Abstract
Interleukin-12 (IL-12) is an anti-tumor cytokine that promotes biological actions through the IL-12/STAT4 axis. Genetic variation and tumor microenvironment dynamics have been identified as critical elements for impaired immune anti-tumor responses. Breast cancer (BC) is a heterogeneous disease classified at the molecular level in several subtypes, each having unique biological and clinical traits. Despite research identifying the relevance of IL-12 in many cancer types, no studies have assessed the role of the IL-12/STAT4 axis in BC. The goal of this study was to evaluate the correlation of the IL-12/STAT4 signaling axis and BC patients’ survival in general and in the context of the BC molecular subtypes. Bioinformatics analyses using TCGA data were completed to evaluate the correlation of the IL-12/STAT4 axis and BC. A high expression of important IL-12/STAT4 axis molecules such as the IL-12 receptor genes (IL12RB1 and IL12RB2), STAT4, IFNG and TBX21 were found to significantly increase BC patients’ survival rates, especially in the most aggressive BC subtypes such as the luminal B (LumB), HER-2+ and basal like (BL). A possible relevant role of the IL-12/STAT4 axis in BC is suggested by this bioinformatics-study, which might also be subtype-specific. Further studies such as molecular and tumor microenvironment analyses will be required to clarify better the specific role of the IL-12 /STAT4 axis in BC. The results from these additional analyses may potentially improve IL-12-related immunotherapeutic approaches to BC.
Keywords: Breast cancer, breast cancer subtypes, cytokines, immunotherapy, Interleukin-12, IL-12/STAT4 axis, tumor microenvironment
1. Introduction
Cytokines are essential modulators of the immune system’s responses against cancer cells [1–3]. Interleukin-12 (IL-12) is a heterodimeric cytokine which promotes relevant immune responses against tumors [4,5]. IL-12 triggers a signaling event that requires the activation of the interleukin-12 receptor (IL-12R) and eventually of the Signal Transducer and Activator of Transcription 4 (STAT4) transcription factor to promote the production of interferon gamma (IFNG) and other anti-tumor effectors [6,7]. Macrophages, dendritic cells, and B cells are among the primary immune cell populations that secrete IL-12 [8]. Also, the presence of IL-12-related immune cells such as cytotoxic T cells (CD8+), Th1 CD4 cells and NK cells in the tumor microenvironment relates to a better prognosis in several cancer types, including breast cancer (BC) [9,10]. BC is still one of the most significant chronic diseases that negatively affect women worldwide [11,12]. The biology of BC is unique, and modern classification systems acknowledge the existence of several molecular subtypes [13,14]. The most common BC subtypes are the luminal A (LumA), luminal B (LumB), HER-2 enriched (HER-2+) and the basal-like (BL) which is part of the triple-negative (TN) breast cancers. BC subtypes are defined primarily by the expression of the estrogen receptor (ER) and the HER-2 oncogene. LumA BC tumors are the most common and typically have the best prognosis of all subtypes [15].
In contrast to the LumA BC, the majority of LumB tumors express the HER-2 oncogene, although a sub-group is HER-2 negative. LumB tumors are more proliferative than LumA, and represents 10–20% of BC [16]. HER-2+ BC shows no expression of the ER and progesterone (PR) receptors, but high expression of the HER-2 oncogene [17].
HER2-enriched accounts for 5–15% of all BC and is typically more aggressive than luminal BC. BL BC, also known as a TN because it lacks the expression of ER, PR, and HER-2 receptors, is highly proliferative and very difficult to treat, making it one of poor prognosis [18]. This BL molecular subtype accounts for 15–20% of the BC cases. BC heterogeneity represents a challenge for targeted therapy. As the traditional “one size fits all” medical approach to fight cancer has proved itself inefficient, personalized therapeutic strategies to BC are currently emerging at a fast rate. Immunotherapy, in particular, has arisen as a potentially useful option for BC treatment [19]. IL-12 induction of tumor infiltration by CD8 + T cells, CD4+ Th1 T cells, and NK cells have resulted in a better prognosis for BC patients [20–22]. Similarly, the direct delivery of IL-12 to the breast tumor microenvironment has shown promising anti-tumor actions in animal and human models [23,24]. However, although research has demonstrated the effectiveness of IL-12 in BC tumor size reduction and elimination, minimal studies are assessing the role of the IL-12/STAT4 axis in BC. Moreover, there are no specific analyses that focus on the role of this IL-12/STAT4 axis and the major BC molecular subtypes.
The primary goal of this study was to evaluate the relationship of IL-12 signaling in BC, and notably, if this axis correlated with better survival in BC patients. Here we assessed the correlation of the expression of the main molecules of the IL-12/STAT4 signaling event in BC and BC subtypes by bioinformatics analyses as an initial step to clarify the role of the IL-12/STAT4 axis in BC.
2. Materials and Methods
2.1. Kaplan Meier bioinformatics analysis
KM Plotter online database (http://www.kmplot.com) was used for assessing the prognostic significance of the mRNA expression of the IL-12/STAT4 axis genes and BC [25]. BC patients’ data was divided into high and low expression cohorts using the median expression of eleven IL-12/STAT4 axis genes (IL12A, IL12B, IL12RB1, IL12RB2, TYK2, JAK2, STAT4, TBX21, IFNG, PIAS2 and SOCS3). A description of the selected eleven molecules pertaining to the IL-12/STAT4 axis is summarized on Table 1. The overall survival of BC patients from the TCGA database was analyzed by Kaplan-Meier (KM) plots focusing on all BC patients (n=1764) and also stratified by BC molecular subtypes such as LumA (n=841), LumB (n=407), HER-2+ (n=156) and BL (n=360). Hazard ratios (HR) and log rank p values were calculated on the web platform. Array quality control was set selecting “remove redundant samples” and “exclude biased arrays”. P values <0.05 were considered statistically significant.
Table 1:
Components of the IL-12/STAT4 axis
| Molecule | Genetic Location | Function |
|---|---|---|
| Interleukin-12 alpha subunit or p35 subunit (IL12A) | Chromosome 3 (3q25.33) | It is the p35 subunit of the dimeric IL-12 and its unique to IL-12. In conjunction with the p40 subunit activates the IL-12 receptor. |
| Interleukin-12 beta subunit or p40 subunit (IL12B) | Chromosome 5 (5q33.3) | It is the p40 subunit of the dimeric IL-12, it is shared between other IL-12 family members such as IL-23A. In conjunction with p35, activates the IL-12 receptor. |
| IL-12 receptor beta 1 subunit (IL12RB1) | Chromosome 19 (19p 13.1) | Type I transmembrane protein that belongs to the hemopoietin receptor Superfamily. This protein binds to IL-12 with a low affinity and is described as part of IL12 receptor complex. Upon joining IL12RB2, the IL-12 high affinity receptor is formed. Interacts with TYK2. |
| IL-12 receptor beta 2 subunit (IL12RB2) | Chromosome 1 (1p31.3) | This subunit is the signaling component that couples the JAK2-STAT4 activating message. It has been described as promoting proliferation of T-cells and NK cells. Additionally, it enhances the Th1 phenotype by mediating the production of interferon gamma. |
| Tyrosine kinase 2 (TYK2) | Chromosome 19 (19p13.2) | Tyrosine kinase belonging to the Janus kinases (JAKs) protein family. Promotes cytokine signaling by phosphorylation of specific cytokine-receptor subunits. |
| JANUS Kinase 2 (JAK2) | Chromosome 9 (9p24.1) | Protein tyrosine kinase (JAK-STAT pathway) related with specific cytokine signaling events. Associated to the prolactin receptor and it is important for interferon gamma-related responses. |
| Signal Transducer and Activator of Transcription 4 (STAT4) | Chromosome 2 (2q32.3) | Protein which is part of the JAK/STAT signaling. This transcription factor is phosphorylated by Janus kinases (Jak2 and Tyk2), and form homo- or heterodimers that translocate to the cell nucleus where they act as transcription activators. It is essential for mediating responses to IL-12 in lymphocytes and regulating the differentiation of T helper cells. |
| TBET (TBX21) | Chromosome 17 (17q21.32) | Transcription factor that controls the expression of interferon-gamma. Also, promotes Th1 cell lineage development and antagonizes Th2 cell programs. |
| Interferon gamma (IFNG) | Chromosome 12 (12q15) | Cytokine with antiviral, immunoregulatory and anti-tumor properties. It has been described as a potent activator of macrophages and other immune cells. |
| Protein Inhibitor of Activated STAT, 2 (PIAS2) | Chromosome 18 (18q21.1) | Member of the protein inhibitor of activated STAT (PIAS) family. It functions as SUMO E3 ligase and plays important roles in many cellular processes by mediating the sumoylation of target proteins. Binds and inhibits STAT4. |
| Suppressor of Cytokine Signaling, 3 (SOCS3) | Chromosome 17 (17q25.3) | It is involved in negative regulation of cytokines that signal through the JAK/STAT pathway. Inhibits cytokine signal transduction by binding to tyrosine kinase receptors including gp130, erythropoietin, insulin, IL12, GCSF and leptin receptors. |
Source: GeneCards
Abbreviations: Th1: T helper cells phenotype 1; Th2: T helper cells phenotype 2; GCSF: Granulocyte colony stimulating factor
2.1. cBioportal database bioinformatics analysis
The open-access tool cBioPortal for cancer genomics (http://www.cbioportal.org/) was used to explore the genetic alterations on the eleven IL-12/STAT4 axis genes [26]. Six different studies were selected with the search parameters for a total of 3,875 breast tumor samples included. Overall survival (OS) and the log rank p values with the 95% CI were calculated following cBioPortal’s online instructions. P <0.05 was considered statistically significant.
3. Results
The IL-12/STAT4 axis signaling event starts with the functional IL-12 molecule (composed of two subunits), binding to the dimeric receptor IL-12 receptor (IL12R) and the activation of specific kinases (JAK2 and TYK2) that in turn activate STAT4. Activated STAT4 molecules translocate into the cell nucleus and promote the transcription of IFNG and other anti-tumor effectors. A bioinformatics analysis was performed using data from the TCGA repository to have a better idea of how the expression of the IL-12/STAT4 axis correlated with overall survival for BC patients. In general, a higher expression of major components of the IL-12 signaling axis genes such as IL12RB1, IL12RB2, STAT4 and IFNG correlated with higher survival rates in BC patients (Fig. 1). There was a significantly high survival rate observed for the upstream IL-12/STAT4 axis components in BC overall, except for IL12B and JAK2 (Fig. 1). Higher IL12A expression also correlated with longer survival rates (HR:0.75; 95% CI: 0.67–0.83; P<0.0001). Conversely, IL12B gene expression did not correlate with higher survival rates among BC patients (Fig. 1). In BC in general, high expression of the IL12R genes correlated with higher survival rates for both of the IL-12R subunits IL12RB1 (HR: 0.70; 95% CI: 0.62–0.78; P<0.0001) and IL12RB2 (HR:0.73, 95% CI: 0.66–0.82; P<0.0001), as shown in Fig. 1.
Figure 1: Survival analysis of BC patients by gene expression of the IL-12/STAT4 axis.
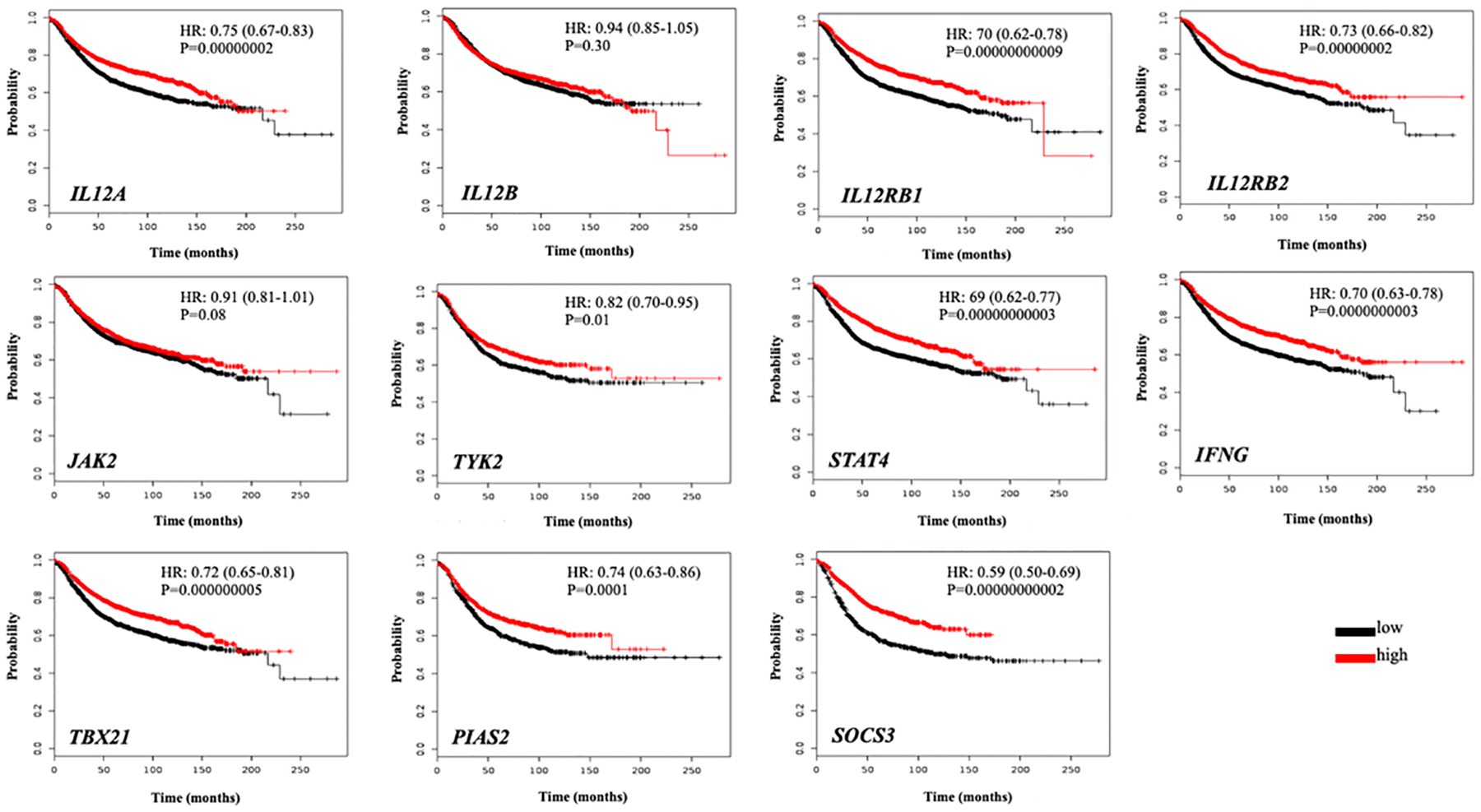
Kaplan Meier plots of the IL-12/STAT4 axis genes expression and overall BC survival are presented. The red line represents a higher expression, whereas the black line represents lower gene expression. Y-axis represents survival probability whereas x-axis the time (in months). HR: Hazards ratio; P: p-value.
Other upstream IL-12/STAT4 axis components such as TYK2 and JAK2 were also evaluated. A high JAK2 expression did not correlate with long survival rates among BC patients (Fig. 1). TYK2 on the other hand was found to be associated with long survival rates, but less robustly, when compared to other upstream IL-12/STAT4 axis molecules (HR: 0.82; 95% CI: 0.70–0.95; P=0.01) as depicted in Fig. 1. STAT4 is the main transcription factor of the IL-12 message.
In this analysis, a higher expression of STAT4 correlated with a significant clear increase in survival rates in BC patients (HR: 0.69; 95% CI: 0.62–0.71; P<0.0001) as presented in Fig. 1. Other IL-12 axis related transcription factor, the TBX21gene, also correlated with high survival rates among BC patients (HR: 0.72; 95% CI: 0.65–0.81; P<0.0001). Interferon gamma is one of the main downstream effectors of the IL-12/STAT4 axis. Overall, a noticeable significant correlation for elevated IFNG expression and long BC patient’s survival rates was also observed for this gene (HR: 0.70; 95% CI: 0.63–0.78; P <0.0001) as shown in Fig. 1. The IL-12/STAT4 regulator molecules PIAS2 and SOCS3 also had a significant correlation with a high expression of these associating with longer survival rates among BC patients (Fig. 1).
Stratification analysis for the BC molecular subtypes was also conducted. A high expression of the IL12 genes, IL12A (HR: 0.76; 95% CI: 0.63–0.92; P=0.005) and IL12B (HR: 0.67; 95% CI: 0.55–0.81; P<0.0001) correlated with longer survival rates for the BC subtypes. This correlation was more evident for the upstream genes IL12A and IL12B in the HER-2+ and BL patients when compared to Luminal BC (Fig. 2). Similarly, the IL12R genes expression was evaluated, and for LumB patients a high expression of IL12RB1 correlated with longer survival rates (HR: 0.57; 95% CI: 0.42–0.78; P=0.0004) when compared to the LumA cases (Fig. 2). For this same gene, a more robust correlation of high expression with longer survival rates was observed for the HER-2+ and BL cases (Fig. 2). There was no correlation of the IL12RB2 expression and survival rates for the LumB cases as opposed to what was found for the LumA ones (Fig. 2). In addition, and consistent to what was observed for all BC patients, there was a significant correlation of higher expression of STAT4 (HR: 0.56; 95% CI: 0.46–0.68; P<0.0001), IFNG (HR: 0.64; 95% CI: 0.53–0.78; P<0.0001) and TBX21 (HR: 0.65; 95% CI: 0.54–0.79; P<0.0001) and longer survival rates for LumA and LumB patients, although this was more evident for the LumB BC (Fig. 3 and 4). Poor or no correlations were observed for TYK2 and JAK2 genes in the luminal BC patients (Fig. 3).
Figure 2: Survival analysis of BC patients by gene expression of the IL-12/STAT4 axis upstream components.

Kaplan Meier plots of the IL12A, IL12B, IL!2RB1 and IL12RB2 genes expression and BC survival stratified by BC subtypes (Luminal A, Luminal B, HER2+ and Basal-like). Red line represents a higher expression, whereas black line represents lower gene expression. Y-axis represents survival probability whereas x-axis the time (in months). HR: Hazards ratio; P: p-value.
Figure 3: Survival analysis of BC patients by gene expression of the IL-12/STAT4 axis upstream and transcription factors components.
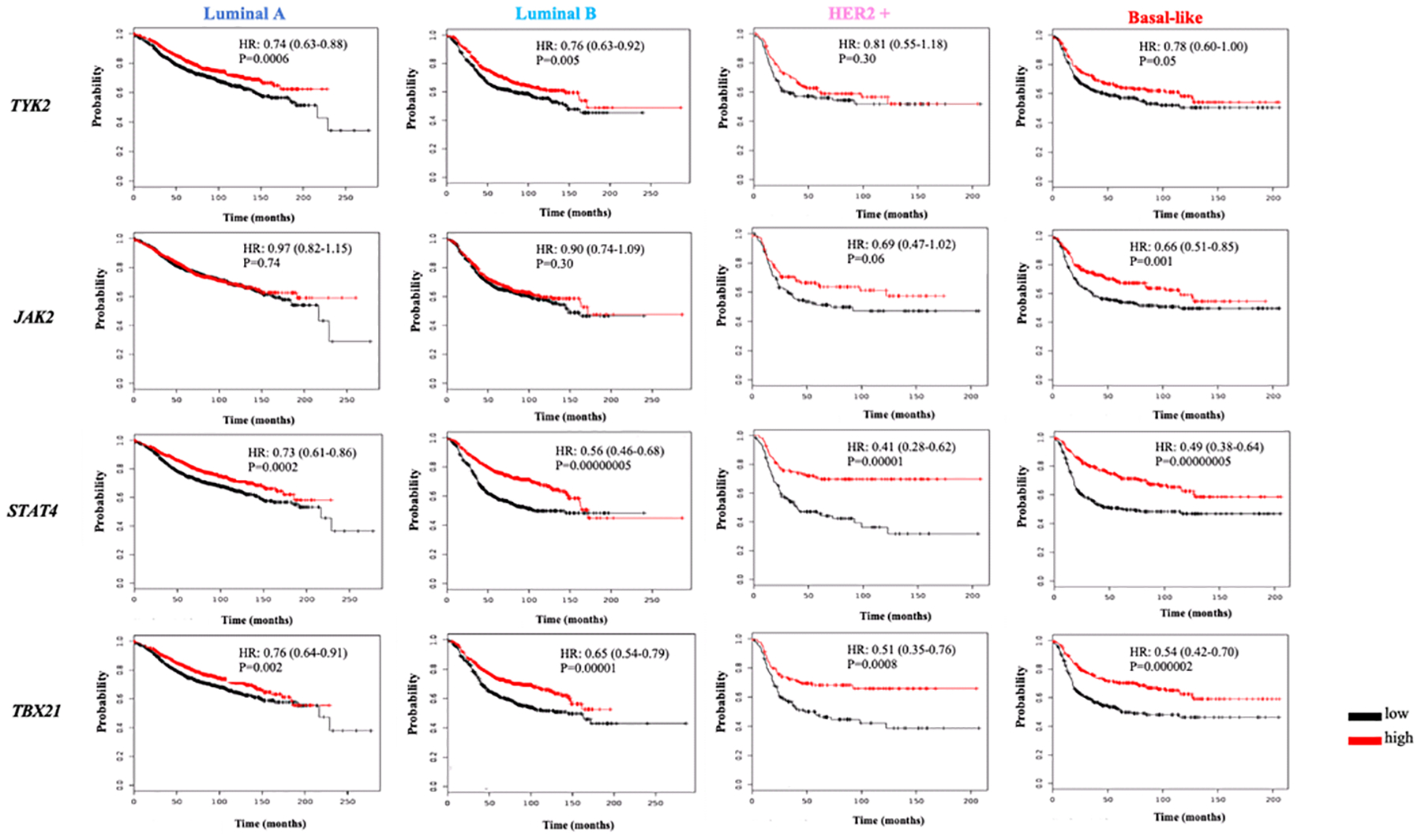
Kaplan Meier plots of the TYK2, JAK2, STAT4 and TBX21 genes expression and BC survival stratified by BC subtypes (Luminal A, Luminal B, HER2+ and Basal-like). The red line represents a higher expression, whereas the black line represents lower gene expression. Y-axis represents survival probability whereas x-axis the time (in months). HR: Hazards ratio; P: p-value.
Figure 4: Survival analysis of BC patients by gene expression of the IL-12/STAT4 axis downstream and regulators components.
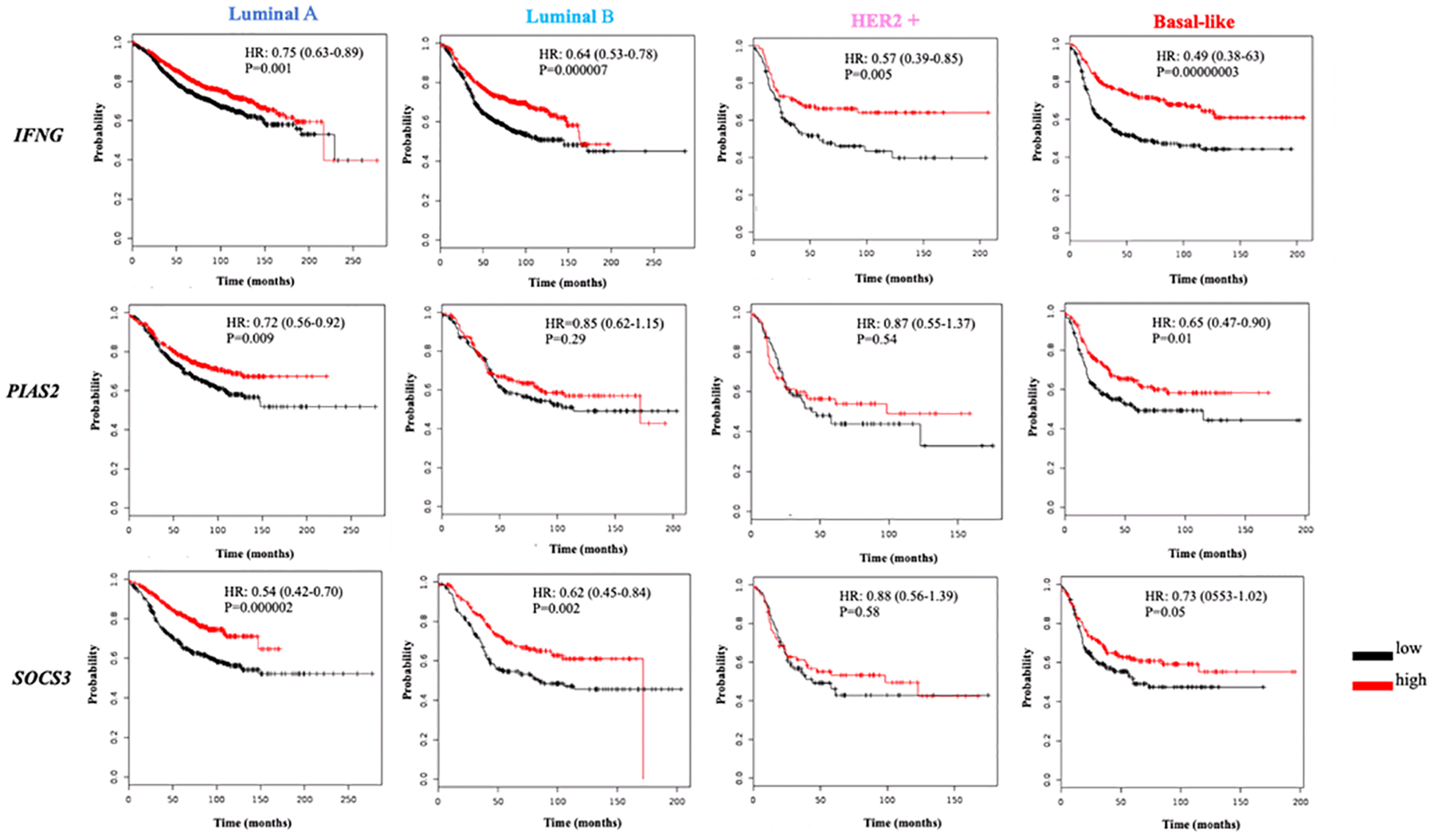
Kaplan Meier plots of the IFNG, PIAS2 and SOCS3 genes expression and BC survival stratified by BC subtypes (Luminal A, Luminal B, HER2+ and Basal-like). The red line represents a higher expression, whereas the black line represents lower gene expression. Y-axis represents survival probability whereas x-axis the time (in months). HR: Hazards ratio; P: p-value.
The HER2-enriched BC subtype has only HER-2 expression. Although IL12A and IL12B genes high expression correlated with longer survival rates (Fig. 2), the most notable correlations for the HER-2+ subtype were observed for STAT4 (HR: 0.41; 95% CI: 0.28–0.62; P<0.0001), IFNG (HR: 0.57; 95% CI: 0.39–0.85; P=0.0008) and TBX21 (HR: 0.51; 95% CI: 0.35–0.76; P=0.005) as presented in Fig. 3 and 4. The IL-12R and the IL-12/STAT4 axis tyrosine kinases genes, except for the IL12RB2, did not correlate with longer survivals for this BC subtype (Fig. 2 and 3). The regulators PIAS2 and SOCS3 also did not correlate with BC patients’ longer survival rates (Fig. 4). The BL subtype lacks expression of any of the hormone and oncogene receptors. The bioinformatics analysis with the TCGA data revealed that higher IL12A (HR:0.65; 95% CI: 0.50–0.84; P<0.0001) and IL12B (HR:0.58; 95% CI: 0.45–0.75; P<0.0001) expression correlated with longer BC patient’s survival (Fig. 2). It also showed that both IL-12R genes’ high expression, for IL12RB1 (HR: 0.68; 95% CI: 0.49–0.94; P=0.02) and IL12RB2 (HR: 0.71; 95% CI: 0.55–0.92; P=0.008) were correlated with longer survival rates of BC patients (Fig. 2). For the LB subtype there was also a stronger correlation of STAT4 (HR:0.49; 95% CI: 0.38–0.64; P<0.0001), IFNG (HR: 0.49; 95% CI: 0.28–0.63; P<0.0001) and TBX21 (HR: 0.54; 95% CI: 0.42–0.70; P<0.0001) high expression and longer survival (Fig. 3 and 4). These were stronger than the overall BC correlations.
In summary, when comparing the correlations of expression and survival rates by KM analyses, the major IL-12/STAT4 axis components that were found to be significantly correlated for all the BC subtypes, and including all BC patients, were STAT4 and IFNG. The luminal BC (LumA and LumB) had a pattern similar to the observed for overall BC, with high expression of STAT4 and SOCS3 being the most significantly correlated genes with long survival rates. The more aggressive BC subtypes (HER-2+ and BL) had a very similar pattern in which in addition to STAT4 and IFNG, a high TBX21 expression also was importantly correlated with more prolonged survival rates for these BC patients.
The KM plot analyses were based on mRNA obtained systemically from patients. To further verify the correlation of the selected IL-12/STAT4 gene-alterations in data from breast tumors, with overall survival among BC patients, an analysis was performed retrieving samples from six different studies using the cBioportal database. Overall, longer survival rates for BC patients whose tumors did not have genetic alterations on the major IL-12/STAT4 axis-genes was observed when compared to those who had the gene alterations, as presented in Fig. 5A. Although not abundant, most of the genetic changes found in the IL-12/STAT4 axis genes within these patients were amplifications, followed by deletions and missense mutations (Fig. 5B). Figure 5C shows the distribution frequencies of genetic alterations per each of the selected studies in this analysis as obtained from the cBioportal website.
Figure 5: IL-12/STAT4 axis-genes tumor mutation analysis in BC samples (cBioportal tool).

(A) Kaplan-Meier plot comparing overall survival in cases with/without IL-12/STAT4-genes alterations. (B) Oncoprint output from the cBioPortal representing the proportion and distribution of samples with alterations in IL-12/STAT4 axis genes. The figure was cropped on the right to exclude samples without alterations. (C) The alteration frequencies from each of the selected studies are shown stratified by synonymous mutations (Mutation), amplifications, deep deletions and multiple alterations.
4. Discussion
IL-12 is an important proinflammatory cytokine which induces a signaling event that triggers STAT4 to eventually generate IFNG and other key anti-tumor effectors [9,27]. IL-12 promote anti-tumor actions of NK, CD8+ T cells, macrophages and CD4+ Th1 cells [21,28]. Despite abundant evidence on the anti-tumor properties of IL-12 in several cancer models, very little is known regarding the role of this cytokine in BC. Moreover, there are no specific studies that have assessed the IL-12 in a pathway-oriented manner, nor the context of the BC molecular subtypes. In this study, high gene expression of key IL-12/STAT4 axis molecules was found to be correlated with better overall prognosis among BC patients. This prolonged survival was especially evident for key players of this signaling event such as STAT4 and IFNG. Overall, lower expression of key IL-12/STAT4 axis molecules such as IL12B, IL12RB1, STAT4, TBX21, IFNG, and SOCS3 was found to be correlated with poorer prognosis in BC, regardless of subtype classification. When a stratification analysis by BC molecular subtypes was completed, a similar pattern was observed for STAT4 and IFNG. In addition, using data from tumors, a longer survival was observed for those BC patients who had no genetic alterations in the IL-12/STAT4 axis components, suggesting the positive role of this IL-12 axis in anti-tumor responses.
BC is heterogeneous, and specific gene signatures drive the unique biological traits of the major BC subtypes [29,30]. Despite the fact that STAT4 and IFNG correlations were similar for all BC subtypes in this study, specific patterns of IL-12/STAT4 axis expression and BC survival were observed for the molecular subtypes. The LumA subtype had strong correlations for the IL12A and the regulators PIAS2 and SOCS3 genes. LumB BC subtype resulted in strong associations for the IL12B, the IL12RB1and SOCS3 genes. LumB BC is more proliferative than LumA, so although not as clear from this analysis, a stronger IL-12 related response may be observed for these LumB tumors. The more proliferative subtypes, the HER-2+, and the BL, also had very similar correlation patterns, as in addition to STAT4 and IFNG; also, TBX21 was strongly associated with prolonged survival in the patients with these types of tumors. TBX21 works closely with STAT4 to induce Th1 T cells, which are involved in anti-tumor responses mediated by IL-12 [28,31]. As several studies have shown, tumor-infiltrating Th1 CD4+ cells promote prolonged survival of cancer patients as these enhance anti-tumor reactions [32]. Thus, it may be possible that while the IL-12/STAT4 axis may be relevant for BC in general, specific dynamics about this signaling event may promote unique immune actions in the context of the BC subtypes. However, one limitation of this study is that with the bioinformatics approach alone although useful to identify significant correlations, it is not possible to adequately address if indeed this is the case.
Analyzing the gene expression of the IL-12/STAT4 axis and its relationship to BC is a good starting point to assess the role of this axis in BC. However, it may not be enough to provide answers to the specific dynamics that may influence this. Some studies have emphasized the difficulty of having a precise BC subtyping classification [33,34]. This limitation may affect targeted approaches to diagnose or treat BC. Noticeably this challenge was not present in this bioinformatics study since the available data assessed belonged to the TCGA repository, which has validated information regarding BC subtyping. Therefore, the results obtained in this study are representative of the real BC subtypes.
Several factors may alter the normal functioning of anti-tumor cytokines including genetics, epigenetics, and environmental dynamics. Genetic variation such as Single Nucleotide Polymorphisms (SNPs) may change normal gene expression patterns and predispose to inefficient immune responses that eventually increase cancer risk [35]. SNPs in the IL-12 genes, IL12Aand IL12B, have been commonly published as related to cancer risk [36,37]. Reduced IL-12 signaling may relate to less effective anti-tumor actions in BC patients. Therefore, a low expression of key IL-12/STAT4 axis key players such as the ones identified by this study might be downregulated in BC. Meta-analyses, including the ones conducted by Xiao and his colleagues in 2018 and Zheng and his colleagues in 2017 have found a relationship between SNPs on the IL12 genes and cancer risk [38,39]. Specific SNPs are believed to induce less-effective IL-12 signaling event, by destabilizing the normal genetic expression process for the cytokine and therefore reducing IL-12-related anti-tumor effectors. The particular way this may happen is not well understood, but it may also include the effects of genetic alteration of the IL-12 signaling axis components.
SNPs in specific IL-12/STAT4 axis have been related to colorectal, cervical, gastric, hepatic, and prostate cancers[40,41]. STAT4 gene alterations, for example, have been implicated with an increased risk to several cancers, such as those of the stomach, colon, lung, and the breasts [42–44]. In this study, a lower STAT4 expression correlated with poorer BC patient’s survival for all BC subtypes, with a noticeable correlation when analyzing the more aggressive subtypes. Similarly, genetic alterations of the IFNG gene have been associated with prostate, colorectal, cervical, and gastric cancers [45,46]. As it has been established by previous research, high IFNG expression relates to better anti-tumor responses [2]. A higher expression of IFNG also correlated with longer survival rates of BC in the present study. Noteworthy this was more evident for the HER-2+ and BL subtypes, as stronger correlations were obtained even when the cohorts for such subtypes were small compared to the overall study population. However, assessing the genetic expression of these critical IL-12/STAT4 axis molecules is only the starting point of a necessary more in-depth evaluation.
SNPs are not the only possible cause of decreased anti-tumor immune responses. Tumor microenvironment dynamics also help shape the extent by which immune cells can target and eradicate cancer cells [47, 48]. Impaired anti-tumor immunity can predispose to cancer and also promote a pro-tumor tumor microenvironment. Overall, the tumor microenvironment is a complex environment in which immune cells and cancer cells interact [49]. The responses of adaptive immune cells to BC cells may be impaired at the tumor microenvironment by a combination of several factors, such as suppressive immune cells and cytokines. This suppression may reduce the capacity of anti-tumor cytokines, such as IL-12, to promote tumor elimination, which eventually facilitates pro tumor dynamics.
Assessing peripheral immune cells and other IL-12 related markers may also aid to better understand the changes in overall systemic immunity for a BC patient, and if indeed these correlates to what it is observed at the gene expression level. In addition, it will be required to evaluate the IL-12/STAT4 axis at the protein level, not only systemically but also at the tumor zones. As it has been the case in other studies for anti-tumor cytokines, the IL-12 axis may be downregulated at the breast tumor microenvironment. The exact possible causes of IL-12/STAT4 impairment in BC could be explained by SNPs and by the altered breast tumor microenvironment. The specific dynamics related to this are not precise yet, but downregulation of the IL-12/STAT4 axis may occur, predisposing to BC, in a way that may also be subtype-specific.
5. Conclusion
The results of this study suggest that the IL-12/STAT4 axis has a role in BC. Moreover, this role may be BC subtype-specific. This bioinformatics approach is a useful tool to generate specific hypotheses regarding the role of the IL-12 axis in BC. However, further strategies targeting genetics and the tumor microenvironment would be necessary to elucidate the role of the IL-12/STAT4 axis in BC better. By doing so, this may enhance IL-12related immunotherapeutic approaches to BC.
6. Future studies
It would be useful for futures studies to assess the IL-12/STAT4 axis in the context of the breast tumor microenvironment. Immunohistochemistry of tumor tissue and co-culture systems between BC cell lines and IL-12 related immune cells could be valuable tools to better evaluate the IL-12/STAT4 axis in BC and BC molecular subtypes.
Acknowledgements
The author acknowledges Dr. Julie Dutil and Dr. Patricia Casbas. The author also acknowledges the NIH-NIGMS fellowship (R25GM082406).
References
- [1].Lippitz BE, “Cytokine patterns in patients with cancer: A systematic review,” The Lancet Oncology. 2013. [DOI] [PubMed] [Google Scholar]
- [2].Duluc D et al. , “Interferon-γ reverses the immunosuppressive and protumoral properties and prevents the generation of human tumor-associated macrophages,” Int. J. Cancer, 2009. [DOI] [PubMed] [Google Scholar]
- [3].Floros T and Tarhini AA, “Anticancer Cytokines: Biology and Clinical Effects of Interferon-α2, Interleukin (IL)-2, IL-15, IL-21, and IL-12,” Semin. Oncol, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Lasek W, Zagozdzon R, and Jakobisiak M, “Interleukin 12: Still a promising candidate for tumor immunotherapy?,” Cancer Immunology, Immunotherapy. 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Watford WT, Moriguchi M, Morinobu A, and O’Shea JJ, “The biology of IL-12: Coordinating innate and adaptive immune responses,” Cytokine and Growth Factor Reviews. 2003. [DOI] [PubMed] [Google Scholar]
- [6].Kerkar SP and Restifo NP, “The power and pitfalls of IL-12,” Blood. 2012. [DOI] [PubMed] [Google Scholar]
- [7].Zundler S and Neurath MF, “Interleukin-12: Functional activities and implications for disease,” Cytokine and Growth Factor Reviews. 2015. [DOI] [PubMed] [Google Scholar]
- [8].Vignali DAA and Kuchroo VK, “IL-12 family cytokines: immunological playmakers,” Nature Immunology. 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Tugues S et al. , “New insights into IL-12-mediated tumor suppression,” Cell Death and Differentiation. 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Kerkar SP et al. , “Tumor-specific CD8+T cells expressing interleukin-12 eradicate established cancers in lymphodepleted hosts,” Cancer Res, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Vogel VG, “Epidemiology of breast cancer,” in The Breast: Comprehensive Management of Benign and Malignant Diseases, 2017. [Google Scholar]
- [12].DeSantis CE, Ma J, Goding Sauer A, Newman LA, and Jemal A, “Breast cancer statistics, 2017, racial disparity in mortality by state,” CA. Cancer J. Clin, 2017. [DOI] [PubMed] [Google Scholar]
- [13].Cancer T and Atlas G, “Comprehensive molecular portraits of human breast tumours.,” Nature, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Dai X et al. , “Breast cancer intrinsic subtype classification, clinical use and future trends,” American Journal of Cancer Research. 2015. [PMC free article] [PubMed] [Google Scholar]
- [15].Ignatiadis M and Sotiriou C, “Luminal breast cancer: From biology to treatment,” Nature Reviews Clinical Oncology. 2013. [DOI] [PubMed] [Google Scholar]
- [16].Ades F et al. , “Luminal B breast cancer: Molecular characterization, clinical management, and future perspectives,” Journal of Clinical Oncology. 2014. [DOI] [PubMed] [Google Scholar]
- [17].Russnes HG, Lingjærde OC, Børresen-Dale AL, and Caldas C, “Breast Cancer Molecular Stratification: From Intrinsic Subtypes to Integrative Clusters,” American Journal of Pathology. 2017. [DOI] [PubMed] [Google Scholar]
- [18].Ahn SG, Kim SJ, Kim C, and Jeong J, “Molecular Classification of Triple-Negative Breast Cancer,” J. Breast Cancer, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Emens LA, “Breast cancer immunotherapy: Facts and hopes,” Clinical Cancer Research. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Hamza T, Barnett JB, and Li B, “Interleukin 12 a key immunoregulatory cytokine in infection applications,” International Journal of Molecular Sciences. 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Ohs I, Ducimetière L, Marinho J, Kulig P, Becher B, and Tugues S, “Restoration of natural killer cell antimetastatic activity by IL12 and checkpoint blockade,” Cancer Res, 2017. [DOI] [PubMed] [Google Scholar]
- [22].Yang S-X et al. , “Interleukin-12 activated CD8+ T cells induces apoptosis in breast cancer cells and reduces tumor growth,” Biomed. Pharmacother, 2016. [DOI] [PubMed] [Google Scholar]
- [23].Kerkar SP et al. , “IL-12 triggers a programmatic change in dysfunctional myeloid-derived cells within mouse tumors,” J. Clin. Invest, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Chinnasamy D et al. , “Local delivery of interleukin-12 using T cells targeting VEGF receptor-2 eradicates multiple vascularized tumors in mice,” Clin. Cancer Res, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Casper J et al. , “The UCSC Genome Browser database: 2018 update,” Nucleic Acids Res, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Gao J et al. , “Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal,” Sci. Signal, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Watford WT, Hissong BD, Bream JH, Kanno Y, Muul L, and O’Shea JJ, “Signaling by IL-12 and IL-23 and the immunoregulatory roles of STAT4,” Immunological Reviews. 2004. [DOI] [PubMed] [Google Scholar]
- [28].Qu Y, Chen L, Lowe DB, Storkus WJ, and Taylor JL, “Combined Tbet and IL12 gene therapy elicits and recruits superior antitumor immunity in vivo,” Mol. Ther, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Casbas-Hernandez P et al. , “Tumor intrinsic subtype is reflected in cancer-adjacent tissue,” Cancer Epidemiol. Biomarkers Prev, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Koboldt DC et al. , “Comprehensive molecular portraits of human breast tumours,” Nature, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Andreev K et al. , “Impaired T-bet-pSTAT1α and perforin-mediated immune responses in the tumoral region of lung adenocarcinoma,” Br. J. Cancer, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Gu-Trantien C et al. , “CD4+follicular helper T cell infiltration predicts breast cancer survival,” J. Clin. Invest, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Rivenbark AG, O’Connor SM, and Coleman WB, “Molecular and cellular heterogeneity in breast cancer: Challenges for personalized medicine,” Am. J. Pathol, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Dai X et al. , “Breast cancer intrinsic subtype classification, clinical use and future trends,” Am J Cancer Res, 2015. [PMC free article] [PubMed] [Google Scholar]
- [35].Deng N, Zhou H, Fan H, and Yuan Y, “Single nucleotide polymorphisms and cancer susceptibility,” Oncotarget, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Chang JS et al. , “Genetic polymorphisms in adaptive immunity genes and childhood acute lymphoblastic leukemia,” Cancer Epidemiol. Biomarkers Prev, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Chang SW, Xu GQ, and Fan YL, “Association of interleukin-12 gene polymorphisms with cancer susceptibility: A meta-analysis,” Int. J. Clin. Exp. Med, 2015. [PMC free article] [PubMed] [Google Scholar]
- [38].Xiao Y, Liu G, and Gong L, “Systematic review and meta-analysis on the association between polymorphisms in genes of IL-12 signaling pathway and hepatocellular carcinoma risk,” J. Cancer, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Zheng Y et al. , “Role of interleukin-12 gene polymorphisms in the onset risk of cancer: a meta-analysis,” Oncotarget, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Slattery ML et al. , “Genetic variation in the JAK/STAT/SOCS signaling pathway influences breast cancer-specific mortality through interaction with cigarette smoking and use of aspirin/NSAIDs: The Breast Cancer Health Disparities Study,” Breast Cancer Res. Treat, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Hussain SK et al. , “Nucleotide variation in IL-10 and IL-12 and their receptors and cervical and vulvar cancer risk: A hybrid case-parent triad and case-control study,” Int. J. Cancer, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Slattery ML et al. , “Genetic variation in the JAK/STAT/SOCS signaling pathway influences breast cancer-specific mortality through interaction with cigarette smoking and use of aspirin/NSAIDs: The Breast Cancer Health Disparities Study,” Breast Cancer Res. Treat, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Moon CM et al. , “Association of signal transducer and activator of transcription 4 genetic variants with extra-intestinal manifestations in inflammatory bowel disease,” Life Sci, 2010. [DOI] [PubMed] [Google Scholar]
- [44].Chanthra N et al. , “Single nucleotide polymorphisms in STAT3 and STAT4 and risk of hepatocellular carcinoma in Thai patients with chronic hepatitis B,” Asian Pacific J. Cancer Prev, 2016. [DOI] [PubMed] [Google Scholar]
- [45].Zhang Z, Fye S, Borecki IB, and Rader JS, “Polymorphisms in immune mediators associate with risk of cervical cancer,” Gynecol. Oncol, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Gonzalez B, Flores-A H, Sanchez D, Alaez C, and Gorodezky C, “IL2 polymorphisms have no impact on risk, but genetic variants in IFNG have a protective role in acute lymphoblastic leukemia and are associated with gender in Mexican children,” Hum. Immunol, 2015. [Google Scholar]
- [47].Larsen SK, Gao Y, and Basse PH, “NK Cells in the Tumor Microenvironment,” Crit Rev Oncog, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Thorsson V et al. , “The Immune Landscape of Cancer,” Immunity, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Rahir G and Moser M, “Tumor microenvironment and lymphocyte infiltration,” Cancer Immunology, Immunotherapy. 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]


