Abstract
Aromatic prenyltransferases (PTases), including ABBA-type and dimethylallyl tryptophan synthase (DMATS)-type enzymes from bacteria and fungi, play important role for diversification of the natural products and improvement of the biological activities. For a decade, the characterization of enzymes and enzymatic synthesis of prenylated compounds by using ABBA-type and DMATS-type PTases have been demonstrated. Here, I introduce several examples of the studies on chemoenzymatic synthesis of unnatural prenylated compounds and the enzyme engineering of ABBA-type and DMATS-type PTases.
Keywords: Biosynthesis, Enzyme engineering, Aromatic prenyltransferase, Prenylated compounds
Introduction
The prenylated indole alkaloids and prenylated aromatic compounds isolated from plants and microorganisms show broad structural diversity and various biological activities [1–6]. The prenylation may increase the lipophilicity and/or binding ability to target protein that directly influences the biological activity [7, 8]. The prenylation to aromatic compounds is catalyzed by the several enzyme groups of prenyltransferases (PTases), including membrane-embedded UbiA-type, bacterial and fungal ABBA-type, and fungal dimethylallyl tryptophan synthase (DMATS)-type PTases [9–20].
UbiA-type PTases possess a conserved (N/D)DXXD motif for binding of Mg2+ ion and diphosphate that is also conserved in the isoprenyl diphosphate synthases [9, 10]. The enzymes in this group are observed in the ubiquinone and menaquinone biosynthesis [10], membrane lipids biosynthesis in archaea [21], in the biosynthesis of prenylated aromatic secondary metabolites in plants [1], and fungal meroterpenoid biosynthesis [22]. On the other hand, ABBA-type and DMATS-type PTases from microorganisms are soluble proteins and do not contain diphosphate and metal ion binding motif [11–18, 20]. The soluble aromatic PTases are involved in the biosynthesis of secondary metabolites in bacteria and fungi.
In the present review, several examples of the recent studies on chemoenzymatic synthesis and the enzyme engineering of soluble ABBA-type and DMATS-type PTases to generate unnatural prenylated aromatic compounds are provided.
Soluble aromatic PTases
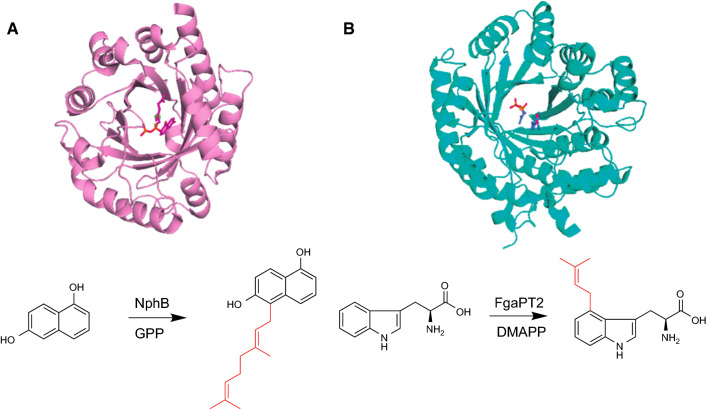
ABBA-type and DMATS-type aromatic PTases catalyze prenylation of dimethylallyl diphosphate (DMAPP) and/or geranyl diphosphate (GPP) to aromatic compounds in bacteria and fungi. The ABBA-type PTases are identified from both of bacteria and fungi, and the CloQ from Streptomyces roseochromogenes var. oscitans is a first characterized ABBA-type of PTases in 2003, which is involved in the biosynthesis of clorobiocin [23]. Different from membrane-bound UbiA-type PTases, these enzyme reactions, except for NphB, are metal-independent enzymes [14]. The first crystal structure of ABBA-type PTase was solved with NphB in biosynthesis of the naphterpin derivatives [24]. The X-ray crystal structure of NphB showed the characteristic β/α barrel fold with antiparallel strands, which is completely distinct from UbiA-type PTases [25]. This enzyme group was later called as ABBA PTases due to their α-β-β-α PT folds (Fig. 1A) [13].
Fig. 1.
The overall structures of ABBA-type and DMATS-type PTases. The crystal structure and reaction of a NphB and b FgaPT2
On the other hand, DMATS-type PTases, identified in fungi, catalyze the prenylation reactions mainly toward indole derivatives, including tryptophan-containing cyclic dipeptides, indole terpenoids, and tryptophan itself [11, 12, 15, 16, 18]. DMATS PTases are also metal-independent enzymes, which do not have aspartate-rich motifs as in the case of ABBA PTases. However, in several cases, the addition of metal ions such as Ca2+ and Mg2+ enhances their activities [12]. So far, the DMATS enzymes that catalyzed at all positions of the indole ring have been identified (N-1, C-2, C-3, C-4, C-5, C-6, and C-7 prenylation DMATS). The structural analysis of DMATS enzymes revealed that the overall structures share the similar α-β-β-α PT folds as in the case of ABBA-type PTases (Fig. 1b) [26]. In many cases, both of ABBA-type and DMATS-type show broad substrate flexibility towards aromatic substrates [13, 27–41] while these enzymes show narrow specificity toward length of prenyl donors [11, 13, 14, 26–42].
Chemoenzymatic syntheses of various prenylated compounds
Specificity for aromatic compounds
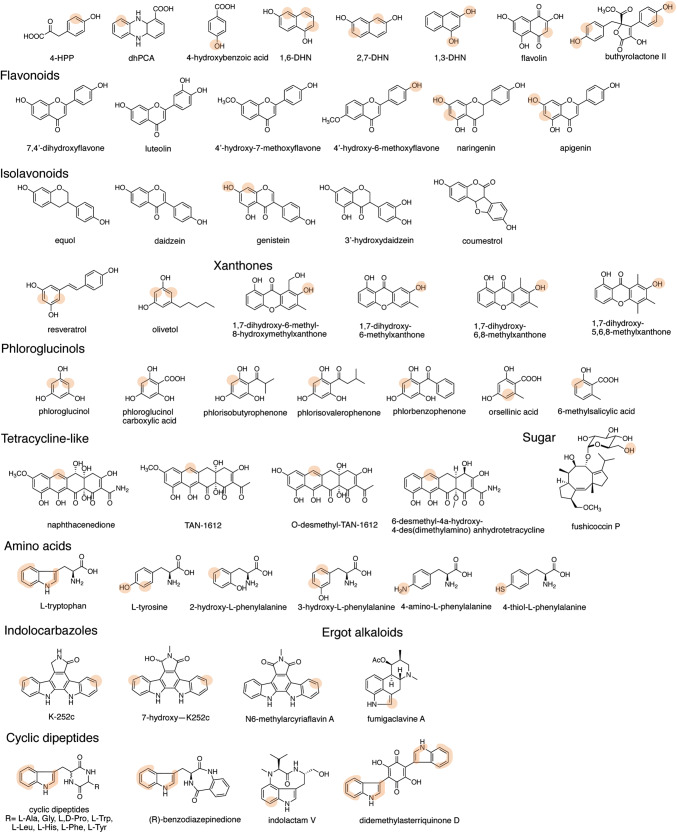
Based on the broad substrate specificity of aromatic prenyltansferases, the chemoenzymatic syntheses of prenylated aromatic derivatives have been performed using the soluble PTases (Table 1). The 4-hydroxyphenylpyruvic acid (4-HPP) derivatives, flavonoids, isoflavonoids, phenylpropanoids, dihydronaphthalenes, and stilbenoids were converted to corresponding dimethylallyl or geranyl group attached products using ABBA-type PTases such as CloQ, NovQ, NphB, SCO7190 and so on [23, 36, 41, 43–46]. The prenylated compounds at different position are obtained using enzymes with different regiospecificity (Fig. 2).
Table 1.
Examples of prenyltransferases and their substrates
| Enzyme | Organism | Prenyl acceptor (examples) | Prenyl donor | References |
|---|---|---|---|---|
| ABBA-type PTases | ||||
| CloQ | Streptomyces roseochromogenes | 4-HPP, flavonoids, isoflavonoids, stilbenoid | DMAPP | [46] |
| NovQ | Streptomyces spheroids | Phenylpropanoids, flavonoids, DHNs | DMAPP | [45] |
| NphB | Streptomyces sp. CL190 | 4-HPP, plant polyketides, DHNs | GPP | [24, 38, 47] |
| Fnq26 | Streptomyces cinnamonensis DSM 1042 | DHNs, flavolin, 4-hydroxybenzoic acid | GPP | [43] |
| SCO7190 | Streptomyces coelicolor A3 | Plant polyketids, DHNs | DMAPP | [24, 38, 47] |
| XptB | Aspergillus nidulans | Xanthone | DMAPP | [48] |
| VrtC | Penicillium aethiapicum | Tetracycline-like naphthacenediones | DMAPP, GPP | [49] |
| PaPT | Phomopsis amygdali | Fusicoccin P | [50] | |
| TleC | Streptomyces blastmyceticus | Indolactam V | DMAPP, GPP, FPP | [83–87] |
| MpnD | Marinactinospora thermotolerans | Indolactam V | DMAPP, GPP, FPP, GGPP, GFPP | [83–87] |
| AtaPT | Aspergillus terreus | Lignanoids, xanthones, quinoline alkaloids, coumarins, benzophenones, curcuminoid, hydroxynaphthalenes | DMAPP, GPP, FPP, GGPP, PPP | [88] |
| Cyanobactin PTases | ||||
| LynF | Lyngbya aestuarii | Tyr residue in cyclic peptides, N-boc-tyrosine | DMAPP | [51] |
| PagF | Oscillatoria agardhii | L-Tyr, N-acetyl-L-Tyr, N-boc-L-Tyr, Tyr-Tyr-Tyr, Tyr4 residue in cyclic[INPYLYP] | DMAPP | [59] |
| TruF | Ser and Thr residues in cyclic peptides | DMAPP | [52, 57, 62] | |
| KgpF |
Microcystis aeruginosa NIES-88 |
Trp residue in kawaguchipeptine B, Fmoc-Trp | DMAPP | [58] |
| TyrPT | Aspergillus niger | Tyr and Trp derivatives | DMAPP, alkyl-PPs | [31, 63] |
| SirD | Leptosphaeria maculans | Tyr and Trp derivatives | DMAPP, alkyl-PPs | [29, 31, 41, 63, 92] |
| DMAT-type PTases | ||||
| 5-DMAT |
Aspergillus clavatus, Streptomyces coelicolor |
Tyr and Trp derivatives, indolocarbazoles | DMAPP, alkyl-PPs | [63, 70, 89] |
| 6-DMAT |
Streptomyces ambofaciens, Streptomyces violaceusniger, Streptomyces sp. SN-593 |
Tyr and Trp derivatives, naphthalene derivatives | DMAPP, GPP, alkyl-PPs | [63, 72, 73] |
| 7-DMAT |
Aspergillus fumigatus, Neosartorya sp. |
Tyr and Trp derivative, naphthalene derivatives, acylphloroglucinols, flavonoids | DMAPP, alkyl-PPs | [44, 63, 66] |
| AnaPT | Neosartorya fischeri | Tyr and Trp derivative, Trp-containing cyclic dipeptides, acylphloroglucinols, flavonoids | DMAPP, alkyl-PPs | [44, 67, 82, 91] |
| FgaPT2 | Aspergillus fumigatus | Tyr and Trp derivatives, Trp-containing cyclic dipeptides, indolocarbazoles | DMAPP, alkyl-PPs | [26, 36, 42, 63, 90, 93] |
| CdpC3PT | Neosartorya fischeri | Trp-containing cyclic dipeptides, acylphloroglucinols | DMAPP, alkyl-PPs | [67, 77, 91] |
| CdpNPT | Aspergillus fumigatus | Trp-containing cyclic dipeptides, naphthalene derivatives | DMAPP, alkyl-PPs | [37, 67, 79, 91] |
| BrePT | Aspergillus versicolor | Trp-containing cyclic dipeptides | DMAPP, alkyl-PPs | [30, 91] |
| FtmPT1 | Aspergillus fumigatus | Trp-containing cyclic dipeptides, indolylbutenone | DMAPP, alkyl-PPs | [27, 28, 91] |
Fig. 2.
Examples of the prenylation substrates of ABBA-type and DMATS-type PTases. The highlighted atoms are the major prenylation points. The prenylation positions of non-highlighted compounds are not determined
First characterized CloQ from Streptomyces roseochromogenes was thought to be specific for 4-HPP [23]. However, recent study on the substrate tolerance of CloQ for various phenolic acceptor revealed that the enzyme accepts flavonoids; 7,4′-dihydroflavone, luteolin, 4′-hydroxy-7-methoxyflavone, and 4′-hydroxy-6-methoxyflavone, isoflavonoids; equol, daidzein, genisein, 3′-hydroxydaidzein, and coumestrol, and stilbenoid; resveratrol to produce corresponding dimethylallyl group attached products (less than 10% yield) [46]. Furthermore, the phenylpropanoids; caffeic acid and p-coumaric acid, the (iso)flavonoids; naringenin, apigenin, and the dihydronaphthalenes (DHNs); 1,6-DHN and 2,7-DHN were prenylated with DMAPP by NovQ from Streptomyces spheroids, involved in the biosynthesis of novobiocin [45]. Interestingly, the yield of prenylated phenylpropanoids, flavonoids, and DHNs are much better compared to the enzyme reaction of CloQ (15–90% by NovQ). Similarly, NphB, which catalyzes geranylation reaction, also shows a broad substrate tolerance toward aromatic compounds, and this enzyme accepts 4-HPP, plant polyketides, and DHNs, including olivetol, olivetolic acid, resveratrol, apigenin, naringenin, genistein, daidzein, 1,6-DHN, and 2,7-DHN to generate corresponding geranylated products [24, 38, 47]. Fnq26 from Streptomyces cinnamonensis DSM 1042 shows slightly different substrate specificity from NphB, whereas Fnq26 shares ~ 40% identity with NphB [43]. Fnq26 catalyzes regular O-prenylation and reverse and regular C-prenylation toward flaviolin, 1,3-DHN, and 4-hydroxybenzoic acid. Further, the NphB homologue SCO7190 from Streptomyces coelicolor A3 shows similar substrate specificity to NphB and generates dimethylallyl attached naringenin, olivetol, resveratrol, 1,6-DHN, and 2,7-DHN [24, 38, 47].
Some of fungal ABBA superfamily enzymes accept different type of polyketides and terpenoids. For example, the hydroxylated and methylated xanthone compounds are prenylated by XptB from Aspergillus nidulans [48]. VrtC from Penicillium aethiapicum and its homologs catalyze prenylation of DMAPP and GPP to tetracycline-like naphthacenedione compounds such as phthacenedione, TAN-1612 (2-acetyl-2-decarboxamido-anthrotainin), and 6-desmethyl-4a-hydroxy-4-des-(dimethylamino)anhydrotetracycline [49]. Moreover, PaPT from Phomopsis amygdali accept glycosylated terpenoid fusicoccin P to generate an O-prenylated compound fusicoccin J in the fusicoccin A biosynthesis [50].
The cyanobactin prenyltransferases LynF, PagF, KgpF, TruF, TyrPT, and SirD form a group of small ABBA-type proteins that catalyze the prenylation of tryptophan, tyrosine, threonine, or serine residues in ribosomally synthesized and post-translationally modified peptides (RiPPs) [50–61]. In these, LynF and PagF also accept N-boc-tyrosine and various tyrosine-containing cyclic peptides [51, 59]. Moreover, PagF used linear peptides with different amino acid sequences as substrates. TruF is encoded in the biosynthetic gene cluster of trunkamide and catalyzes the prenylation of DMAPP on serine or threonine residues of core peptide [62]. Although the substrate selectivity of purified enzyme of TruF has not been elucidated, the expression of tru biosynthetic genes in E. coli created quite large numbers of prenylated peptide derivatives [52, 57]. The tyrosine and tryptophan derivatives, including 4-amino- and 4-thiol-phenylalanine and methyl- or methoxylated tryptophans were accepted by tyrosine O-prenyltransferase SirD from Leptosphaeria maculans, involved in the biosynthesis of sirodesmin PL, to give O-, C-, N-, and S- prenylated compounds [29, 41, 63].
DMATS superfamily enzymes were used for the production of various prenylated indole-containing compounds. The different DMATSs catalyze prenylation toward different position of indole ring. The catalytic reaction of DMATS are well introduced by Winkelblech et al. in 2015 [12]. Using DMATS enzymes, tryptophan derivatives, L-tryptophan-containing cyclic dipeptides, isomers of L-tyrosine, indolocarbazoles, and phenolic molecules were prenylated [26, 29, 41, 63–81]. Some DMATSs also accept aromatic compounds such as acylphloroglucinols and related compounds. AnaPT from Neosartorya fischeri and 7-DMATS from Aspergillus fumigatus, and CdpC3PT from Neosartorya fischeri catalyze prenylation toward phloroglucinol, orsellinic acid, 6-methylsalicylic acid, phloroglucinol carboxylic acid, phlorisobutyrophenone, phlorisovalerophenone, and phlorbenzophenone [82].
Specificity for prenyl donors
In contrast to aforementioned PTases that show broad specificity toward a variety of prenyl acceptors, TleC from Streptomyces blastmyceticus and MpnD from Marinactinospora thermotolerans, in the biosynthesis of teleocidin and pendolmycin, accept only indolactam V as a prenyl acceptor [83–87]. Instead, these enzymes exhibit broad substrate specificity toward prenyl donors (Fig. 3a). The DMAPP (C5), GPP (C10), farnesyl diphosphate (FPP) (C15), geranylgeranyl diphosphate (GGPP) (C20), and geranylfarnesyl diphosphate (GFPP) (C25) were accepted by the enzymes to generate C-7 position or C-5 position prenylated indolactam V. Moreover, the extremely promiscuous AtaPT from Aspergillus terreus was reported to produce 72 prenylated aromatic compounds, including lignanoids, xanthones, quinoline alkaloids, coumarins, benzophenones, curcuminoid, and hydroxynaphthalenes using DMAPP, GPP, and FPP as prenyl donors [88]. The formation of mono-, di-, and/or tri-prenylated compounds were demonstrated. AtaPT also accepts GGPP and phytyl diphosphate (PPP) as prenyl donors to attach geranylgeranyl or phytyl group on (+)-butyrolactone II (Fig. 3b).
Fig. 3.
The substrate specificity of a TleC, MpnD, and b AtaPT toward prenyl donors
The aromatic PTases can also accept synthetic unnatural alkyl donors, e.g. methylallyl diphosphate (MAPP), 2-pentenyl diphosphate (2-pen-PP), and benzyl diphosphate (benzyl-PP) (Fig. 4). When 2-pen-PP were used as a alkyl donor of FgaPT2 and 5-DMATS, the enzymes generated regular C5-alkylated and C6-alkylated L-tryptophan, respectively [89, 90]. Similarly, the regular C4- and C5-alkylated and regular C5- and C6-alkylated L-tryptophan were produced by FgaPT2 and 5-DMAT, respectively, with MAPP as an allyl donor. Furthermore, C2- and C3- reversely alkylated cyclic dipeptides using MAPP and 2-pen-PP as alkyl donors were delivered by C3-prenyltransferases (CdpC3PT, CdpNPT, and AnaPT) and C2-prenyltransferases (BrePT and FtmPT1) [91]. Interestingly, BrePT and FtmPT1 afforded a mixture of C2- and C3-alkylated diastereomers. C5-, C6-, and C7-benzyl-L-tryptophan derivatives were enzymatically synthesized with 5-DMATS, 6-DMATS, TyrPT, and FgaPT2.
Fig. 4.
Structures of the synthetic unnatural prenyl donors tested for FgaPT2 and SirD. The substrates highlighted in purple, red, and blue are tested for the both of FgaPT2 and SirD, only SirD, and only FgaPT2, respectively. The substrates enclosed in red and blue frames are accepted by SirD and FgaPT2, respectively
SirD and FgaPT2 were used for further alkyl-diversification of L-Tyr and indole-containing compounds, respectively. Bandari et al. synthesized various alkyl-PP analogues, including allylic, conjugated diene analogues and benzylic substituted substrates shown in Fig. 4 [92, 93]. The 33 unnatural compounds were used as substrates of FgaPT2 and the 20 unnatural compounds were tested for SirD. SirD accepted 15 out of the 20 unnatural alkyl-PP to deliver the corresponding alkylated L-tyrosine. In these, single corresponding O-monoalkylated L-tyrosine were mainly produced, while two monoalkylated products were given from cinnamyl-PP and (2E,4E)-5-phenylpenta-2,4-dien-1-yl-PP. On the other hand, FgaPT2 used 24 out of the 33 synthetic alkyl-PPs. The C4 and C5-regular alkylated and C3 and N1-reversely alkylated L-tryptophan were synthesized through in vitro enzyme reactions. Additionally, C3 position of 7-hydroxy-indolocarbazole was able to be alkylated by FgaPT2 with (E)-3-methylpent-2-en-1-yl-PP, 3-ethylpent-2-en-1-yl-PP, cyclohex-1-en-1-ylmethyl-PP, 2-cyclopentylideneethyl-PP, 2-cyclohexylideneethyl-PP (Fig. 4).
Structure-based engineering of ABBA and DMATS PTases
So far, more than 15 crystal structures of aromatic PTases have been reported [23, 58, 66, 78, 84, 87, 93–102]. The structure-based engineering of PTases were also performed for several enzymes.
The EpzP and PpzP from Streptomyces cinnamonensis DSM 104 and Streptomyces annulatus 9663, respectively, catalyze prenylation toward phenazine [101]. The crystal structures of EzpP, the docking model with a substrate 5,10-dihydrophenazine-1-carboxylate (dhPCA), and the mutagenesis analysis provided the intimate structural details of the prenylation reaction mechanism. Based on these information together with the sequential comparison between EpzP and PpzP, the catalytic velocity of EpzP was improved by site-directed mutagenesis. V270F mutation was introduced to form π-stacking between dhPCA and Phe residue. As a result, the enzymatic activity of V270F mutant was increased five-times compared to wild type. Furthermore, the substitution of Ala285 with Gln residue to interact with a water molecule in the active site showed ~ 14-fold higher enzymatic activity than wild type.
The substrate specificity of FgaPT2 was altered by structure-based mutagenesis experiment. Lys174 residue in FgaPT2, proposed to abstract a proton from prenyl-attached arenium intermediate, was substituted with phenylalanine to stabilize the arenium intermediate and increase the interaction with benzene ring of non-genuine substrate L-tyrosine [26, 103]. The K174F exhibited 4.9-times higher catalytic efficiency toward L-tyrosine than that of wild type, while the activity toward L-tryptophan was almost abolished. Interestingly, the K174F mutant catalyzes C3-prenylation reaction toward L-tyrosine and its analog 4-amino-L-phenylalanine, and N-prenylation reaction toward 4-amino-L-phenylalanine as a minor reaction [56]. The specificity for the prenylation of L-tyrosine and L-tryptophan was changed from 1:31 (wild type) to 208:1 (K174F mutant). Furthermore, saturation mutagenesis was performed at Arg244, interacting with carboxylate group of substrates [104]. The prenylation activities of 13 Arg244 mutants toward tryptophan-containing cyclic dipeptides were increased up to 76-times compared to wild type. Interestingly, the preferences for tryptophan-containing cyclic dipeptides, including cyclo-L-Trp-L-Leu, cyclo-L-Trp-D-Pro, cyclo-L-Trp-L-Pro, cyclo-L-Trp-Gly, cyclo-L-Trp-L-Trp, and cyclo-L-Trp-L-Phe of these mutants were also changed. For example, the wild-type, R244A, R244T, and R244Q prefer cyclo-L-Trp-D-Pro and cyclo-L-Trp-L-Leu, while R244G utilizes cyclo-L-Trp-L-Leu and cyclo-L-Trp-L-Trp as preferable substrates. The combination of the K174F and R244X mutations succeeded to alter the regiospecificity of prenylation from C4-regular prenylation to C3-reverse prenylation toward tryptophan-containing cyclic dipeptides (Fig. 5a) [105].
Fig. 5.
The enzyme reactions of engineered ABBA-type and DMATS-type PTases. The enzyme reaction of a the FgaPT2 and its K174F/K244X mutants, b the FtmPT1 and its Y205X mutants, c the TleC W97Y/A173M and W97Y/F170W/A173M mutants, and d FgaPT2 and its M328X mutants
The structure-based engineering of DMATS to alter the product specificity was also achieved using FtmPT1 from Aspergillus fumigatus in fumitremorgins biosynthesis. FtmPT1 originally catalyzes C2 prenylation reaction to bevianamide F (cyclo-L-Trp-L-Pro). The structure analysis of FtmPT1 suggested that Tyr205 residue in FtmPT1 interacts with ketone-group of bevianamide F. The saturation-mutagenesis at Tyr205 revealed that the 15 mutants generate regularly C3-prenylated brevianamide F, but not at C2 position [106]. The substrate specificity analysis of the two selected mutants Y205N and Y205L revealed that these mutants generated C3-reverse prenylated compounds as predominant products when cyclo-D-Trp-D-Pro, cyclo-D-Trp-L-Pro, and cyclo-L-Trp-D-Pro were used as substrates (Fig. 5b).
On the other hand, the engineering of substrate specificity toward prenyl donors was demonstrated using MpnD, and TleC. MpnD and TleC prefer to utilize C5 DMAPP and C10 GPP, respectively, and catalyze attachment of prenyl donor at the C-7 position of indolactam V in a reverse fashion [85]. The structural analysis of MpnD and TleC complexed with substrates suggested that the three amino acid residues Trp97, Phe170, and Ala173 in TleC and Tyr80, Trp157, and Met159 in MpnD regulate the selectivity of the length of prenyl donor and regiospecificity of prenylation position (Fig. 5c). Based on these observations, TleC A173M, TleC W97Y/A173M, and TleC W97Y/F170W/A173M, MpnD M159A were constructed and analyzed. The preference for prenyl donors of TleC A173M switched from GPP to the smaller DMAPP. On the contrary, M159A substitution in MpnD improved the GPP prenylation activity to generate lyngbyatoxin A, while DMAPP prenylation activity was decreased. Moreover, the TleC W97Y/A173M and TleC W97Y/F170W/ A173M mutants newly produced teleocidin A-2, C-19-epimer of lyngbyatoxin A, and 5-geranylindolactam V in addition to lyngbyatoxin A.
Similar manipulation of prenyl donor substrates was also performed using FgaPT2. The structure-based modeling of FgaPT2 with substrate suggested that the side chain of Met328 protrudes toward the active site and would decrease the size of active site. Thus, Met328 was thought to regulate the substrate specificity of the length of prenyl donor. The substitution with smaller side chain, including M328A, M328C, M328S, and M328G significantly increased the activity for GPP and FPP prenylation (Fig. 5d) [107]. Furthermore, the model also suggested that the active site residues Lue263 and Tyr398 could also interfere with terminal isoprene unit of FPP. The large-to-small substitution of Leu263 and Tyr398 with Ala and Phe, respectively, improved the FPP prenylation activity.
Conclusion
The development of the sequencing technology and the improvement of methodology to characterize the enzymes have accelerated the understanding of the biosynthesis of secondary metabolites. By using these techniques, dozens of ABBA-type and DMATS-type aromatic PTases were functionally and structurally characterized. The accumulation of the knowledge in enzymes provided the chance for the application and engineering of these aromatic PTases. The results presented in this review would be the model cases toward utilization of the secondary metabolite enzymes to generate structurally diversified and biologically active unnatural novel molecular scaffolds for drug discovery.
Acknowledgements
The works in the authors’ laboratory were supported in part by a Grant-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science, and Technology of Japan.
Footnotes
The original version of this article was revised due to a retrospective Open Access order.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
5/28/2020
The article Enzymatic studies on aromatic prenyltransferases, written by Takahiro��Mori.
References
- 1.Yazaki K, Sasaki K, Tsurumaru Y. Prenylation of aromatic compounds, a key diversification of plant secondary metabolites. Phytochemistry. 2009;70:1739–1745. doi: 10.1016/j.phytochem.2009.08.023. [DOI] [PubMed] [Google Scholar]
- 2.Botta B, Vitali A, Menendez P, et al. Prenylated flavonoids: pharmacology and biotechnology. Curr Med Chem. 2010;12:713–739. doi: 10.2174/0929867053202241. [DOI] [PubMed] [Google Scholar]
- 3.Alhassan AM, Abdullahi MI, Uba A, Umar A. Prenylation of aromatic secondary metabolites: a new frontier for development of novel drugs. Trop J Pharm Res. 2014;13:307–314. doi: 10.4314/tjpr.v13i2.22. [DOI] [Google Scholar]
- 4.Miranda CL, Aponso GLM, Stevens JF, et al. Prenylated chalcones and flavanones as inducers of quinone reductase in mouse Hepa 1c1c7 cells. Cancer Lett. 2000;149:21–29. doi: 10.1016/S0304-3835(99)00328-6. [DOI] [PubMed] [Google Scholar]
- 5.Dong X, Fan Y, Yu L, Hu Y. Synthesis of four natural prenylflavonoids and their estrogen-like activities. Arch Pharm (Weinheim) 2007;340:372–376. doi: 10.1002/ardp.200700057. [DOI] [PubMed] [Google Scholar]
- 6.Appendino G, Gibbons S, Giana A, et al. Antibacterial cannabinoids from Cannabis sativa: a structure-activity study. J Nat Prod. 2008;71:1427–1430. doi: 10.1021/np8002673. [DOI] [PubMed] [Google Scholar]
- 7.Militão GCG, Pinheiro SM, Dantas INF, et al. Bioassay-guided fractionation of pterocarpans from roots of Harpalyce brasiliana Benth. Bioorganic Med Chem. 2007;15:6687–6691. doi: 10.1016/j.bmc.2007.08.011. [DOI] [PubMed] [Google Scholar]
- 8.Mukai R. Prenylation enhances the biological activity of dietary flavonoids by altering their bioavailability. Biosci Biotechnol Biochem. 2018;82:207–215. doi: 10.1080/09168451.2017.1415750. [DOI] [PubMed] [Google Scholar]
- 9.Ashby MN, Kutsunai SY, Ackerman S, et al. COQ2 is a candidate for the structural gene encoding para-hydroxybenzoate:polyprenyltransferase. J Biol Chem. 1992;267:4128–4136. [PubMed] [Google Scholar]
- 10.Suvarna K, Stevenson D, Meganathan R, Hudspeth MES. Menaquinone (vitamin K2) biosynthesis: localization and characterization of the menA gene from Escherichia coli. J Bacteriol. 1998;180:2782–2787. doi: 10.1128/JB.180.10.2782-2787.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tanner ME. Mechanistic studies on the indole prenyltransferases. Nat Prod Rep. 2015;32:88–101. doi: 10.1039/C4NP00099D. [DOI] [PubMed] [Google Scholar]
- 12.Winkelblech J, Fan A, Li SM. Prenyltransferases as key enzymes in primary and secondary metabolism. Appl Microbiol Biotechnol. 2015;99:7379–7397. doi: 10.1007/s00253-015-6811-y. [DOI] [PubMed] [Google Scholar]
- 13.Tello M, Kuzuyama T, Heide L, et al. The ABBA family of aromatic prenyltransferases: broadening natural product diversity. Cell Mol Life Sci. 2008;65:1459–1463. doi: 10.1007/s00018-008-7579-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heide L. Prenyl transfer to aromatic substrates: genetics and enzymology. Curr Opin Chem Biol. 2009;13:171–179. doi: 10.1016/j.cbpa.2009.02.020. [DOI] [PubMed] [Google Scholar]
- 15.Li SM. Applications of dimethylallyltryptophan synthases and other indole prenyltransferases for structural modification of natural products. Appl Microbiol Biotechnol. 2009;84:631–639. doi: 10.1007/s00253-009-2128-z. [DOI] [PubMed] [Google Scholar]
- 16.Li SM. Evolution of aromatic prenyltransferases in the biosynthesis of indole derivatives. Phytochemistry. 2009;70:1746–1757. doi: 10.1016/j.phytochem.2009.03.019. [DOI] [PubMed] [Google Scholar]
- 17.Saleh O, Haagen Y, Seeger K, Heide L. Prenyl transfer to aromatic substrates in the biosynthesis of aminocoumarins, meroterpenoids and phenazines: the ABBA prenyltransferase family. Phytochemistry. 2009;70:1728–1738. doi: 10.1016/j.phytochem.2009.05.009. [DOI] [PubMed] [Google Scholar]
- 18.Li SM. Prenylated indole derivatives from fungi: structure diversity, biological activities, biosynthesis and chemoenzymatic synthesis. Nat Prod Rep. 2010;27:57–78. doi: 10.1039/B909987P. [DOI] [PubMed] [Google Scholar]
- 19.Nakagawa K, Hirota Y, Sawada N, et al. Identification of UBIAD1 as a novel human menaquinone-4 biosynthetic enzyme. Nature. 2010;468:117–121. doi: 10.1038/nature09464. [DOI] [PubMed] [Google Scholar]
- 20.Bonitz T, Alva V, Saleh O, et al. Evolutionary relationships of microbial aromatic prenyltransferases. PLoS ONE. 2011;6:e0143237. doi: 10.1371/journal.pone.0027336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jain S, Caforio A, Driessen AJM. Biosynthesis of archaeal membrane ether lipids. Front Microbiol. 2014;5:641. doi: 10.3389/fmicb.2014.00641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Matsuda Y, Abe I. Biosynthesis of fungal meroterpenoids. Nat Prod Rep. 2016;33:26–53. doi: 10.1039/C5NP00090D. [DOI] [PubMed] [Google Scholar]
- 23.Pojer F, Wemakor E, Kammerer B, et al. CloQ, a prenyltransferase involved in clorobiocin biosynthesis. Proc Natl Acad Sci U S A. 2003;100:2316–2321. doi: 10.1073/pnas.0337708100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuzuyama T, Noel JP, Richard SB. Structural basis for the promiscuous biosynthetic prenylation of aromatic natural products. Nature. 2005;435:983–987. doi: 10.1038/nature03668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang H, Levin EJ, Liu S, et al. Structure of a membrane-embedded prenyltransferase homologous to UBIAD1. PLoS Biol. 2014;12:e1001911. doi: 10.1371/journal.pbio.1001911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Metzger U, Schall C, Zocher G, et al. The structure of dimethylallyl tryptophan synthase reveals a common architecture of aromatic prenyltransferases in fungi and bacteria. Proc Natl Acad Sci U S A. 2009;106:14309–14314. doi: 10.1073/pnas.0904897106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grundmann A, Li SM. Overproduction, purification and characterization of FtmPT1, a brevianamide F prenyltransferase from Aspergillus fumigatus. Microbiology. 2005;151:2199–2207. doi: 10.1099/mic.0.27962-0. [DOI] [PubMed] [Google Scholar]
- 28.Chen J, Morita H, Wakimoto T, et al. Prenylation of a nonaromatic carbon of indolylbutenone by a fungal indole prenyltransferase. Org Lett. 2012;14:3080–3083. doi: 10.1021/ol301129x. [DOI] [PubMed] [Google Scholar]
- 29.Rudolf JD, Poulter CD. Tyrosine O -prenyltransferase SirD catalyzes S -, C -, and N -prenylations on tyrosine and tryptophan derivatives. ACS Chem Biol. 2013;8:2707–2714. doi: 10.1021/cb400691z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yin S, Yu X, Wang Q, et al. Identification of a brevianamide F reverse prenyltransferase BrePT from Aspergillus versicolor with a broad substrate specificity towards tryptophan-containing cyclic dipeptides. Appl Microbiol Biotechnol. 2013;97:1649–1660. doi: 10.1007/s00253-012-4130-0. [DOI] [PubMed] [Google Scholar]
- 31.Yu H, Liebhold M, Xie X, Li SM. Tyrosine O-prenyltransferases TyrPT and SirD displaying similar behavior toward unnatural alkyl or benzyl diphosphate as their natural prenyl donor dimethylallyl diphosphate. Appl Microbiol Biotechnol. 2015;99:7115–7124. doi: 10.1007/s00253-015-6452-1. [DOI] [PubMed] [Google Scholar]
- 32.Leipoldt F, Zeyhle P, Kulik A, et al. Diversity of ABBA Prenyltransferases in marine streptomyces sp. CNQ-509: promiscuous enzymes for the biosynthesis of mixed terpenoid compounds. PLoS ONE. 2015;10:e143237. doi: 10.1371/journal.pone.0143237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang X, Yang J, Jiang Y, et al. Regiospecific synthesis of prenylated flavonoids by a prenyltransferase cloned from Fusarium oxysporum. Sci Rep. 2016;6:24819. doi: 10.1038/srep24819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kremer A, Li SM. Tryptophan aminopeptidase activity of several indole prenyltransferases from Aspergillus fumigatus. Chem Biol. 2008;15:729–738. doi: 10.1016/j.chembiol.2008.05.019. [DOI] [PubMed] [Google Scholar]
- 35.Ruan HL, Yin WB, Wu JZ, Li SM. Reinvestigation of a cyclic dipeptide N-prenyltransferase reveals rearrangement of N-1 prenylated indole derivatives. ChemBioChem. 2008;9:1044–1047. doi: 10.1002/cbic.200700723. [DOI] [PubMed] [Google Scholar]
- 36.Steffan N, Unsöld IA, Li SM. Chemoenzymatic synthesis of prenylated indole derivatives by using a 4-dimethylallyltryptophan synthase from Aspergillus fumigatus. ChemBioChem. 2007;8:1298–1307. doi: 10.1002/cbic.200700107. [DOI] [PubMed] [Google Scholar]
- 37.Yin WB, Ruan HL, Westrich L, et al. CdpNPT, an N-prenyltransferase from Aspergillus fumigatus: overproduction, purification and biochemical characterisation. ChemBioChem. 2007;8:1154–1161. doi: 10.1002/cbic.200700079. [DOI] [PubMed] [Google Scholar]
- 38.Kumano T, Richard SB, Noel JP, et al. Chemoenzymatic syntheses of prenylated aromatic small molecules using Streptomyces prenyltransferases with relaxed substrate specificities. Bioorganic Med Chem. 2008;16:8117–8126. doi: 10.1016/j.bmc.2008.07.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xiao Y, MacHacek M, Lee K, et al. Prenyltransferase substrate binding pocket flexibility and its application in isoprenoid profiling. Mol Biosyst. 2009;5:913–917. doi: 10.1039/b902370d. [DOI] [PubMed] [Google Scholar]
- 40.Haug-Schifferdecker E, Arican D, Brückner R, Heide L. A new group of aromatic prenyltransferases in fungi, catalyzing a 2,7-dihydroxynaphthalene 3-dimethylallyl-transferase reaction. J Biol Chem. 2010;285:16487–16494. doi: 10.1074/jbc.M110.113720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yu X, Xie X, Li SM. Substrate promiscuity of secondary metabolite enzymes: prenylation of hydroxynaphthalenes by fungal indole prenyltransferases. Appl Microbiol Biotechnol. 2011;92:737–748. doi: 10.1007/s00253-011-3351-y. [DOI] [PubMed] [Google Scholar]
- 42.Unsöld IA, Li SM. Overproduction, purification and characterization of FgaPT2, a dimethylallyltryptophan synthase from Aspergillus fumigatus. Microbiology. 2005;151:1499–1505. doi: 10.1099/mic.0.27759-0. [DOI] [PubMed] [Google Scholar]
- 43.Haagen Y, Unsöld I, Westrich L, et al. A soluble, magnesium-independent prenyltransferase catalyzes reverse and regular C-prenylations and O-prenylations of aromatic substrates. FEBS Lett. 2007;581:2889–2893. doi: 10.1016/j.febslet.2007.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhou K, Yu X, Xie X, Li SM. Complementary flavonoid prenylations by fungal indole prenyltransferases. J Nat Prod. 2015;78:2229–2235. doi: 10.1021/acs.jnatprod.5b00422. [DOI] [PubMed] [Google Scholar]
- 45.Ozaki T, Mishima S, Nishiyama M, Kuzuyama T. NovQ is a prenyltransferase capable of catalyzing the addition of a dimethylallyl group to both phenylpropanoids and flavonoids. J Antibiot (Tokyo) 2009;62:385–392. doi: 10.1038/ja.2009.48. [DOI] [PubMed] [Google Scholar]
- 46.Araya-Cloutier C, Martens B, Schaftenaar G, et al. Structural basis for non-genuine phenolic acceptor substrate specificity of Streptomyces roseochromogenes prenyltransferase CloQ from the ABBA/PT-barrel superfamily. PLoS ONE. 2017;12:e0174665. doi: 10.1371/journal.pone.0174665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sugiyama A, Linley PJ, Sasaki K, et al. Metabolic engineering for the production of prenylated polyphenols in transgenic legume plants using bacterial and plant prenyltransferases. Metab Eng. 2011;13:629–637. doi: 10.1016/j.ymben.2011.07.003. [DOI] [PubMed] [Google Scholar]
- 48.Pockrandt D, Ludwig L, Fan A, et al. New insights into the biosynthesis of prenylated Xanthones: Xptb from Aspergillus nidulans catalyses an O-prenylation of Xanthones. ChemBioChem. 2012;13:2764–2771. doi: 10.1002/cbic.201200545. [DOI] [PubMed] [Google Scholar]
- 49.Chooi YH, Wang P, Fang J, et al. Discovery and characterization of a group of fungal polycyclic polyketide prenyltransferases. J Am Chem Soc. 2012;134:9428–9437. doi: 10.1021/ja3028636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Noike M, Liu C, Ono Y, et al. An enzyme catalyzing o-prenylation of the glucose moiety of Fusicoccin A, a Diterpene Glucoside produced by the fungus Phomopsis amygdali. ChemBioChem. 2012;13:566–573. doi: 10.1002/cbic.201100725. [DOI] [PubMed] [Google Scholar]
- 51.McIntosh JA, Donia MS, Nair SK, Schmidt EW. Enzymatic basis of ribosomal peptide prenylation in cyanobacteria. J Am Chem Soc. 2011;133:13698–13705. doi: 10.1021/ja205458h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tianero MDB, Donia MS, Young TS, et al. Ribosomal route to small-molecule diversity. J Am Chem Soc. 2012;134:418–425. doi: 10.1021/ja208278k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gu W, Dong SH, Sarkar S, et al. The biochemistry and structural biology of cyanobactin pathways enabling combinatorial biosynthesis. Methods Enzymol. 2018;604:113–163. doi: 10.1016/bs.mie.2018.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dalponte L, Parajuli A, Younger E, et al. N-prenylation of tryptophan by an aromatic prenyltransferase from the cyanobactin biosynthetic pathway. Biochemistry. 2018;57:6860–6867. doi: 10.1021/acs.biochem.8b00879. [DOI] [PubMed] [Google Scholar]
- 55.Morita M, Hao Y, Jokela JK, et al. Post-translational tyrosine geranylation in cyanobactin biosynthesis. J Am Chem Soc. 2018;140:6044–6048. doi: 10.1021/jacs.8b03137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fan A, Zocher G, Stec E, et al. Site-directed mutagenesis switching a dimethylallyl tryptophan synthase to a specific tyrosine C3-prenylating enzyme. J Biol Chem. 2015;290:1364–1373. doi: 10.1074/jbc.M114.623413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ruffner DE, Schmidt EW, Heemstra JR. Assessing the combinatorial potential of the RiPP cyanobactin tru pathway. ACS Synth Biol. 2015;4:482–492. doi: 10.1021/sb500267d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Okada M, Sugita T, Akita K, et al. Stereospecific prenylation of tryptophan by a cyanobacterial post-translational modification enzyme. Org Biomol Chem. 2016;14:9639–9644. doi: 10.1039/C6OB01759B. [DOI] [PubMed] [Google Scholar]
- 59.Hao Y, Pierce E, Roe D, et al. Molecular basis for the broad substrate selectivity of a peptide prenyltransferase. Proc Natl Acad Sci U S A. 2016;113:14037–14042. doi: 10.1073/pnas.1609869113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Okada M, Sugita T, Abe I. Posttranslational isoprenylation of tryptophan in bacteria. Beilstein J Org Chem. 2017;13:338–346. doi: 10.3762/bjoc.13.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Estrada P, Morita M, Hao Y, et al. A single amino acid switch alters the isoprene donor specificity in ribosomally synthesized and post-translationally modified peptide prenyltransferases. J Am Chem Soc. 2018;140:8124–8127. doi: 10.1021/jacs.8b05187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Donia MS, Ravel J, Schmidt EW. A global assembly line for cyanobactins. Nat Chem Biol. 2008;4:341–343. doi: 10.1038/nchembio.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fan A, Xie X, Li SM. Tryptophan prenyltransferases showing higher catalytic activities for Friedel-Crafts alkylation of o- and m-tyrosines than tyrosine prenyltransferases. Org Biomol Chem. 2015;13:7551–7557. doi: 10.1039/C5OB01040C. [DOI] [PubMed] [Google Scholar]
- 64.Mundt K, Li SM. CdpC2PT, a reverse prenyltransferase from Neosartorya fischeri with a distinct substrate preference from known C2-prenyltransferases. Microbiol (United Kingdom) 2013;159:2169–2179. doi: 10.1099/mic.0.069542-0. [DOI] [PubMed] [Google Scholar]
- 65.Tsai HF, Wang H, Gebler JC, et al. The claviceps purpurea gene encoding dimethylallyltryptophan Synthase, the Committed Step for Ergot Alkaloid Biosynthesis. Biochem Biophys Res Commun. 1995;216:119–125. doi: 10.1006/bbrc.1995.2599. [DOI] [PubMed] [Google Scholar]
- 66.Miyamoto K, Ishikawa F, Nakamura S, et al. A 7-dimethylallyl tryptophan synthase from a fungal Neosartorya sp.: biochemical characterization and structural insight into the regioselective prenylation. Bioorganic Med Chem. 2014;22:2517–2528. doi: 10.1016/j.bmc.2014.02.031. [DOI] [PubMed] [Google Scholar]
- 67.Yu X, Zocher G, Xie X, et al. Catalytic mechanism of stereospecific formation of cis-configured prenylated pyrroloindoline diketopiperazines by indole prenyltransferases. Chem Biol. 2013;20:1492–1501. doi: 10.1016/j.chembiol.2013.10.007. [DOI] [PubMed] [Google Scholar]
- 68.Markert A, Steffan N, Ploss K, et al. Biosynthesis and accumulation of ergoline alkaloids in a mutualistic association between Ipomoea asarifolia (Convolvulaceae) and a clavicipitalean fungus. Plant Physiol. 2008;147:296–305. doi: 10.1104/pp.108.116699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zou HX, Xie XL, Linne U, et al. Simultaneous C7- and N1-prenylation of cyclo-l-Trp-l-Trp catalyzed by a prenyltransferase from Aspergillus oryzae. Org Biomol Chem. 2010;8:3037–3044. doi: 10.1039/c002850a. [DOI] [PubMed] [Google Scholar]
- 70.Yu X, Liu Y, Xie X, et al. Biochemical characterization of indole prenyltransferases: Filling the lastgapofprenylation positions bya5-dimethylallyltryptophan synthase from Aspergillus clavatus. J Biol Chem. 2012;287:1371–1380. doi: 10.1074/jbc.M111.317982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ding Y, Wet JRD, Cavalcoli J, et al. Genome-based characterization of two prenylation steps in the assembly of the stephacidin and notoamide anticancer agents in a marine-derived Aspergillus sp. J Am Chem Soc. 2010;132:12733–12740. doi: 10.1021/ja1049302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Takahashi S, Takagi H, Toyoda A, et al. Biochemical characterization of a novel indole prenyltransferase from streptomyces sp. SN-593. J Bacteriol. 2010;192:2839–2851. doi: 10.1128/JB.01557-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Winkelblech J, Li SM. Biochemical investigations of two 6-DMATS enzymes from streptomyces reveal new features of l -tryptophan prenyltransferases. ChemBioChem. 2014;15:1030–1039. doi: 10.1002/cbic.201400046. [DOI] [PubMed] [Google Scholar]
- 74.Schultz AW, Lewis CA, Luzung MR, et al. Functional characterization of the cyclomarin/cyclomarazine prenyltransferase cymd directs the biosynthesis of unnatural cyclic peptides. J Nat Prod. 2010;73:373–377. doi: 10.1021/np9006876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tarcz S, Ludwig L, Li SM. AstPT catalyses both reverse N1- and regular C2 prenylation of a methylated bisindolyl benzoquinone. ChemBioChem. 2014;15:108–116. doi: 10.1002/cbic.201300610. [DOI] [PubMed] [Google Scholar]
- 76.Schneider P, Weber M, Hoffmeister D. The Aspergillus nidulans enzyme TdiB catalyzes prenyltransfer to the precursor of bioactive asterriquinones. Fungal Genet Biol. 2008;45:302–309. doi: 10.1016/j.fgb.2007.09.004. [DOI] [PubMed] [Google Scholar]
- 77.Yin WB, Yu X, Xie XL, Li SM. Preparation of pyrrolo[2,3-b]indoles carrying a β-configured reverse C3-dimethylallyl moiety by using a recombinant prenyltransferase CdpC3PT. Org Biomol Chem. 2010;8:2430–2438. doi: 10.1039/c000587h. [DOI] [PubMed] [Google Scholar]
- 78.Haynes SW, Gao X, Tang Y, Walsh CT. Complexity generation in fungal peptidyl alkaloid biosynthesis: a two-enzyme pathway to the hexacyclic MDR export pump inhibitor ardeemin. ACS Chem Biol. 2013;8:741–748. doi: 10.1021/cb3006787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Schuller JM, Zocher G, Liebhold M, et al. Structure and catalytic mechanism of a cyclic dipeptide prenyltransferase with broad substrate promiscuity. J Mol Biol. 2012;422:87–99. doi: 10.1016/j.jmb.2012.05.033. [DOI] [PubMed] [Google Scholar]
- 80.Wunsch C, Zou HX, Linne U, Li SM. C7-prenylation of tryptophanyl and O-prenylation of tyrosyl residues in dipeptides by an Aspergillus terreus prenyltransferase. Appl Microbiol Biotechnol. 2015;99:1719–1730. doi: 10.1007/s00253-014-5999-6. [DOI] [PubMed] [Google Scholar]
- 81.Liu X, Walsh CT. Characterization of cyclo-acetoacetyl-l-tryptophan dimethylallyltransferase in cyclopiazonic acid biosynthesis: substrate promiscuity and site directed mutagenesis studies. Biochemistry. 2009;48:11032–11044. doi: 10.1021/bi901597j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhou K, Ludwig L, Li SM. Friedel-Crafts alkylation of acylphloroglucinols catalyzed by a fungal indole prenyltransferase. J Nat Prod. 2015;78:929–933. doi: 10.1021/np5009784. [DOI] [PubMed] [Google Scholar]
- 83.Edwards DJ, Gerwick WH. Lyngbyatoxin biosynthesis: Sequence of biosynthetic gene cluster and identification of a novel aromatic prenyltransferase. J Am Chem Soc. 2004;126:11432–11433. doi: 10.1021/ja047876g. [DOI] [PubMed] [Google Scholar]
- 84.Awakawa T, Zhang L, Wakimoto T, et al. A methyltransferase initiates terpene cyclization in teleocidin B biosynthesis. J Am Chem Soc. 2014;136:9910–9913. doi: 10.1021/ja505224r. [DOI] [PubMed] [Google Scholar]
- 85.Mori T, Zhang L, Awakawa T, et al. Manipulation of prenylation reactions by structure-based engineering of bacterial indolactam prenyltransferases. Nat Commun. 2016;7:10849. doi: 10.1038/ncomms10849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Awakawa T, Abe I. Biosynthesis of the teleocidin-type terpenoid indole alkaloids. Org Biomol Chem. 2018;16:4746–4752. doi: 10.1039/C8OB00803E. [DOI] [PubMed] [Google Scholar]
- 87.Abe I. Biosynthetic studies on teleocidins in Streptomyces. J Antibiot (Tokyo) 2018;71:763–768. doi: 10.1038/s41429-018-0069-4. [DOI] [PubMed] [Google Scholar]
- 88.Chen R, Gao B, Liu X, et al. Molecular insights into the enzyme promiscuity of an aromatic prenyltransferase. Nat Chem Biol. 2017;13:226–234. doi: 10.1038/nchembio.2263. [DOI] [PubMed] [Google Scholar]
- 89.Liebhold M, Xie X, Li SM. Expansion of enzymatic Friedel-Crafts alkylation on indoles: Acceptance of unnatural β-unsaturated allyl diphospates by dimethylallyl-tryptophan synthases. Org Lett. 2012;14:4882–4885. doi: 10.1021/ol302207r. [DOI] [PubMed] [Google Scholar]
- 90.Liebhold M, Li SM. Regiospecific benzylation of tryptophan and derivatives catalyzed by a fungal dimethylallyl transferase. Org Lett. 2013;15:5834–5837. doi: 10.1021/ol4029012. [DOI] [PubMed] [Google Scholar]
- 91.Liebhold M, Xie X, Li SM. Breaking cyclic dipeptide prenyltransferase regioselectivity by unnatural alkyl donors. Org Lett. 2013;15:3062–3065. doi: 10.1021/ol401247s. [DOI] [PubMed] [Google Scholar]
- 92.Bandari C, Scull EM, Masterson JM, et al. Determination of alkyl-donor promiscuity of tyrosine-o-prenyltransferase sird from Leptosphaeria maculans. ChemBioChem. 2017;18:2323–2327. doi: 10.1002/cbic.201700469. [DOI] [PubMed] [Google Scholar]
- 93.Bandari C, Scull EM, Bavineni T, et al. FgaPT2, a biocatalytic tool for alkyl-diversification of indole natural products. Medchemcomm. 2019;10:1465–1475. doi: 10.1039/C9MD00177H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Elshahawi SI, Cao H, Shaaban KA, et al. Structure and specificity of a permissive bacterial C-prenyltransferase. Nat Chem Biol. 2017;13:366–368. doi: 10.1038/nchembio.2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wang J, Chen CC, Yang Y, et al. Structural insight into a novel indole prenyltransferase in hapalindole-type alkaloid biosynthesis. Biochem Biophys Res Commun. 2017;495:1782–1788. doi: 10.1016/j.bbrc.2017.12.039. [DOI] [PubMed] [Google Scholar]
- 96.Awakawa T, Mori T, Nakashima Y, et al. Molecular insight into the Mg2+-dependent allosteric control of indole prenylation by aromatic prenyltransferase AmbP1. Angew Chemie - Int Ed. 2018;57:6810–6813. doi: 10.1002/anie.201800855. [DOI] [PubMed] [Google Scholar]
- 97.Wong CP, Awakawa T, Nakashima Y, et al. Two distinct substrate binding modes for the normal and reverse prenylation of hapalindoles by the prenyltransferase AmbP3. Angew Chemie - Int Ed. 2018;57:560–563. doi: 10.1002/anie.201710682. [DOI] [PubMed] [Google Scholar]
- 98.Roose BW, Christianson DW. Structural basis of tryptophan reverse N-prenylation catalyzed by CymD. Biochemistry. 2019;58:3232–3242. doi: 10.1021/acs.biochem.9b00399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Jost M, Zocher G, Tarcz S, et al. Structure—function analysis of an enzymatic prenyl transfer specificity. J Am Chem Soc. 2010;132:17849–17858. doi: 10.1021/ja106817c. [DOI] [PubMed] [Google Scholar]
- 100.Metzger U, Keller S, Stevenson CEM, et al. Structure and mechanism of the magnesium-independent aromatic prenyltransferase CloQ from the clorobiocin biosynthetic pathway. J Mol Biol. 2010;404:611–626. doi: 10.1016/j.jmb.2010.09.067. [DOI] [PubMed] [Google Scholar]
- 101.Zocher G, Saleh O, Heim JB, et al. Structure-based engineering increased the catalytic turnover rate of a novel phenazine prenyltransferase. PLoS ONE. 2012;7:e48427. doi: 10.1371/journal.pone.0048427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Bent AF, Koehnke J, Houssen WE, et al. Structure of PatF from Prochloron didemni. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2013;69:618–623. doi: 10.1107/S1744309113012931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Luk LYP, Qian Q, Tanner ME. A cope rearrangement in the reaction catalyzed by dimethylallyltryptophan synthase? J Am Chem Soc. 2011;133:12342–12345. doi: 10.1021/ja2034969. [DOI] [PubMed] [Google Scholar]
- 104.Fan A, Li SM. Saturation mutagenesis on Arg244 of the tryptophan C4-prenyltransferase FgaPT2 leads to enhanced catalytic ability and different preferences for tryptophan-containing cyclic dipeptides. Appl Microbiol Biotechnol. 2016;100:5389–5399. doi: 10.1007/s00253-016-7365-3. [DOI] [PubMed] [Google Scholar]
- 105.Zheng L, Mai P, Fan A, Li SM. Switching a regular tryptophan C4-prenyltransferase to a reverse tryptophan-containing cyclic dipeptide C3-prenyltransferase by sequential site-directed mutagenesis. Org Biomol Chem. 2018;16:6688–6694. doi: 10.1039/C8OB01735B. [DOI] [PubMed] [Google Scholar]
- 106.Zhou K, Zhao W, Liu XQ, Li SM. Saturation mutagenesis on Tyr205 of the cyclic dipeptide C2-prenyltransferase FtmPT1 results in mutants with strongly increased C3-prenylating activity. Appl Microbiol Biotechnol. 2016;100:9943–9953. doi: 10.1007/s00253-016-7663-9. [DOI] [PubMed] [Google Scholar]
- 107.Mai P, Zocher G, Stehle T, Li SM. Structure-based protein engineering enables prenyl donor switching of a fungal aromatic prenyltransferase. Org Biomol Chem. 2018;16:7461–7469. doi: 10.1039/C8OB02037J. [DOI] [PubMed] [Google Scholar]