Abstract
Bacterial biofilms are complex surface attached communities of bacteria held together by self-produced polymer matrixs mainly composed of polysaccharides, secreted proteins, and extracellular DNAs. Bacterial biofilm formation is a complex process and can be described in five main phases: (i) reversible attachment phase, where bacteria non-specifically attach to surfaces; (ii) irreversible attachment phase, which involves interaction between bacterial cells and a surface using bacterial adhesins such as fimbriae and lipopolysaccharide (LPS); (iii) production of extracellular polymeric substances (EPS) by the resident bacterial cells; (iv) biofilm maturation phase, in which bacterial cells synthesize and release signaling molecules to sense the presence of each other, conducing to the formation of microcolony and maturation of biofilms; and (v) dispersal/detachment phase, where the bacterial cells depart biofilms and comeback to independent planktonic lifestyle. Biofilm formation is detrimental in healthcare, drinking water distribution systems, food, and marine industries, etc. As a result, current studies have been focused toward control and prevention of biofilms. In an effort to get rid of harmful biofilms, various techniques and approaches have been employed that interfere with bacterial attachment, bacterial communication systems (quorum sensing, QS), and biofilm matrixs. Biofilms, however, also offer beneficial roles in a variety of fields including applications in plant protection, bioremediation, wastewater treatment, and corrosion inhibition amongst others. Development of beneficial biofilms can be promoted through manipulation of adhesion surfaces, QS and environmental conditions. This review describes the events involved in bacterial biofilm formation, lists the negative and positive aspects associated with bacterial biofilms, elaborates the main strategies currently used to regulate establishment of harmful bacterial biofilms as well as certain strategies employed to encourage formation of beneficial bacterial biofilms, and highlights the future perspectives of bacterial biofilms.
Keywords: bacterial biofilm, biofilm formation, biofilm risk, biofilm promotion, regulation strategy
Introduction
It is now understood that about 40–80% of bacterial cells on earth can form biofilms (Flemming and Wuertz, 2019). The formation of biofilms was detrimental in several situations (Donlan and Costerton, 2002; Dobretsov et al., 2006; Coughlan et al., 2016). For example, in food industries, pathogenic bacteria are able to form biofilms inside of processing facilities, leading to food spoilage, and endangering consumer’s health (Galie et al., 2018). In hospital settings, biofilms have also been shown to persist on medical device surfaces and on patient’s tissues causing persistent infections (Dongari-Bagtzoglou, 2008; Percival et al., 2015). In view of the serious impact of biofilms on human health and other aspects, researchers and the public have long focused on prevention and control of the harmful biofilms.
Despite the negative impacts, bacterial biofilms may also have beneficial effects (Rosche et al., 2009). That is, the formation of bacterial biofilms is often important in agricultural and other industrial settings (Bogino et al., 2013; Berlanga and Guerrero, 2016). These beneficial biofilms are currently used as biological control agents against phytopathogens and biofertilizers to enhance crop production (Timmusk et al., 2017), for bioremediation treatment of hazardous pollutants (Irankhah et al., 2019), for wastewater treatment (Ali et al., 2018), for protection of marine ecosystem (Naidoo and Olaniran, 2013), and for prevention of corrosion (Jayaraman et al., 1997; Martinez et al., 2015). Although biofilms can be beneficial to agriculture and industry, people’s understanding of the harmfulside of biofilms has been far better than the benefits for decades. Therefore, the beneficial aspects of biofilms will have great development prospects in the future.
Biofilms are complex surface attached communities of microorganisms held together by self-produced polymer matrixs mainly composed of polysaccharides, secreted proteins, and extracellular DNAs (Tremblay et al., 2013). A biofilm can consist of a single microbial species or a combination of different species of bacteria, protozoa, archaea, algae, filamentous fungi, and yeast that strongly attach to each other and to biotic or abiotic surfaces (Tomaras et al., 2003; Bogino et al., 2013; Silva et al., 2014; Costa-Orlandi et al., 2017; Raghupathi et al., 2017). The ability of microorganisms to develop biofilms has been shown to be an adaptable attribute of microbes (Koczan et al., 2011). The formation of biofilm appears to be an age-old survival mechanism that provides microorganisms with better options compared to their planktonic cells (Dang and Lovell, 2016), including stronger ability to grow in oligotrophic environments (Bowden and Li, 1997), greater access to nutritional resources (Dang and Lovell, 2016), improved survival to biocides (Flemming et al., 2016), enhanced organism productivity and interactions (Roder et al., 2018), as well as greater environmental stability (Dang and Lovell, 2016). It can be seen that biofilms provide protection for bacteria and make them more suitable for the external environment under certain conditions.
Generally, bacterial biofilm formation relies on the interaction between the bacterial cells, the substrates and the surrounding media (Van Houdt and Michiels, 2010). And the formation of bacterial biofilms is a multi-step process starting with reversible attachment to surfaces aided by intermolecular forces and hydrophobicity, and then progress to extracellular polymeric substances (EPS) production which enable the cells to permanently adhere to a surface (Dunne, 2002; Bogino et al., 2013; Caruso et al., 2018). More specially, there are five main phases involved in the biofilm formation process: reversible attachment, irreversible attachment, EPS production, maturation of biofilm, and dispersal/detachment (Stoodley et al., 2002; Toyofuku et al., 2016). However, the expression and regulation mechanisms of different species of bacteria on various phases of biofilms are quite diverse. Before fully understanding the formation process of all bacterial biofilms, researchers still have a long way to walk.
Nowadays, a variety of approaches, which were mostly concerned with interference against bacterial attachment, signal transduction (quorum sensing interference), and disruption of biofilm architecture, have been applied to inhibit formation of harmful biofilms (Chung and Toh, 2014; Galie et al., 2018). In addition, formation of beneficial biofilms can be encouraged through manipulation of adhesion surfaces, quorum sensing (QS) signals and environmental conditions (Upadhyayula and Gadhamshetty, 2010; Renner and Weibel, 2011; Mangwani et al., 2016). Compared with the researches that promote the formation of beneficial biofilms, the investigation on the prevention and control of harmful biofilms is much deeper.
In order to have a comprehensive understanding of bacterial biofilms beyond risk, this review describes the events involved in bacterial biofilm formation, lists the negative and positive aspects associated with bacterial biofilms, elaborates the main strategies currently used to regulate establishment of harmful bacterial biofilms as well as certain strategies employed to encourage formation of beneficial bacterial biofilms, and highlights the future perspectives of bacterial biofilms.
Risks of Bacterial Biofilms
Bacteria are able to colonize and form biofilms on virtually all kinds of surfaces, including natural and synthetic surfaces (Hall-Stoodley et al., 2004; Sweet et al., 2011). Biofilms are responsible for chronic illness and nosocomial infections, industrial pipe fouling, spoilage of foods, contamination of sea food, and dairy products as well as ship hull fouling (Zottola and Sasahara, 1994; Schultz et al., 2011; Abdallah et al., 2014; Khatoon et al., 2018). Therefore, the harmful effects of biofilms on human society are manifold.
Healthcare Issues
In the healthcare settings, biofilms have been shown to develop on medical device surfaces, dead tissues (e.g., sequestra of bones), and inside living tissues (e.g., lung tissue, teeth surfaces; Alav et al., 2018). They may develop on the surface of biomedical devices such as catheters, prosthetic heart valves, pacemakers, breast implants, contact lenses, and cerebrospinal fluid shunts (Table 1; Hall-Stoodley et al., 2004; Wu et al., 2015). Both Gram-positive and Gram-negative bacteria may attach to and develop biofilms on the surfaces of these devices, but the most frequently reported biofilm forming bacteria are Staphylococcus aureus, Staphylococcus epidermidis and Pseudomonas aeruginosa (Hall-Stoodley et al., 2004; Shokouhfard et al., 2015; Khatoon et al., 2018; Pakharukova et al., 2018). It is estimated that about two-thirds of indwelling devices related infections are caused by the staphylococcal species (Khatoon et al., 2018). Bacterial biofilms can also develop in health care water distribution systems. P. aeruginosa can form biofilms on inner surfaces of metal pipes in hospital water system (Loveday et al., 2014). In addition, biofilm forming bacteria contribute to a lot of life-threatening infections and diseases in humans such as cystic fibrosis (CF), otitis media, periodontitis, infective endocarditis (IE), chronic wounds, and osteomyelitis (Southey-Pillig et al., 2005; Akyildiz et al., 2013; Masters et al., 2019). More specially, P. aeruginosa biofilm can cause severe pulmonary infections in patients with CF (Southey-Pillig et al., 2005; Rabin et al., 2015); Haemophilus influenza biofilm is among the causative agents of otitis media (Akyildiz et al., 2013; Bjarnsholt, 2013); Periodontitis, an infection of the gums that damages the soft tissues as well as bones supporting the teeth, is normally caused by the biofilms of Pseudomonas aerobicus and Fusobacterium nucleatum (Jamal et al., 2018); The hypothesis that IE, notoriously difficult to treat, is a biofilm infection explains its resistance to antimicrobials and why surgical disruption and removal of the biofilm improves the chance of cure (Elgharably et al., 2016); P. aeruginosa biofilm is also usually formed on chronic wound (Rabin et al., 2015); and Chronic osteomyelitis is a biofilm infection, where microorganisms adhere to dead bone (Zimmerli and Sendi, 2017). It is believed that biofilm-related organisms account for more than 65% of all microbial infections and exhibit high resistance to antimicrobial agents and components of the host defense system (both innate and adaptive; Jamal et al., 2018; Ciofu and Tolker-Nielsen, 2019). Herein, biofilms have huge impacts on human healthcare.
TABLE 1.
Biofilm forming bacteria on medical devices.
| Medical devices | Biofilm-forming bacteria | References |
| Contact lenses | P. aeruginosa, S. aureus, S. epidermidis, S. saprophyticus, Klebsiella spp. | El-Ganiny et al., 2017 |
| Central venous catheters | Coagulase-negative Staphylococci, S. aureus, Enteric Gram-negative Bacilli | Gominet et al., 2017 |
| Urinary catheters | S. aureus, Enterococcus faecalis, P. aeruginosa | Murugan et al., 2016 |
| Peritoneal dialysis catheters | S. epidermidis, P. acnes, S. warneri, S. lugdunensis, R. mucilaginosa | Pihl et al., 2013 |
| Mechanical heart valves | Streptococcus spp., S. aureus, S. epidermidis, Gram-negative Bacillus, Enterococcus | Jamal et al., 2018 |
| Cerebrospinal fluid shunts | S. aureus, S. epidermidis, Enterococcus faecalis, Enterococcus faecium | Bayston et al., 2012 |
| Breast implants | S. epidermidis, Coagulase-negative Staphylococci, Propionibacterium acnes | Pajkos et al., 2003; Rieger et al., 2013 |
| Orthopaedic implants | S. aureus, S. epidermidis, P. aeruginosa, E. coli, S. haemolyticus | Arciola et al., 2015 |
| Dental implants | Gram-positive cocci, Actinomyces spp., Gram-negative anaerobic oral bacteria | Dhir, 2013; Veerachamy et al., 2014 |
| Voice prostheses | S. aureus, P. aeruginosa, Klebsiella spp., Enterobacterspp., R. dentocariosa, and Proteus spp. | Somogyi-Ganss et al., 2017 |
| Cardiac pacemakers | S. aureus, S. epidermidis | Santos et al., 2011 |
| Intrauterine devices | E. coli, Streptococcus agalactie, S. aureus, Enterococcus faecalis, Lactobacillus spp., Prevotella spp., Porphyromonas spp., Bacteroides, Fusobacterium spp. | Pal et al., 2005 |
| Biliary stents | Pseudomonas, Citrobacter, Klebsiella, Staphylococcus, Enterococcus, Aeromonas, Proteus, Enterobacter | Vaishnavi et al., 2018 |
Plant Diseases
Biofilm-related diseases have also been reported in agricultural settings. Xanthomonas citri biofilm can cause plant diseases like pierce’s disease of grapevines and citrus canker (Ference et al., 2018; Kyrkou et al., 2018). Strong biofilm producer Xylella fastidiosa can also cause pierce’s disease of grapevines by blocking the plant vasculature (Rudrappa et al., 2008; Kyrkou et al., 2018). Biofilms have also been implicated in brown spot disease of bean leaves caused by P. syringae pv. syringae (Monier and Lindow, 2004; Danhorn and Fuqua, 2007). Similarly, P. aeruginosa biofilm on the roots of Arabidopsis and Ocimum basilicum (sweet basil) can cause mortality in a short time. Ralstonia solanacearum, an important plant pathogenic bacterium reported to form biofilms on the surfaces of xylem vessels, cause bacterial wilt disease in plants (Yao and Allen, 2007; Mori et al., 2016). It is becoming obvious that bacteria can form biofilms when colonizing different plant surfaces.
Food Safety and the Food Industry
Within the food industry, biofilms can occur on surfaces contacting with, or without foods (Zottola and Sasahara, 1994; Kumar and Anand, 1998). Biofilms are responsible for about 60% of foodborne outbreaks (Han et al., 2017). Therefore, the presence of biofilms in food processing environments poses significant risk to food safety and the food industry (Galie et al., 2018). In the food processing environments, contaminants mostly come from the surrounding air, equipments, or food surfaces (Kumar and Anand, 1998). Then biofilms growing in food processing environments may lead to spoilage of food, which in turn can cause serious public health risk to consumers and serious economic consequences (Coughlan et al., 2016; Galie et al., 2018). The most common biofilm forming foodborne pathogens and spoilage organisms are Listeria monocytogenes (a ubiquitous species that can cause abortion in pregnant women and other complications in immunocompromised individuals; Galie et al., 2018), Salmonella spp. (a major cause of foodborne diseases which can lead to Reiter’s syndrome or even death; Ajene et al., 2013; Wirtanen and Salo, 2016), Escherichia coli 0157:H7 (a strain which is responsible for hemorrhagic colitis; Wirtanen and Salo, 2016), Pseudomonas spp. (a ubiquitous spoilage organism which produces proteases with negative impacts on foods; Rajmohan et al., 2002), Vibrio parahaemolyticus (its infection most commonly associated with consumption of undercooked seafood; Yeung and Boor, 2004), Clostridium perfringens (a species producing different toxins; Wirtanen and Salo, 2016), Campylobacter jejuni (a major cause of human bacterial gastroenteritis; Wirtanen and Salo, 2016), Bacillus spp. (a species secreting toxins that can cause diarrhea and emetic syndrome; Galie et al., 2018), S. aureus (a species secreting enteric toxins that cause foodborne intoxications; Argudin et al., 2010), Shewanella putrefaciens (a species producing volatile sulfides, amines, and trimethylamine; Bagge et al., 2001), Cronobacter spp. (a genus mostly causing infections in infants and immunocompromised individuals; Wirtanen and Salo, 2016), and Geobacillus stearothermophilus (a common contaminant of dairy products; Table 2; Burgess et al., 2017). These organisms even can establish multi-species biofilms, which are more stable and difficult to control (Bagge et al., 2001; Coughlan et al., 2016; Han et al., 2016; Wirtanen and Salo, 2016; Galie et al., 2018). Biofilms are also responsible for serious technical challenges of food industry in that they may prevent the flow of heat across equipment surfaces, increase the fluid frictional resistance at the surfaces, and promote the corrosion rate of the surfaces, leading to loss of production efficiency (Chmielewski and Frank, 2003; Meesilp and Mesil, 2019). In a word, biofilms have the danger of direct contamination with pathogenic bacteria in the food industries, as well as the risk of contamination of instruments and equipment.
TABLE 2.
Representative of foodborne bacteria that can form biofilms.
| Foodborne bacteria | Growing substrate | Spoiledfood | References |
| Listeria monocytogenes | Wastewater pipes, floors, conveyor belts, rubber seals, elastomers, and stainless steel | Dairy products, melons, coleslaw, ready to eat meat products and ready to eat fish products | Wirtanen and Salo, 2016 |
| Pseudomonas spp. | Conveyor belts, floors, drains, slicing, and milking machine | Dairy products, red meat, and poultry | Korber et al., 2009; Møretrø and Langsrud, 2017 |
| Bacillus cereus | Stainless steel, plastic, soil, and glass wool | Sprouted seeds, fruit juices, fried rice, pasta dishes, meat products, vegetables, and milk products | Korber et al., 2009; Wirtanen and Salo, 2016 |
| Salmonella | Stainless steel, elastomers, concrete, glass, and food surfaces (like lettuce and tomato) | Poultry, pig, cow meats, and dairy products | Wirtanen and Salo, 2016 |
| Escherichia coli | Stainless steel surfaces, food contact surfaces | Dairy products, fermented meat sausage, meat, poultry, fish products, drinks, and vegetables | Wirtanen and Salo, 2016 |
| Clostridium | Multi-species biofilm | Dairy products, fish, cattle meat, poultry, vegetables, honey, and canned food | Wirtanen and Salo, 2016 |
| Cronobacter spp. | Powder service and powder packaging rooms, spray-drying areas, and evaporator rooms | Dairy products, vegetables, grains, bread, herbs, sausages, spices, and meat | Wirtanen and Salo, 2016 |
| Staphylococcus | Stainless steel, plastics (such as polystyrene and polypropylene), and glass | Dairy products, ready to eat meat products, ready to eat fish and seafood products, and ready to eat dairy products | Wirtanen and Salo, 2016 |
Drinking Water Distribution Systems
Biofilms are the predominant mode of microbial growth within the drinking water distribution systems (Mahapatra et al., 2015; Liu et al., 2016). It is well documented that biofilms represent one of the major problems in drinking water distribution systems (Douterelo et al., 2016; Prest et al., 2016). The consumption of contaminated water with pathogenic biofilms has been linked to human infections and waterborne outbreak (Angles et al., 2007; Prest et al., 2016). And the major biofilm producing bacteria in drinking water are P. aeruginosa, Campylobacter jejuni, Legionella pneumophila, Mycobacteria, Aeromonas hydrophila, and Klebsiella pneuminiae (Prest et al., 2016; Chan et al., 2019). Since bacterial cells can attach and develop biofilms on the inner surfaces of piping systems from which cells could be detached into the bulk water, they may cause biocorrosion of pipes, undesirable water quality changes affecting color, taste, turbidity and odors, and reduction of heat exchange efficiency (Prest et al., 2016). More specially, the major biofilm producing bacteria known to promote corrosion of metals are sulfate-reducing bacteria, sulfur-oxidizing bacteria, iron-oxidizers, iron-reducers, and manganese-oxidizers (Kip and van Veen, 2015). All in all, biofilms can affect the safety of drinking water and adversely affect water pipelines.
Marine Biofouling
Marine biofouling portrays the undesirable accumulation of organisms on any natural or man-made objects exposed to seawater (Dobretsov et al., 2013). Common examples of marine substrates include ship hulls and oil or gas installations. Biofouling has been a major challenge in the naval industry and for civilian oceangoing ships (Hopkins and Forrest, 2010; Schultz et al., 2011). Bacteria are among the early microorganisms to settle and colonize substrates in the marine environment and may subsequently facilitate attachment and colonization of larger fouling organisms, such as algae, mussels, and barnacles. Herein, marine biofilms cause biofouling (de Carvalho, 2018). Generally, accumulation of biofoulers by biofilms on ship hulls can increase the hydrodynamic drag of the ships, which causes challenges for shipping industry, including speed reduction, an increase in cleaning time, and greater fuel consumption (Schultz et al., 2011; Demirel et al., 2017). In addition, biofouling of ship hulls has been considered as an important vector for the spread of invasive marine species to new habitats. These transported organisms can adversely affect native species through competition and predation (Minchin and Gollasch, 2003). Therefore, biofilms will affect the cost of ship usage and the balance of marine environment.
Benefits of Bacterial Biofilms
Despite their negative impacts in ecosystems, biofilms have positive effects in agricultural, and other industrial settings (Bogino et al., 2013; Berlanga and Guerrero, 2016). That is, they could be used for plant protection, bioremediation, wastewater treatment, prevention of corrosion, and other useful applications (Table 3; Morikawa, 2006; Singh et al., 2006; Edwards and Kjellerup, 2013; Naidoo and Olaniran, 2013; Singh et al., 2019). As researches progress, the beneficial aspects of biofilms will receive more attention.
TABLE 3.
Examples of beneficial applications of bacterial biofilms.
| Applications | Purposes | References |
| Biofertilizer/biocontrol | Plant growth promotion and protection against phytopathogens | Das et al., 2017 |
| Bioremediation | Transformation of hazardous pollutants to harmless substances | van Dillewijn et al., 2009 |
| Wastewater treatment | Removal of contaminants from wastewater | Yamashita and Yamamoto-Ikemoto, 2014 |
| Microbial fuel cells (MFCs) | Electricity generation, biohydrogen production, and wastewater treatment | Ali et al., 2018 |
| Anticorrosion | Corrosion inhibition for metals | Zuo, 2007 |
| Bioleaching | Extraction of metals from their ores e.g., copper, nickel, cobalt, zinc | Siezen and Wilson, 2009 |
| Biofilm reactor | Production of fermented products and wastewater treatment | Morikawa, 2006 |
| Human gut microbiome | Production of vitamins, degradation of toxic compounds and conversion of complex sugar polymers into short-chain fatty acids | de Vos, 2015 |
Plant Protection Agents
Biofilm formation triggers a number of beneficial effects such as biocontrol and symbiosis. In plants, bacterial biofilms can be formed on the surfaces of leaves, roots, and stems (Morikawa, 2006; Bogino et al., 2013; Schirawski and Perlin, 2018). Biofilm-forming rhizobacteria can act as biocontrol agents due to their successful colonization of plants surfaces (Bais et al., 2004; Vejan et al., 2016). Such rhizobacteria belong to Bacillus, Pseudomonas, Streptomyces, Serratia, and Stenotrophomonas (Arrebola et al., 2019). Beneficial bacteria could also be used as biofertilizers to promote plant growth through nitrogen fixation, mineral nutrient uptake, phytohormone production, and disease suppression as well as protection from both biotic and abiotic stresses (Bais et al., 2004; Khan et al., 2018).
The genus Bacillus consists of important plant-associated strains employed for both biocontrol and plant growth promotion (Morikawa, 2006). For example, Bacillus subtilis is a prominent rhizobacterium which is used as an efficient biocontrol and growth promotion agent to protect plants from bacterial and fungal pathogens due to the formation of robust biofilms and the production of several antagonistic metabolites (Bais et al., 2004; Morikawa, 2006). These metabolites mainly include lipopeptides (such as surfactin, iturin, and fengycins), bacteriocins and siderophores (Meena and Kanwar, 2015; Fira et al., 2018). The colonization by B. subtilis in plant roots is associated with surfactin production and biofilm formation, and the surfactin confers protection of plants from pathogen Pseudomonas syringae infection (Bais et al., 2004; Stein, 2005).
A large number of root-associated Pseudomonas spp. can act as biocontrol agents. They can produce a wide range of antagonistic compounds, including cyclic lypopeptides, pyrrolnitrin, and phenazines, to prevent proliferation of plant pathogens (Arrebola et al., 2019). For example, priming the seeds with phenazine-producing Pseudomonas chlororaphis can provide protection of barley and oats against seed borne diseases (Chin-A-Woeng et al., 2003); Pseudomonas putida 06909 attaches and colonizes the hyphae of citrus root rotting fungus Phytophthora parasitica by feeding on its exudates and then develop a biofilm around the citrus roots, which prevents further proliferation of the fungus (Steddom et al., 2002; Pandin et al., 2017); and Peanut rhizosphere biofilm formation by Paenibacillus polymyxa provides protection of peanut plants against crown root rot disease caused by Aspergillus niger (Haggag and Timmusk, 2008). Rhizhosphere colonization of beneficial biofilms usually offer excellent plant growth promotion and protection against phytopathogens (Das et al., 2017).
Bioremediation
Bioremediation is a process that employs living organisms or their derivatives for treatment of hazardous substances from the environment (soil, water, and air) into lesser or harmless compounds (van Dillewijn et al., 2009). It is thought to be a better option than conventional physical and chemical remediation measures with regard to cost and environmental safety (Singh et al., 2006). Moreover, biofilm-mediated remediation methods exhibit higher efficiency in transforming toxic wastes because of improved bioavailability of the pollutants to degrading organisms and enhanced adaptability of degrading microorganisms to different toxic compounds (Upadhyayula and Gadhamshetty, 2010). The process usually occurs as part of microbial metabolism and relies on the enzymatic attack by microbes to convert environmental pollutants into innocuous products (Karigar and Rao, 2011). Numerous microorganisms are capable of transforming wide varieties of environmental pollutants into non-toxic forms (van Dillewijn et al., 2009). Microbial bioremediation can be at the site of contamination (in situ) or off the place of contamination (ex situ; Kapley and Purohit, 2009). It can be achieved through the incorporation of limiting nutrients and electrons (biostimulation) or by the addition of microbes at the polluted sites (bioaugmentation) to promote the transformation process (Mangwani et al., 2016).
Compared with their planktonic counterparts, microorganisms living in biofilms display greater tolerance to contaminants, higher chance of survival and adaptation as well as stronger abilities to decompose different pollutants through catabolic pathways (van Dillewijn et al., 2009). Biofilm forming bacteria can efficiently be used in the remediation process as cells are encased within a matrix of EPS, which offers protection against several environmental hazards (Mangwani et al., 2016). In addition, biofilms provide an essential habitat which encourages intercellular gene transfer, cellular communication with QS, cohesion and metabolite diffusion as well as bacterial chemotaxis characteristic (Santos et al., 2018).
Biofilm mediated remediation can harbor diverse species of both aerobic and anaerobic bacteria that often use the degradation of pollutants as an energy source (Rodriguez-Martinez et al., 2006). During aerobic degradation, bacteria can use oxygen as final electron acceptor to breakdown toxic contaminants into innocuous products, mainly carbon dioxide, and water (Edwards and Kjellerup, 2013; Azubuike et al., 2016). In anaerobic conditions, electron acceptors such as nitrate and sulfate can play the role of oxygen to transform contaminants into less toxic or harmless substances and the byproduct may depend on the electron acceptor (Rodriguez-Martinez et al., 2006).
Currently, there is an increasing interest in the use of bacterial biofilms mediated remediation for removal of different kinds of environmental pollutants like oil spills, persistent organic pollutants (such as polycyclic aromatic hydrocarbons, polychlorinated biphenyls, and polychlorinated ethenes), heavy metals, dyes, explosives, pesticides, and pharmaceutical products (Edwards and Kjellerup, 2013). Hence, biofilm-mediated bioremediation is employed in the industry for remediation of contaminated soil and groundwater (Edwards and Kjellerup, 2013). Pseudomonas, Dehalococcides, Arthrobacter, Bacillus, Alcanivorax, Cycloclasticus, Burkholderia, and Rhodococcus can remediate these pollutants (Dasgupta et al., 2013; Yoshikawa et al., 2017). It is likely that more and more bacterial biofilms will be applied to bioremediation.
Wastewater Treatment
Nowadays, water contamination caused by industrialization, population growth, and urbanization has become a major global threat (Daud et al., 2017). Wastewater is composed of a broad range of organic and inorganic contaminants originating from storm water, agriculture, industry, domestic, and commercial sewage (Naidoo and Olaniran, 2013). The treatment of wastewater is essential to the protection of aquatic ecosystems and public health (Naidoo and Olaniran, 2013). There are several physicochemical processes for wastewater treatment such as coagulation/flocculation, membrane filtration systems, and electrochemical treatment (Kobya et al., 2009; Francis et al., 2016; Favero et al., 2018). Even though these processes provide effectiveness, they experienced difficulty in removing organic matters in that the main components in the conventional water treatment systems are disinfection and filtration (Hlihor et al., 2017). Bacterial communities have been employed to neutralize and degrade organic and inorganic compounds in wastewater through the use of biofilm-based wastewater treatment technology. Removal of excess nutrients from wastewater is also imperative to avoid aquatic eutrophication which leads to anoxia (Yamashita and Yamamoto-Ikemoto, 2014). The basic nutrients present in wastewater are mostly nitrogen and phosphorous (Yamashita and Yamamoto-Ikemoto, 2014). Hence, among the bacterial species used in wastewater treatment are often denitrifying species or those capable of neutralizing phosphorous (Zielinska et al., 2016).
Biologically active carbon (BAC) process, one of the water treatment biotechnologies, uses granular activated carbon (GAC) as a water filtration media to physically remove water-borne disease causing microorganisms, organic matter and in organic substances (Shirey et al., 2012). After the GAC media particles became exhausted, the rough porous surfaces of this GAC are amenable to colonization of bacteria and formation of bacterial biofilms, which degrade phosphorous and nitrogen-containing compounds, organic carbon as well as other entrapped contaminants in the influent water (Simpson, 2008). Currently, biofilm reactors are developed for wastewater treatment such as membrane reactors, moving beds, fluidized beds, and rotating contactors (Huang et al., 2019).
Biofilms can also be used in bioelectrochemical systems (BESs; Upadhyayula and Gadhamshetty, 2010). BESs are bioreactors that utilize microorganisms as catalysts to convert the energy present in organic wastes into electrical energy (Bajracharya et al., 2016). BESs can facilitate wastewater treatment, bioremediation as well as production of power, fuels and chemicals (Ren et al., 2019). BES electrode surface remodeling has been considered an effective technique to improve the performance of BESs (Ren et al., 2019). Microbial fuel cells (MFCs) are a type of BESs that offer another approach for wastewater treatment in an inexpensive way (Ren et al., 2008; Mei et al., 2017). All sorts of wastewater containing compounds degradable by bacteria can be treated by MFCs, including brewery effluent, petroleum contaminants, domestic wastes, food processing waste, swine manure slurry, landfill leachate, and so on (Franks and Nevin, 2010; Gude, 2016a). MFC uses bacteria in the waste as a biocatalyst to convert the chemical energy present in the wastes to electrical energy using oxidation-reduction reactions (Franks et al., 2010; Angelaalincy et al., 2018). MFCs are primarily made of an anode and a cathode separated by a semi-permeable membrane (Franks et al., 2010). The use of MFCs for wastewater treatment needs a design which permits the passage of wastewater through the cell over the anode surface. Bacterial attachment, colonization and biofilm development occur on the anode surface, the bacteria then oxidize the substrate in wastewater to produce electrons and protons. The electrons released during oxidation flow to the cathode via an electrical circuit to generate current (Ali et al., 2018; Singh et al., 2019). At the cathode, electron acceptors (usually oxygen) react with protons, and electrons to generate water vapor-like reduced compounds (Cao et al., 2019). Most of the MFCs configurations can achieve chemical oxygen demand (COD) removal efficiencies at wastewater treatment (Liu et al., 2004; Min et al., 2005; Gude, 2016b). Liu et al. (2004) were the first to initiate the application of MFCs, which reached 80% of COD removal efficiency from real domestic wastewater with a maximum electrical power generation of 26 mW/m2 using a single-chamber MFC. Min et al. (2005) had demonstrated that COD and ammoniacal nitrogen (NH+4-N) removal are 86% and 83% with a maximum power output of 45 mW/m2 when a swine wastewater is treated with a dual-chambered MFC.
Prevention and Control of Corrosion
Corrosion has now been widely acknowledged as a big problem in drinking water distribution systems, medical, marine, and food processing industry (Prest et al., 2016; Jia et al., 2017; Guo et al., 2018). Both chemical and biological factors can accelerate the rate of corrosion (Kip and van Veen, 2015). Obviously, the activities of microbes on surfaces of metallic materials can either inhibit or promote corrosion (Zuo, 2007). Different strategies, including protective coatings, biocides, cathodic protection and corrosion inhibitors, have been developed to prevent corrosion (Zuo, 2007). However, more recently there has been increased interest in the use of beneficial bacterial biofilms to prevent corrosion because of their effectiveness, cost effective and nature friendly behavior (Zuo, 2007; Guo et al., 2018). The potential strategies may involve: (i) removal of corrosive substances such as oxygen by aerobic bacteria through respiration; (ii) inactivation of corrosive inducing bacteria like sulfate reducing bacteria by inhibitory antimicrobial compounds secreted within biofilms; (iii) production of protective coats such as γ-polyglutamate by biofilms; and (iv) biofilms formation serving as a diffusion barrier to hinder dissolution of metals (Zuo, 2007; Guo et al., 2018). A gramicidin-S-producing Bacillus brevis biofilm has been reported to curtail the rate of corrosion in mild steel by suppressing the growth of sulfate-reducing bacterium Desulfosporosinus orientis and the iron-oxidizing bacterium Leptothrix discophora SP-6 (Zuo et al., 2004). Also, the antimicrobial compounds indolicidin, bactenecin and probactenecin produced by genetically engineered B. subtilis biofilm can suppress metal corrosion by inhibiting the growth of D. vulgaris and D. gigas (sulfate-reducing bacteria; Zuo, 2007). Although, both aerobic and anaerobic biofilms are able to reduce corrosion rates on the surfaces of different materials, the aerobic biofilms remarkably suppress metal corrosion, which suggests that oxygen consumption can further enhance corrosion protection (Kip and van Veen, 2015). Anticorrosive approach via beneficial biofilms has been successfully reported for stainless steel, carbon steel, copper, and aluminum (Guo et al., 2018). The use of bacterial biofilms for prevention and control of corrosion is a relatively new direction and deserves special attention.
Biofilm Formation Process
Bacteria form biofilms in response to environmental stresses such as UV radiation, desiccation, limited nutrients, extreme pH, extreme temperature, high salt concentrations, high pressure, and antimicrobial agents. Herein, the events leading to bacterial biofilm formation are complex (O’Toole et al., 2000; Hall-Stoodley et al., 2004; Lopez et al., 2010; Galie et al., 2018). It is generally believed that biofilm formation starts with a reversible attachment of bacteria onto a surface, followed by the irreversible attachment, usually aided by adhesive structures of bacteria and short-range interactions. Their reversible attachment is progressed through the production of EPS. Later, they develop into an organized structure entrapped in an EPS matrix. Finally, bacterial cells can escape from the mature biofilm and disperse into the environment to colonize new niches (Berne et al., 2015; Hoffman et al., 2015; Limoli et al., 2015; Toyofuku et al., 2016). These phases of biofilm formation are illustrated in Figure 1. Five main phases leading to the development of free-living planktonic life form into a sedentary “biofilm” lifestyle are discussed below.
FIGURE 1.
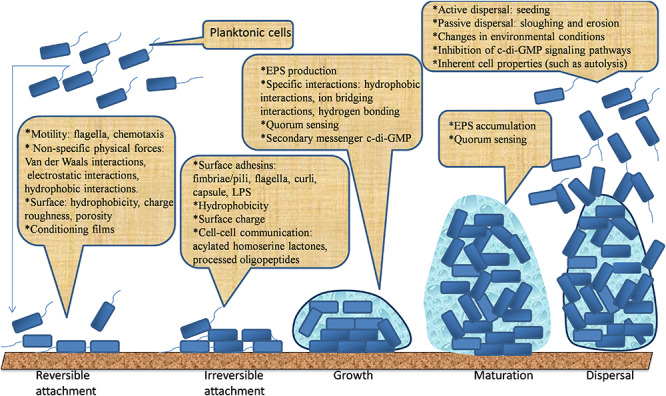
The five main phases leading to the development and dispersal of biofilm.
Reversible Attachment
Bacterial attachment is the initial step of biofilm formation. It begins with the favorable interaction between a few planktonic cells and substrate surfaces. The bacteria must be transported to the surfaces by Brownian motion, sedimentation, or convection (Palmer et al., 2007). Chemotaxis is the directed movement of bacterial cells toward a nutrient source or chemoattractants (e.g., amino acids and sugars) along a concentration gradient in mobile fluids. It occurs in virtually all microorganisms and can facilitate bacterial growth on surfaces by enabling cell-surface interactions (Vladimirov and Sourjik, 2009; Porter et al., 2011). Once the cells reach a surface, the interaction between the cell surfaces and the conditioned surface depends on the net sum of repulsive or attractive forces generated between the two surfaces. If the attractive forces are greater than the repulsive forces, the bacteria will attach to the surface and vice versa (Dunne, 2002; Carniello et al., 2018). This initial attachment is achieved through the effects of non-specific physical forces such as electrostatic forces, hydrophobic interactions and Lifshitz–van der Waals interactions (Dunne, 2002; Carniello et al., 2018). Bacterial attachment has been interpreted within the scope of the classical Derjaguin, Verwey, Landau, and Overbeek (DVLO) DVLO theory, the extended DVLO model, and the thermodynamic approaches (Perni et al., 2014; Zhang et al., 2015; Carniello et al., 2018). These theories describe attachment as the result of a balance between attractive Lifshitz–van der Waals interactions and repulsive forces, based upon electrostatic forces (Morra and Cassinelli, 1997; Rijnaarts et al., 1999), in addition to hydration forces (Jucker et al., 1998; Hermansson, 1999). In general, the reversible bacterial attachment to a surface involves deposition of a bacteria to a substrate in such a way that the bacteria remain in a two dimensional Brownian motion and can be easily detached from the surface by either bacterial mobility or shearing effects of a fluid flowing over the surface (Li and Tang, 2009; Carniello et al., 2018).
Both inert and biological surfaces can be used for initial bacterial attachment. In fact, any substance coming into contact with bacterial suspension is considered to be a substrate for biofilm growth (Donlan, 2002; Tuson and Weibel, 2013). The physicochemical properties of a substratum surface can affect bacterial attachment and how quickly biofilms develop, including surface roughness, hydrophobicity, surface charge, and presence of conditioning films (Donlan, 2002; Srey et al., 2013).
The relationship between bacterial attachment and surface roughness has been reported for years. However, opinion is divided regarding the effect of roughness on bacterial attachment and biofilm formation. Some studies revealed that the irregularities of abiotic surfaces promote bacterial attachment and biofilm development due to lower shear forces and larger surface area to which bacterial cells can attach on rougher surfaces (Pedersen, 1990; Bollen et al., 1997; Donlan, 2002; Yu et al., 2016), whereas a contradictory result showed that surface roughness had no influence on bacterial attachment (Vanhaecke et al., 1990; Flint et al., 2000; Zhao et al., 2014). The opposite results may be due to different extracellular structures and physicochemical properties of different bacteria as well as the diverse physicochemical properties of a substratum surface with varied hydrophobicity, surface charge and conditioning films.
Surface hydrophobicity, the strongest long range non-covalent interactions in biological systems, has been thought to play an important role in bacterial attachment. Hydrophobic surfaces seem to be easier for bacteria to colonize than hydrophilic materials (Teixeira and Oliveira, 1999; Donlan and Costerton, 2002; Sousa et al., 2011). This is probably because hydrophobicity reduces repulsive forces between the bacterial surface and colonization substratum. Yu et al. (2016) attributed hydrophobicity and surface roughness of substratum to the early attachment of Streptococcus mutans. Teixeira and Oliveira (1999) also reported the positive correlation between the degree of hydrophobicity of polymeric substrate materials and the number of attached Alcaligenes denetrificans. A notable exception, however, is that L. monocytogenes is likely to attach to hydrophilic substrates such as stainless steel than hydrophobic surfaces like polytetrafluoroethylene (PTFE; Chavant et al., 2002). This might be due to the fact that attachment of bacterial cells is also influenced by bacterial surface hydrophobicity, which in turn depends on bacterial growth rate, bacterial species, and growth medium (Vacheethasanee et al., 1998; Katsikogianni and Missirlis, 2004). Vacheethasanee et al. (1998) observed that S. epidermidis strains with higher surface hydrophobicity attached to a greater extent than the ones with less surface hydrophobicity to polyethylene (PE). Studies have shown that hydrophobicity affects attachment of spores to surfaces, and that the more hydrophobic a surface or bacterium, the stronger the attachment (Husmark and Rönner, 1992; Faille et al., 2002). Husmark and Rönner (1992) reported that the hydrophobicity of B. cereus spores and hair like appendages surrounding the spores influenced attachment to inert surfaces. Hydrophobicity of bacteria can be determined by bacterial adherence to hydrocarbons (BATH) also currently known as microbial adherence to hydrocarbons (MATH), hydrophobic interaction chromatography (HIC), and contact angle measurements (Ukuku and Fett, 2002; Palmer et al., 2007). The choice of bacteria to attach to hydrophobic or hydrophilic surfaces depends on the structures and complex physiological and biochemical characteristics of both bacteria and their contacting surfaces.
Surface charge is another physical factor that affects the adhesion of bacteria to substratum. It is widely believed that most bacterial cells have a net negative surface charge due to the presence of considerable amount of carboxyl, amino, and phosphate groups (Dziubakiewicz et al., 2013). Thus, surface that is positively charged promotes bacterial attachment while a negatively charged surface will encourage resistance to bacterial attachment (Tuson and Weibel, 2013). It should be further noted that the surface charge of bacteria differs between bacterial species and is influenced by growth medium, bacterial age, pH, and ionic strength (Katsikogianni and Missirlis, 2004). It is often describes by the zeta potential (Palmer et al., 2007). Studies investigating the influence of surface charge on the adhesion ability of E. coli to inert surfaces have shown positive relationship in some cases (Dickson and Koohmaraie, 1989; Ukuku and Fett, 2002) and no correlation in the other (Rivas et al., 2007). The discrepancies in these studies could be due to the employed methods, which utilized different growth media and buffers, to demine bacterial surface charge. Similarly, QS in E. coli causes an increase in the negative charge on cell surfaces, which in turn promote the association of bacteria with surfaces during the early phases of biofilm formation (Tuson and Weibel, 2013). Electrostatic interaction chromatography (ESIC) has been widely used to measure bacterial surface charge (Ukuku and Fett, 2002).
Nearly all bacteria moving from liquid media toward surfaces make their first contact with conditioning films. The films are essential in the bacterial adhesion process and are formed as a result of adsorption of nutrient molecules onto the material surfaces which lead to changes in physicochemical characteristics of the surfaces and in turn affect the bacterial attachment (Lorite et al., 2011). These films are formed within minutes of exposure with concomitant growth for several hours (Donlan, 2002).
Irreversible Adhesion
The irreversible attachment is attained through the effects of short range interactions such as dipole-dipole interactions, hydrogen, ionic and covalent bonding, and hydrophobic interactions with involvement of bacterial structural adhesions (Bos et al., 1999). The surface of bacteria is gifted with different adhesins that are projected away from the cell surface into the extracellular environment (Berne et al., 2015). So far, adhesive structures of bacteria, including flagella, pili/fimbriae, and non-fimbrial adhesions, were identified to be involved in the development of biofilms (Berne et al., 2015). The presence of these surface organelles help bacterial cells to make first physical contact with substrates (Petrova et al., 2012; Berne et al., 2015; Carniello et al., 2018). Flagellum is a whip like filamentous appendage concerned with bacterial locomotion (Haiko and Westerlund-Wikstrom, 2013). Flagella driven motility can either be swimming (in liquids) or swarming (on solid moist surfaces). Various species of bacteria exhibit both type of movements to navigate bacterial cells toward a favorable environment and to attach onto a surface (Kearns, 2010; Hintsche et al., 2017). Numerous studies have reported the importance of flagella mediated motility in early attachment and subsequent biofilm formation. Flagella can initiate the adhesion of cells to surfaces by overcoming the repulsive forces that might hinder cell to surface interactions (Van Houdt and Michiels, 2005; Terashima et al., 2008; Lemon et al., 2007; Haiko and Westerlund-Wikstrom, 2013; Wood, 2013). Non-flagellated mutants of L. monocytogenes were impaired in surface adhesion compared to the wild type with short incubation periods. However, with longer times of incubation, surface coverage by non-flagellated mutant cells almost reach the same level as flagellated cells, suggesting that the presence of flagella is crucial for initial and early attachment (Vatanyoopaisarn et al., 2000).
Pili/fimbriae are also filamentous appendages used for bacterial attachment to each other and early cell-surface attachment (Konto-Ghiorghi et al., 2009; Maldarelli et al., 2016). For example, P. aeruginosa can employ a pilus mediated form of bacterial surface movement called twitching motility (Alarcon et al., 2009). In K. pneumoniae, Streptococcus agalactiae, Clostridium difficile, and Acinetobacter baumannii, pili play important roles in their early attachment to surfaces (Konto-Ghiorghi et al., 2009; Maldarelli et al., 2016; Pakharukova et al., 2018). Type 1 and type 3 fimbriae on the surface of K. pneumoniae facilitate attachment on abiotic surfaces and formation of mature biofilm, while only type 1 fimbriae initiate attachment of E. amylovora on abiotic surfaces and biofilm formation (Di Martino et al., 2003; Murphy et al., 2013). And the wild type of E. amylovora attached in greater numbers to surfaces than the mutant type with a deletion in type I fimbriae, which suggests the importance of adhesion structures in the formation of mature biofilms (Koczan et al., 2011). Additionally, thin aggregative fimbriae, also called curli fimbriae and antigen 43, are found to enhance initial surface attachment of bacteria (Heras et al., 2014; Carter et al., 2016). Moreover, distinct adhesins in some bacteria might be used to mediate transition from transient to permanent surface attachment. For example, formation of the monolayer in Caulobacter crescentus is mediated by a strong adhesive polysaccharide called the holdfast (Karatan and Watnick, 2009). Another example is polysaccharide intercellular adhesin (PIA) produced by S. epidermidis that is essential for cell to cell attachment and subsequent biofilm development (Rohde et al., 2010).
Bacterial pathogens also generate special adhesins that enable them not only adhere to receptors on eukaryotic cell surface but also facilitate their internalization. For instance, Yersinia pseudotuberculosis and Yersinia enterocolitica produce a protein invasin which adheres to β1 integrins on the surface of M-cells and causes crossing of Yersinia into M-cells (Bonazzi et al., 2009; Karatan and Watnick, 2009).
A cell-to-cell signaling mechanism called QS also coordinate individual cells to initiate formation of bacterial biofilms (Abraham, 2016). Using QS, bacteria synthesize and release first messengers like chemical signals (autoinducers, AIs) to enable cell-to-cell communication within bacterial population (Li and Tian, 2012; Papenfort and Bassler, 2016). Both Gram-negative and Gram-positive bacteria employ cell-to-cell signaling mechanisms to regulate biofilm formation. Gram-negative bacteria primarily used acyl homoserine lactones (AHLs), whereas Gram-positive bacteria used oligopeptides, universal AIs that can be utilized by both Gram-negative and Gram-positive bacteria (Miller and Bassler, 2001; Sperandio et al., 2001; Sun et al., 2004).
EPS Production
Irreversible adhesion is progressed through the production of EPS regulated by QS of the resident bacterial cells. Bacteria synthesize and secrete EPSs which are an essential component of biofilm extracellular matrix. EPS can mediate both cohesion of bacteria and adhesion of biofilms to surfaces via hydrophobic interactions and ion bridging interactions (Fahs et al., 2014; Costa et al., 2018). Overall, EPS plays critical roles in adherence to surfaces, cell–cell recognition, biofilm formation, biofilm structure, retention of water, signaling, protection of cells, symbiosis with plants, trap of nutrients, and genetic exchange (Dogsa et al., 2005; Limoli et al., 2015; Flemming, 2016; Costa et al., 2018). In addition, secondary messenger c-di-GMP is regarded as one of the stimuli for the transition from reversible to irreversible adhesion through the production of EPS and cell surface structures (Toyofuku et al., 2016).
The main constituents of EPS, including polysaccharides, proteins, DNAs, lipids and other polymeric compounds, depend on the bacterial species, and the environmental conditions (Myszka and Czaczyk, 2009; Kostakioti et al., 2013; Limoli et al., 2015; Jayathilake et al., 2017; Bacosa et al., 2018; Costa et al., 2018). Polysaccharides are a major constituent of the EPS matrix and necessary for biofilm development and growth in most bacteria (Flemming et al., 2016). In Gram-negative bacteria, the polysaccharides are usually neutral or polyanionic. The anionic property is considered to be as a result of the presence of uronic acids or ketal-linked pyruvates. This is thought to facilitates association of divalent cations such as magnesium and calcium, which are very important for cross-linking of polymer strands leading to greater binding force in a developed biofilm formation (Donlan, 2002). In Gram-positive bacteria such as Staphylococci, however, the EPS is mainly cationic (Donlan, 2002). EPS matrix also contains considerable amounts of proteins such as enzymes and proteinaceous structures like pili and fimbriae. Besides, DNA is an integral part of EPS matrix which acts as an intercellular connector (Flemming et al., 2016). Lipids found in the matrix also play important roles for the attachment of Thiobacillus ferrooxidans (Flemming et al., 2016).
Biofilm Maturation
At this phase, the genetic machineries of EPS such as a 15 gene-long epsA-O cluster concerning biofilm formation in Bacillus subtilis become activated when intensity of the AIs exceed certain threshold. Bacteria continue to multiply within embedded EPS matrix by using the AIs signals, and conduct to formation of microcolonies and maturation of biofilms (Lopez et al., 2010; Toyofuku et al., 2016). Following microcolony formation and EPS accumulation, changes in gene expressions are induced, and the products of these genes are utilized for the production of EPS that act as biological “glue” between embedded bacterial cells (Frederick et al., 2011; Karimi et al., 2015). The formation of matrix is followed by formation of water-filled channels which act like circulatory systems, conveying nutrients to the cells communities and removing unwanted products (Garnett and Matthews, 2012). Structural analysis of the microcolonies often shows a pyramid/mushroom-shaped multicellular structure (Garnett and Matthews, 2012). During the process of maturation, motility is restricted within the microcolonies as the production of bacterial surface structures is inhibited, and the gene expression pattern of the sessile cells differs significantly from the planktonic cells. For example, more than 57 biofilm associated proteins, that were not present in the planktonic cells, have been detected in P. aeruginosa microcolony (Hall-Stoodley and Stoodley, 2002). Moreover, QS enables communication among bacteria of the same or different species through secretion and detection of AIs. Bacteria use these signaling molecules to sense the presence of each other and to regulate gene expression in response to changes of their population density (Kaplan, 2010; Guttenplan and Kearns, 2013; Wei and Ma, 2013; Berlanga and Guerrero, 2016). Herein, AIs have an important role in maintaining existing biofilms.
Dispersal/Detachment
The biofilm detachment process, also known as dispersal, represents the terminal process of biofilm development. It is regarded as a strategy of bacterial cells to leave biofilms and continue another biofilm life cycle (Singh et al., 2017). That is, dispersal of surface attached cells from biofilms is a naturally program phenomenon which allows bacterial cells to form new microcolonies on other fresh substrates in response to particular physiological or environmental conditions (Diaz-Salazar et al., 2017). Dispersal is a complex process regulated by environmental signals, signal transduction pathways, and effectors (Kaplan, 2010).
Although the dispersal mechanisms vary among bacteria, the whole process can still be divided into three common stages: detachment of cells from the microcolonies, movement of cell to a fresh substrate, and adhesion of the cells to the new substrate (Kaplan, 2010; Shen et al., 2018). Furthermore, the detachment can be an active action (i.e., seeding) that cells in biofilms initiate the detachment of themselves in response to changes in their environment such as antimicrobial stress, matrix-degrading enzymes and nutrient starvation, or passive behaviors (i.e., sloughing and erosion) mediated by external forces such as shear forces (Kaplan, 2010; Fleming and Rumbaugh, 2017; Lee and Yoon, 2017). In other words, seeding dispersal is the active detachment mechanism associated with rapid release of microcolonies or planktonic cells from the center of the biofilm, leaving an empty hollow cavity; Sloughing is the sudden detachment of a large portion of a biofilm; Erosion is a release of small portion bacteria from the biofilm. Aggregatibacter actinomycetemcomitans, P. aeruginosa, Serratia marcescens, and S. aureus can exhibit seeding dispersal of biofilms (Kaplan, 2010; Lee and Yoon, 2017).
During active dispersal, genes involved in cell motility, such as flagella synthesis and EPS degradation are usually up-regulated, while genes related to EPS production (i.e., polysaccharide synthesis), attachment, and fimbriae synthesis are often down-regulated (Kostakioti et al., 2013). Another effective way to disperse biofilm is to inhibit the c-di-GMP signaling pathways because reduction of intracellular c-di-GMP levels will either inhibit biofilm development or enhance biofilm dispersal (Kaplan, 2010). Furthermore, environmental factors like temperature change, pH, nutrients, and oxygen deficiency can contribute to biofilm dispersal (Kostakioti et al., 2013). For example, limited oxygen supply facilitates biofilm detachment by promoting c-di-GMP degradation. An increase in glucose supply can decrease intracellular c-di-GMP, resulting in the raise of flagella synthesis that eases detachment process (Lee and Yoon, 2017). Moreover, there are various physicochemical parameters and inherent cell properties such as autolysis that facilitate biofilm dispersal (Kaplan, 2010; Kostakioti et al., 2013; Lee and Yoon, 2017).
Regulating Approaches for Bacterial Biofilms
Unlike the planktonic bacteria, biofilms are not effectively eliminated by ordinary cleaning, washing and disinfection methods (LeChevallier et al., 1988; Somers and Wong, 2004). The formation of biofilm, however, can also play beneficial roles (Morikawa, 2006; Singh et al., 2006; Edwards and Kjellerup, 2013; Naidoo and Olaniran, 2013; Singh et al., 2019). Therefore, multiple factors have also been explored to promote formation of beneficial biofilms (Ansari et al., 2012). Herein, there are different strategies developed to prevent, control or promote bacterial biofilm development, which are closely related to the regulation of bacterial attachment, signal transduction (quorum sensing interference), and bacterial biofilm matrix (Table 4; Chung and Toh, 2014).
TABLE 4.
The regulating approaches for bacterial biofilms.
| Strategy | Mechanism | References |
| 1. Prevention and control as well as promotion of bacterial attachment | ||
| 1.1 Antifouling surfaces | ||
| Poly ethylene glycol (PEG) | Bacteria repelling coatings | Roosjen et al., 2003; Roosjen et al., 2005 |
| 1.2 Antimicrobial surfaces | ||
| Silver | Antimicrobial releasing coatings | Bazaka et al., 2012; Francolini et al., 2017 |
| quaternary ammoniumcompounds (QACs) | Contact killing coatings | Hasan et al., 2013; Achinas et al., 2019 |
| 1.3 Small molecules | ||
| aryl rhodanines | Anti-adhesion | Cegelski et al., 2009; Chung and Toh, 2014 |
| Pilicides and curlicides | Anti-adhesion | Cegelski et al., 2009; Chorell et al., 2012 |
| 1.4 Surface modification | ||
| Oxygen plasma on carbon based materials | Promotion of bacterial attachment, biofilm formation and electricity generation in BESs | Flexer et al., 2013 |
| Nitrogen plasma on carbon anode | Promotion of biofilm formation and electricity production in MFCs | He et al., 2012 |
| Polyethylene membrane (PE) modified with positively charged graft polymer chains (diethylamino) | High adhesiveness for nitrifying bacteria than original unmodified membrane and rapiddevelopment of nitrifying biofilms | Hibiya et al., 2000 |
| Methoxy-PEG-amine (-PEG-NH2) modification on a rough PP surface and the smooth PE surface | Enhancement in biofilm formation | Lackner et al., 2009 |
| 2. Control or promotion of bacterial signal transduction (quorum sensing interference) | ||
| 2.1 Quorum quenchers (QQs) | ||
| Enzymes includinglactonase, acylase, oxidoreductase, and paraoxonase | Enzymatic degradation of signal molecules | Sadekuzzaman et al., 2015 |
| 2.2 Quorum sensing inhibitors (QSIs) | ||
| N-octanoyl-L-HSL (C8-HSL) | Inhibition of the synthesis of signal molecules | Hirakawa and Tomita, 2013 |
| 2.3 Natural agents | ||
| Furanone, ajoene, naringin, musaceae, andcurcumin | Prevention of bacterial biofilm | Ponnusamy et al., 2010; Musthafa et al., 2010; Jakobsen et al., 2012; Truchado et al., 2012; Packiavathy et al., 2014 |
| Honey | Restriction to biofilm development | Sharahi et al., 2019 |
| 2.4 AIs and QS genes | ||
| 10 μM acyl homoserine lactones | Encouragement of beneficial biofilm formation | Chen et al., 2017 |
| 100 μM quinolone | Enhancement in biofilm mass | Chen et al., 2017 |
| increased expression of QS genes lasI and rhlI | Improvement of biofilm formation and EPS production | Mangwani et al., 2016 |
| 3. Disruption of bacterial biofilm matrix | ||
| 3.1 Matrix targeting enzymes | ||
| DNase I, restriction endonucleases, glycoside hydrolases, proteases, and dispersin B | EPS degradation | Kaplan, 2014; Parrino et al., 2019 |
| 3.2 Bacteriophages | ||
| phage SAP-26 | EPS degradation | Lu and Collins, 2007 |
| 3.3 Small molecules | ||
| Cis-2 decenoic acid (C2DA) | Biofilm dispersal | Jennings et al., 2012; Chung and Toh, 2014 |
Prevention and Control as Well as Promotion of Bacterial Attachment
Inhibition of cell attachment is an ideal approach to prevent biofilm formation at an initial phase. Therefore, remodeling the surface or coating the surface with the substances that do not encourage the bacterial adhesion could probably impede establishment of bacterial biofilm (Rogers et al., 1994; Chung and Toh, 2014).
Antibiofilm surfaces can mainly be divided into antifouling surfaces and antibacterial ones. The former prevents bacterial attachment onto the surfaces while the latter kills bacteria on the surfaces (Xu et al., 2005; Li et al., 2018). Coating agents and paints such as silver, titanium oxide, grapheme, arsenic, mercury oxide, copper oxide, and zinc oxide nanoparticles have been developed and used effectively as antifoulants (Kuang et al., 2018). Recently, poly(ethylene glycol) (PEG) has been the most widely used antifouling coatings in the marine and biomedical industries (Zhang et al., 2017). Surfaces covered with PEG have been shown to resist the adhesion of bacteria, because of the hydrophilic surface property. PEG coatings are able to repel quite a number of bacterial species like S. aureus, S. epidermidis, P. aeruginosa, and E. coli (Roosjen et al., 2003; Roosjen et al., 2005). Antibacterial surfaces are designed for indwelling medical devices (e.g., catheters and endotracheal tube), which can be colonized by biofilm forming bacteria, to release antibiotics, bacteriocins, metal ions, plant extracts or nanoparticles against pathogens such as S. aureus, Candida albicans, P. aeruginosa, and E. faecalis (Dror et al., 2009; Chung and Toh, 2014; Sanchez et al., 2016; Kuang et al., 2018; Vasilev et al., 2018). Inhibition of biofilm formation can be achieved on medical device surfaces through coating with silver (Bazaka et al., 2012; Francolini et al., 2017). The mechanism of action of silver-based materials is mainly related with the release of silver ion (Ag+) from the surface, and the required amount for an optimal effect ranging from 10 μM to 10 μM (Bazaka et al., 2012; Francolini et al., 2017). Similarly, quaternary ammonium compounds (QACs) are widely used as antibacterial agents for contact killing coatings. Contrary to the antibiotic-release mechanism of silver ions, QACs coatings have a long-lasting contact based antimicrobial mechanism (Hasan et al., 2013; Achinas et al., 2019). Unfortunately, contact killing surfaces have the drawback that some microorganisms are able to develop resistance against these surfaces (Hasan et al., 2013). What’s more, small molecules like aryl rhodanines can prevent the early phases of biofilms formed by Gram-positive pathogens by inhibiting bacterial adhesion to surfaces (Cegelski et al., 2009; Chung and Toh, 2014). And small synthetic compounds pillicides and curlicides can interfere with bacterial adhesion by inhibiting production of bacterial pili/fimbriae and curli (Cegelski et al., 2009). Natural products honey and tea can also inhibit bacterial attachment (Kuang et al., 2018; Sharahi et al., 2019).
On the other hand, attachment of beneficial bacteria can be promoted by surface modification of materials, which includes both physical and chemical based modification, or electrochemical oxidation treatment (Berlowska et al., 2013; Flexer et al., 2013; Kang et al., 2014).
The surface characteristics can be designed and altered to enhance bacterial attachment and formation of beneficial biofilms for BESs and yeast fermentation industry (Upadhyayula and Gadhamshetty, 2010; Berlowska et al., 2013). The use of nitrogen or oxygen plasma on carbon based materials such as graphite electrodes has been shown to increase surface energy and hydrophilicity which in turn promotes bacterial attachment, biofilm formation and electricity generation in BESs (Flexer et al., 2013). Also, the use of nitrogen plasma on carbon anode can alter surface roughness and hydrophobicity to promote biofilm formation and electricity production in MFCs (He et al., 2012). In addition, carbon felt electrodes treated with UV/O3 can enhance Shewanella oneidensis MR-1 attachment and biofilm formation, leading to increased electron transfer rate and greater current density production in MFCs (Cornejo et al., 2015).
The conversion of ammonium to nitrate (nitrification) is an essential process in wastewater treatment (Lackner et al., 2009). However, the organisms responsible for nitrification have very low growth rates and do not form strong biofilms (Lackner et al., 2009). Hence, efforts need to be made to maintain these nitrifiers in reactor systems (Busscher et al., 1995). It has been proposed that the bond between the attaching bacteria and the materials surface is determinant on biofilm strength and shear resistance (Busscher et al., 1995). Various approaches have been used to promote attachment of nitrifying bacteria onto a membrane surface (Hibiya et al., 2000; Terada et al., 2004; Lackner et al., 2009). Hibiya et al. (2000) observed that the PE membrane whose surface is modified with positively charged graft polymer chains (diethylamino) exhibited a high adhesiveness for nitrifying bacteria than original unmodified membrane, and nitrifying biofilms develop rapidly. Lackner et al. (2009) modified PE and polypropylene (PP) membranes to improve the attachment and shear resistance of nitrifying biofilms. They used a combination of plasma polymerization and wet chemistry to introduce chains of PEG containing two different functional groups (-PEG-NH2 and -PEG-CH3) to the membrane surfaces. They demonstrated that the methoxy-PEG-amine (-PEG-NH2) modification on a rough PP surface and the smooth PE surface had a clear enhancement in biofilm formation. The amino group of methoxy-PEG-amine acts as an attractive force for nitrifiers like Nitrosomonas europea and Nitrobacter winogradskyi, which enhanced formation of biofilm.
Control or Promotion of Bacterial Signal Transduction (Quorum Sensing Interference)
Bacterial QS depends upon a series of events such as production of signal, signal dissemination, signal receptors, signal detection, gene expression, and signaling response. Therefore, quorum quenchers (QQs), or quorum sensing inhibitors (QSIs) that interfere with these processes might potentially inhibit bacterial QS and ultimately biofilm formation (Li and Tian, 2012; Remy et al., 2018). The quorum-quenching approach employing quorum quenching enzymes to inactivate quorum sensing signals is important in healthcare settings and medicine as well as industrial membrane bioreactors, crop production and aquaculture (Fong et al., 2018). Quorum quenching enzymes, lactonase, acylase, oxidoreductase, and paraoxonase, have been discovered in various species of bacteria (Chen et al., 2013). The well-known mechanism of action of QQs is the inactivation of acyl homoserine lactone molecules (Sadekuzzaman et al., 2015). Another mechanism is the inhibition of the synthesis of signal molecules (e.g., AHLs) by QSIs such as N-octanoyl-L-HSL (C8-HSL) that prevents the enzymatic activity of Lux operon proteins (Hirakawa and Tomita, 2013). The natural QSIs known to prevent bacterial biofilm mainly include furanone (Ponnusamy et al., 2010), ajoene (Jakobsen et al., 2012), naringin (Truchado et al., 2012), musaceae (Musthafa et al., 2010), and curcumin (Packiavathy et al., 2014). Moreover, a natural ingredient honey at elevated amount can interfere with genes involved in bacterial communications such as AI-2 and LsrA, thereby limiting biofilm development (Sharahi et al., 2019). Furthermore, the presence of a secondary messenger c-di-GMP in elevated amount promotes biofilm formation in bacteria. Therefore, inhibiting the c-di-GMP pathway may be effective method to prevent biofilm formation (Sharahi et al., 2019).
On the other hand, genetic engineering of AIs can encourage beneficial biofilm formation, which plays critical roles in power generation, wastewater treatment and bioremediation (Mangwani et al., 2016; Chen et al., 2017). For example, a decrease in start-up time from 10 days to 4 days in dual chamber MFC was achieved by adding a kind of AIs (10 μM AHLs; Chen et al., 2017). An AI (100 μM quinolone) enhances biofilm mass of extremophile Halanaerobium praevalence on the anode of MFC, leading to effective treatment of high salinity wastewater and improved power generation (Chen et al., 2017). Furthermore, QS bacteria can degrade a broad range of pollutants (Mangwani et al., 2015). The marine P. aeruginosa N6P6 biofilm formation and EPS production can be improved by increasing the expression of QS genes lasI and rhlI, which contributes to increase rate of polycyclic aromatic hydrocarbon degradation (Mangwani et al., 2016).
Disruption of Bacterial Biofilm Matrix
To disperse bacterial biofilms, it’s essential to destroy the structural components of EPS (Flemming and Wingender, 2010; Wei and Ma, 2013). Thus, degradation of the EPS matrix can be effective method to interfere with bacterial biofilm formation.
EPS matrix-degrading enzymes, including deoxyribonuclease I (DNase I), restriction endonucleases, glycoside hydrolases, proteases, and dispersin B, can inhibit bacterial biofilm formation and facilitate dispersion of established biofilm colonies (Kaplan, 2014). As soon as the biofilm matrix is enzymatically degraded, the bacterial cells are then released as planktonic cells which are easily eliminated by various antibacterial agents, disinfectants, phages, or immune systems (Kaplan, 2014; Parrino et al., 2019).
Phages can cross the EPS matrix by either diffusion or with the assist of phage-derived enzymes (Sao-Jose, 2018; Simmons et al., 2018). A genetically engineered lytic phage having a biofilm degrading enzyme showed more efficient eradication of biofilm than non-enzymatic phage (Lu and Collins, 2007). Combined with antibiotic rifampicin, the phage SAP-26 was able to cross the biofilm matrix leading to disruption of biofilm architectures (Hughes et al., 1998; Rahman et al., 2011). Cis-2 decenoic acid (C2DA) is a medium-chain fatty acid chemical messenger produced by Paeruginosa to initiate the dispersion of established bacterial biofilms C2DA not only effectively induced biofilm dispersal but may also inhibits initiation of biofilm formation (Jennings et al., 2012; Chung and Toh, 2014).
Therefore, EPS matrix can be destructed with enzymes and phages. As the research progresses, more EPS degradation methods will be found.
Conclusion
Bacterial biofilm formation occurs in sequential and well-regulated events and is the predominant bacterial lifestyle in most natural and man-made environments. The ability of bacteria to colonize surfaces and to establish biofilms are considered serious issues and has been associated with detrimental consequences in many branches related to food, water, pharmacy and healthcare. In an effort to get rid of harmful biofilms, various techniques and approaches have been developed which were mostly concerned with interference against bacterial attachment and QS as well as biofilm matrix destruction. However, bacterial biofilms affect the environments beyond risk. There are numerous beneficial applications of bacterial biofilms. Biofilm-associated bacteria play essential roles in the transformation of hazardous pollutants to harmless substances, the protection of plants against phytopathogens, the plant growth promotion, as well as the removal of excess nutrients from wastewater. Moreover, beneficial biofilms formation can be encouraged in many industrial and environmental areas through surface modification and QS signals.
Future Perspectives
Researches on the harmful effects of biofilms on healthcare, agriculture, food industry, drinking water, and oceans will continue to dominate the research field in the near future. However, with the deepening of people’s understanding of the dual-sidedness of the role of biofilms more than risks, the researches focusing on plant protection, bioremediation, wastewater treatment, and corrosion control of biofilms will be increasing. In addition, with the application of next-generation technologies such as various omics to biofilm researches, new bacterial biofilm regulation mechanisms are expected to be discovered. Therefore, researches of biofilms may be carried out in the following aspects in the future:
-
(1)
Control of bacterial biofilms that are harmful to human society;
-
(2)
Utilization of beneficial bacteria with high-production biofilms;
-
(3)
In-depth investigation of regulatory mechanisms for the formation and dispersion of bacterial biofilms, especially researches related to beneficial biofilms;
-
(4)
Elucidation of interaction mechanisms of bacterial biofilms with inanimate or living body interfaces;
-
(5)
Development of new commercial products based on bacterial biofilms;
-
(6)
Exploration of application schemes for bacterial biofilm products.
It is believed that through the unremitting efforts of researchers, biofilms will play more and more important roles in both basic researches and practical applications in recent years.
Author Contributions
The idea of this manuscript was conceived by TH. MM wrote the manuscript. AI, XF, YG, YY, and XJ helped to analyze the literatures. TH, XG, and JQ reviewed the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank the teachers and the students of the Biopesticide Research Center for their help. Thanks were also extended to the referees for their conductive comments. The content is solely the responsibility of the authors and does not necessarily represent the official views of the Ministry of Science and Technology, China, the National Natural Science Foundation of China, and the State Key Laboratory of Ecological Pest Control for Fujian and Taiwan Crops, China.
Footnotes
Funding. This project was supported by the National Key R&D Program of China (Nos. 2017YFD0200400, 2017YFE0121700, and 2017YFE0122000), the National Natural Science Foundation of China (No. 31672084), and the State Key Laboratory of Ecological Pest Control for Fujian and Taiwan Crops (No. SKL2018010).
References
- Abdallah M., Benoliel C., Drider D., Dhulster P., Chihib N. E. (2014). Biofilm formation and persistence on abiotic surfaces in the context of food and medical environments. Arch. Microbiol. 196 453–472. 10.1007/s00203-014-0983-1 [DOI] [PubMed] [Google Scholar]
- Abraham W. R. (2016). Going beyond the control of quorum-sensing to combat biofilm infections. Antibiotics 5:3. 10.3390/antibiotics5010003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Achinas S., Charalampogiannis N., Euverink G. J. W. (2019). A brief recap of microbial adhesion and biofilms. Appl. Sci. 9:2801. [Google Scholar]
- Ajene A. N., Fischer Walker C. L., Black R. E. (2013). Enteric pathogens and reactive arthritis: a systematic review of Campylobacter, salmonella and Shigella-associated reactive arthritis. J. Health Popul. Nutr. 31 299–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akyildiz I., Take G., Uygur K., Kizil Y., Aydil U. (2013). Bacterial biofilm formation in the middle-ear mucosa of chronic otitis media patients. Indian J. Otolaryngol. Head Neck Surg. 65 557–561. 10.1007/s12070-012-0513-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alarcon I., Evans D. J., Fleiszig S. M. (2009). The role of twitching motility in Pseudomonas aeruginosa exit from and translocation of corneal epithelial cells. Invest. Ophthalmol. Vis. Sci. 50 2237–2244. 10.1167/iovs.08-2785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alav I., Sutton J. M., Rahman K. M. (2018). Role of bacterial efflux pumps in biofilm formation. J. Antimicrob. Chemother. 73 2003–2020. 10.1093/jac/dky042 [DOI] [PubMed] [Google Scholar]
- Ali J., Sohail A., Wang L., Rizwan Haider M., Mulk S., Pan G. (2018). Electro-microbiology as a promising approach towards renewable energy and environmental sustainability. Energies 11:1822. [Google Scholar]
- Angelaalincy M. J., Navanietha K. R., Shakambari G., Ashokkumar B., Kathiresan S., Varalakshmi P. (2018). Biofilm engineering approaches for improving the performance of microbial fuel cells and bioelectrochemical systems. Front. Energy Res. 6:63. 10.1016/j.biotechadv.2019.107468 [DOI] [PubMed] [Google Scholar]
- Angles M. L., Chandy J. P., Cox P. T., Fisher I. H., Warnecke M. R. (2007). Implications of biofilm-associated waterborne Cryptosporidium oocysts for the water industry. Trends Parasitol. 23 352–356. [DOI] [PubMed] [Google Scholar]
- Ansari M. I., Schiwon K., Malik A., Grohmann E. (2012). “Biofilm formation by environmental bacteria,” in Environmental Protection Strategies for Sustainable Development, eds Malik A., Grohmann E. (Dordrecht: Springer Netherlands; ). [Google Scholar]
- Arciola C. R., Campoccia D., Ehrlich G. D., Montanaro L. (2015). Biofilm-based implant infections in orthopaedics. Adv. Exp. Med. Biol. 830 29–46. 10.1007/978-3-319-11038-7_2 [DOI] [PubMed] [Google Scholar]
- Argudin M. A., Mendoza M. C., Rodicio M. R. (2010). Food poisoning and Staphylococcus aureus enterotoxins. Toxins 2 1751–1773. 10.3390/toxins2071751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrebola E., Tienda S., Vida C., De Vicente A., Cazorla F. M. (2019). Fitness features involved in the biocontrol interaction of Pseudomonas chlororaphis with host plants: the case study of PcPCL1606. Front. Microbiol. 10:719. 10.3389/fmicb.2019.00719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azubuike C. C., Chikere C. B., Okpokwasili G. C. (2016). Bioremediation techniques-classification based on site of application: principles, advantages, limitations and prospects. World J. Microbiol. Biotechnol. 32:180. 10.1007/s11274-016-2137-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacosa H. P., Kamalanathan M., Chiu M. H., Tsai S. M., Sun L., Labonte J. M. (2018). Extracellular polymeric substances (EPS) producing and oil degrading bacteria isolated from the northern Gulf of Mexico. PLoS One 13:e0208406. 10.1371/journal.pone.0208406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagge D., Hjelm M., Johansen C., Huber I., Gram L. (2001). Shewanella putrefaciens adhesion and biofilm formation on food processing surfaces. Appl. Environ. Microbiol. 67 2319–2325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bais H. P., Fall R., Vivanco J. M. (2004). Biocontrol of Bacillus subtilis against infection of Arabidopsis roots by Pseudomonas syringae is facilitated by biofilm formation and surfactin production. Plant Physiol. 134 307–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajracharya S., Sharma M., Mohanakrishna G., Dominguez Benneton X., Strik D. P. B. T. B., Sarma P. M., et al. (2016). An overview on emerging bioelectrochemical systems (BESs): technology for sustainable electricity, waste remediation, resource recovery, chemical production and beyond. Renew. Energy 98 153–170. [Google Scholar]
- Bayston R., Ullas G., Ashraf W. (2012). Action of linezolid or vancomycin on biofilms in ventriculoperitoneal shunts in vitro. Antimicrob. Agents Chemother. 56 2842–2845. 10.1128/AAC.06326-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazaka K., Jacob M. V., Crawford R. J., Ivanova E. P. (2012). Efficient surface modification of biomaterial to prevent biofilm formation and the attachment of microorganisms. Appl. Microbiol. Biotechnol. 95 299–311. 10.1007/s00253-012-4144-7 [DOI] [PubMed] [Google Scholar]
- Berlanga M., Guerrero R. (2016). Living together in biofilms: the microbial cell factory and its biotechnological implications. Microb. Cell Fact. 15:165. 10.2210/pdb4bhu/pdb) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berlowska J., Kregiel D., Ambroziak W. (2013). Enhancing adhesion of yeast brewery strains to chamotte carriers through aminosilane surface modification. World J. Microbiol. Biotechnol. 29 1307–1316. 10.1007/s11274-013-1294-4 [DOI] [PubMed] [Google Scholar]
- Berne C., Ducret A., Hardy G. G., Brun Y. V. (2015). Adhesins involved in attachment to abiotic surfaces by gram-negative bacteria. Microbiol. Spectr. 3 10.1128/microbiolsec.MB-0018-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjarnsholt T. (2013). The role of bacterial biofilms in chronic infections. APMIS Suppl. 121 1–51. [DOI] [PubMed] [Google Scholar]
- Bogino P. C., Oliva Mde L., Sorroche F. G., Giordano W. (2013). The role of bacterial biofilms and surface components in plant-bacterial associations. Int. J. Mol. Sci. 14 15838–15859. 10.3390/ijms140815838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollen C. M., Lambrechts P., Quirynen M. (1997). Comparison of surface roughness of oral hard materials to the threshold surface roughness for bacterial plaque retention: a review of the literature. Dent. Mater. 13 258–269. [DOI] [PubMed] [Google Scholar]
- Bonazzi M., Lecuit M., Cossart P. (2009). Listeria monocytogenes internalin and E-cadherin: from bench to bedside. Cold Spring Harb. Perspect. Biol. 1:a003087. 10.1101/cshperspect.a003087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bos R., Van Der Mei H. C., Busscher H. J. (1999). Physico-chemistry of initial microbial adhesive interactions–its mechanisms and methods for study. FEMS Microbiol. Rev. 23 179–230. [DOI] [PubMed] [Google Scholar]
- Bowden G. H., Li Y. H. (1997). Nutritional influences on biofilm development. Adv. Dent. Res. 11 81–99. [DOI] [PubMed] [Google Scholar]
- Burgess S. A., Flint S. H., Lindsay D., Cox M. P., Biggs P. J. (2017). Insights into the Geobacillus stearothermophilus species based on phylogenomic principles. BMC Microbiol. 17:140. 10.1186/s12866-017-1047-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busscher H. J., Bos R., Van Der Mei H. C. (1995). Initial microbial adhesion is a determinant for the strength of biofilm adhesion. Fems Microbiol. Lett. 128 229–234. [DOI] [PubMed] [Google Scholar]
- Cao Y., Mu H., Liu W., Zhang R., Guo J., Xian M., et al. (2019). Electricigens in the anode of microbial fuel cells: pure cultures versus mixed communities. Microb. Cell Fact. 18:39. 10.1186/s12934-019-1087-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carniello V., Peterson B. W., Van Der Mei H. C., Busscher H. J. (2018). Physico-chemistry from initial bacterial adhesion to surface-programmed biofilm growth. Adv. Colloid Interface Sci. 261 1–14. 10.1016/j.cis.2018.10.005 [DOI] [PubMed] [Google Scholar]
- Carter M. Q., Louie J. W., Feng D., Zhong W., Brandl M. T. (2016). Curli fimbriae are conditionally required in Escherichia coli O157:H7 for initial attachment and biofilm formation. Food Microbiol. 57 81–89. 10.1016/j.fm.2016.01.006 [DOI] [PubMed] [Google Scholar]
- Caruso C., Rizzo C., Mangano S., Poli A., Di Donato P., Finore I., et al. (2018). Production and biotechnological potential of extracellular polymeric substances from sponge-associated antarctic bacteria. Appl. Environ. Microbiol. 84:e01624-17. 10.1128/AEM.01624-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cegelski L., Pinkner J. S., Hammer N. D., Cusumano C. K., Hung C. S., Chorell E., et al. (2009). Small-molecule inhibitors target Escherichia coli amyloid biogenesis and biofilm formation. Nat. Chem. Biol. 5 913–919. 10.1038/nchembio.242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan S., Pullerits K., Keucken A., Persson K. M., Paul C. J., Radstrom P. (2019). Bacterial release from pipe biofilm in a full-scale drinking water distribution system. NPJ Biofilms Microbiomes 5:9. 10.1038/s41522-019-0082-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavant P., Martinie B., Meylheuc T., Bellon-Fontaine M. N., Hebraud M. (2002). Listeria monocytogenes LO28: surface physicochemical properties and ability to form biofilms at different temperatures and growth phases. Appl. Environ. Microbiol. 68 728–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F., Gao Y., Chen X., Yu Z., Li X. (2013). Quorum quenching enzymes and their application in degrading signal molecules to block quorum sensing-dependent infection. Int. J. Mol. Sci. 14 17477–17500. 10.3390/ijms140917477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S., Jing X., Tang J., Fang Y., Zhou S. (2017). Quorum sensing signals enhance the electrochemical activity and energy recovery of mixed-culture electroactive biofilms. Biosens. Bioelectron. 97 369–376. 10.1016/j.bios.2017.06.024 [DOI] [PubMed] [Google Scholar]
- Chin-A-Woeng T. F. C., Bloemberg G. V., Lugtenberg B. J. J. (2003). Phenazines and their role in biocontrol by Pseudomonas bacteria. New Phytologist 157 503–523. [DOI] [PubMed] [Google Scholar]
- Chmielewski R. A. N., Frank J. F. (2003). Biofilm formation and control in food processing facilities. Compr. Rev. Food Sci. Food Safety 2 22–32. [DOI] [PubMed] [Google Scholar]
- Chorell E., Pinkner J. S., Bengtsson C., Edvinsson S., Cusumano C. K., Rosenbaum E., et al. (2012). Design and synthesis of fluorescent pilicides and curlicides: bioactive tools to study bacterial virulence mechanisms. Chemistry 18 4522–4532. 10.1002/chem.201103936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung P. Y., Toh Y. S. (2014). Anti-biofilm agents: recent breakthrough against multi-drug resistant Staphylococcus aureus. Pathog. Dis. 70 231–239. 10.1111/2049-632X.12141 [DOI] [PubMed] [Google Scholar]
- Ciofu O., Tolker-Nielsen T. (2019). Tolerance and resistance of Pseudomonas aeruginosa biofilms to antimicrobial agents-How P. aeruginosa can escape antibiotics. Front. Microbiol. 10:913. 10.3389/fmicb.2019.00913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornejo J. A., Lopez C., Babanova S., Santoro C., Artyushkova K., Ista L., et al. (2015). Surface modi?cation for enhanced bio?lm formation and electron transport in shewanella anodes. J. Electrochem. Soc. 162 597–603. [Google Scholar]
- Costa O. Y. A., Raaijmakers J. M., Kuramae E. E. (2018). Microbial extracellular polymeric substances: ecological function and impact on soil aggregation. Front. Microbiol. 9:1636. 10.3389/fmicb.2018.01636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa-Orlandi C. B., Sardi J. C. O., Pitangui N. S., De Oliveira H. C., Scorzoni L., Galeane M. C., et al. (2017). Fungal biofilms and polymicrobial diseases. J. Fungi (Basel) 3:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coughlan L. M., Cotter P. D., Hill C., Alvarez-Ordonez A. (2016). New weapons to fight old enemies: novel strategies for the (Bio)control of bacterial biofilms in the food industry. Front. Microbiol. 7:1641. 10.3389/fmicb.2016.01641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang H., Lovell C. R. (2016). Microbial surface colonization and biofilm development in marine environments. Microbiol. Mol. Biol. Rev. 80 91–138. 10.1128/MMBR.00037-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danhorn T., Fuqua C. (2007). Biofilm formation by plant-associated bacteria. Annu. Rev. Microbiol. 61 401–422. [DOI] [PubMed] [Google Scholar]
- Das K., Rajawat M. V. S., Saxena A. K., Prasanna R. (2017). Development of Mesorhizobium ciceri-based biofilms and analyses of their antifungal and plant growth promoting activity in chickpea challenged by Fusarium Wilt. Indian J. Microbiol. 57 48–59. 10.1007/s12088-016-0610-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasgupta D., Ghosh R., Sengupta T. K. (2013). Biofilm-mediated enhanced crude oil degradation by newly isolated pseudomonas species. ISRN Biotechnol. 2013:250749. 10.5402/2013/250749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daud M. K., Nafees M., Ali S., Rizwan M., Bajwa R. A., Shakoor M. B., et al. (2017). Drinking water quality status and contamination in Pakistan. Biomed. Res. Int. 2017:7908183. 10.1155/2017/7908183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Carvalho C. C. C. R. (2018). Marine biofilms: a successful microbial strategy with economic implications. Front. Mar. Sci. 5:126 10.3389/fmars.2018.00126 [DOI] [Google Scholar]
- de Vos W. M. (2015). Microbial biofilms and the human intestinal microbiome. NPJ Biofilms Microbiomes 1:15005. 10.1038/npjbiofilms.2015.5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demirel Y. K., Uzun D., Zhang Y., Fang H. C., Day A. H., Turan O. (2017). Effect of barnacle fouling on ship resistance and powering. Biofouling 33 819–834. 10.1080/08927014.2017.1373279 [DOI] [PubMed] [Google Scholar]
- Dhir S. (2013). Biofilm and dental implant: the microbial link. J. Indian Soc. Periodontol. 17 5–11. 10.4103/0972-124X.107466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Martino P., Cafferini N., Joly B., Darfeuille-Michaud A. (2003). Klebsiella pneumoniae type 3 pili facilitate adherence and biofilm formation on abiotic surfaces. Res. Microbiol. 154 9–16. [DOI] [PubMed] [Google Scholar]
- Diaz-Salazar C., Calero P., Espinosa-Portero R., Jimenez-Fernandez A., Wirebrand L., Velasco-Dominguez M. G., et al. (2017). The stringent response promotes biofilm dispersal in Pseudomonas putida. Sci. Rep. 7:18055. 10.1038/s41598-017-18518-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson J. S., Koohmaraie M. (1989). Cell surface charge characteristics and their relationship to bacterial attachment to meat surfaces. Appl. Environ. Microbiol. 55 832–836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobretsov S., Abed R. M., Teplitski M. (2013). Mini-review: inhibition of biofouling by marine microorganisms. Biofouling 29 423–441. 10.1080/08927014.2013.776042 [DOI] [PubMed] [Google Scholar]
- Dobretsov S., Dahms H. U., Qian P. Y. (2006). Inhibition of biofouling by marine microorganisms and their metabolites. Biofouling 22 43–54. [DOI] [PubMed] [Google Scholar]
- Dogsa I., Kriechbaum M., Stopar D., Laggner P. (2005). Structure of bacterial extracellular polymeric substances at different pH values as determined by SAXS. Biophys. J. 89 2711–2720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dongari-Bagtzoglou A. (2008). Pathogenesis of mucosal biofilm infections: challenges and progress. Expert Rev. Anti Infect. Ther. 6 201–208. 10.1586/14787210.6.2.201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donlan R. M. (2002). Biofilms: microbial life on surfaces. Emerg. Infect. Dis. 8 881–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donlan R. M., Costerton J. W. (2002). Biofilms: survival mechanisms of clinically relevant microorganisms. Clin. Microbiol. Rev. 15 167–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douterelo I., Husband S., Loza V., Boxall J. (2016). Dynamics of biofilm regrowth in drinking water distribution systems. Appl. Environ. Microbiol. 82 4155–4168. 10.1128/AEM.00109-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dror N., Mandel M., Hazan Z., Lavie G. (2009). Advances in microbial biofilm prevention on indwelling medical devices with emphasis on usage of acoustic energy. Sensors (Basel) 9 2538–2554. 10.3390/s90402538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunne W. M., Jr. (2002). Bacterial adhesion: seen any good biofilms lately? Clin. Microbiol. Rev. 15 155–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dziubakiewicz E., Hrynkiewicz K., Walczyk M., Buszewski B. (2013). Study of charge distribution on the surface of biocolloids. Colloids Surf. B Biointerfaces 104 122–127. 10.1016/j.colsurfb.2012.11.018 [DOI] [PubMed] [Google Scholar]
- Edwards S. J., Kjellerup B. V. (2013). Applications of biofilms in bioremediation and biotransformation of persistent organic pollutants, pharmaceuticals/personal care products, and heavy metals. Appl. Microbiol. Biotechnol. 97 9909–9921. 10.1007/s00253-013-5216-z [DOI] [PubMed] [Google Scholar]
- El-Ganiny A. M., Shaker G. H., Aboelazm A. A., El-Dash H. A. (2017). Prevention of bacterial biofilm formation on soft contact lenses using natural compounds. J. Ophthalmic. Inflamm. Infect. 7:11. 10.1186/s12348-017-0129-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elgharably H., Hussain S. T., Shrestha N. K., Blackstone E. H., Pettersson G. B. (2016). Current hypotheses in cardiac surgery: biofilm in infective endocarditis. Semin. Thorac. Cardiovasc. Surg. 28, 56–59. 10.1053/j.semtcvs.2015.12.005 [DOI] [PubMed] [Google Scholar]
- Fahs A., Quiles F., Jamal D., Humbert F., Francius G. (2014). In situ analysis of bacterial extracellular polymeric substances from a Pseudomonas fluorescens biofilm by combined vibrational and single molecule force spectroscopies. J. Phys. Chem. B 118 6702–6713. 10.1021/jp5030872 [DOI] [PubMed] [Google Scholar]
- Faille C., Jullien C., Fontaine F., Bellon-Fontaine M. N., Slomianny C., Benezech T. (2002). Adhesion of Bacillus spores and Escherichia coli cells to inert surfaces: role of surface hydrophobicity. Can. J. Microbiol. 48 728–738. [DOI] [PubMed] [Google Scholar]
- Favero B. M., Favero A. C., Taffarel S. R., Souza F. S. (2018). Evaluation of the efficiency of coagulation/flocculation and Fenton process in reduction of colour, turbidity and COD of a textile effluent. Environ. Technol. 41 1580–1589. [DOI] [PubMed] [Google Scholar]
- Ference C. M., Gochez A. M., Behlau F., Wang N., Graham J. H., Jones J. B. (2018). Recent advances in the understanding of Xanthomonas citri ssp. citri pathogenesis and citrus canker disease management. Mol. Plant Pathol. 19 1302–1318. 10.1111/mpp.12638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fira D., Dimkic I., Beric T., Lozo J., Stankovic S. (2018). Biological control of plant pathogens by Bacillus species. J. Biotechnol. 285 44–55. [DOI] [PubMed] [Google Scholar]
- Fleming D., Rumbaugh K. P. (2017). Approaches to dispersing medical biofilms. Microorganisms 5:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flemming H. C. (2016). EPS-then and now. Microorganisms 4:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flemming H. C., Wingender J. (2010). The biofilm matrix. Nat. Rev. Microbiol. 8 623–633. [DOI] [PubMed] [Google Scholar]
- Flemming H. C., Wingender J., Szewzyk U., Steinberg P., Rice S. A., Kjelleberg S. (2016). Biofilms: an emergent form of bacterial life. Nat. Rev. Microbiol. 14 563–575. [DOI] [PubMed] [Google Scholar]
- Flemming H. C., Wuertz S. (2019). Bacteria and archaea on Earth and their abundance in biofilms. Nat. Rev. Microbiol. 17 247–260. [DOI] [PubMed] [Google Scholar]
- Flexer V., Marque M., Donose B. C., Virdis B., Keller J. (2013). Plasma treatment of electrodes significantly enhances the development of anodic electrochemically active biofilms. Electrochim. Acta 108 566–574. [Google Scholar]
- Flint S. H., Brooks J. D., Bremer P. J. (2000). Properties of the stainless steel substrate, influencing the adhesion of thermo-resistant streptococci. J. Food Eng. 43 235–242. [Google Scholar]
- Fong J., Zhang C., Yang R., Boo Z. Z., Tan S. K., Nielsen T. E., et al. (2018). Combination therapy strategy of quorum quenching enzyme and quorum sensing inhibitor in suppressing multiple quorum sensing pathways of P. aeruginosa. Sci. Rep. 8:1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis M. R., Sarkar R., Roy S., Jaffar S., Mohan V. R., Kang G., et al. (2016). Effectiveness of membrane filtration to improve drinking water: a quasi-experimental study from rural Southern India. Am. J. Trop. Med. Hyg. 95 1192–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francolini I., Vuotto C., Piozzi A., Donelli G. (2017). Antifouling and antimicrobial biomaterials: an overview. APMIS 125 392–417. [DOI] [PubMed] [Google Scholar]
- Franks A. E., Malvankar N., Nevin K. P. (2010). Bacterial biofilms: the powerhouse of a microbial fuel cell. Biofuels 1 589–604. [Google Scholar]
- Franks A. E., Nevin K. P. (2010). Microbial fuel cells. Curr. Rev. Energies 3 899–919. [Google Scholar]
- Frederick M. R., Kuttler C., Hense B. A., Eberl H. J. (2011). A mathematical model of quorum sensing regulated EPS production in biofilm communities. Theor. Biol. Med. Model. 8:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galie S., Garcia-Gutierrez C., Miguelez E. M., Villar C. J., Lombo F. (2018). Biofilms in the food industry: health aspects and control methods. Front. Microbiol. 9:898 10.3389/fmicb.2018.00898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garnett J. A., Matthews S. (2012). Interactions in bacterial biofilm development: a structural perspective. Curr. Protein Pept. Sci. 13 739–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gominet M., Compain F., Beloin C., Lebeaux D. (2017). Central venous catheters and biofilms: where do we stand in 2017? APMIS 125 365–375. [DOI] [PubMed] [Google Scholar]
- Gude V. G. (2016a). “8 – microbial fuel cells for wastewater treatment and energy generation,” in Microbial Electrochemical and Fuel Cells, eds Scott K., Yu E. H. (Boston, MA: Woodhead Publishing; ). [Google Scholar]
- Gude V. G. (2016b). Wastewater treatment in microbial fuel cells – an overview. J. Clean. Prod. 122 287–307. [Google Scholar]
- Guo J., Yuan S., Jiang W., Lv L., Liang B., Pehkonen S. O. (2018). Polymers for combating biocorrosion. Front. Mater. 5:10 10.3389/fmats.2018.00010 [DOI] [Google Scholar]
- Guttenplan S. B., Kearns D. B. (2013). Regulation of flagellar motility during biofilm formation. FEMS Microbiol. Rev. 37 849–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haggag W. M., Timmusk S. (2008). Colonization of peanut roots by biofilm-forming Paenibacillus polymyxa initiates biocontrol against crown rot disease. J. Appl. Microbiol. 104 961–969. [DOI] [PubMed] [Google Scholar]
- Haiko J., Westerlund-Wikstrom B. (2013). The role of the bacterial flagellum in adhesion and virulence. Biology (Basel) 2 1242–1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall-Stoodley L., Costerton J. W., Stoodley P. (2004). Bacterial biofilms: from the natural environment to infectious diseases. Nat. Rev. Microbiol. 2 95–108. [DOI] [PubMed] [Google Scholar]
- Hall-Stoodley L., Stoodley P. (2002). Developmental regulation of microbial biofilms. Curr. Opin. Biotechnol. 13 228–233. [DOI] [PubMed] [Google Scholar]
- Han N., Mizan M. F. R., Jahid I. K., Ha S.-D. (2016). Biofilm formation by Vibrio parahaemolyticus on food and food contact surfaces increases with rise in temperature. Food Control 70 161–166. [Google Scholar]
- Han Q., Song X., Zhang Z., Fu J., Wang X., Malakar P. K., et al. (2017). Removal of foodborne pathogen biofilms by acidic electrolyzed water. Front. Microbiol. 8:988 10.3389/fmicb.2017.00988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasan J., Crawford R. J., Ivanova E. P. (2013). Antibacterial surfaces: the quest for a new generation of biomaterials. Trends Biotechnol. 31 295–304. [DOI] [PubMed] [Google Scholar]
- He Y. R., Xiao X., Li W. W., Sheng G. P., Yan F. F., Yu H. Q., et al. (2012). Enhanced electricity production from microbial fuel cells with plasma-modified carbon paper anode. Phys. Chem. Chem. Phys. 14 9966–9971. [DOI] [PubMed] [Google Scholar]
- Heras B., Totsika M., Peters K. M., Paxman J. J., Gee C. L., Jarrott R. J., et al. (2014). The antigen 43 structure reveals a molecular Velcro-like mechanism of autotransporter-mediated bacterial clumping. Proc. Natl. Acad. Sci. U.S.A. 111 457–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermansson M. (1999). The DLVO theory in microbial adhesion. Colloids Surf. B Biointerfaces 14 105–119. [Google Scholar]
- Hibiya K., Tsuneda S., Hirata A. (2000). Formation and characteristics of nitrifying biofilm on a membrane modified with positively-charged polymer chains. Colloids Surf. B Biointerfaces 18 105–112. [Google Scholar]
- Hintsche M., Waljor V., Grossmann R., Kuhn M. J., Thormann K. M., Peruani F., et al. (2017). A polar bundle of flagella can drive bacterial swimming by pushing, pulling, or coiling around the cell body. Sci. Rep. 7:16771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirakawa H., Tomita H. (2013). Interference of bacterial cell-to-cell communication: a new concept of antimicrobial chemotherapy breaks antibiotic resistance. Front. Microbiol. 4:114 10.3389/fmicb.2013.00114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hlihor R. M., Gavrilescu M., Tavares T., Favier L., Olivieri G. (2017). Bioremediation: an overview on current practices, advances, and new perspectives in environmental pollution treatment. Biomed. Res. Int. 2017:6327610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman M. D., Zucker L. I., Brown P. J., Kysela D. T., Brun Y. V., Jacobson S. C. (2015). Timescales and frequencies of reversible and irreversible adhesion events of single bacterial cells. Anal. Chem. 87 12032–12039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkins G. A., Forrest B. M. (2010). A preliminary assessment of biofouling and non-indigenous marine species associated with commercial slow-moving vessels arriving in New Zealand. Biofouling 26 613–621. [DOI] [PubMed] [Google Scholar]
- Huang H., Peng C., Peng P., Lin Y., Zhang X., Ren H. (2019). Towards the biofilm characterization and regulation in biological wastewater treatment. Appl. Microbiol. Biotechnol. 103 1115–1129. [DOI] [PubMed] [Google Scholar]
- Hughes K. A., Sutherland I. W., Jones M. V. (1998). Biofilm susceptibility to bacteriophage attack: the role of phage-borne polysaccharide depolymerase. Microbiology 144(Pt 11) 3039–3047. [DOI] [PubMed] [Google Scholar]
- Husmark U., Rönner U. (1992). The influence of hydrophobic, electrostatic and morphologic properties on the adhesion of Bacillus spores. Biofouling 5 335–344. [Google Scholar]
- Irankhah S., Abdi Ali A., Mallavarapu M., Soudi M. R., Subashchandrabose S., Gharavi S., et al. (2019). Ecological role of Acinetobacter calcoaceticus GSN3 in natural biofilm formation and its advantages in bioremediation. Biofouling 35 377–391. [DOI] [PubMed] [Google Scholar]
- Jakobsen T. H., Van Gennip M., Phipps R. K., Shanmugham M. S., Christensen L. D., Alhede M., et al. (2012). Ajoene, a sulfur-rich molecule from garlic, inhibits genes controlled by quorum sensing. Antimicrob. Agents Chemother. 56 2314–2325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamal M., Ahmad W., Andleeb S., Jalil F., Imran M., Nawaz M. A., et al. (2018). Bacterial biofilm and associated infections. J. Chin. Med. Assoc. 81 7–11. [DOI] [PubMed] [Google Scholar]
- Jayaraman A., Cheng E. T., Earthman J. C., Wood T. K. (1997). Importance of biofilm formation for corrosion inhibition of SAE 1018 steel by axenic aerobic biofilms. J. Ind. Microbiol. Biotechnol. 18 396–401. [DOI] [PubMed] [Google Scholar]
- Jayathilake P. G., Jana S., Rushton S., Swailes D., Bridgens B., Curtis T., et al. (2017). Extracellular polymeric substance production and aggregated bacteria colonization influence the competition of microbes in biofilms. Front. Microbiol. 8:1865. 10.3389/fmicb.2017.01865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jennings J. A., Courtney H. S., Haggard W. O. (2012). Cis-2-decenoic acid inhibits S. aureus growth and biofilm in vitro: a pilot study. Clin. Orthop. Relat. Res. 470 2663–2670. 10.1007/s11999-012-2388-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia R., Yang D., Xu D., Gu T. (2017). Mitigation of a nitrate reducing Pseudomonas aeruginosa biofilm and anaerobic biocorrosion using ciprofloxacin enhanced by D-tyrosine. Sci. Rep. 7:6946. 10.1038/s41598-017-07312-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jucker B. A., Zehnder A. J. B., Harms H. (1998). Quantification of polymer interactions in bacterial adhesion. Environ. Sci. Technol. 32 2909–2915. [Google Scholar]
- Kang C. S., Eaktasang N., Kwon D. Y., Kim H. S. (2014). Enhanced current production by Desulfovibrio desulfuricans biofilm in a mediator-less microbial fuel cell. Bioresour. Technol. 165 27–30. 10.1016/j.biortech.2014.03.148 [DOI] [PubMed] [Google Scholar]
- Kaplan J. B. (2010). Biofilm dispersal: mechanisms, clinical implications, and potential therapeutic uses. J. Dent. Res. 89 205–218. 10.1177/0022034509359403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan J. B. (2014). Biofilm matrix-degrading enzymes. Methods Mol. Biol. 1147 203–213. 10.1007/978-1-4939-0467-9_14 [DOI] [PubMed] [Google Scholar]
- Kapley A., Purohit H. J. (2009). Genomic tools in bioremediation. Indian J. Microbiol. 49 108–113. 10.1007/s12088-009-0012-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karatan E., Watnick P. (2009). Signals, regulatory networks, and materials that build and break bacterial biofilms. Microbiol. Mol. Biol. Rev. 73 310–347. 10.1128/MMBR.00041-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karigar C. S., Rao S. S. (2011). Role of microbial enzymes in the bioremediation of pollutants: a review. Enzyme Res. 2011:805187. 10.4061/2011/805187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi A., Karig D., Kumar A., Ardekani A. M. (2015). Interplay of physical mechanisms and biofilm processes: review of microfluidic methods. Lab Chip 15 23–42. 10.1039/c4lc01095g [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsikogianni M., Missirlis Y. F. (2004). Concise review of mechanisms of bacterial adhesion to biomaterials and of techniques used in estimating bacteria-material interactions. Eur. Cell Mater. 8 37–57. [DOI] [PubMed] [Google Scholar]
- Kearns D. B. (2010). A field guide to bacterial swarming motility. Nat. Rev. Microbiol. 8 634–644. 10.1038/nrmicro2405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan N., Martinez-Hidalgo P., Ice T. A., Maymon M., Humm E. A., Nejat N., et al. (2018). Antifungal activity of Bacillus species against fusarium and analysis of the potential mechanisms used in biocontrol. Front. Microbiol. 9:2363. 10.3389/fmicb.2018.02363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khatoon Z., Mctiernan C. D., Suuronen E. J., Mah T. F., Alarcon E. I. (2018). Bacterial biofilm formation on implantable devices and approaches to its treatment and prevention. Heliyon 4:e01067. 10.1016/j.heliyon.2018.e01067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kip N., van Veen J. A. (2015). The dual role of microbes in corrosion. ISME J. 9 542–551. 10.1038/ismej.2014.169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobya M., Demirbas E., Akyol A. (2009). Electrochemical treatment and operating cost analysis of textile wastewater using sacrificial iron electrodes. Water Sci. Technol. 60 2261–2270. 10.2166/wst.2009.672 [DOI] [PubMed] [Google Scholar]
- Koczan J. M., Lenneman B. R., Mcgrath M. J., Sundin G. W. (2011). Cell surface attachment structures contribute to biofilm formation and xylem colonization by Erwinia amylovora. Appl. Environ. Microbiol. 77 7031–7039. 10.1128/AEM.05138-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konto-Ghiorghi Y., Mairey E., Mallet A., Dumenil G., Caliot E., Trieu-Cuot P., et al. (2009). Dual role for pilus in adherence to epithelial cells and biofilm formation in Streptococcus agalactiae. PLoS Pathog. 5:e1000422. 10.1371/journal.ppat.1000422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korber D. R., Mangalappalli-Illathu A. K., Vidoviæ S. (2009). “6 – biofilm formation by food spoilage microorganisms in food processing environments,” in Biofilms in the Food and Beverage Industries, eds Fratamico P. M., Annous B. A., Gunther N. W. (Sawston: Woodhead Publishing; ). [Google Scholar]
- Kostakioti M., Hadjifrangiskou M., Hultgren S. J. (2013). Bacterial biofilms: development, dispersal, and therapeutic strategies in the dawn of the postantibiotic era. Cold Spring Harb. Perspect. Med. 3:a010306. 10.1101/cshperspect.a010306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuang X., Chen V., Xu X. (2018). Novel approaches to the control of oral microbial biofilms. Biomed. Res. Int. 2018:6498932. 10.1155/2018/6498932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar C. G., Anand S. K. (1998). Significance of microbial biofilms in food industry: a review. Int. J. Food Microbiol. 42 9–27. [DOI] [PubMed] [Google Scholar]
- Kyrkou I., Pusa T., Ellegaard-Jensen L., Sagot M. F., Hansen L. H. (2018). Pierce’s disease of grapevines: a review of control strategies and an outline of an epidemiological model. Front. Microbiol. 9:2141. 10.3389/fmicb.2018.02141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lackner S., Holmberg M., Terada A., Kingshott P., Smets B. F. (2009). Enhancing the formation and shear resistance of nitrifying biofilms on membranes by surface modification. Water Res. 43 3469–3478. 10.1016/j.watres.2009.05.011 [DOI] [PubMed] [Google Scholar]
- LeChevallier M. W., Cawthon C. D., Lee R. G. (1988). Inactivation of biofilm bacteria. Appl. Environ. Microbiol. 54 2492–2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K., Yoon S. S. (2017). Pseudomonas aeruginosa biofilm, a programmed bacterial life for fitness. J. Microbiol. Biotechnol. 27 1053–1064. 10.4014/jmb.1611.11056 [DOI] [PubMed] [Google Scholar]
- Lemon K. P., Higgins D. E., Kolter R. (2007). Flagellar motility is critical for Listeria monocytogenes biofilm formation. J. Bacteriol. 189 4418–4424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G., Tang J. X. (2009). Accumulation of microswimmers near a surface mediated by collision and rotational Brownian motion. Phys. Rev. Lett. 103:078101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y. H., Tian X. (2012). Quorum sensing and bacterial social interactions in biofilms. Sensors (Basel) 12 2519–2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Hu W., Zhao Y., Ren L., Yuan X. (2018). Integrated antibacterial and antifouling surfaces via cross-linking chitosan-g-eugenol/zwitterionic copolymer on electrospun membranes. Colloids Surf. B Biointerfaces 169 151–159. 10.1016/j.colsurfb.2018.04.056 [DOI] [PubMed] [Google Scholar]
- Limoli D. H., Jones C. J., Wozniak D. J. (2015). Bacterial extracellular polysaccharides in biofilm formation and function. Microbiol. Spectr. 3:10.1128/microbiolsec.MB–0011–2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Ramnarayanan R., Logan B. E. (2004). Production of electricity during wastewater treatment using a single chamber microbial fuel cell. Environ. Sci. Technol. 38 2281–2285. [DOI] [PubMed] [Google Scholar]
- Liu S., Gunawan C., Barraud N., Rice S. A., Harry E. J., Amal R. (2016). Understanding, monitoring, and controlling biofilm growth in drinking water distribution systems. Environ. Sci. Technol. 50 8954–8976. 10.1021/acs.est.6b00835 [DOI] [PubMed] [Google Scholar]
- Lopez D., Vlamakis H., Kolter R. (2010). Biofilms. Cold Spring Harb. Perspect. Biol. 2:a000398. 10.1101/cshperspect.a000398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorite G. S., Rodrigues C. M., De Souza A. A., Kranz C., Mizaikoff B., Cotta M. A. (2011). The role of conditioning film formation and surface chemical changes on Xylella fastidiosa adhesion and biofilm evolution. J. Colloid Interface Sci. 359 289–295. 10.1016/j.jcis.2011.03.066 [DOI] [PubMed] [Google Scholar]
- Loveday H. P., Wilson J. A., Kerr K., Pitchers R., Walker J. T., Browne J. (2014). Association between healthcare water systems and Pseudomonas aeruginosa infections: a rapid systematic review. J. Hosp. Infect. 86 7–15. 10.1016/j.jhin.2013.09.010 [DOI] [PubMed] [Google Scholar]
- Lu T. K., Collins J. J. (2007). Dispersing biofilms with engineered enzymatic bacteriophage. Proc. Natl. Acad. Sci. U.S.A. 104 11197–11202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahapatra A., Padhi N., Mahapatra D., Bhatt M., Sahoo D., Jena S., et al. (2015). Study of biofilm in bacteria from water pipelines. J. Clin. Diagn. Res. 9 DC09–DC11. 10.7860/JCDR/2015/12415.5715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maldarelli G. A., Piepenbrink K. H., Scott A. J., Freiberg J. A., Song Y., Achermann Y., et al. (2016). Type IV pili promote early biofilm formation by Clostridium difficile. Pathog. Dis. 74:ftw061. 10.1093/femspd/ftw061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangwani N., Kumari S., Das S. (2015). Involvement of quorum sensing genes in biofilm development and degradation of polycyclic aromatic hydrocarbons by a marine bacterium Pseudomonas aeruginosa N6P6. Appl. Microbiol. Biotechnol. 99 10283–10297. 10.1007/s00253-015-6868-7 [DOI] [PubMed] [Google Scholar]
- Mangwani N., Kumari S., Das S. (2016). Bacterial biofilms and quorum sensing: fidelity in bioremediation technology. Biotechnol. Genet. Eng. Rev. 32 43–73. 10.1080/02648725.2016.1196554 [DOI] [PubMed] [Google Scholar]
- Martinez P., Vera M., Bobadilla-Fazzini R. A. (2015). Omics on bioleaching: current and future impacts. Appl. Microbiol. Biotechnol. 99 8337–8350. 10.1007/s00253-015-6903-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masters E. A., Trombetta R. P., De Mesy Bentley K. L., Boyce B. F., Gill A. L., Gill S. R., et al. (2019). Evolving concepts in bone infection: redefining “biofilm”, “acute vs. chronic osteomyelitis”, “the immune proteome” and “local antibiotic therapy”. Bone Res. 7:20. 10.1038/s41413-019-0061-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meena K. R., Kanwar S. S. (2015). Lipopeptides as the antifungal and antibacterial agents: applications in food safety and therapeutics. Biomed. Res. Int. 2015:473050. 10.1155/2015/473050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meesilp N., Mesil N. (2019). Effect of microbial sanitizers for reducing biofilm formation of Staphylococcus aureus and Pseudomonas aeruginosa on stainless steel by cultivation with UHT milk. Food Sci. Biotechnol. 28 289–296. 10.1007/s10068-018-0448-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei X., Xing D., Yang Y., Liu Q., Zhou H., Guo C., et al. (2017). Adaptation of microbial community of the anode biofilm in microbial fuel cells to temperature. Bioelectrochemistry 117 29–33. 10.1016/j.bioelechem.2017.04.005 [DOI] [PubMed] [Google Scholar]
- Miller M. B., Bassler B. L. (2001). Quorum sensing in bacteria. Annu. Rev. Microbiol. 55 165–199. [DOI] [PubMed] [Google Scholar]
- Min B., Kim J., Oh S., Regan J. M., Logan B. E. (2005). Electricity generation from swine wastewater using microbial fuel cells. Water Res. 39 4961–4968. [DOI] [PubMed] [Google Scholar]
- Minchin D., Gollasch S. (2003). Fouling and ships’ hulls: how changing circumstances and spawning events may result in the spread of exotic species. Biofouling 19(Suppl) 111–122. [DOI] [PubMed] [Google Scholar]
- Monier J. M., Lindow S. E. (2004). Frequency, size, and localization of bacterial aggregates on bean leaf surfaces. Appl. Environ. Microbiol. 70 346–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Møretrø T., Langsrud S. (2017). Residential bacteria on surfaces in the food industry and their implications for food safety and quality. Compr. Rev. Food Sci. Food Safety 16 1022–1041. [DOI] [PubMed] [Google Scholar]
- Mori Y., Inoue K., Ikeda K., Nakayashiki H., Higashimoto C., Ohnishi K., et al. (2016). The vascular plant-pathogenic bacterium Ralstonia solanacearum produces biofilms required for its virulence on the surfaces of tomato cells adjacent to intercellular spaces. Mol. Plant Pathol. 17 890–902. 10.1111/mpp.12335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morikawa M. (2006). Beneficial biofilm formation by industrial bacteria Bacillus subtilis and related species. J. Biosci. Bioeng. 101 1–8. [DOI] [PubMed] [Google Scholar]
- Morra M., Cassinelli C. (1997). Bacterial adhesion to polymer surfaces: a critical review of surface thermodynamic approaches. J. Biomater. Sci. Polym. Ed 9 55–74. [DOI] [PubMed] [Google Scholar]
- Murphy C. N., Mortensen M. S., Krogfelt K. A., Clegg S. (2013). Role of Klebsiella pneumoniae type 1 and type 3 fimbriae in colonizing silicone tubes implanted into the bladders of mice as a model of catheter-associated urinary tract infections. Infect. Immun. 81 3009–3017. 10.1128/IAI.00348-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murugan K., Selvanayaki K., Al-Sohaibani S. (2016). Urinary catheter indwelling clinical pathogen biofilm formation, exopolysaccharide characterization and their growth influencing parameters. Saudi J. Biol. Sci. 23 150–159. 10.1016/j.sjbs.2015.04.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musthafa K. S., Ravi A. V., Annapoorani A., Packiavathy I. S., Pandian S. K. (2010). Evaluation of anti-quorum-sensing activity of edible plants and fruits through inhibition of the N-acyl-homoserine lactone system in Chromobacterium violaceum and Pseudomonas aeruginosa. Chemotherapy 56 333–339. 10.1159/000320185 [DOI] [PubMed] [Google Scholar]
- Myszka K., Czaczyk K. (2009). Characterization of adhesive exopolysaccharide (EPS) produced by Pseudomonas aeruginosa under starvation conditions. Curr. Microbiol. 58 541–546. 10.1007/s00284-009-9365-3 [DOI] [PubMed] [Google Scholar]
- Naidoo S., Olaniran A. O. (2013). Treated wastewater effluent as a source of microbial pollution of surface water resources. Int. J. Environ. Res. Public Health 11 249–270. 10.3390/ijerph110100249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Toole G., Kaplan H. B., Kolter R. (2000). Biofilm formation as microbial development. Annu. Rev. Microbiol. 54 49–79. [DOI] [PubMed] [Google Scholar]
- Packiavathy I. A., Priya S., Pandian S. K., Ravi A. V. (2014). Inhibition of biofilm development of uropathogens by curcumin – an anti-quorum sensing agent from Curcuma longa. Food Chem. 148 453–460. 10.1016/j.foodchem.2012.08.002 [DOI] [PubMed] [Google Scholar]
- Pajkos A., Deva A. K., Vickery K., Cope C., Chang L., Cossart Y. E. (2003). Detection of subclinical infection in significant breast implant capsules. Plast. Reconstr. Surg. 111 1605–1611. [DOI] [PubMed] [Google Scholar]
- Pakharukova N., Tuittila M., Paavilainen S., Malmi H., Parilova O., Teneberg S., et al. (2018). Structural basis for Acinetobacter baumannii biofilm formation. Proc. Natl. Acad. Sci. U.S.A. 115 5558–5563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal Z., Urban E., Dosa E., Pal A., Nagy E. (2005). Biofilm formation on intrauterine devices in relation to duration of use. J. Med. Microbiol. 54 1199–1203. [DOI] [PubMed] [Google Scholar]
- Palmer J., Flint S., Brooks J. (2007). Bacterial cell attachment, the beginning of a biofilm. J. Ind. Microbiol. Biotechnol. 34 577–588. [DOI] [PubMed] [Google Scholar]
- Pandin C., Le Coq D., Canette A., Aymerich S., Briandet R. (2017). Should the biofilm mode of life be taken into consideration for microbial biocontrol agents? Microb. Biotechnol. 10 719–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papenfort K., Bassler B. L. (2016). Quorum sensing signal-response systems in Gram-negative bacteria. Nat. Rev. Microbiol. 14 576–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrino B., Schillaci D., Carnevale I., Giovannetti E., Diana P., Cirrincione G., et al. (2019). Synthetic small molecules as anti-biofilm agents in the struggle against antibiotic resistance. Eur. J. Med. Chem. 161 154–178. [DOI] [PubMed] [Google Scholar]
- Pedersen K. (1990). Biofilm development on stainless steel and pvc surfaces in drinking water. Water Res. 24 239–243. [Google Scholar]
- Percival S. L., Suleman L., Vuotto C., Donelli G. (2015). Healthcare-associated infections, medical devices and biofilms: risk, tolerance and control. J. Med. Microbiol. 64 323–334. [DOI] [PubMed] [Google Scholar]
- Perni S., Preedy E. C., Prokopovich P. (2014). Success and failure of colloidal approaches in adhesion of microorganisms to surfaces. Adv. Colloid Interface Sci. 206 265–274. [DOI] [PubMed] [Google Scholar]
- Petrova O. E., Schurr J. R., Schurr M. J., Sauer K. (2012). Microcolony formation by the opportunistic pathogen Pseudomonas aeruginosa requires pyruvate and pyruvate fermentation. Mol. Microbiol. 86 819–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pihl M., Davies J. R., Johansson A. C., Svensater G. (2013). Bacteria on catheters in patients undergoing peritoneal dialysis. Perit. Dial. Int. 33 51–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponnusamy K., Paul D., Sam Kim Y., Kweon J. H. (2010). 2(5H)-Furanone: a prospective strategy for biofouling-control in membrane biofilm bacteria by quorum sensing inhibition. Braz. J. Microbiol. 41 227–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter S. L., Wadhams G. H., Armitage J. P. (2011). Signal processing in complex chemotaxis pathways. Nat. Rev. Microbiol. 9 153–165. [DOI] [PubMed] [Google Scholar]
- Prest E. I., Hammes F., Van Loosdrecht M. C., Vrouwenvelder J. S. (2016). Biological stability of drinking water: controlling factors, methods, and challenges. Front. Microbiol. 7:45 10.3389/fmicb.2016.00045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabin N., Zheng Y., Opoku-Temeng C., Du Y., Bonsu E., Sintim H. O. (2015). Biofilm formation mechanisms and targets for developing antibiofilm agents. Future Med. Chem. 7 493–512. [DOI] [PubMed] [Google Scholar]
- Raghupathi P. K., Liu W., Sabbe K., Houf K., Burmolle M., Sorensen S. J. (2017). Synergistic interactions within a multispecies biofilm enhance individual species protection against grazing by a pelagic protozoan. Front. Microbiol. 8:2649 10.3389/fmicb.2017.02649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman M., Kim S., Kim S. M., Seol S. Y., Kim J. (2011). Characterization of induced Staphylococcus aureus bacteriophage Sap-26 and its anti-biofilm activity with rifampicin. Biofouling 27 1087–1093. [DOI] [PubMed] [Google Scholar]
- Rajmohan S., Dodd C. E., Waites W. M. (2002). Enzymes from isolates of Pseudomonas fluorescens involved in food spoilage. J. Appl. Microbiol. 93 205–213. [DOI] [PubMed] [Google Scholar]
- Remy B., Mion S., Plener L., Elias M., Chabriere E., Daude D. (2018). Interference in bacterial quorum sensing: a biopharmaceutical perspective. Front. Pharmacol. 9:203. 10.3389/fphar.2018.00203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren L., Mccuskey S. R., Moreland A., Bazan G. C., Nguyen T.-Q. (2019). Tuning Geobacter sulfurreducens biofilm with conjugated polyelectrolyte for increased performance in bioelectrochemical system. Biosens. Bioelectron. 144:111630. [DOI] [PubMed] [Google Scholar]
- Ren Z., Steinberg L. M., Regan J. M. (2008). Electricity production and microbial biofilm characterization in cellulose-fed microbial fuel cells. Water Sci. Technol. 58 617–622. [DOI] [PubMed] [Google Scholar]
- Renner L. D., Weibel D. B. (2011). Physicochemical regulation of biofilm formation. MRS Bull. 36 347–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieger U. M., Mesina J., Kalbermatten D. F., Haug M., Frey H. P., Pico R., et al. (2013). Bacterial biofilms and capsular contracture in patients with breast implants. Br. J. Surg. 100 768–774. [DOI] [PubMed] [Google Scholar]
- Rijnaarts H. H. M., Norde W., Lyklema J., Zehnder A. J. B. (1999). DLVO and steric contributions to bacterial deposition in media of different ionic strengths. Colloids Surf. B Biointerfaces 14 179–195. [Google Scholar]
- Rivas L., Fegan N., Dykes G. A. (2007). Attachment of Shiga toxigenic Escherichia coli to stainless steel. Int. J. Food Microbiol. 115 89–94. [DOI] [PubMed] [Google Scholar]
- Roder H. L., Herschend J., Russel J., Andersen M. F., Madsen J. S., Sorensen S. J., et al. (2018). Enhanced bacterial mutualism through an evolved biofilm phenotype. ISME J. 12 2608–2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Martinez E. M., Perez E. X., Schadt C. W., Zhou J., Massol-Deya A. A. (2006). Microbial diversity and bioremediation of a hydrocarbon-contaminated aquifer (Vega Baja, Puerto Rico). Int. J. Environ. Res. Public Health 3 292–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers J., Dowsett A. B., Dennis P. J., Lee J. V., Keevil C. W. (1994). Influence of plumbing materials on biofilm formation and growth of Legionella pneumophila in potable water systems. Appl. Environ. Microbiol. 60 1842–1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohde H., Frankenberger S., Zahringer U., Mack D. (2010). Structure, function and contribution of polysaccharide intercellular adhesin (PIA) to Staphylococcus epidermidis biofilm formation and pathogenesis of biomaterial-associated infections. Eur. J. Cell Biol. 89 103–111. [DOI] [PubMed] [Google Scholar]
- Roosjen A., De Vries J., Van Der Mei H. C., Norde W., Busscher H. J. (2005). Stability and effectiveness against bacterial adhesion of poly(ethylene oxide) coatings in biological fluids. J. Biomed. Mater. Res. B Appl. Biomater. 73 347–354. [DOI] [PubMed] [Google Scholar]
- Roosjen A., Kaper H. J., Van Der Mei H. C., Norde W., Busscher H. J. (2003). Inhibition of adhesion of yeasts and bacteria by poly(ethylene oxide)-brushes on glass in a parallel plate flow chamber. Microbiology 149 3239–3246. [DOI] [PubMed] [Google Scholar]
- Rosche B., Li X. Z., Hauer B., Schmid A., Buehler K. (2009). Microbial biofilms: a concept for industrial catalysis? Trends Biotechnol. 27 636–643. [DOI] [PubMed] [Google Scholar]
- Rudrappa T., Biedrzycki M. L., Bais H. P. (2008). Causes and consequences of plant-associated biofilms. FEMS Microbiol. Ecol. 64 153–166. [DOI] [PubMed] [Google Scholar]
- Sadekuzzaman M., Yang S., Mizan M. F. R., Ha S. D. (2015). Current and recent advanced strategies for combating biofilms. Compr. Rev. Food Sci. Food Safety 14 491–509. [Google Scholar]
- Sanchez E., Rivas Morales C., Castillo S., Leos-Rivas C., Garcia-Becerra L., Ortiz Martinez D. M. (2016). Antibacterial and antibiofilm activity of methanolic plant extracts against nosocomial microorganisms. Evid. Based Complement. Alternat. Med. 2016:1572697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos A., Galdino A. C. M., Mello T. P., Ramos L. S., Branquinha M. H., Bolognese A. M., et al. (2018). What are the advantages of living in a community? A microbial biofilm perspective! Mem. Inst. Oswaldo Cruz. 113:e180212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos A. P., Watanabe E., Andrade D. (2011). Biofilm on artificial pacemaker: fiction or reality? Arq. Bras. Cardiol. 97 e113–e120. [DOI] [PubMed] [Google Scholar]
- Sao-Jose C. (2018). Engineering of phage-derived lytic enzymes: improving their potential as antimicrobials. Antibiotics (Basel) 7:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirawski J., Perlin M. H. (2018). Plant(-)microbe interaction 2017-the good, the bad and the diverse. Int. J. Mol. Sci. 19:1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz M. P., Bendick J. A., Holm E. R., Hertel W. M. (2011). Economic impact of biofouling on a naval surface ship. Biofouling 27 87–98. [DOI] [PubMed] [Google Scholar]
- Sharahi J. Y., Azimi T., Shariati A., Safari H., Tehrani M. K., Hashemi A. (2019). Advanced strategies for combating bacterial biofilms. J. Cell. Physiol. 234 14689–14708. [DOI] [PubMed] [Google Scholar]
- Shen D., Langenheder S., Jurgens K. (2018). Dispersal modifies the diversity and composition of active bacterial communities in response to a salinity disturbance. Front. Microbiol. 9:2188 10.3389/fmicb.2018.02188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirey T. B., Thacker R. W., Olson J. B. (2012). Composition and stability of bacterial communities associated with granular activated carbon and anthracite filters in a pilot scale municipal drinking water treatment facility. J. Water Health 10 244–255. [DOI] [PubMed] [Google Scholar]
- Shokouhfard M., Kermanshahi R. K., Shahandashti R. V., Feizabadi M. M., Teimourian S. (2015). The inhibitory effect of a Lactobacillus acidophilus derived biosurfactant on biofilm producer Serratia marcescens. Iran J. Basic Med. Sci. 18 1001–1007. [PMC free article] [PubMed] [Google Scholar]
- Siezen R. J., Wilson G. (2009). Bioleaching genomics. Microb. Biotechnol. 2 297–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva V. O., Soares L. O., Silva Junior A., Mantovani H. C., Chang Y. F., Moreira M. A. (2014). Biofilm formation on biotic and abiotic surfaces in the presence of antimicrobials by Escherichia coli Isolates from cases of bovine mastitis. Appl. Environ. Microbiol. 80 6136–6145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons M., Drescher K., Nadell C. D., Bucci V. (2018). Phage mobility is a core determinant of phage-bacteria coexistence in biofilms. ISME J. 12 531–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson D. R. (2008). Biofilm processes in biologically active carbon water purification. Water Res. 42 2839–2848. [DOI] [PubMed] [Google Scholar]
- Singh H. M., Pathak A. K., Chopra K., Tyagi V. V., Anand S., Kothari R. (2019). Microbial fuel cells: a sustainable solution for bioelectricity generation and wastewater treatment. Biofuels 10 11–31. [Google Scholar]
- Singh P. K., Bartalomej S., Hartmann R., Jeckel H., Vidakovic L., Nadell C. D., et al. (2017). Vibrio cholerae combines individual and collective sensing to trigger biofilm dispersal. Curr. Biol. 27 3359–3366.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R., Paul D., Jain R. K. (2006). Biofilms: implications in bioremediation. Trends Microbiol. 14 389–397. [DOI] [PubMed] [Google Scholar]
- Somers E. B., Wong A. C. (2004). Efficacy of two cleaning and sanitizing combinations on Listeria monocytogenes biofilms formed at low temperature on a variety of materials in the presence of ready-to-eat meat residue. J. Food Prot. 67 2218–2229. [DOI] [PubMed] [Google Scholar]
- Somogyi-Ganss E., Chambers M. S., Lewin J. S., Tarrand J. J., Hutcheson K. A. (2017). Biofilm on the tracheoesophageal voice prosthesis: considerations for oral decontamination. Eur. Arch. Otorhinolaryngol. 274 405–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa C., Rodrigues D., Oliveira R., Song W., Mano J. F., Azeredo J. (2011). Superhydrophobic poly(L-lactic acid) surface as potential bacterial colonization substrate. AMB Express 1:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southey-Pillig C. J., Davies D. G., Sauer K. (2005). Characterization of temporal protein production in Pseudomonas aeruginosa biofilms. J. Bacteriol. 187 8114–8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperandio V., Torres A. G., Giron J. A., Kaper J. B. (2001). Quorum sensing is a global regulatory mechanism in enterohemorrhagic Escherichia coli O157:H7. J. Bacteriol. 183 5187–5197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srey S., Jahid I. K., Ha S.-D. (2013). Biofilm formation in food industries: a food safety concern. Food Control 31 572–585. [Google Scholar]
- Steddom K., Menge J. A., Crowley D., Borneman J. (2002). Effect of repetitive applications of the biocontrol bacterium Pseudomonas putida 06909-rif/nal on citrus soil microbial communities. Phytopathology 92 857–862. [DOI] [PubMed] [Google Scholar]
- Stein T. (2005). Bacillus subtilis antibiotics: structures, syntheses and specific functions. Mol. Microbiol. 56 845–857. [DOI] [PubMed] [Google Scholar]
- Stoodley P., Sauer K., Davies D. G., Costerton J. W. (2002). Biofilms as complex differentiated communities. Annu. Rev. Microbiol. 56 187–209. [DOI] [PubMed] [Google Scholar]
- Sun J., Daniel R., Wagner-Dobler I., Zeng A. P. (2004). Is autoinducer-2 a universal signal for interspecies communication: a comparative genomic and phylogenetic analysis of the synthesis and signal transduction pathways. BMC Evol. Biol. 4:36. 10.1186/1471-2148-4-36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweet M. J., Croquer A., Bythell J. C. (2011). Development of bacterial biofilms on artificial corals in comparison to surface-associated microbes of hard corals. PLoS One 6:e21195. 10.1371/journal.pone.0021195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira P., Oliveira R. (1999). Influence of surface characteristics on the adhesion of Alcaligenes denitrificans to polymeric substrates. J. Adhes. Sci. Technol. 13 1287–1294. [Google Scholar]
- Terada A., Yamamoto T., Hibiya K., Tsuneda S., Hirata A. (2004). Enhancement of biofilm formation onto surface-modified hollow-fiber membranes and its application to a membrane-aerated biofilm reactor. Water Sci. Technol. 49 263–268. [PubMed] [Google Scholar]
- Terashima H., Kojima S., Homma M. (2008). Flagellar motility in bacteria structure and function of flagellar motor. Int. Rev. Cell Mol. Biol. 270 39–85. 10.1016/S1937-6448(08)01402-0 [DOI] [PubMed] [Google Scholar]
- Timmusk S., Behers L., Muthoni J., Muraya A., Aronsson A. C. (2017). Perspectives and challenges of microbial application for crop improvement. Front. Plant Sci. 8:49. 10.3389/fpls.2017.00049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomaras A. P., Dorsey C. W., Edelmann R. E., Actis L. A. (2003). Attachment to and biofilm formation on abiotic surfaces by Acinetobacter baumannii: involvement of a novel chaperone-usher pili assembly system. Microbiology 149 3473–3484. 10.1099/mic.0.26541-0 [DOI] [PubMed] [Google Scholar]
- Toyofuku M., Inaba T., Kiyokawa T., Obana N., Yawata Y., Nomura N. (2016). Environmental factors that shape biofilm formation. Biosci. Biotechnol. Biochem. 80 7–12. 10.1080/09168451.2015.1058701 [DOI] [PubMed] [Google Scholar]
- Tremblay Y. D., Levesque C., Segers R. P., Jacques M. (2013). Method to grow Actinobacillus pleuropneumoniae biofilm on a biotic surface. BMC Vet. Res. 9:213. 10.1186/1746-6148-9-213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truchado P., Gimenez-Bastida J. A., Larrosa M., Castro-Ibanez I., Espin J. C., Tomas-Barberan F. A., et al. (2012). Inhibition of quorum sensing (QS) in Yersinia enterocolitica by an orange extract rich in glycosylated flavanones. J. Agric. Food Chem. 60 8885–8894. [DOI] [PubMed] [Google Scholar]
- Tuson H. H., Weibel D. B. (2013). Bacteria-surface interactions. Soft Matter 9 4368–4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ukuku D. O., Fett W. F. (2002). Relationship of cell surface charge and hydrophobicity to strength of attachment of bacteria to cantaloupe rind. J. Food Prot. 65 1093–1099. [DOI] [PubMed] [Google Scholar]
- Upadhyayula V. K., Gadhamshetty V. (2010). Appreciating the role of carbon nanotube composites in preventing biofouling and promoting biofilms on material surfaces in environmental engineering: a review. Biotechnol. Adv. 28 802–816. 10.1016/j.biotechadv.2010.06.006 [DOI] [PubMed] [Google Scholar]
- Vacheethasanee K., Temenoff J. S., Higashi J. M., Gary A., Anderson J. M., Bayston R., et al. (1998). Bacterial surface properties of clinically isolated Staphylococcus epidermidis strains determine adhesion on polyethylene. J. Biomed. Mater. Res. 42 425–432. [DOI] [PubMed] [Google Scholar]
- Vaishnavi C., Samanta J., Kochhar R. (2018). Characterization of biofilms in biliary stents and potential factors involved in occlusion. World J. Gastroenterol. 24 112–123. 10.3748/wjg.v24.i1.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dillewijn P., Nojiri H., Van Der Meer J. R., Wood T. K. (2009). Bioremediation, a broad perspective. Microb. Biotechnol. 2 125–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Houdt R., Michiels C. W. (2005). Role of bacterial cell surface structures in Escherichia coli biofilm formation. Res. Microbiol. 156 626–633. [DOI] [PubMed] [Google Scholar]
- Van Houdt R., Michiels C. W. (2010). Biofilm formation and the food industry, a focus on the bacterial outer surface. J. Appl. Microbiol. 109 1117–1131. 10.1111/j.1365-2672.2010.04756.x [DOI] [PubMed] [Google Scholar]
- Vanhaecke E., Remon J. P., Moors M., Raes F., De Rudder D., Van Peteghem A. (1990). Kinetics of Pseudomonas aeruginosa adhesion to 304 and 316-L stainless steel: role of cell surface hydrophobicity. Appl. Environ. Microbiol. 56 788–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasilev K., Cavallaro A., Zilm P. (2018). Special Issue: antibacterial materials and coatings. Molecules 23:585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vatanyoopaisarn S., Nazli A., Dodd C. E., Rees C. E., Waites W. M. (2000). Effect of flagella on initial attachment of Listeria monocytogenes to stainless steel. Appl. Environ. Microbiol. 66 860–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veerachamy S., Yarlagadda T., Manivasagam G., Yarlagadda P. K. (2014). Bacterial adherence and biofilm formation on medical implants: a review. Proc. Inst. Mech. Eng. H 228 1083–1099. 10.1177/0954411914556137 [DOI] [PubMed] [Google Scholar]
- Vejan P., Abdullah R., Khadiran T., Ismail S., Nasrulhaq Boyce A. (2016). Role of plant growth promoting Rhizobacteria in agricultural sustainability-a review. Molecules 21:573. 10.3390/molecules21050573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vladimirov N., Sourjik V. (2009). Chemotaxis: how bacteria use memory. Biol. Chem. 390 1097–1104. 10.1515/BC.2009.130 [DOI] [PubMed] [Google Scholar]
- Wei Q., Ma L. Z. (2013). Biofilm matrix and its regulation in Pseudomonas aeruginosa. Int. J. Mol. Sci. 14 20983–21005. 10.3390/ijms141020983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirtanen G., Salo S. (2016). “Chapter 5 – biofilm risks,” in Handbook of Hygiene Control in the Food Industry (Second Edition), eds Lelieveld H., Holah J., Gabriæ D. (San Diego, CA: Woodhead Publishing; ). [Google Scholar]
- Wood T. K. (2013). Precedence for the structural role of flagella in biofilms. mBio 4:e00225-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H., Moser C., Wang H. Z., Hoiby N., Song Z. J. (2015). Strategies for combating bacterial biofilm infections. Int. J. Oral Sci. 7 1–7. 10.1038/ijos.2014.65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Q., Barrios C. A., Cutright T., Newby B. M. (2005). Assessment of antifouling effectiveness of two natural product antifoulants by attachment study with freshwater bacteria. Environ. Sci. Pollut. Res. Int. 12 278–284. [DOI] [PubMed] [Google Scholar]
- Yamashita T., Yamamoto-Ikemoto R. (2014). Nitrogen and phosphorus removal from wastewater treatment plant effluent via bacterial sulfate reduction in an anoxic bioreactor packed with wood and iron. Int. J. Environ. Res. Public Health 11 9835–9853. 10.3390/ijerph110909835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao J., Allen C. (2007). The plant pathogen Ralstonia solanacearum needs aerotaxis for normal biofilm formation and interactions with its tomato host. J. Bacteriol. 189 6415–6424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung P. S., Boor K. J. (2004). Epidemiology, pathogenesis, and prevention of foodborne Vibrio parahaemolyticus infections. Foodborne Pathog. Dis. 1 74–88. [DOI] [PubMed] [Google Scholar]
- Yoshikawa M., Zhang M., Toyota K. (2017). Integrated anaerobic-aerobic biodegradation of multiple contaminants including chlorinated ethylenes, benzene, toluene, and dichloromethane. Water Air Soil Pollut. 228:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu P., Wang C., Zhou J., Jiang L., Xue J., Li W. (2016). Influence of surface properties on adhesion forces and attachment of Streptococcus mutans to Zirconia In Vitro. Biomed. Res. Int. 2016:8901253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Brodus D., Hollimon V., Hu H. (2017). A brief review of recent developments in the designs that prevent bio-fouling on silicon and silicon-based materials. Chem. Cent. J. 11:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Zhang Q., Yan T., Jiang Z., Zhang X., Zuo Y. Y. (2015). Quantitatively predicting bacterial adhesion using surface free energy determined with a spectrophotometric method. Environ. Sci. Technol. 49 6164–6171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B., Van Der Mei H. C., Subbiahdoss G., De Vries J., Rustema-Abbing M., Kuijer R., et al. (2014). Soft tissue integration versus early biofilm formation on different dental implant materials. Dent. Mater. 30 716–727. [DOI] [PubMed] [Google Scholar]
- Zielinska M., Rusanowska P., Jarzabek J., Nielsen J. L. (2016). Community dynamics of denitrifying bacteria in full-scale wastewater treatment plants. Environ. Technol. 37 2358–2367. [DOI] [PubMed] [Google Scholar]
- Zimmerli W., Sendi P. (2017). Orthopaedic biofilm infections. APMIS 125 353–364. 10.1111/apm.12687 [DOI] [PubMed] [Google Scholar]
- Zottola E. A., Sasahara K. C. (1994). Microbial biofilms in the food processing industry–should they be a concern? Int. J. Food Microbiol. 23 125–148. [DOI] [PubMed] [Google Scholar]
- Zuo R. (2007). Biofilms: strategies for metal corrosion inhibition employing microorganisms. Appl. Microbiol. Biotechnol. 76 1245–1253. [DOI] [PubMed] [Google Scholar]
- Zuo R., Ornek D., Syrett B. C., Green R. M., Hsu C. H., Mansfeld F. B., et al. (2004). Inhibiting mild steel corrosion from sulfate-reducing bacteria using antimicrobial-producing biofilms in Three-Mile-Island process water. Appl. Microbiol. Biotechnol. 64 275–283. [DOI] [PubMed] [Google Scholar]


