Abstract
TBR1, a T-box transcription factor expressed in the cerebral cortex, regulates the expression of several candidate genes for autism spectrum disorders (ASD). Although TBR1 has been reported as a high-confidence risk gene for ASD and intellectual disability (ID) in functional and clinical reports since 2011, TBR1 has only recently been recorded as a human disease gene in the OMIM database. Currently, the neurodevelopmental disorders and structural brain anomalies associated with TBR1 variants are not well characterized. Through international data sharing, we collected data from 25 unreported individuals and compared them with data from the literature. We evaluated structural brain anomalies in seven individuals by analysis of MRI images, and compared these with anomalies observed in TBR1 mutant mice. The phenotype included ID in all individuals, associated to autistic traits in 76% of them. No recognizable facial phenotype could be identified. MRI analysis revealed a reduction of the anterior commissure and suggested new features including dysplastic hippocampus and subtle neocortical dysgenesis. This report supports the role of TBR1 in ID associated with autistic traits and suggests new structural brain malformations in humans. We hope this work will help geneticists to interpret TBR1 variants and diagnose ASD probands.
Subject terms: Genetics research, Genetics
Introduction
TBR1 encodes a brain-specific transcription factor of the T-box family. It is expressed both during embryogenesis and adult life in glutamatergic pyramidal neurons of the cerebral cortex, with the highest expression in the preplate and layer 6, the source of corticothalamic axons [1]. Homozygous TBR1 loss-of-function (LoF) in mice causes abnormal differentiation of early born neocortical neurons, leading to severe cortical lamination anomalies, and impaired corticothalamic projections [2]. TBR1 gain-of-function variations promote differentiation of glutamatergic neurons and oligodendrocytes from murine neural stem cells [3]. TBR1 heterozygous mice display defects in amygdalar axonal projections, and deficiencies in social interaction, cognitive flexibility, and associative memory, similar to the behavioral disorders observed in individuals with autism [4].
At the transcriptional level, TBR1 regulates the expression of several genes mutated in intellectual disability (ID) and autism spectrum disorders (ASD) [5]. It forms a nuclear complex with the synaptic scaffolding protein CASK, inducing the transcription of genes containing TBR1 binding sites, such as Reelin [6]. TBR1 is also a part of the PAX6-TBR2-NEUROD-TBR1 transcription factor cascade, essential for controlling the differentiation and connection of glutamatergic neurons in the cortex, cerebellum, and hippocampus [7]. It activates the expression of AUTS2 in the developing neocortex, and directly controls GRIN2B expression in mature neurons to induce neuronal activation [8, 9]. Conversely, TBR1 directly represses FEZF2 activity and thereby inhibits the formation of corticospinal axon projections [10, 11].
TBR1 was first reported as a candidate gene for ID in a male with severe ID, facial particularities, hypotonia, and joint laxity carrying a de novo 5.3 Mb deletion encompassing 20 genes [12]. To date, 12 single nucleotide variants (SNVs) and few copy number variations (CNVs) involving TBR1 have been reported in the literature with limited phenotypic description [13–18], except for four studies with somewhat more detailed phenotypic assessments [19–22]. The majority of these variants were reported through studies designed to identify new genes by next generation sequencing (NGS) in large cohorts of ID/ASD probands with poor clinical descriptions. Further implicating TBR1 in the pathophysiology of ASD, functional analyses have demonstrated that de novo truncating variants identified in sporadic ASD disrupt transcriptional repression activity, localization and homodimerization, and interactions of TBR1 both with the co-activator CASK and with the transcription factors FOXP1, FOXP2, and BCL11A. De novo missense variants induce milder effects, but clearly impact the function and effective dosage of the protein [23, 24]. TBR1 has very recently been associated with a human neurocognitive disorder in OMIM (#606053) in several individual cases with varying amounts of detail, but no significant cohort of individuals with variants affecting TBR1 function has been published so far.
In order to define the phenotypic and genetic diversity of variants affecting TBR1 function, we report on the detailed clinical and genetic features of 25 new individuals with de novo TBR1 SNVs and CNV, complemented by a review of individuals previously reported in the literature. We also describe the structural brain anomalies in seven new affected individuals and in heterozygous and null mutant mice.
Materials and methods
Patients
We recruited 25 unrelated and unreported individuals with a de novo variant affecting TBR1 function through national and international data sharing. The diagnosis of individuals 3, 8, and 15 was made locally by exome sequencing (ES). Twenty-two additional individuals were gathered from a national collaboration call through the AnDDI-Rares network, an international collaboration, GeneMatcher [25], ClinVar, and Decipher [ID 331488]. We did not include individuals with CNVs encompassing genes, other than TBR1, associated with human diseases in the OMIM database.
Diagnostic assessment of the individuals
Each group used a standardized clinical sheet to provide phenotypic details for individuals from their center (Supplemental data). Appropriate informed consent was obtained for affected individuals, healthy parents, and other family members, in accordance with the local ethics committee. We evaluated developmental and neurological features, medical history, physical signs, electroencephalogram (EEG) patterns, and brain magnetic resonance imaging (MRI) data, when available. ASD were assessed by the Diagnostic and Statistical Manual of Mental Disorders (DSM) criteria or the Autism Diagnostic Observation Schedule and Autism Diagnostic Interview (ADOS/ADI) scales, if available and when possible. For individuals with no specific evaluation, we took into consideration the following autistic traits: communication defects, social interaction disorders, poor eye contact, restrained interests, stereotypic behavior, or rituals. IQ evaluation was not available for all patients, so we defined severe DD/ID as walking acquired after 24 months of age or walking acquired between 18 and 24 months with severe speech delay. Moderate DD/ID was defined as normal walking acquired between 12 and 18 months of age with a severe speech delay, or by walking acquired between 18 and 24 months with a moderate speech delay. An absence of speech was considered a severe speech delay.
Molecular analyses
Gene panel tests
Samples from individuals 1, 2, 4, 6, 7, 13, 14, and 24 underwent gene panel tests that included 16–2308 genes implicated in ID/ASD, using HaloPlex or SureSelect QXT (Agilent Technologies) for capture and hybridization, followed by NGS on MiSeq instrument (Illumina) (Supplemental data).
Exome and Genome sequencing
Solo or trio ES variant filtering and analysis were performed in individuals 3, 5, 8, 9, 10, 11, 12, 15, 16, 18–23, as previously described [26–30]. Individual 17 underwent genome sequencing (GS), which was performed as previously described [31]. Alignment was made on the reference human genome GRCh37/hg19.
Array-CGH
Array-comparative genomic hybridization analysis (CGH) was performed using Oxford Gene Technology CytoSure™ ISCA v2 oligoarray set (Oxford Gene Technology, Oxford, UK) for individual 25, according to the manufacturer’s instructions.
Sanger sequencing and quantitative PCR
SNVs were confirmed by Sanger sequencing and CNV by quantitative polymerase chain reaction (PCR), respectively, as well as parental segregation. Primers and PCR conditions (available on request) were designed using the RefSeq NM_006593.2.
Animal studies
All animal procedures were performed with Institutional Animal Care and Use Committee approval from Seattle Children’s Research Institute. TBR1 mutant mice were maintained as heterozygotes on the CD-1 background, and bred to produce wild-type control, heterozygous, and homozygous mutant pups (see Supplementary Materials and Methods). Postnatal day 14 (P14) pups were perfused with 0.9% sodium chloride solution followed by 4% paraformaldehyde in 0.1 M sodium phosphate buffer. P14 is the ideal age to detect developmental delay (DD) or malformation of the dentate gyrus (DG) because morphogenesis is complete in normal mice at this timepoint. Brains were post-fixed and further processed for cryosectioning and other histological procedures as described in Supplementary Materials and Methods.
Review of the literature
We collected all articles reporting TBR1 SNVs or CNVs associated with a human disorder. We selected the individuals carrying a CNV encompassing no gene other than TBR1 associated to a human disorder in the OMIM database. We identified two other individuals with deletion encompassing TBR1, but also other genes associated with ASD or ID. They presented with a similar phenotype, but it seemed difficult to establish a genotype–phenotype correlation, so we decided not to include them in the cohort. In total, 13 individuals were selected [13–18, 20–22].
Statistical analysis for genotype–phenotype correlation
First, we compared the degree of DD/ID according to the type of variant: premature stop codon against missense, and premature stop codon + CNV against missense + in-frame deletion (Table S3a, b). Variants leading to a premature stop codon located in the last exon of the gene were not taken into account for these comparisons, because transcripts with such variants will potentially not be degraded by nonsense mediated decay. We choose to focus on the degree of DD/ID because there are too many other features missing in the literature to make a genotype–phenotype comparison based on the global phenotype. Secondly, we carried out a genotype–phenotype comparison among the eight individuals with the p.(Thr532Argfs*144) variant and all the other individuals in the cohort (Table S3c). We also compared these eight individuals with only the other individuals carrying a premature stop codon variant (not localized in the last exon) or a CNV (Table S3d). The p.(Thr532Argfs*144) variant is located in the last exon of TBR1, but is a recurrent variant in the cohort, justifying this comparison. The statistical analysis was conducted using the Fisher’s exact test because of the small number of individuals, with correction for multiple testing, leading to a significant outcome p < 0.0125.
Results
The 25 individuals in our cohort included 12 females and 13 males (sex ratio = 1.08), aged 2–29 years old. All individuals were born from healthy nonconsanguineous parents. Individual 1 was conceived through a sperm donation. Clinical features are listed in Tables S1 and S2 (Supplemental data). Phenotypic and genotypic data were anonymously submitted to the ClinVar database (Table 1).
Table 1.
SNVs and CNVs affecting the function of TBR1 newly reported and reported in the literature.
| SNVs’ individuals/references | Sequencing method | cDNA (NM_006593.2) | Inheritance | Amino acid | ClinVar ID |
|---|---|---|---|---|---|
| O’Roak et al. [14] PMID: 22495309 | Trio ES | c.405delC | De novo | p.Ala136Profs*80 | – |
| Individual 1 | Cognitive disorders panel 275 genes | c.471delC | NA | p.(Tyr157*) | SCV000747902.1 |
| Individual 2 | ID panel 44 genes | c.553C>T | De novo | p.(Gln185*) | Submitted |
| Individual 3 | Solo ES | c.673A>T | De novo | p.(Ile225Phe) | SCV000747903.1 |
| O’Roak et al. [15] PMID: 23160955 | MIP 13814.p1 | c.682A>G | De novo | p.Lys228Glu | – |
| Individual 4 | Cognitive disorders panel 275 genes | c.713_719del | De novo | p.(Ser238Thrfs*17) | SCV000747904.1 |
| Individual 5 | Solo ES | c.811T>C | De novo | p.Trp271Arg | SCV000747905.1 |
| Hamdan et al. [18] PMID:25356899 | Trio ES | c.811T>C | De novo | p.Trp271Arg | – |
| Individual 6 | ID panel 285 genes | c.812G>C | De novo | p.(Trp271Ser) | SCV000747906.1 |
| O’Roak et al. [16] PMID: 25418537 | MIP | c.813G>T | De novo | p.Trp271Cys | SCV000863780.2 |
| Individual 7 | ID panel 16 genes | c.844C>T | De novo | p.(Gln282*) | SCV000747907.1 |
| Individual 8 | Solo ES | c.896G>A | De novo | p.(Trp299*) | SCV000747908.1 |
| McDermott et al. [21] PMID 29288087 | Trio ES | c.932T>C | De novo | p.(Leu311Pro) | – |
| Individual 9 | Trio ES | c.933_934insCAAAGGA | De novo | p.(Thr312Glnfs11*) | Submitted |
| McDermott et al. [21] PMID 29288087 | Trio ES | c.946G>T | De novo | p.(Gly316*) | – |
| O’Roak et al. [15] PMID: 23160955 | MIP 13796.p1 | c.1049dup | De novo | p.Ser351* | – |
| Individual 10 | Trio ES | c.1105_1113delGTCACCGCC | De novo | p.(Val369_Ala371del) | Submitted |
| Individual 11 | Trio ES | c.1118A>G | De novo | p.(Gln373Arg) | SCV000747909.1 |
| Gilissen et al. [17] PMID:24896178 | Trio GS | c.1118A>G | De novo | p.(Gln373Arg) | – |
| Neale et al. [13] PMID: 22495311 | Trio ES | c.1120A>C | De novo | p.Asn374His | – |
| Individual 12 | Trio ES | c.1155C>G | De novo | p.(Asn385Lys) | SCV000747910.1 |
| O’Roak et al. [16] PMID: 25418537 | MIP | c.1165A>G | De novo | p.(Lys389Glu) | SCV000863781.1 |
| Individual 13 | Autism/ID panel 2308 genes | c.1177dupG | De novo | p.(Asp393Glyfs*2) | SCV000747911.1 |
| Individual 14 | Cognitive disorders panel 456 genes | c.1369_1371delinsCA | De novo | p.(Thr457Glnfs*30) | SCV000747912.1 |
| Individual 15 | Solo ES | c.1588_1594dup | De novo | p.(Thr532Argfs*144) | SCV000747913.1 |
| Individual 16 | Trio ES | c.1588_1594dup | De novo | p.(Thr532Argfs*144) | SCV000965704.1 |
| Individual 17 | GS | c.1588_1594dup | De novo | p.(Thr532Argfs*144) | SCV000747914.1 |
| Individual 18 | Trio ES | c.1588_1594dup | De novo | p.(Thr532Argfs*144) | Submitted |
| Individual 19 | Trio ES | c.1588_1594dup | De novo | p.(Thr532Argfs*144) | SCV000747915.1 |
| Individual 20 | Trio ES | c.1588_1594dup | De novo | p.(Thr532Argfs*144) | Submitted |
| Gilissen et al. [17] PMID:24896178 | Trio GS | c.1588_1594dup | De novo | p.(Thr532Argfs*144) | – |
| Vegas et al. [22] PMID 30268909 | Trio ES | c.1588_1594dup | De novo | p.(Thr532Argfs*144) | SCV000863782.1 |
| Individual 21 | Trio ES | c.1635_1644dup | De novo | p.(Ser549Glyfs*128) | Submitted |
| Individual 22 | Solo ES | c.1639_1648dup | De novo | p.(Pro550fs*127) | SCV000747916.1 |
| Individual 23 | Trio ES | c.1652dup | De novo | p.(Gln552Alafs*122) | SCV000891709.1 |
| Individual 24 | Cognitive disorders panel 456 genes | c.1653_1654del | De novo | p.(Gln552Valfs*121) | SCV000747918.1 |
| CNVs’ Individuals/reference | Method | Size | Coordinates | Band | Genes encompassed in the deletion | Transmission | ClinVar ID |
|---|---|---|---|---|---|---|---|
| Individual 25 | CGH |
Min: 461.24 kb Max: 549.18 kb |
chr2:g.(161927809_161967492)_(162428730_162476985) | 2q24.2 | TANK, PSMD14, TBR1 | De novo | SCV000747921.1 |
| Palumbo et al. [20] PMID:24458984 | SNP | 122 kb | chr2:g.(162269888–162391666) | 2q24.2 | TBR1 | De novo | – |
CNVs copy number variants, ES exome sequencing, GS genome sequencing, ID intellectual deficiency, MIP molecular inversion probe, NA not available, SNVs single nucleotide variants.
Phenotypic description of the new reported individuals
DD and/or ID were present in the 25 individuals; 10 were moderate and 15 were severe. Five individuals acquired independent walking at or after 24 months, four walked with aid only, and two was unable to walk. All other individuals acquired walked independently before 24 months (Tables S1 and S2). Seven individuals had no speech, and the majority of the others had severely delayed speech. Autistic traits were very frequent (19/25 individuals), and five individuals were diagnosed with ASD according to DSM-IV or ADOS/ADI criteria. Stereotypic behavior was frequently reported, sometimes associated with rituals or echolalia. Behavioral disturbances (22/25 individuals) included mainly attention deficit and auto/heteroaggressive behavior.
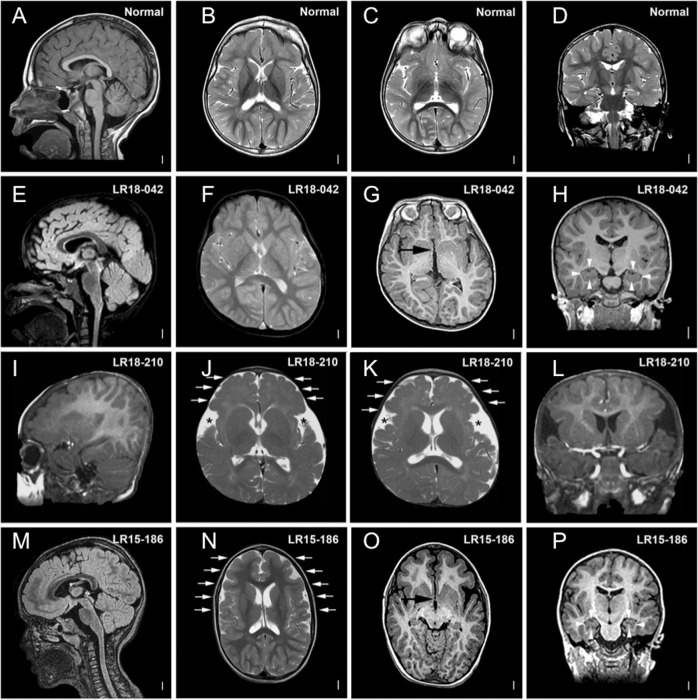
The main neurological signs included hypotonia and fine motor delay (15/21 individuals each), abnormal movements (7/21 individuals) such as dystonia, chorea, or tremors, and gait disorders (6/21 individuals). Five individuals presented seizures: one at 6 months old with tonic-clonic seizures and absences; one with one episode of seizures at 1 year old and several episodes of tonic seizures from 28 years old; and one at 2 years old with generalized refractory epilepsy sensitive to geometric patterns and light. Of the 15 individuals who had an EEG, nine displayed an abnormal unspecific pattern. MRI data were initially provided by collaborators for 22 individuals, with 10 reporting abnormal imaging. However, when possible, in-depth analysis of brain MRI imaging was performed by trained readers. In total, MRI imaging was reexamined for seven individuals. We found anomalies in each case, three of which were initially considered normal. The anterior commissure (AC) was thin or absent in all individuals, the hippocampi were dysplastic for three of them and two presented cortical dysplasia with severe gyral anomalies (Fig. 1 and Fig. S1). Only two individuals presented microcephaly.
Fig. 1. Brain MRIs of individuals 1, 19 and 23 compared with MRIs of noncarrier TBR1 variants.
Brain MRIs of individuals with TBR1 variants affecting function in comparison with MRIs of noncarrier individuals (a–d) showing: g, o anomalies of the anterior commissure (AC) (black arrowheads); h, p hippocampal anomalies (white arrowheads); j, k, n cortical gyral anomalies (white arrows and black stars). LR18-042 = individual 1; LR15-186 = individual 23; LR18–210 = individual 19.
Heterogeneous facial features were frequent, mainly high or large forehead (6/18 individuals) and long philtrum (6/18 individuals) (Fig. S2), as well as variable skeletal anomalies (13/24 individuals) with joint laxity (8/13 individuals), pes planus (5/13 individuals), and scoliosis (4/13 individuals). Infrequent features included intrauterine growth retardation (IUGR) (7/23 individuals) with persistent short stature ≤ −2 standards deviations (SD) (3/25 individuals) and mild-to-severe constipation (7/25 individuals). Feeding difficulties, potentially leading to severe complications, or surgeries such as Nissen fundoplication, were also reported in a few cases (4/25 individuals).
TBR1 molecular results of the newly and previously reported individuals
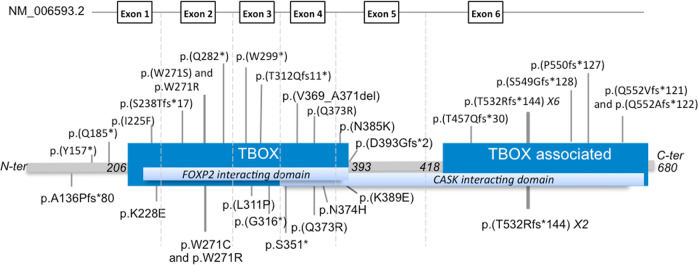
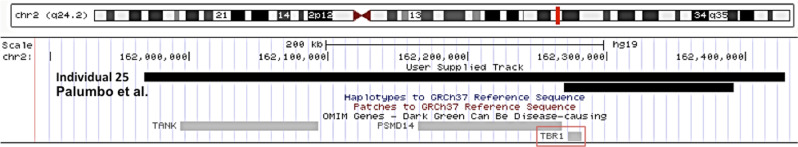
All the newly reported and previously published variants were de novo, except for one variant, which could not be tested in father because of a sperm donation (individual 1). Twenty-nine different SNVs, including ten frameshift, six nonsense, one in-frame deletion and ten missense and two deletions were reported (Table 1, Fig. 2). One deletion encompassed TBR1 and two genes, PSMD14 and TANK, not associated with a human disorder in OMIM, and one deletion only included TBR1 [20] (Table 1 and Fig. 3).
Fig. 2. Schematic representation of TBR1 transcript (NM_006593.2) and TBR1 protein with localization of the different SNVs newly reported in this study on the top, and previously reported in the literature below.
The protein contains a DNA-binding motif or T-box domain: amino acids 206–393 (dark blue box), a dimerization domain or T-box associated domain: amino acids 418–680 (dark blue box), a CASK interacting domain: amino acids 342–681 (light blue box), and a FOXP2 interacting domain: amino acids 213–393 (light blue box).
Fig. 3.
Schematic representation of the two CNVs discussed in this study, the newly reported CNV of individual 25, and that previously reported by Palumbo et al. [20].
Twenty SNVs were located in the T-box domain of the protein, 13 in the T-box-associated domain, and three were out of a functional domain defined according to the Pfam database (Fig. 2). It should be noted that only frameshift variants affected the T-box associated domain.
Genotype–phenotype correlation
There was no significant difference in the severity of DD/ID according to the type of variant. The p was superior to 0.0125 for the comparison between the individuals with SNV leading to a premature stop codon and missense variant, and between the individuals with SNV leading to a premature stop codon or CNV and those with a missense or in-frame deletion (p = 0.99 and 0.73, respectively) (Table S3a, b). We also focused on the p.(Thr532Argfs*144) variant because eight individuals were carrying it. We found no significant difference in the severity of DD/ID when we compared the individuals carrying this variant with the entire cohort or with the individuals carrying an SNV leading to a premature stop codon or CNV (p = 0.36 and 0.56, respectively) (Table S3c, d).
Four individuals, ranging in age from 5 to 10 years, carried three different missense variants impacting the same amino acid (p.Trp271). Two had p.(Trp271Arg) and presented with a similar phenotype including severe DD/ID, autistic traits, and seizures. One carried p.(Trp271Ser) and had moderate DD/ID but no autistic traits or seizures. The other carried p.(Trp271Cys) and presented with ID and autistic traits, but a detailed description was not provided [16].
Cortical malformations in postnatal Tbr1 null mice
In previous studies of mice with a targeted TBR1 null variant, homozygotes showed severe malformations of the neocortex and neonatal lethality [2], while heterozygotes showed behavioral anomalies and defective amygdalar axon connections, but no neocortical malformation [4]. The genetic background of mice in the previous studies was mostly C57BL/6, an inbred strain with relatively high phenotypic susceptibility to genetic variations [4]. In the present study, we bred the TBR1 null allele onto the outbred CD-1 strain, and found that the majority of homozygous TBR1 mutants survived to postnatal ages, allowing for assessment of later stages of cortical development.
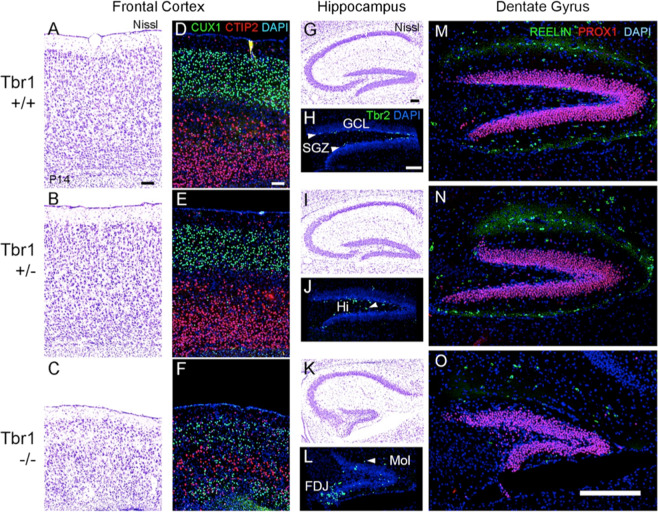
Histology of P14 brains suggested thinning and disorganization of the neocortex in TBR1 homozygous mutants, but no obvious neocortical phenotype in heterozygous mice (Fig. 4a–f). Two-color immunofluorescence, used to detect layer-specific markers, showed that deep layer neurons (CTIP2+) were sandwiched between upper layer neurons (CUX1+) in TBR1 null homozygotes, creating a “mirror-image” laminar phenotype that has also been described in reeler mice [32]. The reeler-like disorganization of TBR1 null neocortex in P14 resembled that in neonatal TBR1 mutants [2], while more severe thinning indicated a postnatal growth defect, atrophy, or apoptosis.
Fig. 4. Neocortical dyslamination and hippocampal dysgenesis in Tbr1 mutant mice (sagittal sections, P14).
a–c Nissl stains of frontal cortex. The cortex was thin and disorganized in TBR1 null cortex (c) compared with wild-type (a) and heterozygous (b) mice. d–f Immunofluorescence for CUX1 (green; upper layers) and CTIP2 (red; deep layers). Compared with wild-type (d) and heterozygous (e) mice, TBR1 null mice (f) showed thin and disorganized cortical layers, with a “mirror-image” laminar phenotype consisting of deep layers sandwiched between upper layers. g, i, k Nissl stains of hippocampus. In TBR1 null mice (k), the pyramidal layer of CA3 was less compact, and the dentate gyrus (DG) was much smaller than in wild-type (g) and heterozygous (i) mice. h, j, l Immunofluorescence for TBR2 (green), a marker of neurogenic intermediate progenitors (IPs; Hodge et al. [33, 34], revealed an immature pattern in TBR1 heterozygous and homozygous mutants. In wild-type mice, TBR2 + IPs were restricted almost exclusively to the subgranular zone (SGZ) of the DG (arrowheads, h). In TBR1 heterozygotes, IPs were dispersed outside the SGZ, notably in the hilus (Hi) of the DG (arrowhead, j). In TBR1 null mice (l), many IPs were clustered at the fimbriodentate junction, a transient niche normally regressed by P14, while other IPs were dispersed in the SGZ, Hi, and molecular layer (Mol; arrowhead, l) of the DG. Scale bar = 100 µm. m–o Immunofluorescence detection of Reelin (green) and Prox1 (red) in dentate gyrus. Reelin expression was markedly reduced in TBR1 null dentate gyrus and neocortex (not shown). Orientation: rostral left, dorsal up. Orientation: sagittal sections, rostral left, dorsal up. Scale bars (a, d, h) = 100 µm; scale bar (o) = 250 µm. hf hippocampal fissure.
In addition, striking defects were observed in the development of the DG in P14 TBR1 mutants, both homozygous and heterozygous (Fig. 4g–o). In wild-type control mice, DG neurogenesis was restricted to the subgranular zone (SGZ), as revealed by the distribution of TBR2+ neurogenic progenitor cells [33]. In TBR1 heterozygotes, the DG appeared slightly truncated, and TBR2+ progenitor cells were scattered in the hilus and molecular layer of the DG, as normally seen at slightly earlier stages of development [34]. In TBR1 homozygotes, the DG appeared very small, and TBR2+ progenitors were clustered at the fimbriodentate junction as normally observed at much earlier stages around the time of birth (Fig. 4k–l) [34]. Furthermore, expression of Reelin protein seemed severely reduced in P14 TBR1 homozygous knockout mice, and DG granule neurons, although fewer, expressed transcription factor PROX1 as usual (Fig. 4o). Also, pyramidal neurons of cornu Ammonis region 3 (CA3) did not form a compact layer. These results indicate that a TBR1 deficiency causes dose-dependent delay and reduction of DG neurogenesis, reduced Reelin expression, and dispersion of CA3 neurons. These hippocampal phenotypes need to be quantified but have never been reported in TBR1 mutant mice. Interestingly, mice with defective Reelin signaling exhibit some similar hippocampal anomalies [35].
Discussion
We provide here a full clinical and molecular description of 25 new unrelated individuals (average age 10 years) carrying a variant affecting TBR1 function. We present a genotype-first approach based on data gained from international collaboration and detailed retrospective phenotyping. With our review of the literature, we gathered 38 individuals harboring 31 different TBR1 variants, including frameshift, nonsense, in-frame deletion, missense, and CNVs. SNVs were distributed all along the protein. Functional studies in cell model systems showed that both missense variants and variants introducing premature stop codons have a deleterious impact on protein function. A dominant-negative effect on function and the localization of the protein was suggested for the missense variants, and a mechanism of haploinsufficiency was suggested for the variants presenting premature stop codons [23, 24]. All the reported TBR1 variants occurred de novo (except for individual 1). It has been shown that while inherited missense variants in ASD probands do not disturb protein function, de novo heterozygous variants present in sporadic ASD do indeed lead to protein dysfunction [23, 24]. Moreover, biallelic variants affecting TBR1 function have never been reported in living humans, and knockout mouse models died shortly after birth in the absence of hand feeding [2], or (on the more salutary CD-1 background) survived with severe cortical malformations (present study). Also, no LoF variant is reported in the GnomAD database, leading to a pLI equal to 1 (one variant observed/24.5 expected). This gene is thus extremely intolerant to LoF variants, but also to missense variants, seeing as the z-score in GnomAD is 3.64 (172 variants observed/368.4 expected). This supports the theory that de novo heterozygous LoF and missense variants have a deleterious effect and highlights the major role of TBR1 in development.
DD/ID was described in all 38 individuals, ranging from mild to severe, and 61% of the individuals presented a severe disorder. Autistic traits were frequent (76%), similar to previous studies that showed incomplete penetrance [13–16, 19, 20]. Additional behavior disorders were observed in 85% of individuals, mainly attention deficit and aggressive behavior [15, 17, 19–21]. Frequent neurological symptoms (83%) included hypotonia (51%), fine motor delay (46%) and abnormal movements (20%). Seizures were less common (17%), with onset from 6 months to 28 years of age, in contrast with frequent abnormal EEG (50%). Brain structures appeared mostly normal according to routine interpretation of imaging, which is concordant with previous reports showing that TBR1 heterozygosity was not associated with severe structural brain malformations in humans [19] or in animal models [1]. Interestingly, the in-depth analysis of MRI brain imaging from seven affected individuals revealed subtle malformation (thickening) of the neocortex and relatively small brains. This may reflect impaired neuron migration, or may be secondary to decreased cortical surface area as a consequence of reduced gyral complexity. Cortical volume seemed reduced in humans and mice, although this reduction was manifest as reduced gyral complexity and increased sulcal width in human patients (Fig. 1 and Fig. S1), and as cortical thinning in TBR1 knockout mice (Fig. 4). Since mice are naturally lissencephalic, reduced gyral complexity is not phenotypically possible. Brain MRIs analysis also reveals dysplastic hippocampi in three individuals, concordant with the observations made in heterozygous and homozygous mutant mice (Fig. 4). The hippocampal defects have never been reported in TBR1 mutant mice or human, and point to impaired and delayed morphogenesis of the DG. The anomalies of the neocortex and DG could be attributable in part to reduced expression of the RELN gene, as shown in Fig. 4 and as documented in TBR1-deficient mice [2]. Overall, these results in mice correlate well with the neocortex and hippocampus malformations seen by MRI in human patients, and could be attributable in part to reduced Reelin signaling. However, since many other genes are dysregulated in Tbr1 mutants, we cannot affirm that Reelin deficiency is the only cause of the malformations. The AC was thin or nearly absent in all seven individuals studied by MRI, concordant with the TBR1± mice studies that showed the absence of the posterior part of the AC [4]. In the literature, brain imaging was available for five individuals. One individual exhibited increased extra-axial spaces [18] and two individuals presented irregular cortical gray and white matter suggestive of pachygyria, a thick frontal cortex, and a thin corpus callosum with large lateral ventricles for one of them [17, 22], but no anomalies of the AC or the hippocampus were reported.
Additional clinical features reported in the 38 individuals included facial particularities (68%), and skeletal anomalies (56%), but no specific pattern was detected. Affected individuals also presented IUGR (30%), short stature (21%), microcephaly (16%), constipation (26%), and feeding difficulties (18%) (Table 2). Though tall stature has never been reported in the literature, our cohort included four individuals whose height was between +2 and +3 SD. Therefore, aside from DD/ID and autistic traits, most of the other TBR1-associated features were either nonspecific or infrequent, providing a strong argument in favor of the genotype-first strategy. This approach provides essential genotype–phenotype and natural history data for use in genetic counseling.
Table 2.
Frequencies of the main clinical features of all individuals (study and literature) according to the variant type.
| Variants | Truncating | Missense | In-frame deletion | CNV | Total | Total in percentage |
|---|---|---|---|---|---|---|
| Number and gender of individuals | 11M/12F | 8M/4F | 1F | 2M | 21M/17F | |
| Mean age at last follow-up | 11 years | 9 years | 3 years | 9.5 years | 10 years | |
| Growth parameters | ||||||
| IUGR (birth weight ≤ 10th per) | 3/18 | 3/6 | 1/1 | 1/2 | 8/27 | 30% |
| Short stature (length ≤ −2 SD) | 2/19 | 3/7 | 0/1 | 1/2 | 6/28 | 21% |
| Tall stature (length ≥ −2 SD) | 4/19 | 0/7 | 0/1 | 0/2 | 4/28 | 14% |
| Microcephaly (OFC ≤ −3 SD) | 4/21 | 0/7 | 0/1 | 1/2 | 5/31 | 16% |
| DD/ID | 23/23 | 12/12 | 1/1 | 2/2 | 38/38 | 100% |
| Mild | 1/23 | 2/12 | 0/1 | 0/2 | 3/38 | 8% |
| Moderate | 6/23 | 3/12 | 0/1 | 1/2 | 10/38 | 26% |
| Severe | 16/23 | 5/12 | 1/1 | 1/2 | 23/38 | 61% |
| NA | 0/23 | 2/12 | 0/1 | 0/2 | 2/38 | 5% |
| ASD | 18/23 | 9/12 | 1/1 | 1/2 | 29/38 | 76% |
| Behavior disorders | 19/23 | 7/8 | 1/1 | 2/2 | 29/34 | 85% |
| Neurological features | 20/23 | 7/9 | 1/1 | 1/2 | 29/35 | 83% |
| Hypotonia | 15/23 | 3/9 | 0/1 | 0/2 | 18/35 | 51% |
| FMD | 12/23 | 3/9 | 0/1 | 1/2 | 16/35 | 46% |
| Abnormal movements | 4/23 | 2/9 | 0/1 | 1/2 | 7/35 | 20% |
| Epilepsy | 2/23 | 4/9 | 0/1 | 0/2 | 6/35 | 17% |
| Brain MRI anomalies | 10/25 | 3/7 | ND | 0/1 | 13/33 | 39% |
| EEG anomalies | 5/15 | 4/4 | ND | 1/1 | 10/20 | 50% |
| Facial particularities | 13/21 | 5/7 | 1/1 | 2/2 | 21/31 | 68% |
| Skeletal anomalies | 11/22 | 4/7 | 1/1 | 2/2 | 18/32 | 56% |
| Feeding difficulties | 6/23 | 0/8 | 0/1 | 0/2 | 6/34 | 18% |
| Constipation | 6/21 | 2/7 | 0/1 | 0/2 | 8/31 | 26% |
Only one facial particularity was needed for the sign to be counted.
ASD autism spectrum disorders, CNV copy number variation, DD developmental delay, FMD fine motor delay, ID intellectual deficiency, IUGR intrauterine growth retardation, ND not done.
Thorough phenotypic analysis and statistical analyses highlight no significant difference in the severity of DD/ID according to the variant type. Among the eight individuals carrying the p.(Thr532Argfs*144) variant, seven presented severe DD/ID, but no significant differences were found with the other individuals of the cohort (Table S3c, d). It has been shown that de novo TBR1 variants introducing a premature stop codon lead to a lack of CASK redistribution and a loss of TBR1–CASK co-localization [23]. Variants that affect this interaction domain are more likely to severely alter the developing cerebral cortex, and so to be deleterious with a remarkable phenotype. Moreover, the TBR1–CASK protein complex induces the transcription of RELN. The consequences of reduced RELN protein expression have been previously mentioned. However, we cannot affirm that this is the case with the p.(Thr532Argfs*144) variant because the functional studies were focused on two other variants, both not located in the last exon (p.Ala136Profs*80 and p.Ser351*). Also, these studies have been realized in vitro and results are not likely to relate to the situation in vivo with these truncating variants, as NMD will most likely prevent the truncated protein to be expressed.
The severity of the cognitive phenotype and the associated features appear to vary in individuals carrying a variation that affects the same amino acid p.(Trp271). In vitro studies showed that p.Trp271Cys had deleterious effects on protein function, contrary to p.Trp271Arg. However, protein levels were lower in the cells expressing this variant, and the authors did not exclude a contribution to the neurodevelopmental phenotype [24]. This idea is supported by our observation that the two individuals carrying the p.Trp271Arg variant presented severe DD/ID in association with other anomalies. The only in-frame deletion reported in this study affects the T-box domain, which is involved in DNA-binding and protein–protein interactions, and the three deleted amino acids are highly conserved in the different species. Even if these biological arguments and the phenotype of individual 10 are convincing, functional analyses would be necessary to confirm the pathogenicity of this variant. The two reported CNVs ranged from 122 to 461.24 kb, including from one to three genes (Fig. 3). In an 18-year-old male with mild ID and short stature, Burrage et al. detected a mosaicism for a 0.422 Mb deletion, present in ~ 61% of cells, including the same three genes as our individual, TBR1 PSMD14, and TANK [36]. TBR1 was the most likely candidate for the ID phenotype. PSMD14 (non-atpase proteasome 26s subunit 14) was also a good candidate because knockdown of this gene in postmitotic neurons resulted in apoptosis. The authors suggested that the association of PSMD14 with decreased cell viability and cell cycle arrest makes it an interesting candidate gene for short stature. This association is only hypothetical considering that our individual did not present short stature and that some patients without the deletion of PSMD14 do [12, 20]. Lastly, TANK (Traf family member-associated NF-Kappa-B activator) is a ubiquitously expressed gene whose protein product activates NF-kb with TRAF2 (tumor necrosis factor receptor-associated factor 2). It was very recently suggested that TANK haploinsufficiency could be responsible for nonsyndromic ID in two siblings [37].
To conclude, de novo variants affecting TBR1 function in humans are responsible for a syndromic nonrecognizable but recurrent neurocognitive disorder that includes DD/ID, autistic traits, and cerebral anomalies. Brain structures in human and mutant mice appeared to present similar anomalies, particularly dysplasia of the cortex and of the hippocampi linked to the regulation of RELN by TBR1. Interestingly, neocortical malformations were seen with heterozygous loss of TBR1 function in some human patients (Fig. 1 and Fig. S1), but not in TBR1 heterozygous knockout mice (Fig. 4 and [2]). This distinction fits with other examples, such as DCX [38], where brain morphogenesis is more sensitive to specific gene variants in humans than in mice. Phenotypic differences may reflect different variant types or effects even for the same type, as indicated by in vitro studies [24]. Hypoplasia/absence of the AC and hippocampal dysplasia were not previously reported in mice or humans with TBR1 alterations, and could be useful features, in association with the core signs of the TBR1 phenotype, for guiding clinicians and biologists in the diagnostic process and variant interpretation. A genotype-first approach seems suitable for diagnosing such patients, using large ID panels or ES/GS; data sharing is a powerful means to validate new genes discovered by NGS. Combining these two methods improves the likelihood of delineating the complex phenotypes that occur in genetic diseases, thus enhancing the clinician’s ability to provide the most suitable medical care and accurate genetic counseling to patients and families.
Web resources
AnDDI-Rares: http://www.anddi-rares.org/
ClinVar: http://www.ncbi.nlm.nih.gov/clinvar
Decipher: http://www.decipher.sanger.ac.uk
GeneMatcher: genematcher.org
gnomAD: http://gnomad.broadinstitute.org/
OMIM: http://www.omim.org/
Pfam: http://pfam.xfam.org/
Supplementary information
Acknowledgements
This study makes use of data shared through the PhenomeCentral repository. Funding for PhenomeCentral was provided by Genome Canada and Canadian Institute of Health Research (CIHR). Research reported in this publication was supported by the French Ministry of Health (PHRC national 2008/2008-A00515-50), the Regional Council of Burgundy/Dijon University hospital (Plan d’Actions Régionales pour l’Innovation PARI 2012, AOI 2013), the European Union through the PO FEDER-FSE Bourgogne 2014/2020 programs (FEDER 2015), the National Institute of Neurological Disorders and Stroke (NINDS) under award number K08NS092898 and Jordan’s Guardian Angels (to GM), by NIH Grants R01 NS092339 and R01 NS085081 (to RFH), and by NHGRI grant UM1HG007301 to GMC. The CAUSES Study is funded by Mining for Miracles, British Columbia Children’s Hospital Foundation and Genome British Columbia. Several authors of this publication are members of the European Reference Network for Developmental Anomalies and Intellectual Disability (ERN-ITHACA). Proofreading of the manuscript was provided by Suzanne Rankin from the Dijon University Hospital.
CAUSES Study
Shelin Adam, Christèle du Souich, Alison M. Elliott, Anna Lehman, Jill Mwenifumbo, Tanya N. Nelson, Clara van Karnebeek, Jan M. Friedman.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Christel Thauvin-Robinet, Email: christel.thauvin@chu-dijon.fr.
CAUSES Study:
Shelin Adam, Christèle du Souich, Alison M. Elliott, Anna Lehman, Jill Mwenifumbo, Tanya N. Nelson, Clara van Karnebeek, and Jan M. Friedman
Supplementary information
The online version of this article (10.1038/s41431-020-0571-6) contains supplementary material, which is available to authorized users.
References
- 1.Bulfone A, Smiga SM, Shimamura K, Peterson A, Puelles L, Rubenstein JLR. T-brain-1: a homolog of Brachyury whose expression defines molecularly distinct domains within the cerebral cortex. Neuron. 1995;15:63–78. doi: 10.1016/0896-6273(95)90065-9. [DOI] [PubMed] [Google Scholar]
- 2.Hevner RF, Shi L, Justice N, Hsueh YP, Sheng M, Smiga S, et al. Tbr1 regulates differentiation of the preplate and layer 6. Neuron. 2001;29:353–66. doi: 10.1016/S0896-6273(01)00211-2. [DOI] [PubMed] [Google Scholar]
- 3.Méndez-Gómez HR1, Vergaño-Vera E, Abad JL, Bulfone A, Moratalla R, de Pablo F, et al. The T-box brain 1 (Tbr1) transcription factor inhibits astrocyte formation in the olfactory bulb and regulates neural stem cell fate. Mol Cell Neurosci. 2011;46:108–21. doi: 10.1016/j.mcn.2010.08.011. [DOI] [PubMed] [Google Scholar]
- 4.Huang TN, Chuang HC, Chou WH, Chen CY, Wang HF, Chou SJ, et al. Tbr1 haploinsufficiency impairs amygdalar axonal projections and results in cognitive abnormality. Nat Neurosci. 2014;17:240–7. doi: 10.1038/nn.3626. [DOI] [PubMed] [Google Scholar]
- 5.Notwell JH, Heavner WE, Darbandi SF, Katzman S, McKenna WL, Ortiz-Londono CF, et al. TBR1 regulates autism risk genes in the developing neocortex. Genome Res. 2016;26:1013–22. doi: 10.1101/gr.203612.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hsueh YP, Wang TF, Yang FC, Sheng M. Nuclear translocation and transcription regulation by the membrane-associated guanylate kinase CASK/LIN-2. Nature. 2000;404:298–302. doi: 10.1038/35005118. [DOI] [PubMed] [Google Scholar]
- 7.Hevner RF, Hodge RD, Daza RA, Englund C. Transcription factors in glutamatergic neurogenesis: conserved programs in neocortex, cerebellum, and adult hippocampus. Neurosci Res. 2006;55:223–33. doi: 10.1016/j.neures.2006.03.004. [DOI] [PubMed] [Google Scholar]
- 8.Bedogni F, Hodge RD, Elsen GE, Nelson BR, Daza RA, Beyer RP, et al. Tbr1 regulates regional and laminar identity of postmitotic neurons in developing neocortex. Proc Natl Acad Sci USA. 2010;107:13129–34. doi: 10.1073/pnas.1002285107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chuang HC, Huang TN, Hsueh YP. Neuronal excitation upregulates Tbr1, a high-confidence risk gene of autism, mediating Grin2b expression in the adult brain. Front Cell Neurosci. 2014;8:280. doi: 10.3389/fncel.2014.00280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Han W, Kwan KY, Shim S, Lam MM, Shin Y, Xu X, et al. TBR1 directly represses Fezf2 to control the laminar origin and development of the corticospinal tract. Proc Natl Acad Sci USA. 2011;108:3041–6. doi: 10.1073/pnas.1016723108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McKenna WL, Betancourt J, Larkin KA, Abrams B, Guo C, Rubenstein JL, et al. Tbr1 and Fezf2 regulate alternate corticofugal neuronal identities during neocortical development. J Neurosci. 2011;31:549–64. doi: 10.1523/JNEUROSCI.4131-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Magri C, Piovani G, Pilotta A, Michele T, Buzi F, Barlati S. De novo deletion of chromosome 2q24.2 region in a mentally retarded boy with muscular hypotonia. Eur J Med Genet. 2011;54:361–4. doi: 10.1016/j.ejmg.2010.12.011. [DOI] [PubMed] [Google Scholar]
- 13.Neale BM, Kou Y, Liu L, Ma’ayan A, Samocha E, Sabo A, et al. Patterns and rates of exonic de novo mutations in autism spectrum disorders. Nature. 2012;485:242–5. doi: 10.1038/nature11011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.O’Roak BJ, Vives L, Girirajan S, Karakoc E, Krumm N, Coe BP, et al. Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature. 2012;485:246–50. doi: 10.1038/nature10989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.O’Roak BJ, Vives L, Fu W, Egertson JD, Stanaway IB, Phelps IG, et al. Multiplex targeted sequencing identifies recurrently mutated genes in autism spectrum disorders. Science. 2012;338:1619–22. doi: 10.1126/science.1227764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.O’Roak BJ, Stessman HA, Boyle EA, Witherspoon KT, Martin B, Lee C, et al. Recurrent de novo mutations implicate novel genes underlying simplex autism risk. Nat Commun. 2014;5:5595. doi: 10.1038/ncomms6595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gilissen C, Hehir-Kwa JY, Thung DT, Witherspoon KT, Martin B, Lee C, et al. Genome sequencing identifies major causes of severe intellectual disability. Nature. 2014;511:344–7. doi: 10.1038/nature13394. [DOI] [PubMed] [Google Scholar]
- 18.Hamdan FF, Srour M, Capo-Chichi JM, Daoud H, Nassif C, Patry L, et al. De novo mutations in moderate or severe intellectual disability. PLoS Genet. 2014;10:e1004772. doi: 10.1371/journal.pgen.1004772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Traylor RN, Dobyns WB, Rosenfeld JA, Wheeler P, Spence JE, Bandholz AM, et al. Investigation of TBR1 hemizygosity: four individuals with 2q24 microdeletions. Mol Syndromol. 2012;3:102–12. doi: 10.1159/000342008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Palumbo O, Fichera M, Palumbo P, Rizzo R, Mazzolla E, Cocuzza DM, et al. TBR1 is the candidate gene for intellectual disability in patients with a 2q24.2 interstitial deletion. Am J Med Genet A. 2014;164A:828–33. doi: 10.1002/ajmg.a.36363. [DOI] [PubMed] [Google Scholar]
- 21.McDermott JH, Study DDD, Clayton-Smith J, Briggs TA. The TBR1-related autistic-spectrum-disorder phenotype and its clinical spectrum. Eur J Med Genet. 2017;17:30427–5. doi: 10.1016/j.ejmg.2017.12.009. [DOI] [PubMed] [Google Scholar]
- 22.Vegas Nancy, Cavallin Mara, Kleefstra Tjitske, de Boer Lonneke, Philbert Marion, Maillard Camille, Boddaert Nathalie, Munnich Arnold, Hubert Laurence, Bery Amandine, Besmond Claude, Bahi-Buisson Nadia. Mutations in TBR1 gene leads to cortical malformations and intellectual disability. European Journal of Medical Genetics. 2018;61(12):759–764. doi: 10.1016/j.ejmg.2018.09.012. [DOI] [PubMed] [Google Scholar]
- 23.Deriziotis P, O’Roak BJ, Graham SA, Estruch SB, Dimitropoulou D, Bernier RA, et al. De novo TBR1 mutations in sporadic autism disrupt protein functions. Nat Commun. 2014;5:4954. [DOI] [PMC free article] [PubMed]
- 24.Den Hoed J, Sollis E, Venselaar H, Estruch SB, Deriziotis P, Fisher SE. Functional characterization of TBR1 variants in neurodevelopmental disorder. Sci Rep. 2018;8:14279. [DOI] [PMC free article] [PubMed]
- 25.Sobreira N, Schiettecatte F, Valle D, Hamosh A. GeneMatcher: a matching tool for connecting investigators with an interest in the same gene. Hum Mutat. 2015;36:928–30. doi: 10.1002/humu.22844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tanaka AJ, Cho MT, Millan F, Juusola J, Retterer K, Joshi C, et al. Mutations in SPATA5 are associated with microcephaly, intellectual disability, seizures, and hearing loss. Am J Hum Genet. 2015;97:457–64. doi: 10.1016/j.ajhg.2015.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thevenon J, Duffourd Y, Masurel-Paulet A, Lefebvre M, Feillet F, El Chehadeh-Djebbar S, et al. Diagnostic odyssey in severe neurodevelopmental disorders: toward clinical whole-exome sequencing as a first-line diagnostic test. Clin Genet. 2016;89:700–7. doi: 10.1111/cge.12732. [DOI] [PubMed] [Google Scholar]
- 28.Yang Y, Muzny DM, Reid JG, Bainbridge MN, Willis A, Ward PA, et al. Clinical whole-exome sequencing for the diagnosis of mendelian disorders. N. Engl J Med. 2013;369:1502–11. doi: 10.1056/NEJMoa1306555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tarailo-Graovac M, Shyr C, Ross CJ, Horvath GA, Salvarinova R, Ye XC, et al. Exome Sequencing and the Management of Neurometabolic Disorders. N. Engl J Med. 2016;374:2246–55. doi: 10.1056/NEJMoa1515792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ruggeri G, Timms AE, Cheng C, Weiss A, Kollros P, Chapman T, et al. Bi-allelic mutations of CCDC88C are a rare cause of severe congenital hydrocephalus. Am J Med Genet A. 2018;176:676–81. doi: 10.1002/ajmg.a.38592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bowling KM, Thompson ML, Amaral MD, Finnila CR, Hiatt SM, Engel KL, et al. Genomic diagnosis for children with intellectual disability and/or developmental delay. Genome Med. 2017;9:43. doi: 10.1186/s13073-017-0433-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Boyle MP, Bernard A, Thompson CL, Ng L, Boe A, Mortrud M, et al. Cell-type-specific consequences of Reelin deficiency in the mouse neocortex, hippocampus, and amygdala. J Comp Neurol. 2011;519:2061–89. doi: 10.1002/cne.22655. [DOI] [PubMed] [Google Scholar]
- 33.Hodge RD, Kowalczyk TD, Wolf SA, Encinas JM, Rippey C, Enikolopov G, et al. Intermediate progenitors in adult hippocampal neurogenesis: Tbr2 expression and coordinate regulation of neuronal output. J Neurosci. 2008;28:3707–17. doi: 10.1523/JNEUROSCI.4280-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hodge RD, Garcia AJ, Elsen GE, Nelson BR, Mussar KE, Reiner SL, et al. Tbr2 expression in Cajal-Retzius cells and intermediate neuronal progenitors is required for morphogenesis of the dentate gyrus. J Neurosci. 2013;33:4165–80. doi: 10.1523/JNEUROSCI.4185-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ha S, Tripathi PP, Mihalas AB, Hevner RF, Beier DR. C-terminal region truncation of RELN disrupts an interaction with VLDLR, causing abnormal development of the cerebral cortex and hippocampus. J Neurosci. 2017;37:960–71. doi: 10.1523/JNEUROSCI.1826-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Burrage LC, Eble TN, Hixson PM, Roney EK, Cheung SW, Franco LM. A mosaic 2q24.2 deletion narrows the critical region to a 0.4 Mb interval that includes TBR1, TANK, and PSMD14. Am J Med Genet A. 2013;161A:841–4. doi: 10.1002/ajmg.a.35751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Karoglan A, Schanze D, Bär C, Muschke P, Zenker M, Schanze I. A 2q24.2 microdeletion containing TANK as novel candidate gene for intellectual disability. Am J Med Genet A. 2019;179:832–6. doi: 10.1002/ajmg.a.61093. [DOI] [PubMed] [Google Scholar]
- 38.Nosten-Bertrand M, Kappeler C, Dinocourt C, Denis C, Germain J, Phan Dinh Tuy F, et al. Epilepsy in Dcx knockout mice associated with discrete lamination defects and enhanced excitability in the hippocampus. PLoS ONE. 2008;3:e2473. doi: 10.1371/journal.pone.0002473. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.