Abstract
Background: CD37, a member of the transmembrane 4 superfamilies (TM4SF), has been proved to be abnormally expressed in a range of malignancies. Herein, we investigate the effects of CD37 expression and analyze its clinical outcome in acute myeloid leukemia (AML) patients.
Methods: The RNA-seq and clinical data of AML patients were obtained from cBioPortal database. CD37 correlated genes, the expression prolife and survival curve of eight key genes were acquired from Gene Expression Profiling Interactive Analysis (GEPIA) and UALCAN. Pathway enrichment and protein–protein interaction (PPI) network analysis were performed based on metascape databases.
Results: Our results showed that CD37 mRNA expression level was significantly up-regulated in patients with AML compared with healthy persons. Patients with high CD37 expression had shorter overall survival (OS) and disease-free survival (DFS). Pathway analysis data showed that CD37 is involved in DNA replication, RNA transport, Salmonella infection, ribonucleoprotein complex biogenesis, cell cycle phase transition and so on. Furthermore, we found eight genes correlated with CD37 are all highly expressed in AML patients, and high expression is associated with poor prognosis.
Conclusion: Our study described systematical expression profiles and the prognostic values of CD37 in AML; our data suggested CD37 might be novel therapeutic target and promising prognostic biomarker in the patients.
Keywords: acute myeloid leukaemia, CD37, gene expression, prognosis
Background
It started a new era when the first monoclonal antibody (mAb), known as rituximab, for treatment of low-grade non-Hodgkins lymphoma was approved by the Food and Drug Administration (FDA) in 1997 [1]. Since then, many other monoclonal antibodies like anti-CD19, anti-CD22, anti-CD23 and anti-CD80 have also been studied. In the late 1980s and early 1990s, the anti-CD37 antibody was used for radioimmunotherapy [2,3].
CD37 belonged to TM4SF (transmembrane 4 superfamily), which was characterized by four potential membrane-spanning regions [4,5]. The CD37 antigen was strongly expressed on B lymphocytes with positive surface immunoglobulin, but it was weakly expressed on a subpopulation of monocytes, granulocytes and T cells, whereas neither platelets nor red blood cells expressed CD37 [6,7]. In terms of cancers, deficiency of CD37 induced the development of B-cell lymphoma [8]. In addition, anti-CD37 agents were clinically active and well tolerated in patients with chronic lymphocytic leukemia (CLL) according to preclinical and early clinical studies [9]. However, the research about CD37 in acute myeloid leukemia (AML) was very limited.
AML is a highly heterogeneous group of diseases characterized by infiltration of the myeloid blasts in bone marrow, peripheral blood or other tissues [10]. It is the most common form of acute leukemia among adults. In 1990, there were 0.96 deaths per 100000 because of AML worldwide, and by 2017 it has increased to 1.31 [11]. The general therapeutic strategy of AML includes induction chemotherapy and consolidation therapy. But if patients do not get poster-mission therapy, they will experience relapse within 6–9 months [12]. Besides, toxicity and morbidity caused by chemotherapy are fatal, especially for elderly patients. Therefore, it is of great urgency to identify reliable biomarkers, enable early diagnosis, improve prognosis, develop new therapeutic strategies.
Methods
TCGA data and the cBioPortal
The cBioPortal is an open-access resource that provides a visual tool for researching and analyzing cancer gene data. Up to now, it offers data on more than 5000 tumor samples from 20 studies [13]. In this study, we obtained CD37 mRNA expression z-scores (RNA Seq V2 RSEM) and detailed information of patients with AML.
The Gene Expression Profiling Interactive Analysis dataset
Gene Expression Profiling Interactive Analysis (GEPIA) is a tool which delivers integrated information to end users, including differential expression analysis, profiling plotting, correlation analysis, patient survival analysis, similar gene detection and dimensionality reduction analysis [14]. We performed CD37 mRNA expression levels in various cancers, the differential expression of CD37 genes in AML and healthy donor samples, survival analysis between high CD37 TPM (transcripts per million) and low CD37 TPM.
UALCAN
UALCAN is an online platform that makes an in-depth analysis of TCGA gene expression data [15]. It is able to assess the effect of gene expression levels and clinicopathological features on patient survival. In the current study, UALCAN was applied to analyze the association between the transcriptional levels of CD37 and clinical outcome. Genes co-expressed with CD37 were also analyzed via UALCAN.
The Cancer Cell Line Encyclopedia dataset
The Cancer Cell Line Encyclopedia (CCLE) assembles 947 human cancer cell lines, covered gene expression, chromosomal copy number, abundant parallel sequencing data and pharmacologic profiles of 24 anticancer drugs across 479 cancer cell lines [16]. By using it, we gained the CD37 expression levels in 14 AML cell lines.
Metascape
Metascape is a powerful and efficient tool, the purpose of which is designed to supply a comprehensive gene list annotation as well as analysis resource for users [17]. Metascape can help users to apply the current popular bioinformatics analysis methods to realize the understanding of genes or protein functions.
DiseaseMeth
DiseaseMeth collects experimental information from over 14000 entries and 175 high-throughput datasets from a wide number of sources. It provides integrated gene methylation data based on cross-dataset analysis for disease and normal samples [18].
Results
CD37 expression in patients with AML
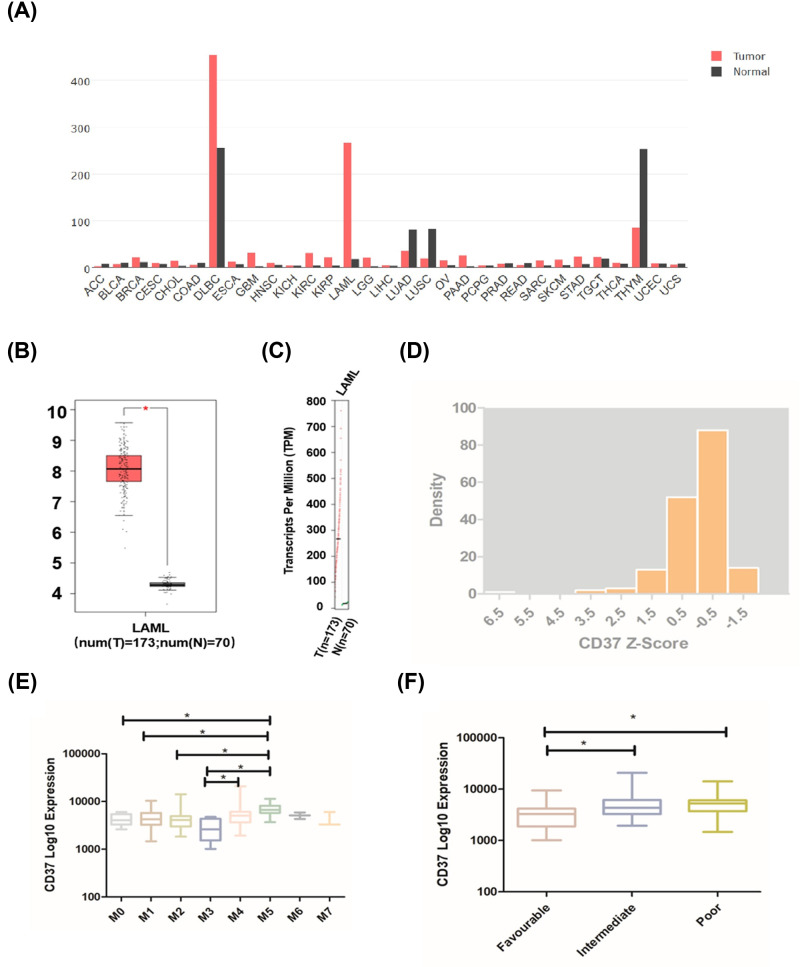
The mRNA levels of CD37 in different cancers were obtained from GEPIA database. As shown in Figure 1A, CD37 has the highest mRNA level in diffuse large B-cell lymphoma (DLBCL) and ranks second in AML. Furthermore, the results showed that CD37 is highly expressed in patients with AML compared with a healthy person (Figure 1B,C). These data suggested that CD37 plays a potential role in AML. We next downloaded CD37 expression data from cBioPortal. When CD37 Z score is between 0 and −1, the number of patients is the largest, and between 6 and 7 is the least (Figure 1D). To figure out whether there was significant difference in CD37 expression among patients classified by The French–American–British (FAB) classification systems, we summarized the expression level of each subgroup (Figure 1E). CD37 expression is relatively higher in M5, which is significantly different from that in M0, M1, M2 and M3 (M0: P=0.017; M1: P=0.008; M2: P=0.007; M3: P=0.000013). We also observed that CD37 expression is relatively lower in M3 but it is only significantly different from M5 and M3 (M3 vs M4: P=0.016). We analyzed CD37 expression in different risk groups based on NCCN AML classification (Figure 1F), and found that the CD37 expression is significantly lower in favorable risk group than that of intermediate and poor risk group (intermediate: P=0.002; poor: P=0.000344). The patients from TCGA datasets were further divided into CD37 high (Z score ≥ 1, n=19) and low (Z score < 1, n=154) group. No significant difference was observed in the two groups on patients’ age, white blood cell counts, percentages of bone marrow blasts, etc. (Table 1). Also, no significant association was identified between the level of CD37 expression and the mutations of IDH1, IDH2, RUNX1, TET2, NRAS, CEBPA, WT1, DNMT3A, NPM1 and TP53 (Table 2).
Figure 1. CD37 expression levels in different types of cancers.
(A) The expression of CD37 in pan-cancer by GEPIA. (B) CD37 expression in AML compared with healthy control. (C) The expression level of CD37 in AML. (D)The distribution of CD37 Z-score. The distribution of CD37 log10 mRNA expression classified by (E) FAB norm; (F) risk status. *P<0.05.
Table 1. The clinical characteristics of patients with AML from cBioPortal.
| Parameter | Total | CD37 low (Z < 1) | CD37 high (Z > 1) | P-value |
|---|---|---|---|---|
| Male (%) | 92 (53.1) | 84 (91.3) | 8 (8.70) | 0.338 |
| Median age (y) | 58 | 57 | 63 | 0.147 |
| Median WBC | 17 | 14.9 | 34.8 | 0.116 |
| Median PB | 39 | 41 | 22 | 0.171 |
| Median BM | 72 | 73 | 63 | 0.129 |
| NCCN subtype | vs favorable | |||
| Favorable | 33 | 32 | 1 | |
| Intermediate | 92 | 82 | 11 | 0.182 |
| Poor | 45 | 37 | 7 | 0.128 |
| FAB subtype, number | ||||
| M0 | 16 | 16 | 0 | |
| M1 | 44 | 40 | 4 | |
| M2 | 38 | 35 | 3 | |
| M3 | 16 | 16 | 0 | |
| M4 | 34 | 29 | 5 | |
| M5 | 18 | 11 | 7 | |
| M6 | 2 | 2 | 0 | |
| M7 | 3 | 3 | 0 |
Table 2. The relation of CD37 expression and common mutation in AML.
| Total | CD37 low (Z < 1) | CD37 high (Z > 1) | P-value | ||
|---|---|---|---|---|---|
| CD37 low vs. high | Fisher exact test | ||||
| FLT3, number (%) | 0.837 | 1 | |||
| Present | 49 (28.32) | 44 (89.80) | 5 (10.20) | ||
| Absent | 124 (71.67) | 110 (88.71) | 14 (11.29) | ||
| IDH1, number (%) | 0.526 | 1 | |||
| Mutated | 16 (9.25) | 15 (93.75) | 1 (6.25) | ||
| Wild-type | 157 (90.75) | 139 (88.54) | 18 (11.46) | ||
| IDH2, number (%) | 0.914 | 1 | |||
| Mutated | 17 (9.83) | 15 (88.24) | 2 (11.76) | ||
| Wild-type | 156 (90.17) | 139 (89.10) | 17 (10.90) | ||
| RUNX1, number (%) | 0.914 | 1 | |||
| Mutated | 17 (9.83) | 15 (88.24) | 2 (11.76) | ||
| Wild-type | 156 (90.17) | 139 (89.10) | 17 (10.90) | ||
| TET2, number (%) | 0.577 | 1 | |||
| Mutated | 15 (8.67) | 14 (93.33) | 1 (6.67) | ||
| Wild type | 158 (91.33) | 140 (88.61) | 18 (11.39) | ||
| NRAS, number (%) | 0.075 | 0.13 | |||
| Mutated | 12 (6.94) | 9 (75) | 3 (25) | ||
| Wild-type | 161 (93.06) | 145 (90.06) | 16 (9.94) | ||
| CEBPA, number (%) | 0.599 | 0.638 | |||
| Mutated | 13 (7.51) | 11 (84.62) | 2 (15.38) | ||
| Wild-type | 160 (92.49) | 143 (89.38) | 17 (10.63) | ||
| WT1, number (%) | 0.254 | 0.604 | |||
| Mutated | 10 (5.78) | 10 (100) | 0 (0) | ||
| Wild-type | 163 (94.22) | 144 (88.34) | 19 (11.66) | ||
| DNMT3A, number (%) | 0.603 | 0.784 | |||
| Mutated | 45 (26.01) | 41 (91.11) | 4 (8.89) | ||
| Wild-type | 128 (73.99) | 113 (88.28) | 15 (11.72) | ||
| NPM1, number (%) | 0.349 | 0.416 | |||
| Mutated | 48 (27.75) | 41 (85.42) | 7 (14.58) | ||
| Wild-type | 125 (72.25) | 113 (90.4) | 12 (9.6) | ||
| TP53, number (%) | 0.633 | 1 | |||
| Mutated | 14 (8.09) | 13 (92.86) | 1 (7.14) | ||
| Wild-type | 159 (91.91) | 141 (88.68) | 18 (11.32) | ||
CD37 expression in AML cell lines
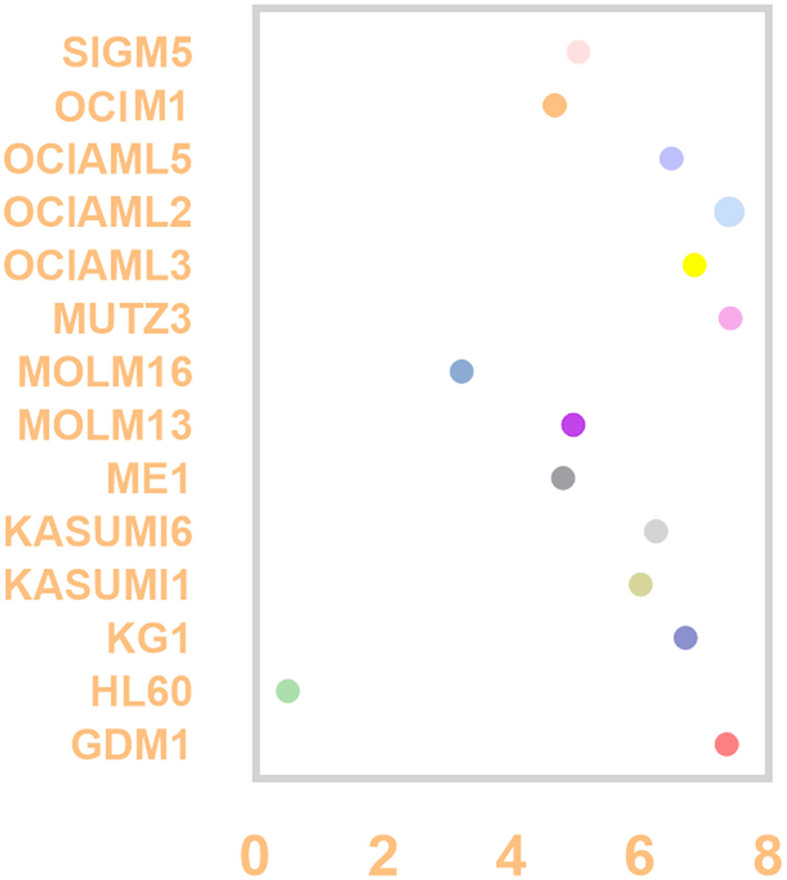
Using the CCLE dataset, we compared the CD37 mRNA level identified by RNA-seq in AML cell lines which include GDM-1, HL-60, KG-1, Kasumi-1, Kasumi-6, ME-1, MOLM-13, MOLM-16, MUTZ-3, OCI-AML3, OCI-AML2, OCI-AML5, OCI-M1 and SIG-M5. The top three cell AML lines with the highest expression are MUTZ-3, OCI-AML2, GDM-1, and the lowest is HL-60 (Figure 2).
Figure 2. CD37 expression level in AML cell lines by CCLE.
Prognostic values of CD37 in AML
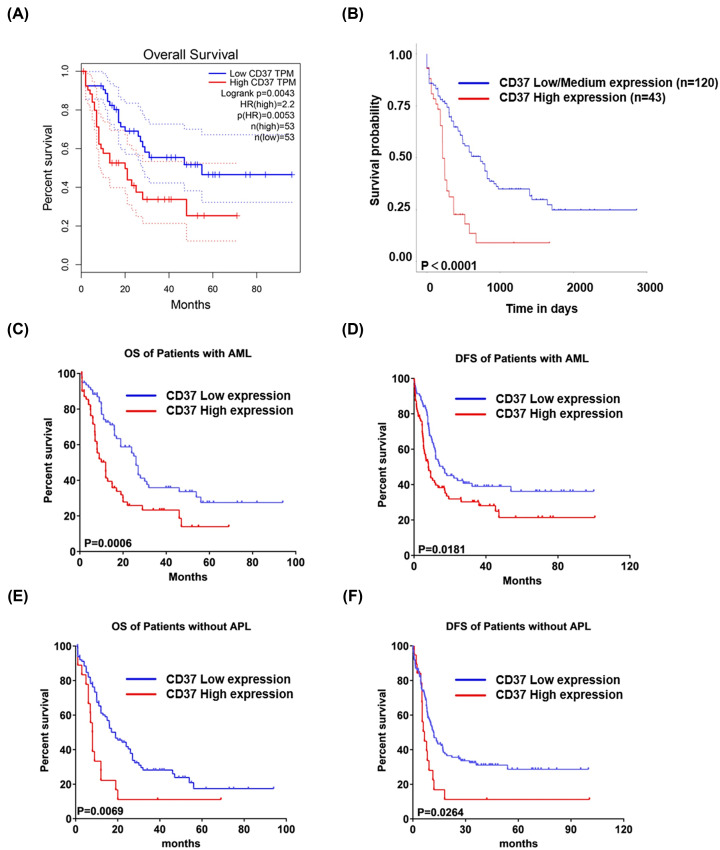
The prognostic role of CD37 in AML was further explored by means of GEPIA and UALCAN databases, and the significant association of CD37 high expression with poor overall survival (OS) in AML was observed in both databases (Figure 3A,B). To further validate the association between CD37 expression level and clinical outcome, we also dichotomized patients into high and low CD37 expression groups by CD37 median expression. A similar trend was observed that patients with high CD37 expression had significantly shorter OS as well as disease-free survival (DFS) than CD37 low expression patients (OS-median: 11.99 vs. 26.02 months, P=0.0006; DFS-median: 8 vs.16.1 months, P=0.0181; Figure 3C,D). As is known to all, acute promyelocytic leukemia (APL) is a particular subtype in AML, characterized by a balanced reciprocal translocation between chromosomes 15 and 17, which leads to the fusion between promyelocytic leukemia (PML) gene and retinoic acid receptor α (RARα) [19]. The good news is the complete remission (CR) rate was increased to 90–95% and 5-year DFS to 74% because of all-trans-retinoic acid (ATRA) or the ATRA-based regimens [20]. For these reasons, we excluded the patients of M3 from the survival analysis. Yet, the OS and DFS of CD37 high group (Z ≥ 1) is still shorter than the low group (OS-median: 7.965 vs. 18.96 months, P=0.0069; DFS-median: 6.6 vs. 11.9 months, P=0.0264; Figure 3E,F). Univariate analysis showed that high CD37 mRNA levels were associated with increasing age, poor molecular risk, transplant status, white blood cell count. This association was confirmed in multivariate analyses except for transplant status (Table 3).
Figure 3. The survival analysis of CD37 in patients with AML.
(A,B) The survival curves of OS in AML patients with regard to CD37 expression by GEPIA and UALCAN, respectively. (C,D) The OS and DFS of patients with AML categorized by CD37 median expression. (E,F) According to CD37 Z-score, the OS and DFS of patients without APL.
Table 3. Multivariate Cox regression model between OS in AML and other clinical parameters.
| Characteristic | CD37 high expression (n=19) | CD37 low expression (n=154) | Univariate analyses (Chi-square tests) | Multivariate analyses (Multivariate Cox model) | |
|---|---|---|---|---|---|
| P-value | P-value | HR (95% CI) | |||
| Age (y; median; range) | 63 (31–88) | 57 (18-82) | <0.001 | <0.001 | 1.041 (1.023–1.058) |
| Male (%) | 42.1 | 53.2 | 0.858 | 0.793 | 0.948 (0.637–1.412) |
| WBC, ×109/l (median; range) | 34.8 (1.6–99.2) | 14.9 (0.4–297.4) | 0.049 | 0.026 | 1.005 (1.001–1.009) |
| Platelets, ×109/l (median; range) | 57 (11–257) | 47 (8–351) | 0.549 | 0.917 | 1.000 (0.996–1.004) |
| Bone marrow blast percentage (median; range) | 63 (30–98) | 73 (30–100) | 0.659 | 0.528 | 1.003 (0.993–1.013) |
| Transplant (%) | 21.1 | 44.8 | <0.001 | 0.089 | 0.671 (0.423–1.063) |
| Poor molecular risk (%) | 36.8 | 24.0 | 0.002 | <0.001 | 1.620 (1.266–2.072) |
Methylation analysis of CD37 gene in AML
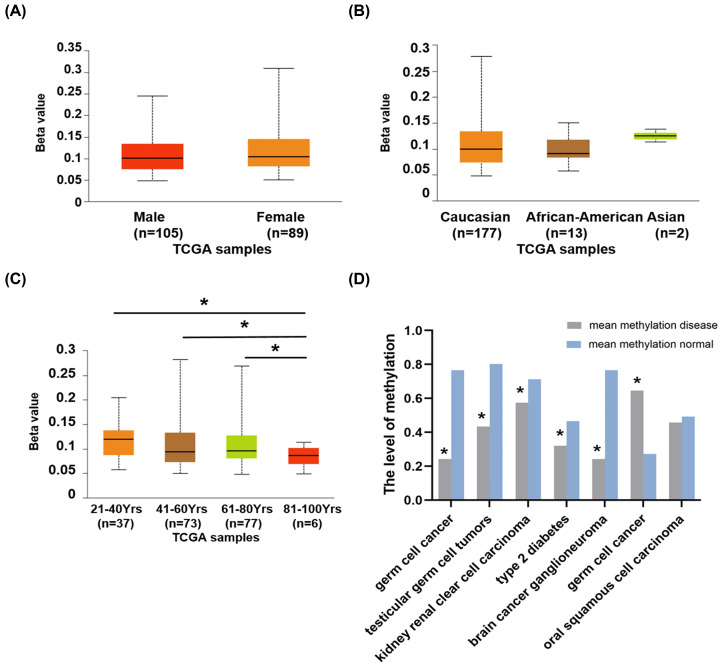
In order to find the main causes of aberrant expression of CD37 in AML, we used DiseaseMeth database as well as UALCAN. We found that CD37 gene is hypermethlated in germ cell cancer, but hypomethlated in testicular germ cell tumors, kidney renal clear cell carcinoma, type 2 diabetes and brain cancer ganglioneuroma (Table 4). In AML, patients aged 81–100 have significantly lower levels of CD37 promoter methylation compared with patients aged 21–40, 41–60 and 61–80 (P=7.80E-04, P=3.02E-03, P=1.18E-03). No significant differences in gender and race (Figure 4).
Table 4. Methylation of CD37 in various cancers.
| Disease | Mean methyldisease | Mean methylnormal | P-value | Methyltype |
|---|---|---|---|---|
| Germ cell cancer | 0.645 | 0.273 | 6.958e-04 | Hypermethylation |
| Kidney renal clear cellcarcinoma | 0.575 | 0.713 | 0.000e+00 | Hypomethylation |
| Trisomy 18 | 0.479 | 0.417 | 1.396e-04 | - |
| Oral squamous cell carcinoma | 0.458 | 0.492 | 1.560e-01 | - |
| Testicular germ tumors | 0.434 | 0.802 | 0.000e+00 | Hypomethylation |
| Type 2 diabetes | 0.321 | 0.465 | 2.218e-10 | Hypomethylation |
| Brain cancer ganglioneuroma | 0.242 | 0.766 | 4.115e-05 | Hypomethylation |
Figure 4. The methylation level of CD37 in AML.
CD37 promoter methylation profile based on (A) gender, (B) race, (C) age. (D) The level of CD37 methylation in different cancers.
CD37 co-expression genes in AML
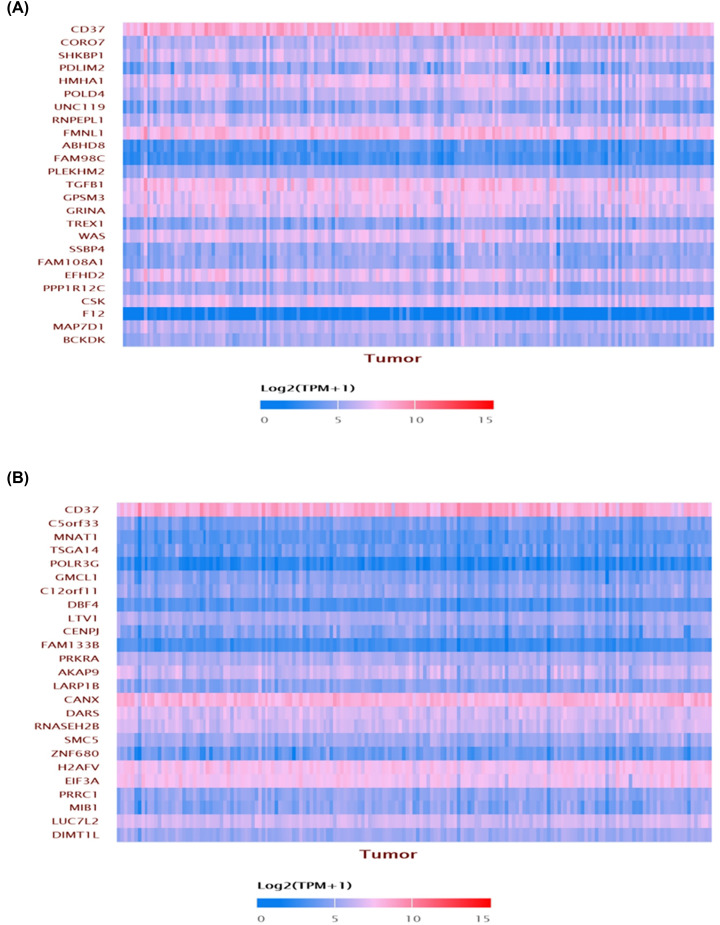
The genes co-expressed with CD37 were obtained in AML from the UALCAN database. CD37 is positively co-expressed with CORO7, SHKBPI, PDLIM2, HMHA1, POLD4, UNC119, RNPEPL1, FMNL1, ABHD8, FAM98C etc. (Figure 5A), and negatively with C5orf33, MNAT1, TSGA14, POLR3G, GMCL1, C12orf11, DBF4, LTV1, CENPJ, FAM133B etc. (Figure 5B).
Figure 5. The CD37 co-expression genes by UALCAN.
(A) Genes positively correlated with CD37 expression in AML. (B) Genes negatively correlated with CD37 in AML.
CD37 function prediction and pathway analysis in AML
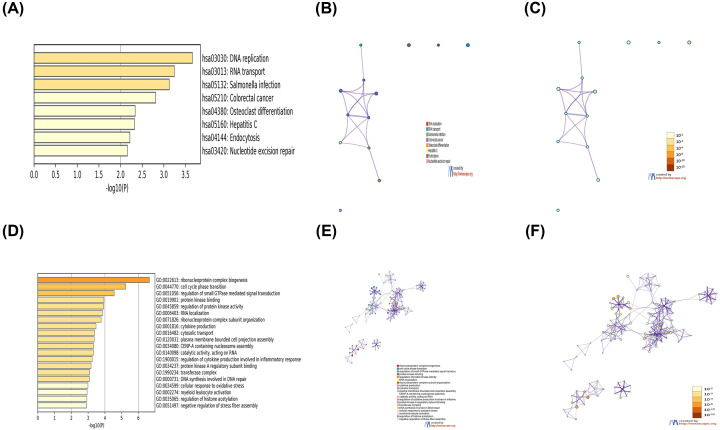
To further understand the role of CD37 in AML, we made a list of genes which included 200 genes that were the most correlated with CD37 according to Pearson correlation coefficient in UALCAN. Subsequently, we analyzed the list using the Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) tools in Metascape (Figure 6). The top 20 GO enrichment items were classified into biological process group, molecular function group and cellular component group. The biological process that these genes were involved in were ribonucleoprotein complex biogenesis, cell cycle phase transition, regulation of small GTPase-mediated signal transduction and so on. The molecular function for these genes were enriched in protein kinase binding, catalytic activity, protein kinase A regulatory subunit binding. The cellular component was mainly associated with terms related to transferase complex. The top eight KEGG pathways for these genes are DNA replication, RNA transport, Salmonella infection, colorectal cancer, osteoclast differentiation, hepatitis C, endocytosis and nucleotide excision repair.
Figure 6. The functions of CD37 and genes significantly associated with CD37 gene alterations.
(A) Heatmap of KEGG enriched terms colored by P-values. (B) Network of KEGG enriched terms colored by cluster ID. (C) Network of KEGG enriched terms colored by P-value. (D) Heatmap of GO enriched terms colored by P-values. (E) Network of KEGG enriched terms colored by cluster ID. (F) Network of GO enriched terms colored by P-value, where terms containing more genes tend to have a more significant P-value.
Protein–protein interaction network analysis
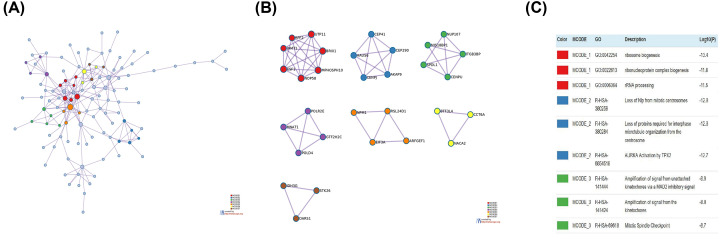
The protein–protein interaction (PPI) topology analysis was performed in order to systemically understand the functions of CD37 gene. The PPI network and Molecular Complex Detection (MCODE) components identified in the gene lists are shown in Figure 7. The seven most significant MCODE components were extracted from the PPI network. After pathway and process enrichment analysis was independently applied to each MCODE component, the results showed that biological function was mainly related to ribosome biogenesis, loss of NIp from mitotic centrosomes, loss of proteins required for interphase microtubule organization from the centrosome, AURKA activation by TPX2, ribonucleoprotein complex biogenesis, rRNA processing.
Figure 7. PPI network and MCODE components identified related to genes significantly associated with CD37 gene alterations.
(A) PPI network of proteins. (B) Modules selected from PPI network using MCODE. (C) The description of each MCODE component.
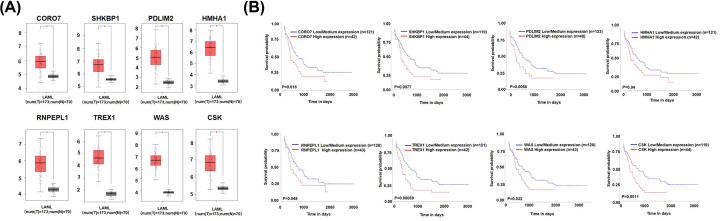
Survival analysis and expression profile of key genes
Based on these correlation genes, eight genes were significantly associated with disease prognosis including CORO7, SHKBP1, PDLM2, HMHA1, RNPEPL1, TREX1, WAS, CSK (Figure 8). Furthermore, they are all highly expressed in AML patients, and high expression is associated with poor prognosis.
Figure 8. The expression level and survival analysis of key genes.
(A) The expression profile of key genes. (B) Survival analysis of key genes. *P<0.05 versus control.
Discussion
CD37 belongs to the members of tetraspanin superfamily, which have reached nearly 20 genes since its discovery in 1990 [4]. CD37 is absent or weakly expressed on normal T cells, monocytes and neutrophils, and is not detected on natural killer (NK) cells, platelets and erythrocytes. Thus, CD37 is considered to be a lineage-specific marker of human B cells and stands for a worthy therapeutic target in NHL. Additionally, the preclinical and early clinical studies suggested that anti-CD37 agents were useful in the treatment of CLL [21,22]. In this study, we found that CD37 is highly expressed in AML and CD37 high expression is associated with poor outcome in the patients.
We observed the expression profiling of CD37 in different types of cancers, and CD37 expression is higher in breast invasive carcinoma, cervical squamous cell carcinoma and endocervical adenocarcinoma, cholangiocarcinoma and DLBCL, but lower in adrenocortical carcinoma, bladder urothelial carcinoma, colon adenocarcinoma compared with normal adjacent tissues (Figure 1A). CD37 is significantly higher in AML. Analysis of the TCGA data showed that high CD37 expression is associated with shorter OS and DFS in AML, even if M3 is removed. These results indicated that high expression of CD37 might be a predictor of poor prognosis.
In line with these observations, Pereira et al. reported that CD37 was overexpressed in AML, and anti-CD37 IgG2 antibody exhibited potent antitumor efficacy in preclinical models of AML as well as patient-derived models [23]. However, another finding published by de Winde et al. implicated that loss of CD37 drives tumor progression via constitutive activation of the IL-6 signaling pathway [8]. In previous study, Debio 1562, a potent antibody–drug conjugate directed at CD37, was demonstrated to suppress the proliferative capacities of AML cell lines, including OCI-AML3, THP-1 and primary AML samples. Moreover, the mice engrafted with the THP-1 cell line showed a survival advantage and elimination of the disease on the basis of histopathology. Although the mechanism is unclear so far, our findings may help to provide some clues. According to our KEGG and GO enrichment results, we speculated that high CD37 blocks cell cycle progression by affecting DNA replication, thereby inhibiting AML cell proliferation.
In recent decades, mAbs have emerged as a potent and effective therapy for human malignancies [24]. For instance, the chimeric anti-CD20 antibody, IDEC-C2B8, also known as rituximab, has favorable clinical responses and became the first mAb approved by the FDA for use in human cancers [25,26]. In AML, CD33, CD123, CD44 and so on are potential targets [27–29]. Pagel et al. have reported a phase I study of an anti-CD37 smip protein in relapsed and/or refractory NHL patients [30]. Stilgenbauer et al. has reported human experience with anti-CD37 antibody in CLL, and indicated that it was a valid treatment with acceptable tolerability and notable efficacy, especially in difficult-to-treat patients with poor-risk features including del(17p) or TP53 mutations [31]. More importantly, Pereira et al. found that CD37 was also well expressed in AML, and an antibody–drug conjugate that targeted CD37-induced cytotoxicity, apoptosis and cell cycle arrest in AML cell lines [23]. This property makes the CD37 molecule an attractive target for immunotherapy. Besides, further analyses of its clinical effects and its combination with other therapeutics would bring about new insights into the mechanism of its action and strategies to improve its effects.
Additionally, our study has its limitations. First, all patients came from one single institution, which means further validations from multicenter and large sample size cohorts are needed. Second, the lack of original data to confirm our findings. Third, the mechanism about CD37 high expression in AML was associated with poor prognosis is unclear, but we believe it deserves further study in the future.
In summary, improving outcomes for AML patients, in particular those with refractory/relapsed AML, remains an ongoing challenge. As evidenced by the clinical success of rituximab, antibody-based therapies are of great importance in the management of a number of hematological malignancies. Our data suggested the oncogenic effect of CD37 high expression in AML and targeting CD37 might be a potential approach for AML therapy, which is valuable to be further explored in future.
Conclusion
Our current study revealed the important role of CD37 gene in clinical outcomes in AML patients, and provided the rationale to identify CD37 as a valuable biomarker and develop strategies to target CD37 for AML therapy.
Abbreviations
- AML
acute myeloid leukemia
- ATRA
all-trans retinoic acid
- AURKA
aurora kinase A
- CCLE
The Cancer Cell Line Encyclopedia
- CLL
chronic lymphocytic leukemia
- DFS
disease-free survival
- DLBCL
diffuse large B-cell lymphoma
- FDA
Food and Drug Administration
- GEPIA
Gene Expression Profiling Interactive Analysis
- GO
Gene Ontology
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- MCODE
Molecular Complex Detection
- NHL
non-Hodgkin's lymphoma
- OS
overall survival
- PPI
protein–protein interaction
- RARα
retinoic acid receptor α
- TCGA
The Cancer Genome Atlas
- TPM
transcript per million
Data Availability
All data generated or analyzed during the present study are included in this published article.
Competing Interests
The authors declare that there are no competing interest associated with the manuscript.
Funding
This work is supported in part by The National Natural Science Foundation of China [grant number 81770172]; Jiangsu Provincial Special Program of Medical Science [grant number BE2017747]; Jiangsu Province “333” project [grant number BRA2019103]; The Fundamental Research Funds for the Central Universities [grant number 2242019K3DZ02]; Milstein Medical Asian American Partnership (MMAAP) Foundation Research Project Award in Hematology (2017); Key Medical of Jiangsu Province [grant number ZDXKB2016020].
Author Contribution
Q.Z., Q.H. and J.Z. performed experiments and analyzed data. Z.G. and C.S. designed and supervised data analysis. Q.Z., C.S. and Z.G. wrote the manuscript.
References
- 1.Marwick C. (1997) Monoclonal antibody to treat lymphoma. JAMA 278, 616–618 10.1001/jama.1997.03550080022011 [DOI] [PubMed] [Google Scholar]
- 2.Press O.W., Eary J.F., Badger C.C., Martin P.J., Appelbaum F.R. and Levy R. (1989) Treatment of refractory non-Hodgkin's lymphoma with radiolabeled MB-1 (anti-CD37) antibody. J. Clin. Oncol. 7, 1027–1038 10.1200/JCO.1989.7.8.1027 [DOI] [PubMed] [Google Scholar]
- 3.Kaminski M.S., Fig L.M., Zasadny K.R., Koral K.F., DelRosario R.B. and Francis I.R. (1992) Imaging, dosimetry, and radioimmunotherapy with iodine 131-labeled anti-CD37 antibody in B-cell lymphoma. J. Clin. Oncol. 10, 1696–1711 10.1200/JCO.1992.10.11.1696 [DOI] [PubMed] [Google Scholar]
- 4.Maecker H.T., Todd S.C. and Levy S. (1997) The tetraspanin superfamily: molecular facilitators. FASEB J. 11, 428–442 10.1096/fasebj.11.6.9194523 [DOI] [PubMed] [Google Scholar]
- 5.Classon B.J., Williams A.F., Willis A.C., Seed B. and Stamenkovic I. (1989) The primary structure of the human leukocyte antigen CD37, a species homologue of the rat MRC OX-44 antigen. J. Exp. Med. 169, 1497–1502 10.1084/jem.169.4.1497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schwartz-Albiez R., Dorken B., Hofmann W. and Moldenhauer G. (1988) The B cell-associated CD37 antigen (gp40-52). Structure and subcellular expression of an extensively glycosylated glycoprotein. J. Immunol. 140, 905–914 [PubMed] [Google Scholar]
- 7.van Spriel A.B., Puls K.L., Sofi M., Pouniotis D., Hochrein H., Orinska Z. et al. (2004) A regulatory role for CD37 in T cell proliferation. J. Immunol. 172, 2953–2961 10.4049/jimmunol.172.5.2953 [DOI] [PubMed] [Google Scholar]
- 8.de Winde C.M., Veenbergen S., Young K.H., Xu-Monette Z.Y. and van Spriel A.B. (2016) Tetraspanin CD37 protects against the development of B cell lymphoma. J. Clin. Invest. 126, 653–666 10.1172/JCI81041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Robak T. and Robak P. (2014) Anti-CD37 antibodies for chronic lymphocytic leukemia. Expert Opin. Biol. Ther. 14, 651–661 10.1517/14712598.2014.890182 [DOI] [PubMed] [Google Scholar]
- 10.Dohner H., Weisdorf D.J. and Bloomfield C.D. (2015) Acute myeloid leukemia. N. Engl. J. Med. 373, 1136–1152 10.1056/NEJMra1406184 [DOI] [PubMed] [Google Scholar]
- 11.Lozano R., Naghavi M., Foreman K. and Lim S. (2012) Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet 380, 2095–2128 10.1016/S0140-6736(12)61728-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.O’Donnell M.R., Abboud C.N., Altman J. and Gregory K.M. (2012) NCCN Clinical Practice Guidelines acute myeloid leukemia. J. Natl. Compr. Canc. Netw. 10, 984–1021 10.6004/jnccn.2012.0103 [DOI] [PubMed] [Google Scholar]
- 13.Cerami E., Gao J., Dogrusoz U., Gross B.E., Sumer S.O. and Aksoy B.A. (2012) The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2, 401–404 10.1158/2159-8290.CD-12-0095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tang Z., Li C., Kang B., Gao G., Li C. and Zhang Z. (2017) GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 45, W98–W102 10.1093/nar/gkx247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chandrashekar D.S., Bashel B., Balasubramanya S., Creighton C.J., Ponce-Rodriguez I., Chakravarthi B. et al. (2017) UALCAN: a portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia 19, 649–658 10.1016/j.neo.2017.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barretina J., Caponigro G., Stransky N. and Venkatesan K. (2012) The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 483, 603–607 10.1038/nature11003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou Y., Zhou B., Pache L., Chang M., Khodabakhshi A.H., Tanaseichuk O. et al. (2019) Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 10, 1523 10.1038/s41467-019-09234-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xiong Y., Wei Y., Gu Y., Zhang S., Lyu J., Zhang B. et al. (2017) DiseaseMeth version 2.0: a major expansion and update of the human disease methylation database. Nucleic Acids Res. 45, D888–D895 10.1093/nar/gkw1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Piazza F., Gurrieri C. and Pandolfi P.P. (2001) The theory of APL. Oncogene 20, 7216–7222 10.1038/sj.onc.1204855 [DOI] [PubMed] [Google Scholar]
- 20.Wang Z.Y. and Chen Z. (2008) Acute promyelocytic leukemia: from highly fatal to highly curable. Blood 111, 2505–2515 10.1182/blood-2007-07-102798 [DOI] [PubMed] [Google Scholar]
- 21.Barrena S., Almeida J., Yunta M., Lopez A., Fernandez-Mosteirin N., Giralt M. et al. (2005) Aberrant expression of tetraspanin molecules in B-cell chronic lymphoproliferative disorders and its correlation with normal B-cell maturation. Leukemia 19, 1376–1383 10.1038/sj.leu.2403822 [DOI] [PubMed] [Google Scholar]
- 22.Byrd J.C., Pagel J.M., Awan F.T., Forero A., Flinn I.W., Deauna-Limayo D.P. et al. (2014) A phase 1 study evaluating the safety and tolerability of otlertuzumab, an anti-CD37 mono-specific ADAPTIR therapeutic protein in chronic lymphocytic leukemia. Blood 123, 1302–1308 10.1182/blood-2013-07-512137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pereira D.S., Guevara C.I., Jin L., Mbong N., Verlinsky A., Hsu S.J. et al. (2015) AGS67E, an anti-CD37 monomethyl auristatin E antibody-drug conjugate as a potential therapeutic for B/T-cell malignancies and AML: a new role for CD37 in AML. Mol. Cancer Ther. 14, 1650–1660 10.1158/1535-7163.MCT-15-0067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Adams G.P. and Weiner L.M. (2005) Monoclonal antibody therapy of cancer. Nat. Biotechnol. 23, 1147–1157 10.1038/nbt1137 [DOI] [PubMed] [Google Scholar]
- 25.Lozano R., Naghavi M., Foreman K. and Lim S. (2012) Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet 380, 2095–2128 10.1016/S0140-6736(12)61728-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maloney D.G., GrilloLopez A.J., White C.A., Bodkin D., Schilder R.J., Neidhart J.A. et al. (1997) IDEC-C2B8 (Rituximab) anti-CD20 monoclonal antibody therapy in patients with relapsed low-grade non-Hodgkin’s lymphoma. Blood 90, 2188–2195 10.1182/blood.V90.6.2188 [DOI] [PubMed] [Google Scholar]
- 27.Sievers E.L., Larson R.A., Stadtmauer E.A., Estey E., Lowenberg B., Dombret H. et al. (2001) Efficacy and safety of gemtuzumab ozogamicin in patients with CD33-positive acute myeloid leukemia in first relapse. J. Clin. Oncol. 19, 3244–3254 10.1200/JCO.2001.19.13.3244 [DOI] [PubMed] [Google Scholar]
- 28.Jin L., Lee E.M., Ramshaw H.S., Busfield S.J., Peoppl A.G., Wilkinson L. et al. (2009) Monoclonal antibody-mediated targeting of CD123, IL-3 receptor alpha chain, eliminates human acute myeloid leukemic stem cells. Cell Stem Cell 5, 31–42 10.1016/j.stem.2009.04.018 [DOI] [PubMed] [Google Scholar]
- 29.Jin L., Hope K.J., Zhai Q., Smadja-Joffe F. and Dick J.E. (2006) Targeting of CD44 eradicates human acute myeloid leukemic stem cells. Nat. Med. 12, 1167–1174 10.1038/nm1483 [DOI] [PubMed] [Google Scholar]
- 30.Pagel J.M., Spurgeon S., Awan F., Blum K.A., Lanasa M.C., Flinn I.W. et al. (2011) Phase 1 study of TRU-016, an anti-CD37 SMIP (TM) protein in relapsed and/or refractory NHL patients. Blood 118, 711 10.1182/blood.V118.21.1636.1636 [DOI] [Google Scholar]
- 31.Stilgenbauer S., Schleinitz T.A., Eichhorst B., Lang F., Offner F., Rossi J. et al. (2019) Phase 1 first-in-human trial of the anti-CD37 antibody BI 836826 in relapsed/refractory chronic lymphocytic leukemia. Leukemia 33, 2531–2535 10.1038/s41375-019-0475-z [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during the present study are included in this published article.