Abstract
Visual impairment due to inherited ophthalmic disorders is amongst the most common disabilities observed in populations practicing consanguineous marriages. Here we investigated the molecular genetic basis of an unselected broad range of ophthalmic disorders in 20 consanguineous families from Arab villages of Israel and the Palestinian Authority. Most patients had little or very poor prior clinical workup and were recruited in a field study. Homozygosity mapping followed by candidate gene sequencing applying conventional Sanger sequencing or targeted next generation sequencing was performed in six families. In the remaining 14 families, one affected subject per family was chosen for whole exome sequencing. We discovered likely disease-causing variants, all homozygous, in 19 of 20 independent families (95%) including a previously reported novel disease gene for congenital nystagmus associated with foveal hypoplasia. Moreover, we found a family in which disease-causing variants for two collagenopathies — Stickler and Knobloch syndrome — segregate within a large sibship. Nine of the 19 distinct variants observed in this study were novel. Our study demonstrated a very high molecular diagnostic yield for a highly diverse spectrum of rare ophthalmic disorders in Arab patients from Israel and the Palestinian Authority, even with very limited prior clinical investigation. We conclude that ‘genetic testing first' may be an economic way to direct clinical care and to support proper genetic counseling and risk assessment in these families.
Subject terms: Neurodegenerative diseases, Genetics research
Introduction
Hereditary ophthalmic diseases such as retinal dystrophies, congenital cataract, and other developmental malformations of the eye have considerable impact on social autonomy and quality of life. In particular, infants with congenital visual impairment show significant delays and difficulties regarding their motor, cognitive, language, educational, social, and emotional development compared with sighted children [1]. Molecular testing can help to provide a patient with early diagnosis and potential treatment, and is further required for accurate genetic counseling. Detection of disease-causing variants in genetic diagnostic and research testing has significantly improved with the introduction of next-generation-sequencing technologies (NGS), especially for rare diseases with a high level of clinical and genetic heterogeneity [2].
Within the last two decades, major progress has been made in our understanding of hereditary ophthalmic diseases. Inherited retinal dystrophies (IRDs), a major cause of childhood vision loss, are a heterogenic group of blinding diseases with more than 225 genes known to be mutated in the various clinical subtypes (https://sph.uth.edu/retnet/). Besides the genetic complexity, phenotypic overlaps and considerable variation in clinical expression may hamper a distinct clinical diagnosis [3]. The different disease entities result from variants in genes critical for development and maintenance of retinal photoreceptors or their supporting cells (for review, see ref. [4]). IRDs can be stationary or progressive (for review, see ref. [5]), and in the latter, vision worsens over time usually to the point of legal blindness, which can occur in early infancy (i.e. Leber congenital amaurosis) or later in life (i.e. retinitis pigmentosa or cone–rod dystrophy).
Congenital eye malformations result from defects in genes expressed during normal embryonic development, and are a significant cause of blindness worldwide [6]. Such developmental ocular defects comprise congenital cataract, congenital corneal anomalies (such as microcornea and sclerocornea), coloboma, microphthalmia (including nanophthalmos), anophthalmia and others, invariably leading to poor or no visual function and cosmetic disfigurement.
Most of these inherited ocular diseases are rare and a proper diagnosis requires expert physicians or multidisciplinary consultations and high-end specialized equipment for a differential diagnosis. However, high level medical infrastructure is expensive and not available in many parts of the world, leaving many patients without appropriate diagnosis and thus precludes proper counseling or knowledge of potential treatment options. Dropping prices for genome-wide genetic analysis such as exome sequencing may offer an economic alternative as a primary diagnostic measure in such scenarios.
Here we present the results of a genetic study on an unselected highly diverse spectrum of rare ophthalmic disorders in consanguineous Arab families from the Palestinian Authority and mainland Israel. Our results demonstrate the high sensitivity of current DNA-based diagnostics for ophthalmic disorders in this population even in families with poor prior clinical workup.
Materials and methods
Subjects
Study participants were recruited in Israel and the Palestinian Authority. In most instances, the patients and family members were recruited based on their self-reported symptoms and visual impairments upon visits of a non-ophthalmologist at the families’ home. Peripheral venous blood was taken from all consenting participants and used to isolate total genomic DNA applying standard procedures according to the manufacturer’s protocol (FlexiGene DNA Kit, QIAGEN, Hilden, Germany). DNA samples were sent to the University of Tuebingen (Germany) for genetic investigation. This study was performed according to the tenets of the Declaration of Helsinki and written informed consent was obtained from each study participant or from the parents in case of minor study subjects.
SNP genotyping and homozygosity mapping
SNP genotyping was done applying Affymetrix 250K NspI or CytoScan HD arrays (Affymetrix, Inc., Santa Clara, CA, USA). SNP chip genotypes were called with the Affymetrix Genotyping Console (GTC) Software v. 2.1 and used to determine homozygous regions by applying the online version of the HomozygosityMapper Software [7]. The threshold was set to a minimum of 2 Mb in length.
Exome sequencing
One affected patient per family underwent whole exome sequencing (WES). Exonic regions were enriched using the Agilent’s SureSelect XT Human All Exon v5 system (and in one case by using the SeqCap EZ Human Exome Library v2.0) according to the manufacturer's protocol (Illumina, San Diego, CA, USA). Sequencing was performed on Illumina HiSeq 2500 or NextSeq 500 instruments, and in one case on an Illumina Genome Analyzer IIx (GAIIx) instrument, in paired-end mode. WES was performed in 14 families (AQ10, AQ20, AQ21, AQ23, AQ24, AQ33, AQ34, AQ46, AQ48, AQ52, AQ53, AQ55, TR16, and MA08). For the (distantly) related families AQ33 and AQ34, WES was performed on one affected patient per family. In two families (AQ09 and TR21), variant detection was done by targeted NGS of >4800 genes applying the Illumina TruSight One Sequencing Panel (“clinical exome”) on Illumina MiSeq and HiSeq instruments. Raw NGS data were processed through an in-house pipeline (megSAP). Alignment was performed with BWA-MEM (https://arxiv.org/abs/1303.3997), freebayes (https://arxiv.org/abs/1207.3907) was used for variant calling, and SnpEff [8] was used for variant annotation. Variants were filtered for changes that are rare (minor allele frequency <1% due to The Genome Aggregation Database (gnomAD)), in homozygous state and in genes known/suggested to be associated with human diseases (“TruSight One Sequencing Panel filter”; 4813 genes).
Variant interpretation, Sanger sequencing, and segregation analysis
Variants identified by NGS were validated by PCR and subsequent Sanger sequencing using BigDye Terminator v. 1.1 chemistry and separation of sequencing products on a 3130xl Genetic Analyzer (Applied Biosystems, Weiterstadt, Germany). Primer sequences are listed in Table S1. Segregation analysis was done by Sanger sequencing with samples from all available family members. Minor allele frequency (MAF) data were obtained from The Genome Aggregation Database (gnomAD) (http://gnomad.broadinstitute.org/; accessed November 10, 2017). Variants were evaluated by the following in silico tools: MutationTaster (https://www.mutationtaster.org/) [9], PolyPhen-2 (https://genetics.bwh.harvard.edu/pph2/) [10], and SIFT (https://sift.jcvi.org/) [11], and classified in accordance with the standards and guidelines provided by the American College of Medical Genetics and Genomics (ACMG) and the Association for Molecular Pathology (AMP) [12] with the help of the Genetic Variant Interpretation Tool https://www.medschool.umaryland.edu/genetic_variant_interpretation_tool2.html/). Variants were submitted to ClinVar.
Results
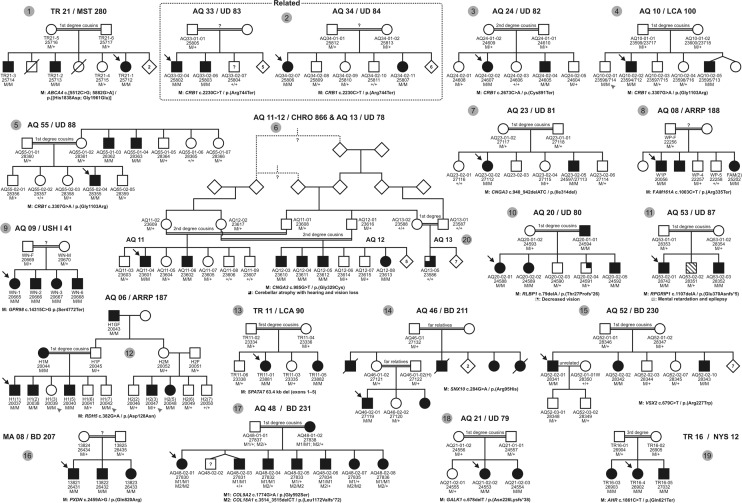
The inclusion criteria for subjects and families were defined as follows: (1) bilateral ophthalmic disease or a disorder with predominant visual symptomology, (2) at least two affected subjects per family, and (3) likely autosomal recessive inheritance with parental consanguinity. The study finally comprised 20 independent Arab families from Israel and the Palestinian Authority. In two instances we merged independently recruited families (AQ11 and AQ12 as well as AQ33 and AQ34), because we could establish familial relationship and affected patients presented with very similar conditions or symptoms. All families met the inclusion criteria with the exception of AQ13. In this family, distantly related to family AQ11/12, there was only a single affected individual suffering from a complex phenotype with ocular and neurodevelopmental symptoms. Only few families had prior undergone a thorough clinical work up. In fact, the majority of families were recruited through personal contacts within their communities by visits at their home places in order to inform them about the study, to interview the patients and family members about their visual and other health complaints and to collect blood samples. Therefore, mostly very limited knowledge on the condition (i.e. 'night blindness and gradual vision loss' or 'decreased vision and sensitivity to light') was available prior to the genetic investigation. Information on the origin, clinical diagnosis, and genetic analysis are provided in Table 1 and all pedigrees and genotypes are shown in Fig. 1.
Table 1.
Overview of the cohort.
| No. | Family ID Index Subject | Origin | First (clinical) diagnosis | Molecular diagnostics procedure | Disease causing variant (reference) | Zygosity | Genetic diagnosis |
|---|---|---|---|---|---|---|---|
| 1 | TR21 MGL#25712 | Israeli Arab | Stargardt disease | Homozygosity Mapping; Clinical exome | ABCA4 c.[5512C > G; c.5882G > A]; p.[(His1838Asp; p.Gly1961Glu)], (complex allele; [20]) | Homozygous | Stargardt disease, autosomal recessive |
| 2 | AQ33a MGL#25802 | Palestinian Arab | Night blindness followed by gradual/total vision loss | Whole exome sequencing | CRB1 c.2230C > T; p.(Arg744Ter) [14] | Homozygous | Leber congenital amaurosis, retinitis pigmentosa, or other early-onset retinal dystrophy, autosomal recessive |
| AQ34a MGL#25806 | Palestinian Arab | Total vision loss | Whole exome sequencing | CRB1 c.2230C > T; p.(Arg744Ter) [14] | Homozygous | Leber congenital amaurosis, retinitis pigmentosa, or other early-onset retinal dystrophy, autosomal recessive | |
| 3 | AQ24 MGL#24607 | Palestinian Arab | Vision loss | Whole exome sequencing | CRB1 c.2673C > A; p.(Cys891Ter) (this study) | Homozygous | Leber congenital amaurosis, retinitis pigmentosa, or other early-onset retinal dystrophy, autosomal recessive |
| 4 | AQ10 MGL#23594 | Palestinian Arab | Decreased vision since birth and color blindness | Homozygosity Mapping; Whole exome sequencing | CRB1 c.3307G > A; p.(Gly1103Arg) [56] | Homozygous | Leber congenital amaurosis, retinitis pigmentosa, or other early-onset retinal dystrophy, autosomal recessive |
| 5 | AQ55 MGL#28355 | Palestinian Arab | Night blindness and gradual vision loss | Whole exome sequencing | CRB1 c.3307G > A; p.(Gly1103Arg) [56] | Homozygous | Leber congenital amaurosis, retinitis pigmentosa, or other early-onset retinal dystrophy, autosomal recessive |
| 6 | AQ11/12 MGL#23601 | Palestinian Arab | Decreased vision and sensitivity to light | Homozygosity Mapping; Candidate gene screening | CNGA3 c.985G > T; p.(Gly329Cys) [57] | Homozygous | Achromatopsia, autosomal recessive |
| 7 | AQ23 MGL#27112 | Palestinian Arab | Retinal dystrophy | Whole exome sequencing | CNGA3 c.940_942delATC; p.(Ile314del) [24] | Homozygous | Achromatopsia, or achromatopsia-like disorder, autosomal recessive |
| 8 | AQ08 MGL#20056 | Palestinian Arab | Retinitis pigmentosa | Homozygosity Mapping; Candidate gene screening | FAM161A c.1003C > T; p.(Arg335Ter) (this study, published elsewhere [18]) | Homozygous | Retinitis pigmentosa, autosomal recessive |
| 9 | AQ09 MGL#20665 | Palestinian Arab | Usher syndrome | Homozygosity Mapping; Clinical exome | GPR98 c.14315C > G; p.(Ser4772Ter) (this study) | Homozygous | Usher syndrome, autosomal recessive |
| 10 | AQ20 MGL#24588 | Palestinian Arab | Visual problems including night blindness | Whole exome sequencing | RLBP1 c.79delA; p.(Thr27Profs*26) (this study) | Homozygous | Retinitis pigmentosa, or retinitis punctata albescens, autosomal recessive |
| 11 | AQ53 MGL#28742 | Palestinian Arab | Vision reduction since birth | Whole exome sequencing | RPGRIP1 c.1107delA; p.(Glu370Asnfs*5) [34] | Homozygous | Leber congenital amaurosis, autosomal recessive |
| 12 | AQ06 MGL#20037 | Palestinian Arab | Ophthalmic disorder | Homozygosity Mapping; Candidate gene screening | RDH5 c.382G > A; p.(Asp128Asn) [29] | Homozygous | Fundus albipunctatus, autosomal recessive |
| 13 | TR11 MGL#23881 | Israeli Arab | Leber congenital amaurosis | Homozygosity Mapping; Candidate gene screening | SPATA7 c.1–23706_373–709delinsTGG (this study, published elsewhere [19]) | Homozygous | Leber congenital amaurosis, or juvenile retinitis pigmentosa, autosomal recessive |
| 14 | AQ46 MGL#27119 | Palestinian Arab | Osteopetrosis with complete vision loss | Whole exome sequencing | SNX10 c.284G > A; p.(Arg95His) [38] | Homozygous | Infantile osteopetrosis, autosomal recessive |
| 15 | AQ52 MGL#28341 | Palestinian Arab | Atrophic eye balls, complete vision loss since birth | Whole exome sequencing | VSX2 c.679C > T; p.(Arg227Trp) [39] | Homozygous | VSX2-based anophthalmia/ microphthalmia, autosomal recessive |
| 16 | MA08 MGL#26431 | Palestinian Arab | Congenital nanophthalmos, microsclerocornea | Whole exome sequencing | PXDN c.2459A > G; p.(Gln820Arg) [this study] | Homozygous | PXDN-based anterior segment dysgenesis, autosomal recessive |
| 17 | AQ48 MGL#27830 | Palestinian Arab | Vision loss and hearing problems of various degrees | Whole exome sequencing | COL9A2 c.1774G > A; p.(Gly592Ser) [this study] COL18A1 c.3514_3515delCT; p.(Leu1172Valfs*72) [45] | Homozygous, triallelic | Stickler syndrome and Knobloch syndrome, autosomal recessive |
| 18 | AQ21 MGL#24553 | Palestinian Arab | Congenital cataract | Whole exome sequencing | GALK1 c.678delT; p.(Asn226Lysfs*38) [this study] | Homozygous | Early cataract formation due to galactokinase deficiency, autosomal recessive |
| 19 | TR16 MGL#26902 | Israeli Arab | (Idiopathic) infantile nystagmus | Whole exome sequencing | AHR c.1861C > T; p.(Gln621Ter) (this study, published elsewhere [13]) | Homozygous | New disease entity, autosomal recessive |
| 20 | AQ13b MGL#23586 | Palestinian Arab | Cerebellar atrophy with hearing loss and decreased vision | Homozygosity Mapping | Unsolved | None |
aThe families AQ33 and AQ34 were found to be distantly related and therefore considered as a single family (No. 2) in this study
bFamily AQ13 (No. 20) was related to AQ11/12, but showed a distinct phenotype
Fig. 1. Pedigrees of all studied families.
Black arrows indicate index patients; gray arrowheads indicate genotype–phenotype discrepancies. The families AQ33 and AQ34 were distantly related and segregate the same mutation and therefore merged to a single family (AQ33/34). Numbering (no. 1−20) of the families corresponds to the numbering shown in Table 1.
Applying homozygosity mapping and subsequent candidate gene analysis or — during the later stage of this project — WES of a single affected patient per family, we identified the most likely disease-causing gene variants in 19 of the 20 (95%) independent families (Table 2). One family (AQ13) remained genetically unsolved.
Table 2.
Disease-causing variants: evaluation and classification.
| Family ID | Gene | cDNA change | Protein change | Pathogenicity | Minor allele frequency (gnomAD) | ClinVar | HGMD accession no. | ACMG-based classification | ||
|---|---|---|---|---|---|---|---|---|---|---|
| MutationTaster | SIFT | PolyPhen-2 | ||||||||
| Retinal dystrophies (isolated and syndromic forms) | ||||||||||
| TR21 | ABCA4 | NM_000350.2:c.[5512C > G;5882G > A] | p.[(His1838Asp;Gly1961Glu)] | Deleterious (0.999);(0.999)a | Deleterious (0.02);(0.00)a | Deleterious (1.00);(1.00)a | 1/246168 (4.062e−6); 1292/277186 (0.004661)a | SCV000924646.1 | CM1311140; CM970016a | n.d. (complex allele) |
| AQ33/34 | CRB1 | NM_201253.3:c.2230C > T | p.(Arg744Ter) | Deleterious (1.000) | – | – | 3/245674 (1.221e−5) | SCV000924647.1 | BM1462770 | Pathogenic |
| AQ24 | CRB1 | NM_201253.3:c.2673C > A | p.(Cys891Ter) | Deleterious (1.000) | – | – | 1/120058 (ExAC) (0.000008329) | SCV000924648.1 | Novel | Pathogenic |
| AQ10 & AQ55 | CRB1 | NM_201253.3:c.3307G > A | p.(Gly1103Arg) | Deleterious (0.204) | Tolerated (0.12) | Deleterious (0.998) | 7/245136 (2.856e−5) | SCV000924649.1 | CM040718 | Pathogenic |
| AQ11/12 | CNGA3 | NM_001298.2:c.985G > T | p.(Gly329Cys) | Deleterious (0.999) | Deleterious (0.00) | Deleterious (1.000) | – | SCV000924650.1 | CM1110706 | Pathogenic |
| AQ23 | CNGA3 | NM_001298.2:c.940_942delATCb | p.(Ile314del) | Deleterious (0.999) | – | – | 6/246266 (2.436e−5) | SCV000924651.1 | CD014652 | Likely pathogenic |
| AQ08 | FAM161A | NM_001201543.2:c.1003C > T | p.(Arg335Ter) | Deleterious (1.000) | – | – | 4/277166 (1.443e−5) | SCV000924652.1 | CM142945 | Pathogenic |
| AQ09 | ADGRV1c | NM_032119.4:c.14315C > G | p.(Ser4772Ter) | Deleterious (1.000) | – | – | – | SCV000924653.1 | Novel | Pathogenic |
| AQ20 | RLBP1 | NM_000326.4:c.79delA | p.(Thr27Profs*26) | Deleterious (1.000) | – | – | – | SCV000924654.1 | Novel | Pathogenic |
| AQ53 | RPGRIP1 | NM_020366.3:c.1107delA | p.(Glu370Asnfs*5) | Deleterious (1.000) | – | – | – | SCV000924655.1 | CD012276 | Pathogenic |
| AQ06 | RDH5 | NM_002905.4:c.382G > A | p.(Asp128Asn) | Deleterious (0.999) | Tolerated (0.09) | Deleterious (1.000) | 2/243954 (8.198e−6) | SCV000924656.1 | CM077621 | Pathogenic |
| TR11 | SPATA7 | NM_018418.4:c.1–23706_373-709delinsTGG NC_000014.9:88,362,113_88,425,523delinsTGG | – | – | – | – | – | SCV000924657.1 | CG152189 | Pathogenic |
| Osteopetrosis (with complete vision loss) | ||||||||||
| AQ46 | SNX10 | NM_001199835.1:c.284G > A | p.(Arg95His) | Tolerated (0.968) | Tolerated (0.55) | Deleterious (1.000) | – | SCV000924658.1 | CM152582 | Likely pathogenic |
| Developmental malformations of the eye | ||||||||||
| AQ52 | VSX2 | NM_182894.2:c.679C > T | p.(Arg227Trp) | Deleterious (0.999) | Deleterious (0.01) | Deleterious (1.000) | 3/245904 (1.22e−5) | SCV000924659.1 | CM042327 | Pathogenic |
| MA08 | PXDN | NM_012293.3:c.2459A > G | p.(Gln820Arg) | Deleterious (0.999) | Deleterious (0.01) | Deleterious (0.992) | – | SCV000924660.1 | Novel | Likely pathogenic |
| Collagenopathies | ||||||||||
| AQ48 | COL9A2 | NM_001852.4:c.1774G > A | p.(Gly592Ser) | Deleterious (0.999) | Deleterious (0.00) | Deleterious (1.000) | 2/227220 (8.802e−6) | SCV000924661.1 | Novel | Likely pathogenic |
| COL18A1 | NM_130445.2:c.3514_3515delCT | p.(Leu1172Valfs*72) | Deleterious (1.000) | – | – | 66/222428 (0.0002967) | SCV000924662.1 | CD023246 | Pathogenic | |
| Congenital cataract | ||||||||||
| AQ21 | GALK1 | NM_000154.1:c.678delT | p.(Asn226Lysfs*38) | Deleterious (1.000) | – | – | – | SCV000924663.1 | Novel | Pathogenic |
| Foveal hypoplasia and infantile nystagmus (novel disease entity) | ||||||||||
| TR16 | AHR | NM_001621.5:c.1861C > T | p.(Gln621Ter) | Deleterious (1.000) | – | – | – | SCV000924664.1 | Novel | Pathogenic |
aPathogenicity score, minor allele frequency, and HGMD accession no. provided for each substitution in ABCA4 individually
bCorresponds to c. 934-936delATC in ref. [24]
cAlternative symbol: GPR98
Most likely disease-causing variants were observed in 15 different already known disease-associated genes (Table 2). This includes several ultrarare disease genes such as PXDN in ANTERIOR SEGMENT DYSGENESIS 7 (ASGD7; OMIM 269400), VSX2 in MICROPHTHAMLIA (OMIM 610093), or SNX10 in autosomal recessive OSTEOPETROSIS (OMIM 615085) for which very few affected families have been reported hitherto. Moreover, in family TR16 presenting with foveal hypoplasia and infantile nystagmus, we identified a nonsense variant in a novel disease candidate gene (AHR) as reported in detail elsewhere [13]. Of 19 different disease-associated variants (one of which representing a complex allele) identified in the present study, 10 were novel at the time of identification (Tables 1 and 2). However, the variant (c.2230C > T;p.(Arg744Ter)) introducing a premature stop codon in CRB1 was meanwhile reported [14]. Disease-associated variants comprised seven different missense changes, five different variants introducing a premature stop codon, five different indel variants (small insertions and/or deletions; four of them causing a frameshift), and one large deletion encompassing five consecutive exons of the SPATA7 gene as well as upstream sequences. The Stargardt disease patients of one family (TR21) were shown to carry a complex ABCA4 allele (c.[5512C > G;c.5882G > A]). All disease-associated variants were present in homozygous state in affecteds.
Surprisingly, we found that deleterious variants in two collagen genes, COL9A2 and COL18A1, which are associated with Stickler and Knobloch syndrome, respectively, are segregating in family AQ48. COL9A2-linked Stickler syndrome and COL18A1-linked Knobloch syndrome are two ultrarare disorders with overlapping ophthalmic phenotype which—to the best of our knowledge—have never been described together in a single patient in the scientific literature before.
Discussion
In 19 of 20 consanguineous Arab families from Israel and the Palestinian Authority with diverse ophthalmic conditions and mostly no or only rudimentary clinical data, we identified disease-causing variants in 16 different genes, including a novel candidate gene for foveal hypoplasia and infantile nystagmus. The prevalence of hereditary recessive diseases in Arab societies is high, mainly because of a long tradition of consanguinity due to socio-cultural factors [15]. Consanguineous marriages in the Israeli Arab society were shown to account for up to 40% of all marriages, and most frequently occurred among first cousins [16, 17]. Therefore, complex relationships within individual families and affected family members in multiple generations are often observed. Although consanguinity and the application of homozygosity mapping, or filtering for homozygous variants in exome sequencing data, likely facilitated the identification of the disease-causing variants, the high diagnostic yield (95%) was remarkable given the heterogeneity of conditions and the mostly poor or absent prior clinical examination. Recruitment of the families was carried out over a period of 6 years, mostly by visits of a non-ophthalmologist at the families’ home (field study), and the patients and family members were enrolled according to their self-reported visual problems. Therefore we argue that a concept of 'reverse diagnostics' by WES-based genetic testing as a first tier approach can be highly efficient, and further represent a model for countries or people with limited access to specialized ophthalmic clinical services.
Genotyping of available family members by PCR and subsequent Sanger sequencing confirmed concordant segregation in all but three families (AQ06, AQ10, and AQ20; Fig. 1). False disease status information for some family members may be the most likely explanation, as well as accidental mislabeling or mix-up of samples during the field study. Other, less likely possibilities includes reduced penetrance or non-causality of the identified variants, either in the entire family or in individual subjects.
Five of the 12 distinct disease-associated variants in nine different genes associated with retinal dystrophies (isolated and syndromic) were novel. To of them — a nonsense variant in FAM161A in family AQ08 and a large deletion in SPATA7 in family TR11 — have already been described in detail elsewhere [18, 19].
In family TR21, three siblings had clinically been diagnosed with Stargardt disease, which was genetically confirmed by the identification of a homozygous ABCA4 variant. The ABCA4 complex allele (c.[5512C > G;5882G > A]) seen in this family has previously been observed in two siblings of a consanguineous Jordanian family, reported to have a severe, early-onset phenotype of Stargardt disease with severe macular atrophy and additional peripheral bone spicule-like pigmentary deposits as seen in retinitis pigmentosa [20].
Retinal dystrophy in four families (AQ33/AQ34, AQ24, AQ10, and AQ55) was CRB1-related. This is in line with a study by Beryozkin et al., showing that CRB1 variants are a relatively frequent cause of autosomal recessive early-onset retinal degeneration in the Israeli and Palestinian populations [21]. Furthermore, the only recurrent disease-associated variant in this study, CRB1: c.3307G > A; p.(Gly1103Arg), observed in two unrelated families (AQ10 and AQ55), was demonstrated to be a true founder variant in this population [21]. Disease-associated variants in CRB1 imply that the affected subjects in our families most likely suffer from severe retinitis pigmentosa (RP12; ref. [22]) or Leber congenital amaurosis [23], which is well compatible with the symptomology described by the families. However, a definite diagnosis would require a detailed clinical workup by an ophthalmologist in these cases.
Two families (AQ11/12 and AQ23) were shown to carry known disease-causing variants in CNGA3, a gene associated with achromatopsia or cone dystrophy [24]. Notably, clinical trials to evaluate safety and efficacy of gene supplementation therapy for CNGA3-linked achromatopsia are currently underway including one multi-site trial, NCT02935517, with a study site in Israel. This further emphasizes the value of this study and testing strategy to inform patients about clinical trials (usually at no costs for the patient) and identify patients with rare conditions as potential participants.
Family AQ11/12 has a further family branch (AQ13) with a single affected individual (AQ13-05). This subject had different symptoms (cerebellar atrophy with hearing loss and decreased vision) compared to his affected cousins in family branches AQ11 and AQ12, and did not carry the CNGA3 variant. AQ13-05 remained the only unsolved patient of this study although a separate WES analysis was done.
A novel nonsense variant in GPR98 (also designated as ADGRV1) was identified in a family with four children suffering from both visual and hearing impairment (AQ09). Variants in GPR98 account for a small but significant proportion of subjects suffering from Usher syndrome type 2 (6.4%). Most of these GPR98 variants, as in our case, result in a truncated protein product [25]. Of note, three of four affected siblings in AQ09 were shown to be heterozygous carriers of an additional complex USH2A allele (c.[3959C > T;12052G > A]), which was inherited from the mother). Both missense USH2A variants have already been reported in Arab-Muslim families in the Jerusalem region [26].
In family AQ20 with multiple affected patients reporting a defect of vision and night blindness, a novel frameshift variant (c.79delA; p.(Thr27Profs*26)) was identified in RLBP1 encoding the cellular retinaldehyde-binding protein (CRALBP). Variants in this gene have been reported to cause autosomal recessive retinitis pigmentosa [27] or retinitis punctata albescens [28], a subtype of retinitis pigmentosa characterized by night blindness, delayed dark adaptation, and the aggregation of small yellow-white dots in the posterior pole.
Another gene involved in the intraretinal regeneration of 11-cis-retinal, RDH5, was mutated in family AQ06. The observed missense variant (c.382G > A; p.(Asp128Asn)) has been reported in several prior studies [29–31] and was shown to strongly reduce protein stability in COS-1 cells [32]. Variants in RDH5 are associated with autosomal recessive Fundus albipunctatus (FA), characterized by night blindness and subretinal yellow-white spots [33]. In FA, night blindness is usually stationary and visual acuity is often normal which may compromise the ascertainment of the clinical status in a field study and explain non-concordant genotypes in this family.
Disease-associated variants in RPGRIP1 were first described in 2001 in a study involving 57 unrelated patients with Leber congenital amaurosis [34], and were also reported to cause a clinically somewhat milder cone–rod dystrophy [35]. In family AQ53, we observed a known frameshift variant (c.1107delA; p.(Glu370Asnfs*5)) (ref. [34]). The reported early disease onset in the affected family members is in accordance with a Leber congenital amaurosis-type of retinal dystrophy in family AQ53.
Affected family members of family AQ46 suffered from osteopetrosis with complete vision loss and were shown to carry a homozygous missense variant in the SNX10 gene. Variants in SNX10 are reported to cause autosomal recessive osteopetrosis type 8. Complete visual loss occurs invariably in all untreated patients [36]. The disease is frequently fatal during infancy unless treated by hematopoietic stem cell transplantation [37]. The SNX10 variant (c.284G > A; p.(Arg95His)) observed in this study has already been described in homozygous state in a female patient from consanguineous parents with autism spectrum disorder, developmental delay, and osteopetrosis [38].
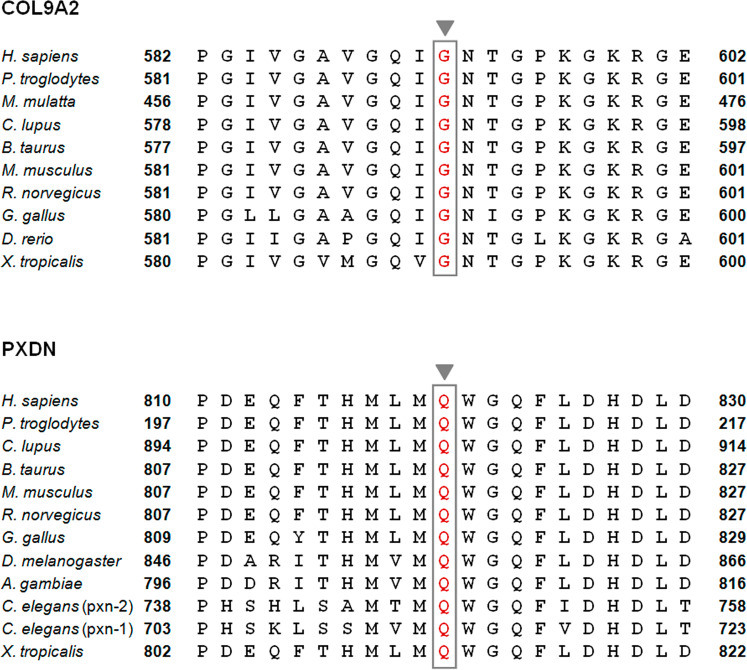
Our study included two families with affected subjects presenting with distinct ocular malformations, who were found to carry homozygous missense variants in VSX2 and PXDN, respectively. The VSX2 variant c.679C > T; p.(Arg227Trp) has previously been reported in a large, consanguineous Arab family with four affected patients suffering from isolated microphthalmia/anophthalmia [39], consistent with the findings (atrophic eye balls, complete vision loss since birth) in family AQ52. Notably, VSX2-associated ocular malformations are almost exclusively reported from the Middle-East in families with parental consanguinity [40]. The novel PXDN missense variant (c.2459A > G; p.(Gln820Arg)) identified in family MA08 with congenital nanophthalmos and corneal opacification affects a highly conserved amino acid as shown in Fig. 2. It is also predicted to be disease-causing by MutationTaster, SIFT, and PolyPhen-2, and further absent from the gnomAD data set (Table 2). Prior reports describe similar ocular anomalies (cataract-microcornea with corneal opacity, microphthalmia, and anterior segment dysgenesis) due to variants in PXDN [41, 42].
Fig. 2.
Amino acid conservation of protein sequences affected by novel missense variants.
Stickler syndrome type V and Knobloch syndrome type I are caused by variants in the two different collagen genes COL9A2 and COL18A1, respectively. Besides a single 5-generation consanguineous family of Asian-Indian origin segregating autosomal recessive Stickler syndrome that has been identified to carry a biallelic 8-bp deletion in the COL9A2 gene leading to a premature termination codon (PTC) [43], only three more patients in two unrelated families have been shown to harbor biallelic disease-associated variants in this gene [44]. In the present study, we now identified a novel missense variant c.1774G > A; p.(Gly592Ser) affecting a highly conserved amino acid in the collagen type IX alpha 2 chain (Fig. 2) in family AQ48, likely representing the fourth case with Stickler syndrome type V due to a biallelic COL9A2 variant. The variant was predicted to be deleterious by all three in silico tools (MutationTaster, SIFT, and PolyPhen-2) and observed with a minor allele frequency of 8.802e−6 in gnomAD (Table 2). The mother (AQ48-01-02) was found to be homozygous for the COL9A2 variant and upon re-interview reported to suffer from severe hearing impairment since birth, requiring assistive devices. Note that neurosensory hearing loss is a consistent feature in Stickler syndrome type V. Family AQ48 additionally segregates a known 2-bp deletion (c.3514_3515delCT; p.(Leu1172Valfs*72)) in COL18A1 repeatedly reported in subjects with autosomal recessive Knobloch syndrome [45–47]. The index patient in AQ48 was homozygous for the COL9A2 as well as the 2-bp deletion in COL18A1, which is unique, as—to the best of our knowledge—Knobloch and Stickler syndrome have never been observed together in a single individual before. Accordingly, her phenotype was reported to be slightly more severe compared to other affected family members that were homozygous for either the COL9A2 or the COL18A1 variant, or were triallelic.
Defects in GALK1 were shown to be responsible for autosomal recessive congenital cataracts [48]. Galactokinase (GALK1) is an enzyme involved in the first step of galactose metabolism. Cataract formation in galactokinase-deficient individuals is the result of osmotic phenomena caused by the accumulation of galactitol, the reduction product of galactose through an alternative pathway, in the lens [49]. In family AQ21 with congenital cataract and a novel 1-bp deletion (c.678delT; p.(Asn226Lysfs*38)) in GALK1, extra-ocular symptoms (speech delay, hearing loss, late walking, huge tongue, and hypothyroidism) have been noted in the index patient. Severe speech delay was also reported in two galactokinase-deficient brothers further showing mental retardation and cataract [50]. In classic galactosemia (linked to defects in GALT) it has been shown that speech, motor coordination, and strength disorders, likely associated with diffuse cerebellar damage, co-occur [51]. Hypothyroidism can be congenital and accompanied by sensorineural hearing loss [52] and a large protruding tongue [53]. This triad of symptoms is unlikely to be caused by the 1-bp deletion in GALK1 given their absence in the affected sister.
In family TR16, a novel disease gene for autosomal recessive infantile nystagmus was identified as described in detail elsewhere [13]. The three affected family members were shown to be homozygous for a nonsense variant (c.1861C > T; p.(Gln621Ter)) in the aryl hydrocarbon receptor (AHR) gene encoding a ligand activated transcription factor involved in various physiological and developmental processes including neurogenesis. A knockout mouse model showing similar disease expression supports our hypothesis [54, 55]. This finding represents a novel disease entity characterized by foveal hypoplasia and infantile nystagmus.
In this study, we demonstrated that the genetic causes of highly heterogeneous ophthalmic disorders can be identified with high efficacy in consanguineous Arab families, even with no or sparse initial clinical data. Especially, exome sequencing could serve as an economic diagnostic approach, enabling efficient 'reverse diagnostics' and the opportunity for tailored care, rehabilitation, and treatment.
Acknowledgements
We are grateful to the patients and family members who participated in this study. Further, we are indebted to Dr. Ibrahim Yehya, the Scientific Director of the Triangle Regional Research and Development Center.
Funding
This work was supported by grants from the German Research Foundation (SCHO 754/5-2, WI 1189/8-2, and BA 2417/2-2) to LS, BW, and PB (principle applicants), and to GB, RS, SA, and AA (co-applicants). LS is a member of the European Network for Rare Neurological Diseases (Project ID No. 739510). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Dale N, Salt A. Early support developmental journal for children with visual impairment: the case for a new developmental framework for early intervention. Child Care Health Dev. 2007;33:684–90. doi: 10.1111/j.1365-2214.2007.00798.x. [DOI] [PubMed] [Google Scholar]
- 2.Lohmann K, Klein C. Next generation sequencing and the future of genetic diagnosis. Neurotherapeutics. 2014;11:699–707. doi: 10.1007/s13311-014-0288-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weisschuh N, Mayer AK, Strom TM, Kohl S, Glöckle N, Schubach M, et al. Mutation detection in patients with retinal dystrophies using targeted next generation sequencing. PLoS One. 2016;11:e0145951. doi: 10.1371/journal.pone.0145951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Veleri S, Lazar CH, Chang B, Sieving PA, Banin E, Swaroop A. Biology and therapy of inherited retinal degenerative disease: insights from mouse models. Dis Model Mech. 2015;8:109–29. doi: 10.1242/dmm.017913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berger W, Kloeckener-Gruissem B, Neidhardt J. The molecular basis of human retinal and vitreoretinal diseases. Prog Retin Eye Res. 2010;29:335–75. doi: 10.1016/j.preteyeres.2010.03.004. [DOI] [PubMed] [Google Scholar]
- 6.Graw J. The genetic and molecular basis of congenital eye defects. Nat Rev Genet. 2003;4:876–88. doi: 10.1038/nrg1202. [DOI] [PubMed] [Google Scholar]
- 7.Seelow D, Schuelke M, Hildebrandt F, Nürnberg P. HomozygosityMapper-an interactive approach to homozygosity mapping. Nucleic Acids Res. 2009;37:W593–99. doi: 10.1093/nar/gkp369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cingolani P, Platts A, Wang le L, Coon M, Nguyen T, Wang L, et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strainw1118; iso-2; iso-3. Fly (Austin) 2012;6:80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schwarz JM, Cooper DN, Schuelke M, Seelow D. MutationTaster2: mutation prediction for the deep-sequencing age. Nat Methods. 2014;11:361–62. doi: 10.1038/nmeth.2890. [DOI] [PubMed] [Google Scholar]
- 10.Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–49. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4:1073–81. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 12.Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. ACMG Laboratory Quality Assurance Committee. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–24. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mayer AK, Mahajnah M, Thomas MG, Cohen Y, Habib A, Schulze M, et al. Homozygous stop mutation in AHR causes autosomal recessive foveal hypoplasia and infantile nystagmus. Brain. 2019;142:1528–34. doi: 10.1093/brain/awz098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kuniyoshi K, Ikeo K, Sakuramoto H, Furuno M, Yoshitake K, Hatsukawa Y, et al. Novel nonsense and splice site mutations in CRB1 gene in two Japanese patients with early-onset retinal dystrophy. Doc Ophthalmol. 2015;130:49–55. doi: 10.1007/s10633-014-9464-8. [DOI] [PubMed] [Google Scholar]
- 15.Tadmouri GO, Nair P, Obeid T, Al Ali MT, Al Khaja N, Hamamy HA. Consanguinity and reproductive health among Arabs. Reprod Health. 2009;6:17. doi: 10.1186/1742-4755-6-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jaber L, Bailey-Wilson JE, Haj-Yehia M, Hernandez J, Shohat M. Consanguineous matings in an Israeli-Arab community. Arch Pediatr Adolesc Med. 1994;148:412–5. doi: 10.1001/archpedi.1994.02170040078013. [DOI] [PubMed] [Google Scholar]
- 17.Sharkia R, Zaid M, Athamna A, Cohen D, Azem A, Zalan A. The changing pattern of consanguinity in a selected region of the Israeli Arab community. Am J Hum Biol. 2008;20:72–7. doi: 10.1002/ajhb.20678. [DOI] [PubMed] [Google Scholar]
- 18.Zobor D, Balousha G, Baumann B, Wissinger B. Homozygosity mapping reveals new nonsense mutation in the FAM161A gene causing autosomal recessive retinitis pigmentosa in a Palestinian family. Mol Vis. 2014;20:178–82. [PMC free article] [PubMed] [Google Scholar]
- 19.Mayer AK, Mahajnah M, Zobor D, Bonin M, Sharkia R, Wissinger B. Novel homozygous large deletion including the 5' part of the SPATA7 gene in a consanguineous Israeli Muslim Arab family. Mol Vis. 2015;21:306–15. [PMC free article] [PubMed] [Google Scholar]
- 20.Burke TR, Fishman GA, Zernant J, Schubert C, Tsang SH, Smith RT, et al. Retinal phenotypes in patients homozygous for the G1961E mutation in the ABCA4 gene. Invest Ophthalmol Vis Sci. 2012;53:4458–67. doi: 10.1167/iovs.11-9166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Beryozkin A, Zelinger L, Bandah-Rozenfeld D, Harel A, Strom TA, Merin S, et al. Mutations in CRB1 are a relatively common cause of autosomal recessive early-onset retinal degeneration in the Israeli and Palestinian populations. Invest Ophthalmol Vis Sci. 2013;54:2068–75. doi: 10.1167/iovs.12-11419. [DOI] [PubMed] [Google Scholar]
- 22.den Hollander AI, ten Brink JB, de Kok YJ, van Soest S, van den Born LI, van Driel MA, et al. Mutations in a human homologue of Drosophila crumbs cause retinitis pigmentosa (RP12) Nat Genet. 1999;23:217–21. doi: 10.1038/13848. [DOI] [PubMed] [Google Scholar]
- 23.Lotery AJ, Jacobson SG, Fishman GA, Weleber RG, Fulton AB, Namperumalsamy P, et al. Mutations in the CRB1 gene cause Leber congenital amaurosis. Arch Ophthalmol. 2001;119:415–20. doi: 10.1001/archopht.119.3.415. [DOI] [PubMed] [Google Scholar]
- 24.Wissinger B, Gamer D, Jägle H, Giorda R, Marx T, Mayer S, et al. CNGA3 mutations in hereditary cone photoreceptor disorders. Am J Hum Genet. 2001;69:722–37. doi: 10.1086/323613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Besnard T, Vaché C, Baux D, Larrieu L, Abadie C, Blanchet C, et al. Non-USH2A mutations in USH2 patients. Hum Mutat. 2012;33:504–10. doi: 10.1002/humu.22004. [DOI] [PubMed] [Google Scholar]
- 26.Sharon D, Banin E. Nonsyndromic retinitis pigmentosa is highly prevalent in the Jerusalem region with a high frequency of founder mutations. Mol Vis. 2015;21:783–92. [PMC free article] [PubMed] [Google Scholar]
- 27.Maw MA, Kennedy B, Knight A, Bridges R, Roth KE, Mani EJ, et al. Mutation of the gene encoding cellular retinaldehyde-binding protein in autosomal recessive retinitis pigmentosa. Nat Genet. 1997;17:198–200. doi: 10.1038/ng1097-198. [DOI] [PubMed] [Google Scholar]
- 28.Morimura H, Berson EL, Dryja TP. Recessive mutations in the RLBP1 gene encoding cellular retinaldehyde-binding protein in a form of retinitis punctata albescens. Invest Ophthalmol Vis Sci. 1999;40:1000–4. [PubMed] [Google Scholar]
- 29.Iannaccone A, Tedesco SA, Gallaher KT, Yamamoto H, Charles S, Dryja TP. Fundus albipunctatus in a 6-year old girl due to compound heterozygous mutations in the RDH5 gene. Doc Ophthalmol. 2007;115:111–6. doi: 10.1007/s10633-007-9054-0. [DOI] [PubMed] [Google Scholar]
- 30.Schatz P, Preising M, Lorenz B, Sander B, Larsen M, Eckstein C, et al. Lack of autofluorescence in fundus albipunctatus associated with mutations in RDH5. Retina. 2010;30:1704–13. doi: 10.1097/IAE.0b013e3181dc050a. [DOI] [PubMed] [Google Scholar]
- 31.Pras E, Pras E, Reznik-Wolf H, Sharon D, Raivech S, Barkana Y, et al. Fundus albipunctatus: novel mutations and phenotypic description of Israeli patients. Mol Vis. 2012;18:1712–8. [PMC free article] [PubMed] [Google Scholar]
- 32.Lidén M, Romert A, Tryggvason K, Persson B, Eriksson U. Biochemical defects in 11-cis-retinol dehydrogenase mutants associated with fundus albipunctatus. J Biol Chem. 2001;276:49251–7. doi: 10.1074/jbc.M107337200. [DOI] [PubMed] [Google Scholar]
- 33.Yamamoto H, Simon A, Eriksson U, Harris E, Berson EL, Dryja TP. Mutations in the gene encoding 11-cis retinol dehydrogenase cause delayed dark adaptation and fundus albipunctatus. Nat Genet. 1999;22:188–91. doi: 10.1038/9707. [DOI] [PubMed] [Google Scholar]
- 34.Dryja TP, Adams SM, Grimsby JL, McGee TL, Hong DH, Li T, et al. Null RPGRIP1 alleles in patients with Leber congenital amaurosis. Am J Hum Genet. 2001;68:1295–8. doi: 10.1086/320113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hameed A, Abid A, Aziz A, Ismail M, Mehdi SQ, Khaliq S. Evidence of RPGRIP1 gene mutations associated with recessive cone-rod dystrophy. J Med Genet. 2003;40:616–9. doi: 10.1136/jmg.40.8.616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Aker M, Rouvinski A, Hashavia S, Ta-Shma A, Shaag A, Zenvirt S, et al. An SNX10 mutation causes malignant osteopetrosis of infancy. J Med Genet. 2012;49:221–6. doi: 10.1136/jmedgenet-2011-100520. [DOI] [PubMed] [Google Scholar]
- 37.Pangrazio A, Fasth A, Sbardellati A, Orchard PJ, Kasow KA, Raza J, et al. SNX10 mutations define a subgroup of human autosomal recessive osteopetrosis with variable clinical severity. J Bone Min Res. 2013;28:1041–9. doi: 10.1002/jbmr.1849. [DOI] [PubMed] [Google Scholar]
- 38.Zhu X, Petrovski S, Xie P, Ruzzo EK, Lu YF, McSweeney KM, et al. Whole-exome sequencing in undiagnosed genetic diseases: interpreting 119 trios. Genet Med. 2015;17:774–81. doi: 10.1038/gim.2014.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bar-Yosef U, Abuelaish I, Harel T, Hendler N, Ofir R, Birk OS. CHX10 mutations cause non-syndromic microphthalmia/ anophthalmia in Arab and Jewish kindreds. Hum Genet. 2004;115:302–9. doi: 10.1007/s00439-004-1154-2. [DOI] [PubMed] [Google Scholar]
- 40.Williamson KA, FitzPatrick DR. The genetic architecture of microphthalmia, anophthalmia and coloboma. Eur J Med Genet. 2014;57:369–80. doi: 10.1016/j.ejmg.2014.05.002. [DOI] [PubMed] [Google Scholar]
- 41.Khan K, Rudkin A, Parry DA, Burdon KP, McKibbin M, Logan CV, et al. Homozygous mutations in PXDN cause congenital cataract, corneal opacity, and developmental glaucoma. Am J Hum Genet. 2011;89:464–73. doi: 10.1016/j.ajhg.2011.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Choi A, Lao R, Ling-Fung Tang P, Wan E, Mayer W, Bardakjian T, et al. Novel mutations in PXDN cause microphthalmia and anterior segment dysgenesis. Eur J Hum Genet. 2015;23:337–41. doi: 10.1038/ejhg.2014.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Baker S, Booth C, Fillman C, Shapiro M, Blair MP, Hyland JC, et al. A loss of function mutation in the COL9A2 gene causes autosomal recessive Stickler syndrome. Am J Med Genet A. 2011;155A:1668–72. doi: 10.1002/ajmg.a.34071. [DOI] [PubMed] [Google Scholar]
- 44.Nixon TRW, Alexander P, Richards A, McNinch A, Bearcrift PWP, Cobben J, et al. Homozygous Type IX collagen variants (COL9A1, COL9A2, and COL9A3) causing recessive Stickler syndrome-expanding the phenotype. Am J Med Genet A. 2019;179:1498–506. doi: 10.1002/ajmg.a.61191. [DOI] [PubMed] [Google Scholar]
- 45.Suzuki OT, Sertié AL, Der Kaloustian VM, Kok F, Carpenter M, Murray J, et al. Molecular analysis of collagen XVIII reveals novel mutations, presence of a third isoform, and possible genetic heterogeneity in Knobloch syndrome. Am J Hum Genet. 2002;71:1320–9. doi: 10.1086/344695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Charsar BA, Goldberg EM. Polymicrogyria and intractable epilepsy in siblings With Knobloch syndrome and homozygous mutation of COL18A1. Pediatr Neurol. 2017;76:91–2. doi: 10.1016/j.pediatrneurol.2017.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang LS, Li HB, Zeng J, Yang Y, Ding C. Knobloch syndrome caused by homozygous frameshift mutation of the COL18A1 gene in a Chinese pedigree. Int J Ophthalmol. 2018;11:918–22. doi: 10.18240/ijo.2018.06.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stambolian D, Ai Y, Sidjanin D, Nesburn K, Sathe G, Rosenberg M, et al. Cloning of the galactokinase cDNA and identification of mutations in two families with cataracts. Nat Genet. 1995;10:307–12. doi: 10.1038/ng0795-307. [DOI] [PubMed] [Google Scholar]
- 49.Asada M, Okano Y, Imamura T, Suyama I, Hase Y, Isshiki G. Molecular characterization of galactokinase deficiency in Japanese patients. J Hum Genet. 1999;44:377–82. doi: 10.1007/s100380050182. [DOI] [PubMed] [Google Scholar]
- 50.Segal S, Rutman JY, Frimpter GW. Galactokinase deficiency and mental retardation. J Pediatr. 1979;95:750–2. doi: 10.1016/s0022-3476(79)80729-5. [DOI] [PubMed] [Google Scholar]
- 51.Potter NL, Nievergelt Y, Shriberg LD. Motor and speech disorders in classic galactosemia. JIMD Rep. 2013;11:31–41. doi: 10.1007/8904_2013_219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vanderschueren-Lodeweyckx M, Debruyne F, Dooms L, Eggermont E, Eeckels R. Sensorineural hearing loss in sporadic congenital hypothyroidism. Arch Dis Child. 1983;58:419–22. doi: 10.1136/adc.58.6.419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Virtanen M. Manifestations of congenital hypothyroidism during the 1st week of life. Eur J Pediatr. 1988;147:270–4. doi: 10.1007/BF00442693. [DOI] [PubMed] [Google Scholar]
- 54.Chevallier A, Mialot A, Petit JM, Fernandez-Salguero P, Barouki R, Coumoul X, et al. Oculomotor deficits in aryl hydrocarbon receptor null mouse. PLoS One. 2013;8:e53520. doi: 10.1371/journal.pone.0053520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Juricek L, Carcaud J, Pelhaitre A, et al. AhR-deficiency as a cause of demyelinating disease and inflammation. Sci Rep. 2017;7:9794. doi: 10.1038/s41598-017-09621-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hanein S, Perrault I, Gerber S, Tanguy G, Barbet F, Ducroq D, et al. Leber congenital amaurosis: comprehensive survey of the genetic heterogeneity, refinement of the clinical definition, and genotype–phenotype correlations as a strategy for molecular diagnosis. Hum Mutat. 2004;23:306–17. doi: 10.1002/humu.20010. [DOI] [PubMed] [Google Scholar]
- 57.Genead MA, Fishman GA, Rha J, Dubis AM, Bonci DM, Dubra A, et al. Photoreceptor structure and function in patients with congenital achromatopsia. Invest Ophthalmol Vis Sci. 2011;52:7298–308. doi: 10.1167/iovs.11-7762. [DOI] [PMC free article] [PubMed] [Google Scholar]