program: Obeticholic acid protocols. figshare. Collection.
- Coleman-Hulbert AL, Johnson E, Sedore CA, Banse SA, Guo M, Driscoll M, Lithgow GJ, Phillips PC. Caenorhabditis Intervention Testing Program: the tyrosine kinase inhibitor imatinib mesylate does not extend lifespan in nematodes. MicroPubl Biol. 2019 Jul 01;2019 doi: 10.17912/micropub.biology.000131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Held JM, White MP, Fisher AL, Gibson BW, Lithgow GJ, Gill MS. DAF-12-dependent rescue of dauer formation in Caenorhabditis elegans by (25S)-cholestenoic acid. Aging Cell. 2006 Aug 01;5(4):283–291. doi: 10.1111/j.1474-9726.2006.00218.x. [DOI] [PubMed] [Google Scholar]
- Lucanic M, Plummer WT, Chen E, Harke J, Foulger AC, Onken B, Coleman-Hulbert AL, Dumas KJ, Guo S, Johnson E, Bhaumik D, Xue J, Crist AB, Presley MP, Harinath G, Sedore CA, Chamoli M, Kamat S, Chen MK, Angeli S, Chang C, Willis JH, Edgar D, Royal MA, Chao EA, Patel S, Garrett T, Ibanez-Ventoso C, Hope J, Kish JL, Guo M, Lithgow GJ, Driscoll M, Phillips PC. Impact of genetic background and experimental reproducibility on identifying chemical compounds with robust longevity effects. Nat Commun. 2017 Feb 21;8:14256–14256. doi: 10.1038/ncomms14256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuschwander-Tetri BA, Loomba R, Sanyal AJ, Lavine JE, Van Natta ML, Abdelmalek MF, Chalasani N, Dasarathy S, Diehl AM, Hameed B, Kowdley KV, McCullough A, Terrault N, Clark JM, Tonascia J, Brunt EM, Kleiner DE, Doo E, NASH Clinical Research Network. Farnesoid X nuclear receptor ligand obeticholic acid for non-cirrhotic, non-alcoholic steatohepatitis (FLINT): a multicentre, randomised, placebo-controlled trial. Lancet. 2014 Nov 01;385(9972):956–965. doi: 10.1016/S0140-6736(14)61933-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team (2019). R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria.
- Stroustrup N, Ulmschneider BE, Nash ZM, López-Moyado IF, Apfeld J, Fontana W. The Caenorhabditis elegans Lifespan Machine. Nat Methods. 2013 May 12;10(7):665–670. doi: 10.1038/nmeth.2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Therneau T.M. (2018). coxme: Mixed Effects Cox Models R. R package version 2.2–10.
- Zhi X, Zhou XE, Melcher K, Motola DL, Gelmedin V, Hawdon J, Kliewer SA, Mangelsdorf DJ, Xu HE. Structural conservation of ligand binding reveals a bile acid-like signaling pathway in nematodes. J Biol Chem. 2011 Dec 14;287(7):4894–4903. doi: 10.1074/jbc.M111.315242. [DOI] [PMC free article] [PubMed] [Google Scholar]