Abstract
Recent studies have used genome-wide single-nucleotide polymorphisms (SNPs) to investigate relationships among various Jewish populations and their non-Jewish historical neighbors, often focusing on small subsets of populations from a limited geographic range or relatively small samples within populations. Here, building on the significant progress that has emerged from genomic SNP studies in the placement of Jewish populations in relation to non-Jewish populations, we focus on population structure among Jewish populations. In particular, we examine Jewish population-genetic structure in samples that span much of the historical range of Jewish populations in Europe, the Middle East, North Africa, and South Asia. Combining 429 newly genotyped samples from 29 Jewish and 3 non-Jewish populations with previously reported genotypes on Jewish and non-Jewish populations, we investigate variation in 2789 individuals from 114 populations at 486,592 genome-wide autosomal SNPs. Using multidimensional scaling analysis, unsupervised model-based clustering, and population trees, we find that, genetically, most Jewish samples fall into four major clusters that largely represent four culturally defined groupings, namely the Ashkenazi, Mizrahi, North African, and Sephardi subdivisions of the Jewish population. We detect high-resolution population structure, including separation of the Ashkenazi and Sephardi groups and distinctions among populations within the Mizrahi and North African groups. Our results refine knowledge of Jewish population-genetic structure and contribute to a growing understanding of the distinctive genetic ancestry evident in closely related but historically separate Jewish communities.
Subject terms: Computational biology and bioinformatics, Genetic variation
Introduction
Relationships among Jewish populations and between Jewish groups and their non-Jewish historical neighbors have long been a topic of great interest in human population genetics. Studies of Jewish populations have examined many forms of variation, from blood groups and immunological markers to mitochondrial and Y-chromosomal DNA variants and autosomal restriction fragment length polymorphisms, microsatellites, and single-nucleotide polymorphisms (SNPs) [1–8].
Until recently, relationships among Jewish population groups were difficult to assess robustly, due to the close relationships among the populations and the relatively small number of markers that had historically been available. During the genomic era, however, progress in Jewish population genetics has been significant. In particular, following a period of emphasis in Jewish population genetics on uniparental Y-chromosomal and mitochondrial loci that follow male and female lineages, respectively—often with differing patterns observed (reviewed in [7])—recent analyses of genome-wide autosomal SNPs have introduced a new era in which it has been possible to clarify population relationships at a fine scale from genome-wide averages. Indeed, some of the first genomic studies spanning large numbers of Jewish populations represented considerable advances in placing Jewish in relation to non-Jewish populations [9, 10]. In population structure inference from autosomal genomes, studies of large numbers of genome-wide polymorphisms in Jewish and closely related non-Jewish populations [9–22] have produced agreement on two main patterns. (1) Genetic clustering of many Jewish populations places them as intermediate between non-Jewish European and Middle Eastern groups. (2) With notable exceptions, most Jewish populations cluster genetically with other Jewish populations, indicating genetic similarity that often exceeds that with non-Jewish historical neighbors.
In this study, we examine Jewish population structure using genome-wide SNPs. Our work builds primarily upon five studies [9, 10, 15, 19, 20], each including a variety of Jewish populations, but each having less geographic coverage or smaller sample sizes per population than the current investigation, or both, and with generally more emphasis on relationships of Jewish and non-Jewish groups. Kopelman et al. [15] studied four Jewish populations, finding that they could be distinguished from neighboring European and Middle Eastern non-Jewish populations, and that the Tunisian Jewish group was the most distinctive of the four. Atzmon et al. [9] identified two clusters among seven Jewish populations, one containing Mizrahi Jewish populations and the other containing European and Syrian Jewish groups. Behar et al. [10] sampled a larger number of populations, but few individuals from most of them, and also uncovered two clusters, again one for Mizrahi populations and the other including Ashkenazi, North African, and Sephardi groups. The study by Campbell et al. [19], which also discerned a Mizrahi cluster, emphasized the analysis of the North African Jewish populations, identifying a North African cluster that was separate from Ashkenazi and some Sephardi populations. That study refined earlier results on distinctiveness of the Tunisian and Libyan Jewish populations [15, 23]. In the largest study to date, Behar et al. [20] analyzed a broad sample from across western Eurasia and North Africa, but focused primarily on one aspect, the Ashkenazi Jews in relation to non-Jewish populations.
With more individuals and populations, our aim is a more comprehensive analysis of Jewish population-genetic substructure. We emphasize relationships among Jewish populations, considering relationships of Jewish populations to non-Jewish groups in order to place the Jewish populations into context. We analyze new data from 429 individuals, representing 29 Jewish and 3 non-Jewish populations, at 486,592 SNPs. Unlike most recent studies on the genetics of Jewish populations, we identify countries of origin in labeling the Ashkenazi samples. We analyze our new data together with previously reported Jewish population-genetic data [10] and with samples from diverse worldwide populations [24, 25], examining a total of 2789 individuals from 114 populations.
Results
We assembled the data (Table S1, Fig. S1, see “Materials and methods” section), classified populations by regional group, and analyzed 12 population subsets (Tables 1 and S2). Because studies of population structure can reveal finer-scale relationships as the analysis narrows to restrict attention to more closely related groups [24, 26, 27], we adopted a nested data analysis strategy. We proceeded from a broad geographic context for Jewish populations to local comparisons of specific Jewish groups and of specific groups of Jewish and non-Jewish populations.
Table 1.
Twelve population sets used in population structure analysis. The study analyzes 12 sets of populations, numbered 1–12. An “X” entry in the table indicates that the group of populations listed in the associated row is included in the set for the associated column.
| Population group | Number of populations | Population set | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | ||
| Africa | 12 | X | |||||||||||
| East Asia | 22 | X | |||||||||||
| Central/South Asia | 13 | X | X | ||||||||||
| Middle East (Armenian, Georgian, Iranian) | 3 | X | X | X | X | ||||||||
| Middle East (Egyptian, Moroccan, Mozabite) | 3 | X | X | X | X | ||||||||
| Middle East (Cypriot, Turkish) | 2 | X | X | X | X | ||||||||
| Middle East (other populations) | 10 | X | X | X | |||||||||
| Europe (Adygei) | 1 | X | X | X | X | X | |||||||
| Europe (Italian, Romanian, Spanish, Tuscan) | 4 | X | X | X | X | X | |||||||
| Europe (other populations) | 13 | X | X | X | X | ||||||||
| Ashkenazi Jewish | 13 | X | X | X | X | X | X | ||||||
| Mizrahi Jewish | 6 | X | X | X | X | X | X | ||||||
| North African Jewish | 5 | X | X | X | X | X | X | ||||||
| Sephardi Jewish | 3 | X | X | X | X | X | X | ||||||
| Yemenite Jewish | 1 | X | X | X | |||||||||
| Indian Jewish | 2 | X | X | ||||||||||
| Ethiopian Jewish | 1 | X | |||||||||||
| Total number of populations | 114 | 114 | 79 | 64 | 27 | 31 | 10 | 8 | 9 | 13 | 6 | 5 | 3 |
We first examined relationships between Jewish and non-Jewish samples from Africa, Asia, and Europe (set 1); Europe, the Middle East, and Central and South Asia only (set 2); Europe and the Middle East only (set 3). Following these initial analyses that aimed at placing the Jewish populations into a broader geographic context and identifying genetically proximate non-Jewish populations for refined analysis, we then focused on Jewish populations only (set 4).
Most Jewish populations largely fall into four major cultural groups: Ashkenazi, Mizrahi, North African, and Sephardi. These groupings represent generally distinct geographic ancestries: Central and Eastern Europe for the Ashkenazi group; the Middle East, Caucasus, and Central Asia for the Mizrahi group; North Africa for the North African group; and Mediterranean regions inhabited by descendants of Jewish populations expelled from Iberia in the late 1400 s for the Sephardi group. Population classifications according to these groupings appear in Table S1. Together with non-Jewish samples from regions historically inhabited by specific Jewish groups, we examined Ashkenazi (set 5), Mizrahi (set 6), North African (set 7), and Sephardi Jewish samples (set 8). Omitting the non-Jewish samples, we also separately analyzed Ashkenazi (set 9), Mizrahi (set 10), North African (set 11), and Sephardi samples (set 12). Four Jewish populations included in the study—Ethiopian Jews, Indian Jews from Cochin, Indian Jews from Mumbai, and Yemenite Jews—are considered to be culturally distinct and not part of the Ashkenazi, Mizrahi, North African, or Sephardi groups; they are therefore not analyzed in sets (5)–(12).
Jewish populations in relation to non-Jewish populations of Africa, Asia, and Europe
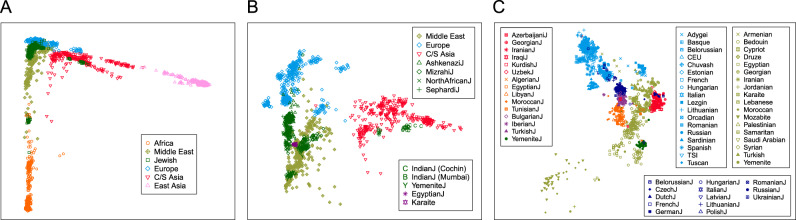
We performed multidimensional scaling (MDS) using the allele-sharing distance [28] between pairs of individuals. Figure 1a shows an MDS plot for 2789 individuals from 114 populations from Africa, Asia, and Europe (population set 1). The plot shows a separation of individuals by geographic region with Jewish samples largely forming a cluster overlapping the European and Middle Eastern samples. Some Jewish samples lie outside this cluster: the two Indian Jewish populations, from Cochin and Mumbai, and the Ethiopian Jewish population lie among non-Jewish populations of Central and South Asia and Africa, respectively.
Fig. 1. Multidimensional scaling analysis of population structure for Jewish populations and non-Jewish populations from specific regions.
a Africa, Asia, and Europe (2789 individuals, 114 populations). b Europe, Middle East, and Central and South Asia (1656 individuals, 79 populations; Ethiopian Jews are excluded). c Europe and Middle East (1288 individuals, 64 populations; Ethiopian and Indian Jews from Cochin and Mumbai are excluded). In c, groups are color-coded: Europe, light blue; Middle East, olive green; Ashkenazi Jewish, dark blue; Mizrahi Jewish, red; North African Jewish, orange; Sephardi Jewish, purple; Yemenite Jewish, green. Population symbols often overlap due to similar placement.
We then narrowed the sample set, excluding non-Jewish populations from Africa and East Asia as well as Ethiopian Jews. This analysis included 1,656 individuals from 79 populations, producing the MDS plot of the Jewish populations together with non-Jewish populations of Europe, the Middle East, and Central and South Asia shown in Fig. 1b (population set 2). The Jewish populations continue to fall among the European and Middle Eastern samples with the exception that the two Indian Jewish populations are found among samples from Central and South Asia. Figure 1b reveals a distinctive position for the Yemenite Jewish samples in relation to other Jewish populations. Mizrahi Jewish samples can be distinguished from Ashkenazi, North African, and Sephardi Jewish samples with most Mizrahi samples clustering among Middle Eastern non-Jewish samples, and the other Jewish samples generally lying between European and Middle Eastern non-Jewish populations.
We further narrowed the sample set, excluding Central and South Asia and the Indian Jewish populations from Cochin and Mumbai, leaving 1288 individuals from 64 populations, with a focus on Jewish populations in relation to European and Middle Eastern non-Jewish populations (population set 3). The resulting MDS plot (Fig. 1c) places the Yemenite Jews near Bedouin, Saudi Arabian, and Yemenite non-Jewish populations. It accentuates the differentiation between the Mizrahi Jews (red) and Ashkenazi (dark blue), North African (orange), and Sephardi Jews (purple). Interestingly, some Russian Jewish samples and one Ukrainian Jewish sample cluster among the Mizrahi Jews. Figure 1c also distinguishes the Ashkenazi and North African populations with the Sephardi populations lying intermediate between these two groups. The Ashkenazi populations appear closer to European populations than do other Jewish populations.
Jewish populations in relation to non-Jewish populations of Europe and the Middle East
Because the MDS analysis of Jewish and non-Jewish populations of Europe and the Middle East separated major Jewish population groups and produced informative placements of Jewish samples in relation to non-Jewish populations, we undertook additional analyses with this same set of populations (population set 3). We first examined genetic diversity, and then conducted additional analyses of population structure.
Heterozygosity
Table S3 shows the mean expected heterozygosity across loci for all Jewish populations (including Ethiopian Jews and Indian Jews from Cochin and Mumbai) and non-Jewish populations of Europe and the Middle East. The mean heterozygosities across 31 Jewish, 18 European, and 18 Middle Eastern populations are 0.3237, 0.3235, and 0.3252, respectively. For the 27 Jewish populations classified in larger regional groups (excluding Ethiopian Jews, Indian Jews from Cochin and Mumbai, and Yemenite Jews), the means are 0.3249 for 13 Ashkenazi populations, 0.3224 for 6 Mizrahi populations, 0.3221 for 5 North African populations, and 0.3259 for 3 Sephardi populations, respectively (Table S4).
Genetic diversity measures in human populations typically vary within a narrow range and largely follow a pattern in which within-population genetic diversity decreases with increasing geographic distance from Sub-Saharan Africa over land-based routes [29, 30]. In accord with this pattern, we find that although their values are similar, European non-Jewish populations, at a greater land-based distance from Sub-Saharan Africa than Middle Eastern populations, have lower heterozygosities (p = 0.0030, two-tailed Wilcoxon two-sample test). The heterozygosity of 0.3291 for the full pooled set of 504 Jewish individuals is intermediate, between 0.3259 for non-Jewish individuals from Europe and 0.3311 for those from the Middle East. Considering paired lists of loci, Jewish heterozygosities in the pooled sample have an intermediate position, greater than European heterozygosities (p < 2.2 × 10−16, two-tailed Wilcoxon signed-rank test) and smaller than those of the non-Jewish Middle Eastern populations (p < 2.2 × 10−16).
Using population-level values in Table S3, comparisons show intermediate heterozygosities for Jewish subgroups between those of European and Middle Eastern non-Jewish populations. For example, Ashkenazi Jewish populations have slightly higher values than non-Jewish European populations (p = 0.0681, two-tailed Wilcoxon two-sample test), and Mizrahi Jewish populations have lower heterozygosities than non-Jewish Middle Eastern populations (p = 0.0191).
Unsupervised model-based clustering
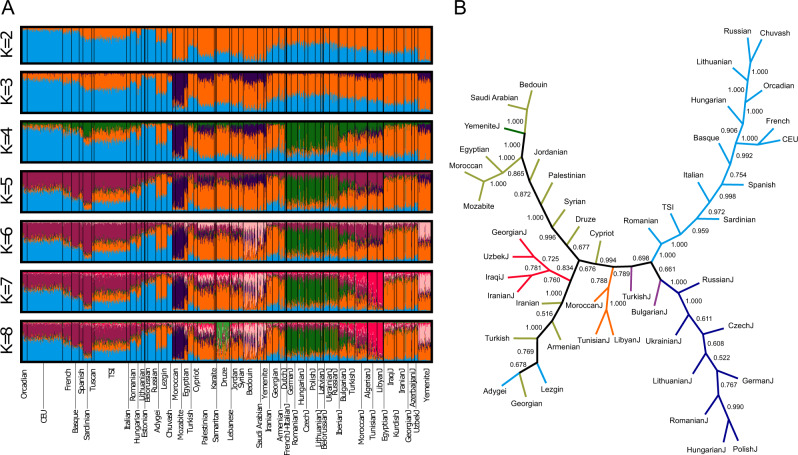
We next performed unsupervised model-based clustering of the set of European, Middle Eastern, and Jewish populations, employing Structure [31]. For each value of the number of clusters K from 2 to 8, we performed 20 replicate runs, identifying modes among these runs using Clumpak [32].
Figure 2a illustrates the major clustering solutions for each K from 2 to 8. For K = 2, one cluster has higher membership in European populations and the other has higher membership in Middle Eastern populations. Jewish populations have mixed membership in the two clusters, with the exception of the Yemenite Jews, who are placed primarily in the main cluster among Middle Eastern populations. For K = 3, the third cluster (dark blue) separates the Mozabite and Moroccan populations. Non-Jewish populations from the Levant generally have substantial membership in this cluster, as do North African and Yemenite Jews.
Fig. 2. Population structure of Jewish, European, and Middle Eastern populations.
a Unsupervised clustering using Structure, for the 1288 individuals in Fig. 1c. For each value of K, the number of predefined clusters, each individual is represented by a vertical line partitioned into K colored components according to inferred membership in K genetic clusters. For each K, the major mode identified by Clumpak is shown; among 20 replicates, this mode represents 20, 20, 18, 17, 11, 13, and 14 runs for K = 2, 3, 4, 5, 6, 7, and 8, respectively. b Neighbor-joining population tree. The plot includes the 47 populations in Fig. 1c for which sample sizes were ten or greater. Numbers indicate bootstrap support out of 1000 replicates. Color codes for labels and external branches follow Fig. 1c. An internal branch is colored black if it subtends a set of two or more populations that appears with at least two colors.
Ashkenazi Jews are largely assigned to the new cluster for K = 4 (green), which also contains sizeable membership from southern Europe, particularly the Sardinians, as well as from Sephardi and North African Jews. For K = 5, the new cluster largely subdivides the non-Jewish European populations (purple), so that the similarity of Sephardi and North African Jews to Ashkenazi Jews seen at K = 4 is partially absorbed in a cluster anchored by Sardinians.
For K = 6, Yemenite Jews have relatively high membership in the new cluster, which also has substantial membership from Middle Eastern populations such as Bedouins and Saudi Arabians (pink). For K = 7, Libyan and Tunisian Jews fall into a new cluster (red). For K = 8, the new cluster is centered on the Druze (light green).
Population tree
For the European, Middle Eastern, and Jewish populations, we carried out a neighbor-joining tree-based clustering. Restricting attention to 47 populations with sample size 10 or more, we obtained population-based distance matrices for the allele-sharing distance, using 1000 bootstrap resamples of the set of loci (Fig. 2b).
In the resulting population tree, one subtree with bootstrap support 1000 of 1000 replicates corresponds to European (non-Caucasus) populations; another with support 996 corresponds to most of the Middle Eastern populations plus the Yemenite Jews. A third subtree contains Mizrahi Jewish populations and non-Jewish populations from nearby regions, with support 760 for a grouping of the Mizrahi populations. Ashkenazi and North African Jewish populations each lie in separate subtrees, with bootstrap support values 1000 and 788, respectively. The general pattern is similar to that seen with Structure (Fig. 2a).
Relationships among Jewish populations
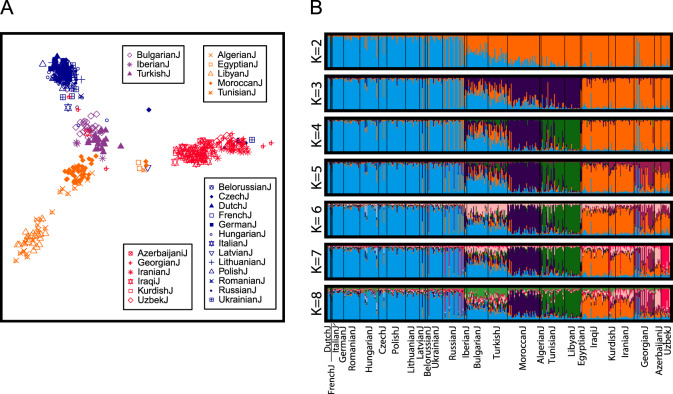
We further reduced the population set, exploring structure among Jewish populations, continuing to exclude Ethiopian and Indian Jews, and also excluding the relatively dissimilar Yemenite Jews (population set 4). We performed MDS and Structure analyses for this subset, which included 420 individuals from 27 Jewish populations.
The MDS plot in Fig. 3a contains clusters representing Ashkenazi, Mizrahi, North African, and Sephardi populations. The Mizrahi cluster is relatively distant from the other three and most distant from the Ashkenazi and North African groups. Notably, the Sephardi populations form a cluster separate from the Ashkenazi and North African populations. The average linkage distance L0, measuring the distance between pairs of individuals, one from each group, is 0.0233 for Sephardi and Ashkenazi populations. This distance gives a significant separation; with group labels permuted (see “Materials and methods” section, multidimensional scaling), Sephardi–Ashkenazi L0 is always smaller than the unpermuted L0 (p < 0.001, 1000 permutations). Sephardi–North African L0 also indicates significant separation (L0 = 0.0250, p < 0.001).
Fig. 3. Population structure for Jewish populations.
a Multidimensional scaling. Color codes follow Fig. 1c. b Unsupervised clustering using Structure. Among 20 replicates, the major mode shown appears in 20, 20, 18, 15, 9, 14, and 10 runs for K = 2, 3, 4, 5, 6, 7, and 8, respectively. Both plots include 420 individuals from 27 populations and exclude Ethiopian, Indian, and Yemenite Jewish populations.
Within the Mizrahi, North African, and Sephardi clusters, but not the Ashkenazi cluster, populations can be differentiated. For North Africans, the Libyan and Tunisian populations lie more distant from the center of the plot than do the other populations. In the Sephardi cluster, the Bulgarian Jews are closer to the Ashkenazi populations and the Turkish Jews are closer to the North African populations. L0 between the Bulgarian Jews and Turkish Jews is 0.0064; this value is significant by a permutation test in which L0 between the populations is compared with the corresponding distance when the population labels of samples from these groups are permuted (p < 0.001). Similarly, Bulgarian–Ashkenazi L0, 0.0207, is significantly smaller than the Turkish–Ashkenazi L0 of 0.0257, by a test in which labels of the Bulgarian and Turkish Jews are permuted and the distance to Ashkenazi Jews is recomputed (p < 0.001).
Structure (Fig. 3b) confirms many of the distinctions observed with MDS. For K = 2, Ashkenazi and Mizrahi Jews are largely assigned to separate clusters, with North African and Sephardi Jews having intermediate membership. For K = 3, North African Jews split into a new cluster that partially contains Sephardi Jews (blue). The new cluster at K = 4 separates Moroccan Jews from the other North African Jewish populations (green). For K = 5, Georgian Jews are assigned mostly to a new cluster that partly contains other Mizrahi populations (purple). At K = 6, the new cluster contains Sephardi Jews and contributions from some Mizrahi populations (pink). At K = 7, the Sephardi separation is less noticeable, with separation visible for Azerbaijani and Uzbek Jews (red). At K = 8, the partial Sephardi separation reappears (light green).
Major subgroups of Jewish populations
Figure 3 distinguishes the Ashkenazi, Mizrahi, North African, and Sephardi populations. We next analyzed these groups separately, using MDS and Structure for each group, and MDS for each group in combination with geographically associated non-Jewish populations (Figs. 4, 5).
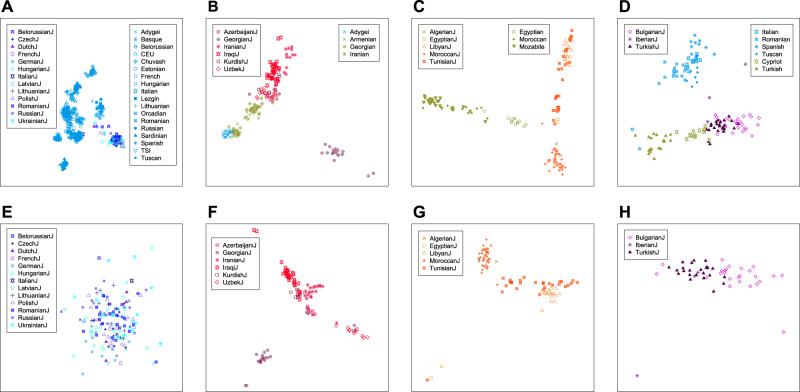
Fig. 4. Multidimensional scaling analysis of population structure for Jewish subgroups.
a Ashkenazi Jewish and nearby non-Jewish populations (31 populations, 632 individuals). b Mizrahi Jewish and nearby non-Jewish populations (ten populations, 179 individuals). c North African Jewish and nearby non-Jewish populations (eight populations, 140 individuals). d Sephardi Jewish and nearby non-Jewish populations (nine populations, 131 individuals). e Ashkenazi Jews (13 populations, 159 individuals). f Mizrahi Jews (six populations, 104 individuals). g North African Jews (five populations, 91 individuals). h Sephardi Jews (three populations, 53 individuals). Non-Jewish comparison populations were chosen from regions near locations historically inhabited by the Jewish populations.
Fig. 5. Unsupervised clustering of subgroups of Jewish populations, using Structure.
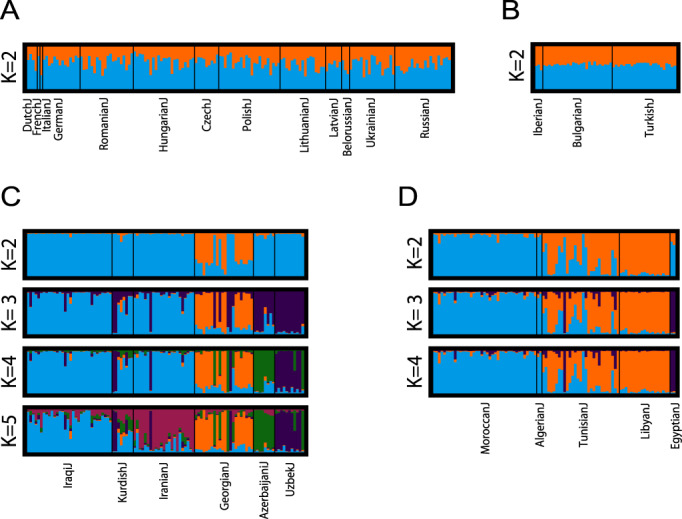
a Ashkenazi. Among 20 replicates, the major mode shown appears in 13 replicates for K = 2. b Sephardi. The major mode appears in 20 replicates for K = 2. c Mizrahi. The major mode appears in 14, 9, 11, and 16 replicates for K = 2, 3, 4, and 5, respectively. d North African. The major mode appears in 20, 15, and 18 replicates for K = 2, 3, and 4, respectively. The analysis uses the individual sets from Figs. 4e, h, f, and g. In each panel, the largest value of K displayed was selected so that no additional structure was observed for larger K values.
Ashkenazi Jewish populations
Unlike most studies of Ashkenazi Jews, we used separate population labels within the Ashkenazi sample to search for structure among different Ashkenazi Jewish populations. However, at the finest-level analysis, neither MDS analysis of Ashkenazi populations together with European non-Jewish populations (population set 5), nor MDS and Structure analyses of Ashkenazi populations alone (population set 9) produced evidence of substantial structure within our Ashkenazi samples (Fig. 4a, e and 5a).
Mizrahi Jewish populations
Structure among Mizrahi populations is shown by both MDS (Fig. 4b, f) and Structure (Fig. 5c). In both types of analysis, the Georgian Jews are relatively distinct, clustering separately from the other Mizrahi populations in MDS analysis both with (Fig. 4b) and without the non-Jewish populations (Fig. 4f), and forming a distinct Structure cluster at K = 2 (Fig. 5c). The remaining Mizrahi Jewish populations all produce largely separate clusters in MDS analysis and in Structure analysis, with the exception that the Kurdish Jews largely overlap Iranian Jews with MDS (Fig. 4f) and share substantial membership with Iraqi Jews in Structure analysis (Fig. 5c). Among non-Jewish populations, the Iranian population falls closest to the Mizrahi groups with MDS (Fig. 4b).
North African Jewish populations
The Libyan and Tunisian Jewish populations separate in MDS plots from Moroccan and Algerian Jews, with the Tunisian Jews placed over a wide area within the plots (Figs. 4c, g). This distinction is also evident using Structure, which identifies a cluster with high membership for Libyan Jews (Fig. 5d); the Algerian and Tunisian populations are spread across multiple clusters. Among non-Jewish populations, the MDS placement of the Egyptians is the closest to the North African Jewish populations, though still somewhat separate (Fig. 4c).
Sephardi Jewish populations
Supporting the separation between Bulgarian and Turkish Jews in Fig. 3a, most Bulgarian and Turkish Jews are placed in distinct locations in MDS analysis (Fig. 4h). Two Iberian Jews from Spain cluster with the Turkish Jews, whereas the third Iberian sample, from Portugal, is placed separately. In relation to non-Jewish populations, the Sephardi Jews, and particularly the Turkish Jews, are placed near the Cypriot population (Fig. 4d). No structure among the Sephardi individuals was detected by Structure (Fig. 5b).
Discussion
Using genome-wide SNPs, we investigated population structure in Jewish and non-Jewish populations. Compared with previous genomic studies of Jewish populations, the large number of markers and populations, combined with larger sample sizes for many populations, enabled a more detailed resolution of Jewish population structure, particularly in regard to divisions among Ashkenazi, Mizrahi, and Sephardi Jewish populations.
Our analyses consistently subdivide most Jewish populations into four major groups, corresponding to Ashkenazi, Mizrahi, North African, and Sephardi populations (Figs. 2, 3), with the Ashkenazi, North African, and Sephardi groups aggregating together in several analyses (Fig. 1b, c). The placement of the Jewish populations follows geography, with Ashkenazi Jews closer than other Jewish populations to non-Jewish Europeans and Mizrahi Jews closer than other Jewish populations to non-Jewish populations of the Middle East and the Caucasus region. North African and Sephardi Jewish populations appear to be intermediate between Ashkenazi Jews and non-Jewish Middle Eastern populations.
The patterns we have detected accord with and refine previous genomic analyses. Behar et al. [10] observed a cluster that included Ashkenazi, Moroccan, and Sephardi Jews and was separate from Mizrahi Jews. Similar population structure was seen in further analysis with additional non-Jewish populations [20]. With a different sample set, Atzmon et al. [9] also found that two Mizrahi Jewish populations—Iranian and Iraqi Jews—clustered separately from Ashkenazi, Sephardi, and Syrian Jews. Campbell et al. [19] augmented the data of Atzmon et al. [9] with additional Mizrahi and North African Jewish samples and confirmed separate placement of Mizrahi Jewish populations while distinguishing some North African populations from the Ashkenazi and Sephardi populations. This distinctiveness of North Africans had been suggested in earlier analyses with fewer populations [15] and markers [23].
Our analysis reveals a new clustering feature: the Sephardi populations, represented by Bulgarian, Iberian, and Turkish Jews, are largely distinguished from Ashkenazi and North African samples (Fig. 3a). Sephardi populations from Bulgaria and Turkey descend from Iberian Jews who were expelled from Spain and Portugal at the end of the 15th century and who settled in the Ottoman Empire. In the Sephardi group, the Bulgarian Jewish population is slightly closer to the Ashkenazi Jews than is the Turkish Jewish population, the latter being closer to the North African Jews (Fig. 3a). This result might reflect admixture of Iberian Jewish exiles with Ashkenazi descendants who arrived in Bulgaria via Hungary and Bavaria.
In agreement with previous studies of European and Middle Eastern non-Jewish populations, our study finds Ashkenazi populations genetically intermediate between southern Europe and the Middle East (Figs. 1c, 2), with heterozygosity slightly greater than in European populations and smaller than in Middle Eastern populations. Unlike most previous studies, our Ashkenazi samples were identified by location. However, we found little evidence of difference by location (Figs. 4e and 5a), suggesting that among the four main groups of Jewish populations, the Ashkenazi group is less genetically structured than the others.
MDS and Structure distinguish the Mizrahi populations from other Jewish regional groups. Mizrahi populations group together in multiple computations (Figs. 1c, 2, and 3), providing a similar signal of shared ancestry to that seen in the within-group clustering of the populations in the Ashkenazi, North African, and Sephardi groups in the same analyses. Unlike the other Jewish groups, however, the Mizrahi populations appear close to the Middle Eastern non-Jewish populations, and not to European non-Jewish populations. In the finest-scale analyses, each Mizrahi population can be distinguished (Figs. 4f and 5c), particularly the Georgian Jews; a possible exception is the Kurdish Jews, who overlap the Iranian and Iraqi Jewish populations. With Structure, the Armenian, Georgian, and Iranian non-Jewish populations are similar to the Mizrahi populations, though with partial membership in a cluster represented in Europe and not in the Mizrahi Jews. Notably, some Mizrahi individuals clustered with Ashkenazi populations, and vice versa, a pattern seen primarily among populations from the former Soviet Union and possibly reflecting internal migration during the Soviet period.
The North African Jewish populations were distinct from Ashkenazi and Sephardi populations, with Moroccan and Algerian Jews clustering closer to Sephardi populations, and Libyan and Tunisian Jews being more distinctive (Figs. 2 and 3a). The pattern, seen with MDS, Structure, and neighbor-joining, accords with the distinctiveness of the Libyan and Tunisian populations found in previous studies [15, 19, 23], and perhaps reflects greater influence of the Iberian exile on Moroccan and Algerian populations than on Libyan and Tunisian Jews [33], stronger and older founder effects in the Libyan and Tunisian Jews, or both.
Among non-Jewish populations, we find several populations relatively close to sets of Jewish groups. For example, in MDS analysis (Fig. 1c), the Cypriots appear near Sephardi Jews; this affinity was also evident in Structure plots (Fig. 2a). Ashkenazi Jews were placed near southern European populations such as Italians, North African Jews were closest to Egyptians, and Mizrahi Jewish populations were within a cluster of non-Jewish Middle Eastern populations, in proximity to such groups as Armenians and Iranians. These observations generally accord with previous studies, which have identified some of these same similarities [9, 10, 15, 19, 20]. No single non-Jewish group consistently overlapped any Jewish population across all analyses.
We note that although coverage in the sample of Jewish populations was relatively broad, coverage of proximate non-Jewish populations was less comprehensive, and might have omitted some of the most relevant non-Jewish groups for particular comparisons. It is possible that the nature of the sample has affected placements of Jewish in relation to non-Jewish populations; for example, Structure separations that occur at low K when a group has a large sample size might occur in a different sequence when sample sizes are matched. Thus, the large combined sample size of the Ashkenazi groups, with relatively little internal structure, could have contributed to the early Ashkenazi separation from non-Jewish and other Jewish populations (Fig. 2a, K = 4). Precise attention to the composition of the sample would be important in future work targeting hypotheses about relationships of Jewish and specific non-Jewish populations.
Our results augment previous studies and refine the understanding of the population structure of Jewish populations, particularly in distinguishing four major subgroups corresponding to the Ashkenazi, Mizrahi, North African, and Sephardi populations, and in identifying subdivisions within all except the Ashkenazi subgroup. Some of the previous studies of Jewish populations have demonstrated the potential of analyses of genomic identity-by-descent (IBD) sharing between pairs of individuals from different populations to reveal fine-scale structure [9, 19–22]. The observation that in a number of cases, non-IBD methods such as MDS, Structure, and neighbor-joining clarify structure not evident in IBD studies suggests that further information might be uncovered by joint use of IBD and non-IBD methods. In addition, although we considered more populations and a finer classification of Ashkenazi Jews than had been used previously, many sample sizes were too small to permit confident population placement. We hypothesize that with larger samples and use of IBD methods, further refinement will be possible, in particular for populations such as the Dutch, Egyptian, and Italian Jews, whose histories are notably distinct from the broader population groups with which they were combined for our data analysis.
Materials and methods
Dataset
The study design involved high-resolution genotyping of an initial sample set at genome-wide SNPs. Informed consent for participation in population-genetic studies was obtained, under ethics approvals provided by Barzilai Medical Center. Individuals included in a sample for a population satisfied a criterion that all four grandparents were members of the population (Supplementary Materials and Methods). Following quality control, which included the exclusion of duplicates and close relatives, Hardy–Weinberg testing, and exclusion of monomorphic SNPs and those with substantial missing data, the collection was reduced to 438 samples and 557,772 SNPs (Supplementary Materials and Methods). This set was merged with data from HGDP-CEPH, HapMap, and Behar et al. [10], producing a final dataset of 2789 individuals—including the 429 of the 438 new samples that did not overlap with the other datasets—114 populations, and 486,592 autosomal SNPs.
Genetic variability
Mean expected heterozygosity across the 486,592 SNPs was computed from the sample-size-corrected estimator [34]. Means across populations of the population-level mean expected heterozygosities were compared for various population sets.
Multidimensional scaling (MDS)
Pairwise distances between individuals were calculated using allele-sharing distance [28] for the 486,592 SNPs. We performed classical MDS for each matrix of individual distances—population sets 1–12—using cmdscale in R, rotating the resulting coordinates to align with approximate geographic coordinates for the populations.
In two-dimensional MDS plots, we evaluated distances between pairs of groups of individuals using the average linkage distance L0 [35, 36]: the mean Euclidean distance between the location in the plot of a randomly chosen member of the first group and a randomly chosen member of the second group. To evaluate the significance of the group separation, the probability that a random permutation of group labels gives rise to a larger L0 for two groups than that seen using the actual labels was obtained from the distribution of L0 across permutations. For three-population comparisons, we tested whether the difference between the L0 distances of each of two populations to a third population increased when labels of individuals in the first two populations were permuted.
Structure
The Structure 2.2.3 [31] admixture model assuming correlated allele frequencies among clusters was used to assess population structure, with a pruned set of 5233 widely separated markers (Supplementary Materials and Methods) and population sets 3, 4, 9, 10, 11, and 12. We modulated K from 2 to 8 for datasets 3 and 4, and from 2 to 6 for datasets 9–12. For each K, we performed 20 replicates with burn-in 10,000 iterations followed by 20,000 additional iterations. We used Clumpak [32] to identify clustering modes for each K and to align clusters across K values (default parameters: LargeKGreedy algorithm with 2000 repetitions, dynamic threshold for similarity scores). Multimodality in clustering solutions was observed for some datasets and some choices of K (Table S5); we present the mode with the most replicates (the “major mode”), reporting its associated number of replicates.
Some individuals in Fig. 3b have Structure memberships that differ considerably from other individuals in their populations: among Ashkenazi and Mizrahi individuals, some have cluster memberships typical of the other of the two groups. We excluded 13 outliers (1 Czech Jewish, 3 Georgian Jewish, 1 Iraqi Jewish, 1 Latvian Jewish, 6 Russian Jewish, 1 Ukrainian Jewish) from fine-scale analysis of Jewish populations with sample sets 5–12 (Table S6).
Population trees
Neighbor-joining trees [37] were produced from allele frequencies for populations with sample size ≥10, using the Phylip 3.65 Neighbor program [38]. Distance matrices computed as one minus the proportion of shared alleles under Hardy–Weinberg proportions [39] were obtained with Microsat [40], bootstrapping across loci. We constructed a majority-rule consensus tree, resolving multifurcations by sequentially incorporating the groupings that had the highest frequencies in the set of bootstraps and that were compatible with groupings already incorporated. Trees were edited using FigTree (http://tree.bio.ed.ac.uk/software/figtree/).
Supplementary information
Acknowledgements
We acknowledge support from the CNRS-PICS program on genetic diversity in Central Asian populations and the University of Michigan Life Sciences Institute/Israeli Universities Partnership.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version of this article (10.1038/s41431-019-0542-y) contains supplementary material, which is available to authorized users.
References
- 1.Mourant AE, Kopec AC, Dmaniewska-Sobczak K. The Genetics of the Jews. Oxford: Clarendon Press; 1978. [Google Scholar]
- 2.Carmelli D, Cavalli-Sforza LL. The genetic origin of the Jews: a multivariate approach. Hum Biol. 1979;51:41–61. [PubMed] [Google Scholar]
- 3.Goodman RM. Genetic disorders among the Jewish people. Baltimore: Johns Hopkins University Press; 1979. [Google Scholar]
- 4.Karlin S, Kenett R, Bonné-Tamir B. Analysis of biochemical genetic data on Jewish populations: II. Results and interpretations of heterogeneity indices and genetic distance measures with respect to standards. Am J Hum Genet. 1979;31:341–65. [PMC free article] [PubMed] [Google Scholar]
- 5.Ostrer H. A genetic profile of contemporary Jewish populations. Nat Rev Genet. 2001;2:891–8. doi: 10.1038/35098506. [DOI] [PubMed] [Google Scholar]
- 6.Klitz W, Gragert L, Maiers M, Fernandez-Vina M, Ben-Naeh Y, Benedek G, et al. Genetic differentiation of Jewish populations. Tissue Antigens. 2010;76:442–58. doi: 10.1111/j.1399-0039.2010.01549.x. [DOI] [PubMed] [Google Scholar]
- 7.Ostrer H, Skorecki K. The population genetics of the Jewish people. Hum Genet. 2013;132:119–27. doi: 10.1007/s00439-012-1235-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rosenberg NA, Weitzman SP. From generation to generation: the genetics of Jewish populations. Hum Biol. 2013;85:817–23. doi: 10.3378/027.085.0603. [DOI] [PubMed] [Google Scholar]
- 9.Atzmon G, Hao L, Pe’er I, Velez C, Pearlman A, Palamara PF, et al. Abraham’s children in the genome era: major Jewish diaspora populations comprise distinct genetic clusters with shared Middle Eastern ancestry. Am J Hum Genet. 2010;86:850–9. doi: 10.1016/j.ajhg.2010.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Behar DM, Yunusbayev B, Metspalu M, Metspalu E, Rosset S, Parik J, et al. The genome-wide structure of the Jewish people. Nature. 2010;466:238–42. doi: 10.1038/nature09103. [DOI] [PubMed] [Google Scholar]
- 11.Seldin MF, Shigeta R, Villoslada P, Selmi C, Tuomilehto J, Silva G, et al. European population substructure: clustering of northern and southern populations. PLoS Genet. 2006;2:e143. doi: 10.1371/journal.pgen.0020143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bauchet M, McEvoy B, Pearson LN, Quillen EE, Sarkisian T, Hovhannesyan K, et al. Measuring European population stratification with microarray genotype data. Am J Hum Genet. 2007;80:948–56. doi: 10.1086/513477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Price AL, Butler J, Patterson N, Capelli C, Pascali VL, Scarnicci F, et al. Discerning the ancestry of European Americans in genetic association studies. PLoS Genet. 2008;4:e236. doi: 10.1371/journal.pgen.0030236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tian C, Plenge RM, Ransom M, Lee A, Villoslada P, Selmi C, et al. Analysis and application of European genetic substructure using 300 K SNP information. PLoS Genet. 2008;4:e4. doi: 10.1371/journal.pgen.0040004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kopelman NM, Stone L, Wang C, Gefel D, Feldman MW, Hillel J, et al. Genomic microsatellites identify shared Jewish ancestry intermediate between Middle Eastern and European populations. BMC Genet. 2009;10:80. doi: 10.1186/1471-2156-10-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Need AC, Kasperaviciute D, Cirulli ET, Goldstein DB. A genome-wide genetic signature of Jewish ancestry perfectly separates individuals with and without full Jewish ancestry in a large random sample of European Americans. Genome Biol. 2009;10:R7. doi: 10.1186/gb-2009-10-1-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bray SM, Mulle JG, Dodd AF, Pulver AE, Wooding S, Warren ST. Signatures of founder effects, admixture, and selection in the Ashkenazi Jewish population. Proc Natl Acad Sci USA. 2010;107:16222–7. doi: 10.1073/pnas.1004381107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Listman JB, Hasin D, Kranzler HR, Malison RT, Mutirangura A, Sughondhabirom A, et al. Identification of population substruture among Jews using STR markers and dependence on reference populations studied. BMC Genet. 2010;11:48. doi: 10.1186/1471-2156-11-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Campbell CL, Palamara PF, Dubrovsky M, Botigué LR, Fellous M, Atzmon G, et al. North African Jewish and non-Jewish populations form distinctive, orthogonal clusters. Proc Natl Acad Sci USA. 2012;109:13865–70. doi: 10.1073/pnas.1204840109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Behar DM, Metspalu M, Baran Y, Kopelman NM, Yunusbayev B, Gladstein A, et al. No evidence from genome-wide data of a Khazar origin for the Ashkenazi Jews. Hum Biol. 2013;85:859–900. doi: 10.3378/027.085.0604. [DOI] [PubMed] [Google Scholar]
- 21.Waldman YY, Biddanda A, Davidson NR, Billing-Ross P, Dubrovsky M, Campbell CL, et al. The genetics of Bene Israel from India reveals both substantial Jewish and Indian ancestry. PLoS ONE. 2016;11:e0152056. doi: 10.1371/journal.pone.0152056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Waldman YY, Biddanda A, Dubrovsky M, Campbell CL, Oddoux C, Friedman E, et al. The genetic history of Cochin Jews from India. Hum Genet. 2016;135:1127–43. doi: 10.1007/s00439-016-1698-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rosenberg NA, Woolf E, Pritchard JK, Schaap T, Gefel D, Shprier I, et al. Distinctive genetic signatures in the Libyan Jews. Proc Natl Acad Sci USA. 2001;98:858–63. doi: 10.1073/pnas.98.3.858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li JZ, Absher DM, Tang H, Southwick AM, Casto AM, Ramachandran S, et al. Worldwide human relationships inferred from genome-wide patterns of variation. Science. 2008;319:1100–4. doi: 10.1126/science.1153717. [DOI] [PubMed] [Google Scholar]
- 25.International HapMap 3 Consortium. Integrating common and rare genetic variation in diverse human populations. Nature. 2010;467:52–58. doi: 10.1038/nature09298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rosenberg NA, Pritchard JK, Weber JL, Cann HM, Kidd KK, Zhivotovsky LA, et al. Genetic structure of human populations. Science. 2002;298:2381–5. doi: 10.1126/science.1078311. [DOI] [PubMed] [Google Scholar]
- 27.Jakobsson M, Scholz SW, Schet P, Gibbs JR, VanLiere JM, Fund H-C, et al. Genotype, haplotype and copy-number variation in worldwide human populations. Nature. 2008;451:998–1003. doi: 10.1038/nature06742. [DOI] [PubMed] [Google Scholar]
- 28.Mountain JL, Cavalli-Sforza LL. Multilocus genotypes, a tree of individuals, and human evolutionary history. Am J Hum Genet. 1997;61:705–18. doi: 10.1086/515510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Prugnolle F, Manica A, Balloux F. Geography predicts neutral genetic diversity of human populations. Curr Biol. 2005;15:R159–R160. doi: 10.1016/j.cub.2005.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ramachandran S, Deshpande O, Roseman CC, Rosenberg NA, Feldman MW, Cavalli-Sforza LL. Support from the relationship of genetic and geographic distance in human populations for a serial founder effect originating in Africa. Proc Natl Acad Sci USA. 2005;102:15942–7. doi: 10.1073/pnas.0507611102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics. 2003;164:1567–87. doi: 10.1093/genetics/164.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kopelman NM, Mayzel J, Jakobsson M, Rosenberg NA, Mayrose I. CLUMPAK: a program for identifying clustering modes and packaging population structure inferences across K. Mol Ecol Resour. 2015;15:1179–91. doi: 10.1111/1755-0998.12387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chouraqui AN. Between east and west: a history of the Jews of North Africa. Philadelphia: Jewish Publication Society of America; 1968. [Google Scholar]
- 34.Nei M. Molecular evolutionary genetics. New York: Columbia University Press; 1987. [Google Scholar]
- 35.Timm NH. Applied multivariate analysis. New York: Springer; 2002. [Google Scholar]
- 36.Le Roux B, Rouanet H. Geometric data analysis: from correspondence analysis to structured data analysis. Dordrecht: Kluwer; 2004. [Google Scholar]
- 37.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–25. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 38.Felsenstein J. PHYLIP (Phylogeny Inference Package) version 3.6. Seattle, WA: Department of Genome Sciences, University of Washington; 2005.
- 39.Bowcock AM, Ruiz-Linares A, Tomfohrde J, Minch E, Kidd JR, Cavalli-Sforza LL. High resolution of human evolutionary trees with polymorphic microsatellites. Nature. 1994;368:455–7. doi: 10.1038/368455a0. [DOI] [PubMed] [Google Scholar]
- 40.Minch E, Ruiz Linares A, Goldstein DB, Feldman MW, Cavalli-Sforza LL. MICROSAT version 1.5d: a program for calculating statistics on microsatellite data. Stanford, CA: Department of Genetics, Stanford University; 1998.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.