Since its initial discovery in mouse and human cells as nuclear protein 95 (Np95; [1]) and inverted CCAAT box-binding protein 90 (ICBP90; [2]), respectively, ubiquitin-like PHD and RING finger domain-containing protein 1 (UHRF1) has quickly risen to current prominence as a key epigenetic regulator in diverse cellular and developmental processes (reviewed in [3]). A recent in vivo study further revealed a critical role of UHRF1 during mouse spermatogenesis [4].
Much like the famed Swiss Army knife, the multidomain UHRF1 protein functions through its five arms, including from N-terminus to C-terminus, a ubiquitin-like (UBL) domain, a tandem Tudor domain (TTD), a plant homeodomain (PHD), a SET- and RING-associated (SRA) domain, and a really interesting new gene (RING) domain (Figure 1A). Initial studies focused on the characterization of individual domains and their interacting partners in vitro and ex vivo. Later studies uncovered the incredible amount of coordination and crosstalk among these domains, as UHRF1 carries out its multifaceted functions through precise interactions with DNA, histones, and other effector proteins.
Figure 1.
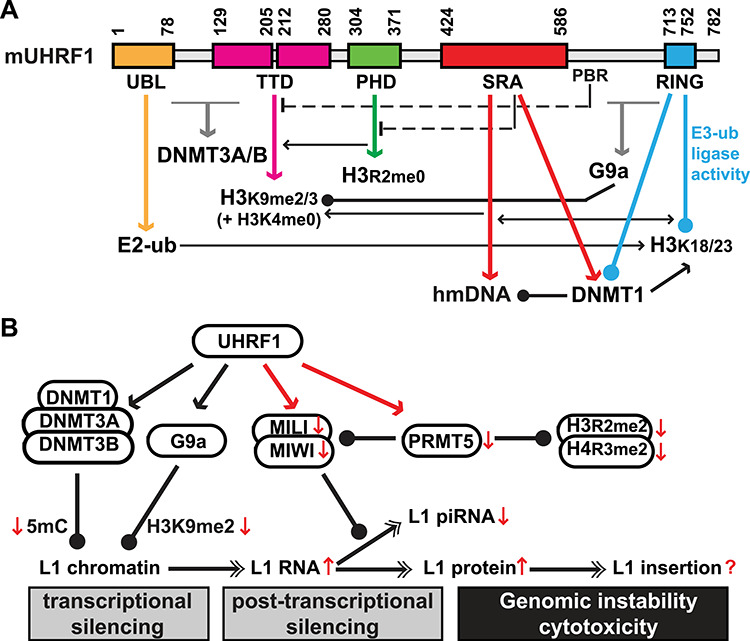
UHRF1 regulates multiple epigenetic pathways during spermatogenesis. (A) Mouse UHRF1 domains and interacting partners. Domain boundaries are labeled above the domain structure as amino acid coordinates per UniProt (https://www.uniprot.org/uniprot/Q8VDF2). Drawn to scale. Six domains/regions are labeled below the domain structure: UBL, ubiquitin like; TTD, tandem Tudor domain; PHD, plant homeodomain; SRA, SET- and RING-associated; PBR, polybasic region; and RING, really interesting new gene. Domains and corresponding interactions (down arrows) are color coded. Interactions with DNMT3A/3B and G9a have not been mapped to defined domains, thus in gray arrows. Ball-headed lines indicate enzymatic modifications. Crosstalk is annotated as either stimulatory (arrow) or inhibitory (T-end dashed lines for intramolecular interactions between TTD and PBR, and between PHD and SRA). hmDNA, hemi-methylated DNA. E2-ub, E2 ubiquitin conjugating enzyme. (B) Model for UHRF1-mediated epigenetic silencing of L1 during spermatogenesis. In postnatal testes, L1 retrotransposons are transcriptionally silenced by DNA methylation and H3K9me2/3, and post-transcriptionally by the pachytene piRNA pathway. In the light of the recent work by Dong and colleagues [4], UHRF1 has emerged as the master regulator of these epigenetic pathways. Newly reported interactions of UHRF1 with PIWI proteins and PRMT5 are highlighted (in red). The conditional loss of UHRF1 function causes meiotic catastrophe and germ cell death. Many changes are observed at molecular levels (↑, upregulation; ↓, downregulation). In particular, L1s are de-repressed at both RNA and protein levels. However, whether there is an increase in L1 insertion and to which degree L1 retrotransposition contributes to genomic instability in Uhrf1-deficient germ cells remain unknown.
UHRF1 is best known for its function in DNA methylation maintenance. It is a critical partner for the maintenance DNA methyltransferase DNMT1, which methylates the newly synthesized daughter strand following semi-conservative DNA replication. UHRF1 preferentially binds to hemi-methylated DNA through its SRA domain [5–7]. The SRA domain also interacts directly with DNMT1’s replication focus targeting sequence (RFTS) [7, 8], thus tethering DNMT1 to newly replicated DNA. The physical interaction between UHRF1 and DNMT1 increases the activity and specificity of DNMT1 for methylating hemi-methylated CG sites [9, 10]. In addition, UHRF1 may regulate do novo DNA methylation through interactions with DNMT3A and DNMT3B under specific cellular contexts [11].
Hemi-methylated DNA is not the only cue that UHRF1 takes from the genome. To guide DNMT1 to the right spot in the nucleus, UHRF1 also understands the highly complex language of chromatin and employs its other domains to decipher the combinatorial state of post-translational histone modifications (i.e., the histone code). Its TTD domain preferentially binds to a single histone H3 N-terminal tail with di- or tri-methylated lysine 9 (H3K9me2/3) and unmethylated lysine 4 (H3K4me0) [12–15]. The importance of this interaction has been shown in human HeLa cells [15] and mouse embryonic stem cells (ESCs) [16]. There is also evidence that UHRF1 interacts with G9a, one of the histone methyltransferases responsible for H3K9me2 [17]. There are additional layers of crosstalk among the domains. The PHD domain, which has an affinity to unmodified arginine 2 on the H3 tail (H3R2me0) [14, 18–20], facilitates the interaction between TTD and H3K9me3 [21]. Importantly, structural studies illustrate that the full-length UHRF1 protein adopts a closed conformation due to intramolecular interactions in the absence of ligands. Binding to H3K9me3 is blocked by an intramolecular interaction of TTD with a polybasic region (PBR) between the SRA and RING domains [22, 23]. Meanwhile, binding to H3R2me0 is inhibited by an intramolecular interaction of the PHD domain with the SRA domain [22]. Binding to hemi-methylated DNA shifts UHRF1 to an open state, which promotes H3K9me3 recognition by UHRF1 [22, 23]. Thus, UHRF1 integrates the two major epigenetic silencing pathways by its dynamic interactions with hemi-methylated DNA and H3K9me2/3.
UHRF1’s role in maintenance DNA methylation is also dependent on two other domains: the RING finger domain and the UBL domain. Its RING finger domain has E3 ubiquitin ligase activity toward histone H3 [24], DNMT1 [25, 26], and UHRF1 itself [27]. UHRF1-dependent H3 ubiquitination is a prerequisite for DNMT1 binding to DNA replication sites [28]. Hemi-methylated DNA stimulates UHRF1 ubiquitin ligase activity on H3 by directing TTD-PHD-bound H3 substrate to the active site of E2 ubiquitin ligase [29]. The tandem monoubiquitin marks on H3 are recognized by DNMT1 via a ubiquitin interaction motif (part of the RFTS binding module) [30, 31], which enhances DNMT1 recruitment and high-fidelity maintenance of DNA methylation. Indeed, it has been demonstrated that UHRF1’s E3 ubiquitin ligase activity is required for maintenance DNA methylation at retrotransposons and satellite repeats [28, 31]. In contrast, the ubiquitination of DNMT1 by UHRF1 regulates DNMT1 stability and promotes its degradation [25, 26]. The role of the UBL domain was only recently revealed. It is required for H3 ubiquitination by UHRF1 RING E3 ligase in stabilizing the E2-E3-chromatin complex, and also for maintenance DNA methylation on retrotransposons [32, 33].
So far, only a few studies have explored UHRF1’s function in vivo, in sharp contrast to aforementioned numerous in vitro and ex vivo studies that helped to illuminate UHRF1’s biochemical and cellular functions. UHRF1 is essential for early development. Global loss of function causes early developmental arrest shortly after gastrulation in mice [6, 34]. Thus, conditional knockout (cKO) strategies have been adopted to investigate the role of UHRF1 in other stages of development, including during the proliferation and maturation of colonic regulatory T cells [35], chondrocyte differentiation and limb growth [36], neuronal differentiation and survival [37], oocyte growth [38], and, most recently, spermatogenesis [4].
DNA methylation in the germline of mammalian genomes is not static. Globally, the germline genome undergoes two rounds of DNA methylation reprogramming during development. The first round occurs in preimplantation embryos and the second in migrating/post-migratory primordial germ cells (PGCs). Recent data indicate the erasure of global methylation in PGCs is largely due to replication-coupled passive DNA demethylation (reviewed in [39]). In rapidly proliferating PGCs between embryonic day (E) 9.5 and E11.5, both UHRF1 and de novo methyltransferases DNMT3A/3B are downregulated and become undetectable [40, 41]. Later, in the female germline, an oocyte-specific methylation pattern is established in growing oocytes (GOs) and completed in fully grown oocytes (FGOs) via de novo DNA methyltransferases DNMT3a/DNMT3L [42, 43]. To study the role of UHRF1 in oocyte and preimplantation embryonic development, the Zp3-Cre line was used, which is expressed exclusively in GOs [38]. In Uhrf1 cKO females, oogenesis is unaffected. At the molecular level, there is 20% reduction in global CG methylation and 15% reduction in global non-CG methylation in FGOs. As oocyte growth does not involve maintenance DNA methylation (no DNA replication in meiotic prophase I), these data suggest that UHRF1 participates in de novo CG and non-CG methylation in oogenesis. Importantly, in vitro fertilization of Uhrf1 cKO oocytes with wild-type sperm leads to the developmental arrest in 80% of the embryos [38]. Thus, the maternally derived UHRF1 protein is also required for preimplantation embryonic development. The loss of maternal UHRF1 leads to a global reduction in CG methylation, encompassing genic, intergenic, tandem repeats, retrotransposons (L1, B1, and IAP), and imprinted sequences, suggesting maternal UHRF1 is required for maintenance CG methylation in preimplantation embryos. However, its impact on retrotransposon expression was not assessed by the study [38].
DNA methylation is an important mechanism of controlling the proliferation and differentiation of stem cells, not only ESCs but also adult stem cells. Indeed, in the mouse brain, UHRF1 is expressed specifically in fetal and adult neural stem cells (NSCs) [37, 44]. To investigate whether early epigenetic mechanisms impact the long-term behavior of NSCs and derivatives, a recent study used Emx1-Cre to specifically abolish UHRF1 function in the dorsal telencephalon [37]. The Emx1 promoter is expressed exclusively in the dorsal telencephalon from embryo to adulthood. The conditional deletion of UHRF1 starts as early as E10 in the NSCs of the developing cerebral cortex. However, no gross morphological defect is observed during the embryonic development. The first morphological phenotype becomes detectable at postnatal day (P) 7 as the thickness of the cerebral cortex is reduced. The delayed neurodegeneration appears to be the result of increased cell death at the onset of embryonic neurogenesis and the subsequent escalated cell death during the neuronal maturation at postnatal stages. At the molecular level, reflecting an essential role of UHRF1 in maintaining DNA methylation, global DNA methylation, is significantly reduced, including 35% loss on L1, B1, and IAP retrotransposons. Surprisingly, there is little change in transcription of L1, B1, and annotated genes. The only exception is for IAP retrotransposons, more specifically from the IAPEz family, which shows ~130-fold increase in the cKO cortices. The extraordinary magnitude of change contrasts sharply with more modest alterations in Uhrf1 KO ESCs (2-fold) and embryos (4~8-fold) [6, 45]. These results indicate that UHRF1 is critical for IAP repression during neurogenesis [37].
Until recently, the physiological role of UHRF1 in spermatogenesis has not been explored. The testis boasts the highest level of UHRF1 RNA expression among a panel of mouse tissues when examined by northern blot [1]. Indeed, UHRF1 protein is present not only in proliferating spermatogonia but also in meiotic spermatocytes and differentiating spermatids [46], implicating a potential function during spermatogenesis. In a landmark paper, Dong and colleagues conditionally abrogated UHRF1 in the differentiating spermatogonia and spermatocytes of postnatal testes using a Stra8-Cre mouse line [4]. UHRF1 protein abundance and subcellular localization during male germ cell development were thoroughly analyzed. Interestingly, wild-type UHRF1 protein is detected in nearly all developmental stages, encompassing fetal, neonatal, and adult stages, from prospermatogonia to early round spermatids. Notably, there is a dynamic change in subcellular localization, beginning as mainly cytoplasmic in fetal prospermatogonia, nuclear in mitotic spermatogonia, back to cytoplasmic at the onset of meiosis (preleptotene, leptotene, zygotene, and early pachytene spermatocytes), and then nuclear again (late pachytene and round spermatids), although the regulation and significance of the cytoplasmic/nuclear shuttling remain unclear (see later discussions in the context of UHRF1’s interactions with PIWI proteins and PRMT5).
Phenotypically, the conditional deletion of UHRF1 in differentiating spermatogonia leads to a spermatogenic arrest at the pachytene spermatocyte stage and ultimately male infertility [4]. At the histological level, abnormality in seminiferous tubules becomes apparent as early as P14, when the first cohort of pachytene spermatocytes is formed. Cell marker analyses indicate that both spermatogonial differentiation and meiotic initiation are unaffected in cKO testes. However, staining of phosphorylated histone H2A.X (γ-H2A.X), a chromatin marker for adjacent DNA double-strand breaks (DSBs), shows an unusual nucleus-wide distribution in cKO pachytene spermatocytes, instead of being confined to the XY body. Such a pattern is indicative of a systemic failure in DNA damage repair and the persistence of genome-wide unrepaired DSBs. Many meiotic mouse mutants are accompanied with extensive asynapsis and failure in meiotic sex chromosome inactivation (MSCI), which is thought to trigger germ cell apoptosis at the mid-pachytene stage [47]. However, despite widespread γ-H2A.X signals, the autosomal synapsis appears to be complete in the residual Uhrf1 cKO pachytene spermatocytes. Additionally, the number of RPA2 foci is reduced in the Uhrf1 cKO spermatocytes, indicating a defect in meiotic recombination in meiotic prophase I. Regardless of the checkpoint pathways, the developmental defect in Uhrf1 cKO spermatocytes can be attributed to elevated apoptosis (approximately 3-fold higher than the wild-type control at P14), which persisted until at least P35 [4].
Given the role of UHRF1 in DNA methylation, Dong and colleagues first examined methylation levels by immunostaining of methylated cytosines (5mCs) [4]. DNA methylation in the Uhrf1 cKO appears normal at P10 but reduced globally in P14 testes. No apparent change is found in differentiating spermatogonia but 5mC signals become nearly undetectable in leptotene, zygotene, and pachytene spermatocytes. Bisulfite sequencing shows a substantial reduction in DNA methylation in the L1 5’UTR promoter region from 86 to 45% at P18, which is comparable to that seen in Uhrf1 KO embryos (85 to 40%) [6]. DNA methylation is also reduced from 90 to 70% at IAP elements, although the magnitude is much less than that in Uhrf1 KO embryos (95 to 20%) [6]. Mild reduction in methylation is observed in the typically paternally methylated intergenic differentially methylated region (DMR) between the imprinted genes Dlk1 and Gtl2 (93 to 68%). These data suggest that UHRF1 is required for DNA methylation maintenance during the postnatal male germ cell development. The current study did not examine methylation for other genes or heterochromatic regions, such as major or minor microsatellites. The magnitude of methylation loss in germ cells might have been substantially underestimated due to the presence of testicular somatic cells. To fully understand the role of UHRF1-mediated methylation in postnatal germ cells, it is imperative to pinpoint the precise timing of the methylation loss as well as to identify the genomic regions that are affected, preferably using a methylomics approach on stage-specific germ cell populations.
Following DNA methylation reprogramming in PGCs, in the postnatal testis, the male germline genome undergoes at least two additional episodes of dynamic DNA methylation. The first episode occurs as spermatogonia transition from undifferentiated (Kit−) to differentiating (Kit+) spermatogonia [48, 49]. Among the reported DMRs from whole-genome bisulfite sequencing analyses, more genomic regions lose methylation than gain methylation at this transition. Only 40 genomic regions gain >30% CG methylation, while 1352 regions lose >30% CG methylation [49]. The second wave is at the onset of meiosis, which features a transient reduction in DNA methylation (TRDM), equivalent to a genome-wide 12% of CG methylation loss, primarily in preleptotene spermatocytes [50]. The TRDM is thought to occur via DNA replication-dependent DNA demethylation due to a delay in maintenance DNA methylation. The global loss in CG methylation is fully regained in pachytene spermatocytes [50].
UHRF1 has been shown to play a role in spermatogonial differentiation in a Uhrf1 inducible KO (iKO) model [51]. In this iKO model, no Kit+ cells are identified, suggesting the loss of UHRF1 completely blocks spermatogonial differentiation and/or required for the survival of differentiating spermatogonia. The phenotype in the iKO model is much more severe than that observed in the cKO model by Dong and colleagues. However, the interpretation of the iKO results may be confounded by an unexpected adverse effect of tamoxifen itself on spermatogenesis [52]. In the paper of Dong and colleagues, Stra8-Cre is used to ablate Uhrf1 function genetically. Endogenous STRA8 protein is expressed only in differentiating spermatogonia through leptotene spermatocytes [53, 54]. However, the Stra8-Cre clearly functions in a subset of undifferentiated spermatogonia in the transgenic mice [55], potentially due to the lack of some endogenous regulatory elements in the promoter sequence used in the transgene. Thus, it is reasonable to suspect that Stra8-Cre-mediated deletion of UHRF1 may occur prior to the type A to A1 transition, which marks the beginning of spermatogonial differentiation [56]. Therefore, the impact from a loss of UHRF1, in the current model, points to a role of maintenance DNA methylation in differentiating spermatogonia as well as in meiotic prophase. Again, further analysis of DNA methylation changes is warranted to dissect the differential contribution of UHRF1 during these two developmental stages.
What is the impact of reduced methylation on gene expression? To this end, the authors examined the effects on gene expression by RNA-seq [4]. Few genes show deregulation at P9, but 200 genes are upregulated by >2-fold at P12. Additionally, many retrotransposon families are upregulated. The upregulation of L1 and IAP is also confirmed by qPCR, ranging from 3–7-fold in P18 and P56 testes. As retrotransposon expression is regulated in germ cells by the PIWI-interacting RNA (piRNA) pathway (reviewed by [57, 58]), the authors checked PIWI protein abundance and piRNA production [4]. Unexpectedly, both MILI and MIWI, two PIWI proteins that are expressed in spermatocytes, are downregulated in Uhrf1 cKO spermatocytes. In addition, the piRNA biogenesis is compromised. How does UHRF1 crosstalk with the piRNA pathway? In this regard, the authors offered novel evidence for the physical interactions of UHRF1 with MILI and MIWI [4]. Therefore, it is likely that such interactions in normal germ cells help to stabilize MILI and MIWI and facilitate the generation of pachytene piRNAs. What process might mediate this stabilization? Based on the involvement of PRMT5 (see below), the data support a model in which the stabilization of MILI and MIWI by UHRF1 is mediated via its recruitment of PRMT5 and subsequent symmetric dimethylation of these PIWI proteins (Figure 1B).
Indeed, the authors presented compelling evidence for an interaction between UHRF1’s TTD domain and PRMT5’s SAM domain [4]. In wild-type leptotene and zygotene spermatocytes, both PRMT5 and UHRF1 are enriched in the cytoplasm. In pachytene spermatocytes, PRMT5 and UHRF1 colocalize in the nucleus. In P18 Uhrf1 cKO testes, both Prmt5 mRNA and protein are downregulated, so are the PRMT5-mediated symmetrically dimethylated H3R2me2 and H4R3me2 marks. The impact of altered histone modifications in the Uhrf1 cKO testis is unclear. Interestingly, Prmt5 cKO, mediated by Stra8-Cre, phenocopies the Uhrf1 cKO, showing a similar meiotic arrest [59]. At P10, loss of PRMT5 has no impact on H3R2me2, although H4R2me2 is lost. L1 and IAP expression are also unaffected at P10. The authors of the Prmt5 cKO study did not examine PRMT5-mediated histone modifications and retrotransposon expression at later stages [59]. Therefore, it is still unclear whether the downregulation of PRMT5 in developing germ cells directly contributes to derepression of retrotransposons. It is also noteworthy that UHRF1 has a preference for unmethylated H3R2 [14, 18–20]. Both symmetrically and asymmetrically dimethylated H3R2s impede the interaction between UHRF1 and H3 tail in vitro [19]. In addition, H3R2me2 is only present in the cytoplasm of wild-type spermatocytes [4], suggesting H3R2 is not a substrate for PRMT5 in these cells. Taken together, it is more likely that the reduction of PRMT5 impacts substrates other than H3, including PIWI proteins and spliceosomal Sm proteins [60, 61]. The proposed model further predicts that the symmetrical dimethylation of MILI and MIWI is compromised in the Uhrf1 cKO spermatocytes, which should be experimentally validated in future studies (Figure 1B).
In summary, the recent study by Dong and colleagues [4] adds to our knowledge about the essential roles that UHRF1 plays in vivo. Already recognized as a jack of all trades, this study establishes UHRF1 as a master regulator of multiple epigenetic pathways during male germ cell development. Importantly, the observed infertility phenotype is highly relevant to human male reproduction as genetic polymorphisms in the human UHRF1 gene have been associated with oligospermia [62]. As discussed above, there remain many unanswered questions. In particular, the role of retrotransposon activation in the Uhrf1 cKO model awaits further elaboration. Deregulation of L1 retrotransposons can potentially impact germ cell well-being at three discrete stages of the L1 life cycle: transcription, translation, and retrotransposition [63] (Figure 1B). Previous studies have identified illegitimate meiotic recombination in Dnmt3l KO spermatocytes [64] and an increase in retrotransposition in Mov10l1 KO spermatocytes [63]. Thus, it will be informative to examine levels of retrotransposition by either NextGen sequencing approaches or the new L1 reporter mouse [63, 65]. In parallel, many domain-specific Uhrf1 mutants have been characterized in vitro and ex vivo. At least two knock-in (KI) mouse models have been established, including the SRA KI [45] and TTD KI [66]. Interestingly, the Uhrf1 TTD KI (Y191A/P192A) mouse model displays no overt phenotype, despite a failure in binding to H3K9me2/3 by the mutant UHRF1 protein, suggesting the binding of UHRF1 to H3K9me2/3 is not essential for animal development and reproduction [66]. Nevertheless, there is a modest global reduction in DNA methylation in several somatic tissues examined, including retrotransposons [66]. These and additional new KI models should allow the molecular dissection of the function of UHRF1’s individual domain module(s) during mammalian reproduction.
Grant Support: The research in the An lab is supported by grants from National Institutes of Health (R21HD080143, R21OD017965, R15GM131263, and R03HD099412) and the Markl Faculty Scholar Fund.
References
- 1. Fujimori A, Matsuda Y, Takemoto Y, Hashimoto Y, Kubo E, Araki R, Fukumura R, Mita K, Tatsumi K, Muto M. Cloning and mapping of Np95 gene which encodes a novel nuclear protein associated with cell proliferation. Mamm Genome 1998; 9:1032–1035. [DOI] [PubMed] [Google Scholar]
- 2. Hopfner R, Mousli M, Jeltsch JM, Voulgaris A, Lutz Y, Marin C, Bellocq JP, Oudet P, Bronner C. ICBP90, a novel human CCAAT binding protein, involved in the regulation of topoisomerase IIalpha expression. Cancer Res 2000; 60:121–128. [PubMed] [Google Scholar]
- 3. Bronner C, Alhosin M, Hamiche A, Mousli M. Coordinated dialogue between UHRF1 and DNMT1 to ensure faithful inheritance of methylated DNA patterns. Genes (Basel) 2019; 10:65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dong J, Wang X, Cao C, Wen Y, Sakashita A, Chen S, Zhang J, Zhang Y, Zhou L, Luo M, Liu M, Liao A et al. . UHRF1 suppresses retrotransposons and cooperates with PRMT5 and PIWI proteins in male germ cells. Nat Commun 2019; 10:4705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Unoki M, Nishidate T, Nakamura Y. ICBP90, an E2F-1 target, recruits HDAC1 and binds to methyl-CpG through its SRA domain. Oncogene 2004; 23:7601–7610. [DOI] [PubMed] [Google Scholar]
- 6. Sharif J, Muto M, Takebayashi S, Suetake I, Iwamatsu A, Endo TA, Shinga J, Mizutani-Koseki Y, Toyoda T, Okamura K, Tajima S, Mitsuya K et al. . The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA. Nature 2007; 450:908–912. [DOI] [PubMed] [Google Scholar]
- 7. Bostick M, Kim JK, Esteve PO, Clark A, Pradhan S, Jacobsen SE. UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science 2007; 317:1760–1764. [DOI] [PubMed] [Google Scholar]
- 8. Achour M, Jacq X, Ronde P, Alhosin M, Charlot C, Chataigneau T, Jeanblanc M, Macaluso M, Giordano A, Hughes AD, Schini-Kerth VB, Bronner C. The interaction of the SRA domain of ICBP90 with a novel domain of DNMT1 is involved in the regulation of VEGF gene expression. Oncogene 2008; 27:2187–2197. [DOI] [PubMed] [Google Scholar]
- 9. Bashtrykov P, Jankevicius G, Jurkowska RZ, Ragozin S, Jeltsch A. The UHRF1 protein stimulates the activity and specificity of the maintenance DNA methyltransferase DNMT1 by an allosteric mechanism. J Biol Chem 2014; 289:4106–4115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Berkyurek AC, Suetake I, Arita K, Takeshita K, Nakagawa A, Shirakawa M, Tajima S. The DNA methyltransferase Dnmt1 directly interacts with the SET and RING finger-associated (SRA) domain of the multifunctional protein Uhrf1 to facilitate accession of the catalytic center to hemi-methylated DNA. J Biol Chem 2014; 289:379–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Meilinger D, Fellinger K, Bultmann S, Rothbauer U, Bonapace IM, Klinkert WE, Spada F, Leonhardt H. Np95 interacts with de novo DNA methyltransferases, Dnmt3a and Dnmt3b, and mediates epigenetic silencing of the viral CMV promoter in embryonic stem cells. EMBO Rep 2009; 10:1259–1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Rottach A, Frauer C, Pichler G, Bonapace IM, Spada F, Leonhardt H. The multi-domain protein Np95 connects DNA methylation and histone modification. Nucleic Acids Res 2010; 38:1796–1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Nady N, Lemak A, Walker JR, Avvakumov GV, Kareta MS, Achour M, Xue S, Duan S, Allali-Hassani A, Zuo X, Wang YX, Bronner C et al. . Recognition of multivalent histone states associated with heterochromatin by UHRF1 protein. J Biol Chem 2011; 286:24300–24311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Arita K, Isogai S, Oda T, Unoki M, Sugita K, Sekiyama N, Kuwata K, Hamamoto R, Tochio H, Sato M, Ariyoshi M, Shirakawa M. Recognition of modification status on a histone H3 tail by linked histone reader modules of the epigenetic regulator UHRF1. Proc Natl Acad Sci USA 2012; 109:12950–12955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rothbart SB, Krajewski K, Nady N, Tempel W, Xue S, Badeaux AI, Barsyte-Lovejoy D, Martinez JY, Bedford MT, Fuchs SM, Arrowsmith CH, Strahl BD. Association of UHRF1 with methylated H3K9 directs the maintenance of DNA methylation. Nat Struct Mol Biol 2012; 19:1155–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Liu X, Gao Q, Li P, Zhao Q, Zhang J, Li J, Koseki H, Wong J. UHRF1 targets DNMT1 for DNA methylation through cooperative binding of hemi-methylated DNA and methylated H3K9. Nat Commun 2013; 4:1563. [DOI] [PubMed] [Google Scholar]
- 17. Kim JK, Esteve PO, Jacobsen SE, Pradhan S. UHRF1 binds G9a and participates in p21 transcriptional regulation in mammalian cells. Nucleic Acids Res 2009; 37:493–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hu L, Li Z, Wang P, Lin Y, Xu Y. Crystal structure of PHD domain of UHRF1 and insights into recognition of unmodified histone H3 arginine residue 2. Cell Res 2011; 21:1374–1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wang C, Shen J, Yang Z, Chen P, Zhao B, Hu W, Lan W, Tong X, Wu H, Li G, Cao C. Structural basis for site-specific reading of unmodified R2 of histone H3 tail by UHRF1 PHD finger. Cell Res 2011; 21:1379–1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rajakumara E, Wang Z, Ma H, Hu L, Chen H, Lin Y, Guo R, Wu F, Li H, Lan F, Shi YG, Xu Y et al. . PHD finger recognition of unmodified histone H3R2 links UHRF1 to regulation of euchromatic gene expression. Mol Cell 2011; 43:275–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Xie S, Jakoncic J, Qian C. UHRF1 double tudor domain and the adjacent PHD finger act together to recognize K9me3-containing histone H3 tail. J Mol Biol 2012; 415:318–328. [DOI] [PubMed] [Google Scholar]
- 22. Fang J, Cheng J, Wang J, Zhang Q, Liu M, Gong R, Wang P, Zhang X, Feng Y, Lan W, Gong Z, Tang C et al. . Hemi-methylated DNA opens a closed conformation of UHRF1 to facilitate its histone recognition. Nat Commun 2016; 7:11197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gao L, Tan XF, Zhang S, Wu T, Zhang ZM, Ai HW, Song J. An Intramolecular interaction of UHRF1 reveals dual control for its histone association. Structure 2018; 26:304–311e303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Citterio E, Papait R, Nicassio F, Vecchi M, Gomiero P, Mantovani R, Di Fiore PP, Bonapace IM. Np95 is a histone-binding protein endowed with ubiquitin ligase activity. Mol Cell Biol 2004; 24:2526–2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Qin W, Leonhardt H, Spada F. Usp7 and Uhrf1 control ubiquitination and stability of the maintenance DNA methyltransferase Dnmt1. J Cell Biochem 2011; 112:439–444. [DOI] [PubMed] [Google Scholar]
- 26. Du Z, Song J, Wang Y, Zhao Y, Guda K, Yang S, Kao HY, Xu Y, Willis J, Markowitz SD, Sedwick D, Ewing RM et al. . DNMT1 stability is regulated by proteins coordinating deubiquitination and acetylation-driven ubiquitination. Sci Signal 2010; 3:ra80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Jenkins Y, Markovtsov V, Lang W, Sharma P, Pearsall D, Warner J, Franci C, Huang B, Huang J, Yam GC, Vistan JP, Pali E et al. . Critical role of the ubiquitin ligase activity of UHRF1, a nuclear RING finger protein, in tumor cell growth. Mol Biol Cell 2005; 16:5621–5629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Nishiyama A, Yamaguchi L, Sharif J, Johmura Y, Kawamura T, Nakanishi K, Shimamura S, Arita K, Kodama T, Ishikawa F, Koseki H, Nakanishi M. Uhrf1-dependent H3K23 ubiquitylation couples maintenance DNA methylation and replication. Nature 2013; 502:249–253. [DOI] [PubMed] [Google Scholar]
- 29. Harrison JS, Cornett EM, Goldfarb D, DaRosa PA, Li ZM, Yan F, Dickson BM, Guo AH, Cantu DV, Kaustov L, Brown PJ, Arrowsmith CH et al. . Hemi-methylated DNA regulates DNA methylation inheritance through allosteric activation of H3 ubiquitylation by UHRF1. Elife 2016; 5:e17101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ishiyama S, Nishiyama A, Saeki Y, Moritsugu K, Morimoto D, Yamaguchi L, Arai N, Matsumura R, Kawakami T, Mishima Y, Hojo H, Shimamura S et al. . Structure of the Dnmt1 reader module complexed with a unique two-mono-ubiquitin mark on histone H3 reveals the basis for DNA methylation maintenance. Mol Cell 2017; 68:350–360e357. [DOI] [PubMed] [Google Scholar]
- 31. Qin W, Wolf P, Liu N, Link S, Smets M, La Mastra F, Forne I, Pichler G, Horl D, Fellinger K, Spada F, Bonapace IM et al. . DNA methylation requires a DNMT1 ubiquitin interacting motif (UIM) and histone ubiquitination. Cell Res 2015; 25:911–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. DaRosa PA, Harrison JS, Zelter A, Davis TN, Brzovic P, Kuhlman B, Klevit RE. A Bifunctional role for the UHRF1 UBL domain in the control of hemi-methylated DNA-dependent histone ubiquitylation. Mol Cell 2018; 72:753–765e756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Foster BM, Stolz P, Mulholland CB, Montoya A, Kramer H, Bultmann S, Bartke T. Critical role of the UBL domain in stimulating the E3 ubiquitin ligase activity of UHRF1 toward chromatin. Mol Cell 2018; 72:739–752e739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Muto M, Kanari Y, Kubo E, Takabe T, Kurihara T, Fujimori A, Tatsumi K. Targeted disruption of Np95 gene renders murine embryonic stem cells hypersensitive to DNA damaging agents and DNA replication blocks. J Biol Chem 2002; 277:34549–34555. [DOI] [PubMed] [Google Scholar]
- 35. Obata Y, Furusawa Y, Endo TA, Sharif J, Takahashi D, Atarashi K, Nakayama M, Onawa S, Fujimura Y, Takahashi M, Ikawa T, Otsubo T et al. . The epigenetic regulator Uhrf1 facilitates the proliferation and maturation of colonic regulatory T cells. Nat Immunol 2014; 15:571–579. [DOI] [PubMed] [Google Scholar]
- 36. Yamashita M, Inoue K, Saeki N, Ideta-Otsuka M, Yanagihara Y, Sawada Y, Sakakibara I, Lee J, Ichikawa K, Kamei Y, Iimura T, Igarashi K et al. . Uhrf1 is indispensable for normal limb growth by regulating chondrocyte differentiation through specific gene expression. Development 2018; 145:dev157412. [DOI] [PubMed] [Google Scholar]
- 37. Ramesh V, Bayam E, Cernilogar FM, Bonapace IM, Schulze M, Riemenschneider MJ, Schotta G, Gotz M. Loss of Uhrf1 in neural stem cells leads to activation of retroviral elements and delayed neurodegeneration. Genes Dev 2016; 30:2199–2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Maenohara S, Unoki M, Toh H, Ohishi H, Sharif J, Koseki H, Sasaki H. Role of UHRF1 in de novo DNA methylation in oocytes and maintenance methylation in preimplantation embryos. PLoS Genet 2017; 13:e1007042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Messerschmidt DM, Knowles BB, Solter D. DNA methylation dynamics during epigenetic reprogramming in the germline and preimplantation embryos. Genes Dev 2014; 28:812–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kagiwada S, Kurimoto K, Hirota T, Yamaji M, Saitou M. Replication-coupled passive DNA demethylation for the erasure of genome imprints in mice. EMBO J 2013; 32:340–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ohno R, Nakayama M, Naruse C, Okashita N, Takano O, Tachibana M, Asano M, Saitou M, Seki Y. A replication-dependent passive mechanism modulates DNA demethylation in mouse primordial germ cells. Development 2013; 140:2892–2903. [DOI] [PubMed] [Google Scholar]
- 42. Smallwood SA, Tomizawa S, Krueger F, Ruf N, Carli N, Segonds-Pichon A, Sato S, Hata K, Andrews SR, Kelsey G. Dynamic CpG island methylation landscape in oocytes and preimplantation embryos. Nat Genet 2011; 43:811–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Shirane K, Toh H, Kobayashi H, Miura F, Chiba H, Ito T, Kono T, Sasaki H. Mouse oocyte methylomes at base resolution reveal genome-wide accumulation of non-CpG methylation and role of DNA methyltransferases. PLoS Genet 2013; 9:e1003439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Murao N, Matsuda T, Noguchi H, Koseki H, Namihira M, Nakashima K. Characterization of Np95 expression in mouse brain from embryo to adult: A novel marker for proliferating neural stem/precursor cells. Neurogenesis (Austin) 2014; 1:e976026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sharif J, Endo TA, Nakayama M, Karimi MM, Shimada M, Katsuyama K, Goyal P, Brind'Amour J, Sun MA, Sun Z, Ishikura T, Mizutani-Koseki Y et al. . Activation of endogenous retroviruses in Dnmt1(−/−) ESCs involves disruption of SETDB1-mediated repression by NP95 binding to Hemimethylated DNA. Cell Stem Cell 2016; 19:81–94. [DOI] [PubMed] [Google Scholar]
- 46. Uemura T, Kubo E, Kanari Y, Ikemura T, Tatsumi K, Muto M. Temporal and spatial localization of novel nuclear protein NP95 in mitotic and meiotic cells. Cell Struct Funct 2000; 25:149–159. [DOI] [PubMed] [Google Scholar]
- 47. Burgoyne PS, Mahadevaiah SK, Turner JM. The consequences of asynapsis for mammalian meiosis. Nat Rev Genet 2009; 10:207–216. [DOI] [PubMed] [Google Scholar]
- 48. Hammoud SS, Low DH, Yi C, Lee CL, Oatley JM, Payne CJ, Carrell DT, Guccione E, Cairns BR. Transcription and imprinting dynamics in developing postnatal male germline stem cells. Genes Dev 2015; 29:2312–2324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kubo N, Toh H, Shirane K, Shirakawa T, Kobayashi H, Sato T, Sone H, Sato Y, Tomizawa S, Tsurusaki Y, Shibata H, Saitsu H et al. . DNA methylation and gene expression dynamics during spermatogonial stem cell differentiation in the early postnatal mouse testis. BMC Genomics 2015; 16:624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Gaysinskaya V, Miller BF, De Luca C, van der Heijden GW, Hansen KD, Bortvin A. Transient reduction of DNA methylation at the onset of meiosis in male mice. Epigenetics Chromatin 2018; 11:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Shirakawa T, Yaman-Deveci R, Tomizawa S, Kamizato Y, Nakajima K, Sone H, Sato Y, Sharif J, Yamashita A, Takada-Horisawa Y, Yoshida S, Ura K et al. . An epigenetic switch is crucial for spermatogonia to exit the undifferentiated state toward a kit-positive identity. Development 2013; 140:3565–3576. [DOI] [PubMed] [Google Scholar]
- 52. Patel SH, O'Hara L, Atanassova N, Smith SE, Curley MK, Rebourcet D, Darbey AL, Gannon AL, Sharpe RM, Smith LB. Low-dose tamoxifen treatment in juvenile males has long-term adverse effects on the reproductive system: Implications for inducible transgenics. Sci Rep 2017; 7:8991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Zhou Q, Nie R, Li Y, Friel P, Mitchell D, Hess RA, Small C, Griswold MD. Expression of stimulated by retinoic acid gene 8 (Stra8) in spermatogenic cells induced by retinoic acid: An in vivo study in vitamin A-sufficient postnatal murine testes. Biol Reprod 2008; 79:35–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Hogarth CA, Arnold S, Kent T, Mitchell D, Isoherranen N, Griswold MD. Processive pulses of retinoic acid propel asynchronous and continuous murine sperm production. Biol Reprod 2015; 92:37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Sadate-Ngatchou PI, Payne CJ, Dearth AT, Braun RE. Cre recombinase activity specific to postnatal, premeiotic male germ cells in transgenic mice. Genesis 2008; 46:738–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Rooij DG, Russell LD. All you wanted to know about spermatogonia but were afraid to ask. J Androl 2000; 21:776–798. [PubMed] [Google Scholar]
- 57. Newkirk SJ, An W. L1 regulation in mouse and human germ cells In: Cristofari G. (ed.), Human Retrotransposons in Health and Disease. Springer International Publishing: Gewerbestrasse, Switzerland; 2017: 29–61. [Google Scholar]
- 58. Bao J, Yan W. Male germline control of transposable elements. Biol Reprod 2012; 86:161–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Wang Y, Zhu T, Li Q, Liu C, Han F, Chen M, Zhang L, Cui X, Qin Y, Bao S, Gao F. Prmt5 is required for germ cell survival during spermatogenesis in mice. Sci Rep 2015; 5:11031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Vagin VV, Wohlschlegel J, Qu J, Jonsson Z, Huang X, Chuma S, Girard A, Sachidanandam R, Hannon GJ, Aravin AA. Proteomic analysis of murine Piwi proteins reveals a role for arginine methylation in specifying interaction with Tudor family members. Genes Dev 2009; 23:1749–1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Kirino Y, Kim N, de Planell-Saguer M, Khandros E, Chiorean S, Klein PS, Rigoutsos I, Jongens TA, Mourelatos Z. Arginine methylation of Piwi proteins catalysed by dPRMT5 is required for Ago3 and Aub stability. Nat Cell Biol 2009; 11:652–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Zhu W, Du J, Chen Q, Zhang Z, Wu B, Xu J, Li T, Bi Y, Shi H, Li R. Association of UHRF1 gene polymorphisms with oligospermia in Chinese males. J Assist Reprod Genet 2019; 36:2563–2573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Newkirk SJ, Lee S, Grandi FC, Gaysinskaya V, Rosser JM, Vanden N, Hogarth CA, Marchetto MCN, Muotri AR, Griswold MD, Ye P, Bortvin A et al. . Intact piRNA pathway prevents L1 mobilization in male meiosis. Proc Natl Acad Sci U S A 2017; 114:E5635–E5644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Zamudio N, Barau J, Teissandier A, Walter M, Borsos M, Servant N, Bourc'his D. DNA methylation restrains transposons from adopting a chromatin signature permissive for meiotic recombination. Genes Dev 2015; 29:1256–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Chen C. A novel reporter mouse to monitor in vivo retrotransposition in the germline. Biol Reprod 2017; 97:335–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Zhao Q, Zhang J, Chen R, Wang L, Li B, Cheng H, Duan X, Zhu H, Wei W, Li J, Wu Q, Han JD et al. . Dissecting the precise role of H3K9 methylation in crosstalk with DNA maintenance methylation in mammals. Nat Commun 2016; 7:12464. [DOI] [PMC free article] [PubMed] [Google Scholar]


